Abstract
Background
The injured liver loses normal function, with concomitant decrease of key identity genes. Super-enhancers contribute to mammalian cell identity. Here, we identified core transcription factors (TFs) that are active in hepatocytes, using genome-wide analysis and hierarchical ordering of super-enhancer distribution.
Methods
Expression of core TFs was assessed in a cohort of patients with hepatitis or cirrhosis and animal models. Quantitative PCR, chromatin immunoprecipitation assays, and hydrodynamic gene delivery methods were used to assess gene regulation and hepatocyte viability. RNA-sequencing data were generated to investigate the role of LRH1 in hepatocyte protection from injury.
Results
Network analysis of super-enhancer-associated gene interactions and expression arrays for cohorts of patients with hepatitis and cirrhosis enabled us to identify a super-enhancer-associated network, and LRH1, HNF4α, PPARα, and RXRα as core TFs. In mouse models, expression of core TFs was robustly inhibited by single and multiple challenge(s) with liver toxicant. RNA-seq analysis revealed changes in expression in the super-enhancer-associated genes sensitively biased toward repression by intoxication. LRH1 gene delivery prevented the loss of hepatic super-enhancer-associated signaling circuitry in toxicant-challenged mice, and protected the liver from injury, indicating the role of LRH1 in hepatocyte identity and viability. In hepatocytes, overexpression of each core TF promoted induction of other TFs.
Conclusion
Overall, this study identified LRH1-driven pathway as a circuitry responsible for hepatocyte identity by using cistromic analysis, improving our understanding of liver pathophysiology and identifying novel therapeutic targets.
Keywords: Super-enhancer, Liver disease, LRH1, Acute liver injury, Acetaminophen
Abbreviation: APAP, acetaminophen; ChIP, chromatin immunoprecipitation; ChIP-seq, chromatin immunoprecipitation-sequencing; DMEM, Dulbecco's modified Eagle's medium; FPKM, Fragments Per Kilobase Million; GEO, Gene Expression Omnibus; GO, gene ontology; H3K27Ac, K27-acetylated histone H3; HSC, hepatic stellate cell; ITSX, insulin-transferrin-selenium X; qRT-PCR, quantitative reverse-transcription polymerase chain reaction; RNA-seq, RNA-sequencing; SEM, standard error of mean; TF, transcription factor; TSS, transcription start site; TUNEL, terminal deoxynucleotidyl transferase dUTP nick end labeling
Research in context.
Evidence before this study
-
►
A recent genome-wide study demonstrated the existence of “super-enhancers”, which are large genomic regions containing clusters of closely spaced transcription factor binding sites. Identity of the cell may be determined by the ranks and distributions of super-enhancers in the genome.
-
►
Transcription factors regulate the maintenance of normal liver functions. No attempt had been made to integrate and stratify the core transcriptional network in the liver.
-
►
The expression of liver-specific genes (e.g., albumin) decreases during the state of chronic liver injury, which is accompanied by progressive loss of liver functions. Super-enhancer-associated network was not studied in this event.
Added value of this study
-
►
Genome-wide analysis of super-enhancers and the associated transcriptional network enabled us to identify LRH1, HNF4α, PPARα and RXRα as the core transcription factors in hepatocytes.
-
►
Expression of the core transcription factors in the liver is robustly suppressed in patients with hepatitis or cirrhosis caused by various etiologies, and in animal models.
-
►
LRH1 regulates the interconnected feed-forward loop for LRH1, HNF4α, PPARα and RXRα expression, and the maintenance of hepatocyte identity and normal liver function.
-
►
LRH1 levels correlate with liver-specific gene expression levels in patients or animals with liver cirrhosis (or fibrosis). LRH1 overexpression protects the liver from injurious stimuli.
Implications of all the available evidence
-
►
The information on hepatocyte-specific super-enhancers, LRH1-mediated super-enhancer circuitry, and the associated novel transcriptional network may be of great value to understand liver pathophysiology and design therapeutic targets and strategies for the treatment of liver diseases.
Alt-text: Unlabelled Box
1. Introduction
A recent genome-wide study demonstrated the existence of ‘super-enhancers’, which cover large genomic regions (several kilobases) containing clusters of closely spaced transcription factor (TF)-binding regions [1,2]. Because super-enhancers have the potential to recruit large numbers of transcriptional complexes, super-enhancer-associated genes are highly sensitive to decreases in the levels of enhancer-bound factors and cofactors. Thus, we hypothesized that TFs associated with super-enhancers regulate many subsets of genes. As key regulators, super-enhancers are cell type-specific. They are closely associated with binding of various TFs, and concentrated K27-acetylated histone H3 (H3K27Ac) modification around particular subsets of genes [1,2]. Recently, a few studies have shown the role of super-enhancers in liver physiology [[3], [4], [5]]. However, no attempt has been made to stratify and integrate the core transcriptional network under normal or disease conditions although certain TFs have been implicated as regulators that maintain normal liver function [[6], [7], [8]].
Accumulating evidence indicates that, in chronic liver diseases, expression of liver-specific genes is down-regulated, with progressive loss of parenchymal function [[9], [10], [11], [12]]. Following disease progression and subsequent functional failure, parenchymal cells, which are primarily responsible for maintaining organ function, lose their key identity genes [9,10,13]. Moreover, clinical and experimental observations indicate that loss of hepatocyte identity may be associated with the development of cirrhosis of various etiologies, or its advanced complications (e.g. hepatocellular carcinoma) [11,14,15]. However, the mechanism of parenchymal dedifferentiation and the underlying molecular basis have yet to be defined.
A genome-wide study of super-enhancer patterns is a promising way to identify master transcriptional regulators and to clarify downstream gene transcription patterns. Super-enhancers are distinguished from typical enhancers by their size, ability to recruit TFs, and transcriptional activity [1,2]. The present study was designed to identify transcriptional regulators that are highly expressed in the healthy liver, and to hierarchize them using a genome-wide analysis of super-enhancer distribution according to their interacting molecules. Thus, identifying the super-enhancer-associated network enabled us to predict LRH1, HNF4α, PPARα, and RXRα as core super-enhancer-associated TFs. Liver receptor homolog 1 (LRH1, also known as NR5A2) was sensitively and specifically inhibited as liver function worsened, whereas restoration of LRH1 level using a liver-specific hydrodynamic delivery method allowed hepatocytes to resume their normal phenotype and functions. Moreover, the results of RNA-sequencing (RNA-seq) analysis on liver of animals treated with acetaminophen (APAP), alone or in combination with LRH1 overexpression, demonstrated that a large number of clusters of hepatic identity genes are under the control of LRH1. Our results also showed that LRH1 serves a strong master TF that is necessary for the hepatocyte-specific super-enhancer circuitry, implying its potential therapeutic value for the maintenance of cell identity in response to injurious stimuli and/or during repair processes. Finally, we also examined the super-enhancer signature in hepatic stellate cells (HSCs).
2. Materials and methods
2.1. Bioinformatic analysis
H3K27Ac ChIP-seq (GSE31039) and DNase-seq (GSE83169 for mouse liver, and GSE32970 for human hepatic stellate cell) data were employed to search for enhancer regions. The peak list processed and deposited to UCSC genome browser (RRID: SCR_005780), ENCODE (RRID: SCR_015482), and GEO (RRID: SCR_005012) under the accession numbers of wgEncodeEM002500, ENCSR000CDH, and GSE31039 by the original depositor were used for our super-enhancer analysis. H3K27Ac peaks and scores (from 1 to 67) representing fold-enrichments over background values in the peak data extracted using MACS v1 by the original depositor were used for our analysis. Constituent enhancers were defined as H3K27Ac peaks with amplitude above 5. According to the previous report that firstly suggested the concept of “super-enhancer”, our analysis using super-enhancer proximity assignments method showed highly consistent results (95% agreement) with enhancer-promoter unit assignments, as supported by empirical experiments using Hi-C (i.e., 93% of the super-enhancer-promoter pairs identified by proximity occur within the same topological domains defined by Hi-C) [2]. Thus, we used the super-enhancer proximity assignments method to assess super-enhancer-associated genes in liver. Peaks adjacent within 12.5 kb distance were grouped into a single larger enhancer domain. Then, all of the enhancers were plotted in the order of enhancer magnitude. The enhancers above the contact between a line with a slope of 2 and the enhancer plotting curve were defined as super-enhancers as studied previously [1,2]. The genes in closest vicinity to super-enhancers were clustered by gene ontology (GO) using DAVID 6.7 software (https://david-d.ncifcrf.gov/, RRID: SCR_001881). The GO term ‘Molecular Function-FAT’ was used to assess the most popular functions among genes regulated by super-enhancers, and then GO terms having P-values <0.05 were displayed. Interactions of the genes having transcriptional regulatory activity were analyzed using STRING v9.1 database (http://string91.embl.de/, RRID: SCR_005223). BiNGO analysis (RRID: SCR_005736) was done using the Cytoscape 3.4.0 software application (http://www.cytoscape.org/, RRID: SCR_003032). Publicly accessible patient or mouse gene expression data were downloaded from GEO database (https://www.ncbi.nlm.nih.gov/geo/; GSE74000, GSE28619, GSE38941, GSE25097, GSE17649 and GSE68718). Motif analysis was done using JASPAR 2018 database (RRID: SCR_003030) [16]. dbCoRC database was used to generate core regulatory circuitry in the liver of mice [17]. H3K27Ac ChIP-seq data for human liver were downloaded from ENCODE (accession number: ENCSR230IMS). DNase-seq (GSE32970) data were analyzed to find super-enhancers in HSCs.
For RNA-seq analysis, the raw ‘Fragments Per Kilobase Million’ (FPKM) values were processed and normalized by logarithm and quantile normalization method. Differentially expressed genes (DEGs) by APAP were then identified using independent t-test: DEGs were selected as the genes with P-values < 0.05 with a fold-change of >2. Principal component analysis (PCA) and hierarchical clustering with Pearson's correlation of the DEGs were analyzed using R software (Bioconductor; http://www.bioconductor.org in the public domain, RRID: SCR_001905). Statistically enriched signaling pathways of clustered DEGs were ranked and categorized according to KEGG pathway using DAVID 6.8 software (https://david.ncifcrf.gov/). Cytoscape 3.4.0 software (http://www.cytoscape.org/) visualized gene interaction networks overlapped with gene expression changes from RNA-seq data.
2.2. RNA-sequencing analysis
For RNA-sequencing (RNA-seq) analysis, liver tissues from BALB/c mice treated with APAP were used (n = 3 each) as stated in the animal experiments section. Total RNA concentration was calculated by Quant-IT RiboGreen (Invitrogen). To assess the integrity of the total RNA, samples were run on the TapeStation RNA screentape (Agilent). Only high-quality RNA preparations, with RIN >7.0, were used for RNA library construction. cDNA library preparation is described in the Supporting Information. The libraries were quantified using qPCR according to the qPCR Quantification Protocol Guide (KAPA Library Quantification kits for Illumina Sequencing platforms) and qualified using the TapeStation D1000 ScreenTape (Agilent). Indexed libraries were sequenced using the HiSeq2500 platform (Illumina).
2.3. Animal experiments
Animal experiments were conducted in accordance with the guidelines of the Institutional Animal Use and Care Committee at Seoul National University. Male C57BL/6 or BALB/c mice were purchased from Charles River Orient (Seoul, Korea), and housed at 20 ± 2 °C with 12 h light/dark cycles and a relative humidity of 50 ± 5% under filtered, pathogen-free air, with food and water available ad libitum. C57BL/6 or BALB/c mice were used for CCl4- or APAP-induced injury models, respectively. Briefly, C57BL/6 mice at six weeks of age were intraperitoneally injected with a single dose of CCl4 (0.6 mL in corn oil/kg body weight), and were subjected to analyses 48 h afterward. For the multiple exposure model, mice were injected with CCl4 (0.6 mL/kg, i.p.) twice a week for six weeks. Liver tissues were excised 24 h after last dose of CCl4. For hydrodynamic gene delivery, mice were injected with the empty vector (pcDNA3.1) or a plasmid encoding LRH1 (pCMV-HA backbone) through tail vein (8.3 μg/mL plasmid in saline; 10% of body weight in volume).
It has been shown that blood ALT activity returned to normal four days after a hydrodynamic gene injection [18]. Hence, mice were exposed to a single dose of CCl4 (0.6 mL/kg, i.p.) or vehicle four days after gene delivery, and were sacrificed 48 h thereafter. Previously, ALT activities were elevated to maximum 48 h after CCl4 treatment [19]. So, the time point was chosen to assess LRH1 effect on CCl4-induced toxicity. For the APAP model, male BALB/c mice were hydrodynamically injected with the empty vector (pcDNA3.1) or a plasmid encoding LRH1 (pCMV-HA backbone) through tail vein (8.3 μg/mL plasmid in saline; 10% of body weight in volume). Four days after the injection, the mice were fasted overnight, followed by a single intraperitoneal dose of APAP (300 mg in warmed phosphate-buffered saline/kg body weight), and were subjected to analyses six h afterward. Because ALT activity peaked six h after 300 mg/kg APAP treatment [20], the time-point was selected.
