Abstract
Influenza vaccines are the most effective intervention to prevent the substantial public health burden of seasonal and pandemic influenza. Hemagglutinin (HA), as the main antigen in inactivated influenza vaccines (IIVs), elicits functional neutralizing antibodies and largely determines IIV effectiveness. HA potency has been evaluated by single-radial immunodiffusion (SRID), the standard in vitro potency assay for IIVs, to predict vaccine immunogenicity with a correlation to protective efficacy. We previously reported that limited trypsin digestion (LTD) selectively degraded stressed HA, so that an otherwise conformationally insensitive biophysical quantification technique could specifically quantify trypsin-resistant, immunologically active HA. Here, we demonstrate that isotope dilution mass spectrometry (IDMS), a method capable of quantifying the absolute HA concentration without reference antigen use, can be further expanded by adding LTD followed with precipitation to selectively quantify the active HA. We test the LTD-IDMS assay on H7N9 vaccines stressed by low pH, raised temperature, or freeze/ thaw cycles. This method, unlike SRID, has no requirement for strain-specific reference antigens or antibodies and can generate potency values that correlate with SRID. Thus, LTD-IDMS is a promising alternative in vitro potency assay for influenza vaccines to complement and potentially replace SRID in a pandemic when strain specific reagents may not be readily available.
Keywords: Hemagglutinin, Influenza, Isotope dilution mass spectrometry, Limited trypsin digestion, Potency, Precipitation, Single-radial immunodiffusion, Vaccine
1. Introduction
Influenza viruses cause both seasonal, epidemic infections and periodic, unpredictable pandemics. Vaccination is the most effective means to reduce the substantial morbidity and mortality caused by influenza infection [1,2]. Hemagglutinin (HA), the major influenza surface antigen, binds host cell surface receptors and mediates viral entry by mediating membrane fusion [3,4]. HA is the major target for virus-neutralizing antibodies and the most important antigen in subunit or split influenza vaccines (inactivated influenza vaccine-IIV) [5].
IIV potency is determined primarily by the quantification of immunologically active HA (able to elicit robust neutralizing or hemagglutination inhibiting [HI] antibody responses) that a dose contains. For IIV formulation, release, and stability testing, an in vitro potency test is used, single-radial immunodiffusion (SRID) [6,7]. This modified Ouchterlony test quantifies HA based on the relative diameters of immunoprecipitin rings that form when vaccine antigen or homologous antigen standards diffuse radially from a circular well punched into an agarose gel that has been cast with a matched strain-specific sheep antiserum. The immunoprecipitin ring is detected by Coomassie blue staining after non-complexed antigen and antibody are removed by blotting with filter paper and washing with water. Although HA in IIVs form rosettes and other complexes, zwittergent is added to the antigen to disperse HA to smaller complexes such that the ring size is expected to be proportional to the HA concentration [7]. A weak correlation has been shown between SRID-measured vaccine potency and vaccine immunogenicity in clinical trials [8–10]. SRID has been accepted by international regulatory agencies and used by influenza vaccine manufacturers for IIV formulation, release, and stability testing for four decades.
Despite its selectivity for immunologically active HA, SRID has shortcomings. The most obvious shortcoming is the need for large quantities of strain-specific reference reagents, calibrated reference antigens and antisera. Generation and calibration of these strain-specific reagents is a time-consuming process that can delay vaccine release. The strain-specific HA reference antigens are produced by growing, inactivating, and purifying whole influenza viruses. The corresponding strain-specific antisera are generated by immunizing sheep multiple times with HA cleaved from purified whole virus by bromelain [6,11]. For highly pathogenic pandemic strains, safety concerns can slow the production of both immunological reference reagents. Reference antigens and antisera are calibrated and distributed by the World Health Organization (WHO) Essential Regulatory Laboratories (ERLs). Primary liquid standards (PLS) are generated by assigning a value with physicochemical methods to inactivated whole virus preparations. A conformationally independent total protein assay (BCA, Lowry, or Kjel-dahl) is used to quantify the standard’s total protein content. In addition, sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) is used to determine the percent of HA contained in the sample through densitometry of the Coomassie blue stained bands [12,13]. At this point, SRID is used to calibrate lyophilized standards which manufacturers will use against the relevant characterized PLS. PLS and reference standard are calibrated by the four ERLs in parallel. Thus, material shipment between geographically dispersed organizations, with diverse import regulations, can impose a significant barrier for timely pandemic responses [14]. The time required to generate and calibrate the reagents needed for SRID was evident during the early days of the 2009 pandemic, prompting efforts to develop more practical, alternative in vitro potency assays that do not require strain-specific immunological reagents [13].
HA forms a membrane-bound trimer of disulfide linked HA1/ HA2 heterodimers [15]. HA generally maintains a “metastable”, pre-fusion conformation at neutral pH, which elicits influenza neutralizing and HI antibodies. Once an energy barrier is overcome by an endosomal low pH drop, HA refolds extensively and irreversibly to a post-fusion conformation that is more stable, but significantly less immunologically active [16–19]. HA conformation is also correlated with HA susceptibility to proteolysis [20,21]. HA1 in prefusion HA, the more immunogenic form, is trypsin-resistant whereas HA1 in the less immunogenic, post-fusion HA form is trypsin-sensitive. This is consistent with the observation that native, well-folded protein domains are often protease resistant and stressed, denatured proteins are trypsin sensitive. Limited trypsin digestion (LTD) has been shown to be able to differentiate HA conformation [22,23] and confer functional specificity to biophysical techniques that are generally insensitive to conformation without requiring strain-specific antibodies.
