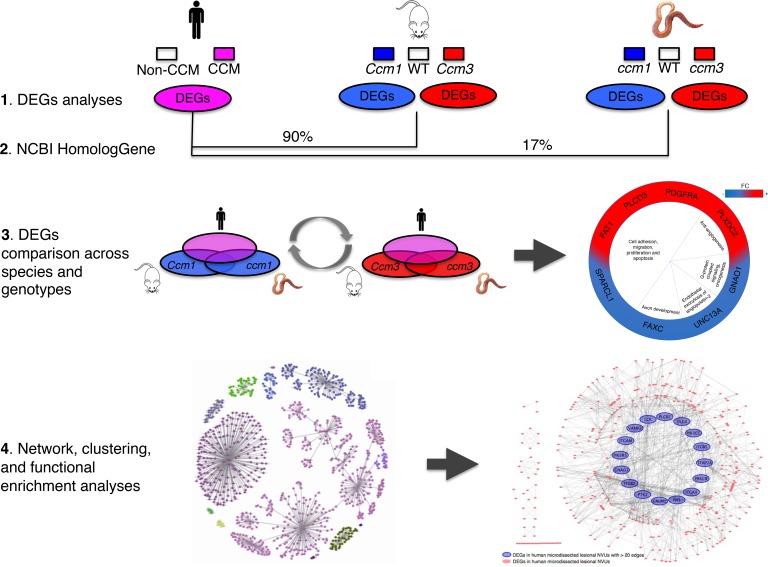
Abstract
The purpose of this study was to determine important genes, functions, and networks contributing to the pathobiology of cerebral cavernous malformation (CCM) from transcriptomic analyses across 3 species and 2 disease genotypes. Sequencing of RNA from laser microdissected neurovascular units of 5 human surgically resected CCM lesions, mouse brain microvascular endothelial cells, Caenorhabditis elegans with induced Ccm gene loss, and their respective controls provided differentially expressed genes (DEGs). DEGs from mouse and C. elegans were annotated into human homologous genes. Cross-comparisons of DEGs between species and genotypes, as well as network and gene ontology (GO) enrichment analyses, were performed. Among hundreds of DEGs identified in each model, common genes and 1 GO term (GO:0051656, establishment of organelle localization) were commonly identified across the different species and genotypes. In addition, 24 GO functions were present in 4 of 5 models and were related to cell-to-cell adhesion, neutrophil-mediated immunity, ion transmembrane transporter activity, and responses to oxidative stress. We have provided a comprehensive transcriptome library of CCM disease across species and for the first time to our knowledge in Ccm1/Krit1 versus Ccm3/Pdcd10 genotypes. We have provided examples of how results can be used in hypothesis generation or mechanistic confirmatory studies.
Keywords: Neuroscience, Vascular Biology
Keywords: Molecular genetics, Transcription, endothelial cells
A cross-comparison of the transcriptomes of human and 4 animal models identifies important genes, functions, and networks contributing to the pathobiology of cerebral cavernous malformation.
Introduction
Cerebral cavernous malformations (CCMs) are hemorrhagic vascular brain lesions predisposing 0.5% of the worldwide population to a lifetime risk of intracerebral hemorrhage and seizures. CCMs are clusters of caverns lined with endothelium lacking mature blood vessel architecture (1). Two forms of the disease exist, one of which is a sporadic form accounting for 70% of cases manifesting a solitary lesion (2). The familial form is inherited in an autosomal dominant pattern with mutations in 1 of 3 documented genes (CCM1/KRIT1, CCM2/MGC4607, or CCM3/PDCD10) causing multifocal lesions to develop stochastically throughout the brain during the patient’s life (2). Familial and sporadic CCMs are histologically indistinguishable and harbor somatic mutations in the same genes, suggesting a common pathological mechanism (3). However, cases with CCM3/PDCD10 mutations exhibit exceptional disease aggressiveness, the cause of which remains speculative (2).
CCM genes encode proteins responsible for maintaining integrity of endothelial permeability by inhibiting RhoA-associated kinase (ROCK) activation (4). Greater ROCK activity leads to dysregulated endothelial junctional proteins resulting in increased blood vessel permeability (4). In addition, in vivo and in vitro mechanistic studies have reported a complex interplay of angiogenic and inflammatory processes in the genesis and maturation of CCM lesions, including signaling aberrations related to endothelial MEKK3-KLF2/4 signaling (5, 6), angiogenic activity (6–8), antigen-mediated immune response (9), and endothelial-mesenchymal transition (5).
Previous studies on the differential transcriptome of resected CCM lesions were performed on the whole lesional tissue, which includes many cell types (10, 11). However, experimental evidence supports that CCM physiopathology arises primarily in endothelium (5, 12). In addition, it is clear that CCM3 engages distinct signaling pathways from CCM1/CCM2, yet patients with mutations in any of the 3 genes develop the same lesions, suggesting that they likely converge on similar biological processes. Therefore, it is important to specifically study the transcriptomic differences between in vivo and in vitro lesional endothelial cells (ECs) and between different genotypes. Caenorhabditis elegans has a well-characterized genome but lacks a circulatory system, yet it manifests a discrete phenotype with Ccm homologous gene loss (13, 14). Recently, Lant and coworkers conducted a genome-wide RNA interference screen that uncovered several genes with human homologs that induced a CCM-like phenotype in C. elegans (13). The remarkable conservation of CCM gene function from nematode to human prompted us to ask if we could construct a refined network by systematically analyzing DEGs across species and genotypes.
Herein, we leveraged models and data from different laboratories, to cross-compare the differentially expressed genes (DEGs), their networks, their pathways, and their functions in human neurovascular units (NVUs) from surgically resected lesions, and with the induced loss of 2 disease genes (Ccm1/Krit1 and Ccm3/Pdcd10) in cultured mouse brain microvascular endothelial cells (BMECs) and in C. elegans. We hypothesized that common DEGs across species may reflect conserved and important pathological processes relevant in the human disease. We also postulated that DEGs between the 2 different genotypes in mouse BMEC and C. elegans models would reflect differences in disease manifestations and aggressiveness.
Results
DEGs and gene ontologies in lesional human NVUs, mouse BMECs, and C. elegans.
After sequencing, 915 DEGs (fold change [FC] ≥ 1.2; P < 0.05, FDR corrected) were identified in human microdissected lesional NVUs compared with normal control capillaries (Supplemental Figure 2A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.126167DS1). In addition, the individual expression profile for each patient (Supplemental Figure 3) suggests a downregulation of at least 1 of the CCM genes (CCM1, CCM2, and CCM3) in the microdissected lesional NVUs.
In mouse BMECs, 1,932 and 524 DEGs (FC ≥ 1.2; P < 0.05, FDR corrected) were observed in Ccm1/Krit1ECKO and Ccm3/Pdcd10ECKO, respectively (Supplemental Figure 2, B and D). In C. elegans ccm1/kri-1 and ccm3/pdcd10 mutants, 1,643 and 1,581 DEGs (FC ≥ 1.2; P < 0.05, FDR corrected) were identified, respectively (Supplemental Figure 2, C and E).