2.4. Cell culture
Hepatocytes were isolated from C57BL/6 mice. The isolation of primary hepatocytes is described in the Supporting Information. AML12 cells (RRID: CVCL_0140) were purchased from American Type Culture Collection (RockvilleMD). The cells were cultured in the DMEM/F-12 containing 10% FBS, insulin-transferrin-selenium X (ITSX), dexamethasone (40 ng/mL; Sigma), and the antibiotics. The cells with <20 passage numbers were used.
2.5. Quantitative reverse-transcription polymerase chain reaction (qRT-PCR)
Total RNA was isolated from cells using TRIzol reagent (Thermo Fisher Scientific) and was reverse-transcribed. QRT-PCR was performed using a StepOne real-time polymerase chain reaction instrument (Thermo Fisher Scientific) and SYBR Premix Ex Taq II kit (Takara Bio, Shiga, Japan) according to the manufacturer's instructions. The relative levels of reverse-transcribed mRNAs were normalized based on β-actin levels. After PCR amplification, a melting curve of each amplicon was determined to verify its accuracy. For the analysis of gene expression data, the relative fold changes were normalized to the control using the 2–∆∆Ct method.
2.6. Histopathology and immunohistochemistry
The liver tissues were fixed in 10% buffered neutral formalin for six h. The samples were stained with H&E or using TUNEL method. Images of liver morphology were obtained by light microscopy or VECTRA according to the manufacturer's instruction. Liver tissue specimens were fixed in 10% formalin, embedded in paraffin, cut into four-micrometer-thick sections, and mounted on slides. Tissue sections were immunostained for HA-LRH1 using an antibody directed against HA-tag (Cell Signaling Technology, Beverly, MA, Cat# 2367, RRID:AB_10691311). The paraffin-embedded tissue sections were deparaffinized with xylene and rehydrated with alcohols series. After antigen retrieval was performed, the endogenous peroxidase activity was quenched. The sections were pretreated with 10% normal donkey serum for 40 min to block nonspecific antibody binding and were incubated with the antibody for overnight at four degrees Celsius. The sections were then treated with 2% normal donkey serum for 15 min and incubated with biotin-SP-conjugated affinity pure donkey anti-mouse IgG. The labeling was done using 3,3′-diaminobenzidine.
2.7. Transient transfection and luciferase assays
The plasmids encoding mouse LRH1 (#16342, RRID: Addgene_16,342) and mouse PPARα (#22751, RRID: Addgene_22,751) were supplied from Addgene (Cambridge, MA). The human HNF4α expression plasmid (pDGT26.1-HNF4α) was provided by Dr. T. Leff (Wayne State University, Detroit, MI). The human RXRα expression plasmid (PECE-RXRα) was provided by Dr. M. O. Lee (Seoul National University). To increase RXRα expression, the coding region of RXRα derived from PECE-RXRα (HindIII and EcoRI digestion) was subcloned into pcDNA3.1 (Invitrogen, Carlsbad, CA; i.e., pcDNA-RXRα) [21]. The cells were plated in six-well plates overnight and transiently transfected with the indicated plasmid in the presence of lipofectamine 2000 Reagent (Invitrogen, Carlsbad, CA). Super-enhancer elements were cloned into pGL3 vector containing 45-base pair minimal mouse hsp70 promoter. Luciferase activity assays were done with Luciferase assay system (Promega) according to the manufacturers' protocols.
2.8. Chromatin immunoprecipitation (ChIP) assay
Primary hepatocytes isolated from mice treated with CCl4 or vehicle for three weeks (1.2 mL/kg body weight, twice a week) were fixed with 1% formaldehyde for cross-linking of chromatin. The ChIP assay was done using anti-H3K27Ac antibody (Cell Signaling Technology, Beverly, MA, Cat# 8173, RRID: AB_10949503) according to the EZ ChIP kit protocol (Upstate Biotechnology, Lake Placid, NY). One tenth of cross-linked lysates served as the input control.
2.9. Statistics
Student's t-test was performed to assess the significance of differences among experimental groups. Pearson's coefficients with associated P-values were used for correlation analysis. Chi-squared test was done to determine P-values for comparison of proportions.
3. Results
3.1. Identification of core TFs in the mouse liver
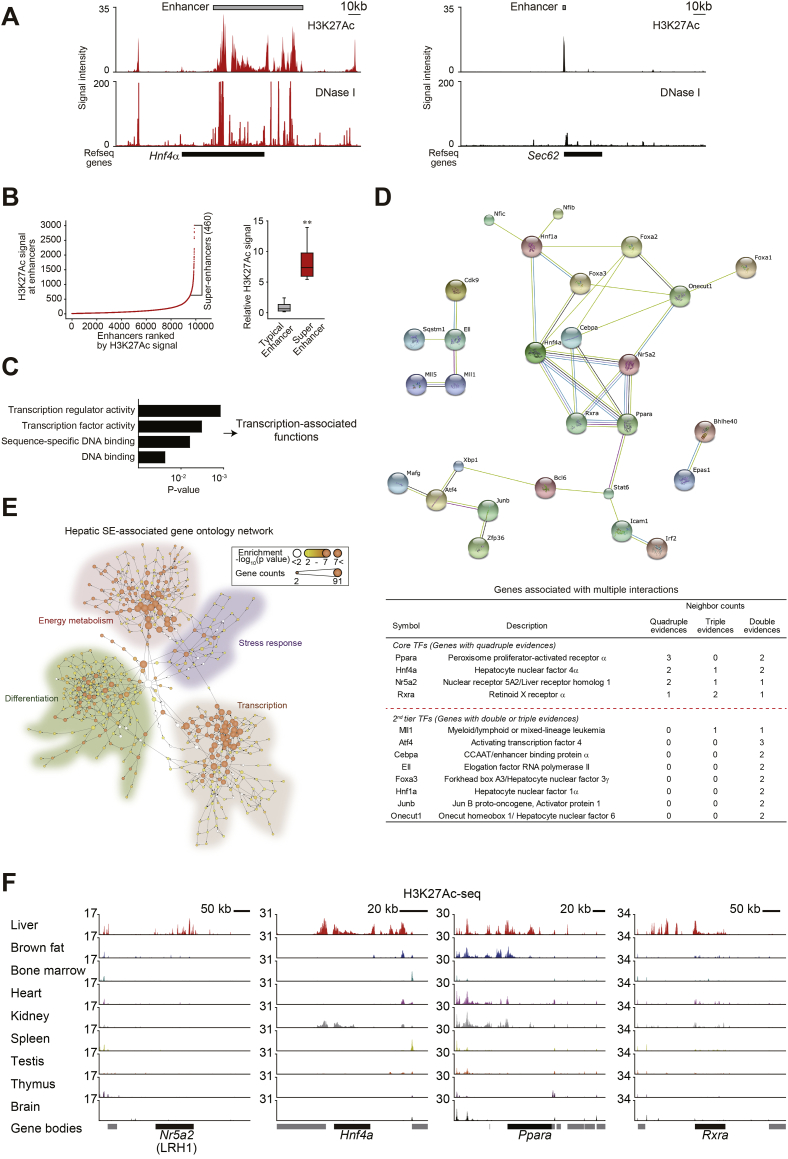
To identify super-enhancer-associated master regulators in the liver, we analyzed the mouse H3K27Ac ChIP-seq and DNase-seq data available in the ENCODE databases (GSE31039 and GSE83169). The enhancer signals proximal to the coding genes in the liver were separated into two distinct groups. Of note, some portions of the genome contained clusters of enhancers spanning as much as 50 kb (i.e. a super-enhancer feature) although a vast majority of enhancers spanned DNA segments of a few hundred base pairs (i.e. typical enhancers) (Fig. 1A). A similar characteristic division was also observed in cells of other organs such as embryonic stem cells, myotubes, and blood cells (ref. [2]). To classify genes associated with hepatic super-enhancers, distribution of enhancer signals across the mouse liver genome was plotted according to signal intensity (Fig. 1B, left and Supporting Table S1). Average signal intensity of the super-enhancers was clearly distinguishable from that of typical enhancers (Fig. 1B, right). Further analysis of GO suggested that genes encoding TFs were enriched among 460 hepatic super-enhancer-associated genes (Fig. 1C).
Fig. 1.
The core TFs identified in the liver.
(A) H3K27Ac ChIP-seq and DNase-seq profiles at the Hnf4a (left) and the Sec62 (right) loci in mouse liver. Gray bars indicate enhancer regions.
(B) Distribution of H3K27Ac ChIP-seq signal intensities across 9891 enhancers in the liver. H3K27Ac occupancy was not evenly distributed across the enhancer regions, with a subset of 460 enhancers containing exceptionally high amounts of H3K27Ac (i.e., super-enhancers) (left). A box plot of H3K27Ac ChIP-seq densities at constituent enhancers within 9431 typical enhancers or 460 super-enhancers (right).
(C) Gene ontology (GO) functional categories regarding molecular functions for super-enhancer-associated genes. Genes encoding for the factors controlling transcription were enriched.
(D) A protein-protein interaction network of super-enhancer-associated transcription factors (TFs) according to STRING database. LRH1 (also known as Nr5a2), HNF4α, PPARα, and RXRα make a core network. The TFs were divided into two groups (multiple interactions with quadruple evidences and multiple interactions with double or triple evidences) according to the number of evidences in the above network. The red dotted line designates the cutoff dividing core and second-tier TFs.
(E) A network displaying interactions between GO categories. Each node indicates GO term. The thickness of node colour represents the degree of statistical significance for enrichment. Node sizes show the number of gene counts assigned to each GO term. The network was generated by analysis of Cytoscape plugin BiNGO.
(F) H3K27Ac ChIP-seq data at the loci of Nr5a2 (LRH1), Hnf4a, Ppara, and Rxra. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
With the aim of identifying key TFs associated with the super-enhancers, a protein-protein interaction network of TFs was produced using the STRING database, and TFs were then grouped according to number of interactions: 1) core TFs having four recorded interactions, and 2) second-tier TFs having three recorded interactions with another TF(s) or less (Fig. 1D, upper). Analyses of co-expression, protein homology and text-mining using STRING database provide the information for not only physical interaction between proteins, but their functional relevance (ref. [22]). Both core and second-tier TFs included liver-enriched transcriptional proteins. Moreover, our findings showed that LRH1 (also known as Nr5a2), HNF4α, PPARα, and RXRα constituted a core network (Fig. 1D, lower). Putative motif analysis using JASPAR database showed congruent existence of the binding motifs of each core TF in the super-enhancer of each gene (Supplementary Fig. S1A). Mouse liver H3K27Ac data was additionally analyzed using dbCoRC, one of the super-enhancer analysis tools to define core TF circuitry based on ROSE and CRCmapper (Supplementary Fig. S1B). The dbCoRC analysis similarly showed a LRH1-containing core regulatory circuitry.
It has been shown that super-enhancers and target gene expression may define cell identity (ref. [2]). To investigate whether super-enhancer-associated genes in hepatocytes include those related to features of liver function, super-enhancer-associated genes were analyzed for biological process enrichment by functional GO clustering. Genes responsible for metabolism of lipids, carbohydrates, amino acids, and cholesterol belonged to the most strongly enriched clusters of super-enhancer target genes (Supporting Table S2), supporting the notion that super-enhancer target genes may encode the regulators necessary to establish and maintain normal healthy liver function. To better understand the role of the super-enhancer-associated gene network, annotated biological pathways of the genes were hierarchized using BiNGO analysis, and the outcome was visualized (Fig. 1E and Supporting Table S3). The pathways of energy metabolism, stress response, differentiation, and transcription were clustered together, indicating that the super-enhancer-associated gene network might be indispensable for maintaining hepatocyte differentiation and viability. In line with this, the super-enhancer signals at the core TF loci (Nr5a2, Hnf4a, Ppara and Rxra) showed a highly liver-enriched pattern in healthy liver (Fig. 1F), confirmative of their roles in liver specification. We also found that the genomic sequences at the super-enhancer regions residing Nr5a2, Hnf4a, Ppara and Rxra were highly conserved across different species including Homo sapiens (Supplementary Fig. S1C, left). Moreover, analysis of the H3K27Ac-seq data for human adult liver from ENCODE (ENCSR230IMS) displayed signal patterns similar to those in mouse liver (Supplementary Fig. S1C, right).