Biophysical techniques, such as reversed-phase high performance liquid chromatography (RP-HPLC) with either ultraviolet or fluorescence detection, have been developed and used for quantitative analysis of influenza HA [24,25]. These methods measure the peak area of the HA1 and quantify it by comparing it to peak areas from reference standards. Like SRID, RP-HPLC still relies on the availability and reliability of the standardized reference antigens. RP-HPLC separates HA1 from other viral proteins based on their different hydrophobicity, but cannot distinguish HA1 between subtypes with similar hydrophobicity, therefore it cannot reliably quantitate subtype-specific HAs in multivalent vaccines.
Isotope dilution mass spectrometry (IDMS) is another biophysical technology recently developed for absolute quantification of HA in a vaccine mixture [26]. IDMS involves selecting specific target peptides from HA sequences as a stoichiometric representative of the intact protein. A known amount of synthetic reference peptide, in which one amino acid has been isotopically labeled, is spiked into the sample. Quantification is achieved by comparing the peak area of the isotopically labeled reference peptide with that of an endogenous target peptide generated by target protein proteolytic cleavage. Therefore, this approach can provide absolute HA quantification without the need for the time-consuming generation and calibration of strain-specific standardized reference antigens. In addition, since the method is based on sequence-specific peptides from each subtype, IDMS has been shown to be capable of selectively quantifying subtype-specific HAs in multivalent vaccines.
LTD functions as a pre-treatment to differentiate immunologically active HA without the need for sheep antisera. IDMS serves as an accurate biophysical approach to evaluate absolute HA quantification without the need of reference antigens. Here we report that we develop an LTD-IDMS method including LTD, IDMS and an intermediate biophysical step, precipitation, to separate LTD- resistant HA from LTD-digested HA to enable the specific, absolute quantification of immunologically active HA. We demonstrate that this biophysical alternative potency assay is able to provide potency measurements of pre-pandemic A/Shanghai/2/2013 (H7N9) vaccine that are comparable to SRID. Because the IDMS target peptides are conserved through the subtype, no method optimization was required since the peptides were first chosen for H7N2 and H7N7 [27]. Thus, the LTD-IDMS assay was rapidly applied for potency evaluation of the new H7N9 strain, A/Hong Kong/125/2017, before its strain-specific SRID reference reagents became available. The comparability study performed later confirmed that LTD-IDMS led to potency results comparable to SRID.
2. Results
Human infections with A(H7N9) viruses in China were first reported to the WHO in March 2013. Since then six waves of human infection have been reported with a total of 1567 cases and a 39% fatality rate [28]. In 2013, A/Shanghai/2/2013 (H7N9) vaccines were developed and produced for rapid response to the A(H7N9) emerging pandemic threat. In 2017, additional A(H7N9) vaccines, including A/Hong Kong/125/2017 (H7N9), were developed to manage the new strains emerging during the fifth and largest wave of the epidemic, which were less well covered by the original A/Shanghai/2/2013 (H7N9) vaccines [29].
2.1. Potency quantification by RP-HPLC and SRID with LTD as a pretreatment for A/Shanghai/2/2013 (H7N9) vaccine
LTD was developed for the purpose of distinguishing immunologically active HA from inactive HA independent of strain-specific antibodies [22]. To confirm the specific digestion of post-fusion HA in the A/Shanghai/2/2013 (H7N9) monobulk by LTD, the monobulk was stressed transiently with low pH and quantified by RP-HPLC and SRID following LTD. The results showed that RP-HPLC alone measured HA in both pre-fusion and post-fusion conformations (Fig. 1A). However, when LTD was included as pre-treatment step digesting HA1 of HA stressed by low pH, RP-HPLC specifically detected and quantified non-stressed, pre-fusion HA, with potency values correlating with the results from SRID, which independently quantified potency without the need for LTD (Fig. 1A). Reducing and non-reducing SDS-PAGE clearly showed that LTD pre-treatment had little effect on non-stressed HA but it led to HA1 loss in low pH stressed HA (Sup. Fig. 1). HA2 in both low pH stressed and non-stressed samples remained relatively unchanged by LTD.
Fig. 1.
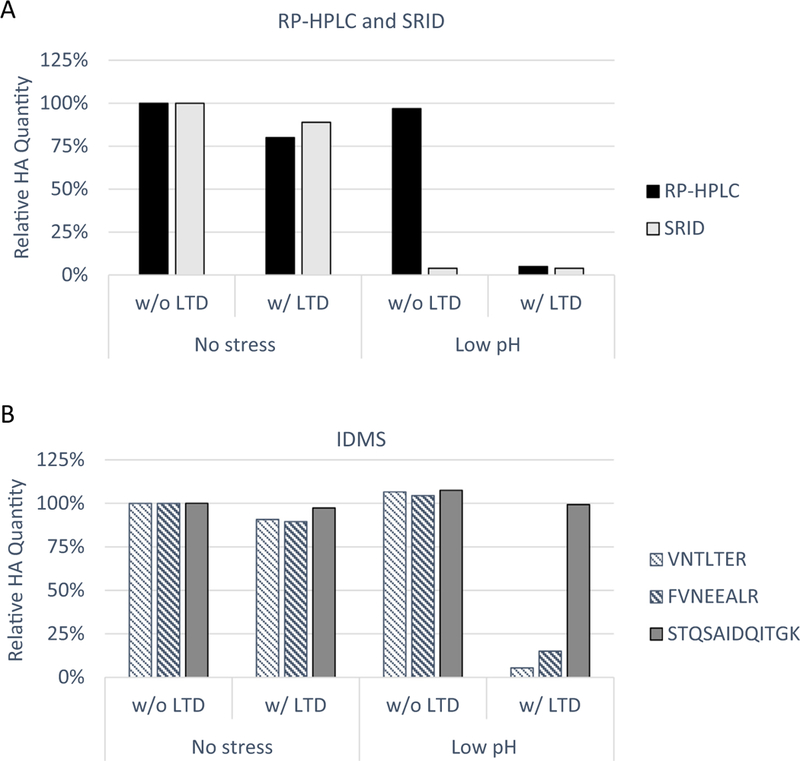
Relative HA quantification for A/Shanghai/2/2013 (H7N9) preparations. Non-stressed and low pH-stressed monobulks were pre-treated with and without LTD as indicated. (A) Relative HA quantification by RP-HPLC and SRID. (B) Relative HA quantification by IDMS following precipitation, reconstitution and analytical digestion using three reference peptides, VNTLTER (striped), FVNEEALR (bold striped) from HA1 sequence and STQSAIDQITGK (solid) from HA2 sequence for measurement.
