Abstract
Caenorhabditis elegans is an exceptionally valuable model for aging research because of many advantages, including its genetic tractability, short lifespan, and clear age‐dependent physiological changes. Aged C. elegans display a decline in their anatomical and functional features, including tissue integrity, motility, learning and memory, and immunity. Caenorhabditis elegans also exhibit many age‐associated changes in the expression of microRNAs and stress‐responsive genes and in RNA and protein quality control systems. Many of these age‐associated changes provide information on the health of the animals and serve as valuable biomarkers for aging research. Here, we review the age‐dependent changes in C. elegans and their utility as aging biomarkers indicative of the physiological status of aging.
1. INTRODUCTION
Living organisms undergo many degenerative and deleterious functional and structural changes with aging. In animals, these changes include reduced motor activity, overall metabolic rates, and cognitive function, as well as impaired digestive function, and increased DNA damage. Delaying or reversing these degenerative changes is critical because one of the important goals of aging research is to prolong the healthy longevity of humans. To achieve this goal, a key initial step is to dene and characterize age‐dependent physiological and molecular changes. However, it takes a very long time for most mammals to display such age‐dependent changes. Therefore, short‐lived organisms have been used as valuable models for aging research.
Caenorhabditis elegans is one of the outstanding model organisms used for aging research because of its very short lifespan and simple physiology. Moreover, experiments with C. elegans are free of ethical concerns. Many breakthrough discoveries in the eld of aging research have been made using C. elegans because of these advantages (Kenyon, 2010). To date, the measurement of lifespan is the most popular method used to study aging in C. elegans. However, this assay still takes a relatively long time because the lifespan of wild‐type C. elegans is approximately 3 weeks. Additionally, chronological age does not always reflect biological (functional and physiological) age; a cohort with the same chronological age undergoes biological changes at variable rates, especially in the later stages of life. Therefore, dening quantitative changes associated with physiological aging, referred to as biomarkers of aging, is crucial for expediting the development of potential anti‐aging therapies.
Legitimate biomarkers of aging should exhibit changes in expression level or activity during physiological aging and therefore serve as a reference to predict the status of aging without harming the subjects. Thus, prominent age‐dependent molecular and physiological changes are reasonable candidates for biomarkers of aging. The characteristics of aging in C. elegans have been described in another review article (Collins, Huang, Hughes, & Kornfeld, 2008). In addition, a recently published book chapter is an outstanding resource for the age‐dependent anatomical changes of C. elegans (Herdon, Wolkow, Driscoll, & Hall, 2017). Here, we summarize the recent ndings regarding potential biomarkers of aging in C. elegans. We classify and describe age‐related changes in C. elegans at the tissue, cellular, and molecular levels (Figure 1). We also review the roles of insulin/IGF‐1 signaling (IIS) and dietary restriction (DR), two evolutionarily conserved aging‐modulating regimes, in age‐related changes. We then discuss the potential utility of these changes as biomarkers of aging in C. elegans. We believe that these biomarkers will provide valuable insights into the aging process and that this information will advance our understanding of aging and age‐related diseases in mammals, including humans.
Figure 1.
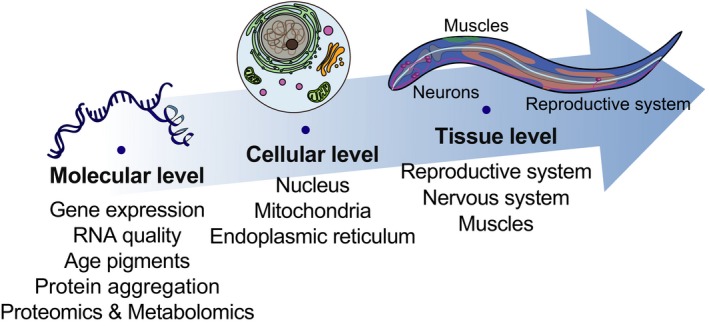
Age‐related changes in Caenorhabditis elegans. A schematic diagram shows age‐related changes that are discussed in this review. We classified age‐related changes into three levels: tissue, cellular, and molecular
2. AGE‐DEPENDENT CHANGES
2.1. Age‐dependent changes at the tissue level
2.1.1. Reproductive system
Like many other physiological systems, the reproductive system of C. elegans undergoes age‐dependent changes (Figure 2). The rate of reproduction signicantly decreases with age, and the structure of the reproductive system deteriorates (Garigan et al., 2002; Hughes, Evason, Xiong, & Kornfeld, 2007; Luo, Kleemann, Ashraf, Shaw, & Murphy, 2010; McGee, Day, Graham, & Melov, 2012; Pickett, Dietrich, Chen, Xiong, & Kornfeld, 2013). As C. elegans is a self‐fertile hermaphrodite with a limited number of sperm (Ward & Carrel, 1979), sperm shortage is one of the reasons for reduced progeny at an early age. This is because the number of progeny produced by unmated (self‐fertilized) C. elegans decreases sharply and relatively early (Day 3) in adulthood (Hughes et al., 2007). In contrast, age‐dependent reductions in the number of progeny are delayed in mated hermaphrodites that receive sperm from other males compared to unmated hermaphrodites (Hughes et al., 2007; Luo et al., 2010). However, mated hermaphrodites also cease reproduction after a certain time point, implying the existence of factors, other than sperm shortage, that limit the reproductive period. Age‐associated deterioration of tissues in the reproductive system may hamper prolonged reproduction, despite the supply of extra sperm at an advanced age. In aged worms, the number of nuclei in the mitotic germline diminishes and the nucleoplasm displays an increased accumulation of grainy material and cavities (Garigan et al., 2002). Oocyte size also decreases during aging, which correlates with their reduced quality, suggesting that an appropriate oocyte size is crucial for reproduction (Andux & Ellis, 2008). In addition, the number of unfertilized oocytes in mated hermaphrodites increases with age (Luo et al., 2010). Thus, the germline of old worms undergoes sperm shortages and structural changes, which eventually render the worms sterile.
Figure 2.
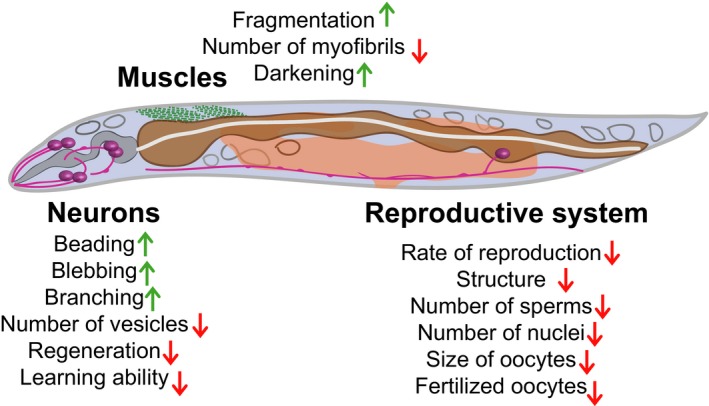
Age‐related changes at the tissue level. As Caenorhabditis elegans ages, the integrity of muscles, neurons, and the reproductive system declines. Muscles become fragmented, and the tissues of the reproductive system degenerate. Neuronal integrity appears to be prolonged compared to other tissues, but neurons also undergo age‐dependent deterioration, such as blebbing
Mechanisms of reduced reproductive potency in aged male C. elegans have also been proposed (Chatterjee et al., 2013; Guo, Navetta, Gualberto, & Garcia, 2012). Although the activity, morphology, and number of sperm are preserved during the rst 3 days of adulthood, 3‐day‐old adults exhibit uncoordinated mating behaviors and a rapid decline in reproductive potency (Chatterjee et al., 2013; Guo et al., 2012). This uncoordinated mating behavior appears to be caused by hyperexcited muscles in the male reproductive system (Guo et al., 2012), as decreasing the activity of these hyperexcited muscles has been shown to improve mating potency. Overall, the reproductive systems of both hermaphrodites and males display age‐related changes, which lead to a reduction in the reproductive potential.