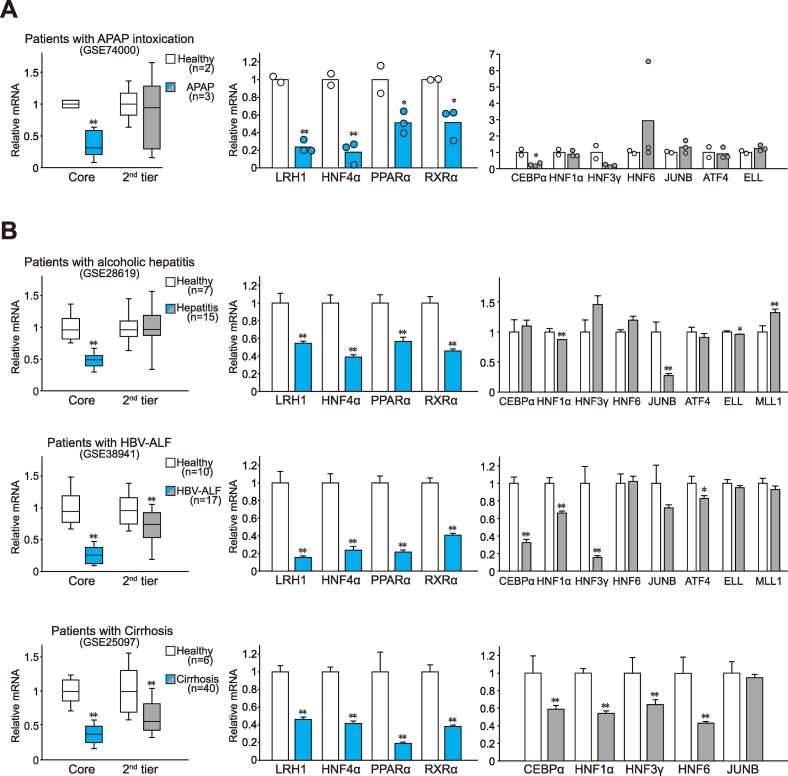
3.2. Repression of hepatic core TFs in patients and mice with hepatitis or cirrhosis
Since genes associated with super-enhancers are in a highly activated state of transcription under basal conditions, we hypothesized that they would also be more sensitive to liver injury elicited by various etiologies. To assess this possibility, we compared the core and the second-tier TFs' transcript levels in healthy individuals with those from patients with APAP intoxication, and found that core TF transcript levels were markedly lower in the patient group (Fig. 2A). However, expression levels of second-tier TFs were in general comparable between groups. Interestingly, analyses of databases for patients with alcoholic hepatitis, HBV-induced acute liver failure or liver cirrhosis robustly reproduced similar patterns (Fig. 2B). The core TF mRNA levels (LRH1, HNF4α, PPARα, and RXRα) were all lowered across different etiologies of liver disease, whereas the second-tier TFs were unchanged or only weakly-to-moderately diminished.
Fig. 2.
Loss of core TFs in the liver of patients with hepatitis or cirrhosis from various etiologies.
(A–B) Averages of the core or the second tier of TF transcript levels in the liver of patients with acetaminophen (APAP) intoxication (A), alcoholic hepatitis, hepatitis B virus-induced acute liver failure (HBV-ALF), or cirrhosis (B). Data were extracted from GSE74000 (quantile normalization), GSE28619 [Gene Chip Robust Multiarray Averaging (GC-RMA) normalization], GSE38941 (RMA normalization), and GSE25097 (RMA normalization). Gene expression changes were calculated relative to the respective healthy liver group included in each dataset.
Data information: For left of A and B, data were shown as box and whisker plot (significantly different as compared with healthy donors: *P < .05; **P < .01). Box, IQR; whiskers, 5–95 percentiles; and horizontal line within box, median. For middle and right of A and B, data represent the means ± SEM (significantly different as compared with healthy donors: *P < .05; **P < .01). Group sizes (n) are denoted in each figure.
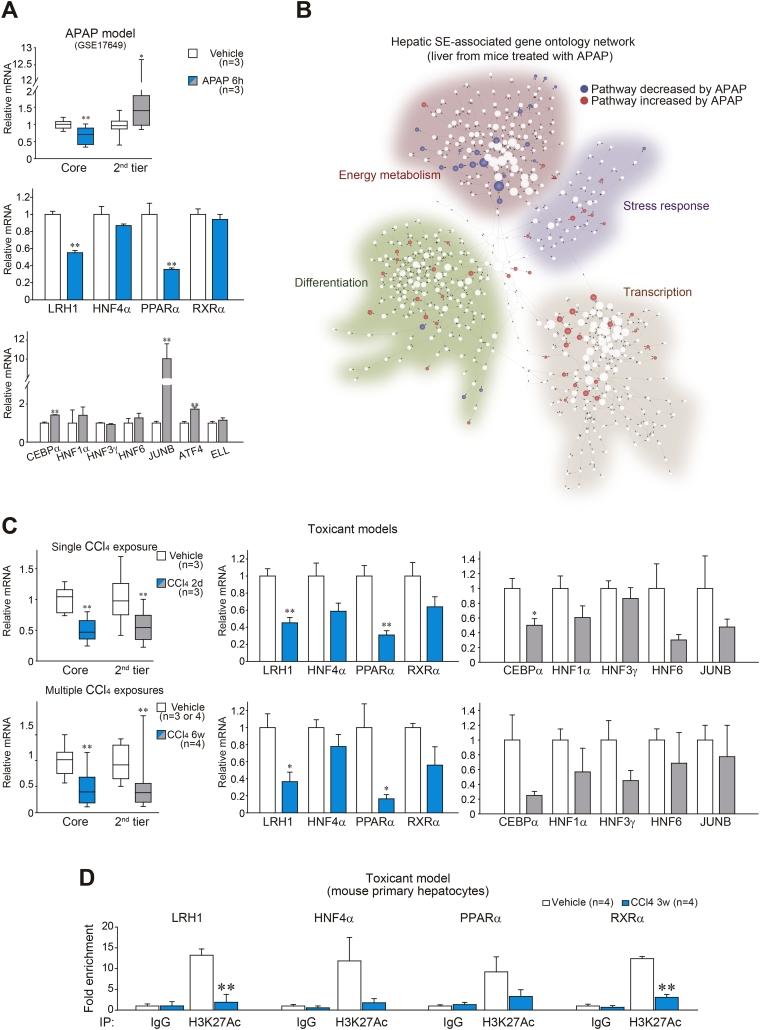
To assess whether this change in expression occurs in a hepatocyte-specific manner, we employed mouse models of liver injury, using liver toxicants (APAP and CCl4), and of liver fibrosis. The toxicants were chosen because they directly and specifically damage hepatocytes after primary biotransformation (ref. [23,24]). In the APAP mouse model, core TF transcript levels were significantly decreased six h after APAP treatment (Fig. 3A), concordant with the outcome obtained from the human database analyses. In this model, only LRH1 and PPARα mRNA levels were significantly repressed, implying that they might have particular sensitivity to drug-induced acute liver injury. To globally assess the genes affected by an acute intoxication of APAP, we performed RNA-seq analysis using the mouse liver tissue, and found notable down- or up-regulated transcriptional alterations in major super-enhancer-associated pathways (Fig. 3B, Supplementary Fig. S2A and B). The pathways related to energy metabolism (a normal function of healthy liver) were markedly suppressed upon liver injury, whereas others related to stress response, differentiation and transcription (necessary functions for cell survival) were adaptively activated. The core TFs' transcript levels were similarly suppressed two days after a single exposure to CCl4 (Fig. 3C, upper). The second-tier TFs were also repressed. Of note, LRH1 and PPARα mRNAs were significantly lowered shortly after a single injection with CCl4. In mice repetitively treated with CCl4 for six weeks, these changes were reproducible (Fig. 3C, lower). All of the outcomes from the analyses of human databases and animal experiments provide strong evidence that hepatocyte injury is accompanied prominently by the inhibition of core TFs.
Fig. 3.
Loss of core TFs in mouse liver disease models.
(A) TF transcript levels in the liver of mice treated with APAP. Data were extracted from GSE17649 (Affymetrix global scaling normalization). Gene expression changes in liver were calculated relative to vehicle-treated group.
(B) Hepatic super-enhancer-associated GO network displaying RNA-seq expression data from the liver of mice treated with APAP. Decreased or increased pathways were depicted as a blue or red node, respectively. RNA-seq data were generated using the mouse liver tissues obtained six h after APAP treatment. RNA-seq data are deposited in the GEO under accession number GSE104302.
(C) TF transcript levels in the injured liver of mice. The mice were sacrificed two days after a single injection of CCl4, or multiple injections of CCl4 for six weeks.
(D) ChIP assays. Crosslinked protein–DNA complexes were immunoprecipitated using anti-H3K27Ac antibody or preimmune-IgG (negative control) in primary hepatocytes isolated from mice treated with vehicle or CCl4 for three weeks. qPCR assays were done to quantify DNAs in the immunoprecipitates using specific primers for each super-enhancer region.
Data information: For upper panel of A and left of C, data were shown as box and whisker plot (significantly different as compared with vehicle-treated control, *P < .05; **P < .01). Box, IQR; whiskers, 5–95 percentiles; and horizontal line within box, median. For lower panels of A and middle and right of C, data represent the means ± SEM (significantly different as compared with vehicle-treated controls, *P < .05; **P < .01). Group sizes (n) are denoted in each figure. For D, data represent the means ± SEM (n = 4 each, significantly different as compared to vehicle control, **P < .01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
To understand whether reductions of core TFs are associated with alterations of super-enhancer activity in hepatocytes, we examined the status of histone acetylation at super-enhancer sites near the transcription start sites (TSSs) of the target genes. In ChIP assays using primary hepatocytes isolated from mice treated with multiple doses of CCl4 for three weeks, chromatin of the enhancer region containing LRH1 significantly changed toward the closed state (Fig. 3D), consistent with a decrease in the level of LRH1 transcript. Likewise, chromatin of the enhancer regions containing RXRα similarly (but to a lesser degree) changed, as indicated by a decrease in H3K27Ac level. A similar tendency was also observed in the chromatin status of the enhancer regions containing HNF4α and PPARα. Our results raised the notion that the identified core TFs may serve as a vanguard contributing to overall transcriptomic perturbations in response to harmful stimuli.
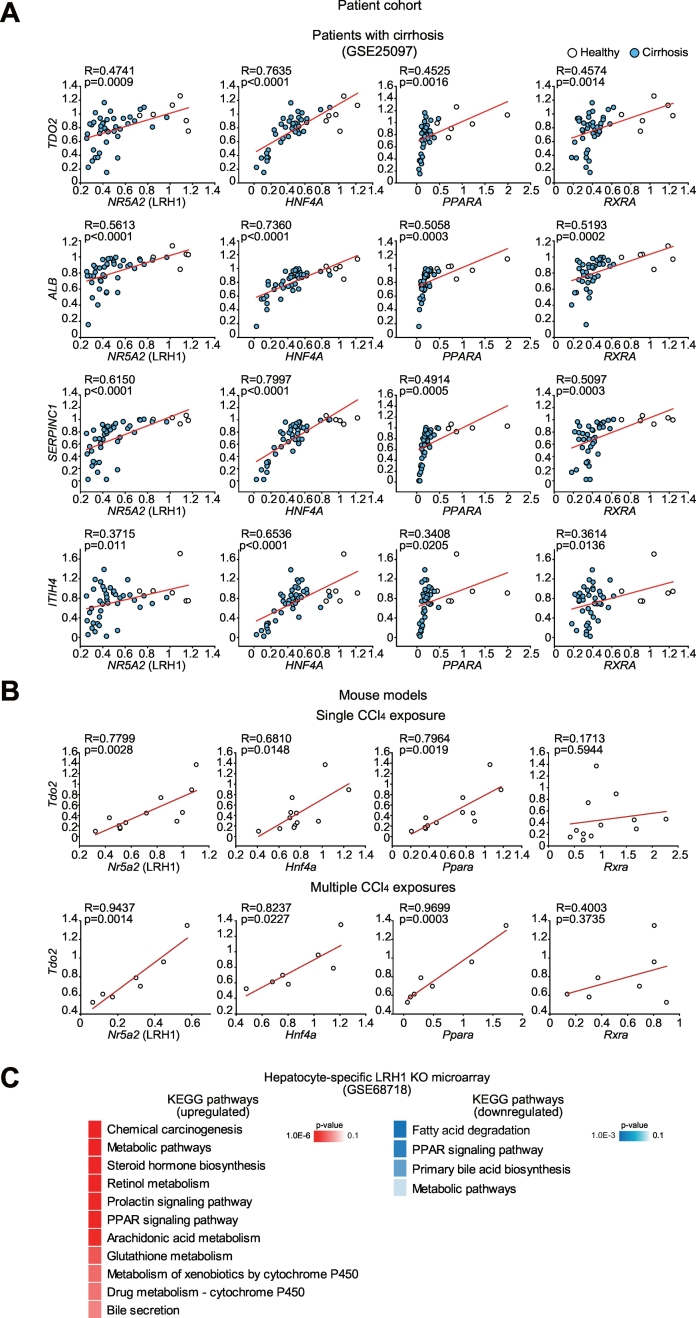
3.3. Correlation of expression between the core TFs and hepatocyte identity genes
In a disease state, hepatocytes lose their normal function, with concomitant decreases in key identity genes such as albumin and anti-thrombin [12,25,26]. Because core TFs were robustly decreased in different conditions of liver diseases, we were tempted to analyze the relationship between core TFs and representative liver identity genes, using a cohort database (GSE25097) for patients with cirrhosis and healthy volunteers. As expected, NR5A2 (LRH1), HNF4A, PPARA, and RXRA mRNA levels each correlated strongly with those of major hepatocyte identity genes (i.e. ALB, albumin; SERPINC1, anti-thrombin; TDO2, tryptophan 2,3-dioxygenase; and ITIH4, inter-alpha-trypsin inhibitor heavy chain family member 4) (Fig. 4A). To confirm the outcomes experimentally, we also used mouse models of liver injury (acute or multiple exposures), and found that Tdo2 (a key liver identity gene) transcript levels strongly correlated with those of Nr5a2, Hnf4a and Ppara, (Fig. 4B). In this approach, however, the correlation between Tdo2 and Rxra mRNA levels was not statistically significant. In the subsequent experiments, emphasis was placed on LRH1 because of its incomplete physiological information despite its high correlation with hepatocyte identity gene expression and specific presence of H3K27Ac signals around the gene in liver (Fig. 1F). Among the core TFs, signals for Ppara were strong in brown adipose tissue, bone marrow, heart, and kidneys as well as liver, presumably because PPARα plays significant roles in normal physiology of these tissues [[27], [28], [29], [30]]. Additional pathway analysis using public microarray data (GSE68718) from hepatocyte-specific LRH1 knockout mouse liver showed that basal functions of LRH1 in hepatocytes mainly associate with hepatocyte-specific functions (Fig. 4C).