2.2. Potency quantification by LTD-IDMS for A/Shanghai/2/2013 (H7N9) vaccine
A similar sample set were quantified by IDMS as it was described previously [26,27,30,31]. Briefly, HA peptides were chosen as the stoichiometric representative of the larger HA protein with the assumption that one mole of that specific peptide represented one mole of the matched protein. For this reason, target peptides needed to not contain modifications that would alter the peptide’s mass and to be in areas of the detergent denatured HA easily accessible for proteolytic digestion. Therefore, peptides with the following characteristics were avoided: potential N-linked glycosylation sites, oxidizable residues (e.g., methionine, tryptophan), containing cysteines or near cysteines. Prior to analysis, HA protein was denatured with RapiGest™, analytically digested with trypsin, and spiked with a labeled peptide standard. After separation by HPLC, individual peptides were quantified using multiple reaction monitoring (MRM) analysis. This process employed the internal peptide standards with one 13C and 15N labeled amino acid giving it the same chemical and chromatographic properties as the target HA peptide, but a different molecular weight that is distinguishable by mass spectrometry. Both labeled and native peptides were assigned a value by amino acid analysis utilizing National Institute of Standards and Technology (NIST) amino acid standards for accuracy and traceability [32]. In order to confirm complete protein digestion, three peptides were chosen and quantified independently. Since it was assumed that there were equal molar amounts of protein and target peptide, the three individual peptides representing the same protein were expected to have similar concentrations.
As an assay control, non-stressed, pre-fusion HA was precipitated directly without LTD pre-treatment, reconstituted in Rapi-Gest™, analytically digested with trypsin, and quantified by IDMS using three target peptides distantly located across the protein, VNTLTER and FVNEEALR on HA1 and STQSAIDQITGK on HA2 (Sup. Fig. 2). Independent measurements obtained from each of the three target peptides agreed within 10%, confirming the complete analytical digestion of the HA protein in the target peptide areas (Fig. 1B). When LTD was added prior to precipitation, the LTD-IDMS assay, including LTD, precipitation, reconstitution, analytical digestion and IDMS quantification, showed similar results suggesting the high recovery of pre-fusion HA. Low pH-stressed, post-fusion HA was precipitated without LTD, and IDMS quantification achieved peptide agreement. The results were similar to those for non-stressed, pre-fusion HA suggesting that IDMS alone quantified HA in all conformations, similar to RP-HPLC. When LTD was added as pre-treatment for this low-pH stressed HA, LTD-IDMS results showed that the two peptides, VNTLTER and FVNEEALR of HA1, substantially decreased in concentration, while the peptide STQSAIDQITGK of HA2 remained similar in amount as in the non-stressed HA. These results indicated the LTD-IDMS specifically quantitated pre-fusion HA represented by the HA1 peptides and total HA represented by the HA2 peptide. The amount of HA1 peptides determined by LTD-IDMS shown in Fig. 1B correlated well with the results from SRID shown in Fig. 1A.
2.3. Selective potency quantification by LTD-IDMS for A/Shanghai/2/ 2013 (H7N9) vaccine in mixed modality
To further evaluate the selectivity of potency quantification, LTD-IDMS was performed to quantify the non-stressed, pre-fusion HA in a mixture of non-stressed and low pH-stressed A/Shanghai/2/2013 (H7N9) monobulk (Fig. 2). The 100% non-stressed (0% low pH-stressed) monobulk was considered as a control. Other mixtures, that maintained the same absolute HA amount, were blended using different percentages of non-stressed and low pH-stressed monobulk with percent of non-stressed HA and low pH-stressed HA shown on the X-axis. LTD-IDMS of the control sample showed good peptide agreement among the three peptides (data not shown). Reduction in HA1 peptides in the remaining samples correlated with the percentage of low pH-stressed HA in each sample, with negligible concentrations in the final 100% low pH-stressed sample. The LTD-IDMS results with HA1 peptides correlated with the percent of non-stressed HA in each sample indicating the selective quantification of pre-fusion, potent HA in the mixture. The HA1 peptide VNTLTER is closer to the N-terminus and was somewhat more susceptible to LTD than the HA1 peptide FVNEEALR. The tendency of HA2 peptide (STQSAIDQITGK) to remain relatively consistent with the control indicated that the HA2 portion of HA is resistant to LTD, as confirmed in the SDS-PAGE shown in Sup. Fig. 1. Therefore, these results confirmed that the degree to which the total HA has been irreversibly shifted into its less immunogenically relevant conformational state can be quantified by LTD-IDMS.
Fig. 2.
