Abstract
Background
Fast growing cancer cells require greater amounts of ATP than normal cells. Although glycolysis was suggested as a source of anabolic metabolism based on lactate production, the main source of ATP to support cancer cell metabolism remains unidentified.
Methods
We have proposed that the oxoglutarate carrier SLC25A11 is important for ATP production in cancer by NADH transportation from the cytosol to mitochondria as a malate. We have examined not only changes of ATP and NADH but also changes of metabolites after SLC25A11 knock down in cancer cells.
Findings
The mitochondrial electron transport chain was functionally active in cancer cells. The cytosolic to mitochondrial NADH ratio was higher in non-small cell lung cancer (NSCLC) and melanoma cells than in normal cells. This was consistent with higher levels of the oxoglutarate carrier SLC25A11. Blocking malate transport by knockdown of SLC25A11 significantly impaired ATP production and inhibited the growth of cancer cells, which was not observed in normal cells. In in vivo experiments, heterozygote of SLC25A11 knock out mice suppressed KRASLA2 lung tumor formation by cross breeding.
Interpretation
Cancer cells critically depended on the oxoglutarate carrier SLC25A11 for transporting NADH from cytosol to mitochondria as a malate form for the purpose of ATP production. Therefore blocking SLC25A11 may have an advantage in stopping cancer growth by reducing ATP production.
Fund
The Basic Science Research Program through the National Research Foundation of Korea funded by the Ministry of Science and ICT to SYK (NRF-2017R1A2B2003428).
Keywords: Cancer metabolism, Oxoglutarate carrier, SLC25A11, Malate aspartate shuttle, Cancer therapeutic target
Research in context.
Evidence before this study
Studies based on Warburg's hypothesis showed that cancer cells depended on glycolysis as the major source of ATP production because cancer mitochondria were damaged. ATP from glycolysis, however, was not enough for supporting vigorous cancer growth. The main source of ATP to support cancer cell metabolism remains unidentified.
Added value of this study
Our study showed that the mitochondrial activity of cancer cells including the oxidative respiratory rates and the mitochondrial membrane potential was similar or higher to that of normal cells. This study revealed that cancer cells significantly depended on the cytosolic NADH for mitochondrial ATP production from oxidative phosphorylation through the malate-aspartate shuttle (MAS).
Implications of all the available evidence
This study showed the first time that blocking cytosolic NADH transportation into mitochondria by knock down of MAS induced cancer cell death with significant decrease of ATP production while the normal cell was not affected at all. Furthermore, a cross-breeding study between spontaneous lung cancer mice and MAS knock out mice showed that the number of mouse tumors was 60% decreased.
Alt-text: Unlabelled Box
1. Introduction
Warburg's hypothesis that cancer cells generate lactate from glucose because of mitochondrial destruction led to the misconception that cancer cells depend on glycolysis as the major source of ATP [1]. However, the respiratory rates of cancer cells are as high as those of normal cells [2,3]. The mitochondrial membrane potential of cancer cells is similar to that of normal cells, whereas the glucose-driven TCA cycle is stalled [[4], [5], [6], [7]]. Zu and Guppy in 2004 reported the pathway contributions to the ATP supply in different cancer cells, which revealed that oxidative phosphorylation (OxPhos) was the main ATP supplier despite enhanced rates of glycolysis [[8], [9], [10]]. However, in cancer cells in which OxPhos is active for ATP synthesis and glucose-driven pyruvate cannot supply NADH through the TCA cycle, the major source of electrons remains unidentified.
Alternative sources of ATP such as glutaminolysis have been suggested [11]. However, metabolic flux analysis revealed that glutamine is catabolised to lactate through the malic enzyme pathway or fatty acid synthesis through acetyl-CoA [4]. Therefore, glutamine is needed mainly for biosynthesis rather than energy metabolism. A recent study showed that human non-small cell lung cancer (NSCLC) tumors can use lactate to support growth, suggesting the contribution of lactate to the TCA cycle [12]. Lactate uptake in cancer cells increases TCA cycle intermediates; however, whether lactate uptake increases ATP production or OxPhos activity remains to be elucidated [12].
In previous work from our group, we showed that blocking one‑carbon metabolism through cytosolic ALDH1L1 knockdown, which decreases ATP production by >35%, reduces the NADH supply by >15% in NSCLC [7,13]. This suggests that cancer cells prefer cytosolic NADH as an electron source [14]. To transfer cytosolic NADH into mitochondria, cancer cells need the malate-aspartate shuttle (MAS), which is composed of two antiporters, the malate-α-ketoglutarate antiporter SLC25A11 (also known as oxoglutarate carrier, OGC) and the glutamate-aspartate antiporter SLC25A12 (also known as aspartate-glutamate carrier isoform 1, AGC1) as well as GOT1 and 2, and MDH1 and 2. Blocking MAS by knocking down the mitochondrial malate dehydrogenase (MDH2) reduces NADH levels by approximately 70% and ATP by 50% [7]. Glutamine contributes to the increase of MAS activity through glutamate production in NSCLC [15,16]. Blocking glutaminolysis through GLS1 knockdown in NSCLC decreases ATP levels by 70%, which is comparable to the levels observed in response to glutamic-oxaloacetic transaminase (GOT)-2 knockdown [16]. This implies that glutaminolysis also contributes to MAS activity despite the role of glutamine in the production of glutathione and α-ketoglutarate [11]. In summary, cancer cells rely on cytosolic NADH transported through the MAS system for OxPhos, whereas normal cells consume mitochondrial NADH produced by the TCA cycle for OxPhos. Lehninger et al. generated metabolic flux diagrams using D-[14C]glucose, and showed that large amounts of cytosolic NADH are oxidised by the mitochondrial respiratory chain via the MAS [17]. This accounts for up to 80% of the total flow of reducing equivalents to oxygen in cancer [18].
Here, we hypothesised that in cancer cells such as NSCLC and melanoma cells the supply of ATP may depend on cytosolic NADH produced by various sources including lipid peroxidation and aldehyde dehydrogenase, which require the MAS system for the transport of electrons into mitochondria. To test this, SLC25A11 was targeted to evaluate the role of the MAS in cancer energy metabolism.
2. Materials and methods
2.1. Cell lines
NSCLC cell lines including A549, H1650, EKVX, HOP62, H322M, H522 and melanoma cell lines including UACC257, UACC62, MDA-MB-435, M14, MALME-3 M, A375, SK-MEL-5, SK-MEL-28 were obtained from the US National Cancer Institute (NCI, Bethesda, MD, USA) (MTA 1-2702-09). A375 (ATCC CCL-1619), H1975 (ATCC CCL-5908), H23 (ATCC CCL-5800), H226 (ATCC CCL-5826), IMR90 (normal lung fibroblast, ATCC CCL-186) and melanocyte (ATCC PCS-200-013) were obtained from American Type Culture Collection (ATCC, Manassas, VA, USA).
2.2. Cell culture
For standard cell culture medium, we used RPMI 1640 medium (SH30027.01, Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS),Cellmaxin (C3314–020, GenDEPOT). IMR-90 cell was grown in DMEM/HIGH GLUCOSE medium (SH30243.01, Hyclone, Logan, UT, USA) containing 10% FBS (SH30084.03, Hyclone, Logan, UT, USA). Melanocyte was grown in dermal cell basal medium (ATCC PCS-200-030) with adult melanocyte growth kit (ATCC PCS-200-020). All cells were cultured at 37 °C with 5% CO2.
2.3. Animal experiments
Generation of SLC25A11 knockout mouse: Mixture of Cas9 protein (100 ng/ul) and gRNA (50 ng/ul) was injected in the cytoplasm of pronuclei. sgRNA sequences, 5’-ACTGCATCCGGTTCTTCACC-3′,5’-CGGATGCAGTTGAGTGGTGA-3′. Indel mutations in F1 mice were identified after TA cloning and sequencing. Cas9 protein (EnGen Cas9 NLS) was purchased from NEB (Ipswich, MA, USA). sgRNAs were generated using T7 in vitro transcription kit (NEB, Ipswich, MA, USA).
This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Cancer Center Research Institute (NCCRI; protocols: NCC-18-429), which is an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC International) accredited facility that abides by the Institute of Laboratory Animal Resources guide. All animal experiments were performed in accordance with the guidelines for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee (IACUC) of the National Cancer Center Research Institute.
2.4. Mouse breeding
Kras-LA2 mouse (B6.129S-Krastm3Tyj/Nci) was received from NCI mouse repository (NCI, Bethesda, MD, USA). The mice were used after backcrossing with C57BL/6.