Fig. 4.
Correlation between identified core TF and each gene transcript levels.
(A) Correlation analyses in a large cohort of cirrhosis patients (GSE25097) (N = 46). Pearson's r correlation coefficients with corresponding P-values for co-variation between each core TF mRNA levels (x axis) and hepatocyte identity gene transcripts (y axis) show robust correlations.
(B) Pearson correlation analyses in mice treated as in Fig. 3C. For single and multiple CCl4 treatment models, group sizes (N) are 12 and 7, respectively.
(C) Results of KEGG pathway analysis of the up- or down-regulated genes after hepatocyte-specific deletion of LRH1 (GSE68718). Enriched signaling pathways of each gene cluster were analyzed using DAVID.
3.4. Effect of LRH1 on toxicant-induced liver injury
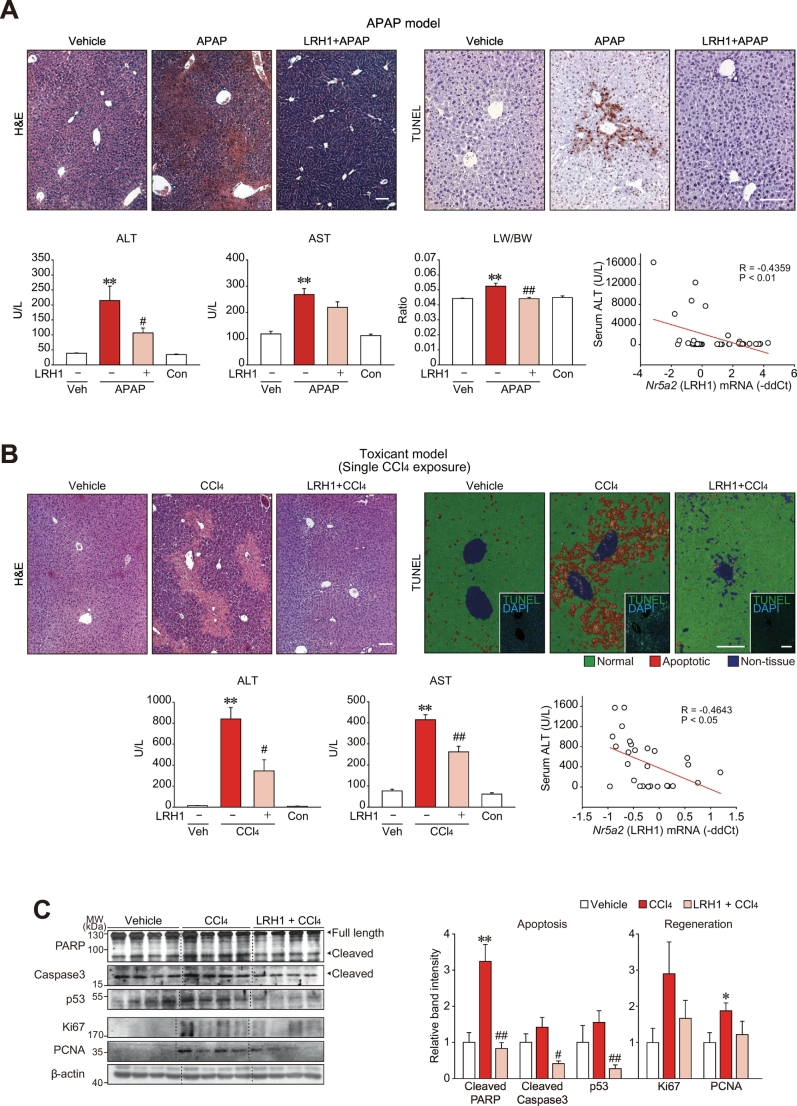
Having identified the strong intensity and liver specificity of enhancer signatures at the Nr5a2 locus, we hypothesized that LRH1 plays a role in the activation of the core TF network, providing a therapeutic advantage for treatment of liver disease. In this approach, we injected mice with a single sublethal dose of APAP after liver-specific delivery of a plasmid encoding LRH1 (or a control vector) in vivo using hydrodynamic injection. Overexpression of LRH1 in the liver was confirmed by immunohistochemistry and qRT-PCR assays (Supplementary Fig. S3A and B). LRH1 overexpression alone without toxicant treatment did not induce histological changes compared to the control liver, nor show toxicity (Supplementary Fig. S3C). In addition, LRH1 overexpression without toxicant treatment did not induce Hnf4a, Ppara, and Rxra expression, albeit enforced expression of LRH1 increased Hnf4a and Rxra transcript levels in mice treated with APAP or CCl4. The lack of effect by LRH1 alone may be due to high basal expression of Nr5a2, Hnf4a, Ppara, and Rxra in healthy liver. Of note, enforced expression of LRH1 protected hepatocytes from APAP challenge, as evidenced by the outcomes of histopathology, TUNEL staining examinations, serum ALT activities, and liver weight to body weight ratios (Fig. 5A). Serum AST activities were not significantly changed, but showed a decreased tendency. An inverse correlation between LRH1 transcript levels and serum ALT activities also supported the physiological impact of LRH1 on hepatocyte survival (Fig. 5A, lower right).
Fig. 5.
LRH1 protection of the liver from toxicant-induced injury.
(A) LRH1 protection of the liver from APAP-induced injury. H&E staining (upper left). At four days after a hydrodynamic injection of the plasmid encoding LRH1 or mock vector (pcDNA3.1), mice were fasted overnight and subjected to a single dose of APAP (300 mg/kg), and the liver tissues were obtained six h afterward. TUNEL staining (upper right). The scale bars represent 100 μm. Serum ALT and AST activities (lower left). Liver weight per body weight ratio (middle). Correlation between serum ALT activities and LRH1 transcript levels in the liver (lower right).
(B) LRH1 protection of the liver from CCl4-induced injury. H&E staining (upper left). Mice were subjected to a single dose of CCl4 (0.6 mL/kg) four days after a hydrodynamic injection of the plasmid encoding LRH1 or mock vector (pcDNA3.1), and the liver tissues were obtained two days afterward. TUNEL staining (upper right). TUNEL-stained tissues were separated to non-tissue, normal and apoptotic areas by blue, green and red colors, respectively. Insets showed true-colour images. The scale bars represent 100 μm. Serum ALT and AST activities (lower left). Correlation between serum ALT activities and LRH1 (Nr5a2) mRNA levels in the liver (lower right).
(C) Immunoblottings for apoptosis or liver regeneration markers (left). Values were obtained using scanning densitometry (right).
Data information: For A, data represent the means ± SEM (Mock+Veh, n = 7; Mock+APAP, n = 8; LRH1 + APAP, n = 13; and non-injected control (Con), n = 5, significantly different as compared to vehicle control, **P < .01; or APAP-treated control, #P < .05; ##P < .01). For B, data represent the means ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; LRH1 + CCl4, n = 4; and non-injected control (Con), n = 4, significantly different as compared to vehicle control, **P < .01; or CCl4-treated control: #P < .05; ##P < .01). For C, data represent the means ± SEM (n = 4 each, significantly different as compared to vehicle control, *P < .05; **P < .01; or CCl4-treated control: #P < .05; ##P < .01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
As an additional liver toxicant, we used CCl4, and obtained almost identical results, which confirmed the hepatoprotective effect of LRH1 against toxicants (Fig. 5B). Ectopic expression of LRH1 primarily at the peri-central region, as shown in Supplementary Fig. S3A, might be sufficient to protect liver from APAP or CCl4 presumably because the toxicities mainly occur at the centrilobular zone. Consistent with histochemical changes and blood biochemical parameters, apoptosis biomarker levels (i.e., cleaved PARP, cleaved caspase-3, and p53) were all normalized (Fig. 5C). In addition, increases of Ki67 and PCNA (regeneration markers) by CCl4 were slightly diminished by LRH1 overexpression, excluding the possibility that LRH1 stimulates cell proliferation. The absence of any effect of LRH1 on CYP2E1, an enzyme responsible for CCl4 bioactivation, confirmed that LRH1 indeed protected liver from injury during unaltered toxicogenic process of CCl4 biotransformation (Supplementary Fig. S3D). All of these results support the contention that overexpression of LRH1 diminished hepatotoxicity, possibly allowing recovery of normal liver function.
3.5. Role of LRH1 in maintenance of hepatocyte viability and function
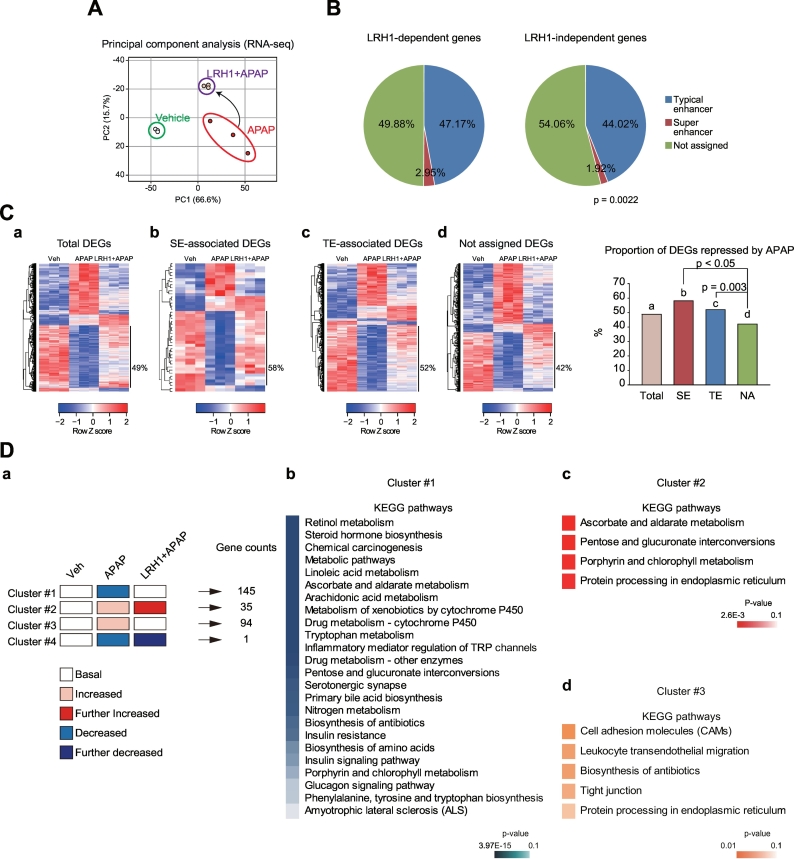
To validate the role of LRH1 in maintenance of normal liver function, we next performed RNA-seq analyses using the liver tissues of mice subjected to liver-specific delivery of LRH1 prior to APAP intoxication. Principal component analysis of the gene expression data revealed that LRH1 overexpression plus APAP treatment elicited major changes in gene expression compared with APAP treatment alone (Fig. 6A). First, we defined LRH1-dependent genes according to the statistical significance of the comparison between LRH1 + APAP and APAP alone groups; the differentially expressed genes (DEGs) were classified into separate groups based on enhancer features in their genomic vicinity (i.e., genes having nearest TSS from each enhancer). All genes not comprised in the LRH1-dependent gene list, including non-expressed genes, were classified as LRH1-independent genes. Of the total number of genes identified, LRH1-dependent super-enhancer-associated genes were ~3.0% in hepatocytes, whereas LRH1-independent super-enhancer-associated genes were only ~1.9% (Fig. 6B). Thus, it is highly likely that super-enhancer-associated genes are sensitively affected by LRH1 in hepatocytes. We further sought to assess global changes in the expression of super-enhancer-associated genes. Among all differentially expressed genes (total DEGs, 1338 genes), APAP intoxication inhibited expression of 653 genes (49%), which was prevented by LRH1 overexpression (Fig. 6Ca). More importantly, a larger portion of the super-enhancer-associated genes were suppressed by APAP (25 out of 43 genes, ~58%), which was also prevented by LRH1 (Fig. 6Cb and Supplementary Fig. S4). Typical enhancer-associated DEGs similarly affected by APAP plus LRH1 were 52% (320 out of 615) (Fig. 6Cc and Supplementary Fig. S5). Among DEGs not assigned to enhancers, the proportion was 42% (286 out of 680) (Fig. 6Cd). Proportions of the gene clusters (LRH1-preventable genes inhibited by APAP) among the groups having different enhancer features in their genomic vicinity were compared in a graph (Fig. 6C, right). These results support the notion that LRH1-dependent changes in expression in the hepatic super-enhancer-associated genes may be biased toward repression after intoxication.