2.1.2. Nervous system
Although it was initially reported that C. elegans neurons remain relatively intact during aging compared with other tissues (Collins et al., 2008; Herndon et al., 2002), subsequent studies have shown that neurons display subtle but reproducible age‐dependent changes (Chen, Chen, Jiang, Chen, & Pan, 2013; Chew, Fan, Gotz, & Nicholas, 2013; Collins et al., 2008; Herndon et al., 2002; Pan, Peng, Chen, & McIntire, 2011; Tank, Rodgers, & Kenyon, 2011; Toth et al., 2012; Figure 1). Touch receptor neurons, such as ALM neurons, display gradual beading, blebbing, and branching during aging (Pan et al., 2011; Tank et al., 2011). In addition, the number of vesicles per synapse in touch receptor neurons is signicantly lower in aged worms than in young worms (Toth et al., 2012). This indicates that the synaptic integrity of worms also degenerates during aging. The extent of aberrant neuronal changes varies among worms of the same age, and neuron integrity shows a positive correlation with the coordinated motility of worms. In addition to the touch receptor neurons, GABAergic motor neurons in aged worms display neurite branching (Tank et al., 2011). Another study, however, suggested that GABAergic motor neurons undergo minimal age‐related changes (Toth et al., 2012). Further studies are needed to resolve these discrepancies, which perhaps originate from dierences in experimental conditions.
In addition to these structural changes, aged C. elegans display a functional deterioration of neurons (Hammarlund, Nix, Hauth, Jorgensen, & Bastiani, 2009; Kauman, Ashraf, Corces‐Zimmerman, Landis, & Murphy, 2010; Liu et al., 2013; Murakami & Murakami, 2005). Caenorhabditis elegans motor neurons undergo functional declines starting at an early age (Liu et al., 2013) and display reduced regeneration capacity (Hammarlund et al., 2009; Kauman et al., 2010; Liu et al., 2013; Murakami & Murakami, 2005). Associative learning ability also decreases with age (Hammarlund et al., 2009; Kauman et al., 2010; Murakami & Murakami, 2005). Thus, dierent neuron types undergo dierent age‐dependent structural and functional declines.
2.1.3. Muscles
An age‐dependent reduction in the muscle integrity of C. elegans has been widely reported (Chow, Glenn, Johnston, Goldberg, & Wolkow, 2006; Garigan et al., 2002; Glenn et al., 2004; Herndon et al., 2002; Johnston, Iser, Chow, Goldberg, & Wolkow, 2008; Shamir, Wolkow, & Goldberg, 2009; Figure 1). Nuclei and sarcomeres in the body wall muscle cells undergo age‐dependent changes. For example, nuclei in the body wall muscle cells undergo redistribution with age. However, the extent of this redistribution varies between as well as within individual worms, suggesting that stochastic factors play a role in this phenomenon (Herndon et al., 2002). Additionally, the size of muscle cell nucleoli tends to increase during aging (Herndon et al., 2002, see also Tiku et al., 2017). Furthermore, sarcomeres in the body wall muscles lose their densely packed structures and regular orientations with increasing age (Glenn et al., 2004; Herndon et al., 2002). The C. elegans pharynx, a neuromuscular organ composed of 20 muscle cells and 20 neurons, also loses its integrity during aging (Albertson & Thomson, 1976; Chow et al., 2006; Garigan et al., 2002; Herndon et al., 2002; Johnston et al., 2008; Podshivalova, Kerr, & Kenyon, 2017; Zhao et al., 2017). In aged worms, the number of myobrils diminishes and the pharyngeal muscles exhibit an abnormal appearance (Chow et al., 2006; Garigan et al., 2002; Herndon et al., 2002; Podshivalova et al., 2017; Zhao et al., 2017). Other age‐dependent degenerative structural changes, such as darkening of the pharyngeal regions, also occur (Chow et al., 2006; Garigan et al., 2002; Herndon et al., 2002). These age‐dependent functional and structural abnormalities of pharyngeal muscles appear to be associated with bacterial accumulation in the terminal bulb of old worms (Podshivalova et al., 2017; Zhao et al., 2017). This is consistent with the susceptibility of old worms to infection by pathogenic bacteria, including Pseudomonas aeruginosa (Youngman, Rogers, & Kim, 2011). Degenerative changes in the muscle morphology of C. elegans have been conrmed via quantitative analytical methods (Johnston et al., 2008; Shamir et al., 2009). Thus, the loss of muscular integrity in old C. elegans results in age‐related changes, including impaired motility and increased susceptibility to infection, as in humans.
2.2. Age‐dependent changes at the cellular level
2.2.1. Nucleus
The integrity of C. elegans nuclei generally diminishes with age, although the degree of deterioration may vary among tissues (Garigan et al., 2002; Golden et al., 2007; Haithcock et al., 2005; Herndon et al., 2002; McGee et al., 2011; Figure 3). The intestine of adult C. elegans has approximately 30–34 cells whose nuclei start to degenerate at Day 12 of adulthood, resulting in a reduction in their number and size (McGee et al., 2011). The nuclei of muscle cells also develop dark patches with age (Haithcock et al., 2005; Herndon et al., 2002), and the relative size of the nucleoli increases with aging (Herndon et al., 2002; Tiku et al., 2017). Likewise, hypodermal and pharyngeal cell nuclei exhibit age‐associated abnormalities (Haithcock et al., 2005). The nuclear lamina, which supports the structure of nucleus, displays irregular peripheral structures in the body wall muscle, hypodermal, and intestinal cells (Haithcock et al., 2005). The space between the nuclei of syncytial germ cells increases, and the number of the nuclei decreases during aging (Garigan et al., 2002). Overall, nuclei in C. elegans undergo age‐dependent structural changes to varying degrees.
Figure 3.
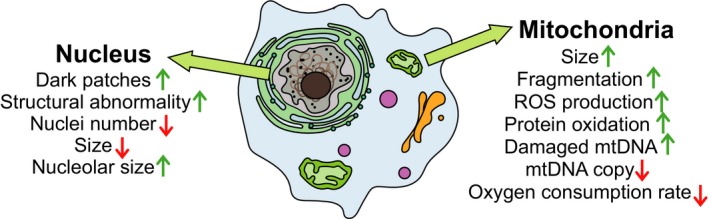
Age‐related changes at the cellular level. A schematic diagram that shows changes in the nuclei and mitochondria of old Caenorhabditis elegans cells
2.2.2. Mitochondria
Mitochondria undergo age‐dependent structural and functional changes as well (Gruber et al., 2011; Yasuda et al., 2006; Figure 3). For example, aged mitochondria in the body wall muscle cells become enlarged and swollen. These changes seem to be caused by mitochondrial fusion rather than an increase in the size of individual mitochondria because the total mitochondrial mass does not change during aging (Yasuda et al., 2006). Old worms also display fragmented mitochondria (Hahm, Kim, DiLoreto, & Shi, 2015; Regmi, Rolland, & Conradt, 2014; Roux, Langhans, Huynh, & Kenyon, 2016), and this correlates with uncoordinated locomotion in aged C. elegans (Hahm et al., 2015). Mitochondria are the major organelles that produce reactive oxygen species (ROS) that oxidize proteins and induce structural DNA damage, thus impairing the function of mitochondria (Birben, Sahiner, Sackesen, Erzurum, & Kalayci, 2012). Increases in the level of ROS, carbonylated proteins, and damaged mitochondrial DNA have been reported during aging in C. elegans (Berlett & Stadtman, 1997; Cui, Kong, & Zhang, 2012; Gruber et al., 2011; Stadtman & Berlett, 1997; Yasuda et al., 2006). Consistent with these changes, mitochondrial DNA copy numbers (Gruber et al., 2011) and oxygen consumption rates (Gruber et al., 2011; Yasuda et al., 2006) decrease during aging. Thus, age‐dependent declines in the integrity of mitochondria appear to be associated with functional impairments.
2.2.3. Endoplasmic reticulum
The endoplasmic reticulum (ER) is an organelle where proteins and lipids are synthesized and modied and serves as a calcium reservoir. The ER is also a site where the unfolded protein response (UPR) or ERUPR occurs (Smith & Wilkinson, 2017). When misfolded proteins accumulate under various stress conditions, the ERUPR is activated to degrade the misfolded proteins through three key pathways including the protein kinase RNA‐like ER kinase (PERK), the inositol‐requiring enzyme‐1 (IRE‐1)/X‐box binding protein‐1 (XBP‐1), and the activating transcription factor 6 (ATF6) pathways. The capacity of the ERUPR seems to be reduced during aging, as has been determined by measuring the levels of HSP‐4, a target chaperone of IRE‐1/XBP‐1, under ER stress conditions (Ben‐Zvi, Miller, & Morimoto, 2009; Maity et al., 2016; Martinez, Duran‐Aniotz, Cabral‐Miranda, Vivar, & Hetz, 2017; Taylor & Dillin, 2013). In addition, the overexpression of XBP‐1 in neurons is sucient for enhancing ER stress resistance and longevity (Taylor & Dillin, 2013). Overall, these results suggest that the ERUPR plays a functional role in aging and lifespan.