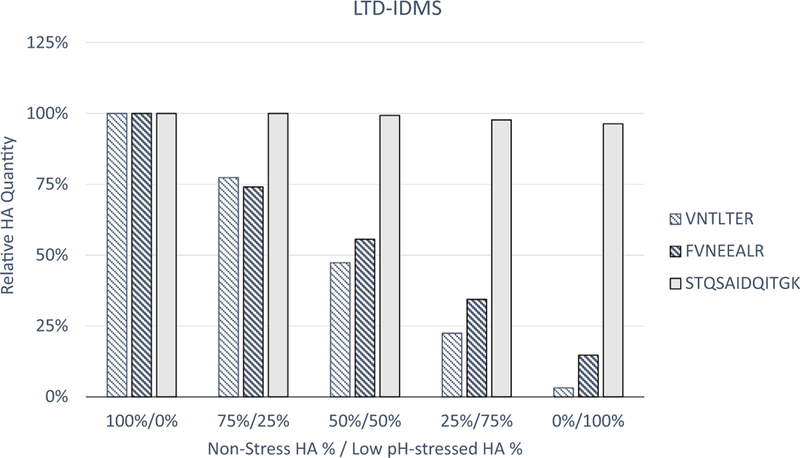
Relative HA potency quantification for A/Shanghai/2/2013 (H7N9) mixed preparations by LTD-IDMS. Non-stressed monobulk was mixed with low pH-stressed monobulk as indicated. LTD-IDMS measured with three reference peptides, VNTLTER (striped), FVNEEALR (bold striped) from HA1 and STQSAIDQITGK (solid) from HA2.
2.4. Potency quantification by RP-HPLC with LTD and LTD-IDMS for A/Hong Kong/125/2017 (H7N9) vaccine
The A/Hong Kong/125/2017 (H7N9) strain was recommended to cover newly emerging strains in 2017. Vaccine production was initiated, but SRID reference reagents did not become available until nine months later. The protein sequence alignment for A/Shanghai/2/2013 (H7N9) HA and A/Hong Kong/125/2017 (H7N9) HA (Sup. Fig. 2) showed the three target peptides used for A/Shanghai/2/2013 (H7N9) were conserved and could be used for analysis of A/Hong Kong/125/2017 (H7N9).
To confirm the selectivity for potent HA by LTD for the A/Hong Kong/125/2017 (H7N9) vaccine, a similar set of mixed non-stressed and low-pH stressed monobulks were prepared (Sup. Fig. 3). While RP-HPLC alone detected both non-stressed and stressed HA in the preparations, RP-HPLC with LTD analyses showed that the method could selectively measure the non-stressed HA (Fig. 3A). RP-HPLC relies on SRID reference antigens as standard for absolute quantification; as such, the A/Shanghai/2/2013 (H7N9) SRID reference antigen was used due to the unavailability of a matched A/Hong Kong/125/2017 SRID reference antigen.
Fig. 3.
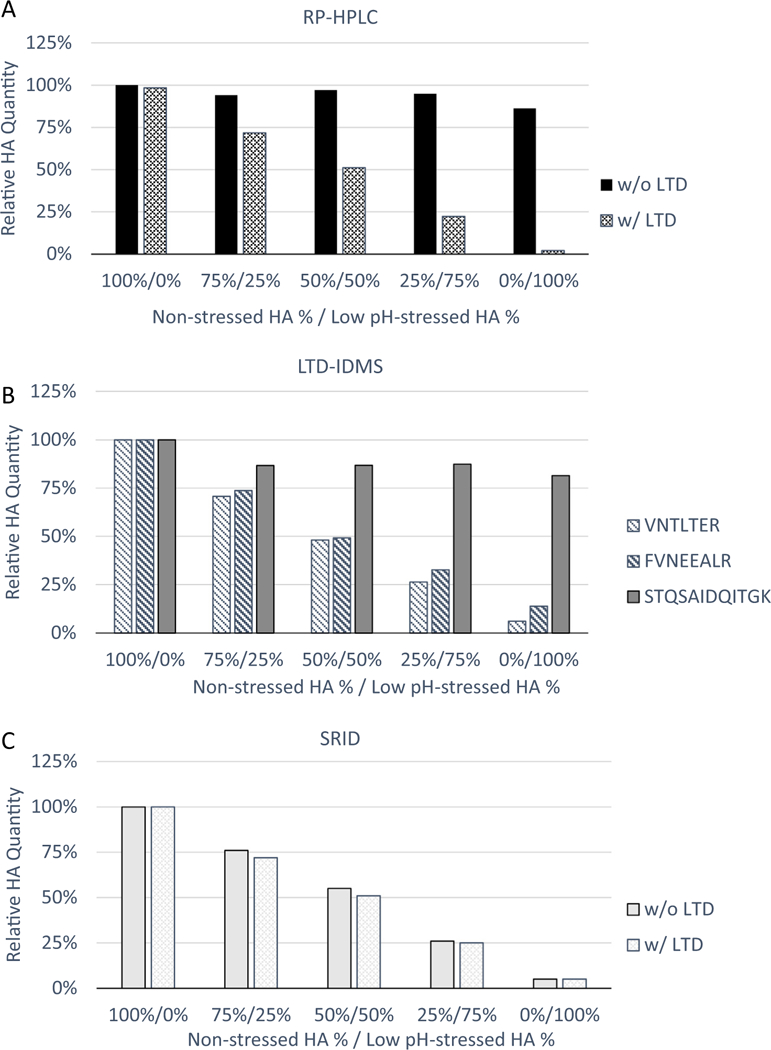
Relative HA potency quantification for A/Hong Kong/125/2017 (H7N9) mixed preparations by RP-HPLC with LTD and LTD-IDMS and correlation with potency quantitation by SRID. Non-stressed monobulk was mixed with low pH-stressed monobulk as indicated. (A) Relative HA quantification by RP-HPLC with and without LTD asa pre-treatment. (B) Relative HA quantification by LTD-IDMS with reference peptides, VNTLTER (striped), FVNEEALR (bold striped) from HA1 and STQSAIDQITGK (solid) from HA2. (C) Relative HA quantification by SRID with and without LTD as a pre-treatment.