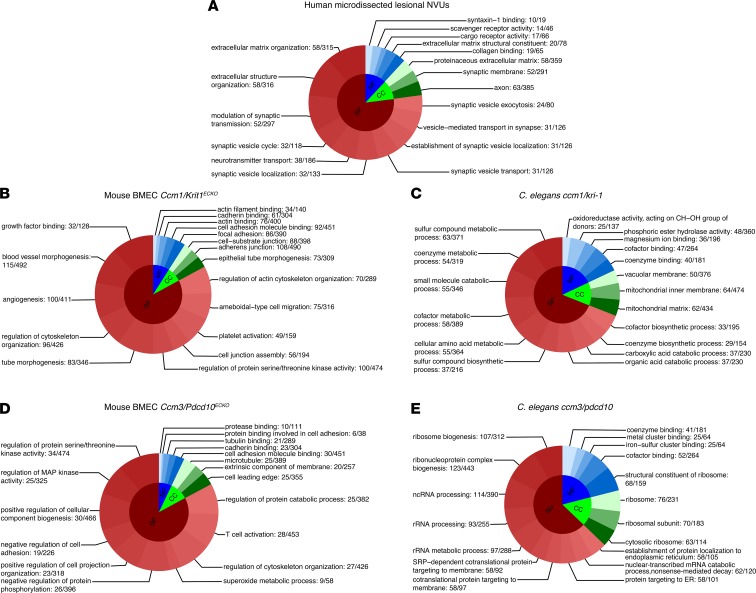
Further analyses showed that 1,307 gene ontology (GO) terms (P < 0.1, FDR corrected) were overrepresented in human lesional NVUs (Figure 1A), 1,565 GO terms in Ccm1/Krit1ECKO BMECs, 283 GO terms in Ccm3/Pdcd10ECKO BMECs (Figure 1, B and D), 519 GO terms in C. elegans ccm1/kri-1 and 685 GO terms in ccm3/pdcd10 (Figure 1, C and E). The GO analyses in the human NVUs reflected neuronal as well vascular processes. The GO analyses across the different species and genotypes revealed functions related to angiogenesis, inflammation, immune cell activity, junctional adhesion, intracellular organelle localization, extracellular matrix and structure remodeling, serine/threonine kinase activity, and apoptosis. One GO term (GO:0051656, establishment of organelle localization) was commonly identified across the different species and genotypes. In addition, 24 GO functions were present in 4 out of 5 models and were related to cell-to-cell adhesion, neutrophil-mediated immunity, ion transmembrane transporter activity, and responses to oxidative stress (Supplemental Figure 4).
Figure 1. Top 18 GO functions of DEGs for each species by genotype.
The gene set enrichment analyses (P < 0.1 for all, FDR corrected) identified (A) 1,307 GO terms in human lesional NVUs, (B) 1,565 in Ccm1/Krit1ECKO BMECs, (C) 519 in C. elegans with a ccm1/kri-1 mutation, (D) 283 in Ccm3/Pdcd10ECKO BMECs, and (E) 685 in C. elegans ccm3/pdcd10. The DEGs were defined as FC greater than or equal to 1.2, with P less than 0.05 (FDR corrected), for all the models. For display purposes, only the top GOs are presented in each category (BP, MF, and CC) by their respective proportion. BP, biological processes; MF, molecular functions; CC, cellular components; ncRNA, noncoding RNA; and SRP, signal recognition particle. Gene ratio is reported at the end of each respective GO.
Network and GO analysis of DEGs in human CCM lesions.
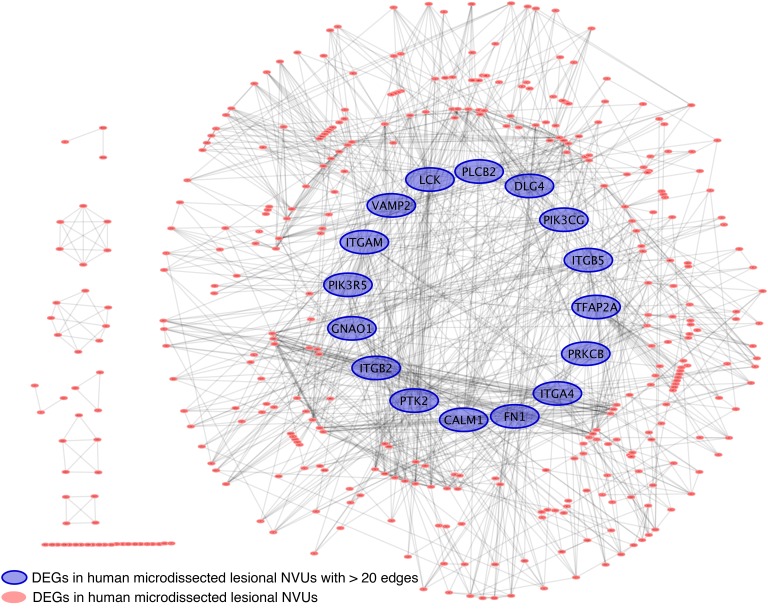
Sixteen genes were highly connected (with at least 20 edges) in the gene network defined for the 915 DEGs identified in the human microdissected lesional NVUs (Figure 2). Among these 16 genes, 4 were related to the expression of integrin subunits, ITGA4, ITGB5, ITGB2, and ITGAM, and 1 to the expression of fibronectin (FN1). Three genes, namely PLCB2, PIK3CG, and PIK3R5, belong to the phosphatidylinositol pathway. PTK2 was also part of this highly connected network and encodes for a focal adhesion kinase (FAK) that is directly related to cell adhesion (15). Furthermore, PRKCB, which belongs to a protein kinase C family; VAMP2, which produces vesicle-associated proteins (16); GNAO1, which encodes an α subunit of the Go protein abundant in the CNS and promotes oncogenesis and TLR signaling (17, 18); DLG4, which encodes postsynaptic density protein 95 and belongs to a membrane-associated guanylate kinase family; and lymphocyte-specific protein kinase were all also identified in this highly connected human network. Finally, 2 other genes were also present in this highly connected network, including CALM1, which encodes for calmodulin 1 with an established role in cellular growth and neurotransmitter production and release (19), and TFAP2A, which codes for the transcription factor AP-2α.
Figure 2. Focused functional interaction network of DEGs in human lesional NVUs.
A total of 915 DEGs (FC ≥ 1.2; P < 0.05, FDR corrected) were identified in human lesional NVUs and used to generate a gene network. Sixteen DEGs were identified with more than 20 edges. Among these highly connected DEGs, the GNAO1 was also conserved in BMECs of Ccm1/Krit1ECKO and C. elegans ccm1/kri-1.
We identified 95 enriched GO terms (P < 0.01, FDR corrected) using the 16 genes with more than 20 edges. The results showed defined enriched functions related to extracellular matrix processes, cellular adhesion, integrin processes, blood vessel morphogenesis, the phosphatidylinositol pathway, the TLR4 signaling pathway, vesicle transportation, and metal ion transportation (Supplemental Table 4).
Common DEGs and gene networks among human NVUs, mouse BMECs, and C.
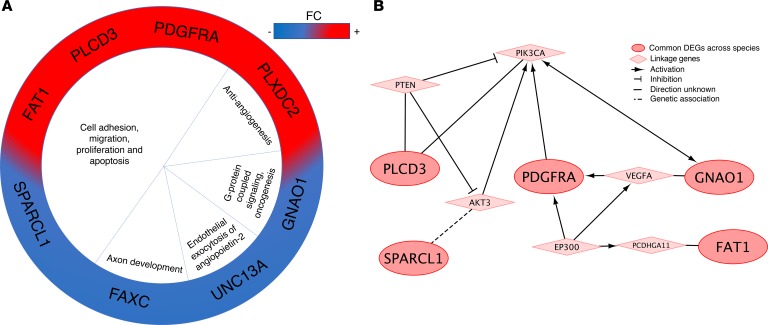
elegans. SPARCL1, PDGRFA, FAXC, and UNC13A (FC ≥ 1.2; P < 0.05, FDR corrected for all) were common between human lesional NVUs and Ccm1/Krit1 models of both the mouse BMECs and C. elegans. We then considered loosening the P value threshold for C. elegans (P < 0.25, FDR corrected) to correct unbalanced restriction in analyses because only 17% of C. elegans genes have corresponding human gene homologs. We then identified 4 additional common DEGs, namely FAT1, PLXDC2, PLCD3, and GNAO1, in both Ccm1/Krit1 models, mouse BMECs, and C. elegans, as well as human lesional NVUs (Supplemental Figure 5). FAT1 was also found to be commonly differentially expressed in human NVUs and in Ccm3/Pdcd10 mouse BMECs and Ccm3/Pdcd10 C. elegans (Figure 3, A and B). PLCD3, FAT1, GNAO1, PDGFRA, and SPARCL1 were identified in the Reactome FIViz database (Figure 3B).