2.5. Tissue array from clinical samples
Tissue microarray (TMA) from formalin-fixed, paraffin-embedded tissue blocks of NSCLC including normal lung tissue (CCN, CC, Super Bio Chips, Seoul, Korea) and malignant melanoma patients were purchased (ME2082b melanoma TMA: US Biomax, Bethesda, MD, USA). The TMA from lung cancer comprised of total 59 NSCLC tumors and 59 normal lung samples. The TMA of malignant melanoma consists of 128 cases of primary tumors and 64 cases of metastatic melanoma including both 8 cases of normal skin from non-cancer individuals and melanoma adjacent skin of primary tumors. Immunohistochemical staining for SLC25A11 was performed according to standard procedures. Immunostained TMAs were scanned with digital Aperio imagescope (Leica, Wetzlar, Germany). Staining pattern and intensity of SLC25A11 were interpreted by two pathologists (E.S.C. & J.I.Y.), and consistency between observers was reasonable (intraclass correlation coefficient = 0.955). The staining intensity was divided into negative, weak, moderate, and strong according to the SLC25A11 abundance. Statistical significance was calculated by Fisher's Exact test.
2.6. Western blot analysis
The cells were harvested, washed in phosphate-buffered saline (PBS) and lysed in lysis buffer (20 mM Tris-HCl (pH 7.4), 150 mM NaCl, 1% (v/v) Triton X-100, 1 mM EDTA, protease, phosphatase inhibitor cocktail. The protein concentration of the cell lysates was quantified by a BCA Protein Assay Kit (Pierce, Rockford, USA). The protein samples were loaded onto 6–15% SDS-PAGE and transferred onto PVDF membranes. After blocking by 5% BSA, and then detected with the following antibodies: GOT1 (ab170950), MDH1 (ab76616), MDH2 (ab135530), SLC25A11 (ab155196), Total OXPHOS human WB antibody Cocktail (ab110411), eIF4A1 (ab31217), Phospho-eIF4B(Ser406) (ab134138), c-Myc (ab32072) were purchased from Abcam (Cambridge, United Kingdom). GOT2 (sc-135,181), eIF4B (sc-3909) and β-actin (SC-47778) were purchased from Santa Cruz Biotechnology (Dallas, TX, USA). mTOR (#2983S), Phospho-mTOR (Ser2481) (#2974S), 4EBP1 (#9644S), Phospho-4EBP1(Thr37/46) (#2855S), P70S6K (#2708), Phospho- P70S6K (Ser371) (#9280) were purchased from Cell Signalling Technology (Danvers, MA, USA). The membranes were incubated in the primary antibodies diluted by 5% BSA buffer for overnight or 1 h at 4 °C and then in the HRP-conjugated secondary antibody for 1 h at room temperature. Finally, the protein band images were captured by a FUSION Solo (VILBER, Collégien, France) with ECL reagent (LF-QC0101, Ab FRONTIER, Seoul, Korea). And the protein band images were quantified by ImageJ.
2.7. Transfections and lentiviral transductions
For loss of function study, siRNAs were transfected by using Lipofector-EXT(AptaBio). Targeted sense sequences of the mRNA are as follows (5′-3′): siSLC25A11#1:GGAAUACAAGAACGGGCUGGACGUGdTdT; siSLC25A11#2: ACAUUGCCAAGACCCGAAUCCAGAAdTdT. Myc-DDK-tagged-SLC25A11 (CAT#:RC200768) was purchased from Origene (Rockville, MD, USA). shRNA vectors against SLC25A11 (TRCN0000322801, TRCN0000044416) was purchased from Sigma (St. Louis, MO, USA). Lentiviruses were produced by transfecting 293T cells. 48 h after transfection, virus particles were filtered (0.45 μm) and used diluted with polybrene to infect cells. Starting 48 h after infection, cells were selected by puromycin for 2 days and then released in fresh medium.
2.8. XF cell mito stress analysis
Cells were transfected with NT siRNA or SLC25A11 siRNA (40 nM) in 60 mm dishes. After 24 h, transfected cells were seeded in XF cell culture microplate at a cellular density of 30,000/well and incubated for 48 h at 37 °C. For oxygen consumption rate (OCR) determination, cells were incubated in XF base medium supplemented with 10 mM glucose, 1 mM sodium pyruvate and 2 mM l-glutamine and equilibrated in non-CO2 incubator for 1 h before starting the assay. The samples were mixed (3 min) and measured (3 min) using XFe96 extracellular flux analyzer (Seahorse Bioscience, North Billerica, Billerica, MA, USA). Oligomycin (1 μM), FCCP (1 μM), and rotenone/antimycin A (0.5 μM) were injected at the indicated time points. Finally, the oxygen consumption rate was normalized by SRB assay.
2.9. Measurement of ATP levels
ATP levels were monitored using an ATP Colorimetric/Fluorometric Assay Kit as per the manufacturer's instructions (K354-100, BioVision, Milpitas, CA, USA) that we reported before [16].
2.10. Measurement of malate levels
Malate levels were monitored using a malate Assay Kit as per the manufacturer's instructions (K637–100, BioVision, Milpitas, CA, USA). The cells (1 × 106) were lysed in 100 μl of assay buffer and then centrifuged under ice-cold conditions at 15,000 rpm for 10 min to pellet the insoluble materials. The supernatant was then collected, and 10–50 μl of this supernatant was added to a 96-well plate; the final volume was topped up to 50 μl/well with assay buffer. Reaction mix was made (assay buffer 38 μl, WST substrate 10 μl and malate enzyme mix 2 μl), and 50 μl of the reaction mix was added to each well containing a test sample. Then, the plate was incubated at 37 °C for 30 min in the dark, and the OD was measured at 450 nm using a microplate reader.
2.11. Measurement of NADH levels
NADH levels were monitored using a NADH Assay Kit as per the manufacturer's instructions (K337–100, BioVision, Milpitas, CA, USA). The cells or mitochondria were lysed in 400 μl of assay buffer and extracted with 400 μl of NADH/NAD Extraction Buffer by freeze/thaw two cycles. Then centrifuged under ice-cold conditions at 15,000 rpm for 10 min to pellet the insoluble materials. The supernatant was then collected. To detect NADH, NAD needs to be decomposed before the reaction. To decompose NAD, aliquot 200 μl of extracted samples into tubes. Heat to 60 °C for 30 min. and 50 μl of this supernatant was added to a 96-well plate; Reaction mix was made (assay buffer 98 μl, NADH developer 10 μl and NAD cycling enzyme mix 2 μl), and 110 μl of the reaction mix was added to each well containing a test sample. Then, the plate was incubated at RT for 60 min in the dark, and the OD was measured at 450 nm using a microplate reader.
2.12. FITC annexin V apoptosis detection
The cells were incubated after SLC25A11 siRNA treatment. The cells were then collected, washed with cold PBS, centrifuged at 1500 rpm for 3 min, and resuspended in 1× binding buffer at a concentration of 5 × 106 cells/ml. The solution (100 μl) was transferred (1 × 105) to a 5 ml polystyrene round-bottom tube, and 5 μl each of annexin V-FITC and PI were added. The cells were gently vortexed and incubated for 15 min at room temperature in the dark. Then, 400 μl of 1× binding buffer was added to each tube, and the samples were analyzed by FACS flow cytometry (BD Falcon, Bedford, MA, USA).
2.13. Clonogenic assay
Cells were plated in 6 well plates at (1000–5000) cells per well in 2 ml media. Media was not changed throughout the course of the experiment. After 14 days, colonies were stained with 0.005% crystal violet staining solution.
2.14. Mitochondrial isolation
Mitochondrial and cytosolic fractions from cells were performed by using the Mitochondria Isolation Kit for Cultured Cells (Thermo Fisher Scientific, Waltham, MA, USA). Cells (2 × 107) were lysed in Mitochondria Isolation Reagent A, and 10 μl reagent B was added and vortexed at maximum speed for 5 s. Tubes were incubated on ice for 5 min, and vortexed at maximum speed every minute. Then 800 μL of Mitochondria Isolation Reagent C was added and mixed by inverting the tubes several times, followed by centrifugation at 700 ×g for 10 min at 4 °C. The supernatant was further centrifuged at 12,000 ×g for 15 min at 4 °C to pellet the mitochondria. The crude mitochondrial fraction was resuspended for washing and centrifuged at 12,000 ×g for 15 min at 4 °C. The pellets were collected as the mitochondrial fraction.
2.15. Immunofluorescence staining
Cells were fixed with 4% (w/v) paraformaldehyde and permeabilized with 0.5% Triton X-100. The cells were then stained with anti-SLC25A11 polyclonal antibody, Mitotracker (M22425, Invitrogen Life Technologies, Carlsbad, CA, USA) and Alexa Fluor 488-conjugated anti-rabbit antibody (A11008, Invitrogen Life Technologies, Carlsbad, CA, USA). Live cell imaging was taken by LSM510 Laser Scanning Microscope and Axio Observer Z1 (Carl Zeiss, Oberkochen, Germany). The relative intensity was normalized by the arithmetic mean intensity (from Zen software 2.6 blue edition).