The LTD-IDMS method was performed on the same set of mixed non-stressed and stressed A/Hong Kong/125/2017 (H7N9) monobulks. A similar trend was observed, with the two HA1 peptides decreasing in concentration in an inverse relation to the amount of low pH-stressed HA in the sample (Fig. 3B), confirming the selective quantification of non-stressed HA by LTD-IDMS. LTD-IDMS results for the HA2 peptide showed a slight decrease in HA2 quantity for the preparations with stressed monobulk added, consistent with SDS-PAGE result shown in Sup. Fig. 3.
2.5. Correlation of LTD-IDMS and SRID results for A/Hong Kong/125/2017 (H7N9) vaccine
SRID reference reagents for A/Hong Kong/125/2017 (H7N9) were available in November 2017 from CBER/FDA. The potency evaluation by SRID and LTD-IDMS was performed on a new set of mixed non-stressed and stressed monobulks to compare SRID and LTD-IDMS potency results. SRID measured relative potency correlating with the percentage of non-stressed HA in each preparation (Fig. 3C), and correlating with LTD-IDMS results (data not shown). This indicated that LTD-IDMS can quantify A/Hong Kong/125/2017 (H7N9) potency in a similar manner as SRID without the need for strain specific antibodies and reference antigens.
2.6. Potency quantification by LTD-IDMS for A/Hong Kong/125/2017 (H7N9) vaccine stressed by heat and freeze/thaw
To evaluate LTD-IDMS robustness for potency quantification, the effect of other stresses on vaccine potency was evaluated by LTD-IDMS with SRID for comparison (Fig. 4). In these experiments, either raised temperature (56 °C for 63 hours) or one cycle of freeze/thaw was applied to A/Hong Kong/125/2017 (H7N9) monobulks prior to LTD-IDMS. Results were reported as relative HA quantity to the non-stressed control by averaging results from the two HA1 peptides. When a freeze/thaw-stress was administered to the sample, both LTD-IDMS and SRID showed reduction in relative HA concentration to ~60% of the control. With the heat stress, a more substantial reduction was quantified by both methods as <11% HA was detected. These results further illustrated that LTD-IDMS has the capability to quantify pre-fusion HA in stressed vaccines.
Fig. 4.
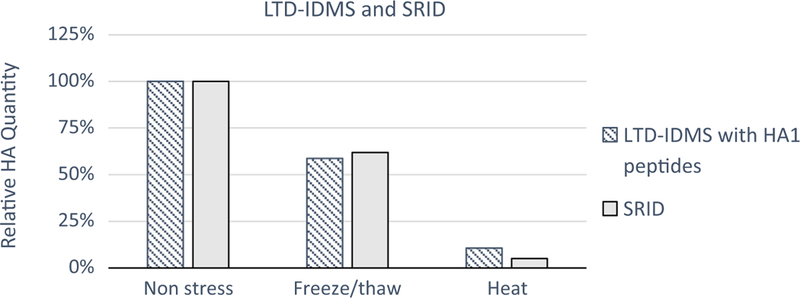
Correlation of relative HA potency quantification for freeze/thaw-stressed and heat-stressed A/Hong Kong/125/2017 (H7N9) preparations by LTD-IDMS (striped) and SRID (grey). Relative HA quantification by LTD-IDMS were the average of results from two HA1 peptides, VNTLTER and FVNEEALR.
3. Discussion
SRID was developed as an in vitro potency assay to quantify the immunologically active HA antigen in influenza vaccines and the measured potency has been shown to correlate with protective clinical responses. SRID has been accepted by international regulatory agencies for IIV release and stability testing for four decades. However, there are significant limitations to the assay. The generation and calibration of SRID sheep antisera and inactivated purified whole virus antigens for each new strain is a lengthy process that can delay vaccine release and a pandemic response. Additionally, differing lots of sheep antisera can change the quantitative results of the SRID assay.
LTD has been shown to selectively digest immunogenicially less active HA and enable biophysical quantification techniques for specific quantification of trypsin-resistant, immunologically active HA as a potential potency assay [22]. LTD eliminates the need to generate strain-specific antibodies for potency determination. LTD followed by RP-HPLC could quantify potent HA in monovalent vaccines. Although LTD selectively degrades stressed HA in multivalent vaccines, RP-HPLC does not separate HA1 from different subtypes well, complicating quantification of each individual vaccine component in multivalent preparations. More importantly, RP-HPLC methods still rely on SRID reference antigens, inactivated purified whole viruses, for potency quantification. IDMS was developed for absolute HA quantification in a vaccine mixture [26] and is an alternative method by which ERLs assign values to SRID reference antigens. This approach uses strain-specific synthetic peptides instead of virus antigen as standards, so IDMS can quantify each HA in the multivalent vaccine without the need to generate and calibrate whole virus standards. The target peptides for IDMS are specific to the subtype so it is a straightforward means of quantifying the amount of each virus strain in a multivalent vaccine. The target peptides are also conserved among subtypes so that it is unaffected by frequent strain updates.
To bridge LTD and IDMS together to form the complete biophysical potency assay, precipitation was introduced post-LTD to remove trypsin-digested HA1 peptides and retain intact HA1 from potent HA, as well as HA2 from all HA for quantification with IDMS. As a result, precipitation enabled IDMS to quantify HA potency and mass in the same test: HA1 peptides quantified potent HA and HA2 peptides quantified total HA.