Figure 3. Common genes conserved across species.
(A) Eight DEGs were commonly identified in human lesional NVUs, both Ccm1/Krit1ECKO and Ccm3/Pdcd10ECKO mouse BMECs, and both ccm1/kri-1 and ccm3/pdcd10 C. elegans models (FC ≥ 1.2; P < 0.05, FDR corrected for human and both genotypes of mouse BMECs. FC ≥ 1.2; P < 0.25, FDR corrected for both genotypes of C. elegans). Functions of the DEGs were related to antiangiogenesis, cell growth, vesicle exocytosis, axon development, G protein–coupled signaling and oncogenesis. Red indicates an increased FC and blue a decreased FC. (B) The focused protein functional interaction network analysis identified 5 of the 8 common DEGs: PLCD3, FAT1, GNAO1, PDGFRA, and PLCD3. PLCD3 is linked with PIK3CA directly but also via PTEN that inhibits PIK3CA. SPARCL1 is associated with AKT3 that activates PIK3CA. GNAO1 is linked to VEGF that activates PDGFRA that further activates PIK3CA. Interestingly, FAT1 is linked to PCDHGA11 that receives activation from EP300. EP300 further activates VEGF and PDGFRA, creating bidirectional activation for PDGFRA that in turn activates PIK3CA.
Further analyses between mouse BMEC models, regardless of the genotype, and human microdissected lesional NVUs identified 36 common DEGs (Supplemental Figure 6 and Supplemental Table 5). In addition, 10 and 109 genes were commonly differently expressed between human microdissected lesional NVUs and Ccm3/Pdcd10ECKO and Ccm1/Krit1ECKO, respectively.
DEGs, enriched GOs, and gene networks of Ccm1/Krit1 and Ccm3/Pdcd10 genotypes.
We finally assessed the unique DEGs of each genotype for Ccm1/Krit1 and Ccm3/Pdcd10 in mouse BMEC and C. elegans models (FC ≥ 1.2; P < 0.05, FDR corrected). We identified 1,541 DEGs only in Ccm1/Krit1ECKO BMECs, and 195 in Ccm3/Pdcd10ECKO BMECs, without comparison to other models. In C. elegans models alone, 676 DEGs were found only in ccm1/krit-1 and 656 only in ccm3/pdcd10. Finally, 71 DEGs were commonly identified only in Ccm1/Krit1-mutant models and 11 only in Ccm3/Pdcd10 using comparisons across the murine and worm models (Supplemental Tables 6 and 7). Notably, we observed that both genotypes showed differential expression of their respective knockout genes, Ccm1/Krit1 and Ccm3/Pdcd10 (Supplemental Tables 2 and 8).
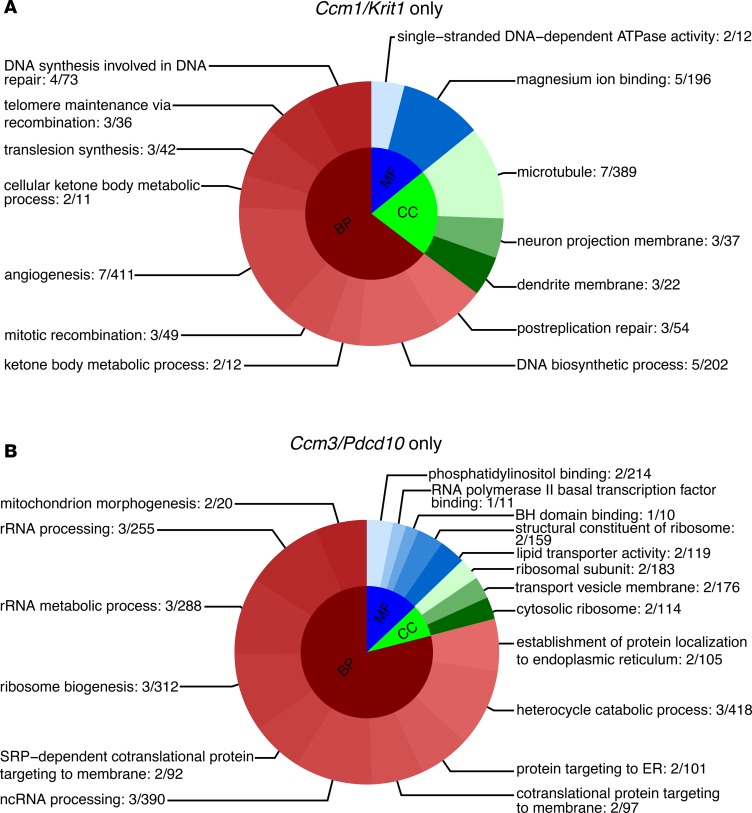
Further analyses identified 14 and 247 (P < 0.1, FDR corrected) enriched GO terms in Ccm1/Krit1 and Ccm3/Pdcd10 genotypes, respectively (Supplemental Tables 9 and 10). Ccm1/Krit1 genotype GO functions were primarily related to DNA repair, angiogenesis, microtubule function, and magnesium ion binding (Figure 4A). Ccm3/Pdcd10 genotype showed fundamental functions for rRNA processing, ribosome biogenesis and its structural components, protein targeting to endoplasmic reticulum, protein intracellular targeting, and vesicle transportation to a cell membrane (Figure 4B).
Figure 4. Top GO functions unique to either Ccm1/Krit1 or Ccm3/Pdcd10 genotypes.
(A) A total of 71 DEGs were identified only in the Ccm1/Krit1 genotype in both mouse BMECs and C. elegans. The gene set enrichment analyses showed 14 enriched GO terms (P < 0.1, FDR corrected) in Ccm1/Krit1 related to DNA repair, angiogenesis, microtubule functions, and magnesium ion binding. (B) Eleven DEGs were identified only in the Ccm3/Pdcd10 genotype and were associated with 247 GO functions. The top 18 GO terms were related to fundamental functions for rRNA processing, ribosome biogenesis and structural constituents, protein targeting to endoplasmic reticulum, protein intracellular targeting, and vesicle transportation to a cell membrane. The DEGs were defined as FC ≥ 1.2 and P < 0.05 (FDR corrected). BH, Bcl-2 homology. Gene ratio is reported at the end of each respective GO.
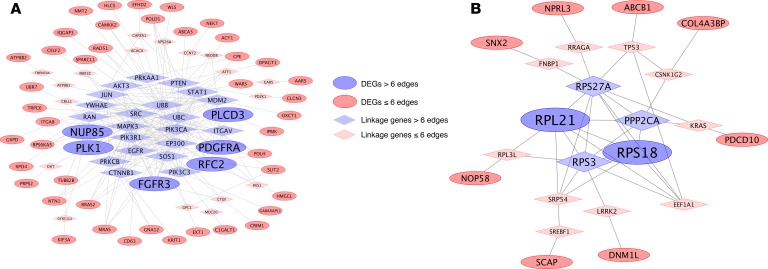
Gene network analyses were performed separately for Ccm1/Krit1 and Ccm3/Pdcd10 (Figure 5). In Ccm1/Krit1, 6 genes with more than 6 edges were identified: FGFR3, PDGFRA, PLK1, RFC2, NUP85, and PLCD3. In the Ccm3/Pdcd10 model, only RPL21 and RPS18 had more than 6 edges.
