Abstract
Polypropylene meshes that are commonly used for inguinal hernia repair may trigger granulomatous foreign body reactions. Here, we show that asymptomatic patients display mesh-associated inflammatory granulomas long after surgery, which are dominated by monocyte-derived macrophages expressing high levels of inflammatory activation markers. In mice, mesh implantation by the onlay technique induced rapid and strong myeloid cell accumulation, without substantial attenuation for up to 90 days. Myeloid cells segregated into distinct macrophage subsets with separate spatial distribution, activation profiles, and functional properties, showing a stable inflammatory phenotype in the tissue surrounding the biomaterial and a mixed, wound-healing phenotype in the surrounding stromal tissue. Protein mass spectrometry confirmed the inflammatory nature of the foreign body reaction, as characterized by cytokines, complement activation, and matrix-modulating factors. Moreover, immunoglobulin deposition increased over time around the implant, arguing for humoral immune responses in association with the cell-driven inflammation. Intravital multiphoton microscopy revealed a high motility and continuous recruitment of myeloid cells, which is partly dependent on the chemokine receptor CCR2. CCR2-dependent macrophages are particular drivers of fibroblast proliferation. Thus, our work functionally characterizes myeloid cell–dependent inflammation following mesh implantation, thereby providing insights into the dynamics and mechanisms of foreign body reactions to implanted biomaterials.
Keywords: Gastroenterology, Inflammation
Keywords: Chemokines, Macrophages, Surgery
Mesh implantation is associated with long lasting accumulation of myeloid cells in patients and mice.
Introduction
Implantation of biomaterials is commonly associated with inflammatory foreign body reaction (FBR) or granuloma formation (1, 2), a rapid, yet highly orchestrated process involving activation of humoral immunity, coagulation, molecular pattern recognition, and danger signal release from damaged tissue (e.g., DNA, ATP, heat shock proteins, S100 proteins, or uric acid) (3). Prolonged inflammation can provoke clinical complications, such as chronic pain, defective wound healing, damage to the implant, and the potential need of surgical explantation (4).
Recruitment of innate immune cells is considered an early event in establishing FBRs against implanted biomaterials (5). CCR2+Ly6C+ monocytes in mice and classical CD14+CD16– monocytes in humans represent the main source for inflammatory tissue macrophages appearing during inflammation, characterized by expression of CD86, NOS2, and TNF (6). Persistence of these cells leads to chronic inflammation, tissue damage, extracellular matrix remodeling (7, 8), and damage to the implant by oxidative stress responses (9). Inflammation and (aberrant) wound healing can also incite macrophages to a phenotypic adaptation favoring tissue remodeling, secreting IL-10, TGF-β, and VEGF (10), as also observed in several murine studies of organ injury and fibrosis (11, 12), emphasizing the high plasticity of myeloid cells in response to their microenvironment.
Biomaterial implantation, a standard surgical procedure, is often used to repair inguinal hernias in adults, for which the lifetime risk is about 27% in men and 3% in women (4). Surgical hernia repair is commonly achieved by the implantation of low-weight, large-pore polypropylene mesh (PP) material (13, 14). Mesh-induced inflammatory responses may lead to postoperative complications and inflammation (15, 16). Previous studies have addressed the general issue of FBR development (2, 17) and proposed different strategies, e.g., coating the implants with isolated extracellular matrix proteins (18), hydrogels (19, 20), or mesenchymal stem cells (21), in order to attenuate mesh-associated inflammation. Nonetheless, the dynamics of underlying cellular and molecular mechanisms leading to the FBR and the modulations by the differential pretreatments have remained largely obscure.
In this study, we comprehensively characterize the inflammatory response in a murine model for hernia mesh implantation. We herein describe a rapid and strong inflammatory foreign body (FB) response following implantation of PP meshes, without substantial attenuation over a time course of up to 90 days. Detailed immunophenotyping revealed a prevailing mixed myeloid cell response involving activated macrophages and conventional DCs, associated with danger signals, inflammasome activation, and inflammatory cytokine release (IL-1b, IL-6, and TNF). Furthermore, protein mass spectrometry identified strong complement activation and immunoglobulin deposition. Intravital multiphoton microscopy demonstrated the high motility and continuous recruitment of myeloid cells, which was partly dependent on the chemokine receptor CCR2. These macrophages were found to be drivers of fibroblast proliferation in response to FB-associated tissue explants.
Results
Monocyte-derived macrophages dominate cellular immune reactions in early and late stages of FB granulomatous disease in men and mice.
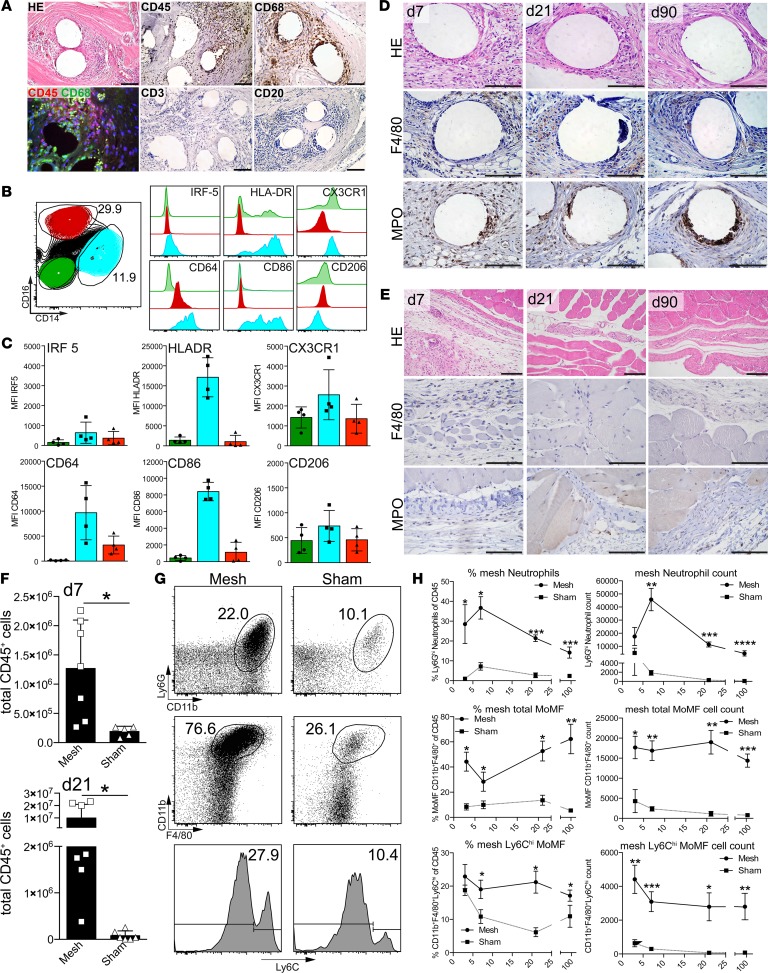
FB-associated inflammation following PP mesh implantation have been implicated in long-term complications after hernia surgery (e.g., hernia recurrence, chronic postherniorrhaphy groin pain, or aberrant scarring) (4). To characterize the potential inflammatory reaction following mesh implantation, tissue samples from asymptomatic patients were retrieved during relaparotomy due to other medical indications >1 year following implantation. Histology revealed dense infiltration of mononuclear cells, giant cells, and connective tissue deposition surrounding mesh fibers. Immunohistochemistry for the pan-leukocyte marker CD45 and macrophage marker CD68 indicated a predominant accumulation and activation of myeloid cells following mesh implantation but no apparent accumulation of CD3+ T cells or CD20+ B cells (Figure 1A). Flow cytometric analysis revealed substantial numbers of both CD14+ and CD16+ myeloid cells (Figure 1B) but only a minor amount of CD3+, CD56+, or CD20+ lymphocytes (data not shown), demonstrating the predominance of monocyte-derived macrophages. CD14+ macrophages expressed high levels of inflammatory activation markers, such as CD64, HLA-DR, CD86, and, also, albeit to a lesser extent, IRF-5 (Figure 1, B and C), while CD16+ cells only showed a slight upregulation of CD64. Furthermore, the macrophage-associated chemokine receptor CX3CR1 was detected on CD14+ cells. CD206, a marker commonly associated with antiinflammatory (“M2-type”) polarization of macrophages, was only detected at low levels on all myeloid populations (Figure 1C). Thus, mesh implantation led to a chronic and predominantly myeloid inflammatory response, regardless of imminent clinical complications associated with the FBR.