Generation of pandemic SRID reagents is challenging due to the specific features of the viruses. Pandemic HAs are usually conformationally less stable [33] than seasonal HAs and, consequently, are suboptimal as immunogens to raise SRID sheep antisera. The safety considerations and import restrictions of pandemic viruses also hinder the growth and purification of whole virus to be used as the SRID reference antigen. LTD-IDMS provides a potential time advantage in a pandemic as it is independent of any immunological reagent. This has been demonstrated in this study on potency quantification of H7N9 vaccines. Because one of the selection criteria for IDMS reference peptides is evolutionary conservation throughout the subtype, the peptides formerly prepared for H7N2 and H7N7 were unchanged in H7N9 HA sequences. LTD-IDMS was applied directly for H7N9 potency determination without the need to generate new strain-specific reference peptides. LTD-IDMS showed with the HA1 peptides the selective quantification of potent HA in H7N9 vaccines stressed by low pH, raised temperature, and freeze/thaw cycle which led to a potency reduction comparable to SRID assessment once SRID reagents were available.
In addition to the relative potency change of stressed material, absolute potency for non-stressed and stressed vaccines was also quantified by LTD-IDMS for comparison with SRID values (data not shown). The absolute potency values by LTD-IDMS analysis were not always consistent with SRID results. Potency absolute values are determined by the calibrated value of standards. Unlike LTD-IDMS, calibration of SRID still relies on inactivated purified whole virus as standards whose HA quantity is determined by combining SDS-PAGE HA proportions characterized with densitometry and total protein determination. The calibrated value is influenced by the accuracy, precision and consistency of the two methods. In addition, potent HA quantification by SRID is also affected by other factors, e.g. diffusion of HA antigen, the formation of immuno-complex by antigens and antibodies [19] and interference by non-HA antibodies such as anti-NA antibodies. Validation of potentially more accurate, precise, and efficient influenza vaccine potency assays includes bridging studies to SRID assays [13]. However, given the inherent issues with SRID, matching SRID with new potency assays may prove impractical and alternate methods for validating the approaches may be needed.
In summary, LTD-IDMS can selectively quantify potent HA in vaccines without the need of sheep antisera nor purified inactivated purified virus antigen. Thus, LTD-IDMS is a promising alternative in vitro potency assay for influenza vaccines with particularly applicability to speedy pandemic responses.
4. Materials and methods
4.1. Influenza reference reagents
SRID reference reagents for A/Anhui/01/2013 (H7N9) were provided by the National Institute for Biological Standards and Control (NIBSC, London, UK): sheep polyclonal reference antiserum A/ Anhui/1/2013 x A/PR/8/34 NIBRG-270 Lot 13/180 and reference antigen A/Anhui/01/2013 (H7N9) NIBRG-268 Lot 14/250. SRID reference reagents for A/Hong Kong/125/2017 (H7N9) were provided by the US Food and Drug Administration’s Center for Biologics Evaluation and Research (FDA CBER, Silver Spring, MD, USA): sheep polyclonal reference antiserum A/Hong Kong/125/2017 (H7N9) Lot 88 and reference antigen A/Hong Kong/125/2017 (H7N9) Lot H7-Ab-1706.
4.2. Influenza vaccines
A/Shanghai/2/2013 IDCDC-RG32A (H7N9) and A/Hong Kong/125/2017 IDCDC-RG56B (H7N9) monobulks were produced from embryonated chicken eggs with the Agrippal® subunit influenza vaccine process by Seqirus. In summary, harvested viruses were inactivated by formaldehyde and purified by centrifugation. After CTAB splitting, the viral surface proteins were further purified and formulated in PBS buffer. These monobulks were prepared for research purposes only.
4.3. Sample stress by low pH
Influenza monobulks were treated with 10% (volume/volume [v/v]) 50 mM citrate, pH 4.0 (Boston BioProducts, Ashland, MA, USA) to reduce the sample pH to 4.0 and then incubated at room temperature for 30 min with shaking at 2500 RPM with an Eppendorf® MixMate®. 12% (v/v) of 1 M Tris, pH 8.4 (Boston BioProducts) was added to neutralize pH to 7.2. Samples were stored at 4 °C until analyzed.
4.4. Sample stress by raised temperature
Raised temperature stressing was performed by incubation of influenza monobulks at 56 °C for 63 hours, followed by a return to 4 °C. Samples were stored at 4 °C until analyzed.
4.5. Sample stress by freeze/thaw
Influenza monobulks were kept at –20 °C for 24 hours, then moved to 4 °C until analyzed.
4.6. Limited trypsin digestion (LTD)
Samples were incubated with bovine trypsin (T7901, Sigma, St. Louis, MO, USA) in 1 mM HCl (Sigma) at 37 °C for 120 min. The trypsin was diluted and sample concentration adjusted so that there was 40 U of trypsin activity per 100 μg of HA. Trypsin activity was quenched by placing the sample on ice. Samples were stored at 4 °C until analyzed.
4.7. SDS-PAGE, RP-HPLC and SRID
SDS-PAGE, RP-HPLC and SRID have been described in [22].
4.8. Precipitation
Samples were diluted 4-fold with cold acetone (Sigma) (–20 °C) and centrifuged at 20,817g for 20 min at 4 °C. The supernatants were separated from the precipitate after centrifugation. Three subsequent washes were performed with cold ethanol (Sigma) (–20 °C). The resulting precipitates were stored at –20 °C for 16 h before being reconstituted in 0.1% RapiGest™ (Waters, Milford, MA, USA) in 50 mM ammonium bicarbonate (Sigma).