Figure 5. Functional interaction network of DEGs conserved in both mouse BMECs and C. elegans unique to either Ccm1/Krit1 or Ccm3/Pdcd10.
(A) The functional interaction network analysis of the 71 common DEGs (FC ≥ 1.2; P < 0.05, FDR corrected) identified only in Ccm1/Krit1 models showed that FGFR3, PDGFRA, PLK1, NUP85, and PLCD3 had more than 6 edges. (B) The functional interaction network analysis of the 11 common DEGs (FC ≥ 1.2; P < 0.05, FDR corrected) identified only in the Ccm3/Pdcd10 genotype revealed that RPL21 and RPS18 were the genes with the most edges (more than 6).
Discussion
The loss of all 3 known CCM genes in endothelium is embryonically lethal in mammals (12, 20), hence highlighting the fundamental biological role of these genes and associated networks and pathways. We here cross-compared the transcriptomic profiles of human resected lesional NVUs, and 2 genotypes, Ccm1/Krit1 and Ccm3/Pdcd10, in both mouse BMECs and C. elegans. The 3 respective models reflected differential transcriptomes of human lesional endothelium and its adjacent milieu and the impact of Ccm gene loss in cultured BMECs as well as in a whole organism. We aimed to identify conserved genes, GO functions, and related networks across these models and those associated commonly or uniquely in Ccm1/Krit1 and Ccm3/Pdcd10 genotypes. Some of the results validate previously published mechanistic links, while others shed new light on disease pathobiology. The data presented herein will be useful to the research community in querying and validating potential mechanistic, biomarker, and therapeutic targets.
The transcriptome profile of the human microdissected lesional NVUs.
The transcriptome profile of the human lesional microdissected NVUs showed 16 highly connected (more than 20 edges) genes. From those genes, GNAO1 was also identified in 2 models: mouse BMEC Ccm1/Krit1ECKO and C. elegans ccm1/kri-1. GNAO1 encodes an α subunit of the Go (Gαo) protein that is abundant in the CNS (17). A mutation of GNAO1 has been shown to promote cellular transformation and oncogenic signaling (18). Similarly, a mutation in GNAQ, a Gαq protein–coding gene, has been reported to cause constitutive subunit activation by attenuating GTPase activity of the α subunit (20). A greater number of ECs with mutations on GNAQ has been shown to be associated with neurovascular disorders, including capillary malformations, Sturge-Weber syndrome, as well as port-wine stains (21, 22). GNAO1 has also been associated with TLR2 receptor downstream signaling (17). TLR2 receptor has been found to be the key molecular sensor for oxidative stress that links oxidative stress, inflammation, innate immunity, and angiogenesis (23). In addition, maturation of CCM lesions has been recently associated with TLR4, signaling which activates the same pathways as TLR2, although the ligands may be different (24).
PIK3CG, PIK3R5, and PLCB2 were also identified to be highly connected in the human DEG network. Phosphoinositide 3-kinase (PI3K) proteins were shown to be involved in vascular disease (20). They are activated through G protein–coupled receptors and tyrosine kinase receptors. Particularly, PIK3CG is a catalytic subunit of class I PI3Ks that produces phosphatidylinositol (3,4)-biphosphate (PtdIns[3,4]P2), phosphatidylinositol(3,4,5)-trisphosphate (PIP3), as well as phosphatidylinositol 3-phosphate (PI3P). PIK3R5 protein belongs to a group of class I PI3Ks functioning as a regulatory subunit for PI3Ks. Phospholipase C β2 (PLCB2) is also activated by G protein–coupled receptor signaling, and PLCB2 protein catalyzes PIP2 breakdown to inositol triphosphate (IP3) and diacylglycerol. Previous work has shown that venous and capillary malformations are associated with mutations of the PIK3CA (20). This gene belongs to the same class of PI3Ks as the PIK3CG gene that is identified to be highly connected in the network. PIK3CA acts as a catalytic subunit for PI3Ks, similar to PIK3CG, inducing phosphorylation of downstream signaling of phosphoinositides. This pathway activates responses related to growth, proliferation, cell survival, and angiogenesis. There is also strong evidence from mouse models supporting a role of the PI3K subunit in the regulation of angiogenesis and activation of RhoA GTPase that contribute to CCM lesion progression (12, 20).
Four genes coding integrin subunits ITGA4, ITGB5, ITGB2, and ITGAM were also highly connected in the human network. Previous studies have reported the importance of integrins, cadherins, and tight junctions in the maintenance of vascular integrity (1). The dysregulation of CCM proteins affects integrin β1-KLF2 activity, which mediates angiogenesis, suggesting a role of integrins as downstream signaling proteins (6). Previous studies in cultured ECs suggest that vascular endothelial growth factor A (VEGF-A) stimulates phosphorylation and recruitment of focal adhesions (15). Recently our group has also shown that VEGF-A levels are altered in plasma of CCM patients (7). In addition, a recent article demonstrated a circuitous role for IL-7, CXCR4, and FAK in B cell development (25). Interestingly, B cell depletion treatments have been shown to blunt lesion burden and hemorrhage in murine models of CCM disease (9). The present study showed upregulation of the IL7R and CXCR4 genes, as well as downregulation and high connectivity of the PTK2 gene, which codes for FAK and is also connected to ITGB5 in our human network. Further mechanistic studies are needed to define the complex role of junctional proteins in CCM disease in further detail.
Fibronectin (FN1) was also a highly connected gene in the human network analysis. Fibronectin has been reported to be involved during the early stages of the coagulation process when endothelium is injured and becomes an antithrombotic agent during the latest stages (26). In addition, our previous studies in vivo have suggested that thrombospondin-1, a natural antiangiogenic protein related to the blood coagulation cascade, affects CCM lesion progression (8). In light of these results, the role of fibronectin in CCM lesion physiopathogenesis should be studied in greater detail.
The common transcriptome profile among human microdissected lesional NVUs, BMECs, and C. elegans.
The cross-comparison of the transcriptomes between the human lesional NVUs and 4 different models identified the FAT1 gene was conserved across the species and genotypes. FAT1 is a protocadherin associated with the hippo pathway and has been shown to regulate cell proliferation, apoptosis, cell polarization, and cell-to-cell adhesion (27). In addition, FAT1 has modulatory activity to β-catenin in vascular smooth muscle cells as it also stabilizes the CCM complex in ECs in Ccm1/Krit1 disease models (27, 28). However, its role in CCM physiopathogenesis remains unclear.
Seven additional genes (UNC13A, SPARCL1, PDGFRA, GNAO1, PLCD3, FAXC, and PLXDC2) were commonly identified in human lesional NVUs and Ccm1/Krit1 genotypes of both BMECs and C. elegans. In normal ECs from the brain of Ccm3/Pdcd10 mice, an inhibition of Unc13b, a paralog of Unc13a, and Vamp3 through Ccm3 protein is associated with a decrease in angiopoietin-2 exocytosis (16). Thus, a loss of the Ccm3/Pdcd10 gene increases exocytosis of angiopoietin-2 (16). These physiopathological processes are associated with a wide range of EC dysfunction promoting CCM disease progression (16). In addition, Vamp2, an important paralog of Vamp3, was identified to be highly connected in the human lesional NVUs network. These results suggest a role of vesicle transportation in the physiopathology of CCM disease, which is supported by observations in C. elegans showing that ccm3 promotes Cdc42 activity and endocytic recycling (14).
