Abstract
Background
Recently, innovative attempts have been made to identify moyamoya disease (MMD) by focusing on the morphological differences in the head of MMD patients. Following the recent revolution in the development of deep learning (DL) algorithms, we designed this study to determine whether DL can distinguish MMD in plain skull radiograph images.
Methods
Three hundred forty-five skull images were collected as an MMD-labeled dataset from patients aged 18 to 50 years with definite MMD. As a control-labeled data set, 408 skull images of trauma patients were selected by age and sex matching. Skull images were partitioned into training and test datasets at a 7:3 ratio using permutation. A total of six convolution layers were designed and trained. The accuracy and area under the receiver operating characteristic (AUROC) curve were evaluated as classifier performance. To identify areas of attention, gradient-weighted class activation mapping was applied. External validation was performed with a new dataset from another hospital.
Findings
For the institutional test set, the classifier predicted the true label with 84·1% accuracy. Sensitivity and specificity were both 0·84. AUROC was 0·91. MMD was predicted by attention to the lower face in most cases. Overall accuracy for external validation data set was 75·9%.
Interpretation
DL can distinguish MMD cases within specific ages from controls in plain skull radiograph images with considerable accuracy and AUROC. The viscerocranium may play a role in MMD-related skull features.
Fund
This work was supported by grant no. 18-2018-029 from the Seoul National University Bundang Hospital Research Fund.
Keywords: Convolutional neural network, Deep learning, Moyamoya, Skull
Research in context.
Evidence before this study
Moyamoya disease is a type of rare cerebrovascular disease, characterized by spontaneous occlusion of the terminal internal carotid artery, proximal anterior cerebral artery, and proximal middle cerebral artery. Innovative attempts have been made to identify novel characteristics of moyamoya disease by focusing on morphological differences in the heads of moyamoya disease patients. Direct measurements show that cephalometric features of moyamoya disease patients differ from those of the normal population. Moreover, network analysis of facial features revealed remarkable differences around the eyes and forehead of Western moyamoya disease patients. Previous three-dimensional morphologic analysis of the internal carotid artery also confirmed differences in vascular tortuosity and related hemodynamics.
Added value of this study
This is the first study to assess whether deep learning can distinguish moyamoya disease in plain skull radiograph images. We add to the previous findings by showing a potential relationship between moyamoya disease and morphological characteristics of the head. Our results showed that deep learning can distinguish moyamoya disease from a control group within specific ages with considerable accuracy. Additionally, gradient-weighted class activation mapping analysis showed that, in most cases, prediction of moyamoya disease could be made by attention to the lower face, from the skull base to the mandibular angle. These results suggest that moyamoya disease patients have distinct features in the viscerocranium, which contains the conduit between the heart and brain.
Implications of all the available evidence
Based on this study and previous reports, moyamoya disease seems to exhibit unique morphologic characteristics in the head. Furthermore, we found that viscerocranium is the most distinctive region, based on deep learning using skull radiographs. The origin of the viscerocranium is the neural crest, whereas the neurocranium is derived from the paraxial mesoderm. Considering that steno-occlusive changes of the cerebral arteries in moyamoya disease occurred only in arteries of neural crest origin, the viscerocranium, sharing an origin with moyamoya disease vessels, might be affected by alteration in neural crest cells. Deep learning may assist in diagnosing moyamoya disease with less invasive tools; moreover, it may also inspire a novel point of view for assessment of skull images. Furthermore, moyamoya disease prediction by deep learning with a non-invasive imaging technique could be a useful tool with further development and validation.
Alt-text: Unlabelled Box
1. Introduction
Moyamoya disease (MMD) is a kind of rare cerebrovascular disease. It is characterized by spontaneous occlusion of the terminal internal carotid artery (ICA), proximal anterior cerebral artery, and proximal middle cerebral artery. As the vessel occlusion progresses, cerebral ischemia develops, leading to a transient ischemic attack or cerebral infarction. Moreover, cerebral ischemia induces proliferation of collateral channels, which mainly arise from choroidal arteries and are responsible for intracerebral hemorrhage [1]. The pathophysiology of MMD is not yet clear, although significant genetic factors have recently been discovered [2]. In terms of treatment, revascularization surgery is accepted as an effective option for preventing both ischemic and hemorrhagic stroke [3,4].
Several investigators have made significant efforts to identify the distinctive features of MMD in order to elucidate its pathophysiology and pathobiology. The reports thus far have been consistent regarding regional differences, with a high incidence and prevalence in East Asia [5]. This epidemiologic trait has been supported by genetic differences [6]. Female predominance is also a well-known epidemiologic characteristic [5].
Typically, invasive examinations, such as cerebral angiography, are required to make a diagnosis of MMD. However, innovative attempts have recently been made to identify novel characteristics of MMD by focusing on the morphological differences in the heads of MMD patients. Direct measurements show that cephalometric features of MMD patients differ from those of the normal population [7]. Moreover, network analysis of facial features revealed remarkable differences around the eyes and forehead of MMD patients [8].