2.16. Measurement of mitochondrial membrane potential (∆ψm)
Mitochondrial membrane potential was analyzed by measuring tetramethylrodamine ester (TMRE) (87,917, Sigma, St. Louis, MO, USA) followed by an established method [19]. Cells were plated 4 well chambered coverglass (155382, Thermo Fisher Scientific) in 0.5 ml culture media. After 24 h, cells were transfected with NT siRNA or SLC25A11 siRNA (40 nM) for 48 h at 37 Co. 100 nM of TMRE and 5 μg/ml of Hoechst 33342 were added to the culture medium for 15 min at 37 Co. Negative control was treated with 50 μM trifluoromethoxy carbonylcyanide phenylhydrazone (FCCP, C2920, Sigma, St. Louis, MO, USA) in serum free media for 15 min at 37 Co before TMRE stanning. 4 well chambered cover glass was placed on LSM510 Laser Scanning Microscope in the presence of the TMRE and Hoechst 33342. Live cell imaging was taken by LSM510 Laser Scanning Microscope and Axio Observer Z1 (Carl Zeiss, Oberkochen, Germany). The relative intensity of TMRE was normalized by the arithmetic mean intensity (from Zen software 2.6 blue edition).
2.17. Puromycin incorporation assay (SUnSET assay)
SUnSET assay was performed as per manufacturer's recommendations (Kerafast, Boston, MA, USA). Cells were incubated with puromycin (2 μg/mL) for 15 min. Post incubation, cells were washed with ice cold PBS and lysed using RIPA lysis buffer. Equal quantity of protein lysates was separated on SDS-PAGE and probed with anti-puromycin antibody. Signals were normalized with probing beta-actin (loading control).
2.18. Sulforhodamine B (SRB) assay
To measure cell proliferation, we used SRB assay [20]. Assay method was followed as we published before [16].
2.19. Relative quantitation of metabolites of energy metabolism using liquid chromatography-tandem mass spectrometry (LC-MS/MS)
Metabolites in energy metabolism were analyzed with LC-MS/MS equipped with 1290 HPLC (Agilent, Santa Clara, CA, USA), Qtrap 5500 (ABSciex, Concord, Ontario, Canada), and a reverse phase column (Synergi fusion RP 50 × 2 mm) followed by method as we reported before [16].
2.20. Pre-clinical xenograft tumor model
Balb/c-nu mice (Central Lab. Animal, Highland Heights, KY, USA) were aged 6–8 weeks. This study was reviewed and approved by the Institutional Animal Care and Use Committee (IACUC) of the National Cancer Center Research Institute (NCCRI; protocols: NCC-17-395), which is an Association for Assessment and Accreditation of Laboratory Animal Care International (AAALAC International) accredited facility that abides by the Institute of Laboratory Animal Resources guide. Cells (5.0 × 106–1.0 × 107) were injected subcutaneously using a 1 ml syringe. Primary tumor size was measured every week using calipers. Tumor volume was calculated using the formula, V = (A × B2)/2, where V is the volume (mm3), A is the long diameter, and B is the short diameter.
2.21. Immunohistochemistry
Formaldehyde (4%) fixed specimens were paraffin-embedded and cut at a thickness of 4 μm. Section were dried for 1 h at 56 °C and immunohistochemical staining performed with the automated instrument Discovery XT (Ventana medical system, Tucson, Arizona, USA) using the Chromomap DAB Detection kit as follow: sections were deparaffinized and rehydrated with EZ prep and washed with Reaction buffer (Ventana medical system, Tucson, Arizona, USA). The antigens were retrieved with heat treatment in pH 6.0 citrate buffer (Ribo CC, Ventana) at 90 °C for 30 min for anti-c-Myc (ab32072). Images were taken by microscope (Axio imagerM2, Carl Zeiss, Oberkochen, Germany).
2.22. Statistical analysis
Statistical analysis was performed using the Student's t-test as appropriate. Tumor growth and tumor nodules of spontaneous mouse model was analyzed statistically by two-way analysis of variance (ANOVA) tests using GraphPad PRISM 5 (GraphPad Software, San Diego, CA, USA).
3. Results
3.1. High SLC25A11 levels increased the mitochondrial membrane potential in cancer cells
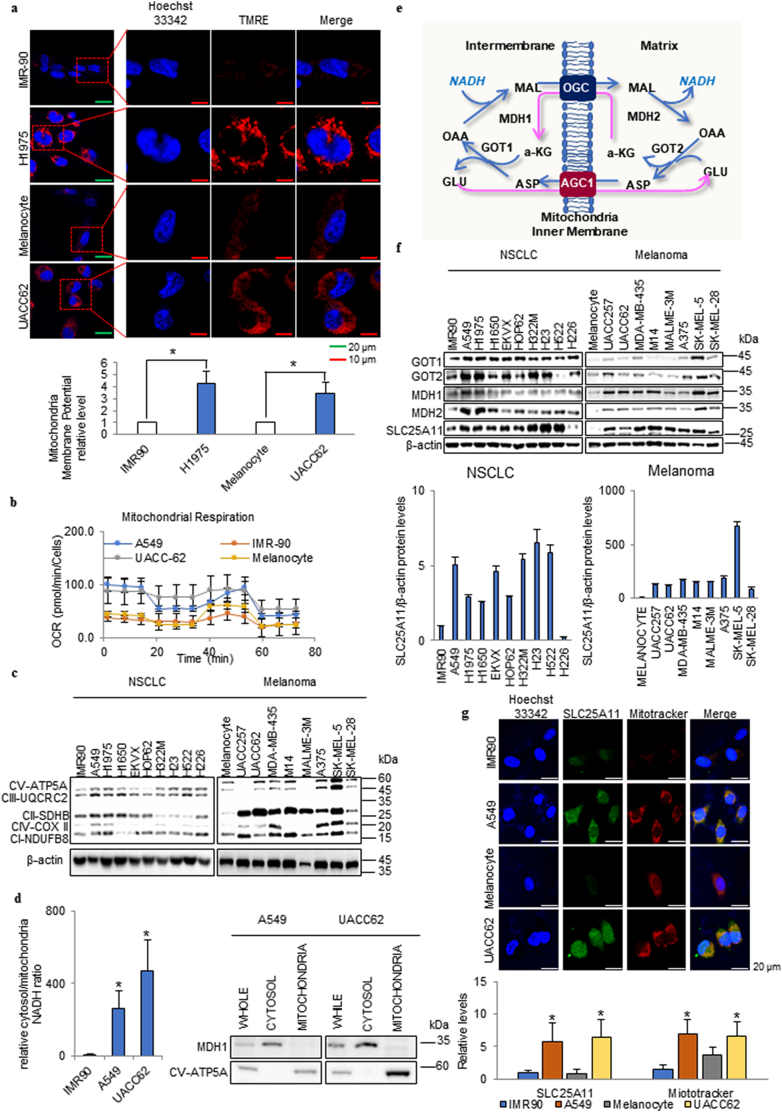
In cancer cells, a highly activated mitochondrial membrane potential is often detected using TMRE staining in comparison with normal cell lines such as the immortalised lung epithelial cell line IMR90 and primary melanocytes under normal culture conditions (normoxia and high glucose) (Fig. 1a). Therefore, we investigated whether OxPhos is increased in cancer, and explored the mechanism underlying the supply of electrons in cancer. Analysis of mitochondrial respiration showed that the oxygen consumption rate (OCR) was higher in cancer than in normal cells (Fig. 1b). This was consistent with the higher expression of OxPhos components in cancer cells than in normal cells, as determined by immunoblotting (Fig. 1c). To supply electrons for OxPhos, cancer cells can obtain them from the cytosol. The cytosolic:mitochondrial NADH ratio was approximately 30-fold higher in cancer cells than in normal cells (Fig. 1d). The transfer of NADH produced in the cytosol into mitochondria depends on the activity of the MAS (Fig. 1e). In the cytoplasm, oxaloacetate is catalysed to malate by malate dehydrogenase (MDH1) with oxidisation of NADH to NAD+. Malate is transported into mitochondria by the OGC SLC25A11 as an ion transporter, and α-ketoglutarate is transported in the opposite direction [21]. Glutamate is transported into mitochondria by the aspartate/glutamate carrier SLC25A12, and aspartate is transported in the opposite direction [22]. Malate is then oxidised to oxaloacetate by the mitochondrial MDH2, resulting in the formation of NADH. The electrons from NADH enter the respiratory chain. The cycle of oxaloacetate to the cytoplasm is mediated by the aspartate aminotransferase GOT2. Aspartate and α-ketoglutarate return to the cytosol and are converted back to oxaloacetate and glutamate, respectively, by GOT1 [16]. The expression of MAS components including GOT1/2, MDH1/2, and SLC25A11 was higher in NSCLC and melanoma cell lines than in normal primary melanocytes or IMR90 cells (Fig. 1f). Especially SLC25A11 level was consistently higher in NSCLC and melanoma cell lines by densitometry analysis (Fig. 1f). The expressions of SLC25A11 (green) and mitrotracker (red) were also higher in NSCLC and melanoma cell lines than in normal primary melanocytes or IMR90 cells (Fig. 1g).