Fig. 6.
LRH1-dependent recovery of hepatocyte identity genes.
(A) Principal component analysis of RNA-seq data. RNA-seq data was generated using liver of mice treated with APAP alone or APAP+LRH1 overexpression vector as in Fig. 5A. RNA-seq data are deposited in the GEO under accession number GSE104302.
(B) Pie graphs showing the enhancer composition. Genes having P-values lower than 0.05 between APAP and LRH1 + APAP groups were defined as LRH1-dependent genes.
(C) Heatmaps and hierarchical correlation analyses of differentially expressed genes (DEGs). DEGs were selected as the genes with independent t-test (P-values < .05 with a fold-change of >1.5). The DEGs were hierarchically clustered and presented as heatmaps according to the row Z score. Super-enhancer- or typical enhancer (SE or TE)-associated DEGs represent significantly altered genes in the APAP group as compared to the vehicle group among the SE-associated or TE-associated genes. Not assigned DEGs are DEGs which are assigned neither to SE nor TE. Heatmaps of total DEGs (a), SE-associated DEGs (b), TE-associated DEGs (c), and not assigned DEGs (d) are presented in left. The proportions of the gene clusters depicted in the heatmaps (left) were shown as a graph (right). (D) Results of KEGG pathway analysis of the clustered DEGs. A schematic description of the gene clusters (a). Enriched signaling pathways of each gene cluster were analyzed using DAVID (b-d).
Next, we performed KEGG analyses using the above liver samples to assess functions of four clusters showing differential expression patterns (Fig. 6Da), and found that cluster #1 (LRH1-preventable genes inhibited by APAP) might represent the pathways characteristic of liver identity (Fig. 6Db, and Supporting Table S4). The genes in cluster #2 (LRH1-augmented genes inducible by APAP) might encode effector molecules related to stress responses for cell survival, which was adaptively enhanced by LRH1 overexpression (Fig. 6Dc, and Supporting Table S5). Moreover, the genes in cluster #3 (i.e., LRH1-preventable genes inducible by APAP) constituted a larger portion, representing inflammation pathways (Fig. 6Dd, and Supporting Table S6). The genes assigned to cluster #4 did not show significantly enriched pathway(s) due to low gene counts. Overall, outcomes of the RNA-seq data analysis enabled us to confirm LRH1 as a key regulator of the genes associated with hepatic identity, adaptive survival pathways, and inhibition of inflammation.
In another intoxication animal model using CCl4, a set of genes including Gclm, Gsta1, Cpt1a, and Cd36 were repressed, whereas others (Tfam and Gsta2) were induced, possibly due to compensatory responses (data not shown). LRH1 overexpression attenuated the inhibitory effect on the genes. In addition, the transcript levels of Ppargc1a, Mt-co1, and Gsta2, which belong to the genes involved in adaption to external stresses, were increased. Our results corroborate the hypothesis that LRH1 plays a role in transcription of the genes responsible for liver identity, and those necessary for maintenance of normal hepatocyte viability and function (e.g., hepatocyte adaptation to external stresses).
3.6. Identification of LRH1-driven TF circuitry
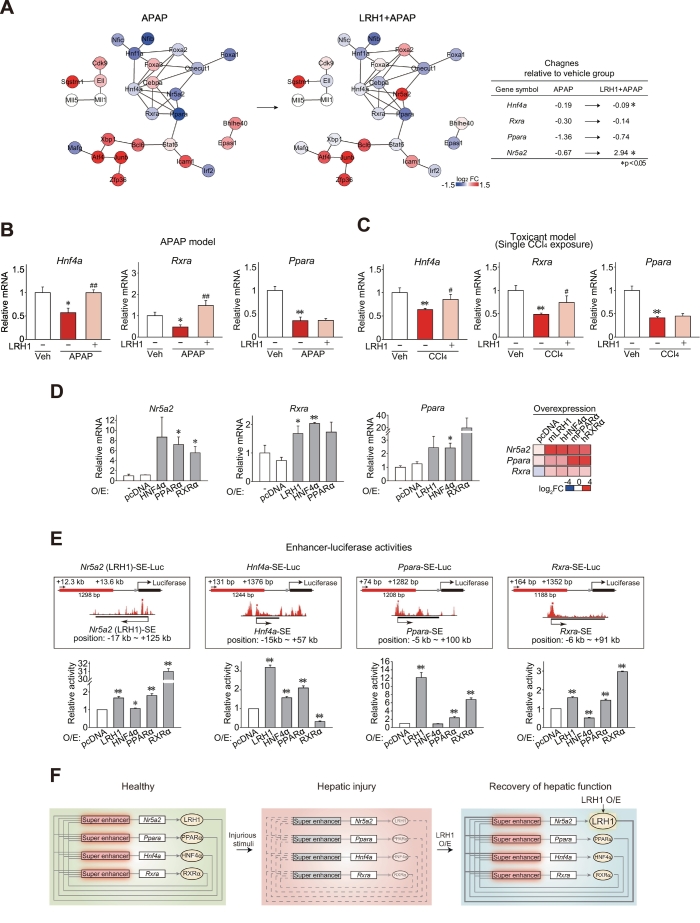
To further delineate the effect of LRH1 on the genes associated with super-enhancers, expression profiles from RNA-seq data were overlapped with those in the hepatocyte super-enhancer TF network presented in Fig. 1D. Similarly to the liver identity genes, APAP treatment inhibited expression of the core TF genes that were directly connected to LRH1 in the network. Moreover, expression of the core TF genes was mostly recovered by LRH1 overexpression (Fig. 7A). This result raised a notion that the identified core TFs (i.e. LRH1, HNF4α, PPARα, and RXRα) might form an auto-regulatory loop for mutual induction to maintain hepatocyte identity and cell-specific functions.
Fig. 7.
LRH1 as a driver gene for the core TF circuitry.
(A) Hepatic super-enhancer-associated TF network with gene expression changes in mice treated with APAP alone or APAP+LRH1 overexpression vector. The node colors reflect log2 gene expression ratio in mice treated with APAP alone (left) or APAP+LRH1 overexpression (right) as compared to vehicle treatment (red, upregulation; blue, downregulation). Log2 fold changes of the core TFs are presented as an inset table.
(B) The core TF mRNA levels from the APAP model.
(C) The core TF mRNA levels from the CCl4 model.
(D) The effect of each core TF overexpression on other core TFs. qRT-PCR assays were done on AML12 cells transfected with pcDNA3.1, LRH1, HNF4α, PPARα or RXRα for 48 h. The first lane of each graph is transfection reagent-treated control. Heatmap presents averages of core TF mRNA levels. O/E, overexpression.
(E) Super-enhancer (SE)-luciferase reporter assays. Luciferase assays were done on AML12 cells co-transfected with each SE-luciferase reporter, and pcDNA3.1, LRH1, HNF4α, PPARα or RXRα overexpression vector for 24 h. Relative luciferase activities represent arbitrary units of luminescence normalized to the pcDNA3.1 group. The schematic illustrations showing each SE-luciferase construct are presented in the upper panel. The ChIP-seq signal peaks in the scheme are also shown in Fig. 1F. Red bars indicate the peaks excised for cloning of each SE-luciferase reporter construct. O/E, overexpression.
(F) A proposed scheme showing auto-regulatory loops for the core TFs. In healthy liver, the core TFs form an interconnected feedback loop for gene expression. Upon injury, the signal circuitry loses its integrity with decrease of hepatocyte identity. LRH1 serves a driver for reconstitution of the signal circuitry.
Data information: For B, data represent the means ± SEM (Mock+Veh, n = 7; Mock+APAP, n = 8; and LRH1 + APAP, n = 13, significantly different as compared to vehicle control, **P < .01; or APAP-treated control, #P < .05; ##P < .01). For C, data represent the means ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; and LRH1 + CCl4, n = 4; significantly different as compared to vehicle control, *P < .05; **P < .01; or CCl4-treated control: #P < .05; ##P < .01). For D and E, data represent the means ± SEM (n = 3 each, significantly different as compared to pcDNA3.1 group, *P < .05; **P < .01). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
We also assessed the effects of LRH1 on core TF transcript levels in vivo using qRT-PCR assays. These showed that enforced expression of LRH1 promoted recovery of Hnf4a and Rxra transcript levels in mice treated with APAP (Fig. 7B). Levels of Ppara mRNA were unaffected. These changes were robustly reproducible in the mouse model of CCl4 (Fig. 7C). In qRT-PCR assays, overexpression of LRH1 in AML12 cells increased the level of RXRα transcript, whereas HNF4α increased RXRα and PPARα transcripts (Fig. 7D, left). PPARα overexpression promoted LRH1 transcript. RXRα increased LRH1 transcript level. Of note, LRH1 mRNA increase was strongest after overexpression of each of the other core TFs, as depicted by relative changes in the heatmap (Fig. 7D, right).
To test whether the core TFs had an auto-regulatory effect, we excised a DNA fragment with the highest peak of H3K27Ac from each of the super-enhancer elements (SEs) of Nr5a2, Ppara, and Rxra, and cloned them into luciferase reporter constructs (i.e., Nr5a2-SE-Luc, Ppara-SE-Luc, and Rxra-SE-Luc) (Fig. 7E, upper). For Hnf4a-SE-Luc, the TSS-proximal H3K27Ac peak was cloned because of the failure to amplify a DNA fragment having the strongest peak of H3K27Ac. Enforced expression of LRH1, PPARα, or RXRα significantly increased luciferase expression from Nr5a2-SE-Luc construct (Fig. 7E, lower left). LRH1, HNF4α, or PPARα overexpression enhanced reporter activity from Hnf4a-SE-Luc, whereas LRH1, PPARα, or RXRα overexpression did so from Ppara-SE-Luc (Fig. 7E, lower middle). Rxra-SE-Luc activities were promoted by LRH1, PPARα, or RXRα (Fig. 7E, lower right). Of the core TFs, LRH1 overexpression stimulated luciferase expression from all of the super enhancer reporter constructs, suggestive of its key role in the super-enhancer-associated auto-regulatory loop in hepatocytes.
Overall, these results show that the core TFs may facilitate the expression of each other, and contribute to recovery of the super-enhancer-associated auto-regulatory loop against injurious stimuli (Fig. 7F).
3.7. Super-enhancers in HSCs
To clarify the hepatocyte-specific features of identity gene expression, we compared the super-enhancer signatures in HSCs. Levels of markers indicating non-parenchymal cell activation did not differ between the LRH1-negative and LRH1-positive groups (i.e. F4/80 for macrophages, and α-SMA for HSCs) (Supplementary Fig. S6A). The super-enhancers as marked by H3K27Ac for the core TFs of mouse liver appeared to be not active in mouse Kupffer cells except for RXRα (Supplementary Fig. S6B). Moreover, HSCs and hepatocytes showed distinct super-enhancer signatures, with different key TF profiles. Analysis of the human HSC DNase-seq database (GSE32970) and the distribution of enhancer signals by magnitude across the genome enabled us to annotate 40,421 enhancers and identify 865 super-enhancers (Supplementary Fig. S6C). In the GO analysis, the genes encoding effector molecules related to cellular responses to retinoic acid and extracellular matrix organization (i.e. well-characterized features of HSCs) were found to be super-enhancer genes in HSCs (Supplementary Fig. S6D).