2.3. Age‐dependent changes at the molecular level
2.3.1. Gene expression
It is not surprising that the expression of many genes is altered during C. elegans aging (Cortes‐Lopez et al., 2018; de Lencastre et al., 2010; Golden, Hubbard, Dando, Herren, & Melov, 2008; Golden & Melov, 2004; Kato, Chen, Inukai, Zhao, & Slack, 2011; Lund et al., 2002; Rangaraju et al., 2015; Vinuela, Snoek, Riksen, & Kammenga, 2012; Youngman et al., 2011). For example, mRNA levels of heat‐shock protein‐encoding genes increase until reaching midlife and then decrease in old age (Golden & Melov, 2004; Golden et al., 2008; Lund et al., 2002). In addition, the expression of many collagen genes, whose overexpression increases lifespan, decreases with age (Ewald, Landis, Porter Abate, Murphy, & Blackwell, 2015; Golden et al., 2008). Many other aging‐associated gene expression changes have been identied, but we still do not fully understand the eects of global gene expression changes on aging. It will be important to determine whether these gene expression changes have causative roles in aging or are the consequences of aging.
The expression of many noncoding RNAs, including microRNAs (miRNAs), Piwi‐interacting RNAs (piRNAs), transfer RNAs (tRNAs), ribosomal RNAs (rRNAs), small nucleolar RNAs (snoRNAs), and circular RNAs (circRNAs), changes with age (Cortes‐Lopez et al., 2018; de Lencastre et al., 2010; Ibanez‐Ventoso et al., 2006; Inukai & Slack, 2013; Kato & Slack, 2013; Kato et al., 2011; Lucanic et al., 2013). The overall levels of miRNAs tend to decrease during aging (de Lencastre et al., 2010; Ibanez‐Ventoso et al., 2006), and this appears to be caused by age‐dependent reductions in the mRNA levels of dcr‐1/Dicer (Mori et al., 2012) and alg‐1/Argonaut (Inukai, Pincus, de Lencastre, & Slack, 2018). The mRNA levels of alg‐1 appear to be decreased by miR‐71, whose levels increase with age (de Lencastre et al., 2010; Inukai et al., 2018). Interestingly, miR‐71 is necessary and sucient for longevity (de Lencastre et al., 2010), suggesting a role for miR‐71 in aging through its regulation of alg‐1. The overall levels of tRNAs, rRNAs, and snoRNAs increase with age, whereas the levels of piRNAs decrease with age (Kato et al., 2011). circRNAs are a recently identied type of noncoding RNAs, whose ends are covalently linked and are implicated in transcriptional regulation (Knupp & Miura, 2018). A recent study indicated that the overall levels of circRNAs increase during C. elegans aging (Cortes‐Lopez et al., 2018). Whether various noncoding RNAs, other than miRNAs, act as biomarkers of aging and/or aect lifespan remains to be determined.
2.3.2. RNA quality
RNA quality control mechanisms decline during aging in C. elegans (Heintz et al., 2017; Son et al., 2017). Nonsense‐mediated mRNA decay (NMD) is a cellular protective pathway that degrades mRNAs containing premature termination codons (PTCs), as these generate potentially toxic truncated proteins (Miller & Pearce, 2014). NMD activity decreases during aging in the tissues of various organs, including the muscles, hypodermis, and intestine (Son et al., 2017). In addition, mRNA splicing delity decreases with age, as shown by the increased levels of introns and unannotated regions in the mRNAs of aged worms (Heintz et al., 2017). Age‐dependent declines in RNA quality control negatively aect longevity, because enhancing NMD activity or overexpressing key splicing factors increases lifespan (Heintz et al., 2017; Son et al., 2017).
mRNAs that are destined to be degraded, such as those targeted by miRNAs, are stored and degraded in processing bodies (P‐bodies; Chantarachot & Bailey‐Serres, 2018). In the P‐body, most mRNAs are degraded by decapping enzymes and exonucleases. DCP1/decapping protein 1 and DCP2/decapping protein 2 remove the 5' cap of target mRNAs, and then the XRN1/5'‐3' exoribonuclease fully degrades the mRNAs (Labno, Tomecki, & Dziembowski, 2016). Interestingly, the inhibition of RNA decay by RNAi targeting xrn‐1/XRN1 increases the number of dcp‐1::gfp granules, which are also increased during aging (Rousakis et al., 2014). These results suggest that RNA decay eciency decreases during aging. Furthermore, perturbation of the RNA decay components aects lifespan; genetic inhibition of dcap‐1/DCP1, dcap‐2/DCP2, or xrn‐1/XRN1 decreases lifespan in C. elegans (Rousakis et al., 2014). In contrast to P‐body granules, aging does not aect the number of stress granules that are induced by various stresses (Chantarachot & Bailey‐Serres, 2018; Rousakis et al., 2014). Overall, the capacity for mRNA decay and proper mRNA metabolism decreases with age.
2.3.3. Age pigments
Organisms contain various types of fluorescent materials, including aromatic amino acids, lipofuscin, advanced glycation end products, and anthranilic acids, several of which accumulate with age. Lipofuscin is well known as an “age pigment,” as its level increases with aging in many organisms (Gray & Woulfe, 2005; Klass, 1977; Xu, Chen, Manivannan, Lois, & Forrester, 2008). Age pigments can be quantitated using in vivo uorescence spectroscopy that detects a broad range of excitation/emission spectra. The accumulation of age pigments is gradual until early adulthood (Days 5–10) and rapid in midlife (Days 10–15; Gerstbrein, Stamatas, Kollias, & Driscoll, 2005). Furthermore, the absolute amount of generated age pigments negatively correlates with health and lifespan, and possibly predicts the lifespan of C. elegans (Gerstbrein et al., 2005; Pincus, Smith‐Vikos, & Slack, 2011). However, a study by the Gems group challenges the view of lipofuscin as an age pigment in C. elegans (Coburn et al., 2013). They have shown that kynurenines, which emit blue uorescence, exhibit characteristics similar to those of lipofuscin. Kynurenine levels do not increase gradually with aging, but they are precipitously elevated upon the death of the organism, resulting in sudden bursts of blue uorescence (Coburn et al., 2013). A recent study proposes that substances emitting red signals increase gradually with age (Pincus, Mazer, & Slack, 2016), whereas those emitting blue signals increase only marginally during aging and dramatically after death (Coburn et al., 2013; Pincus et al., 2016). Further characterization of the chemical nature of age pigments is required to resolve this issue.
2.3.4. Protein aggregation
Protein homeostasis generally declines during aging, and this decline is associated with age‐related diseases. For example, aggregated proteins contribute to the pathology of neurodegenerative diseases, such as Alzheimer's, Parkinson's, and polyglutamine diseases in humans (Nollen et al., 2004; Taylor, Hardy, & Fischbeck, 2002). C. elegans displays an aging‐associated aggregation of transgene‐encoded polyQ and α‐synuclein, which are models for Huntington's and Parkinson's diseases, respectively (Morley, Brignull, Weyers, & Morimoto, 2002; van Ham et al., 2008). Many inherent proteins become insoluble and form aggregates in aged worms (David et al., 2010; Reis‐Rodrigues et al., 2012; Walther et al., 2015). Genetically inhibiting each of many aggregation‐prone protein‐coding genes increases lifespan (Reis‐Rodrigues et al., 2012), suggesting that protein aggregation has a negative eect on longevity. Thus, aging processes are accompanied by an increase in the accumulation of nonfunctional and insoluble protein aggregates.
2.3.5. Proteomic and metabolomic changes
“Omics” techniques allow researchers to examine universal changes in genes, RNAs, proteins, and metabolites during aging. Using stable isotope labeling with amino acids in cell culture (SILAC) followed by LC‐MS/MS methods, C. elegans proteins whose levels increase or decrease with age have been reported (Narayan et al., 2016). During aging, proteins that likely function in nucleosome assembly, ER‐nuclear signaling, and the response to unfolded proteins increase, whereas the abundance of proteins involved in metabolism such as fatty acid, carbohydrate, and amino acid metabolism decreases (Narayan et al., 2016). A similar observation has been reported showing that overall levels of fatty acid metabolic proteins decrease (Copes et al., 2015). Because altering fat metabolism has huge impacts on aging and lifespan (Lee et al., 2015; Lemieux & Ashra, 2016), the proper maintenance of fat metabolism in old age will be benecial for longevity.