With the recent revolution in the development of machine learning algorithms, classification and prediction of large imaging datasets using convolutional neural networks (CNNs) can recognize distinctive features [[9], [10], [11], [12], [13], [14], [15]]. Thus, we designed this study to determine whether MMD could be distinguished in plain skull radiograph images using CNN analysis.
2. Materials and methods
2.1. Study enrollment
This study used electronic healthcare records (EHR) to identify the morphologic features of skulls of MMD patients using deep learning. The facilities are academic teaching hospitals located in South Korea with >1400 beds. This study was approved by the institutional review boards of Seoul National University Bundang Hospital (B-1808-484-128) and Incheon St. Mary's Hospital (OC18REDI0125). The requirement for informed consent was waived.
The inclusion criteria for MMD group were as follows: 1) patients aged 18 to 50 years, 2) patients with confirmed definite bilateral stenosis or occlusion of the terminal ICA, accompanied by basal collateral formation that was verified by cerebral angiography, and 3) patients who did not undergo surgical revascularization at the time of diagnosis. Unilateral MMD was excluded to avoid possible heterogeneity in the MMD group. A total of 623 patients were identified as MMD patients with bilateral angiopathy. Skull radiographs taken outside of the specified age range were excluded because the skull shape changes with age [16,17]. Post-surgical or defective (i.e., seriously rotated) skull images were also not considered. Eligible skull radiograph images were available in 436 patients.
For the control group, patients underwent skull radiography in the emergency department for minor trauma; this excluded any fracture, dislocation, or evidence of a foreign body in skull images. In the same age range, 4258 patients underwent skull radiograph scans for evaluation of trauma. An additional review was performed to exclude skull images taken post-operatively, those with fracture or foreign bodies, an those that were severely defective, in order to exclude traumatic changes in skull radiograph analysis.
2.2. Data partition and preprocessing
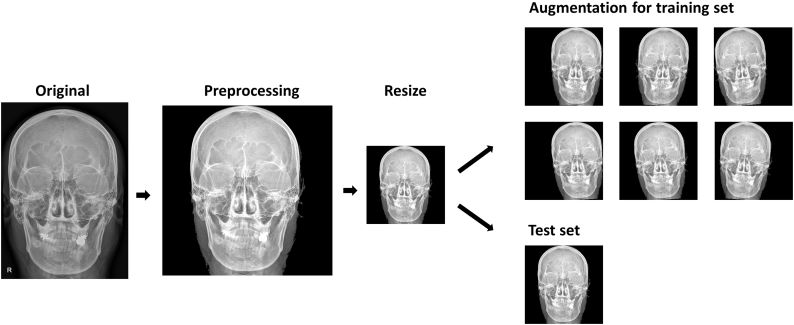
Each group was partitioned into training and test dataset at a 7:3 ratio using permutation. The image preprocessing procedure is summarized as follows: 1) Markers indicating left and right are removed from the original image using morphological operations including image thresholding and connected component labelling. 2) Then, all images are transformed to 256 × 256 square images using zero-padding and image resizing as appropriate. 3) Images in the training dataset undergo data augmentation procedure using a horizontal flip, rotation (within 5°), and horizontal shift (within 15%) producing a total of 50 folds of augmented training dataset. Fig. 1 summarizes the flow of the entire preprocessing.
Fig. 1.
Diagram shows steps in image pre-processing. The skull image is of one of the authors. After resizing, each image is assigned to a training set or test set, although we present same skull image in both sets. The images of the training set are augmented by random application of a horizontal flip, rotation within 5°, and horizontal shift within 15%.
2.3. Convolutional neural network classifier
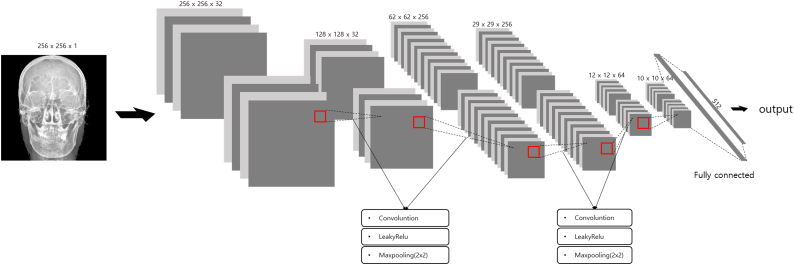
We used Keras (ver. 2.20.0, https://keras.io/) over TensorFlow (ver. 1.8.0, https://www.tensorflow.org/) environment for CNN building, training, and evaluation [18]. Input images were 256 × 256 with a single channel. The first two convolution layers were 3 × 3 kernel size and used zero-padding without stride to maintain feature map size; these were followed by maximum pooling with a 2 × 2 matrix. Using the first two layers, 64 × 64 × 32 feature maps were generated. The next two convolution layers were 3 × 3 kernel size and used zero-padding without stride, followed by maximum pooling with a 2 × 2 matrix. Using these two layers, 14 × 14 × 256 feature maps were made. Through an additional two convolution layers, 10 × 10 × 64 feature maps were produced and connected with 6400 fully connected nodes, which were connected with 512 nodes of a hidden layer. Finally, a single node generated predictive values using a sigmoid function . Each convolution layer was activated by using the LeakyReLU function (f(x) = 0 · 1 ∗ x for x < 0, f(x) = x for x ≥ 0) [19]. Throughout the designed network, 4,135,905 parameters were ready to be trained (Fig. 2).