The protein-protein interaction network of super-enhancer-associated genes in HSCs contained two subsets of core transcriptional regulators: one for nuclear receptors related to retinoic acid, and the other for SMAD-TGFβ signaling. Each cluster comprised a subset of core transcriptional regulators (Supplementary Fig. S6E). In cluster #1, Nr5a1 and Nr2f6 levels were marginally enhanced in activated HSCs, whereas those of Nr2f1, Rarg, Nrfd1, and Nr4a1 were suppressed (Supplementary Fig. S6F). Rara was unchanged. In cluster #2, Smad3 levels significantly increased following suppression of Smad6, an inhibitory SMAD. Smad7 remained unchanged. HSCs showed a distinct super-enhancer profile (i.e. loss of retinoic acid response, and an increase in TGFβ signal), corroborating the cell-type specificity of super-enhancers and the differing roles of non-parenchymal cells in disease progression.
4. Discussion
Genes in close proximity to super-enhancers encode a set of proteins critical to determining cell identity. Super-enhancers are therefore regarded as key regulators of cell identity [1,2]. In previous studies of super-enhancers in embryonic stem cells, myotubes, and blood cells [2], cell identity appeared to be determined by distribution of super-enhancers in the genome. However, whether super-enhancers globally regulate normal liver function, and, if so, whether loss of super-enhancer function precedes progression of liver disease, remained unknown. TFs frequently form an auto-regulatory loop to reinforce their activity [[31], [32], [33]]. Our genome-wide cistromic analysis of super-enhancer distribution showed that LRH1, HNF4α, PPARα, and RXRα are core regulators that are highly expressed and interconnected in healthy hepatocytes. Our results revealed an LRH1-driven hepatocyte-specific auto-regulatory transcription loop, and this loop pathway contributes to maintaining hepatocyte identity.
During the progression of liver diseases, hepatocytes lose their original functions [[9], [10], [11], [12]]. This event is accompanied by a process known as ‘dedifferentiation of hepatocytes’ which is characterized by loss of hepatocyte identity, and is often observed during regenerative repair processes [14,15]. At the terminal stages of liver diseases, the cirrhotic liver loses its function, with repression of hepatocyte-specific genes [12,25,26]. As a well-known example, the rate of albumin production decreases in the liver of patients with cirrhosis, often causing ascite formation [25]. Similarly, coagulation disorder is a common complication of liver diseases [26]. Because the damaged liver cannot produce sufficient quantities of clotting factors, patients with liver diseases often develop gastrointestinal tract bleeding. Here, we found that in liver diseases of various etiologies, core TF levels were markedly decreased, whereas expression of second-tier TFs varied. Our results also demonstrate a strong correlation between core TF levels and hepatocyte identity gene expression (e.g. albumin, anti-thrombin, TDO2, and ITIH4), suggestive of the roles of the core TFs in the maintenance of hepatocyte physiology.
Among the mechanisms proposed to underlie disruption of liver in disease conditions, activation of extrinsic inflammatory pathways could be one of the causes of the descent of super-enhancers during disease progression, since inflammatory cytokines, such as TNF-α, IL-6, and TGF-β, worsen liver diseases across etiologies [34,35]. Accumulating evidence indicates that patients with cirrhosis often display an accompanying endogenous endotoxemic event in the absence of infection [36,37]. In a recent study, LPS/TLR4 signaling rapidly changed super-enhancer distributions in macrophages [38]. Hence, it is likely that deterioration of the inflammatory microenvironment changes the distribution of super-enhancers in hepatocytes, as extrinsic pathways re-establish a new transcriptional network that can no longer compensate for recovery of normal gene expression.
APAP is one of the best-selling drugs in the world. Although it is safe in a therapeutic dose range, when given at an excessively high dose it may produce drug-induced liver injury, which exceeds all other causes of acute liver failure in Western countries [39]. APAP has the unique feature that it can also trigger cell death by covalent binding with macromolecules [23]. In the liver, the majority of a therapeutic dose of APAP is glucuronidated or sulfated for excretion. However, a small percentage is metabolized to the reactive intermediate N-acetyl-p-benzoquinone imine in hepatocytes, causing oxidative damage in the cell. In view of its cell-type specific toxicity, the present study employed an APAP-induced liver injury model to assess the protective role of LRH1 in hepatocytes [40,41]. Specifically, ER stress effector CHOP mediates APAP-induced toxicity to cell death signal in hepatocytes [41]. Since LRH1 is essential for ER stress resolution [42], LRH1 suppression by APAP may contribute to APAP-induced liver toxicity. As another model of toxicant-induced liver injury, we used CCl4, a representative liver toxicant, to more completely understand the role of LRH1 under conditions of hepatocyte injury and regeneration, since CCl4 shares a similar mode of hepatotoxicity with APAP in that it exerts damage only after being metabolized in hepatocytes [24]. In our results, mice with LRH1 overexpression were protected from injury induced by either APAP or CCl4, showing the robust protective effect on hepatocytes of LRH1 supplementation.
LRH1, a TF belonging to the NR5A superfamily of nuclear receptors, is highly expressed in the liver, promoting the expression of CYP7A1 and CYP8B1, bile acid-synthesizing enzymes [43,44]. In another study, LRH1 drove transcription of the genes associated with high-density lipoprotein formation, cholesterol uptake and efflux, and fatty acid synthesis in the liver [45]. Consistently, bile acid metabolism was affected in mice with liver deficiency of LRH1 [46]. In addition, LRH1 suppresses acute phase response genes [47]. LRH1 heterozygous knockout animals display an exacerbated inflammatory response [48].
LRH1 has only recently been implied in the context of its roles against liver homeostasis. Overexpression of LRH1 in mouse liver prevented lipid accumulation under high-fat diet feeding condition [49]. In addition, genetic knockout studies revealed necessity of LRH1 in metabolic homeostasis against atherosclerosis and weight gain [50,51]. While these studies emphasized the role of LRH1 in hepatic energy metabolism, our findings focused on its protective role against liver injury. Our data expands knowledge on the function of LRH1 in hepatocyte physiology, and may be of help in understanding its role in liver pathophysiology and recovery. In addition, as fatty liver disease often involves hepatocyte death, our results strengthen a link between metabolic dysregulation and hepatocyte viability.
The findings that LRH1 was sensitively suppressed under disease conditions, and that enforced expression of LRH1 protected liver from injury, reinforce the concept that LRH1 plays a role in maintenance of hepatocyte identity. This was also verified by the outcome of our gene cluster analysis. Moreover, the results of RNA-seq and super-enhancer-associated TF network analyses enabled us to raise the concept that core TFs linked to LRH1 in the TF network may constitute an inter-connected auto-regulatory loop to maintain their abundant expression, as reinforced by the finding that transient restoration of a core TF permitted hepatocytes to recover normal functions under disease conditions. This concept is consistent with previous reports; genetically engineered mice lacking either PPARα, HNF4α, or RXRα were more vulnerable to liver injury [[52], [53], [54]], whereas HNF4α overexpression reversed terminal hepatic failure by resetting the transcriptional network [11]. Our motif analysis using JASPAR or dbCoRC showing that the upstream super-enhancers of LRH1, HNF4α, PPARα, and RXRα contain motifs for each of the core TFs further supports the idea. However, since the motif analyses are based on in silico prediction methods, direct binding of each core TF on the super-enhancers needs to be confirmed using an empirical experiment (i.e., ChIP assay). Together, our results and others indicate that restoration of a feed-forward auto-regulatory loop of core TFs could permit hepatocytes to recover cell identity.
Nonetheless, there were some inconsistencies across the experiments summarized in Fig. 7. Ppara mRNA levels were unaffected by LRH1 overexpression in liver samples from APAP-treated or CCl4-treated mice. LRH1 increased transcriptional activity of super-enhancer-residing Ppara gene in the luciferase assays, but not in the cell-based assays. HNF4α decreased transcriptional activity of super-enhancer-residing Rxra, and vice versa. However, HNF4α overexpression increased Rxra mRNA. The whole super-enhancer region containing each core TF gene spans >50 kb. However, we used only a part of each super-enhancer containing core TF genes for luciferase assays. Therefore, the discrepancies mentioned above may result from limitations in cloning and transfection of larger DNA fragments. Our data also do not exclude more complex regulatory mechanisms underlying core TF expression (e.g., non-SE-associated TFs could be upstream of the core TFs).
The primary characteristics of liver tissue architecture include a functional parenchymal tissue of hepatocytes and surrounding supportive cells. However, non-parenchymal cells frequently show different, or opposite, characteristics from parenchymal cells, particularly during disease progression. HSCs undergo transdifferentiation from quiescent to myofibroblast-like cells for the production of fibrillar matrix. They are well known to play critical roles in liver disease progression [55], but analysis of the super-enhancer signature of HSCs illuminated a set of key regulators distinct from that of hepatocytes (Note: our analysis on HSCs was based on DNase I HS data using human genome, whereas major analysis on the liver was based on H3K27Ac data using mouse genome). Intriguingly, super-enhancers were concentrated in close proximity to genes that encode the most highly specialized functions of HSCs (i.e. retinoid metabolism and extracellular matrix production). Moreover, expression of key super-enhancer-related genes changed drastically during HSC activation. This supports the contention that cistromic analysis of super-enhancer patterns in other liver cell types (e.g. Kupffer cells or endothelial cells) is warranted.
In summary, our cistromic analysis of super-enhancers in hepatocytes revealed a novel hepatocyte-specific transcriptional network, and core TFs necessary for the maintenance of hepatocyte identity and functions. LRH1 was found to be a driver gene of the positive feed-forward circuitry in hepatocytes. It may serve as a novel target for the recovery of cell identity under conditions of liver injury and regeneration. Given that LRH1 contains a ligand-binding domain, it would be plausible to target this with small molecular ligands. Collectively, our results provide key data for understanding liver pathophysiology and designing new molecules and strategies for treatment of liver disease. Moreover, this approach may be expanded to other cells for identification of a cell-type specific TF network.
The following are the supplementary data related to this article.
Supportive data for super-enhancer analysis in mouse liver. (A) Motif analyses using JASPAR 2018 database. Default cutoff score was used for the analysis (relative profile score threshold 80%). Arrows indicate DNA regions used for the motif analyses. Matrix profiles used for the analysis are Hnf4a (MA0114.3), Hnf4a_1 (PB0030.1), Hnf4a_2 (PB0134.1), Nr5a2 (MA0505.1), PPARA::RXRA (MA1148.1), Rxra (MA0512.1), Rxra (MA0512.2), RxRa_1 (PB0057.1), and Rxra_2 (PB0161.1). (B) The results of dbCoRC analysis. The core transcription regulatory circuitry (CRC) model in mouse liver analyzed by dbCoRC was shown in the left. Potential downstream targets of Nr5a2 (LRH1) within the CRC model were shown in the right. Arrows indicate the regulatory units that one TF at the start of the arrow has putative binding motifs at the super-enhancer region of the target TF gene. The size of node indicates the number of LRH1-binding motifs. (C) Human conservation of super-enhancers at the genomic loci of the core TFs. Snapshot of UCSC genome browser showing the tracks of H3K27Ac ChIP-seq from mouse liver and mammalian conservation data (left). H3K27Ac ChIP-Seq from human liver at the loci of NR5A2 (LRH1), HNF4A, PPARA, and RXRA (right).
Supportive data for RNA-seq data analysis. (A) Quality check data of the RNA-seq data normalization. The raw data from RNA-seq analysis (non-normal data) were processed and normalized by logarithm and quantile normalization method. (B) Volcano plot of RNA-seq data. Red horizontal and blue vertical lines indicate the filtering criteria (fold-change = ±2.0 and P-value = .05, respectively). Blue and yellow dots represent differentially expressed genes (DEGs) downregulated (481 genes) or upregulated (476 genes) by APAP treatment, respectively.