Levels of metabolites such as amino acids change with age as well. A study using ultra performance liquid chromatography (UPLC)‐MS/MS indicated that the levels of all of the amino acids except glycine and aspartic acid increase at an early age (approximately 3 days of adulthood) and then decrease sharply afterward (Gao et al., 2017). Another study using proton NMR spectroscopy reported that the levels of three amino acids (glycine, serine, and tyrosine) increase during aging, whereas the levels of four amino acids (alanine, asparagine, glutamate, and glutamine) decrease during aging (Davies, Bundy, & Leroi, 2015). Importantly, some of these age‐dependent changes reflect the biological age of worms (Davies et al., 2015), and treatment with any amino acids except phenylalanine and aspartate increases lifespan (Edwards et al., 2015). Further studies are required to comprehensively understand the roles of age‐dependent changes in amino acid levels in aging and longevity.
3. THE ROLE OF INSULIN/IGF‐1 SIGNALING AND DIETARY RESTRICTION IN AGE‐ASSOCIATED CHANGES IN C. elegans
Many intrinsic and extrinsic lifespan regulatory factors, including insulin/IGF‐1 signaling (IIS), target of rapamycin (TOR), mitochondrial respiration, reproduction, and dietary restriction (DR), have been identied using C. elegans. Among them, here we review age‐related changes under reduced IIS and DR conditions, which are two representative regimens for extending lifespan and delaying aging. The IIS pathway is an evolutionarily conserved aging regulatory pathway (Altintas, Park, & Lee, 2016; Kenyon, 2010). Mutations in the daf‐2/insulin/IGF‐1 receptor double the lifespan of C. elegans by upregulating various transcription factors, including DAF‐16/FOXO, heat‐shock factor 1 (HSF‐1), and SKN‐1/NRF. DR also promotes longevity across phyla (Kapahi, Kaeberlein, & Hansen, 2017). Caenorhabditis elegans eat‐2 mutants, which display reduced feeding rates and food intake, are a widely used DR model (Lakowski & Hekimi, 1998; Raizen, Lee, & Avery, 1995). Reduced IIS and DR delay various age‐related degenerative changes. The motility of worms, as measured by maximum velocity, is prolonged in daf‐2 and eat‐2 mutants compared to wild‐type animals (Hahm et al., 2015; Huang, Xiong, & Kornfeld, 2004), although daf‐2 mutants display reduced spontaneous movements for a relatively long time (Bansal, Zhu, Yen, & Tissenbaum, 2015). Additionally, daf‐2 mutants are resistant to bacterial colonization (Podshivalova et al., 2017), one of the main causes of early death in C. elegans (Garigan et al., 2002; Zhao et al., 2017). Moreover, the reproductive span of daf‐2 and eat‐2 mutant hermaphrodites (Hughes et al., 2007; Luo et al., 2010; Luo, Shaw, Ashraf, & Murphy, 2009) is longer than that of wild‐type worms, although the total number of progeny produced by daf‐2 mutants is reduced (Hughes et al., 2007). Mutations in daf‐2 also prolong the reproductive period of males (Chatterjee et al., 2013). Furthermore, daf‐2 mutants exhibit reduced uterine mass (McGee et al., 2012) and delayed deterioration of germ cells (Garigan et al., 2002) compared to wild‐type worms. Age‐dependent increases in neuronal abnormalities, including neurite branching and irregularly shaped soma in mechanosensory neurons, are delayed in daf‐2 mutants compared to the wild type (Pan et al., 2011; Tank et al., 2011). Axon regeneration capacity is also prolonged in daf‐2 mutants compared to the wild‐type animals (Byrne et al., 2014). In contrast, neither neurite branching nor axon regeneration appears to be delayed by mutations in eat‐2 (Byrne et al., 2014; Tank et al., 2011). Neuronal functions, such as associative learning ability, are maintained longer in both daf‐2 and eat‐2 mutants than in wild‐type worms (Kauman et al., 2010).
Nuclear integrity in diverse tissues is maintained for a longer period of time in daf‐2 mutants compared to in wild‐type worms, as exemplied by the delayed degeneration of daf‐2 mutant nuclear lamina (Haithcock et al., 2005). Mutations in age‐1, the gene that encodes the phosphoinositide 3‐kinase acting immediately downstream of DAF‐2, delay nuclear degeneration in muscle (Herndon et al., 2002). In addition, the age‐related loss of intestinal nuclei (McGee et al., 2011) and hypodermal nuclei (Golden et al., 2007) is reduced in daf‐2 mutants. Nucleolar size is negatively correlated with longevity and is reduced in daf‐2 and eat‐2 mutants (Tiku et al., 2017). In addition, daf‐2 mutants display higher ATP levels and antioxidant activity and lower ROS levels than wild‐type worms (Zarse et al., 2012). This indicates that mitochondrial activity is increased, while ROS levels are reduced, likely because of an upregulation of antioxidants in daf‐2 mutants (Zarse et al., 2012). RNAi knockdown of daf‐2 during adulthood, however, transiently increases the level of ROS, which acts as a signal for longevity (Zarse et al., 2012), suggesting a stage‐specic regulation of ROS levels by IIS.
Mutations in daf‐2 delay the age‐associated accumulation of abnormal macromolecules, including mRNAs with PTCs (Son et al., 2017) and insoluble proteins (David et al., 2010). Age pigment accumulation is reduced both in daf‐2 mutants and in eat‐2 mutants (Gerstbrein et al., 2005). Overall, the delay in age‐related degenerative changes caused by daf‐2 mutations and DR is consistent with the idea that reduced IIS and DR confer healthy aging with a long lifespan.
4. POTENTIAL BIOMARKERS OF AGING
Biomarkers of aging reflect the physiological and functional age of organisms (Baker & Sprott, 1988). An important way to validate biomarkers of aging is by testing whether they predict the remaining lifespan of an organism (Baker & Sprott, 1988; Johnson, 2006). Several biomarkers predictive of lifespan have been reported in C. elegans (Table 1). First, physiological functions are indicators of the biological age of worms. For example, worms that display fast locomotion during early adulthood (Hahm et al., 2015; Huang et al., 2004; Pincus et al., 2011) or maintain their youth speed in middle age (Hsu, Feng, Hsieh, & Xu, 2009) tend to have longer lifespans compared with slow‐moving worms. The rate of pharyngeal pumping and the number of progeny after mating also positively correlate with remaining lifespan (Huang et al., 2004; Pickett et al., 2013). Second, nucleolar size has a negative correlation with longevity (Tiku et al., 2017). Importantly, nucleolar size is reduced by interventions that extend lifespan in Drosophila melanogaster, mice, and possibly humans as well (Tiku et al., 2017). Third, molecular genetic changes are used as biomarkers of aging. The expression levels of several genes, including a small heat‐shock protein (hsp‐16.2) after transient heat shock (Rea, Wu, Cypser, Vaupel, & Johnson, 2005) and superoxide dismutase 3 (sod‐3; Sanchez‐Blanco & Kim, 2011), are positively correlated with a long lifespan. In addition, the expression levels of certain miRNAs increase or decrease as worms grow old. Changes in these miRNA levels successfully predict the remaining lifespan of C. elegans (Pincus et al., 2011). Fourth, the amount of autofluorescent age pigments has been used as biomarkers of aging. These age pigments accumulate in old worms, and worms with a high accumulation of age pigments early in adulthood tend to live short lives (Pincus et al., 2016, 2011 ). Finally, mitochondrial functional integrity also predicts lifespan. Superoxide bursts (Cheng et al., 2014; Shen et al., 2014) or pH changes (Schwarzlander et al., 2014) in the mitochondria, referred to as “mitoashes,” early in adulthood (Day 3) show a negative correlation with lifespan. However, morphological changes of the mitochondria do not correlate with lifespan (Regmi et al., 2014) and therefore do not seem to be suitable as a biomarker of aging. Overall, physiological and molecular factors, age pigments, and mitochondrial integrity parameters are important biomarkers of aging in C. elegans. Whether these biomarkers are applicable to other organisms needs to be validated. After validation, these universal biomarkers of aging will be very helpful in understanding the mechanistic and physiological causes and consequences of the aging process in multiple species, including mammals.