Fig. 2.
Diagram shows schematic structure of convolutional neural network. The skull image is of one of the authors.
Binary cross-entropy function was adopted as loss function to fit binary classifier, and Adamax was used as optimizer function (learning rate = 0·0001, beta1 = 0·9, and beta2 = 0·999) [20]. Early stopping was set at the point of increment of validation loss with reference to ten epochs.
2.4. Statistical analysis and model evaluation
All continuous variables are presented as mean ± standard deviation and were analyzed by Student's t-test or paired t-test. Categorical variables are presented as number (percent) and were analyzed by Pearson's chi-squared test.
After completion of learning, the performance was evaluated using the institutional test set that the trained classifier had never previously encountered. A confusion matrix was made using the number of true positive (TP, predicts MMD as MMD), true negative (TN, predicts control as control), false positive (FP, predicts control as MMD), and false negative (FN, predicts MMD as control). Accuracy of the classifier was defined as . Area under receiver operating characteristic (AUROC) was calculated with predicted label and related sigmoid output value. Sensitivity, specificity, positive predictive value, and negative predictive value were calculated as per original definition. Confidence interval (CI) for each value was deduced using exact binomial confidence limits [21].
2.5. Class activation map analysis
Each convolutional layer contains spatial information with features deduced by convolutional filter application. Although we could obtain predictive values throughout the established network, we needed to determine the underlying basis for the predictions. Thus, we applied gradient-weighted class activation mapping (Grad-CAM) as a visualization tool for identifying certain features or regions of interest that made the classifier predict [22]. This could be visualized through calculation of the gradient of weights forming each convolution layer using global-average-pooling. Thus, important (larger gradient) features could be calculated and visualized in accordance with the original image.
2.6. External validation
Further validation was planned using skull images obtained from other independent hospitals for the general performance assessment of the classifier. The external validation skull images comprised a new dataset collected by an independent hospital located in another city and blinded to other investigators. We sent verification codes and weights to that hospital. Then, confusion matrices were composed and returned to our team to evaluate the accuracy of the classifier.
3. Results
Among all eligible patients, the sex ratios were statistically different (p < 0·0001, Pearson chi-squared test) between the two groups (64·2% vs. 39·0%, proportions of patients who were female). Thus, coarsened exact matching was performed in this phase using sex and age, and 753 (345 MMD, 408 control) matched patients were selected. The mean age of those patients was 35·2 ± 9·2 years. The proportion of female patients was 64.3% (484 of 753). The mean height and weight were 164·1 ± 8·4 cm and 62·6 ± 14·1 kg, respectively.
The mean ages of the patients in the MMD and control group were 35·2 ± 9·1 years and 35·3 ± 9·1 years, respectively. The distribution of age was not statistically different between the matched groups (p = 0·92, t-test). The proportions of patients who were female were 63·8% (220 of 345) in the MMD group and 64·7% (264 of 408) in the control group. Pearson chi-squared test for sex proportion yielded a p-value of 0·85. The distributions of height and weight were not statistically different between two groups (Table 1).
Table 1.
Basic characteristics of enrolled subjects.
| All | Moyamoya group | Control group | p-value | ||
|---|---|---|---|---|---|
| Age (years) | 35·2 ± 9·2 | 35·2 ± 9·1 | 35·3 ± 9·1 | 0·92a | |
| Sex | Male | 269 (35·7) | 125 (36·2) | 144 (35.3) | 0·85b |
| Female | 484 (64·3) | 220 (63·8) | 264 (64·7) | ||
| Height (cm) | 164·1 ± 8·4 | 164·2 ± 8·2 | 163·5 ± 9·2 | 0·41a | |
| Weight (kg) | 62·6 ± 14·1 | 63·1 ± 14·7 | 61·0 ± 11·9 | 0·16a |
Continuous values are presented as mean ± standard deviation, while categorical values are as number (percent).
Student t-test, bPearson chi-square test.