Fig. 1.
High SLC25A11 levels increased the mitochondrial membrane potential in cancer cells. (a) The mitochondrial membrane potential was analyzed by live cell imaging in cancer cell lines in comparison with normal cells using ZEN software (scale bar, 20 μm, 10 μm). (b) The oxygen consumption rate (OCR) was analyzed using the Seahorse XFe analyzer in cancer cell lines compared to normal cells and then normalized by SRB assay. (c) The expression levels of oxidative phosphorylation (OxPhos) components were analyzed in NSCLC and melanoma cells by immunoblotting. (d) Cytosolic/mitochondrial NADH ratio in cancer cell lines compared with normal cells. Western blot confirming the isolation of mitochondria from cytosol with MDH1 (cytosol marker) and CV-ATP5A (mitochondria marker) antibodies. (e) Malate-aspartate shuttle (MAS) for NADH transportation into the mitochondrial matrix. MAT, malate-α-ketoglutarate transporter; GAT, glutamate-aspartate transporter; OAA, oxaloacetate; α-KG, α-ketoglutarate. (f) The expression levels of SLC25A11, GOT1, GOT2, MDH1, and MDH2 in NSCLC and melanoma cells were analyzed by immunoblotting and quantified by ImageJ. (g) Comparison of SLC25A11 expression in cancer cells and normal cells by immunofluorescence staining and intensity analyzed by ZEN software (scale bar = 20 μm). (Data were presented as mean ± SD. ***p < .001, **p < .01, *p < .05).
3.2. MAS activity was required for ATP production in cancer cells
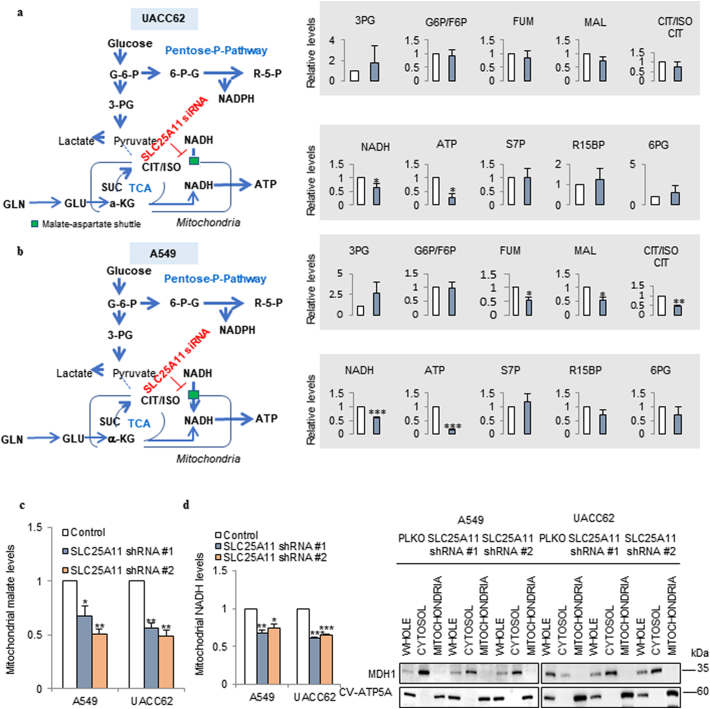
To test whether ATP production depends on cytosolic NADH through the MAS system, metabolite analysis was performed in cells subjected to SLC25A11 knockdown for 24 h (Fig. 2). In UACC62 cells, SLC25A11 knockdown had no significant effect on the levels of glycolysis and pentose phosphate pathway metabolites, whereas it decreased the levels of TCA cycle intermediates by approximately 20% compared with those in the untreated group (Fig. 2a). NADH and ATP production were decreased by approximately 40% and 75%, respectively, by SLC25A11 knockdown in culture media containing 10% FBS and under normal oxygen conditions (Fig. 2a). In A549 cells, SLC25A11 knockdown decreased TCA cycle intermediates by approximately 40% compared with those in the untreated group, and NADH and ATP production by approximately 40% and 80%, respectively (Fig. 2b). By mitochondrial subcellular fraction analysis, SLC25A11 knockdown decreased malate uptake into mitochondria by approximately 50% compared with control levels (Fig. 2c), and this was correlated with a decrease of mitochondrial NADH in NSCLC and melanoma (Fig. 2d). To verify mitochondrial subcellular fraction, we have employed MDH1 for cytosolic marker and CV-ATP5A for mitochondrial marker (Fig. 2d). These results suggest that mitochondria in cancer cells depend on cytosolic NADH to produce ATP through the MAS system for energy supply.
Fig. 2.
Metabolite analysis of SLC25A11 knockdown cancer cells revealed that the MAS is responsible for ATP production in cancer cells. (a, b) Targeted LC-MS/MS metabolite analysis in UACC62 (a) and A549 (b) cells treated with siRNA against SLC25A11 (40 nM) for 24 h. Metabolite levels were measured in triplicate and normalized by BCA assay. (c) Mitochondrial malate levels were measured in SLC25A11-knockdown A549 and UACC62 cells using a mitochondrial fractionation kit and malate assay kit. (d) Mitochondrial NADH levels were measured in SLC25A11-knockdown A549 and UACC62 cells using a mitochondrial fractionation kit and NADH assay kit. Western blot confirming the isolation of mitochondria from cytosol with MDH1 (cytosol marker) and CV-ATP5A (mitochondria marker) antibodies. (Data were presented as mean ± SD. ***p < .001, **p < .01, *p < .05).
3.3. SLC25A11 knockdown decreased cancer cell proliferation
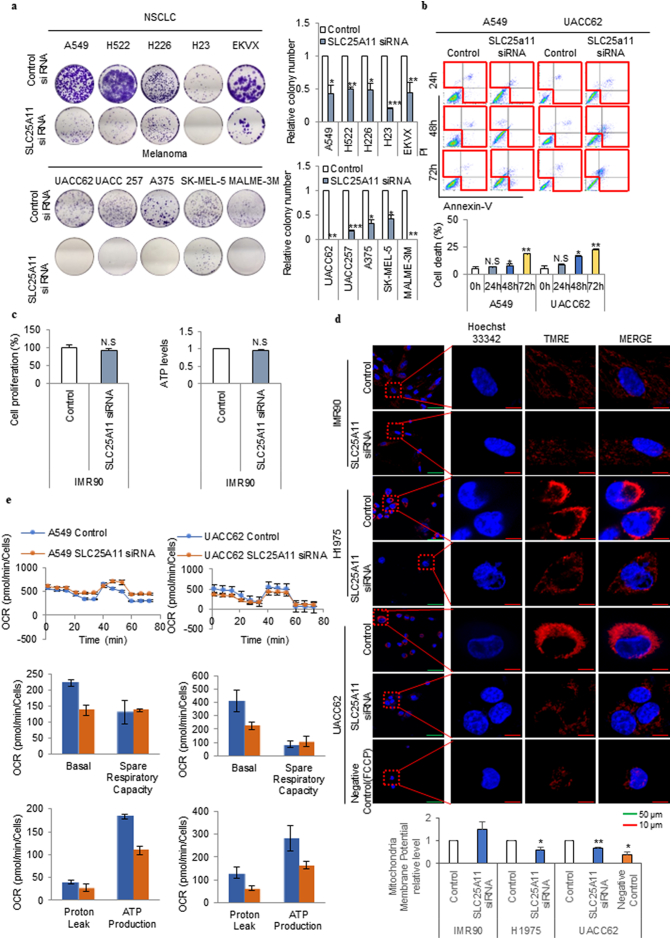
To test the role of SLC25A11 in cancer cell proliferation, colony formation assays were performed in five NSCLC cell lines and five melanoma cell lines with siRNA-mediated (Figs. 3a and S1a) or shRNA-mediated (Fig. S1b and c) SLC25A11 knockdown. SLC25A11 knockdown using siRNA (Fig. 3a) or shRNA (Fig. S1c) decreased colony formation by 50–100% in various cancer cell lines. Cancer cell death following SLC25A11 knockdown was assessed by Annexin V staining at different times (Fig. 3b). SLC25A11 siRNA treatment for 24, 48, and 72 h increased cell death by approximately 20%, 40%, and 400%, respectively, in A549 cells, and by approximately 50%, 300%, and 400%, respectively, in UACC62 cells (Fig. 3b). In IMR90 normal human lung fibroblasts, SLC25A11 knockdown had no effect on ATP production and proliferation under normal culture conditions (Fig. 3c). SLC25A11 knockdown decreased the mitochondrial membrane potential by 40% and 30% in H1975 and UACC62 cells respectively, whereas no changes were observed in IMR90 cells (Fig. 3d). To test whether loss of mitochondrial membrane potential by SLC25A11 knockdown can be rescued by over expression of SLC25A11, UACC62 cell was transfected with SLC25A11 clone for 24 h following to SLC25A11 knock down for 24 h (Fig. S1d-f). The levels of mitochondrial potential (Fig. S1d) and ATP (Fig. S1e) were rescued in UACC62 cell with SLC25A11 transfection (Fig. S1f). SLC25A11 knockdown for 24 h decreased the OCR by approximately 36% in A549 cells and 45% in UACC62 cells (Fig. 3e). These results suggested that SLC25A11 knockdown significantly decreased ATP production in cancer cells at 24 h prior to the occurrence of apoptosis at 48–72 h. We therefore investigated the mechanism by which ATP depletion caused cancer cell death through SLC25A11.