Table 1.
List of potential biomarkers of aging in Caenorhabditis elegans
| Biomarkers of aging | Correlation with lifespan | References | |
|---|---|---|---|
| Physiological markers | Locomotion | Positive | Hahm et al. (2015), Hsu et al. (2009), Huang et al. (2004) and Pincus et al. (2011) |
| Pharyngeal pumping rate | Positive | Huang et al. (2004) | |
| Progeny number | Positive | Pickett et al. (2013) | |
| Cellular markers | Nucleolar size | Negative | Tiku et al. (2017) |
| Molecular markers | sod –3 expression | Positive | Sanchez‐Blanco and Kim (2011) |
| hsp –16 .2 expression | Positive | Rea et al. (2005) | |
| miRNAs | Positive or negative | Pincus et al. (2011) | |
| Age pigments | Negative | Pincus et al. (2011) and Pincus et al. (2016) | |
| Mitochondrial activity | Negative | Cheng et al. (2014) and Shen et al. (2014) |
5. CONCLUSIONS
Risks for many chronic diseases, including cancer, cardiovascular diseases, and neurodegenerative diseases, exponentially increase with age. Research aiming at understanding the mechanisms of aging or delaying aging can be benecial for reducing these risks. The development of biomarkers of aging is crucial for aging research, as they reflect the biological age of an organism. In this review, we described age‐related changes that can potentially be used as biomarkers of aging in C. elegans.
So far, diverse, promising biomarkers of aging have been identied in C. elegans. Several limitations, however, need to be addressed in future research. First, the validity of several potential biomarkers of aging is controversial, and, therefore, further studies resolving these discrepancies are needed. Second, a biomarker of aging that reports one physiological or physical state usually does not reflect the general biological age of an organism. For example, a single worm that displays increased overall motility may have a reduced feeding rate. Therefore, it will be important to use multiple biomarkers of aging collectively, from physiological markers to molecular markers, to precisely estimate the biological age of an organism. In this sense, the standardization of which combinations of biomarkers are the most eective for predicting the biological age of worms will be helpful for the C. elegans aging research eld.
Recent studies have also demonstrated changes in the levels of proteins and metabolites as well as RNAs during aging. As aging is a stochastic process that inuences many dierent biological components, the analysis of these factors globally using omics technologies will be crucial for the progress of the aging research eld. Moreover, the integration of omics data with physiological and molecular markers of aging will provide even more powerful tools toward a better understanding of the mysteries of organismal aging.
CONFLICT OF INTEREST
None declared.
ACKNOWLEDGMENTS
We thank all Lee laboratory members for their help and discussion. This work was supported by the Korean Government (MSIP) through the National Research Foundation of Korea (NRF; NRF‐2016R1E1A1A01941152) to S‐J. V. L.
Son HG, Altintas O, Kim EJE, Kwon S, Lee S‐JV. Age‐dependent changes and biomarkers of aging in Caenorhabditis elegans . Aging Cell. 2019;18:e12853 10.1111/acel.12853
REFERENCES
- Albertson, D. G. , & Thomson, J. N. (1976). The pharynx of Caenorhabditis elegans . Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, 275(938), 299–325. 10.1098/rstb.1976.0085 [DOI] [PubMed] [Google Scholar]
- Altintas, O. , Park, S. , & Lee, S. J. (2016). The role of insulin/IGF‐1 signaling in the longevity of model invertebrates, C. elegans and D. melanogaster . BMB Reports, 49(2), 81–92. 10.5483/BMBRep.2016.49.2.261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andux, S. , & Ellis, R. E. (2008). Apoptosis maintains oocyte quality in aging Caenorhabditis elegans females. PLoS Genetics, 4(12), e1000295 10.1371/journal.pgen.1000295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker, G. T. 3rd , & Sprott, R. L. (1988). Biomarkers of aging. Experimental Gerontology, 23(4–5), 223–239. 10.1016/0531-5565(88)90025-3 [DOI] [PubMed] [Google Scholar]
- Bansal, A. , Zhu, L. J. , Yen, K. , & Tissenbaum, H. A. (2015). Uncoupling lifespan and healthspan in Caenorhabditis elegans longevity mutants. Proceedings of the National Academy of Sciences of the United States of America, 112(3), E277–E286. 10.1073/pnas.1412192112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ben‐Zvi, A. , Miller, E. A. , & Morimoto, R. I. (2009). Collapse of proteostasis represents an early molecular event in Caenorhabditis elegans aging. Proceedings of the National Academy of Sciences of the United States of America, 106(35), 14914–14919. 10.1073/pnas.0902882106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berlett, B. S. , & Stadtman, E. R. (1997). Protein oxidation in aging, disease, and oxidative stress. Journal of Biological Chemistry, 272(33), 20313–20316. 10.1074/jbc.272.33.20313 [DOI] [PubMed] [Google Scholar]
- Birben, E. , Sahiner, U. M. , Sackesen, C. , Erzurum, S. , & Kalayci, O. (2012). Oxidative stress and antioxidant defense. World Allergy Organization Journal, 5(1), 9–19. 10.1097/WOX.0b013e3182439613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Byrne, A. B. , Walradt, T. , Gardner, K. E. , Hubbert, A. , Reinke, V. , & Hammarlund, M. (2014). Insulin/IGF1 signaling inhibits age‐dependent axon regeneration. Neuron, 81(3), 561–573. 10.1016/j.neuron.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chantarachot, T. , & Bailey‐Serres, J. (2018). Polysomes, stress granules, and processing bodies: A dynamic triumvirate controlling cytoplasmic mRNA fate and function. Plant Physiology, 176(1), 254–269. 10.1104/pp.17.01468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chatterjee, I. , Ibanez‐Ventoso, C. , Vijay, P. , Singaravelu, G. , Baldi, C. , Bair, J. ,… Singson, A. (2013). Dramatic fertility decline in aging C. elegans males is associated with mating execution decits rather than diminished sperm quality. Experimental Gerontology, 48(11), 1156–1166. 10.1016/j.exger.2013.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, C. H. , Chen, Y. C. , Jiang, H. C. , Chen, C. K. , & Pan, C. L. (2013). Neuronal aging: Learning from C. elegans . Journal of Molecular Signaling, 8(1), 14 10.1186/1750-2187-8-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, H. , Wang, W. , Wang, X. , Sheu, S. S. , Dirksen, R. T. , & Dong, M. Q. (2014). Cheng et al. reply. Nature, 514(7523), E14–15. 10.1038/nature13859 [DOI] [PubMed] [Google Scholar]
- Chew, Y. L. , Fan, X. , Gotz, J. , & Nicholas, H. R. (2013). Aging in the nervous system of Caenorhabditis elegans . Communicative and Integrative Biology, 6(5), e25288 10.4161/cib.25288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow, D. K. , Glenn, C. F. , Johnston, J. L. , Goldberg, I. G. , & Wolkow, C. A. (2006). Sarcopenia in the Caenorhabditis elegans pharynx correlates with muscle contraction rate over lifespan. Experimental Gerontology, 41(3), 252–260. 10.1016/j.exger.2005.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn, C. , Allman, E. , Mahanti, P. , Benedetto, A. , Cabreiro, F. , Pincus, Z. , … Gems, D. (2013). Anthranilate uorescence marks a calcium‐propagated necrotic wave that promotes organismal death in C. elegans . PLoS Biology, 11(7), e1001613 10.1371/journal.pbio.1001613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins, J. J. , Huang, C. , Hughes, S. , & Kornfeld, K. (2008). The measurement and analysis of age‐related changes in Caenorhabditis elegans . WormBook, 24, 1–21. 10.1895/wormbook.1.137.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copes, N. , Edwards, C. , Chaput, D. , Saifee, M. , Barjuca, I. , Nelson, D. , … Bradshaw, P. C. (2015). Metabolome and proteome changes with aging in Caenorhabditis elegans . Experimental Gerontology, 72, 67–84. 10.1016/j.exger.2015.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortes‐Lopez, M. , Gruner, M. R. , Cooper, D. A. , Gruner, H. N. , Voda, A. I. , van der Linden, A. M. , & Miura, P. (2018). Global accumulation of circRNAs during aging in Caenorhabditis elegans . BMC Genomics, 19(1), 8 10.1186/s12864-017-4386-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui, H. , Kong, Y. , & Zhang, H. (2012). Oxidative stress, mitochondrial dysfunction, and aging. Journal of Signal Transduction, 2012, 646354 10.1155/2012/646354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David, D. C. , Ollikainen, N. , Trinidad, J. C. , Cary, M. P. , Burlingame, A. L. , & Kenyon, C. (2010). Widespread protein aggregation as an inherent part of aging in C. elegans . PLoS Biology, 8(8), e1000450 10.1371/journal.pbio.1000450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies, S. K. , Bundy, J. G. , & Leroi, A. M. (2015). Metabolic youth in middle age: Predicting aging in Caenorhabditis elegans using metabolomics. Journal of Proteome Research, 14(11), 4603–4609. 10.1021/acs.jproteome.5b00442 [DOI] [PubMed] [Google Scholar]