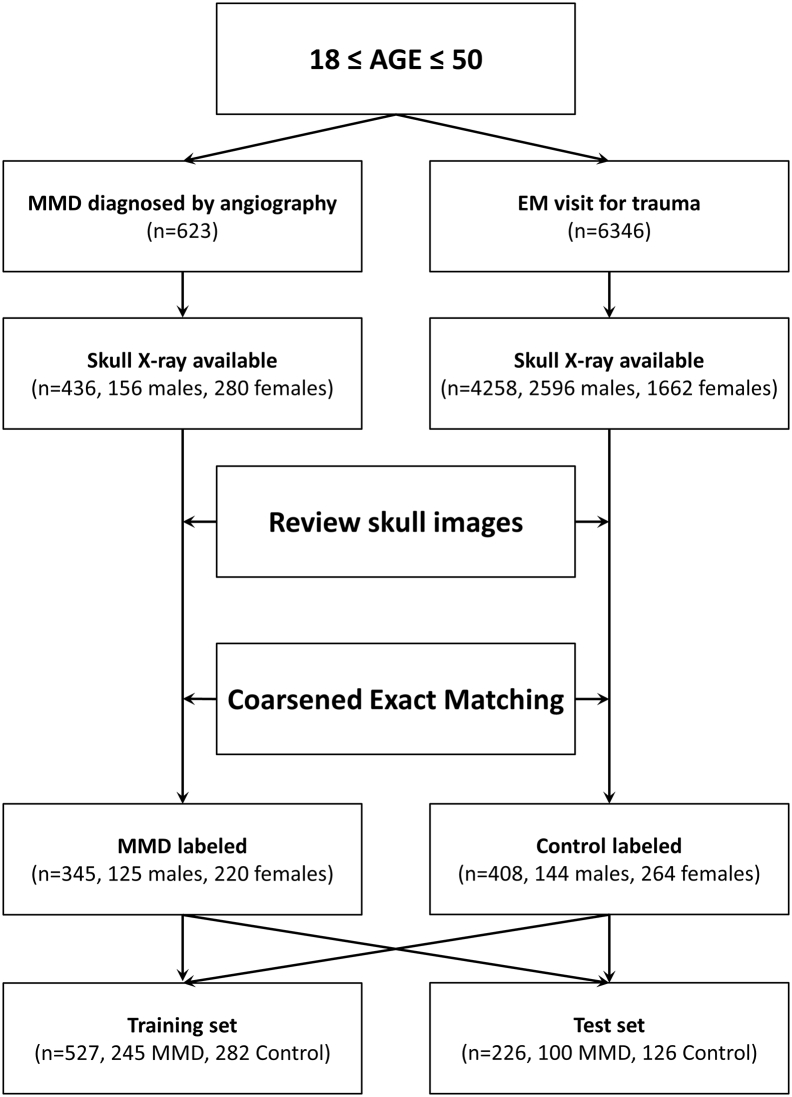
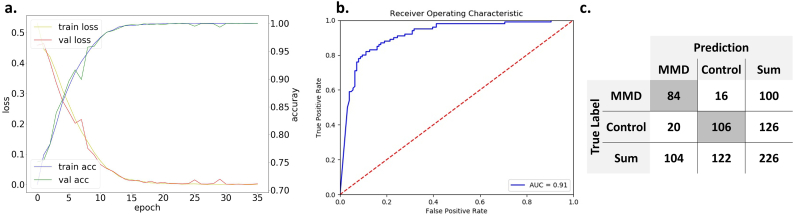
Among the selected images, 226 skull images were separated as the test set (100 for MMD and 126 for control) using random selection (Fig. 3). The remaining 527 images were used in the training process, in which 150 images were partitioned as the validation set. A total of 26,301 augmented images were used. The training process ended in the 36th epoch reaching 99·3% accuracy for the training set and 99·1% for the validation set. In the evaluation process with separated test sets, the classifier predicted the true label for 190 of 226 cases reaching 84·1% accuracy (95% confidence interval [CI], 78·3–88·6%). Sensitivity was 0·84 for MMD (95% CI, 0·78–0·89), and specificity was 0·84 (95% CI, 0·79–0·88). AUROC was 0·91. Positive and negative predictive values of the classifier were 0·81 (95% CI, 0·75–0·86) and 0·87 (95% CI, 0·82–0·91), respectively (Fig. 4).
Fig. 3.
Flow chart shows the schematic process of data collection and partitioning.
Fig. 4.
Charts show training process and evaluation metrics. (a) As epochs proceed, loss of training and validation sets converges to near 0, while accuracy rose to near 0·99. (b) Area under receiver operative characteristic curve is 0·91 for predicting separated test sets. (c) The confusion matrix shows 84·1% accuracy. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
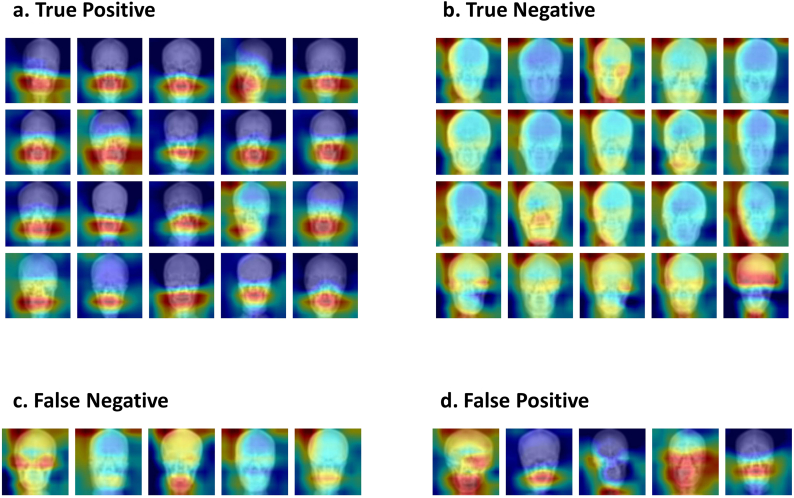
Grad-CAM saliency maps showed that prediction of MMD was made by attention to the lower face, from the skull base to the mandibular angle in most cases. In contrast, attention area for predicting the control group was more scattered and sparser. The ratio between mean weights in saliency maps of lower and upper image halves was 1·25 ± 0·19 in true positive subjects (N = 84), whereas it was 0·98 ± 0·06 in true negative subjects (N = 106); this difference was statistically significant (p 〈10−31). Representative Grad-CAM saliency maps projected for a corresponding test set image are shown in Fig. 5.