Figure 1. Characterization of cellular infiltrates in human and murine mesh samples.
(A) Histological analysis of foreign body granulomatous reaction from a patient explanted >1 year following mesh implantation (H&E). Cells were further subtyped using immunohistochemistry with the indicated antibodies. CD68 was costained with CD45 to estimate the proportion of mesh-infiltrating macrophages. Scale bar: 100 μm. (B) Flow cytometric analysis of myeloid subpopulations and their activation isolated from human mesh explants. Exemplary results are shown. n ≥ 3 samples were analyzed for flow cytometry and histology. (C) Mean fluorescence intensity (MFI) for macrophage markers expressed on the populations described in B. (D–H) Foreign body–induced inflammation was assessed in mice subjected to mesh implantation after 7, 21, and 90 days. Histological analysis of mesh-implanted (D) and sham-operated (E) animals. Scale bar: 100 μm. Cellular infiltration was analyzed by H&E histology and immunohistochemistry. (F) Quantification of total leukocyte infiltrates by flow cytometry 7 and 21 days following implantation. (G) Exemplary gating to characterize myeloid cell populations in the foreign body reaction. (H) Relative and absolute quantification of leukocyte subpopulations of the populations shown in G. MoMF, monocyte-derived macrophages. Statistical analysis was performed with at least 4 animals per group in 2 independent sets of experiments; experimental data were pooled for statistical analysis. Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001, ****P < 0.0001. Error bars represent mean ± SD.
To further gain mechanistic insights into myeloid cell functionality in FBR, we used a mouse model of surgically implanting PP meshes that are commonly used for human abdominal hernia repair (Supplemental Figure 1A; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123862DS1). Meshes were excised after 7, 21, and 90 days, while control animals were subjected to surgical intervention without mesh implantation. Similar to human pathology, mesh implants provoked a dense accumulation of inflammatory cells at all time points after surgery (Figure 1D) in comparison to sham-operated abdominal wall tissue (Figure 1E), characterized by F4/80 as well as myeloperoxidase (MPO) expression, indicating an infiltration of monocytes/macrophages and neutrophils. Quantitative flow cytometry at day 7 and day 21 indicated a massive influx of inflammatory cells followed by sustained inflammation in the mesh-surrounding tissue, compared with only minute amounts of CD45+ leukocytes detectable in abdominal explants recovered from sham-operated animals (Figure 1F). Flow cytometric immune phenotyping revealed an infiltration of neutrophilic granulocytes (characterized as Ly6G+CD11b+ cells) and monocyte-derived macrophages (characterized as Ly6G–CD11b+F4/80+ cells), including increased levels of Ly6Chi monocytes compared with sham-operated animals (Figure 1, G and H).
The amount of macrophages remained almost stable over the time course of the experiment, showing persistently elevated numbers of CD11b+F4/80+Ly6Chi monocyte-derived macrophages (Figure 1H). In contrast, the number of neutrophilic granulocytes peaked at the early time points of the experiment but declined by about 50% at the later stages of chronic inflammation, albeit staying markedly elevated compared with sham-operated control animals. Lymphocytes were only transiently detectable at early stages following mesh implantation, as shown by flow cytometry and immunohistochemistry (Supplemental Figure 1, B–D).
Mesh-infiltrating myeloid cells consist of distinct clusters with monocyte-, macrophage-, and DC-like features.
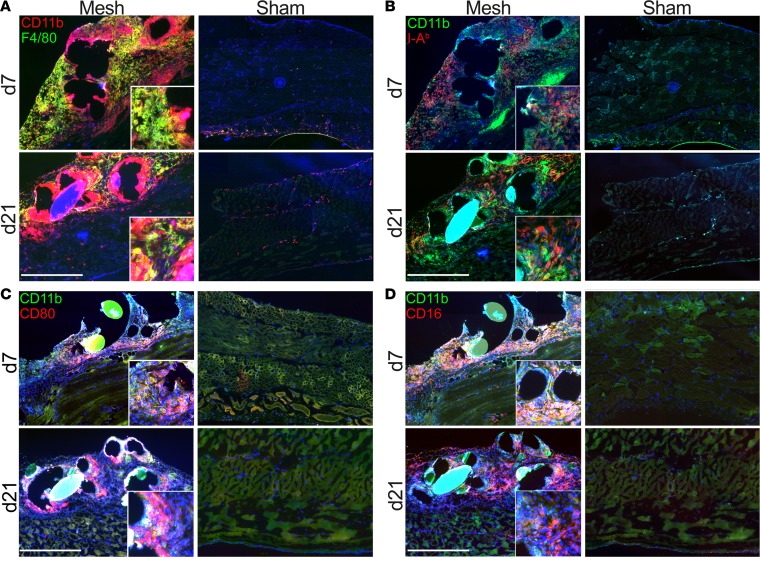
To better understand the myeloid cell response within the mesh-associated infiltrates in the mouse model, we first performed multicolor immunofluorescence histology. Consecutive sections of mesh explants were stained with CD11b and F4/80 to identify macrophages (Figure 2A) and with activation markers, such as MHC-II (I-Ab) (Figure 2B), CD80 (Figure 2C), and CD16 (Figure 2D), showing a zonation between the mesh-surrounding CD11b+ cells (mesh-associated macrophages [MAMs]) and more distant CD11b+F4/80+ cells (stroma-infiltrating macrophages [SIMs]). This was further substantiated by the I-Ab staining, showing a scattered pattern of I-Ab+ cells at day 7 and a more condensed localization at day 21, with an upregulation of I-Ab expression also seen in the CD11b+ cells (Figure 2B). CD80 expression was also detectable at early and late time points in both macrophage populations (Figure 2C), while CD16, though abundantly present in both subtypes, showed higher expression in the CD11b+F4/80+ fraction at day 21 (Figure 2D). Furthermore, the macrophages in the immediate surroundings of the fibers also expressed CD11c at day 7, and this was even more pronounced on day 21 (Supplemental Figure 2A). In addition, Ly6G+ cells were visible in the immediate surroundings of the fibers but were absent in the stroma (Supplemental Figure 2B). CD206 was detected in areas associated with the stroma-infiltrating macrophages but was absent in the immediate surroundings of the fibers, similar to CD301 distribution (Supplemental Figure 2, C and D).
Figure 2. Immunophenotyping of myeloid cells at day 7 and day 21 after mesh implantation in mice.
(A–D) Multicolor immunofluorescence staining of mesh explants and sham-operated animals. Costaining was performed for (A) CD11b and F4/80, (B) CD11b and I-Ab, (C) CD11b and CD80, and (D) CD11b and CD16. Inset images show magnified mesh-surrounding cells in high detail. Scale bar: 500 μm. Original magnification, ×2.5 (inset). Exemplary results are shown in the images; results were verified in at least 4 animals per group from 2 independent sets of experiments.