- de Lencastre, A. , Pincus, Z. , Zhou, K. , Kato, M. , Lee, S. S. , & Slack, F. J. (2010). MicroRNAs both promote and antagonize longevity in C. elegans . Current Biology, 20(24), 2159–2168. 10.1016/j.cub.2010.11.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards, C. , Caneld, J. , Copes, N. , Brito, A. , Rehan, M. , Lipps, D. , … Bradshaw, P. C. (2015). Mechanisms of amino acid‐mediated lifespan extension in Caenorhabditis elegans . BMC Genetics, 16, 8 10.1186/s12863-015-0167-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewald, C. Y. , Landis, J. N. , Porter Abate, J. , Murphy, C. T. , & Blackwell, T. K. (2015). Dauer‐independent insulin/IGF‐1‐signalling implicates collagen remodelling in longevity. Nature, 519(7541), 97–101. 10.1038/nature14021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao, A. W. , Chatzispyrou, I. A. , Kamble, R. , Liu, Y. J. , Herzog, K. , Smith, R. L. , … Houtkooper, R. H. (2017). A sensitive mass spectrometry platform identies metabolic changes of life history traits in C. elegans . Scientic Reports, 7(1), 2408 10.1038/s41598-017-02539-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garigan, D. , Hsu, A. L. , Fraser, A. G. , Kamath, R. S. , Ahringer, J. , & Kenyon, C. (2002). Genetic analysis of tissue aging in Caenorhabditis elegans: A role for heat‐shock factor and bacterial proliferation. Genetics, 161(3), 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstbrein, B. , Stamatas, G. , Kollias, N. , & Driscoll, M. (2005). In vivo spectrouorimetry reveals endogenous biomarkers that report healthspan and dietary restriction in Caenorhabditis elegans . Aging Cell, 4(3), 127–137. 10.1111/j.1474-9726.2005.00153.x [DOI] [PubMed] [Google Scholar]
- Glenn, C. F. , Chow, D. K. , David, L. , Cooke, C. A. , Gami, M. S. , Iser, W. B. , … Wolkow, C. A. (2004). Behavioral decits during early stages of aging in Caenorhabditis elegans result from locomotory decits possibly linked to muscle frailty. Journals of Gerontology. Series A, Biological Sciences and Medical Sciences, 59(12), 1251–1260. 10.1093/gerona/59.12.1251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, T. R. , Beckman, K. B. , Lee, A. H. , Dudek, N. , Hubbard, A. , Samper, E. , & Melov, S. (2007). Dramatic age‐related changes in nuclear and genome copy number in the nematode Caenorhabditis elegans . Aging Cell, 6(2), 179–188. 10.1111/j.1474-9726.2007.00273.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, T. R. , Hubbard, A. , Dando, C. , Herren, M. A. , & Melov, S. (2008). Age‐related behaviors have distinct transcriptional proles in Caenorhabditis elegans . Aging Cell, 7(6), 850–865. 10.1111/j.1474-9726.2008.00433.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golden, T. R. , & Melov, S. (2004). Microarray analysis of gene expression with age in individual nematodes. Aging Cell, 3(3), 111–124. 10.1111/j.1474-9728.2004.00095.x [DOI] [PubMed] [Google Scholar]
- Gray, D. A. , & Woulfe, J. (2005). Lipofuscin and aging: A matter of toxic waste. Science of Aging Knowledge Environment, 2005(5), re1 10.1126/sageke.2005.5.re1 [DOI] [PubMed] [Google Scholar]
- Gruber, J. , Ng, L. F. , Fong, S. , Wong, Y. T. , Koh, S. A. , Chen, C. B. , … Halliwell, B. (2011). Mitochondrial changes in ageing Caenorhabditis elegans–what do we learn from superoxide dismutase knockouts? PLoS One, 6(5), e19444 10.1371/journal.pone.0019444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo, X. , Navetta, A. , Gualberto, D. G. , & Garcia, L. R. (2012). Behavioral decay in aging male C. elegans correlates with increased cell excitability. Neurobiology of Aging, 33(7), 1483.e5–1423.e23. 10.1016/j.neurobiolaging.2011.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahm, J. H. , Kim, S. , DiLoreto, R. , & Shi, C. (2015). C. elegans maximum velocity correlates with healthspan and is maintained in worms with an insulin receptor mutation. Nature Communications, 6, 8919 10.1038/ncomms9919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haithcock, E. , Dayani, Y. , Neufeld, E. , Zahand, A. J. , Feinstein, N. , Mattout, A. , … Liu, J. (2005). Age‐related changes of nuclear architecture in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 102(46), 16690–16695. 10.1073/pnas.0506955102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarlund, M. , Nix, P. , Hauth, L. , Jorgensen, E. M. , & Bastiani, M. (2009). Axon regeneration requires a conserved MAP kinase pathway. Science, 323(5915), 802–806. 10.1126/science.1165527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heintz, C. , Doktor, T. K. , Lanjuin, A. , Escoubas, C. C. , Zhang, Y. , Weir, H. J. , … Mair, W. B. (2017). Splicing factor 1 modulates dietary restriction and TORC1 pathway longevity in C. elegans . Nature, 541(7635), 102–106. 10.1038/nature20789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herdon, L. A. , Wolkow, C. A. , Driscoll, M. , & Hall, D. H. (2017). Eects of ageing on the basic biology and anatomy of C. elegans In Olsen A., & Gill M. S. (Eds.), Ageing: Lessons from C. elegans (pp. 9–39). Cham, Switzerland: Springer International Publishing; 10.1007/978-3-319-44703-2 [DOI] [Google Scholar]
- Herndon, L. A. , Schmeissner, P. J. , Dudaronek, J. M. , Brown, P. A. , Listner, K. M. , Sakano, Y. , … Driscoll, M. (2002). Stochastic and genetic factors inuence tissue‐specic decline in ageing C. elegans . Nature, 419(6909), 808–814. 10.1038/nature01135 [DOI] [PubMed] [Google Scholar]
- Hsu, A. L. , Feng, Z. , Hsieh, M. Y. , & Xu, X. Z. (2009). Identication by machine vision of the rate of motor activity decline as a lifespan predictor in C. elegans . Neurobiology of Aging, 30(9), 1498–1503. 10.1016/j.neurobiolaging.2007.12.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang, C. , Xiong, C. , & Kornfeld, K. (2004). Measurements of age‐related changes of physiological processes that predict lifespan of Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 101(21), 8084–8089. 10.1073/pnas.0400848101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes, S. E. , Evason, K. , Xiong, C. , & Kornfeld, K. (2007). Genetic and pharmacological factors that inuence reproductive aging in nematodes. PLoS Genetics, 3(2), e25 10.1371/journal.pgen.0030025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez‐Ventoso, C. , Yang, M. , Guo, S. , Robins, H. , Padgett, R. W. , & Driscoll, M. (2006). Modulated microRNA expression during adult lifespan in Caenorhabditis elegans . Aging Cell, 5(3), 235–246. 10.1111/j.1474-9726.2006.00210.x [DOI] [PubMed] [Google Scholar]
- Inukai, S. , Pincus, Z. , de Lencastre, A. , & Slack, F. J. (2018). A microRNA feedback loop regulates global microRNA abundance during aging. RNA, 24(2), 159–172. 10.1261/rna.062190.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inukai, S. , & Slack, F. (2013). MicroRNAs and the genetic network in aging. Journal of Molecular Biology, 425(19), 3601–3608. 10.1016/j.jmb.2013.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, T. E. (2006). Recent results: Biomarkers of aging. Experimental Gerontology, 41(12), 1243–1246. 10.1016/j.exger.2006.09.006 [DOI] [PubMed] [Google Scholar]