Fig. 5.
Matrix shows representative saliency maps. (a) and (b) show correctly classified images. Most moyamoya cases were classified by the attention around the lower face. However, attention areas are more scattered and sparser for predicting the control. (c) shows the cases where the classifier predicts moyamoya as control, and (d) shows the opposite.
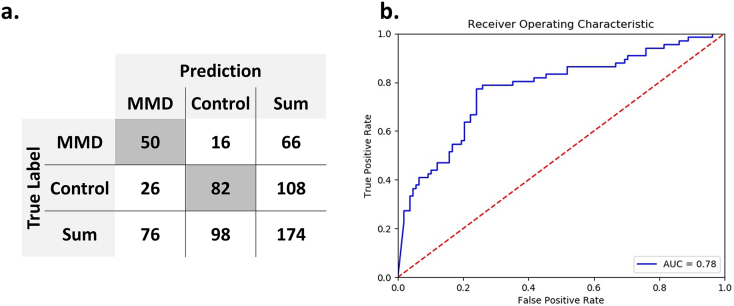
External validation showed overall accuracy to be 75·9% (95% CI, 68·5–81·9%). Sensitivity was 0·76 (95% CI, 0·69–0·82), and specificity was 0·76 (95% CI, 0·70–0·81). Positive and negative predictive values were 0·66 (95% CI, 0·57–0·73) and 0·84 (95% CI, 0·77–0·89), respectively. AUROC was 0·78 (Fig. 6).
Fig. 6.
Charts show the results of external validation. (a) The confusion matrix shows 75·9% accuracy. (b) Area under receiver operative characteristic curve is 0·78 for predicting the external validation set.
4. Discussion
In this study, we determined whether MMD could be distinguished in plain skull radiograph images using deep learning. Our results showed that deep learning can indeed distinguish MMD from a control group of trauma images with considerable accuracy and AUROC within specific ages. We attempted to train our classifier using deeper published networks, such as InceptionResNetV2, DenseNet201, and VGG16 [[23], [24], [25]]. However, there was no particular performance advantage in identifying MMD; indeed, Grad-CAM analysis was more difficult to understand with more scattered attention areas. Conversely, the loss function diverged when four or fewer convolution layers were applied, and the training process was not performed properly. We chose to use a methodology to verify separate test sets, rather than cross-validation where all images were used for learning, because the most important point we considered in designing the classifier was to ensure the accuracy of the classification of radiographs that the classifier had never seen. We separated 226 (30%) images without augmentation as an institutional test set and did not use a training process, in order to prevent institutional over-fitting. Moreover, additional validation tests were performed with a new dataset from a geographically separate hospital for further general performance assessment.
Preceding studies that used deep learning for image diagnosis mostly focused on diagnostic ability while comparing with human-labeled (especially, radiologist-labeled) diagnosis using the same image modalities [9,11,15]. For MMD diagnosis, the golden standard has been cerebral angiography, although MR angiography or CT angiography can show characteristic changes of cerebral vessels in MMD [1]. Plain skull images have not been used as a modality for diagnosing MMD because it has been thought that distinctive features are not detectable. However, in our investigation using plain skull images, the image classifier showed acceptable discrimination power over coincidence through an overall accuracy of 84·1%. Thus, this classifier recognized characteristics that had never been considered before. Machine learning has already shown the possibility of recognizing features that humans are unable to see and distinguish in medical image analysis. For example, non-enhanced cardiac CT images usually did not allow the diagnosis of cardiac abnormalities other than calcifications. However, a recent study reported that machine learning using texture analysis could identify features imperceptible to radiologists in detecting myocardial infarction on non-contrast CT [14].
A few studies have investigated the relationship between MMD and skull images. Qureshi et al. discussed the possible relationship between cephalometric parameters and MMD [7], and they highlighted previous studies showing that cranial synostosis is related to reduced arterial flow velocities and increased pulsatility index, which could improve after surgical correction [26,27]. In contrast, MMD could play a role in alteration of cephalometric parameters through increased expression of various growth factors [28,29]. In short, alteration of skull configuration could affect or could be affected by MMD. More recently, Kraemer et al. investigated the facial landmarks of patients with MMD in comparison with those of a matched control group [8]. They concluded a resemblance among faces of Caucasian patients with idiopathic MMD and a difference from matched controls, especially around the nasion. However, the potential mechanism was not discussed in detail.