Supportive data for in vivo experiments. (A) Immunohistochemistry for HA-LRH1. Ectopically expressed HA-LRH1 in liver sections from mice treated as in Fig. 5A (upper) and Fig. 5B (lower) were immunostained for HA-tag and were colored using 3,3′-diaminobenzidine. Arrows indicate hepatocytes positive for HA-LRH1. The scale bars represent 100 μm. (B) qRT-PCR assays for Nr5a2 (LRH1) in APAP-(left) or CCl4-treated animal models (right). (C) Effects of LRH1 overexpression alone on liver histology and gene expression in mouse liver. H&E or HA-tag staining (left). At four days after a hydrodynamic injection of mock vector (pcDNA3.1) or the plasmid encoding LRH1, mice were sacrificed and the liver tissues were obtained. The scale bar represents 100 μm. qRT-PCR assays for Nr5a2 (LRH1), Hnf4a, Ppara and Rxra (right). (D) Immunoblotting for CYP2E1 (upper). Relative band intensity was obtained using scanning densitometry (lower). Data information: For left of B, data represent the means ± SEM (Mock+Veh, n = 7; Mock+APAP, n = 8; and LRH1 + APAP, n = 13, significantly different as compared to APAP-treated control, ##P < .01). For right of B, data represent the means ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; and LRH1 + CCl4, n = 4, significantly different as compared to vehicle-treated control, **P < .01; or CCl4-treated control, ##P < .01). For right of C, data represent the means ± SEM (Mock, n = 6; and LRH1, n = 4, significantly different as compared to Mock, **P < .01). For D, data represent the means ± SEM (n = 4 each, significantly different as compared to vehicle-treated control, **P < .01; N.S., not significant).
Heatmap and hierarchical correlation analysis of differentially expressed hepatic super-enhancer-associated genes after APAP treatment. RNA-seq data was generated using liver from mice treated with APAP alone or APAP+LRH1 overexpression vector as in Fig. 5A. Differentially expressed super-enhancer-associated genes were selected among the genes with independent t-test (P-values < .05 with a fold-change of >1.5).
Heatmap and hierarchical correlation analysis of differentially expressed hepatic typical-enhancer-associated genes after APAP treatment. RNA-seq data was generated using liver from mice treated with APAP alone and APAP+LRH1 overexpression vector as in Fig. 5A. Differentially expressed genes were selected as those with independent t-test (P-values < .05 with a fold-change of >1.5).
The core TFs in human HSCs. (A) F4/80 and αSMA mRNA levels in the liver of mice treated as in Fig. 5B. (B) H3K27Ac ChIP-seq data at the loci of Nr5a2 (LRH1), Hnf4a, Ppara, and Rxra in mouse Kupffer cells. (C) Distribution of DNase-seq signal intensities across 40,421 enhancers in human HSCs. 865 enhancers were annotated as super-enhancers. (D) GO of biological processes for super-enhancer-associated genes. (E) Protein-protein interaction network of super-enhancer-associated TFs according to STRING database. NR5A1, RARG, NR4A1, NR2F6, NR2F1, RARA and NR1D1 make a core TF cluster of the network (cluster #1). SMAD3, SMAD6 and SMAD7 constitute another core TF cluster (cluster #2). (F) Representative gene transcript levels in cluster #1 or #2 in the quiescent or activated mouse HSCs. Data were extracted from GSE34949. Data information: For A, data represent the mean ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; and LRH1 + CCl4, n = 4, significantly different as compared to vehicle control, **P < .01; N.S., not significant). For F, data represent the means ± SEM (n = 3 each, significantly different as compared to quiescent HSCs, *P < .05; **P < .01).
Genome-wide mapping of hepatic super-enhancers.
Top 5 enriched functional clusters of super-enhancer-associated genes.
BinGO analysis of super-enhancer-associated genes.
KEGG pathway analysis of gene cluster (i).
KEGG pathway analysis of gene cluster (ii).
KEGG pathway analysis of gene cluster (iii).
Supporting Information
Supplementary material 2.
Supplementary material 3.
Transcriptome profiling
RNA-seq data are deposited in the Gene Expression Omnibus (http://www.ncbi.nim.nih.gov/geo) under accession number GSE104302.
Funding sources
This research was supported by the National Research Foundation of Korea (NRF) grants funded by the Korea government (MSIP) (2014M1A3A3A02034698, 2017K1A1A2004511 and 2018R1A2A1A05078694). The funding body had no role in study design, data collection, data analyses, data interpretation, or writing of manuscript.
Author contributions
The overall study was conceived and designed by MS Joo, JH Koo and SG Kim; MS Joo, JH Koo, TH Kim, and YS Kim performed the experiments; MS Joo, JH Koo and SG Kim analyzed data; MS Joo and JH Koo analyzed bioinformatic data; MS Joo, JH Koo, and SG Kim wrote the paper; SG Kim supervised the study and obtained funding.
Disclosures
The authors have nothing to disclose.
References
- 1.Hnisz D., Abraham B.J., Lee T.I., Lau A., Saint-André V., Sigova A.A. Super-enhancers in the control of cell identity and disease. Cell. 2013;155:934–947. doi: 10.1016/j.cell.2013.09.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Whyte W.A., Orlando D.A., Hnisz D., Abraham B.J., Lin C.Y., Kagey M.H. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yan L., Guo H., Hu B., Li R., Yong J., Zhao Y. Epigenomic landscape of human fetal brain, heart, and liver. J Biol Chem. 2016;291:4386–4398. doi: 10.1074/jbc.M115.672931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Z., Zhao M., Joshi P.D., Li P., Zhang Y., Guo W. A class of circadian long non-coding RNAs mark enhancers modulating long-range circadian gene regulation. Nucleic Acids Res. 2017;45:5720–5738. doi: 10.1093/nar/gkx156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matthews B.J., Waxman D.J. Computational prediction of CTCF/cohesin-based intra-TAD loops that insulate chromatin contacts and gene expression in mouse liver. Elife. 2018;7 doi: 10.7554/eLife.34077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schrem H., Klempnauer J., Borlak J. Liver-enriched transcription factors in liver function and development. Part I: the hepatocyte nuclear factor network and liver-specific gene expression. Pharmacol Rev. 2002;54:129–158. doi: 10.1124/pr.54.1.129. [DOI] [PubMed] [Google Scholar]
- 7.Nagy P., Bisgaard H.C., Thorgeirsson S.S. Expression of hepatic transcription factors during liver development and oval cell differentiation. J Cell Biol. 1994;126:223–233. doi: 10.1083/jcb.126.1.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Costa R.H., Kalinichenko V.V., Holterman A.X., Wang X. Transcription factors in liver development, differentiation, and regeneration. Hepatology. 2003;38:1331–1347. doi: 10.1016/j.hep.2003.09.034. [DOI] [PubMed] [Google Scholar]
- 9.Martinez-Hernandez A., Martinez J. The role of capillarization in hepatic failure: studies in carbon tetrachloride-induced cirrhosis. Hepatology. 1991;14:864–874. doi: 10.1002/hep.1840140519. [DOI] [PubMed] [Google Scholar]
- 10.Pessayre D., Lebrec D., Descatoire V., Peignoux M., Benhamou J.P. Mechanism for reduced drug clearance in patients with cirrhosis. Gastroenterology. 1978;74:566–571. [PubMed] [Google Scholar]
- 11.Nishikawa T., Bell A., Brooks J.M., Setoyama K., Melis M., Han B. Resetting the transcription factor network reverses terminal chronic hepatic failure. J Clin Invest. 2015;125:1533–1544. doi: 10.1172/JCI73137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kurumiya Y., Nozawa K., Sakaguchi K., Nagino M., Nimura Y., Yoshida S. Differential suppression of liver-specific genes in regenerating rat liver induced by extended hepatectomy. J Hepatol. 2000;32:636–644. doi: 10.1016/s0168-8278(00)80226-3. [DOI] [PubMed] [Google Scholar]
- 13.Puri S., Folias A.E., Hebrok M. Plasticity and dedifferentiation within the pancreas: development, homeostasis, and disease. Cell Stem Cell. 2015;16:18–31. doi: 10.1016/j.stem.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blaheta R.A., Kronenberger B., Woitaschek D., Auth M.K., Scholz M., Weber S. Dedifferentiation of human hepatocytes by extracellular matrix proteins in vitro: quantitative and qualitative investigation of cytokeratin 7, 8, 18, 19 and vimentin filaments. J Hepatol. 1998;28:677–690. doi: 10.1016/s0168-8278(98)80293-6. [DOI] [PubMed] [Google Scholar]
- 15.Sun W., Ding J., Wu K., Ning B.F., Wen W., Sun H.Y. Gankyrin-mediated dedifferentiation facilitates the tumorigenicity of rat hepatocytes and hepatoma cells. Hepatology. 2011;54:1259–1272. doi: 10.1002/hep.24530. [DOI] [PubMed] [Google Scholar]
- 16.Khan A., Fornes O., Stigliani A., Gheorghe M., Castro-Mondragon J.A., van der Lee R. JASPAR 2018: update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018;4:D1284. doi: 10.1093/nar/gkx1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang M., Chen Y., Yang M., Guo A., Xu Y., Xu L. dbCoRC: a database of core transcriptional regulatory circuitries modeled by H3K27ac ChIP-seq signals. Nucleic Acids Res. 2018;4:D71–D77. doi: 10.1093/nar/gkx796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Higuchi N., Maruyama H., Kuroda T., Kameda S., Iino N., Kawachi H. Hydrodynamics-based delivery of the viral interleukin-10 gene suppresses experimental crescentic glomerulonephritis in Wistar-Kyoto rats. Gene Ther. 2003;10:1297–1310. doi: 10.1038/sj.gt.3301988. [DOI] [PubMed] [Google Scholar]
- 19.Yang P., Han Z., Chen P., Zhu L., Wang S., Hua Z. A contradictory role of A1 adenosine receptor in carbon tetrachloride- and bile duct ligation-induced liver fibrosis in mice. J Pharmacol Exp Ther. 2010;332:747–754. doi: 10.1124/jpet.109.162727. [DOI] [PubMed] [Google Scholar]
- 20.Mohar I., Stamper B.D., Rademacher P.M., White C.C., Nelson S.D., Kavanagh T.J. Acetaminophen-induced liver damage in mice is associated with gender-specific adduction of peroxiredoxin-6. Redox Biol. 2014;2:377–387. doi: 10.1016/j.redox.2014.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park E.Y., Cho I.J., Kim S.G. Transactivation of the PPAR-responsive enhancer module in chemopreventive glutathione S-transferase gene by the peroxisome proliferator-activated receptor-gamma and retinoid X receptor heterodimer. Cancer Res. 2004;64:3701–3713. doi: 10.1158/0008-5472.CAN-03-3924. [DOI] [PubMed] [Google Scholar]
- 22.Szklarczyk D., Morris J.H., Cook H., Kuhn M., Wyder S., Simonovic M. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45:D362–D368. doi: 10.1093/nar/gkw937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hinson J.A., Roberts D.W., James L.P. Mechanisms of acetaminophen-induced liver necrosis. Handb Exp Pharmacol. 2010;196:369–405. doi: 10.1007/978-3-642-00663-0_12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weber L.W., Boll M., Stampfl A. Hepatotoxicity and mechanism of action of haloalkanes: carbon tetrachloride as a toxicological model. Crit Rev Toxicol. 2003;33:105–136. doi: 10.1080/713611034. [DOI] [PubMed] [Google Scholar]
- 25.Gines P., Cardenas A., Arroyo V., Rodés J. Management of cirrhosis and ascites. N Engl J Med. 2004;350:1646–1654. doi: 10.1056/NEJMra035021. [DOI] [PubMed] [Google Scholar]
- 26.Caldwell S.H., Hoffman M., Lisman T., Macik B.G., Northup P.G., Reddy K.R. Coagulation disorders and hemostasis in liver disease: pathophysiology and critical assessment of current management. Hepatology. 2006;44:1039–1046. doi: 10.1002/hep.21303. [DOI] [PubMed] [Google Scholar]
- 27.Benameur T., Tual-Chalot S., Andriantsitohaina R. PPARalpha is essential for microparticle-induced differentiation of mouse bone marrow-derived endothelial progenitor cells and angiogenesis. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Djouadi F., Bastin J. PPARalpha gene expression in the developing rat kidney: role of glucocorticoids. J Am Soc Nephrol. 2001;12:1197–1203. doi: 10.1681/ASN.V1261197. [DOI] [PubMed] [Google Scholar]
- 29.Finck B.N. The PPAR regulatory system in cardiac physiology and disease. Cardiovasc Res. 2007;73:269–277. doi: 10.1016/j.cardiores.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 30.Kelly L.J., Vicario P.P., Thompson G.M. Peroxisome proliferator-activated receptors gamma and alpha mediate in vivo regulation of uncoupling protein (UCP-1, UCP-2, UCP-3) gene expression. Endocrinology. 1998;139:4920–4927. doi: 10.1210/endo.139.12.6384. [DOI] [PubMed] [Google Scholar]