The 5 DEGs (SPARCL1, PLCD3, PDGFRA, PLXDC2, and FAXC) that were common among human lesional NVUs and the Ccm1/Krit1 genotypes of both BMECs and C. elegans do not currently have postulated functions in CCM disease. However, SPARCL1 and PLCD3 have been associated with cellular proliferation, migration, and cancer (29, 30). PLCD3 belongs to a phospholipase C family and may be linked to the phosphatidylinositol signaling pathway (30). PDGFRA is a growth factor also related to cellular growth, differentiation, and tumor vasculogenesis (31). Interestingly, PLXDC2 relays antiangiogenic signaling of pigment epithelium-derived factor (PEDF, SERPINF1) (32). Hypoxic conditions have been shown to downregulate antiangiogenetic activity of PEDF (32).
Last, FAXC is mainly expressed in axon bundles and related to axon development (33). Its role in CCM remains unclear but may reflect a role of the whole NVU and neuronal environment in CCM physiopathogenesis. Interestingly, deletion of ccm3/pdcd10 in glia can induce lesion formation in the neural vasculature of mice (34).
These 8 genes provide potentially novel information of a conserved part of CCM physiopathogenesis and highlight commonalities between disease models and human lesions, providing strong candidate genes for further mechanistic studies. Finally, the common GO terms across the species reflected current knowledge on CCM physiopathogenesis.
Unique transcriptome profiles of Ccm1 versus Ccm3.
The different genotypes in CCM disease have important implications regarding clinical outcomes. Patients with the CCM3 genotype show earlier onset, higher lesion burden, and an increased risk of bleeding compared with patients harboring the CCM1 mutation (2). These physiopathological and phenotypic differences observed between CCM1 and CCM3 genotypes may be explained at the transcriptomic level.
In Ccm1/Krit1, 6 DEGs with more than 6 edges, namely FGFR3, PDGFRA, RFC2, PLK1, NUP85, and PLCD3, were identified. FGFR3 relays cell growth and angiogenesis signals and is broadly associated with tumorigenesis (31). PDGFRA is a growth factor receptor for platelet-derived growth factor that supports cellular growth, survival, and proliferation (31). RFC2 also promotes cell survival by regulating cell cycle progression from G1 to S phase (35). Interestingly, PLK1 takes part in the cell cycle, controlling progression from G2 to M phase, and is considered a protooncogene (36). NUP85 was also only identified in Ccm1/Krit1. It is a subunit of the Nup107-160 complex responsible for formation of normal nuclear pores and is required for mitotic spindle assembly during mitosis (37). A dysregulation of this complex causes deficits in transportation of macromolecules, such as RNA, across the nucleus and cytoplasm (37).
The GO analyses of unique genes for Ccm1/Krit1 were related to DNA repair, angiogenesis, microtubule functions, and magnesium ion binding. Mitotic spindle formation and macromolecule transportation across the nucleus and cytoplasm are important steps in cell cycle control (36). Our results suggest that a dysregulation of the cell cycle control may be an important process in the Ccm1/Krit1 genotype that may explain the differences between the 2 phenotypes.
In the Ccm3/Pdcd10 model, RPL21 and RPS18 were highly connected (more than 6 edges) and were also connected with KRAS, which linked Ccm3/Pdcd10 to the rest of the network. RPL21 encodes a protein component for ribosome 60S subunit, and RPS18 encodes a component for ribosome 40S subunit (38). In addition, recent evidence shows that RPL29, a component of ribosome 60S subunit, is involved in angiogenesis processes (39). A dysfunction of RPL29 affects the integrins of ECs (39). These results may suggest a role of ribosomal dysfunction in CCM physiopathogenesis.
Furthermore, SNX2, ALDH4A1, COL4A3BP, and DNM1L observed only in Ccm3/Pdcd10 are related to apoptosis, ROS signaling, and intracellular endosome trafficking (40–43). SNX2 is linked to intracellular trafficking–sensing membrane curvature and interacts with phosphoinositols PI3P and PtdIns(3,5)P2 in earlier stages of the formation of the endosomes (41). ALDH4A1 codes for a mitochondrial NAD-dependent hydrogenase involved in proline metabolism that maintains important balance of mitochondrial ROS (40). COL4A3BP regulates ceramide transportation from endoplasmic reticulum (42). A dysregulation of this gene causes ceramide accumulation in mitochondria, inducing Bax-dependent apoptosis (42). Interestingly, C. elegans with ccm1/kri-1 mutation are resistant to stress-induced apoptosis (44), suggesting a role for cell death in the pathobiology of CCM. Finally, DNM1L is involved in numerous functions in mitochondria. Its dysregulation causes increased production of ROS, mutations of DNA regulation, as well as dysfunctions in mitochondrial fission and fusion (43).
The enriched GO functions in Ccm3/Pdcd10-specific genes were related to rRNA processing, ribosome biogenesis and structural constituents, protein targeting to endoplasmic reticulum, protein intracellular targeting, and vesicle transportation to a cell membrane. Defects in vesicle transportation and protein intracellular targeting are well-established features in the Ccm3 genotype (13, 14, 16, 45). However, suggested GO functions related to rRNA processing and functioning are not yet understood and may reflect defects in protein construction.
The genes dysregulated in Ccm1/Krit1 genotype are associated with functions of cell cycle control, growth, and proliferation. In contrast, in Ccm3/Pdcd10 the dysregulated genes are associated with apoptosis, ROS metabolism, and intracellular vesicle trafficking. Ribosomal dysfunction in human disease is an interesting concept that remains to be established with the appropriate models. These results provide conserved candidate genes for interrogation that are unique for either genotype in mouse BMECs and C. elegans CCM disease models.
Use of transcriptome library to support molecular mechanisms in CCM disease.
Our transcriptome library provides a database to query DEGs in human lesional NVUs and 2 widely used genetically engineered animal models of CCM disease. Transcriptome data of human lesional NVUs have already been used to support mechanistic findings in animal models, allowing the identification of potential biomarkers to be used in CCM patients (8). Indeed, we have recently reported that loss of brain endothelial thrombospondin-1 (TSP1) in murine Ccm1/Krit1ECKO has been shown to contribute to CCM pathogenesis (8), which is consistent with the downregulation of TSP1 in human lesional NVUs.
Limitations.
It is important to acknowledge the limitations of our study. First, we defined a common FC threshold (≥1.2) to identify the DEGs for all the different models studied. A lower FC threshold applied to RNA-Seq results can increase the false positive discovery. We used an arbitrary FC threshold to allow the comparisons between different experimental designs. In addition, low expressed genes are usually hard to detect. However, we performed a rigorous FDR correction, rather than using the nominal P value, thus decreasing the possibility of false positive discovery in our results. In addition, different databases can be used during the different steps of the analyses and may give slightly different results. Here we used the well-established open-source GO database based on hypergeometric test and Cytoscape with ReactomeFIViz module.
For RNA-Seq analysis, the sequencing depth is another concern. Even though different sequencing depth has been normalized in DEG identification analysis, sequencing depth or library size difference between different species because of samples’ RNA extraction difficulty might be severer in comparison with a study within the same species. The DEG analyses are conducted based on the statistical model. Statistical analysis by itself has its own statistical power to identify DEGs in a small sample size. In addition, when comparing different species’ identified DEGs, the homologous genes between mouse BMECs and C. elegans and humans relies on the homologous genes database. Thus definition of homologous genes between species also needs to be understood. Furthermore, gene comparison of DEGs between species were based solely on the homology of the genes. This explains how FCs of some common genes are observed in the opposite direction (Supplemental Tables 6 and 7). The phenomenon may be due to fundamental differences in models used. Mouse BMECs are specifically ECs, while C. elegans is a whole organism with multiple cell types.