In the current study, Grad-CAM revealed that the classifier paid attention to the viscerocranium rather than the neurocranium. This area contains the common carotid artery and the ICA, which are the conduit between the heart and brain. A previous three dimensional analysis reported that the morphology of extradural ICAs of MMD patients was different from that of normal subjects, particularly in tortuosity [30]. Although the report focused on petrous and cavernous segments of ICAs, it seemed to be straighter along the entire ICA in MMD patients. These different courses might influence the structure of the viscerocranium, including the mandible, petrous bone, and their joints.
The origin of the viscerocranium is the neural crest, whereas the neurocranium is derived from the paraxial mesoderm [31]. The neural crest also is the origin for smooth muscle cells and pericytes of the cerebral arteries of prosencephalon while the arteries of mesencephalon and rhombencephalon are derived from mesodermal origin vessels [32]. Recently, Komiyama introduced a hypothesis stating that MMD could be regarded as a vascular form of neurocristopathy, because the steno-occlusive changes of the cerebral arteries in MMD occurred only in the arteries of neural crest origin [33]. Thus, the viscerocranium, sharing an origin with MMD vessels, might be affected by alteration in neural crest cells.
A crucial limitation of this study was that we selected trauma patients as the control group. This was done for the following reasons. First, most normal subjects would not undergo skull imaging without a particular reason. In our EHR, most skull radiographs belonged to patients who underwent cranial surgery or patients who needed an evaluation of trauma. Second, if we selected patients who underwent cranial surgery, neurosurgical disease (e.g., vascular or tumor) may act as a confounding factor when extracting distinct features. Third, skull radiographs would not present features that differed from those of normal subjects if the trauma did not cause skull damage. Thus, these patients were chosen as a reasonable control group in order to obtain a sufficient number of skull radiographs from patients without skull damage. Nevertheless, the fact that the control group comprised traumatic patients requires a consideration of the potential bias in interpreting the results of the study.
Another consideration when interpreting the results of the current study is that the study was derived from a high prevalence country, South Korea. The large difference in the incidence of MMD, with a marked East–West gradient, is suggestive of a founder effect in East Asia. In Asia, in 95% of familiar and 79% of sporadic MMD, the RNF213 mutation p.R4810K is present as a founder mutation. Previously, the c.14576G > A variant has been identified in 90% of Japanese patients, 79% of Korean patients, and 23% of Chinese patients [[34], [35], [36], [37]]. Considering these prior observations, we cannot guarantee that our classifier will work properly among Western populations with different genetic backgrounds. Indeed, a previous investigation performed in Europe concluded that Western MMD patients were different from matched controls, especially around the nasion. Thus, interesting results may be observed if our training model is applied to skull radiographs of individuals from Western nations with correspondingly different genetic backgrounds.
In addition to the rarity of MMD, the strict inclusion criteria (definite bilateral MMD and age range) to identify representative features of skull images of MMD patients may have been a contributing factor for the modest sample size in the present study. Moreover, some patients did not have pre-operative skull images because this approach is not a routinely used diagnostic modality for MMD. As the control group had a preponderance of men, coarsened exact matching was performed and many eligible control subjects were excluded to meet age and sex ratio restriction. In addition, some images had to be excluded due to factors such as foreign body presence and serious malalignment. Despite the modest sample size, our classifier was trained with acceptable reduction of loss function and could be applied to the institutional test set, showing robust discriminative power. Moreover, the trained classifier could predict MMD using a new dataset from a separate hospital, although overall accuracy dropped slightly. Although multi-centered image collection could produce better performance, the provision of medical images to other institutes as a research resource is strictly prohibited in Korea. However, using a trained classifier with saved weights, follow-up studies can be designed using boosted or transfer learning achieved by other institutes in the future.
In brief, CNN can distinguish plain skull images of MMD from those of a control group within specific age range with particular attention to the viscerocranium, which had never been considered before. We add to the previous findings by showing the potential relationship between MMD and morphological characteristics of the head. Deep learning may assist in diagnosing MMD with less invasive tools, and may inspire a novel point of view for assessment of skull images.
Data sharing statement
The data are not available for public access because of patient privacy concerns but are available from the corresponding author on reasonable request approved by the institutional review boards of Seoul National University Bundang Hospital and Incheon St. Mary's Hospital.
Funding sources
This work was supported by grant no 18–2018-029 from the Seoul National University Bundang Hospital Research Fund. The funding source had no involvement in any phase of the current study.
Declaration of interests
None.
Authors' contributions
TK, JH, DKJ and CWO developed the study concept and design and extracted data from database. TK, JH, and DKJ performed training of convolutional neural network. TK wrote drafted the manuscript. LS, JK, KJL, SHK, SJP, OKK, and CWO critically revised drafted manuscript. CWO had full access to all the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis. All authors reviewed the manuscript.