- Johnston, J. , Iser, W. B. , Chow, D. K. , Goldberg, I. G. , & Wolkow, C. A. (2008). Quantitative image analysis reveals distinct structural transitions during aging in Caenorhabditis elegans tissues. PLoS One, 3(7), e2821 10.1371/journal.pone.0002821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapahi, P. , Kaeberlein, M. , & Hansen, M. (2017). Dietary restriction and lifespan: Lessons from invertebrate models. Ageing Research Reviews, 39, 3–14. 10.1016/j.arr.2016.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , Chen, X. , Inukai, S. , Zhao, H. , & Slack, F. J. (2011). Age‐associated changes in expression of small, noncoding RNAs, including microRNAs, in C. elegans . RNA, 17(10), 1804–1820. 10.1261/rna.2714411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato, M. , & Slack, F. J. (2013). Ageing and the small, non‐coding RNA world. Ageing Research Reviews, 12(1), 429–435. 10.1016/j.arr.2012.03.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauman, A. L. , Ashraf, J. M. , Corces‐Zimmerman, M. R. , Landis, J. N. , & Murphy, C. T. (2010). Insulin signaling and dietary restriction dierentially inuence the decline of learning and memory with age. PLoS Biology, 8(5), e1000372 10.1371/journal.pbio.1000372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenyon, C. J. (2010). The genetics of ageing. Nature, 464(7288), 504–512. 10.1038/nature08980 [DOI] [PubMed] [Google Scholar]
- Klass, M. R. (1977). Aging in the nematode Caenorhabditis elegans: Major biological and environmental factors inuencing life span. Mechanisms of Ageing and Development, 6(6), 413–429. 10.1016/0047-6374(77)90043-4 [DOI] [PubMed] [Google Scholar]
- Knupp, D. , & Miura, P. (2018). CircRNA accumulation: A new hallmark of aging? Mechanisms of Ageing and Development, 173, 71–79. 10.1016/j.mad.2018.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labno, A. , Tomecki, R. , & Dziembowski, A. (2016). Cytoplasmic RNA decay pathways – Enzymes and mechanisms. Biochimica et Biophysica Acta, 1863(12), 3125–3147. 10.1016/j.bbamcr.2016.09.023 [DOI] [PubMed] [Google Scholar]
- Lakowski, B. , & Hekimi, S. (1998). The genetics of caloric restriction in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 95(22), 13091–13096. 10.1073/pnas.95.22.13091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, D. , Jeong, D. E. , Son, H. G. , Yamaoka, Y. , Kim, H. , Seo, K. , … Lee, S. J. (2015). SREBP and MDT‐15 protect C. elegans from glucose‐induced accelerated aging by preventing accumulation of saturated fat. Genes and Development, 29(23), 2490–2503. 10.1101/gad.266304.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemieux, G. A. , & Ashra, K. (2016). Investigating connections between metabolism, longevity, and behavior in Caenorhabditis elegans . Trends in Endocrinology and Metabolism, 27(8), 586–596. 10.1016/j.tem.2016.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, J. , Zhang, B. , Lei, H. , Feng, Z. , Liu, J. , Hsu, A. L. , & Xu, X. Z. (2013). Functional aging in the nervous system contributes to age‐dependent motor activity decline in C. elegans . Cell Metabolism, 18(3), 392–402. 10.1016/j.cmet.2013.08.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucanic, M. , Graham, J. , Scott, G. , Bhaumik, D. , Benz, C. C. , Hubbard, A. , … Melov, S. (2013). Age‐related micro‐RNA abundance in individual C. elegans . Aging (Albany NY), 5(6), 394–411. 10.18632/aging.100564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund, J. , Tedesco, P. , Duke, K. , Wang, J. , Kim, S. K. , & Johnson, T. E. (2002). Transcriptional prole of aging in C. elegans . Current Biology, 12(18), 1566–1573. 10.1016/S0960-9822(02)01146-6 [DOI] [PubMed] [Google Scholar]
- Luo, S. , Kleemann, G. A. , Ashraf, J. M. , Shaw, W. M. , & Murphy, C. T. (2010). TGF‐beta and insulin signaling regulate reproductive aging via oocyte and germline quality maintenance. Cell, 143(2), 299–312. 10.1016/j.cell.2010.09.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo, S. , Shaw, W. M. , Ashraf, J. , & Murphy, C. T. (2009). TGF‐beta Sma/Mab signaling mutations uncouple reproductive aging from somatic aging. PLoS Genetics, 5(12), e1000789 10.1371/journal.pgen.1000789 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity, S. , Rajkumar, A. , Matai, L. , Bhat, A. , Ghosh, A. , Agam, G. , … Chakraborty, K. (2016). Oxidative homeostasis regulates the response to reductive endoplasmic reticulum stress through translation control. Cell Reports, 16(3), 851–865. 10.1016/j.celrep.2016.06.025 [DOI] [PubMed] [Google Scholar]
- Martinez, G. , Duran‐Aniotz, C. , Cabral‐Miranda, F. , Vivar, J. P. , & Hetz, C. (2017). Endoplasmic reticulum proteostasis impairment in aging. Aging Cell, 16(4), 615–623. 10.1111/acel.12599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M. D. , Day, N. , Graham, J. , & Melov, S. (2012). cep‐1/p53‐dependent dysplastic pathology of the aging C. elegans gonad. Aging (Albany NY), 4(4), 256–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGee, M. D. , Weber, D. , Day, N. , Vitelli, C. , Crippen, D. , Herndon, L. A. , … Melov, S. (2011). Loss of intestinal nuclei and intestinal integrity in aging C. elegans . Aging Cell, 10(4), 699–710. 10.1111/j.1474-9726.2011.00713.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller, J. N. , & Pearce, D. A. (2014). Nonsense‐mediated decay in genetic disease: Friend or foe? Mutation Research, Reviews in Mutation Research, 762, 52–64. 10.1016/j.mrrev.2014.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mori, M. A. , Raghavan, P. , Thomou, T. , Boucher, J. , Robida‐Stubbs, S. , Macotela, Y. , … Kahn, C. R. (2012). Role of microRNA processing in adipose tissue in stress defense and longevity. Cell Metabolism, 16(3), 336–347. 10.1016/j.cmet.2012.07.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley, J. F. , Brignull, H. R. , Weyers, J. J. , & Morimoto, R. I. (2002). The threshold for polyglutamine‐expansion protein aggregation and cellular toxicity is dynamic and inuenced by aging in Caenorhabditis elegans . Proceedings of the National Academy of Sciences of the United States of America, 99(16), 10417–10422. 10.1073/pnas.152161099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murakami, S. , & Murakami, H. (2005). The eects of aging and oxidative stress on learning behavior in C. elegans . Neurobiology of Aging, 26(6), 899–905. 10.1016/j.neurobiolaging.2004.08.007 [DOI] [PubMed] [Google Scholar]
- Narayan, V. , Ly, T. , Pourkarimi, E. , Murillo, A. B. , Gartner, A. , Lamond, A. I. , & Kenyon, C. (2016). Deep proteome analysis identies age‐related processes in C. elegans . Cell Systems, 3(2), 144–159. 10.1016/j.cels.2016.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nollen, E. A. , Garcia, S. M. , van Haaften, G. , Kim, S. , Chavez, A. , Morimoto, R. I. , & Plasterk, R. H. (2004). Genome‐wide RNA interference screen identies previously undescribed regulators of polyglutamine aggregation. Proceedings of the National Academy of Sciences of the United States of America, 101(17), 6403–6408. 10.1073/pnas.0307697101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan, C. L. , Peng, C. Y. , Chen, C. H. , & McIntire, S. (2011). Genetic analysis of age‐dependent defects of the Caenorhabditis elegans touch receptor neurons. Proceedings of the National Academy of Sciences of the United States of America, 108(22), 9274–9279. 10.1073/pnas.1011711108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett, C. L. , Dietrich, N. , Chen, J. , Xiong, C. , & Kornfeld, K. (2013). Mated progeny production is a biomarker of aging in Caenorhabditis elegans . G3 (Bethesda), 3(12), 2219–2232. 10.1534/g3.113.008664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus, Z. , Mazer, T. C. , & Slack, F. J. (2016). Autouorescence as a measure of senescence in C. elegans: Look to red, not blue or green. Aging (Albany NY), 8(5), 889–898. 