4.9. Analytical digestion and IDMS
10 μL of sample were combined with 10 μL of 0.2% RapiGest™ and heated for 5 min at 100 °C. After cooling, 5 mL (~86 pmol) of sequencing grade modified porcine trypsin (Promega, Madison, WI, USA) were added and incubated with the sample for 2 h at 37 °C. 10 μL of 0.45 M HCl (Thermo Fisher Scientific, Waltham, MA, USA) were added after incubation to degrade the acid labile RapiGest™. 10 μL of the 0.5 pmol/μL heavy-isotope labeled influenza H7 peptides cocktail (Midwest Bio-Tech, Fishers, IN, USA) in 0.1% formic acid (Thermo Fisher Scientific) was added to serve as the internal standard. 45 μL of 0.1% formic acid were added to yield a final volume of 100 μL. This solution was then transferred to an autosampler vial for LC/MS/MS analysis.
A NanoAcquity (Waters, Milford, MA, USA) was employed for HPLC separation of the target peptides. The system used HPLC- grade water with 0.1% formic acid as its aqueous mobile phase (A) and acetonitrile with 0.1% formic acid (Thermo Fisher Scientific) as its organic phase (B). The analytical column was a 150 mm × 1 mm i.d. Symmetry300 reverse phase C18 (3.5 mm particle size, Waters). The column was conditioned to 99.5% A and ramped up to 20% B within 20 min and finally to 25% B at 25 min. The column was then washed with 98% B for 10 min and then allowed to re-equilibrate to initial conditions for 20 min. The run time for the method was 57 min. The column eluent was introduced into a Thermo Quantum TSQ triple quadrupole tandem mass spectrometer with an electrospray interface (Thermo Fisher Scientific). The instrument, operating in positive ion mode, monitored for the precursor/product ion transition pairs for the native and labeled peptides as shown in Table 1. The data was processed with the Thermo Xcalibur™ Quan software.
Table 1.
Target peptides used for the LTD-IDMS quantification of HA.
| Target peptide | Precursor ion m/z |
Fragment ion m/z |
Fragment ion m/z |
Fragment ion m/z |
Collision energy (eV) |
|---|---|---|---|---|---|
| VNTLTER | 416.7 (+2) | 619.3 | 733.4 | 518.3 | 17 |
| VNTLTER | 420.2 (+2) | 626.4 | 740.4 | 525.3 | 17 |
| FVNEEALR | 489.3 (+2) | 617.3 | 731.4 | 830.4 | 20 |
| FVNEEALR | 492.8 (+2) | 624.3 | 738.4 | 837.5 | 20 |
| STQSAIDQITGK | 624.8 (+2) | 774.4 | 845.5 | 661.4 | 25 |
| STQSAIDQITGK | 626.8 (+2) | 781.5 | 852.5 | 668.4 | 25 |
Supplementary Material
Acknowledgements
We thank those who have participated in helpful discussions at multi-institution meetings on the development of alternative influenza in vitro potency assays, including members of the International Federation of Pharmaceutical Manufacturers and Associations’ Potency Assay Working Group, the National Institute for Biological Standards and Control (NIBSC), the US Food and Drug Administration (FDA) and the Biomedical Advanced Research and Development Authority (BARDA).
Abbreviations:
- HA
hemagglutinin
- IDMS
isotope dilution mass spectrometry
- IIV
inactivated influenza vaccine
- LTD
limited trypsin digestion
- RP-HPLC
reversed-phase high pressure liquid chromatography
- SDS-PAGE
sodium dodecyl sulfate polyacrylamide gel electrophoresis
- SRID
single-radial immunodiffusion
Footnotes
Disclaimer
The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Centers for Disease Control and Prevention. The authors affiliated with Seqirus are employees of Seqirus and shareholders of CSL.
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.vaccine.2018.08.065.
References
- [1].Zambon MC. Epidemiology and pathogenesis of influenza. J Antimicrob Chemother 1999;44(Suppl B):3–9. [DOI] [PubMed] [Google Scholar]
- [2].Poland GA, Rottinghaus ST, Jacobson RM. Influenza vaccines: a review and rationale for use in developed and underdeveloped countries. Vaccine 2001;19 (17–19):2216–20. [DOI] [PubMed] [Google Scholar]
- [3].Wiley DC, Wilson IA, Skehel JJ. Structural identification of the antibody-binding sites of Hong Kong influenza haemagglutinin and their involvement in antigenic variation. Nature 1981;289(5796):373–8. [DOI] [PubMed] [Google Scholar]
- [4].Wilson IA, Skehel JJ, Wiley DC. Structure of the haemagglutinin membrane glycoprotein of influenza virus at 3 A resolution. Nature 1981;289 (5796):366–73. [DOI] [PubMed] [Google Scholar]
- [5].Knossow M et al. Mechanism of neutralization of influenza virus infectivity by antibodies. Virology 2002;302(2):294–8. [DOI] [PubMed] [Google Scholar]
- [6].Wood JM, Schild GC, Newman RW, Seagroatt V. An improved single-radial- immunodiffusion technique for the assay ofinfluenza haemagglutinin antigen: application for potency determinations of inactivated whole virus and subunit vaccines. J Biol Stand 1977;5(3):237–47. [DOI] [PubMed] [Google Scholar]
- [7].Williams MS. Single-radial-immunodiffusion as an in vitro potency assay for human inactivated viral vaccines. Vet Microbiol 1993;37(3–4):253–62. [DOI] [PubMed] [Google Scholar]
- [8].Rowlen K Validation of alternative potency assays for influenza vaccines requires clinical studies. Vaccine 2015. [DOI] [PubMed] [Google Scholar]