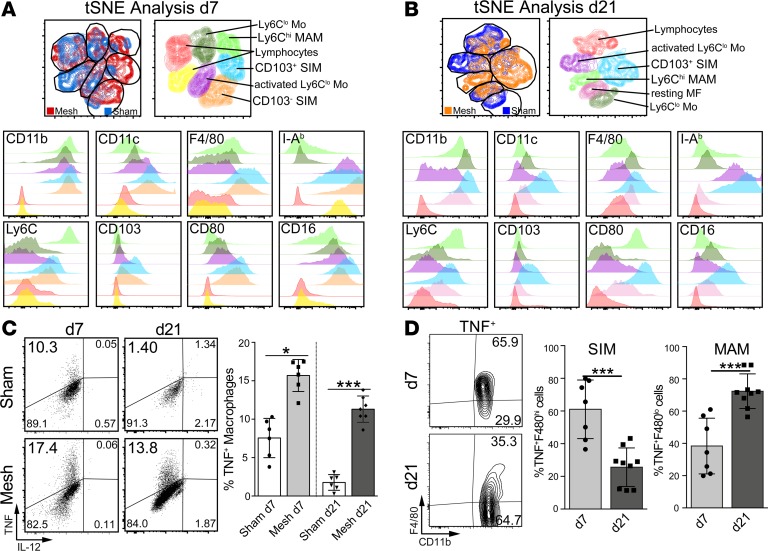
The myeloid cell populations were next characterized using multicolor flow cytometry analysis and an unbiased t-distributed Stochastic Neighbor Embedding (t-SNE) algorithm on CD45+Ly6G– populations derived from both mesh- and sham-operated animals at day 7 (Figure 3A) and day 21 (Figure 3B). In line with the morphological subsets observed by the immunofluorescence histology, two major populations emerged following the cluster analysis at both day 7 and day 21. On day 7, the predominant FBR-associated populations could be characterized as cells expressing CD11bhiCD11cloF4/80loI-AbloLy6ChiCD103negCD80hiCD16in (Figure 3, A and B, light green), likely reflecting the MAMs, and furthermore as CD11bhiCD11chiF4/80hiI-AbhiLy6CloCD80hiCD16hi cells, resembling the SIM population. In the early phase of the FBR, the SIM population further subdivided into two populations expressing different levels of CD103, clustering into CD103lo (Figure 3A, orange) and CD103hi (Figure 3A, light blue) subpopulations. On day 21, two clusters of cells were predominantly present in mesh-operated animals, one being CD11bhiCD11chiF4/80loI-AbloLy6ChiCD103negCD80hiCD16int (Figure 3B, light green), again resembling the MAMs, and the other one being CD11bhiCD11chiF4/80hiI-AbhiLy6ChiCD103hiCD80hiCD16int (Figure 3B, light blue), matching SIMs, with all cells being CD103+ at this time point.
Figure 3. Flow cytometry profiling of myeloid cells at day 7 and day 21 after mesh implantation in mice.
Flow cytometric characterization of mesh explants: t-SNE flow cytometry analysis of murine mesh explants at day 7 (A) and day 21 (B) after implantation. Cells were pregated on CD45+Ly6G– cells and subjected to unbiased t-SNE analysis. Overlays were generated from mesh- and sham-operated animals, and the clusters were then analyzed in detail for their marker expression profiles, as shown in the subsequent histogram overlays. MAM, mesh-associated macrophage; SIM, stroma-infiltrating macrophage. (C) Cytokine production from myeloid cells isolated from mesh-associated tissue. Cells were pregated on CD45+Ly6G–F480+CD11b+. (D) Phenotyping of TNF+ cells pregated on CD45+Ly6G–. Exemplary results are shown in the dot plots; statistical analysis was performed from ≥5 animals per group. Student’s t test: *P < 0.05; ***P < 0.001. Error bars represent mean ± SD.
The macrophages were also functionally analyzed for inflammatory and regulatory cytokine production by intracellular flow cytometry, revealing a strong TNF signal at day 7 and day 21 but no IL-10 (data not shown) or IL-12 signal (Figure 3C). TNF was also detectable following sham operation at day 7, likely reflecting trauma-related inflammation triggered by the sham surgery at the early time point. While in the early phase TNF was produced in a more generalized fashion and was predominantly detected in F4/80hi cells (SIM), chronic FBR was associated with TNF production within the F4/80lo population (MAM) (Figure 3D). In line with the inflammatory macrophage activation especially in the immediate vicinity of the mesh fiber, we could also detect activation of the inflammasome Nlrp3 at both day 7 and day 21 in myeloid cells (Supplemental Figure 2E). Histological TUNEL staining revealed a FBR inflammation–associated cell death in the immediate surrounding of the fibers (Supplemental Figure 2F).
Thus, two major myeloid cell populations were identified by histological and flow cytometry analysis, resembling activated cells with a similarity to inflammatory monocyte-derived macrophages (MAMs) with a mature phenotype and immunological activation, expressing high levels of MHC-II, CD80, and CD16 and TNF (but not IL-12 [SIM]) and, on the other hand, at least to some extent, wound healing–associated markers, such as CD206 and CD301, indicating a dual functionality of the cells promoting inflammation but also antigen presentation and costimulation.
FB implantation provokes the rapid and continuous infiltration of transient BM-derived monocytes.
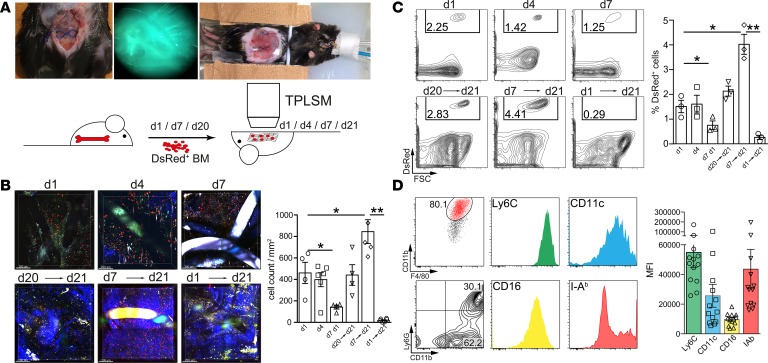
In order to unravel the kinetics of recruitment, accumulation, and persistence of the myeloid cells infiltrating the mesh implants, we performed intravital 2-photon laser-scanning microscopy (TPLSM) in animals receiving an adoptive i.v. transfer of genetically labeled (Actin-DsRed+) BM cells. To assess early infiltration following implantation, we injected DsRed+ cells 12 hours after mesh implantation followed by intravital TPLSM imaging on the following day (day 1), day 4, and day 7 (Figure 4A). DsRed+ cells were already present at day 1 after implantation, indicating a rapid onset of recruitment of BM-derived cells. Infiltrating cell numbers remained stable for several days in the early phase following implantation but showed a significant drop after 7 days (Figure 4, B and C, and Supplemental Video 1).
Figure 4. Intravital 2-photon laser-scanning microscopy imaging of abdominal meshes following adoptive Actin-DsRed+ BM cell transfer.
2 × 107 BM cells from Actin-DsRed donor mice were transferred i.v. into mesh-implanted mice and followed by intravital 2-photon laser-scanning microscopy (TPLSM) imaging (imaging for up to 3 hours). (A) Representative images of the experimental setting and schematic overview of the experimental design. (B) TPLSM images of cellular infiltrates at early stages of foreign body reaction (top row). Cells were injected 12 hours following surgery and analyzed at day 1, day 4, and day 7 (acute FBR). Imaging at day 21 (bottom row) was commenced following cell injection 1, 14, and 20 days before imaging (chronic FBR). Images were composed of 4 adjacent view fields. Mesh fibers are visible as either black or highly fluorescent coherent structures. Blue fibers show collagen fibers visualized by second harmonic generation. Scale bar: 200 μm. Statistical analysis was performed by cell counting in at least 4 view fields of surrounding mesh fibers with a penetration depth of 200 μm. (C) Flow cytometric assessment of cellular infiltration visible in B. Cells were stained for CD45, Ly6G, CD11b, F4/80, CD11c, I-Ab, Ly6C, and CD16. Cells were gated as CD45+Ly6G–DsRed+ cells and for quantification of relative monocyte recruitment efficacy. (D) Flow cytometric subtyping of cellular infiltrates from DsRed+ cells after 14 days (day 7→day 21). Cells were pregated as CD45+Ly6G–DsRed+ and analyzed for their expression of the indicated surface markers. Expression strength was quantified by calculating the MFI of the indicated markers. Exemplary results are shown in the dot plots; statistical analysis was performed from ≥3 animals per group. Student’s t test: *P < 0.05; **P < 0.01. Error bars represent mean ± SD.