We acknowledge the technical limitations of laser capture microdissection to isolate pure capillary ECs from lesions. Our differential transcriptome in humans may be composed of cell material from the extremely close vicinity of ECs. However, the DEGs observed in the lesional human NVUs seem to reflect an EC expression profile strongly (Supplemental Results). In addition, the comparison between the DEGs in human lesional NVUs, mouse BMECs, and C. elegans highlights dysregulated genes specifically expressed in ECs of CCM lesions.
The inclusion of C. elegans results in this study adds a cross-species perspective of analyzing transcriptomic data and in a whole organism. But because only 17% of C. elegans genes have currently corresponding human gene homologs, the ability to perform shared DEG analyses between mammals and C. elegans is limited. In addition, C. elegans does not have ECs. Although C. elegans with a ccm3/pdcd10 mutation exhibits phenotypes in tube development within stress fiber formation and defects in cell-to-cell contacts, there are other phenotypes apparently unrelated to CCM disease, and the ccm1/kri-1–mutated organisms do not seem to exhibit a CCM-like phenotype. Thus, addition of C. elegans data may exclude some key genes involved in CCM pathogenesis while it may narrow down the number of key genes.
We used human lesional microdissected NVUs from mostly sporadic lesions as opposed to familial CCMs with specific germline mutations. Although all CCM lesions likely share common pathway-related signaling aberrations (20), we could not distinguish how the different genotypes might contribute to the human lesional transcriptome results. Although the human lesion transcriptomic libraries allow exploratory comparisons with DEGs in different genotypes and organisms, we would be cautious about drawing further conclusions from individual lesion transcriptomes (Supplemental Figure 3). We also did not use a model of Ccm2 disease for this study. In addition, some studies suggested that the lesional in vivo microenvironment may influence the CCM physiopathogenesis, which is consistent with observations implicating cell-nonautonomous mechanisms for CCM proteins (34, 44). Furthermore, phenotypic differences between engineered animal models suggest that these models recapitulate different features of the development and maturation of CCM lesions (1). We plan to address these gaps by comparing transcriptomes of in vivo microdissected NVUs from murine lesions in comparison with those derived herein by in vitro BMEC gene loss. And we plan to analyze the transcriptome from NVUs of human and murine lesions with various genotypes and from human lesional NVUs with and without recent symptomatic hemorrhage, a clinically relevant disease feature. Finally, our study did not address the effect of loss of CCM gene function in epithelium and other cells (46), which may play secondary roles in disease development, such as influencing gut permeability, which in turn may drive disease development (24). Certainly, our study does not provide direct confirmation of the mechanistic primacy of various DEGs or networks.
Conclusions.
We provide a unique transcriptome library of CCM disease in human lesional NVUs and in 2 genotypes, Ccm1/Krit1 and Ccm3/Pdcd10, of 2 animal models. Our results suggest important conserved genes with a role in G protein–coupled signaling associated with the phosphoinositol pathway, integrin-related signaling, and vesicle transportation. For the first time to our knowledge, we also provide fundamental transcriptomic differences between Ccm1/Krit1 and Ccm3/Pdcd10 genotypes in 2 disease models. Our results validate previously postulated mechanisms and can be used to generate novel hypotheses to gain a greater understanding of CCM disease, its biomarkers, and its therapeutic targeting.
Methods
Supplemental Methods are available online with this article.
Laser-captured NVUs of human surgically resected CCM lesions and normal brain capillaries.
Five surgically resected human CCM lesions (Supplemental Table 1 and Supplemental Figure 7) were embedded in optimal cutting temperature, snap-frozen, and stored at –80°C. Normal brain tissue from 3 subjects free of neurological disease was acquired during autopsy, fixed in formalin, and embedded in paraffin blocks. We mounted 5-μm tissue sections on Leica glass slides (Leica Biosystems Inc) and stained them with HistoGene (Applied Biosystems) for frozen tissue and with Paradise stain (Applied Biosystems) for paraffin-embedded tissue, according to the manufacturers’ protocols. The endothelial layer, including adjacent luminal and extraluminal elements (NVUs) from CCMs and normal brain capillaries, were then collected using laser capture microdissection and stored at –80°C (supplemental material).
RNA was extracted using an RNA isolation kit (RNeasy Micro Kit, Qiagen). cDNA libraries were generated using commercial low-input strand-specific RNA-Seq kits (Clontech) and sequenced on the Illumina HiSeq 4000 platform using single-end 50-bp reads.
Ccm1/Krit1ECKO and Ccm3/Pdcd10ECKO BMEC and corresponding wild-type controls.
Adult male and female 2- to 4-month-old mice were used for isolation and purification of primary BMECs. All experiments were performed using the same C57BL/6 background. We generated 3 endothelial-specific conditional Ccm1/Krit1–null mice (Ccm1/Krit1ECKO) using a Pdgfb promoter–driven, tamoxifen-inducible Cre recombinase, Pdgfb-iCreERT2; Krit1fl/fl (8). We also generated 3 endothelial-specific conditional Ccm3/Pdcd10–null mice (Ccm3/Pdcd10ECKO) using a Pdgfb promoter–driven tamoxifen-inducible Cre recombinase, Pdgfb-iCreERT2; Pdcd10fl/fl (8). BMECs were isolated and purified following a previously described protocol (8). Following purification, BMECs were treated using 5 μM 4-hydroxytamoxifen for 48 hours to induce allelic loss in ECs harboring the Cre recombinase–floxed allelic system (8). We chose to add tamoxifen to cultured cells, to insure full gene knockout, rather than speculate about the efficiency of gene deletion in vivo. After 48 hours, the medium was replaced with medium lacking 4-hydroxytamoxifen, and BMECs were harvested in culture after 72 hours for Ccm1/Krit1ECKO and 168 hours for Ccm3/Pdcd10ECKO. We performed genome-wide RNA-Seq to detect a decrease of greater than 90% of Ccm1/Krit1 or Ccm3/Pdcd10 mRNA after the initial 4-hydroxytamoxifen treatment. After 5 days, a decrease of greater than 90% of Ccm1/Krit1 mRNA was observed in Ccm1/Krit1ECKO BMECs but not for Ccm3/Pdcd10 mRNA in Ccm3/Pdcd10ECKO BMECs. The decrease of greater than 90% was reached after 9 days for Ccm3/Pdcd10 mRNA in Ccm3/Pdcd10ECKO BMECs. For each genotype (Ccm1/Krit1fl/fl and Ccm3/Pdcd10fl/fl), 3 control mice were similarly treated with 5 μM 4-hydroxytamoxifen.
RNA was extracted using the TRIzol protocol (Thermo Fisher Scientific). TruSeq Stranded mRNA Sample Prep Kit (Illumina) was used for library generation. RNA samples were sequenced using 1-yield single-end 100-bp reads for Ccm1/Krit1ECKO and 1-yield single-end 75-bp reads for Ccm3/Pdcd10ECKO.
Ccm1/kri-1 and ccm3/pdcd10 C.
elegans and corresponding wild-type controls. We used 3 biological replicates of C. elegansccm1/kri-1(ok1251)–knockout and ccm3/pdcd10(tm2806)–knockout mutants to isolate total RNA for sequencing. Wild-type C. elegans (N2) were used as controls. Both C. elegans models and controls were harvested 24 hours past the L4 state (young adults). RNA was extracted using the TRIzol protocol (Thermo Fisher Scientific). TruSeq Stranded mRNA Sample Prep Kit (Illumina) was used for library generation. RNA samples were sequenced using 1-yield single-end 50-bp reads.
Statistics.