- [9].Ennis FA et al. Correlation of laboratory studies with clinical responses to A/ New Jersey influenza vaccines. J Infect Dis 1977;136(Suppl):S397–406. [DOI] [PubMed] [Google Scholar]
- [10].La Montagne JR et al. Summary of clinical trials of inactivated influenza vaccine - 1978. Rev Infect Dis 1983;5(4):723–36. [DOI] [PubMed] [Google Scholar]
- [11].Brand CM, Skehel JJ. Crystalline antigen from the influenza virus envelope. Nat New Biol 1972;238(83):145–7. [DOI] [PubMed] [Google Scholar]
- [12].Minor PD. Assaying the Potency of Influenza Vaccines. Vaccines 2015;3:90–104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Wood JM, Weir JP. Standardisation of inactivated influenza vaccines-Learning from history. Influenza Other Respir Viruses 2018;12(2):195–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Hardy S et al. Confronting the next pandemic-workshop on lessons learned from potency testing of pandemic (H1N1) 2009 influenza vaccines and considerations for future potency tests, Ottawa, Canada, July 27–29, 2010. Influenza Other Respir Viruses 2011;5(6):438–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wiley DC, Skehel JJ. Crystallization and x-ray diffraction studies on the haemagglutinin glycoprotein from the membrane of influenza virus. J Mol Biol 1977;112(2):343–7. [DOI] [PubMed] [Google Scholar]
- [16].Bullough pA, Hughson FM, Skehel JJ, Wiley DC. Structure of influenza haemagglutinin at the pH of membrane fusion. Nature 1994;371 (6492):37–43. [DOI] [PubMed] [Google Scholar]
- [17].Skehel JJ, Wiley DC. Receptor binding and membrane fusion in virus entry: the influenza hemagglutinin. Annu Rev Biochem 2000;69:531–69. [DOI] [PubMed] [Google Scholar]
- [18].Ruigrok RW et al. Studies on the structure of the influenza virus haemagglutinin at the pH of membrane fusion. J Gen Virol 1988;69(Pt 11):2785–95. [DOI] [PubMed] [Google Scholar]
- [19].Wen Y et al. Trypsin pre-treatment corrects SRID over-estimation of immunologically active, pre-fusion HA caused by mixed immunoprecipitin rings. Vaccine 2016;34(29):3388–95. [DOI] [PubMed] [Google Scholar]
- [20].Skehel JJ et al. Changes in the conformation of influenza virus hemagglutinin at the pH optimum of virus-mediated membrane fusion. Proc Natl Acad SciUSA 1982;79(4):968–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Ruigrok RW et al. Conformational changes in the hemagglutinin of influenza virus which accompany heat-induced fusion of virus with liposomes. Virology 1986;155(2):484–97. [DOI] [PubMed] [Google Scholar]
- [22].Wen Y et al. Conformationally selective biophysical assay for influenza vaccine potency determination. Vaccine 2015;33(41):5342–9. [DOI] [PubMed] [Google Scholar]
- [23].Wen Y, Palladino G, Xie Y, Ferrari A, Settembre EC. Inactivated influenza vaccine stress can affect in vitro potency assay relationship to immunogenicity. Vaccine 2018;36(21):3010–7. [DOI] [PubMed] [Google Scholar]
- [24].Kapteyn JC et al. Haemagglutinin quantification and identification of influenza A&B strains propagated in PER.C6 cells: a novel RP-HPLC method. Vaccine 2006;24(16):3137–44. [DOI] [PubMed] [Google Scholar]
- [25].Garcia-Canas V, Lorbetskie B, Girard M. Rapid and selective characterization of influenza virus constituents in monovalent and multivalent preparations using non-porous reversed-phase high performance liquid chromatography columns. J Chromatogr A 2006;1123(2):225–32. [DOI] [PubMed] [Google Scholar]
- [26].Williams TL et al. Quantification of influenza virus hemagglutinins in complex mixtures using isotope dilution tandem mass spectrometry. Vaccine 2008;26 (20):2510–20. [DOI] [PubMed] [Google Scholar]
- [27].Santana WI, Williams TL, Winne EK, Pirkle JL, Barr JR. Quantification of viral proteins of the avian H7 subtype of influenza virus: an isotope dilution mass spectrometry method applicable for producing more rapid vaccines in the case of an influenza pandemic. Anal Chem 2014;86(9):4088–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Kile JC et al. Update: increase in human infections with novel Asian lineage avian influenza A(H7N9) viruses during the fifth epidemic - China, October 1, 2016-August 7, 2017. MMWR Morb Mortal Wkly Rep 2017;66(35):928–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].WHO. Zoonotic influenza viruses: antigenic and genetic characteristics and development of candidate vaccine viruses for pandemic preparedness. Wkly Epidemiol Rec 2017;92(12):129–44. [PubMed] [Google Scholar]
- [30].Williams TL, Pirkle JL, Barr JR. Simultaneous quantification of hemagglutinin and neuraminidase of influenza virus using isotope dilution mass spectrometry. Vaccine 2012;30(14):2475–82. [DOI] [PubMed] [Google Scholar]
- [31].Norrgran J et al. Optimization of digestion parameters for protein quantification. Anal Biochem 2009;393(1):48–55. [DOI] [PubMed] [Google Scholar]
- [32].Woolfitt AR, Solano MI, Williams TL, Pirkle JL, Barr JR. Amino acid analysis of peptides using isobaric-tagged isotope dilution LC-MS/MS. Anal Chem 2009;81 (10):3979–85. [DOI] [PubMed] [Google Scholar]
- [33].Xiong X et al. Receptor binding by an H7N9 influenza virus from humans. Nature 2013;499(7459):496–9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