For the chronic inflammatory stage at day 21, we injected DsRed+ BM cells either 1 day before TPLSM imaging (day 20→day 21), 1 week after implantation (day 7→day 21), or 12 hours after implantation (day 1→day 21) (Figure 4A). Cells were readily recruited also 3 weeks following mesh implantation (day 20 → day 21), showing that infiltration of BM cells is a continuous process also at the later stages of FB-induced inflammation. Cells that had been transferred 7 days after mesh implantation were detectable in mesh-induced infiltrates 14 days later (day 7→day 21), while cells injected 1 day after the surgery (day 1→day 21) could not be found anymore in the FB infiltrates (Figure 4, B and C, and Supplemental Videos 2 and 3), demonstrating a continuous turnover of the myeloid cells in vivo as well as a limited life span of the BM-derived cells.
Cells close to the fibers appeared to remain mostly sessile, while the surrounding cells showed high motility and active scanning (Supplemental Videos 4 and 5). Flow cytometry performed on explanted mesh-surrounding tissue confirmed quantification by TPLSM (Figure 4C). Adoptively transferred BM cells infiltrating meshes were mainly CD11b+F4/80+ cells that stained also positive for Ly6C as well as CD16 and had a mixed expression of I-Ab, arguing for different monocyte-derived subsets (Figure 4D). Moreover, we observed that 30% of infiltrating cells were Ly6G+CD11b+DsRed+, indicating a continuous recruitment of neutrophilic granulocytes accompanying the continuous monocytic infiltration (Figure 4D). These experiments indicate a variable half-life time of the infiltrating cells in the inflamed tissue, dependent on early or established FB inflammation.
Molecular characteristics of myeloid cell–driven inflammation in mesh-associated FBR.
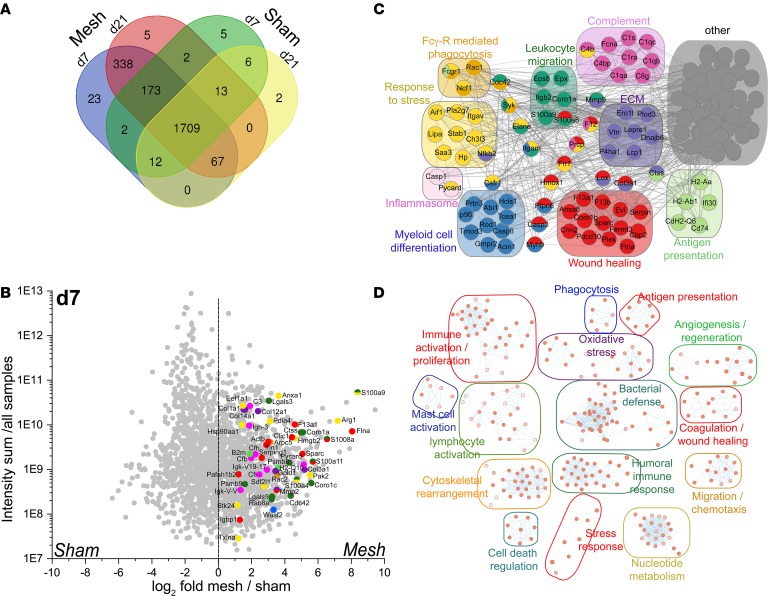
Next, we aimed at elucidating the molecular mechanisms and consequences of myeloid cell accumulation in FBR by transcriptomic and proteomic analyses on murine mesh explants. To address the protein factors associated with the FBR, we performed protein mass spectrometry of abdominal wall explants isolated from mesh-implanted and sham-operated animals at day 7 and day 21 following surgery. An overall comparison between the different conditions revealed a set of 366 proteins (15.5%) unique for the mesh-implanted animals, of which 338 proteins overlapped (92% similarity) between day 7 and day 21 (Figure 5A and Supplemental Table 1). Differential protein quantification revealed a high expression of inflammatory proteins, such as S100 family proteins, stress response proteins (e.g., HMGB2, Hsp90), and wound-healing factors (FLNA, F13A1, SPARC) as well as complement-associated proteins (C3, Cfb, Cfh) and immunoglobulin proteins, e.g., Igh-3 and Igk-V-V (Figure 5B and Supplemental Figure 3C). The abundance of signature proteins for the associated gene ontology (GO) pathways as well their weighted distribution was investigated to further delineate their overall relevance for the mesh-associated inflammation. For the differentially expressed proteins, we performed STRING protein network cluster analysis to identify the protein clusters associated with the FBR. To this end, all proteins with a higher than log2 4-fold abundance were subjected to STRING protein network analysis. The resulting protein network was further filtered, isolating proteins associated with GO pathways related to inflammation, stress responses, wound healing, and general immune system activation (Supplemental Tables 2 and 3). This analysis confirmed myeloid cell activation and differentiation at both time points, as shown by detection of CSFR1, FCGR1, CD74, FCNA, H2, and others. Furthermore, several factors associated with inflammasome activation and alarmin signaling were detected at day 7 and day 21 (S100A8/9, CASP1, PYCARD). Strikingly, protein network clusters indicated a strong presence of complement proteins associated with the classical pathway (C1 complex, C4) in mesh-associated inflammation (Figure 5C and Supplemental Figure 3D), hinting at immunoglobulin deposition in the inflamed areas. Proteins specific for mesh-associated inflammation were then subjected to pathway enrichment for day 7 and day 21, revealing a strong myeloid cell response as well as humoral immune activation at both day 7 and day 21, in combination with oxidative stress, cellular apoptosis, general immune activation, and cell recruitment but also angiogenesis and wound healing (Figure 5D and Supplemental Figure 3B).
Figure 5. Protein expression analysis of inflammation-associated factors from murine mesh explants.
Protein mass spectrometry analysis of tissue isolated from mesh- and sham-operated animals at day 7 and day 21. (A) Venn diagram showing differential protein signatures. Overlapping areas show proteins coidentified at the corresponding conditions. (B) Scatter plot displaying weighted protein expression of mesh and sham isolated tissue. Total protein counts of all samples (sum) were plotted on that y axis, weighted protein expression was displayed as log2-fold expression ratio. Signature proteins of GO pathways described in C were highlighted in the corresponding colors. (C) Cluster network analysis using BinGO (55) to determine clusters enriched in mesh-associated tissue at day 7. Protein expression was calculated against corresponding sham-operated controls with n = 3 animals per group. (D) String protein interaction network analysis. Proteins expressed with a log2 fold ratio of >4 in mesh versus sham samples at day 7 were filtered for generalized pathways relevant for inflammation, immune system activation, and wound healing. Relevant protein networks and exemplary targets are shown.