10.18632/aging.100936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pincus, Z. , Smith‐Vikos, T. , & Slack, F. J. (2011). MicroRNA predictors of longevity in Caenorhabditis elegans . PLoS Genetics, 7(9), e1002306 10.1371/journal.pgen.1002306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podshivalova, K. , Kerr, R. A. , & Kenyon, C. (2017). How a mutation that slows aging can also disproportionately extend end‐of‐life decrepitude. Cell Reports, 19(3), 441–450. 10.1016/j.celrep.2017.03.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raizen, D. M. , Lee, R. Y. , & Avery, L. (1995). Interacting genes required for pharyngeal excitation by motor neuron MC in Caenorhabditis elegans . Genetics, 141(4), 1365–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rangaraju, S. , Solis, G. M. , Thompson, R. C. , Gomez‐Amaro, R. L. , Kurian, L. , Encalada, S. E. , … Petrascheck, M. (2015). Suppression of transcriptional drift extends C. elegans lifespan by postponing the onset of mortality. Elife, 4, e08833 10.7554/eLife.08833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rea, S. L. , Wu, D. , Cypser, J. R. , Vaupel, J. W. , & Johnson, T. E. (2005). A stress‐sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans . Nature Genetics, 37(8), 894–898. 10.1038/ng1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Regmi, S. G. , Rolland, S. G. , & Conradt, B. (2014). Age‐dependent changes in mitochondrial morphology and volume are not predictors of lifespan. Aging (Albany NY), 6(2), 118–130. 10.18632/aging.100639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reis‐Rodrigues, P. , Czerwieniec, G. , Peters, T. W. , Evani, U. S. , Alavez, S. , Gaman, E. A. , … Hughes, R. E. (2012). Proteomic analysis of age‐dependent changes in protein solubility identies genes that modulate lifespan. Aging Cell, 11(1), 120–127. 10.1111/j.1474-9726.2011.00765.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rousakis, A. , Vlanti, A. , Borbolis, F. , Roumelioti, F. , Kapetanou, M. , & Syntichaki, P. (2014). Diverse functions of mRNA metabolism factors in stress defense and aging of Caenorhabditis elegans . PLoS One, 9(7), e103365 10.1371/journal.pone.0103365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux, A. E. , Langhans, K. , Huynh, W. , & Kenyon, C. (2016). Reversible age‐related phenotypes induced during larval quiescence in C. elegans . Cell Metabolism, 23(6), 1113–1126. 10.1016/j.cmet.2016.05.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez‐Blanco, A. , & Kim, S. K. (2011). Variable pathogenicity determines individual lifespan in Caenorhabditis elegans . PLoS Genetics, 7(4), e1002047 10.1371/journal.pgen.1002047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwarzlander, M. , Wagner, S. , Ermakova, Y. G. , Belousov, V. V. , Radi, R. , Beckman, J. S. , … Murphy, M. P. (2014). The 'mitoash' probe cpYFP does not respond to superoxide. Nature, 514(7523), E12–E14. 10.1038/nature13858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shamir, L. , Wolkow, C. A. , & Goldberg, I. G. (2009). Quantitative measurement of aging using image texture entropy. Bioinformatics, 25(23), 3060–3063. 10.1093/bioinformatics/btp571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, E. Z. , Song, C. Q. , Lin, Y. , Zhang, W. H. , Su, P. F. , Liu, W. Y. , … Dong, M. Q. (2014). Mitoash frequency in early adulthood predicts lifespan in Caenorhabditis elegans . Nature, 508(7494), 128–132. 10.1038/nature13012 [DOI] [PubMed] [Google Scholar]
- Smith, M. , & Wilkinson, S. (2017). ER homeostasis and autophagy. Essays in Biochemistry, 61(6), 625–635. 10.1042/ebc20170092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Son, H. G. , Seo, M. , Ham, S. , Hwang, W. , Lee, D. , An, S. W. , … Lee, S. V. (2017). RNA surveillance via nonsense‐mediated mRNA decay is crucial for longevity in daf‐2/insulin/IGF‐1 mutant C. elegans . Nature Communications, 8, 14749 10.1038/ncomms14749 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadtman, E. R. , & Berlett, B. S. (1997). Reactive oxygen‐mediated protein oxidation in aging and disease. Chemical Research in Toxicology, 10(5), 485–494. 10.1021/tx960133r [DOI] [PubMed] [Google Scholar]
- Tank, E. M. , Rodgers, K. E. , & Kenyon, C. (2011). Spontaneous age‐related neurite branching in Caenorhabditis elegans . Journal of Neuroscience, 31(25), 9279–9288. 10.1523/JNEUROSCI.6606-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, R. C. , & Dillin, A. (2013). XBP‐1 is a cell‐nonautonomous regulator of stress resistance and longevity. Cell, 153(7), 1435–1447. 10.1016/j.cell.2013.05.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor, J. P. , Hardy, J. , & Fischbeck, K. H. (2002). Toxic proteins in neurodegenerative disease. Science, 296(5575), 1991–1995. 10.1126/science.1067122 [DOI] [PubMed] [Google Scholar]
- Tiku, V. , Jain, C. , Raz, Y. , Nakamura, S. , Heestand, B. , Liu, W. , … Antebi, A. (2017). Small nucleoli are a cellular hallmark of longevity. Nature Communications, 8, 16083 10.1038/ncomms16083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth, M. L. , Melentijevic, I. , Shah, L. , Bhatia, A. , Lu, K. , Talwar, A. , … Driscoll, M. (2012). Neurite sprouting and synapse deterioration in the aging Caenorhabditis elegans nervous system. Journal of Neuroscience, 32(26), 8778–8790. 10.1523/JNEUROSCI.1494-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham, T. J. , Thijssen, K. L. , Breitling, R. , Hofstra, R. M. , Plasterk, R. H. , & Nollen, E. A. (2008). C. elegans model identies genetic modiers of alpha‐synuclein inclusion formation during aging. PLoS Genetics, 4(3), e1000027 10.1371/journal.pgen.1000027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vinuela, A. , Snoek, L. B. , Riksen, J. A. , & Kammenga, J. E. (2012). Aging uncouples heritability and expression‐QTL in Caenorhabditis elegans . G3 (Bethesda), 2(5), 597–605. 10.1534/g3.112.002212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walther, D. M. , Kasturi, P. , Zheng, M. , Pinkert, S. , Vecchi, G. , Ciryam, P. , … Hartl, F. U. (2015). Widespread proteome remodeling and aggregation in aging C. elegans . Cell, 161(4), 919–932. 10.1016/j.cell.2015.03.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward, S. , & Carrel, J. S. (1979). Fertilization and sperm competition in the nematode Caenorhabditis elegans . Developmental Biology, 73(2), 304–321. 10.1016/0012-1606(79)90069-1 [DOI] [PubMed] [Google Scholar]
- Xu, H. , Chen, M. , Manivannan, A. , Lois, N. , & Forrester, J. V. (2008). Age‐dependent accumulation of lipofuscin in perivascular and subretinal microglia in experimental mice. Aging Cell, 7(1), 58–68. 10.1111/j.1474-9726.2007.00351.x [DOI] [PubMed] [Google Scholar]
- Yasuda, K. , Ishii, T. , Suda, H. , Akatsuka, A. , Hartman, P. S. , Goto, S. , … Ishii, N. (2006). Age‐related changes of mitochondrial structure and function in Caenorhabditis elegans . Mechanisms of Ageing and Development, 127(10), 763–770. 10.1016/j.mad.2006.07.002 [DOI] [PubMed] [Google Scholar]
- Youngman, M. J. , Rogers, Z. N. , & Kim, D. H. (2011). A decline in p38 MAPK signaling underlies immunosenescence in Caenorhabditis elegans . PLoS Genetics, 7(5), e1002082 10.1371/journal.pgen.1002082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zarse, K. , Schmeisser, S. , Groth, M. , Priebe, S. , Beuster, G. , Kuhlow, D. , … Ristow, M. (2012). Impaired insulin/IGF1 signaling extends life span by promoting mitochondrial L‐proline catabolism to induce a transient ROS signal. Cell Metabolism, 15(4), 451–465. 10.1016/j.cmet.2012.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y. , Gilliat, A. F. , Ziehm, M. , Turmaine, M. , Wang, H. , Ezcurra, M. , … Gems, D. (2017). Two forms of death in ageing Caenorhabditis elegans . Nature Communications, 8, 15458 10.1038/ncomms15458 [DOI] [PMC free article] [PubMed] [Google Scholar]