Fig. 3.
SLC25A11 knockdown induced cell death following cell growth arrest through OxPhos reduction in melanoma and NSCLC cells. (a) Clonogenic assay was performed in NSCLC, and melanoma cells were treated with control or SLC25A11 siRNA (40 nM) for 2 weeks. (b) Cell death was measured using an Annexin V staining kit in cells treated with SLC25A11 siRNA (40 nM) for the indicated times. (c) Cell proliferation assay using SRB assay and ATP assay were performed with IMR90 cells by transfection of SLC25A11 siRNA (40 nM) or control siRNA for 48 h. (d) The mitochondrial membrane potential of H1975, UACC62, and IMR90 cells treated with SLC25A11 siRNA for 48 h was analyzed by live cell imaging using ZEN software (scale bar, 50 μm, 10 μm). (e) The oxygen consumption rate was analyzed using the Seahorse XFe analyzer in A549 and UACC62 cells treated with control or SLC25A11 siRNA (40 nM) for 24 h and then normalized by SRB assay. (Data were presented as mean ± SD. ***p < .001, **p < .01, *p < .05).
3.4. SLC25A11 knockdown significantly reduced protein translation by downregulating mTOR
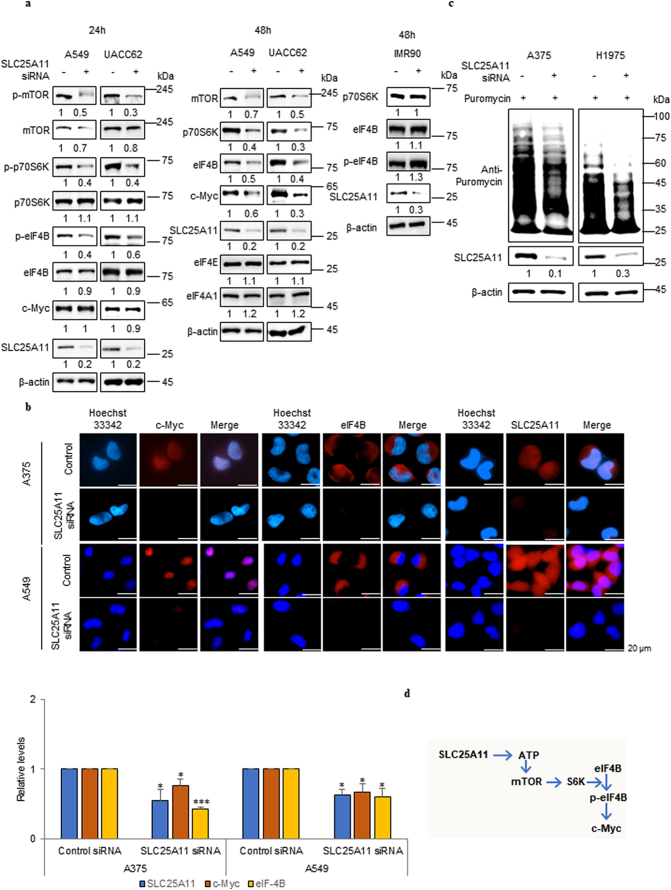
Fig. 2 shows that SLC25A11 knockdown reduced ATP production. The mTOR complex is regulated by ATP levels [23]. To determine whether SLC25A11 expression affects mTOR signalling, the expression of factors downstream of mTOR signalling was measured by immunoblotting at 24 and 48 h after SLC25A11 knockdown in NSCLC and melanoma cell lines (Fig. 4a). The levels of p-mTOR and p-p70SK were critically reduced at 24 h after SLC25A11 knockdown, whereas protein levels were not affected (Fig. 4a). SLC25A11 knockdown downregulated mTOR, eukaryotic translation initiation factor (eIF)-4B, and c-Myc protein expression at 48 h in A549 and UACC62 whereas SLC25A11 knockdown did not affect protein expressions of p70S6K, eIF4B and p-eIF4B at 48 h in IMR90 (Fig. 4a). EIF4B-dependent modulation of c-Myc translation regulates cancer cell proliferation [24]. The effect of SLC25A11 knockdown on eIF4B and c-Myc expression was assessed by immunohistochemistry. The results showed that SLC25A11 knockdown downregulated eIF4B and c-Myc protein expression in NSCLC and melanoma cells (Fig. 4b). We hypothesised that the effect of SLC25A11 knockdown on inhibiting protein translation was mediated by the inactivation of translational elongation factors such as eIF4B through the depletion of the ATP supply, which resulted in a critical reduction of p-eIF4B dependent synthesis of proteins including c-Myc in cancer cells (Fig. 4b). Immunoblot detection of puromycin showed a significant decrease of protein translation in response to SLC25A11 knockdown (Fig. 4c). Taken together, these results suggested that decreased ATP production induced by SLC25A11 knockdown inhibited mTOR signalling by blocking mTOR phosphorylation, which significantly decreased cell proliferation by downregulating c-Myc and eIF4B (Fig. 4d).
Fig. 4.
Knockdown of SLC25A11 inhibited protein translation by inactivating eIF4B. (a) Western blot was performed against phosphorylated mTOR, p70S6K, eIF4B and c-Myc in SLC25A11 knockdown cells and quantification was performed by ImageJ software. (b) Immunofluorescence was performed against eIF4B and c-Myc expression in SLC25A11 knockdown cells and the intensity was analyzed by ZEN software. (scale bar, 20 μm). (c) The indicated cells were treated with SLC25A11 siRNA for 48 h followed by 15 min incubation with puromycin (2 μg/ml). Cell lysates were subjected to immunoblotting using anti-puromycin antibody (SUnSET assay). (d) Possible regulation pathway of SLC25A11. (Data were presented as mean ± SD. ***p < .001, **p < .01, *p < .05).
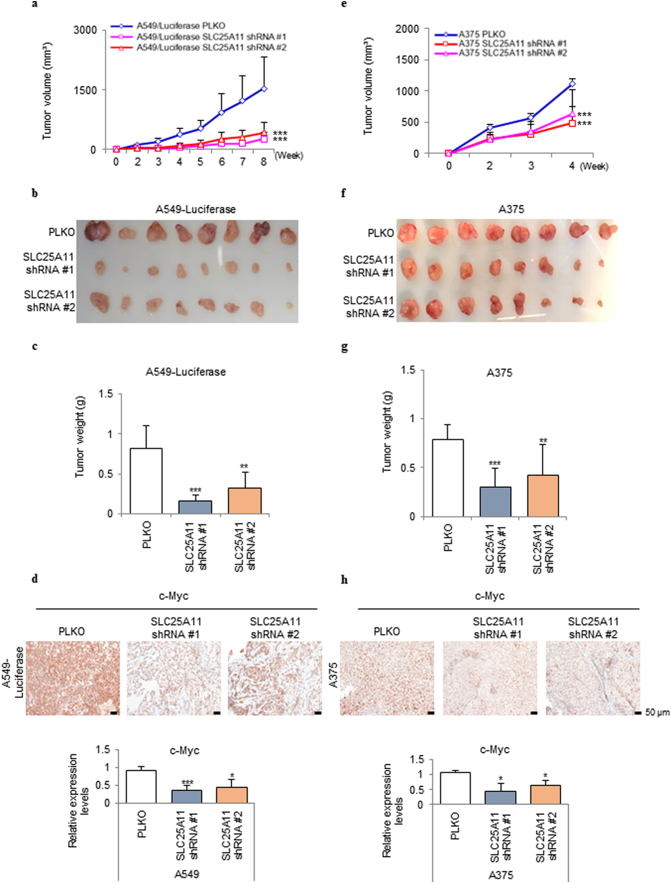
3.5. SLC25A11 knockdown suppressed tumor formation in vivo
To test the contribution of SLC25A11 to tumor growth, SLC25A11 was knocked down by shRNA in the A549 and A375 cell lines (Fig. 5) and the H226 and UACC62 cell lines (Fig. S2), and two knockdown lines were selected from each group for inoculation into mice. Assessment of tumor size over time showed that tumors formed by SLC25A11 knockdown cells were approximately one-fifth the size of control tumors in A549 and A375 cells (Fig. 5a–c and 5e–g respectively) and in H226 and UACC62 cells (Fig. S2a-c and S2d-f respectively). Proliferation activity was assessed by immunohistochemical staining of c-Myc in xenograft tumor tissues. The intensity of c-Myc staining was significantly lower in SLC25A11 knockdown tumors than in the wild-type controls (Fig. 5 d and h).