Bioinformatics analyses and study flow (Supplemental Figure 1) are described in detail in supplemental material. Source data, including full lists of DEGs of human NVUs, mouse BMECs (Ccm1/Krit1ECKO, Ccm3/Pdcd10ECKO), and C. elegans (ccm1/kri-1, ccm3/pdcd10) as well as corresponding GO analyses, are presented in detail in supplemental material (Supplemental Tables 2 and 3). The raw sequencing data used in this study are available in the National Center for Biotechnology Information’s Gene Expression Omnibus (GEO) database and is accessible through GEO series accession number GSE123968.
Study approval.
All patients enrolled in the study gave written informed consent approved by the University of Chicago Institutional Review Board (protocol number 10-295-A). The Declaration of Helsinki and its amendments were followed. The ethical principles guiding the institutional review board are consistent with the Belmont Report and comply with the rules and regulations of the Federal Policy for the Protection of Human Subjects (56 FR 28003). All mice experiments were performed in compliance with animal procedure protocols and approved by Institutional Animal Care and Use Committees at the UCSD.
Author contributions
Studies were designed and conducted by IAA, JK, RG, LS, HAZ, RL, TM, SL, KA, RS, CS, SPP, DZ, JCP, SR, MALR, EMC, EP, ATT, AA, MG, WBD, MLK, and DAM. Samples were sequenced and analyzed by GF, YL, PF, JA, and YC. Results of the study were interpreted and the paper was written by JK, RG, RS, and IAA. The paper was edited by all authors, and all the authors approved the final manuscript.
Supplementary Material
Acknowledgments
This work was partially supported by grants from the NIH (P01 NS092521, to MG, MK, DM, and IAA), to AT (F30NS100252), to MALR (K01HL133530), and to MK (R01HL094326 and R01NS100949); by the University of Chicago Medicine Comprehensive Cancer Center Support Grant (P30 CA14599), the William and Judith Davis Fund in Neurovascular Surgery Research, and the Safadi Translational Fellowship to RG; by the University of Chicago Pritzker School of Medicine to SL; by the Sigrid Juselius Foundation to JK; by a studentship from the National Sciences and Engineering Council to EC; and by the Canadian Institutes of Health Research to WBD (PJT 153000).
Version 1. 02/07/2019
Electronic publication
Funding Statement
to MG, MK, DM and IAA
to AT
to MK
to MK
University of Chicago Medicine Comprehensive Cancer Center Support Grant
to RG
to SL
to JK
studentship to EC
to WBD
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(3):e126167. https://doi.org/10.1172/jci.insight.126167.
Contributor Information
Janne Koskimäki, Email: koskimaki@uchicago.edu.
Romuald Girard, Email: rgirard@surgery.bsd.uchicago.edu.
Yan Li, Email: yli22@bsd.uchicago.edu.
Laleh Saadat, Email: lalehsaadat1215@gmail.com.
Hussein A. Zeineddine, Email: h.a.zeineddine@gmail.com.
Rhonda Lightle, Email: rlightle@surgery.bsd.uchicago.edu.
Thomas Moore, Email: tmoore10@surgery.bsd.uchicago.edu.
Seán Lyne, Email: Sean.Lyne@uchospitals.edu.
Kenneth Avner, Email: Kenneth.Avner@uchospitals.edu.
Robert Shenkar, Email: rshenkar@uchicago.edu.
Ying Cao, Email: ycao2@surgery.bsd.uchicago.edu.
Changbin Shi, Email: changbinshi@hotmail.com.
Sean P. Polster, Email: Sean.Polster@uchospitals.edu.
Dongdong Zhang, Email: dzhang4@surgery.bsd.uchicago.edu.
Julián Carrión-Penagos, Email: jcarrion@surgery.bsd.uchicago.edu.
Sharbel Romanos, Email: sromanos@uchicago.edu.
Miguel A. Lopez-Ramirez, Email: malopezramirez@ucsd.edu.
Eric M. Chapman, Email: echapman.utoronto@gmail.com.
Evelyn Popiel, Email: evelynmpopiel@gmail.com.
Alan T. Tang, Email: tangalan@mail.med.upenn.edu.
Jorge Andrade, Email: jandrade@bsd.uchicago.edu.
Mark Ginsberg, Email: mhginsberg@ucsd.edu.
Mark L. Kahn, Email: markkahn@mail.med.upenn.edu.
Douglas A. Marchuk, Email: douglas.marchuk@duke.edu.
References
- 1.Zeineddine HA, et al. Phenotypic characterization of murine models of cerebral cavernous malformations. Lab Invest. doi: 10.1038/s41374.018.0030-y. doi: 10.1038/s41374.018.0030-y. [published online ahead of print June 26, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shenkar R, et al. Exceptional aggressiveness of cerebral cavernous malformation disease associated with PDCD10 mutations. Genet Med. 2015;17(3):188–196. doi: 10.1038/gim.2014.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McDonald DA, et al. Lesions from patients with sporadic cerebral cavernous malformations harbor somatic mutations in the CCM genes: evidence for a common biochemical pathway for CCM pathogenesis. Hum Mol Genet. 2014;23(16):4357–4370. doi: 10.1093/hmg/ddu153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stockton RA, Shenkar R, Awad IA, Ginsberg MH. Cerebral cavernous malformations proteins inhibit Rho kinase to stabilize vascular integrity. J Exp Med. 2010;207(4):881–896. doi: 10.1084/jem.20091258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhou Z, et al. Cerebral cavernous malformations arise from endothelial gain of MEKK3-KLF2/4 signalling. Nature. 2016;532(7597):122–126. doi: 10.1038/nature17178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Renz M. Regulation of β1 integrin-Klf2-mediated angiogenesis by CCM proteins. Dev Cell. 2015;32(2):181–190. doi: 10.1016/j.devcel.2014.12.016. [DOI] [PubMed] [Google Scholar]
- 7.Girard R, et al. Plasma biomarkers of inflammation and angiogenesis predict cerebral cavernous malformation symptomatic hemorrhage or lesional growth. Circ Res. 2018;122(12):1716–1721. doi: 10.1161/CIRCRESAHA.118.312680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lopez-Ramirez MA, et al. Thrombospondin1 (TSP1) replacement prevents cerebral cavernous malformations. J Exp Med. 2017;214(11):3331–3346. doi: 10.1084/jem.20171178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Shi C, et al. B-cell depletion reduces the maturation of cerebral cavernous malformations in murine models. J Neuroimmune Pharmacol. 2016;11(2):369–377. doi: 10.1007/s11481-016-9670-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shenkar R, et al. Differential gene expression in human cerebrovascular malformations. Neurosurgery. 2003;52(2):465–477. doi: 10.1227/01.NEU.0000044131.03495.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lin X, et al. The differentially expressed genes of human sporadic cerebral cavernous malformations. World Neurosurg. 2018;113:e247–e270. doi: 10.1016/j.wneu.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 12.Whitehead KJ, et al. The cerebral cavernous malformation signaling pathway promotes vascular integrity via Rho GTPases. Nat Med. 2009;15(2):177–184. doi: 10.1038/nm.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lant B, et al. Interrogating the ccm-3 Gene Network. Cell Rep. 2018;24(11):2857–2868.e4. doi: 10.1016/j.celrep.2018.08.039. [DOI] [PubMed] [Google Scholar]
- 14.Lant B, et al. CCM-3/STRIPAK promotes seamless tube extension through endocytic recycling. Nat Commun. 2015;6:6449. doi: 10.1038/ncomms7449. [DOI] [PubMed] [Google Scholar]
- 15.Abedi H, Zachary I. Vascular endothelial growth factor stimulates tyrosine phosphorylation and recruitment to new focal adhesions of focal adhesion kinase and paxillin in endothelial cells. J Biol Chem. 1997;272(24):15442–15451. doi: 10.1074/jbc.272.24.15442. [DOI] [PubMed] [Google Scholar]