Acknowledgments
None.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.043.
Appendix A. Supplementary data
Python codes used in model training
References
- 1.Kim T., Moyamoya Disease Oh.C.W. In: Stroke revisited: diagnosis and treatment of ischemic stroke. Lee S.-H., editor. Springer; Singapore: 2017. pp. 213–219. [Google Scholar]
- 2.Bang O.Y., Fujimura M., Kim S.K. The pathophysiology of moyamoya disease: an update. J Stroke. 2016;18(1):12–20. doi: 10.5853/jos.2015.01760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kim T., Oh C.W., Bang J.S., Kim J.E., Cho W.S. Moyamoya disease: treatment and outcomes. J Stroke. 2016;18(1):21–30. doi: 10.5853/jos.2015.01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee S.U., Oh C.W., Kwon O.K. Surgical treatment of adult moyamoya disease. Curr Treat Options Neurol. 2018;20(7):22. doi: 10.1007/s11940-018-0511-8. [DOI] [PubMed] [Google Scholar]
- 5.Kim T., Lee H., Bang J.S., Kwon O.K., Hwang G., Oh C.W. Epidemiology of moyamoya disease in Korea: based on National health insurance service data. J Korean Neurosurg Soc. 2015;57(6):390–395. doi: 10.3340/jkns.2015.57.6.390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shoemaker L.D., Clark M.J., Patwardhan A. Disease variant landscape of a large multiethnic population of moyamoya patients by exome sequencing. G3 (Bethesda) 2015;6(1):41–49. doi: 10.1534/g3.115.020321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qureshi A.I., Gilani W.I., Gilani S.I., Adil M.M. Cephalometric features of moyamoya disease: a case control study. J Vasc Interv Neurol. 2014;7(4):13–18. [PMC free article] [PubMed] [Google Scholar]
- 8.Kraemer M., Huynh Q.B., Wieczorek D., Balliu B., Mikat B., Boehringer S. Distinctive facial features in idiopathic Moyamoya disease in Caucasians: a first systematic analysis. PeerJ. 2018;6 doi: 10.7717/peerj.4740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim Y., Lee K.J., Sunwoo L. Deep learning in diagnosis of maxillary sinusitis using conventional radiography. Invest Radiol. 2018;54(1):7–15. doi: 10.1097/RLI.0000000000000503. [DOI] [PubMed] [Google Scholar]
- 10.Son J., Park S.J., Jung K.-H. 2017. Retinal vessel segmentation in fundoscopic images with generative adversarial networks. arXiv preprint arXiv:170609318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Becker A.S., Marcon M., Ghafoor S., Wurnig M.C., Frauenfelder T., Boss A. Deep learning in mammography: diagnostic accuracy of a multipurpose image analysis software in the detection of breast cancer. Invest Radiol. 2017;52(7):434–440. doi: 10.1097/RLI.0000000000000358. [DOI] [PubMed] [Google Scholar]
- 12.Cicero M., Bilbily A., Colak E. Training and validating a deep convolutional neural network for computer-aided detection and classification of abnormalities on frontal chest radiographs. Invest Radiol. 2017;52(5):281–287. doi: 10.1097/RLI.0000000000000341. [DOI] [PubMed] [Google Scholar]
- 13.Depeursinge A., Chin A.S., Leung A.N. Automated classification of usual interstitial pneumonia using regional volumetric texture analysis in high-resolution computed tomography. Invest Radiol. 2015;50(4):261–267. doi: 10.1097/RLI.0000000000000127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mannil M., von Spiczak J., Manka R., Alkadhi H. Texture analysis and machine learning for detecting myocardial infarction in noncontrast low-dose computed tomography: unveiling the invisible. Invest Radiol. 2018;53(6):338–343. doi: 10.1097/RLI.0000000000000448. [DOI] [PubMed] [Google Scholar]
- 15.Larson D.B., Chen M.C., Lungren M.P., Halabi S.S., Stence N.V., Langlotz C.P. Performance of a deep-learning neural network model in assessing skeletal maturity on pediatric hand radiographs. Radiology. 2018;287(1):313–322. doi: 10.1148/radiol.2017170236. [DOI] [PubMed] [Google Scholar]
- 16.Lynnerup N. Cranial thickness in relation to age, sex and general body build in a Danish forensic sample. Forensic Sci Int. 2001;117(1–2):45–51. doi: 10.1016/s0379-0738(00)00447-3. [DOI] [PubMed] [Google Scholar]
- 17.Finby N., Kraft E. The aging skull: comparative roentgen study. 25 to 34 year interval. Clin Radiol. 1972;23(4):410–414. doi: 10.1016/s0009-9260(72)80004-7. [DOI] [PubMed] [Google Scholar]