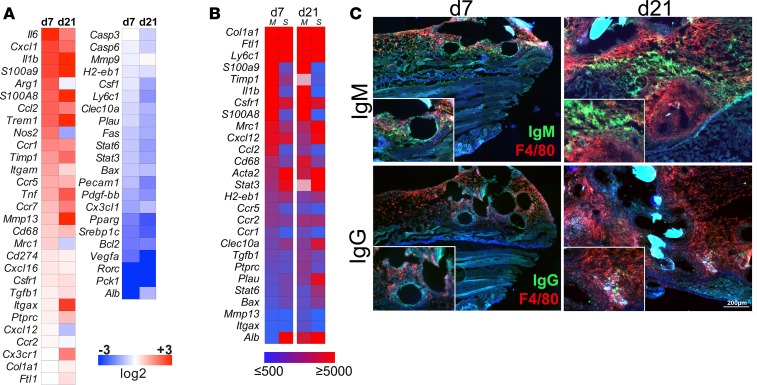
We additionally performed Nanostring-based RNA expression arrays of genes associated with myeloid cell activation from mesh tissue at day 7 and day 21, in comparison with the collated sham-operated animals (Figure 6A). Factors such as Il6, Il1b, Ccl2, Cxcl1, Tnf, S100A8/9, and Nos2 as well as intracellular myeloid activation markers such as Trem1 and Csfr1 where highly upregulated during FBR, while signaling molecules such as Stat3 and Stat6, associated with tolerogenic myeloid cell responses, were markedly downregulated at day 7, a finding that was even more pronounced on day 21. The chemokine receptors Ccr1, Ccr5, Ccr7, and, to a lesser extent, Ccr2 and Cx3cr1 were also increased; Ccr2 and Cx3cr1 were particularly present at the later stages of the FBR. In contrast, some regulatory markers, such as Cday 274 (Pdl1), Mrc1, and Tgfb1, were only slightly upregulated compared with the sham-operated animals. The expression of matrix remodeling factors Mmp9, Mmp13, and Timp1 exhibited a heterogeneous pattern, showing upregulation of Mmp13 but a moderate downregulation of Mmp9. In addition, Timp1 was expressed at both day 7 and day 21, likely by fibroblasts associated with the formation of mesh-surrounding tissue (Figure 6A). The abundance of target RNA was analyzed by the total number of counts for each gene to identify the key regulator genes (Figure 6B). In particular, inflammation-associated markers S100A8/9, Il1b, Csfr1, Ccl2, CD68, Ccr1, Ccr, and Ccr5 as well as the antiinflammatory gene Tgfb1 were detected at high levels. Conventional quantitative RT-PCR for selected genes was performed to validate the Nanostring gene expression, showing similar gene regulation (Supplemental Figure 3A). Thus, gene expression profiling supported a persistent inflammation and a progressive aggravation over time, with higher levels of inflammation-associated mediators as well as myeloid cell maturation and downregulation of inhibitory cellular signaling pathways. To substantiate the hypothesis of immunoglobulin deposition within the FBR tissue, we next performed immunofluorescence microscopy for IgM and IgG, revealing an abundant deposition of IgM at day 7 that was further increased on day 21. In line with these findings, IgG was already present at basal levels after 7 days and was more pronounced at day 21, showing deposition of immune complexes surrounding the mesh fibers (Figure 6C).
Figure 6. Gene expression and protein analysis of inflammation-associated factors from murine mesh explants.
(A) 72-plex Nanostring gene expression array of mesh-surrounding tissue day 7 and day 21 following implantation. Gene expression was normalized against corresponding sham-operated controls. Expression profiles were analyzed with a group size of 3 animals per group and displayed as color-coded heat maps showing log2 fold gene expression. (B) Total count of genes identified by Nanostring gene expression. Absolute counts were measured to validate relative expression data shown in A. Color-coded heat map showing relevant genes with a minimum count threshold of ≥500 total counts. M, mesh; S, sham. (C) Immunofluorescence microscopy of IgM and IgG deposition in mesh-surrounding tissue at day 7 and day 21. Scale bar: 200 μm. Original magnification, ×1.5 (inset). Exemplary results are shown in the images; results were verified in at least 3 animals per group from 2 independent sets of experiments.
Taken together, gene and protein analyses corroborated the persistent inflammatory nature of FBR, with key molecular signals of myeloid cell activation as well as related consequences, such as complement activation and immunoglobulin deposition.
Chemokine receptor CCR2-dependent myeloid cell infiltration determines the inflammatory FB response.
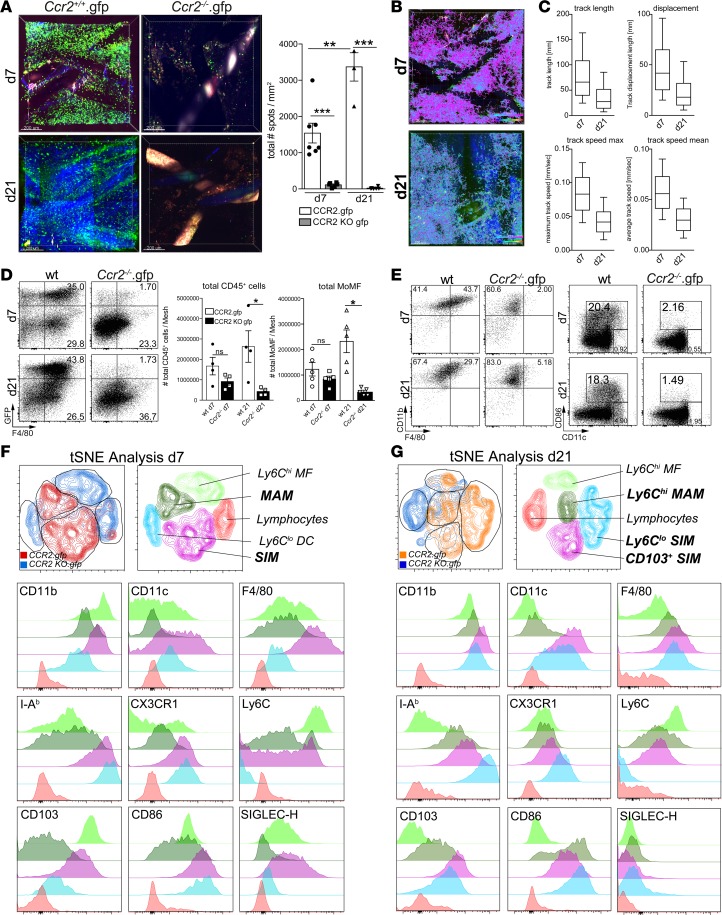
Based on the crucial role of chemokine receptor CCR2 signaling for monocyte recruitment to inflamed tissue (6), we hypothesized that CCR2-dependent recruitment would determine inflammatory myeloid cell responses in FBR. We therefore subjected Ccr2.gfp as well as Ccr2–/–.gfp reporter mice to mesh implantation, followed by intravital TPLSM and flow cytometry analysis at day 7 and day 21 after surgery. While GFP+ cells were detected with high abundance in Ccr2.gfp mice at both time points, virtually no GFP+ cells could be observed in the Ccr2–/–.gfp reporter mice on day 7 or on day 21 using both TPLSM and flow cytometry (Figure 7A, Supplemental Figure 4A, and Supplemental Videos 6 and 7). Ccr2.gfp cells exhibited high motility and showed active scanning of the mesh-surrounding tissue at day 7 and day 21 by intravital TPLSM (Figure 7, B and C, and Supplemental Videos 8 and 9), with a marked accumulation around the fibers at day 21 (Figure 7A). In comparison to day 7, CCR2+ cells displayed a reduction in average migration speed, average track length, and the average cellular displacement at day 21 (Figure 7C), although the maximum track length was similar between both time points (Supplemental Figure 4B). Of note, collagen deposition was also markedly increased on day 21, as evidenced by the increased second harmonic signal. These data demonstrate a continuous infiltration of myeloid cells, which was highly dependent on the ability of monocytes to respond to CCR2-mediated signaling. Furthermore, similar to the adoptive transfer experiments, we noted that infiltrating myeloid cells remained highly motile and were actively migrating through the mesh-surrounding tissue within the mesh-surrounding cell clusters.
Figure 7. Chemokine receptor CCR2-dependent alterations of mesh-associated granulomatous inflammation.