- 16.Zhou HJ, et al. Endothelial exocytosis of angiopoietin-2 resulting from CCM3 deficiency contributes to cerebral cavernous malformation. Nat Med. 2016;22(9):1033–1042. doi: 10.1038/nm.4169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jin M, et al. Toll-like receptor 2-mediated MAPKs and NF-κB activation requires the GNAO1-dependent pathway in human mast cells. Integr Biol (Camb) 2016;8(9):968–975. doi: 10.1039/c6ib00097e. [DOI] [PubMed] [Google Scholar]
- 18.Garcia-Marcos M, Ghosh P, Farquhar MG. Molecular basis of a novel oncogenic mutation in GNAO1. Oncogene. 2011;30(23):2691–2696. doi: 10.1038/onc.2010.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Berchtold MW, Villalobo A. The many faces of calmodulin in cell proliferation, programmed cell death, autophagy, and cancer. Biochim Biophys Acta. 2014;1843(2):398–435. doi: 10.1016/j.bbamcr.2013.10.021. [DOI] [PubMed] [Google Scholar]
- 20.Wetzel-Strong SE, Detter MR, Marchuk DA. The pathobiology of vascular malformations: insights from human and model organism genetics. J Pathol. 2017;241(2):281–293. doi: 10.1002/path.4844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Couto JA, et al. Endothelial cells from capillary malformations are enriched for somatic GNAQ mutations. Plast Reconstr Surg. 2016;137(1):77e–82e. doi: 10.1097/PRS.0000000000001868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shirley MD, et al. Sturge-Weber syndrome and port-wine stains caused by somatic mutation in GNAQ. N Engl J Med. 2013;368(21):1971–1979. doi: 10.1056/NEJMoa1213507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.West XZ, et al. Oxidative stress induces angiogenesis by activating TLR2 with novel endogenous ligands. Nature. 2010;467(7318):972–976. doi: 10.1038/nature09421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang AT, et al. Endothelial TLR4 and the microbiome drive cerebral cavernous malformations. Nature. 2017;545(7654):305–310. doi: 10.1038/nature22075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fistonich C, et al. Cell circuits between B cell progenitors and IL-7+ mesenchymal progenitor cells control B cell development. J Exp Med. 2018;215(10):2586–2599. doi: 10.1084/jem.20180778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang Y, et al. Plasma fibronectin supports hemostasis and regulates thrombosis. J Clin Invest. 2014;124(10):4281–4293. doi: 10.1172/JCI74630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sadeqzadeh E, de Bock CE, Thorne RF. Sleeping giants: emerging roles for the fat cadherins in health and disease. Med Res Rev. 2014;34(1):190–221. doi: 10.1002/med.21286. [DOI] [PubMed] [Google Scholar]
- 28.Glading AJ, Ginsberg MH. Rap1 and its effector KRIT1/CCM1 regulate beta-catenin signaling. Dis Model Mech. 2010;3(1-2):73–83. doi: 10.1242/dmm.003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gagliardi F, Narayanan A, Mortini P. SPARCL1 a novel player in cancer biology. Crit Rev Oncol Hematol. 2017;109:63–68. doi: 10.1016/j.critrevonc.2016.11.013. [DOI] [PubMed] [Google Scholar]
- 30.Liu W, et al. PLCD3, a flotillin2-interacting protein, is involved in proliferation, migration and invasion of nasopharyngeal carcinoma cells. Oncol Rep. 2018;39(1):45–52. doi: 10.3892/or.2017.6080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cao Y, Cao R, Hedlund EM. R Regulation of tumor angiogenesis and metastasis by FGF and PDGF signaling pathways. J Mol Med. 2008;86(7):785–789. doi: 10.1007/s00109-008-0337-z. [DOI] [PubMed] [Google Scholar]
- 32.Gao G, et al. Down-regulation of vascular endothelial growth factor and up-regulation of pigment epithelium-derived factor: a possible mechanism for the anti-angiogenic activity of plasminogen kringle 5. J Biol Chem. 2002;277(11):9492–9497. doi: 10.1074/jbc.M108004200. [DOI] [PubMed] [Google Scholar]
- 33.Hill KK, Bedian V, Juang JL, Hoffmann FM. Genetic interactions between the Drosophila Abelson (Abl) tyrosine kinase and failed axon connections (fax), a novel protein in axon bundles. Genetics. 1995;141(2):595–606. doi: 10.1093/genetics/141.2.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Louvi A, Chen L, Two AM, Zhang H, Min W, Günel M. Loss of cerebral cavernous malformation 3 (Ccm3) in neuroglia leads to CCM and vascular pathology. Proc Natl Acad Sci U S A. 2011;108(9):3737–3742. doi: 10.1073/pnas.1012617108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gupte RS, Pozarowski P, Grabarek J, Traganos F, Darzynkiewicz Z, Lee MY. RIalpha influences cellular proliferation in cancer cells by transporting RFC40 into the nucleus. Cancer Biol Ther. 2005;4(4):429–437. doi: 10.4161/cbt.4.4.1621. [DOI] [PubMed] [Google Scholar]
- 36.Lee SY, Jang C, Lee KA. Polo-like kinases (plks), a key regulator of cell cycle and new potential target for cancer therapy. Dev Reprod. 2014;18(1):65–71. doi: 10.12717/DR.2014.18.1.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harel A, et al. Removal of a single pore subcomplex results in vertebrate nuclei devoid of nuclear pores. Mol Cell. 2003;11(4):853–864. doi: 10.1016/S1097-2765(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 38.Guimaraes JC, Zavolan M. Patterns of ribosomal protein expression specify normal and malignant human cells. Genome Biol. 2016;17(1):236. doi: 10.1186/s13059-016-1104-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jones DT, et al. Endogenous ribosomal protein L29 (RPL29): a newly identified regulator of angiogenesis in mice. Dis Model Mech. 2013;6(1):115–124. doi: 10.1242/dmm.009183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang X, Zhang L, Natarajan SK, Becker DF. Proline mechanisms of stress survival. Antioxid Redox Signal. 2013;19(9):998–1011. doi: 10.1089/ars.2012.5074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Carlton JG, et al. Sorting nexin-2 is associated with tubular elements of the early endosome, but is not essential for retromer-mediated endosome-to-TGN transport. J Cell Sci. 2005;118(pt 19):4527–4539. doi: 10.1242/jcs.02568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jain A, Beutel O, Ebell K, Korneev S, Holthuis JC. Diverting CERT-mediated ceramide transport to mitochondria triggers Bax-dependent apoptosis. J Cell Sci. 2017;130(2):360–371. doi: 10.1242/jcs.194191. [DOI] [PubMed] [Google Scholar]
- 43.Archer SL. Mitochondrial dynamics — mitochondrial fission and fusion in human diseases. N Engl J Med. 2013;369(23):2236–2251. doi: 10.1056/NEJMra1215233. [DOI] [PubMed] [Google Scholar]
- 44.Ito S, Greiss S, Gartner A, Derry WB. Cell-nonautonomous regulation of C. elegans germ cell death by kri-1. Curr Biol. 2010;20(4):333–338. doi: 10.1016/j.cub.2009.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pal S, et al. CCM-3 promotes C. elegans germline development by regulating vesicle trafficking cytokinesis and polarity. Curr Biol. 2017;27(6):868–876. doi: 10.1016/j.cub.2017.02.028. [DOI] [PubMed] [Google Scholar]
- 46.Wang Y, et al. The cerebral cavernous malformation disease causing gene KRIT1 participates in intestinal epithelial barrier maintenance and regulation. FASEB J. doi: 10.1096/fj.201800343R. doi: 10.1096/fj.201800343R. [published online ahead of print September 25, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.