Fig. 5.
SLC25A11 knockdown reduced NSCLC and melanoma tumor growth in vivo. (a) Volume of subcutaneous tumors derived from SLC25A11 shRNA transduced A549 cells. (b) Representative images of tumors derived from SLC25A11 shRNA transduced A549 cells. (c) Weight of subcutaneous tumors derived from SLC25A11 shRNA transduced A549 cells. (d) Immunohistochemical (IHC) analysis of c-Myc in A549 xenograft tumor tissues and quantification by positive cell counting. (e) Volume of subcutaneous tumors derived from SLC25A11 shRNA transduced A375 cells. (f) Representative images of tumors derived from SLC25A11 shRNA transduced A375 cells. (g) Weight of subcutaneous tumors derived from SLC25A11 shRNA transduced A375 cells. (h) IHC analysis of c-Myc in A375 xenograft tumor tissues and quantification by positive cell counting. (scale bar, 50 μm). (Data were analyzed statistically by two-way analysis of variance ANOVA tests using GraphPad PRISM 5 ***p < .001, **p < .01, *p < .05).
3.6. Genetic loss of SLC25A11 in a mouse model suppressed K-ras-mediated lung tumorigenesis
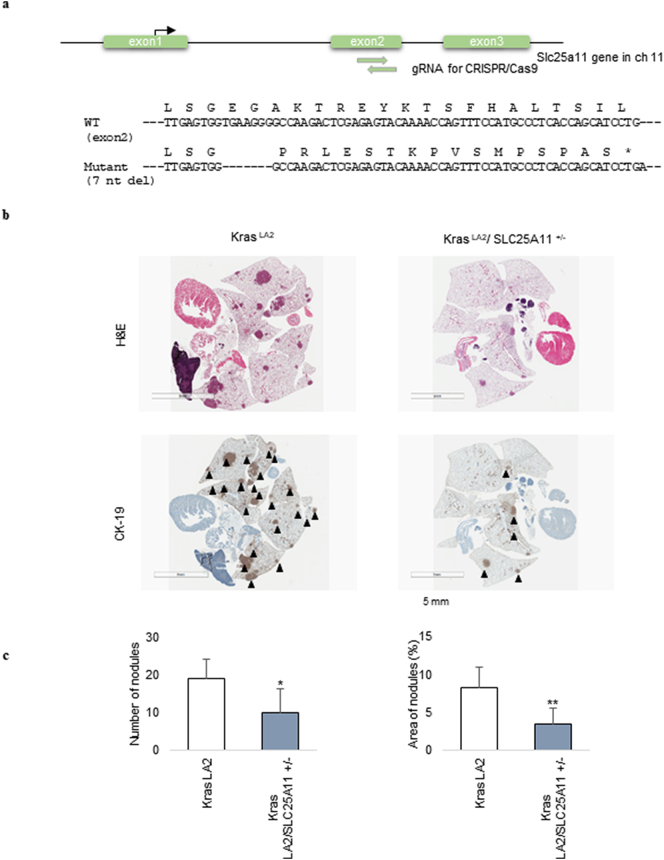
To generate knockout mice with an indel mutation in exon 2 of the SLC25A11 gene, small guide RNA (sgRNA) target sequences were selected using the CRISPR design tool (crispor.tefor.net). A mixture of the Cas9 protein (100 ng/μl) and 2 sgRNAs (50 ng/μl) was injected into the cytoplasm of mouse pronuclei. Indel mutations in F1 mice were identified after TA cloning and sequencing. Mice with a 7 nt deletion in exon 2 of SLC25A11 causing premature translation termination were used for breeding with KrasLA2 mice (Fig. 6a).
Fig. 6.
Genetic loss of SLC25A11 suppressed Kras-mediated lung tumorigenesis. (a) Generation of Slc25a11 knockout mice. A mixture of Cas9 protein and two guide RNAs (gRNA) was injected into the cytoplasm of mouse pronuclei. Mutations in mice were identified by TA cloning and sequencing. A 7 nt deletion in exon 2 of Slc25a11 causes premature translation termination. (b) Representative photomicrographs of tumor nodules detected by haematoxylin and eosin (H&E) and CK-19 staining (scale bar, 5 mm). (c) Quantitative analysis of tumor nodule number and tumor area showed a statistically significant decrease in these parameters in KRASLA2/SLC25A11+/− mice compared with KRASLA2 mice. (Data were analyzed statistically by two-way analysis of variance (ANOVA) tests using GraphPad PRISM 5 ***p < .001, **p < .01, *p < .05).
The effect of SLC25A11 knockout on lung tumorigenesis was investigated by cross-breeding SLC25A11 heterozygous mice to generate SLC25A11 knockout mice. However, SLC25A11 knockout was embryonic lethal in mice (Table. S1). Therefore, paraffin blocks of the lungs of 12-week-old KRASLA2/SLC25A11+/− heterozygous mice and KRASLA2 mice were stained with haematoxylin and eosin and CK-19 to assess lung tumorigenesis (Figs. 6b and S3). Quantitative analysis of tumor nodules showed that the number of KRASLA2/SLC25A11+/− mouse tumors was 40% of that in control KRASLA2 mice, and the tumor area in KRASLA2/SLC25A11+/− mice was 46% of that in control KRASLA2 mice (Fig. 6c).
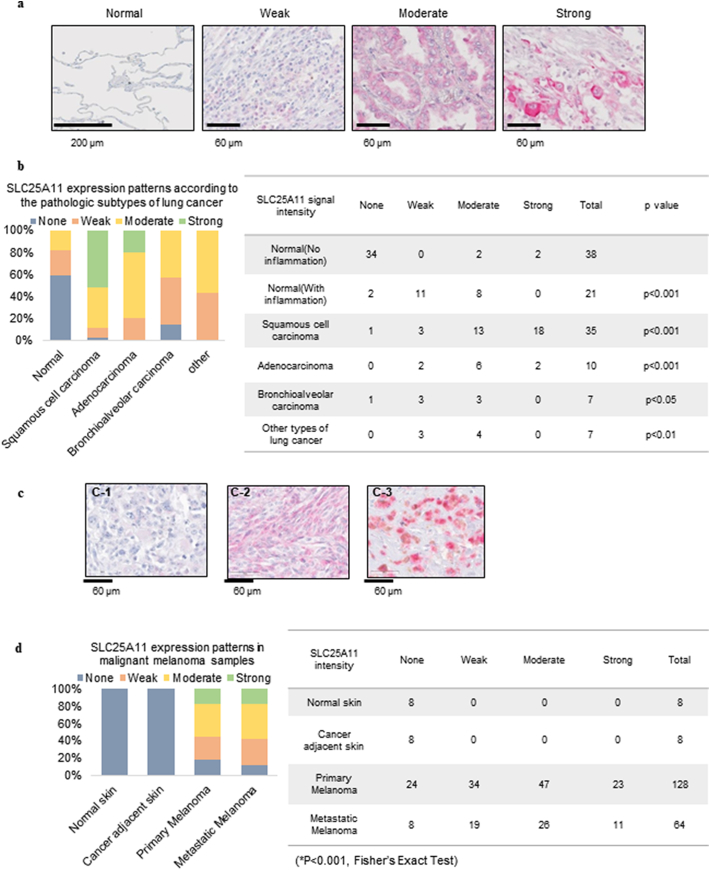
3.7. SLC25A11 expression was increased in tissue microarrays from NSCLC and melanoma patients
SLC25A11 expression in clinical cancer samples was analyzed using tissue arrays from patients with NSCLC and malignant melanoma. Lung cancer tissue microarrays included 59 normal lung tissues with or without chronic inflammation, 35 squamous cell carcinoma samples, ten adenocarcinoma samples, and seven bronchioalveolar carcinoma samples. Malignant melanoma samples included 128 primary melanoma and 64 metastatic melanoma cases, as well as eight normal skin and eight cancer-adjacent normal epithelium samples. SLC25A11 was mostly detected in the cytoplasm of epithelial lung cancer cells (Fig. 7a). In normal lung tissues, SLC25A11 staining was negative or weak (Fig. 7a). Normal alveolar epithelial cells were mostly negative for SLC25A11 expression, whereas non-cancerous epithelial cells with chronic inflammation showed variable SLC25A11 expression (Fig. S4a), indicating that SLC25A11 is expressed in the normal alveolar epithelium in response to chronic inflammation. SLC25A11 expression was higher in NSCLC tissues than in normal lung tissues (Fig. 7b), and it was significantly increased in squamous cell carcinoma and adenocarcinoma, which are major subtypes of lung cancer. SLC25A11 upregulation was not detected in non-tumor cells such as tumor-associated fibroblasts and inflammatory cells (Fig. S4b). SC25A11 upregulation in lung cancer samples was not correlated to TNM stage (Fig. S4c).