(A–C) Ccr.gfp and Ccr2–/–.gfp reporter mice were subjected to abdominal mesh surgery, and cellular infiltration was assessed using intravital TPLSM tracking of GFP+ monocytes at day 7 and day 21 after implantation. (A) TPLSM images of GFP+ monocytes day 7 and day 21 after surgery performed on Ccr2.gfp-competent and Ccr2–/–.gfp mice. Mesh fibers are visible as either black or highly fluorescent coherent structures. Blue fibers show collagen fibers visualized by second harmonic generation. Scale bar: 200 μm. For statistical analysis, cells were counted in 4 adjacent view fields surrounding mesh fibers with a penetration depth of 200 μm. (B) Migration tracking of GFP+ monocytes by intravital TPLSM. Cells were imaged and tracked over time for a period of approximately 2 hours. Migration is shown as color-coded tracks. Pink: fast moving cells ≥ 0.1 μm/sec, turquoise: slow moving cells ≤ 0.01 μm/s. Scale bar: 250 μm. (C) Statistical analysis of cellular migration shown in B, showing overall track length, cellular displacement, maximum track speed, and average track speed at day 7 and day 21. (D–G) Flow cytometry assessment of mesh-infiltrating cells. Total leukocytes and the GFP+ fraction (D) are shown. Statistical data show absolute cell counts of total leukocytes (CD45+) and monocyte-derived macrophages (GFP+Ly6G–CD11b+F4/80+). (E) GFP+ wells were subdivided into classical myeloid cell populations showing macrophage-like cells as CD11b+F4/80+ cells and activated DC-like cells as CD11c+CD86+ cells. (F and G) t-SNE analysis of myeloid cell populations infiltrating murine abdominal meshes isolated from CCR2.gfp and Ccr2–/–.gfp reporter mice at day 7 (F) and day 21 (G) after mesh implantation. Cells were pregated on CD45+GFP+ cells, and 2 × 104 cells of each genotype were concatenated and subjected to t-SNE analysis. CCR2-competent cells are highlighted in red/orange, knockout cells are highlighted in blue. Detailed expression marker profile of cells shown in the color-coded histograms. MAM, mesh-associated macrophage; SIM: stroma-infiltrating macrophage. Error bars represent mean ± SD. Student’s t test: *P < 0.05; **P < 0.01; ***P < 0.001.
Flow cytometry analysis in Ccr2.gfp and Ccr2–/–.gfp animals revealed a steady population of GFP+ cells within the mesh-associated CD45+ population in Ccr2.gfp mice. While overall numbers of CD45+ as well as monocyte-derived macrophages (CD45+Ly6G–CD11b+) were similar in Ccr2–/–.gfp animal compared with the Ccr2.gfp control animals at day 7, there was a significant decrease of macrophages detected in CCR2-deficient mice at day 21, while the CCR2-competent animals showed a massive rise in myeloid cell numbers within the mesh-surrounding tissue (Figure 7, D and E). Myeloid cells retrieved from Ccr2–/–.gfp mice furthermore exhibited a defect in maturation and a retention in their monocytic stage, lacking the expression of F4/80, the activation marker CD86, and, to a lesser extent, CD11c, while still remaining positive for CD11b (Figure 7E).
Cells were further analyzed in an unbiased t-SNE approach by pregating on the GFP+ cells isolated from Ccr2.gfp and Ccr2–/–.gfp mice. Comparing the mesh-implanted WT animals (Figure 3) and the two predominant clusters observed in the Ccr2.gfp animals, MAMs as well as SIMs could be clearly identified in the GFP+ population. Strikingly, both macrophage populations were markedly reduced in Ccr2–/–.gfp mice (Figure 7F). At day 7, the MAM population (Figure 7F, dark green) expressing CD11b+CD11c–F4/80–IAb–CX3CR1–Ly6C+/–CD103–CD86intSIGLEC-H– resembled a more immature monocyte phenotype in CCR2-competent mice, while the more prominent SIM cluster (Figure 7F, pink) expressing CD11b+CD11cintF4/80hiIAb+ CX3CR1+Ly6C+/–CD103+CD86+SIGLEC-H+ appeared to be more mature, with both macrophage and also DC like features. In contrast, the populations most prominent in Ccr2–/–.gfp mice either expressed CD11b+CD11c–F4/80–IAb+CX3CR1+Ly6C–CD103–CD86+SIGLEC-H– (Figure 7F, light blue) and therefore most likely represent partially activated Ly6Clo monocytes or expressed CD11b+CD11cloF4/80loIAbloCX3CR1lo/intLy6C+CD103+ CD86+SIGLEC-H+ (Figure 7F, light green), similar to the matured myeloid cells (SIMs) from the Ccr2.gfp animals, albeit with some clear maturation deficits expressing markedly lower levels of F4/80, IAb, CX3CR1, and CD86.
On day 21 (Figure 7G), the matured SIM population remained the dominant population in the mesh-implanted Ccr2.gfp animals (Figure 7G, pink) but showed a downregulation of SIGLEC-H. Furthermore, a subcluster within the SIM population (Figure 7G, light blue) expressing CD11b+CD11c+F4/80+IAb+CX3CR1+Ly6C–CD103+CD86+SIGLEC-H– appeared, likely representing a further stage of maturation. The MAM macrophages (Figure 7G, dark green) were also more prominent within the GFP+ cells derived from Ccr2.gfp animals compared with Ccr2-deficient mice at day 21, albeit not as clearly as on day 7. In contrast, Ccr2–/–.gfp mice contained mainly cells with a phenotype resembling immature monocytes (Figure 7G, light green). Thus, CCR2 is a key molecule in the recruitment of myeloid cells that give rise to inflammatory cells with a mixed morphology, resembling in part both macrophages and DCs, while, in Ccr2–/–.gfp mice, the myeloid cells appeared less differentiated, with lower levels of CD11c, F4/80, and other maturation markers.
CCR2-dependent myeloid cells promote fibroblast activation and proliferation in FBRs.
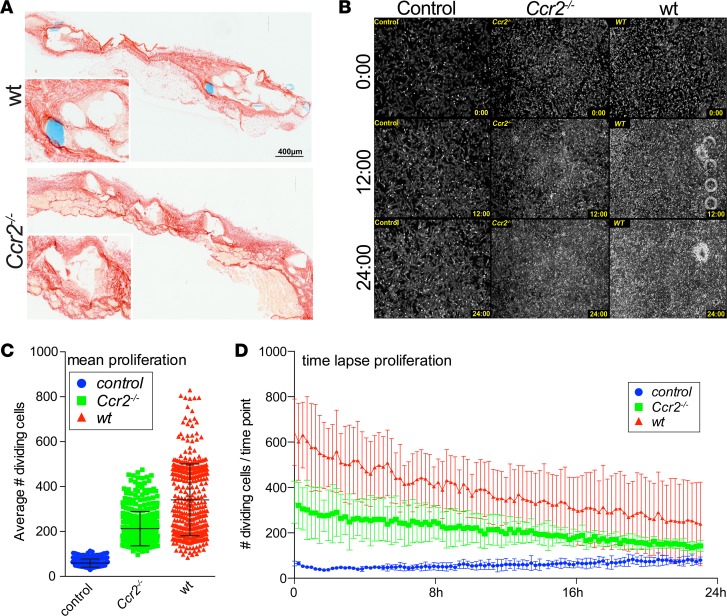
From models of organ fibrosis, it is well established that CCR2-dependent, monocyte-derived macrophages activate fibrogenic cells (10). The activation of fibroblasts is also crucial for wound healing and implant acceptance (22), e.g., via factors such as MMP9, MMP13, and TGF-β. Therefore, we investigated the functional role of CCR2-dependent myeloid cells in the FBR with regard to fibroblast activation. Histological assessment of extracellular matrix deposition revealed no significant difference between Ccr2.gfp and Ccr2–/–.gfp mice at day 21 following mesh implantation, despite an attenuated infiltration of mononuclear cells (Figure 8A and Supplemental Figure 5A), demonstrating that the amount of scarring was not significantly affected by the lack of infiltrating CCR2+ monocytes (Supplemental Figure 5A). However, we hypothesized that Ccr2–/– mice might have an attenuated course of scar deposition due to reduced fibroblast activation. We therefore performed in vitro time-lapse microscopy of NIH/3T3 fibroblasts in coculture with 0.5 cm2 mesh explants (day 21) derived from WT or Ccr2–/– donors to investigate the response to the stimuli secreted by the cells within the explanted tissue. Cell division was determined using phase-contrast microscopy (Supplemental Video 10 and Supplemental Figure 5B). WT mesh explants as well as tissue derived from Ccr2–/–.gfp animals triggered fibroblast proliferation compared with untreated controls, albeit to a different extent. While NIH/3T3 cells became confluent within 12 hours of the imaging when cultured with WT explants, confluency was obtained at around 20–24 hours with the Ccr2‑/– derived tissue (Figure 8, B–D, and Supplemental Video 10). In line, the average fibroblast cell division as well as the cell division over time increased in both coculture conditions, again showing a stronger effect for the WT versus the Ccr2–/–.gfp mesh explants. Thus, our data indicate that myeloid cells strongly promote fibroblast activation and proliferation, as mesh tissue explants derived from CCR2-deficient animals were less capable stimulating fibroblast growth compared with WT explants.