- 31.Boyer L.A., Lee T.I., Cole M.F. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 2005;122:947–956. doi: 10.1016/j.cell.2005.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Loh Y.H., Wu Q., Chew J.L. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 2006;38:431–440. doi: 10.1038/ng1760. [DOI] [PubMed] [Google Scholar]
- 33.Lu X. Tied up in loops: positive and negative autoregulation of p53. Cold Spring Harb Perspect Biol. 2010;2:a000984. doi: 10.1101/cshperspect.a000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamimura S., Tsukamoto H. Cytokine gene expression by Kupffer cells in experimental alcoholic liver disease. Hepatology. 1995;22:1304–1309. [PubMed] [Google Scholar]
- 35.Pellicoro A., Ramachandran P., Iredale J.P., Fallowfield J.A. Liver fibrosis and repair: immune regulation of wound healing in a solid organ. Nat Rev Immunol. 2014;14:181–194. doi: 10.1038/nri3623. [DOI] [PubMed] [Google Scholar]
- 36.Lin R.S., Lee F.Y., Lee S.D., Tsai Y.T., Lin H.C., Lu R.H. Endotoxemia in patients with chronic liver diseases: relationship to severity of liver diseases, presence of esophageal varices, and hyperdynamic circulation. J Hepatol. 1995;22:165–172. doi: 10.1016/0168-8278(95)80424-2. [DOI] [PubMed] [Google Scholar]
- 37.Rao R. Endotoxemia and gut barrier dysfunction in alcoholic liver disease. Hepatology. 2009;50:638–644. doi: 10.1002/hep.23009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hah N., Benner C., Chong L.W., Yu R.T., Downes M., Evans R.M. Inflammation-sensitive super enhancers form domains of coordinately regulated enhancer RNAs. Proc Natl Acad Sci U S A. 2015;112:E297–E302. doi: 10.1073/pnas.1424028112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Larson A.M., Polson J., Fontana R.J., Davern T.J., Lalani E., Hynan L.S. Acetaminophen-induced acute liver failure: results of a United States multicenter, prospective study. Hepatology. 2005;42:1364–1372. doi: 10.1002/hep.20948. [DOI] [PubMed] [Google Scholar]
- 40.Kaplowitz N. Acetaminophen hepatoxicity: what do we know, what don't we know, and what do we do next? Hepatology. 2004;40:23–26. doi: 10.1002/hep.20312. [DOI] [PubMed] [Google Scholar]
- 41.Uzi D., Barda L., Scaiewicz V., Mills M., Mueller T., Gonzalez-Rodriguez A. CHOP is a critical regulator of acetaminophen-induced hepatotoxicity. J Hepatol. 2013;59:495–503. doi: 10.1016/j.jhep.2013.04.024. [DOI] [PubMed] [Google Scholar]
- 42.Mamrosh J.L., Lee J.M., Wagner M., Stambrook P.J., Whitby R.J., Sifers R.N. Nuclear receptor LRH-1/NR5A2 is required and targetable for liver endoplasmic reticulum stress resolution. Elife. 2014;3 doi: 10.7554/eLife.01694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.del Castillo-Olivares A., Gil G. Alpha 1-fetoprotein transcription factor is required for the expression of sterol 12alpha-hydroxylase, the specific enzyme for cholic acid synthesis. Potential role in the bile acid-mediated regulation of gene transcription. J Biol Chem. 2000;275:17793–17799. doi: 10.1074/jbc.M000996200. [DOI] [PubMed] [Google Scholar]
- 44.Nitta M., Ku S., Brown C., Okamoto A.Y., Shan B. CPF: an orphan nuclear receptor that regulates liver-specific expression of the human cholesterol 7alpha-hydroxylase gene. Proc Natl Acad Sci U S A. 1999;96:6660–6665. doi: 10.1073/pnas.96.12.6660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chong H.K., Biesinger J., Seo Y.K., Xie X., Osborne T.F. Genome-wide analysis of hepatic LRH1 reveals a promoter binding preference and suggests a role in regulating genes of lipid metabolism in concert with FXR. BMC Genomics. 2012;13:51. doi: 10.1186/1471-2164-13-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Out C., Hageman J., Bloks V.W., Gerrits H., Sollewijn Gelpke M.D., Bos T. Liver receptor homolog-1 is critical for adequate up-regulation of Cyp7a1 gene transcription and bile salt synthesis during bile salt sequestration. Hepatology. 2011;53:2075–2085. doi: 10.1002/hep.24286. [DOI] [PubMed] [Google Scholar]
- 47.Venteclef N., Smith J.C., Goodwin B., Delerive P. Liver receptor homolog 1 is a negative regulator of the hepatic acute-phase response. Mol Cell Biol. 2006;26:6799–6807. doi: 10.1128/MCB.00579-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lefevre L., Authier H., Stein S., Majorel C., Couderc B., Dardenne C. LRH1 mediates anti-inflammatory and antifungal phenotype of IL-13-activated macrophages through the PPARγ ligand synthesis. Nat Commun. 2015;6:6801. doi: 10.1038/ncomms7801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miranda D.A., Krause W.C., Cazenave-Gassiot A., Suzawa M., Escusa H., Foo J.C. LRH-1 regulates hepatic lipid homeostasis and maintains arachidonoyl phospholipid pools critical for phospholipid diversity. JCI Insight. 2018;3 doi: 10.1172/jci.insight.96151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stein S., Oosterveer M.H., Mataki C., Xu P., Lemos V., Havinga R. SUMOylation-dependent LRH-1/PROX1 interaction promotes atherosclerosis by decreasing hepatic reverse cholesterol transport. Cell Metab. 2014;20:603–613. doi: 10.1016/j.cmet.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 51.Hattori T., Iizuka K., Horikawa Y., Takeda J. LRH-1 heterozygous knockout mice are prone to mild obesity. Endocr J. 2014;61:471–480. doi: 10.1507/endocrj.ej14-0017. [DOI] [PubMed] [Google Scholar]
- 52.Nakajima T., Kamijo Y., Tanaka N., Sugiyama E., Tanaka E., Kiyosawa K. Peroxisome proliferator-activated receptor alpha protects against alcohol-induced liver damage. Hepatology. 2004;40:972–980. doi: 10.1002/hep.20399. [DOI] [PubMed] [Google Scholar]
- 53.Bonzo J.A., Ferry C.H., Matsubara T. Suppression of hepatocyte proliferation by hepatocyte nuclear factor 4α in adult mice. J Biol Chem. 2012;287:7345–7356. doi: 10.1074/jbc.M111.334599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gyamfi M.A., He L., French S.W., Kim J.H., Gonzalez F.J. Hepatocyte retinoid X receptor alpha-dependent regulation of lipid homeostasis and inflammatory cytokine expression contributes to alcohol-induced liver injury. J Pharmacol Exp Ther. 2008;324:443–453. doi: 10.1124/jpet.107.132258. [DOI] [PubMed] [Google Scholar]
- 55.Mederacke I., Hsu C.C., Troeger J.S., Huebener P., Mu X., Dapito D.H. Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology. Nat Commun. 2013;4:2823. doi: 10.1038/ncomms3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supportive data for super-enhancer analysis in mouse liver. (A) Motif analyses using JASPAR 2018 database. Default cutoff score was used for the analysis (relative profile score threshold 80%). Arrows indicate DNA regions used for the motif analyses. Matrix profiles used for the analysis are Hnf4a (MA0114.3), Hnf4a_1 (PB0030.1), Hnf4a_2 (PB0134.1), Nr5a2 (MA0505.1), PPARA::RXRA (MA1148.1), Rxra (MA0512.1), Rxra (MA0512.2), RxRa_1 (PB0057.1), and Rxra_2 (PB0161.1). (B) The results of dbCoRC analysis. The core transcription regulatory circuitry (CRC) model in mouse liver analyzed by dbCoRC was shown in the left. Potential downstream targets of Nr5a2 (LRH1) within the CRC model were shown in the right. Arrows indicate the regulatory units that one TF at the start of the arrow has putative binding motifs at the super-enhancer region of the target TF gene. The size of node indicates the number of LRH1-binding motifs. (C) Human conservation of super-enhancers at the genomic loci of the core TFs. Snapshot of UCSC genome browser showing the tracks of H3K27Ac ChIP-seq from mouse liver and mammalian conservation data (left). H3K27Ac ChIP-Seq from human liver at the loci of NR5A2 (LRH1), HNF4A, PPARA, and RXRA (right).
Supportive data for RNA-seq data analysis. (A) Quality check data of the RNA-seq data normalization. The raw data from RNA-seq analysis (non-normal data) were processed and normalized by logarithm and quantile normalization method. (B) Volcano plot of RNA-seq data. Red horizontal and blue vertical lines indicate the filtering criteria (fold-change = ±2.0 and P-value = .05, respectively). Blue and yellow dots represent differentially expressed genes (DEGs) downregulated (481 genes) or upregulated (476 genes) by APAP treatment, respectively.
Supportive data for in vivo experiments. (A) Immunohistochemistry for HA-LRH1. Ectopically expressed HA-LRH1 in liver sections from mice treated as in Fig. 5A (upper) and Fig. 5B (lower) were immunostained for HA-tag and were colored using 3,3′-diaminobenzidine. Arrows indicate hepatocytes positive for HA-LRH1. The scale bars represent 100 μm. (B) qRT-PCR assays for Nr5a2 (LRH1) in APAP-(left) or CCl4-treated animal models (right). (C) Effects of LRH1 overexpression alone on liver histology and gene expression in mouse liver. H&E or HA-tag staining (left). At four days after a hydrodynamic injection of mock vector (pcDNA3.1) or the plasmid encoding LRH1, mice were sacrificed and the liver tissues were obtained. The scale bar represents 100 μm. qRT-PCR assays for Nr5a2 (LRH1), Hnf4a, Ppara and Rxra (right). (D) Immunoblotting for CYP2E1 (upper). Relative band intensity was obtained using scanning densitometry (lower). Data information: For left of B, data represent the means ± SEM (Mock+Veh, n = 7; Mock+APAP, n = 8; and LRH1 + APAP, n = 13, significantly different as compared to APAP-treated control, ##P < .01). For right of B, data represent the means ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; and LRH1 + CCl4, n = 4, significantly different as compared to vehicle-treated control, **P < .01; or CCl4-treated control, ##P < .01). For right of C, data represent the means ± SEM (Mock, n = 6; and LRH1, n = 4, significantly different as compared to Mock, **P < .01). For D, data represent the means ± SEM (n = 4 each, significantly different as compared to vehicle-treated control, **P < .01; N.S., not significant).
Heatmap and hierarchical correlation analysis of differentially expressed hepatic super-enhancer-associated genes after APAP treatment. RNA-seq data was generated using liver from mice treated with APAP alone or APAP+LRH1 overexpression vector as in Fig. 5A. Differentially expressed super-enhancer-associated genes were selected among the genes with independent t-test (P-values < .05 with a fold-change of >1.5).
Heatmap and hierarchical correlation analysis of differentially expressed hepatic typical-enhancer-associated genes after APAP treatment. RNA-seq data was generated using liver from mice treated with APAP alone and APAP+LRH1 overexpression vector as in Fig. 5A. Differentially expressed genes were selected as those with independent t-test (P-values < .05 with a fold-change of >1.5).
The core TFs in human HSCs. (A) F4/80 and αSMA mRNA levels in the liver of mice treated as in Fig. 5B. (B) H3K27Ac ChIP-seq data at the loci of Nr5a2 (LRH1), Hnf4a, Ppara, and Rxra in mouse Kupffer cells. (C) Distribution of DNase-seq signal intensities across 40,421 enhancers in human HSCs. 865 enhancers were annotated as super-enhancers. (D) GO of biological processes for super-enhancer-associated genes. (E) Protein-protein interaction network of super-enhancer-associated TFs according to STRING database. NR5A1, RARG, NR4A1, NR2F6, NR2F1, RARA and NR1D1 make a core TF cluster of the network (cluster #1). SMAD3, SMAD6 and SMAD7 constitute another core TF cluster (cluster #2). (F) Representative gene transcript levels in cluster #1 or #2 in the quiescent or activated mouse HSCs. Data were extracted from GSE34949. Data information: For A, data represent the mean ± SEM (Mock+Veh, n = 6; Mock+CCl4, n = 14; and LRH1 + CCl4, n = 4, significantly different as compared to vehicle control, **P < .01; N.S., not significant). For F, data represent the means ± SEM (n = 3 each, significantly different as compared to quiescent HSCs, *P < .05; **P < .01).
Genome-wide mapping of hepatic super-enhancers.
Top 5 enriched functional clusters of super-enhancer-associated genes.
BinGO analysis of super-enhancer-associated genes.
KEGG pathway analysis of gene cluster (i).
KEGG pathway analysis of gene cluster (ii).
KEGG pathway analysis of gene cluster (iii).
Supporting Information
Supplementary material 2.
Supplementary material 3.