Fig. 7.
Expression and distribution of SLC25A11 in lung cancer and malignant melanoma tissue samples. (a) Tissue microarray was performed to determine the expression of SLC25A11 in normal lung tissues and lung cancer samples. A-1: Normal lung tissue showing no staining (×200; scale bar, 200 μm); A-2: Weak, A-3: Moderate, and A-4: Strong expression of SLC25A11 in lung cancer samples (×400; scale bar, 60 μm). (b) Percent distribution (graph) and SLC25A11 expression patterns in normal and cancer samples (Table) according to histologic subtypes. Statistical significance compared with normal lung tissues was determined by Fisher's exact test. (c) The expression patterns of SLC25A11 in malignant melanoma were determined by IHC. A-1: Weak, A-2: Moderate, A-3: Strong (×400; scale bar, 60 μm). (d) Percent SLC25A11 expression in normal skin and malignant melanoma samples. (Statistical significance was calculated by Fisher's Exact test. ***p < .001, **p < .01, *p < .05).
In melanoma samples, SLC25A11 expression was weakly detected in keratinocytes of basal and suprabasal layers, and undetectable in basal layer melanocytes with a clear cytoplasm (Fig. S5a). Cytoplasmic SLC25A11 expression was highly increased in primary and metastatic melanoma samples, and its expression did not differ significantly between primary and metastatic lesions (Fig. 7c and d). SLC25A11 expression levels in melanoma were positively correlated to TNM stage (Fig. S5b).
4. Discussion
Warburg observed that in the presence of glucose, tumors acidified the Ringer solution, and this was attributed to the production of lactic acid. When glucose is the only source of nutrients, it can serve both for biosynthesis and energy production. However, studies performed under physiological conditions to identify the sources of ATP in cancer cells indicate that cancer energy metabolism is not as simple as suggested by the Warburg theory. For instance, despite the accelerated glycolysis observed in many cancer cell types, the glycolytic contribution to the total cellular ATP supply is negligible, reaching a maximum of only 10% of the total ATP [25]. Limiting glycolytic ATP production by knocking out pyruvate kinase (PKM2) fails to prevent tumorigenesis, suggesting that PKM2 is not necessary for cancer cell proliferation, and the main role of glycolysis is not to supply ATP [26]. Inactive PKM2 is associated with the proliferating cancer cell population, whereas nonproliferating cancer cells require active pyruvate kinase. An active mitochondrial membrane potential is necessary for cancer cell proliferation and tumorigenesis [27]. Therefore, despite their high glycolytic rates, most cancer cells rely on mitochondrial OxPhos for the production of ATP. Cancer cells adapt mitochondrial function to support survival. In hypoxic cancer cells, mitochondrial OxPhos functions optimally when oxygen levels are reduced to 0.5% [28]. OxPhos is active in cancer cells and was proposed as a therapeutic target despite the stalled TCA cycle activity in cancer cells [29]. However, the source of electrons remains unclear, as OxPhos requires electrons for proton pumping and ATP production. Cytosolic NADH is one of the main NADH sources in cancer [7,16], and via a series of enzymatic and transport steps, MAS is involved in the transfer of electrons from cytosolic NADH to mitochondrial NAD+, which then enter OxPhos in cancer cells under normal culture conditions.
Studies have suggested several possible sources of energy to support cancer cell metabolism. In addition to pyruvate derived from glycolysis or lactate, it is possible that fatty acids and glutamine can supply substrates to the TCA cycle to sustain mitochondrial ATP production in cancer cells. There is a report that certain Ras-driven cancer cells can consume lipids to support ATP production under starvation condition [30]. Cancer cells can use fatty acids from adipocytes to produce mitochondrial ATP [31]. The catabolism of fatty acids (β-oxidation) generates acetyl-CoA and the reducing equivalents NADH and FADH2 in mitochondria, which can be used by TCA cycle and OxPhos to produce mitochondrial ATP [32]. In the early stages of pancreatic adenocarcinoma, the branched-chain amino acids isoleucine, valine, and leucine are highly increased and can be catabolised to acetyl-CoA and TCA cycle intermediates [33]. When pyruvate oxidation to acetyl-CoA is blocked by hypoxia or OxPhos impairment, glutamine can act as an alternative source of acetyl-CoA as a biosynthetic precursor, especially for lipid synthesis [34]. Glutamine can contribute to NADH production through the TCA cycle via glutamate and α-ketoglutarate [11]. Alternative sources of ATP such as glutaminolysis have been suggested [11]. However, metabolic flux analysis shows that glutamine is catabolised to lactate through the malic enzyme pathway or fatty acid synthesis through acetyl-CoA [4], indicating that glutamine is needed for biosynthesis. A recent report showed that human NSCLC tumors can use lactate, suggesting the contribution of lactate to the TCA cycle [12]. Lactate uptake in cancer cells results in increased TCA cycle intermediates, whereas it does not increase ATP production or OxPhos [12]. Nutrient-deficient cancer cells adapt to the microenvironment by activating AMPK and decreasing ATP, inhibiting the anabolic kinase mTOR, which drives the energetic growth of cancer cells [35]. Decreased mTORC1 activity increases autophagy, thereby providing an intracellular glutamine supply to sustain mitochondrial function [36].
MAS is composed of two antiporters, the malate-α-ketoglutarate antiporter SLC25A11 and the glutamate-aspartate antiporter SLC25A12. There is no isotype of SLC25A11 while SLC25A12 has an isotype of calcium-binding aspartate/glutamate carrier as SLC25A13 [37]. Therefore, we have targeted SLC25A11 instead of SLC25A12 to block MAS efficiently in this study. The knock out mice of SLC25A11 showed lethality between embryonic day 10.5 and 14.5 (Table S1). However, knock down of SLC25A11 in normal cell (IMR90) showed no change of mitochondrial potential as well as cell proliferation (Fig. 3c and d) while cancer cells showed decrease of mitochondrial potential as well as increase of cell death (Fig. 3b and d). It implies that SLC25A11 is required for embryogenic development, but it may not be required for proliferation of differentiated normal cells. This suggests that there is a window of targeting SLC25A11 as a therapeutic approach in cancer.
All suggested pathways of energy metabolism are capable of sustaining cancer cell growth under various conditions. However, a conserved pathway for the supply of energy must exist in cancer, as cancer anabolism is typically indicated by lactate production. The results of the present study showed an increase in cytosolic NADH and increased activity of the MAS in NSCLC and melanoma compared with those in normal cells, which is consistent with previous reports showing that blocking cytosolic NADH production decreases ATP production by up to 40%. The mechanisms underlying the regulation of SLC25A11 expression as well as the role of SLC25A11 in various cancers remain to be elucidated. Oncogenes or cancer signalling may also modulate energy metabolism by regulating the MAS system in cancer.
Funding source
This research was supported by the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT to SYK (NRF-2017R1A2B2003428) and by a research grant from the National Cancer Center in Korea to SYK; NCC1910291-1).
Declaration of interests
The authors have no conflict of interests to declare.
Author contributions
S.-Y.K. conceived the project, S.-Y.K. wrote the main manuscript, J.-S.L. and H.L. designed the experiments, J.-S.L., S.L., J.K., S.-H.L., S.-G.K. carried out experiments, E.C., N.K., J.Y performed immunohistochemistry and analyzed the statistics. All authors reviewed and edited the manuscript.
Conflict of interests
The authors declare no competing interests.
Acknowledgments
We thank Ms. Mi Ae Kim in the Microscopy Core and Mr. Tae Sik Kim in the Flow Cytometry Core (National Cancer Center, Korea) for their expert assistance and helpful suggestions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2019.01.036.