- 18.Abadi M. Proceedings of the 12th USENIX Symposium on Operating Systems Design and Implementation (OSDI’16)(Savannah, GA, USA) Vol. 2016. 2016. TensorFlow AABP. Large-scale machine learning on heterogeneous distributed systems; pp. 265–283. [Google Scholar]
- 19.Maas A.L., Hannun A.Y., Ng A.Y. Rectifier nonlinearities improve neural network acoustic models. Proc icml. 2013;2013:3. [Google Scholar]
- 20.Kingma D.P., Ba J. Adam. 2014. A method for stochastic optimization. arXiv preprint arXiv:14126980. [Google Scholar]
- 21.Clopper C.J., Pearson E.S. The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika. 1934;26(4):404–413. [Google Scholar]
- 22.Selvaraju R.R., Das A., Vedantam R., Cogswell M., Parikh D., Batra D. 2016. Grad-CAM: Why did you say that? arXiv preprint arXiv:161107450. [Google Scholar]
- 23.Huang G., Liu Z., Van Der Maaten L., Weinberger K.Q. Densely connected convolutional networks. CVPR. 2017;2017:3. [Google Scholar]
- 24.Szegedy C., Ioffe S., Vanhoucke V., Alemi A.A. Inception-v4, inception-resnet and the impact of residual connections on learning. AAAI. 2017;2017:12. [Google Scholar]
- 25.Simonyan K., AJapa Zisserman. 2014. Very deep convolutional networks for large-scale image recognition. [Google Scholar]
- 26.Wang B., Cheng Z., Mu X., Fan B., Guo Z. Preoperative and postoperative transcranial Doppler sonographic evaluations of the cerebral hemodynamics of craniostenosis. J Craniofac Surg. 2010;21(2):432–435. doi: 10.1097/SCS.0b013e3181cfa7bf. [DOI] [PubMed] [Google Scholar]
- 27.Westra S.J., Stotland M.A., Lazareff J., Anderson C.T., Sayre J.W., Kawamoto H. Perioperative transcranial Doppler US to evaluate intracranial compliance in young children undergoing craniosynostosis repair surgery. Radiology. 2001;218(3):816–823. doi: 10.1148/radiology.218.3.r01mr36816. [DOI] [PubMed] [Google Scholar]
- 28.Hojo M., Hoshimaru M., Miyamoto S. Role of transforming growth factor-beta1 in the pathogenesis of moyamoya disease. J Neurosurg. 1998;89(4):623–629. doi: 10.3171/jns.1998.89.4.0623. [DOI] [PubMed] [Google Scholar]
- 29.Hoshimaru M., Takahashi J.A., Kikuchi H., Nagata I., Hatanaka M. Possible roles of basic fibroblast growth factor in the pathogenesis of moyamoya disease: an immunohistochemical study. J Neurosurg. 1991;75(2):267–270. doi: 10.3171/jns.1991.75.2.0267. [DOI] [PubMed] [Google Scholar]
- 30.Kim T., Bang J.S., Kwon O.K. Morphology and related hemodynamics of the internal carotid arteries of moyamoya patients. Acta Neurochir. 2015;157(5):755–761. doi: 10.1007/s00701-015-2367-y. [DOI] [PubMed] [Google Scholar]
- 31.Bildsoe H., Loebel D.A., Jones V.J. The mesenchymal architecture of the cranial mesoderm of mouse embryos is disrupted by the loss of Twist1 function. Dev Biol. 2013;374(2):295–307. doi: 10.1016/j.ydbio.2012.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Etchevers H.C., Vincent C., Le Douarin N.M., Couly G.F. The cephalic neural crest provides pericytes and smooth muscle cells to all blood vessels of the face and forebrain. Development. 2001;128(7):1059–1068. doi: 10.1242/dev.128.7.1059. [DOI] [PubMed] [Google Scholar]
- 33.Komiyama M. Moyamoya disease is a vascular form of neurocristopathy: disease of the embryologic cephalic neural crest. Childs Nerv Syst. 2017;33(4):567–568. doi: 10.1007/s00381-017-3369-2. [DOI] [PubMed] [Google Scholar]
- 34.Bersano A., Guey S., Bedini G. Research Progresses in Understanding the Pathophysiology of Moyamoya Disease. Cerebrovasc Dis. 2016;41(3–4):105–118. doi: 10.1159/000442298. [DOI] [PubMed] [Google Scholar]
- 35.Miyatake S., Miyake N., Touho H. Homozygous c.14576G>a variant of RNF213 predicts early-onset and severe form of moyamoya disease. Neurology. 2012;78(11):803–810. doi: 10.1212/WNL.0b013e318249f71f. [DOI] [PubMed] [Google Scholar]
- 36.Akagawa H., Mukawa M., Nariai T. Novel and recurrent RNF213 variants in Japanese pediatric patients with moyamoya disease. Hum Genome Var. 2018;5 doi: 10.1038/hgv.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu W., Morito D., Takashima S. Identification of RNF213 as a susceptibility gene for moyamoya disease and its possible role in vascular development. PLoS One. 2011;6(7) doi: 10.1371/journal.pone.0022542. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Python codes used in model training