Figure 8. Loss of CCR2 impairs fibroblast growth but does not hamper wound healing following mesh implantation.
(A) Sirius red staining of mesh explants isolated from WT and Ccr2–/– mice on day 21 after implantation. Inset images show magnified mesh-surrounding extracellular matrix. Scale bar: 400 μm; ×2 (inset). (B–D) In vitro fibroblast activation by mesh-explanted tissue. 5 × 104 NIH/3T3 fibroblasts were plated and grown for 12 hours initially. 0.5 cm2 explanted mesh derived from WT and Ccr2–/– mice isolated on day 21 after implantation was added to the cell cultures, and cells were allowed to grow for another 12 hours. Fibroblast activation was determined by in vitro time-lapse microscopy. (B) Phase-contrast images of fibroblast cell cultures. Cells were imaged continuously for a time period of 24 hours, showing control cells without addition of mesh explants (left) and cells in coculture with mesh tissue derived from Ccr2–/– mice (middle) and from WT control animals (right). (C and D) Statistical analysis of cell division detected by phase-contrast changes visualized in B. Cell division was calculated, determining the statistical mean of dividing cells visible in each time point (C) and also displayed over time (D) for control cells (blue), cells cultured with tissue from Ccr2–/– mice (green), and WT control mesh tissue (red). Error bars represent mean ± SD.
Discussion
FBR is a common complication following biomaterial implantation associated with chronic inflammation (23); it is mainly driven by macrophages orchestrating wound healing but also granulomatous responses (24). Albeit, materials such as the polypropylene meshes have been described to be immunogenic per se (25, 26), long-term inflammatory responses have been mainly linked to infectious complications following the mesh implantation (9). In the present study, we show an active macrophage-driven inflammatory response in patients, even without imminent complications associated to the implant. Histology revealed high numbers of CD68+ cells in the immediate surroundings of the mesh fibers, which contained high levels of CD14+ cells expressing HLA-DR, CD64 (FcgRI), IRF5, and CD86, indicating a myeloid cell–driven chronic inflammation in humans. By investigating the FBR following PP mesh implantation in mice, we could show a high and continuous response of macrophages especially following mesh implantation over a period of up to 90 days. Our study extends the conventional histological descriptions of FBR (27), indicating a strong, chronic innate immune response with mononuclear cell infiltrates at all time points, the lack of a switch to an antiinflammatory response, and little formation of FB-associated giant cells, as revealed by extensive flow cytometric and histological immunophenotyping in mice and humans.
While we noted a clear accumulation of macrophages with an inflammatory phenotype in the immediate vicinity of the mesh fibers, we also identified a second population of macrophages characterized by high expression levels of F4/80, CD16 (FcgRIII), and I-Ab (MHC-II), which marked them as cells specialized for scavenging and antigen presentation. Detailed cell phenotyping showed the expression of activation markers such as CD80 on F4/80hi cells, indicating an immunogenic rather than wound-healing phenotype, as suggested earlier (28). Interestingly, these cells also expressed CD103, a marker commonly associated with antigen cross presentation in myeloid cells, suggesting that the FBR associated with PP mesh implantation leads to a chronic immune reaction driven by inflammatory macrophages, expressing high levels of CD80, CD16, and TNF, without relevant attenuation over time. In support of this interpretation, intravital TPLSM following adoptive transfer of DsRed+ BM cells revealed a high motility of the infiltrating cells at all time points and a half-life time of the murine myeloid cells of between 14 and 20 days, indicating a constant replacement and no long-term survival of the myeloid cells within the FB infiltrates.
Genes associated with cell survival, such as Bcl-2, as well as specific macrophage survival factors, such as Csf-1 (Mcsf) and Cx3cl1 (29), were markedly downregulated, alongside markers, such as Vegf, Pdgf, Stat2, and Stat6 (30), associated with a wound-healing macrophage response. This further supports the hypothesis of a chronic inflammation with high turnover and constant recruitment of myeloid cells, rather than a differentiation of the infiltrating cells into long-lived tissue macrophages.
Chronic inflammatory reactions are generally dependent on danger signals or alarmins (3), e.g., cellular ATP, genomic DNA, uric acid, and factors actively synthesized, such as HMGB-1, IL-33, or S100 proteins, all leading to an activation of downstream intracellular signaling, inflammasome activation, and subsequent release of IL-1b, IL-6, TNF, MCP-1, and other proinflammatory cytokines or chemokines (31). A majority of these mediators was readily detectable in explanted mesh tissue, including S100A8/9 (32–34) and markers of inflammasome activation (35). This suggests that these factors are key drivers of chronic FB inflammation and polarization of inflammatory macrophages, e.g., via interaction with surface receptors, such as CD68, TLR4, and RAGE (32–34). In addition to the inflammatory gene cascades upregulated in the FB response, the deposition of immunoglobulins and subsequent activation of the complement cascade, which has been previously noted in in vitro studies using whole-blood models in contact with various biomaterial surfaces (36), are likely to be key drivers of the chronic inflammatory response perpetuating the myeloid cell response. Our study provides clear in vivo evidence for the deposition and activation of humoral immune factors at solid biomaterial surfaces following implantation.
IL-4/IL-13–dependent formation of FB giant cells is one potential escape mechanism for myeloid cells to prevent cell death associated with anoikis (an impairment of macrophage survival due to the lack of adherence to extracellular matrix) (9, 37). Giant cell formation primarily arises from macrophages undergoing abortive cell cycles (38), rather than cellular fusion, as suggested earlier (9, 37, 39, 40).
CCR2 is a key chemokine in myeloid cell recruitment through the promotion of inflammatory Ly6Chi monocyte egression from the BM in mice (41, 42). Intravital TPSLM as well as flow cytometric analyses revealed a massive recruitment of highly motile CCR2+ cells to the sites of mesh implantation in Ccr2-reporter mice. In contrast, Ccr2–/–.gfp mice showed no substantial infiltration of GFP+ cells, arguing for a critical role of CCR2 in the infiltration of inflammatory monocytes. However, Ly6G–CD11b+ myeloid cells could still be found, even in the absence of CCR2 signaling. While cells were present at much lower numbers, these data indicate alternative pathways bypassing the conventional CCR2-mediated monocyte recruitment. Remarkably, myeloid infiltrates in CCR2-deficient mice exhibited a less activated phenotype, presumably due to the general attenuation of the inflammatory response or to a specific role of CCR2 for macrophage and DC maturation (43).
Even though the myeloid cell infiltration was substantially hampered by the loss of CCR2, there was no apparent difference in the net collagen deposition over 21 days in our model, arguing that the blocking of CCR2 may suffice as a potential therapeutic strategy to specifically limit FB-induced inflammation without disrupting the overall process of wound healing. This view was further supported by the macroscopic observation that the CCR2-knockout animals also did not exhibit prolonged wound healing following the surgical incisions performed for the implantations. However, interactions between macrophages and the fibroblast response have been documented before (44, 45), emphasizing the necessity of myeloid cells to orchestrate the wound healing response (46, 47). As we demonstrate by the in vitro fibroblast activation assay, tissues derived from CCR2-knockout animals exhibited a lower potential for fibroblast activation compared with the corresponding WT controls (8, 10) but still induced enhanced fibroblast activation compared with the untreated control cells. Therefore, CCR2 could potentially be an attractive pharmacological target to attenuate FB-associated inflammatory responses without interfering with the general formation of scar tissue and the healing process, ultimately leading to a stable encapsulation and incorporation of the biomaterial. In conclusion, our study identified cellular and molecular mechanisms of FBR to implanted hernia repair meshes related to the chronic accumulation and activation of myeloid cells, thereby providing the basis for developing either less immunogenic biomaterial or adjunct strategies to dampen persistent inflammation.