Appendix A. Supplementary data
Supplementary material
References
- 1.Warburg O. On respiratory impairment in cancer cells. Science. 1956;124(3215):269–270. [PubMed] [Google Scholar]
- 2.Weinhouse S. On respiratory impairment in cancer cells. Science. 1956;124(3215):267–269. doi: 10.1126/science.124.3215.267. [DOI] [PubMed] [Google Scholar]
- 3.DeBerardinis R.J., Chandel N.S. Fundamentals of cancer metabolism. Sci Adv. 2016;2(5) doi: 10.1126/sciadv.1600200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DeBerardinis R.J., Mancuso A., Daikhin E., Nissim I., Yudkoff M., Wehrli S. Beyond aerobic glycolysis: transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc Natl Acad Sci U S A. 2007;104(49):19345–19350. doi: 10.1073/pnas.0709747104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smolkova K., Bellance N., Scandurra F., Genot E., Gnaiger E., Plecita-Hlavata L. Mitochondrial bioenergetic adaptations of breast cancer cells to aglycemia and hypoxia. J Bioenerg Biomembr. 2010;42(1):55–67. doi: 10.1007/s10863-009-9267-x. [DOI] [PubMed] [Google Scholar]
- 6.Wagner B.A., Venkataraman S., Buettner G.R. The rate of oxygen utilization by cells. Free Radic Biol Med. 2011;51(3):700–712. doi: 10.1016/j.freeradbiomed.2011.05.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang J.H., Lee S.H., Lee J.S., Nam B., Seong T.W., Son J. Aldehyde dehydrogenase inhibition combined with phenformin treatment reversed NSCLC through ATP depletion. Oncotarget. 2016;7(31):49397–49410. doi: 10.18632/oncotarget.10354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Guppy M., Leedman P., Zu X., Russell V. Contribution by different fuels and metabolic pathways to the total ATP turnover of proliferating MCF-7 breast cancer cells. Biochem J. 2002;364(Pt 1):309–315. doi: 10.1042/bj3640309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elwood J.C., Lin Y.C., Cristofalo V.J., Weinhouse S., Morris H.P. Glucose utilization in homogenates of the morris hepatoma 5123 and related tumors. Cancer Res. 1963;23:906–913. [PubMed] [Google Scholar]
- 10.Kallinowski F., Schlenger K.H., Runkel S., Kloes M., Stohrer M., Okunieff P. Blood flow, metabolism, cellular microenvironment, and growth rate of human tumor xenografts. Cancer Res. 1989;49(14):3759–3764. [PubMed] [Google Scholar]
- 11.Hensley C.T., Wasti A.T., DeBerardinis R.J. Glutamine and cancer: cell biology, physiology, and clinical opportunities. J Clin Invest. 2013;123(9):3678–3684. doi: 10.1172/JCI69600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Faubert B., Li K.Y., Cai L., Hensley C.T., Kim J., Zacharias L.G. Lactate metabolism in human lung tumors. Cell. 2017;171(2):358–371. doi: 10.1016/j.cell.2017.09.019. [e9] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kang J.H., Lee S.H., Hong D., Lee J.S., Ahn H.S., Ahn J.H. Aldehyde dehydrogenase is used by cancer cells for energy metabolism. Exp Mol Med. 2016;48(11):e272. doi: 10.1038/emm.2016.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kim S.Y. Cancer Energy Metabolism: Shutting Power off Cancer Factory. Biomol Ther (Seoul) 2018;26(1):39–44. doi: 10.4062/biomolther.2017.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee J.S., Kang J.H., Lee S.H., Lee C.H., Son J., Kim S.Y. Glutaminase 1 inhibition reduces thymidine synthesis in NSCLC. Biochem Biophys Res Commun. 2016;477(3):374–382. doi: 10.1016/j.bbrc.2016.06.095. [DOI] [PubMed] [Google Scholar]
- 16.Lee J.S., Kang J.H., Lee S.H., Hong D., Son J., Hong K.M. Dual targeting of glutaminase 1 and thymidylate synthase elicits death synergistically in NSCLC. Cell Death Dis. 2016;7(12):e2511. doi: 10.1038/cddis.2016.404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Greenhouse W.V., Lehninger A.L. Occurrence of the malate-aspartate shuttle in various tumor types. Cancer Res. 1976;36(4):1392–1396. [PubMed] [Google Scholar]
- 18.Greenhouse W.V., Lehninger A.L. Magnitude of malate-aspartate reduced nicotinamide adenine dinucleotide shuttle activity in intact respiring tumor cells. Cancer Res. 1977;37(11):4173–4181. [PubMed] [Google Scholar]
- 19.Crowley L.C., Christensen M.E., Waterhouse N.J. Measuring mitochondrial transmembrane potential by TMRE staining. Cold Spring Harb Protoc. 2016;2016(12) doi: 10.1101/pdb.prot087361. [DOI] [PubMed] [Google Scholar]
- 20.Skehan P., Storeng R., Scudiero D., Monks A., McMahon J., Vistica D. New colorimetric cytotoxicity assay for anticancer-drug screening. J Natl Cancer Inst. 1990;82(13):1107–1112. doi: 10.1093/jnci/82.13.1107. [DOI] [PubMed] [Google Scholar]
- 21.Piccininni S., Iacobazzi V., Lauria G., Rocchi M., Palmieri F. Assignment of the oxoglutarate carrier gene (SLC20A4) to human chromosome 17p13.3. Cytogenet Cell Genet. 1998;83(3–4):256–257. doi: 10.1159/000015198. [DOI] [PubMed] [Google Scholar]
- 22.Crackower M.A., Sinasac D.S., Lee J.R., Herbrick J.A., Tsui L.C., Scherer S.W. Assignment of the SLC25A12 gene coding for the human calcium-binding mitochondrial solute carrier protein aralar to human chromosome 2q24. Cytogenet Cell Genet. 1999;87(3–4):197–198. doi: 10.1159/000015465. [DOI] [PubMed] [Google Scholar]
- 23.Mihaylova M.M., Shaw R.J. The AMPK signalling pathway coordinates cell growth, autophagy and metabolism. Nat Cell Biol. 2011;13(9):1016–1023. doi: 10.1038/ncb2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Csibi A., Lee G., Yoon S.O., Tong H., Ilter D., Elia I. The mTORC1/S6K1 pathway regulates glutamine metabolism through the eIF4B-dependent control of c-Myc translation. Curr Biol. 2014;24(19):2274–2280. doi: 10.1016/j.cub.2014.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zu X.L., Guppy M. Cancer metabolism: facts, fantasy, and fiction. Biochem Biophys Res Commun. 2004;313(3):459–465. doi: 10.1016/j.bbrc.2003.11.136. [DOI] [PubMed] [Google Scholar]
- 26.Israelsen W.J., Dayton T.L., Davidson S.M., Fiske B.P., Hosios A.M., Bellinger G. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell. 2013;155(2):397–409. doi: 10.1016/j.cell.2013.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Martinez-Reyes I., Diebold L.P., Kong H., Schieber M., Huang H., Hensley C.T. TCA cycle and mitochondrial membrane potential are necessary for diverse biological functions. Mol Cell. 2016;61(2):199–209. doi: 10.1016/j.molcel.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chandel N.S., Budinger G.R., Choe S.H., Schumacker P.T. Cellular respiration during hypoxia. Role of cytochrome oxidase as the oxygen sensor in hepatocytes. J Biol Chem. 1997;272(30):18808–18816. doi: 10.1074/jbc.272.30.18808. [DOI] [PubMed] [Google Scholar]
- 29.Ashton T.M., McKenna W.G., Kunz-Schughart L.A., Higgins G.S. Oxidative phosphorylation as an emerging target in cancer therapy. Clin Cancer Res. 2018;24(11):2482–2490. doi: 10.1158/1078-0432.CCR-17-3070. [DOI] [PubMed] [Google Scholar]
- 30.Kamphorst J.J., Cross J.R., Fan J., de Stanchina E., Mathew R., White E.P. Hypoxic and Ras-transformed cells support growth by scavenging unsaturated fatty acids from lysophospholipids. Proc Natl Acad Sci U S A. 2013;110(22):8882–8887. doi: 10.1073/pnas.1307237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nieman K.M., Kenny H.A., Penicka C.V., Ladanyi A., Buell-Gutbrod R., Zillhardt M.R. Adipocytes promote ovarian cancer metastasis and provide energy for rapid tumor growth. Nat Med. 2011;17(11):1498–1503. doi: 10.1038/nm.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carracedo A., Cantley L.C., Pandolfi P.P. Cancer metabolism: fatty acid oxidation in the limelight. Nat Rev Cancer. 2013;13(4):227–232. doi: 10.1038/nrc3483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mayers J.R., Wu C., Clish C.B., Kraft P., Torrence M.E., Fiske B.P. Elevation of circulating branched-chain amino acids is an early event in human pancreatic adenocarcinoma development. Nat Med. 2014;20(10):1193–1198. doi: 10.1038/nm.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mullen A.R., Wheaton W.W., Jin E.S., Chen P.H., Sullivan L.B., Cheng T. Reductive carboxylation supports growth in tumour cells with defective mitochondria. Nature. 2011;481(7381):385–388. doi: 10.1038/nature10642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guo J.Y., Chen H.Y., Mathew R., Fan J., Strohecker A.M., Karsli-Uzunbas G. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. doi: 10.1101/gad.2016311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thangaratnarajah C., Ruprecht J.J., Kunji E.R. Calcium-induced conformational changes of the regulatory domain of human mitochondrial aspartate/glutamate carriers. Nat Commun. 2014;5:5491. doi: 10.1038/ncomms6491. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material