Methods
Contact for reagent and resource sharing
Information regarding reagents can be found in the Supplemental Methods Reagents. Further information and requests for reagents may be directed to authors Felix Heymann (fheymann@ukaachen.de) or Frank Tacke (frank.tacke@gmx.net).
Experimental model and subject details
Mice.
C57BL/6, Ccr2.BAC-gfp (48), and Ccr2–/– (49) mice crossed with Ccr2.BAC-gfp (Ccr2–/–.gfp) mice were housed in a pathogen-free environment (Supplemental Table 5). Experiments were performed with age- and sex-matched animals at 8–14 weeks of age. Surgery for mesh implantation using the onlay technique was performed following anesthesia using ketamine/xylazine (100 mg/kg and 10 mg/kg i.p.) and prior analgesic treatment using buprenorphine (0.1 mg/kg), followed by subsequent s.c. injections every 6–8 hours over the first 5 days following implantation. For implantation, the abdominal skin was shaved, disinfected, and opened with a 0.5- to 1-cm incision. Subcutaneous connective tissue was carefully removed to separate the skin from the abdominal wall. 2 cm2 of Prolene polypropylene mesh (Ethicon) was instilled on top of the lower abdomen, and the skin was stitched using a 5-0 Prolene surgical thread (Ethicon). To prevent microbial infection following implantation, the mice were treated with 0.1% Borgal (Virbac) in drinking water over a period of 7 days. For explantation, the mice were euthanized, and the skin was reopened at the indicated time points. The mesh was carefully detached from the skin and explanted, including the underlying abdominal tissue. Female mice were used for all experiments because of the sterical hindrance of the preputial gland in male mice. For adoptive cell transfers, BM cells were purified from sex-matched Actin-DsRed donor animals (The Jackson Laboratory), and 2 × 107 cells were injected i.v. into recipient mice before the imaging and flow cytometry experiments described below.
Human tissue samples.
Mesh tissue was collected from patients explanted >1 year following mesh implantation. For histological analysis, explants were stored in 4% formalin solution until further processing and stained either with H&E or for immunofluorescence markers. For flow cytometry, mesh samples were initially digested with collagenase type-IV (Worthington) at 37°C for 30 minutes, following explantation and subsequent mechanical disruption by cutting the tissue in fragments <2 mm2. Cells were filtered using a 70-μm cell strainer (Falcon) and stained for flow cytometric analysis. Cells were pregated on total CD45+CD56– cells to identify leukocytes and exclude NK cells from the analysis. Markers for phenotyping and activation were stained as indicated.
Flow cytometry analysis and t-SNE analysis
Flow cytometric analysis of intrahepatic leukocytes was performed as described previously (7). Total cell numbers were determined by adding fixed numbers of Calibrite APC beads (BD) before measurement as internal reference. Leukocyte isolation from mesh-associated tissues was performed by initial digestion with Collagenase type-IV (Worthington) at 37°C for 30 minutes following explantation and mechanical disruption by cutting the tissue in fragments <2 mm2. The tissue was further homogenized carefully by pipetting and subjected to a second digestion step for 15 minutes. Extracts were filtered using 70-μm cell strainers (Falcon). Staining was performed in PBS with 2 mM EDTA and 0.1% BSA using combinations of the following monoclonal antibodies (Supplemental Table 5): I-Ab and Ly6G (BD Bioscience); CD11b, CD11c, CD16/32, CD45, CD80, CD86, CD103, F4/80, IL-10, IL-12p35, Ly6C, SIGLEC-H, and TNF (eBioscience); and CX3CR1 (Biolegend). If not indicated otherwise, cells were pregated on FSC/SSC and CD45+ to identify leukocytes and those cells that were negative for 7-AAD to exclude dead cells from the analysis. Neutrophils were gated as CD11b+Ly6G+ cell fraction and were gated out for the subsequent analysis of the remaining myeloid subpopulations. Analysis was performed using a FACS Fortessa (BD). Data were analyzed using FlowJo 10.2 (TreeStar). For generation of cluster plots cells were pregated on CD45+Ly6G–, and populations derived from several animals of each group were concatenated and subjected to simultaneous t-SNE analysis. Parameters displayed were as follows: iterations, 1000; perplexity, 50; and Eta, 200. Cluster formation was validated using a different set of t-SNE parameters: iterations, 500; perplexity, 30; and Eta, 200. Cluster sets were compared, and only clusters appearing under both conditions were used for further analysis, depicting their marker expression profiles by generating overlay histograms to assess their phenotype and functional profile.
Intracellular staining of cytokine expression
For intracellular staining, cells were cultured overnight in RPMI-1640 containing 10% FCS and 1:1000 BrefeldinA (eBioscience) to prevent cytokine secretion following explantation and isolation into single cell cultures as described above for the flow cytometric analysis. All buffers used during the staining were kept at 4°C to prevent reactivation of the cells before the fixation. The staining was performed in two steps. Initially, surface antigen staining was performed at 4°C as described above and cells were subsequently fixed using 2% formalin. For permeabilization, cells were washed twice in PBS with 0.5% saponine (MilliporeSigma), stained for intracellularly trapped cytokines, and afterward washed twice using the same buffer.
Histopathology and immunofluorescence histology
Conventional H&E and Sirius Red stainings were performed according to standard protocols using an autostainer. For immunofluorescence histology, mesh explants were cut into consecutive 7-μm cryosections, dried for adhesion, and rehydrated in PBS. Sections were mounted on adhesive slides (Klinipath) to prevent detachment during the antibody staining. Dual immunofluorescence stainings were performed against F4/80, CD11c, I-Ab, Ly6G, CD80, and CD16, all against CD11b either overnight at 4°C or at room temperature for 2 hours in a dark chamber (Supplemental Table 5). Nuclei were stained using Hoechst 33258 (0.1 mg/ml) following the antibody staining. Sections were mounted using a specialized antifading mounting media (Thermo Fisher Scientific). Imaging was carried out using an Axio Observer Z1 (Zeiss). Images were shading corrected and subsequently assembled from stitched tiles using ZEN blue (Zeiss) to allow high-resolution imaging in combination with large fields of view.
Intravital time-lapse laser-scanning microscopy
For intravital TPLSM imaging, animals were initially anesthetized using ketamine/xylazine (100 mg/kg and 10 mg/kg i.p.) in combination with analgesic treatment using buprenorphine (0.1 mg/kg), according to the animal welfare guidelines of The Society for Laboratory Animal Science, followed by tracheotomy (50) and controlled respiration with 2.5% isoflurane in 100% O2 over the time course of the experiment. Analgesia and anesthesia were maintained by i.p. administration of 50 μl/h ketamine (100 mg/kg)/xylazine (2.5 mg/kg)/buprenorphine (0.1 mg/kg) using a syringe pump. For hydratization, 100 μl/h PBS was infused via the same route. Meshes were surgically exposed by removal of the covering skin and were fixed in position using 3% PBS agarose using a custom made tray system. GFP and DsRed were imaged at 900 nm excitation wavelength using 525/50 and a 593/40 band-pass filters. Imaging was performed over a time period of up to 3 hours, and microscopy was performed using a Laviosion Trimscope I (Lavision Biotec), scanning at least 3 view fields per animal over time with a penetration depth between 70 and 150 μm, depending on the tissue, with a spatial resolution of 512 × 512 pixels per view field (resembling 0.5 mm2) and a time increment of approximately 5 minutes per frame. Focal correction was performed manually, if required, though the implant was virtually stable over the period of imaging. Data reconstruction and analysis were performed using Imaris 7.7 (Bitplane), including the XT and the cell tracking package and Fiji version 1.48s. Signal enhancement was performed using Ilastik 1.2. Tracking was performed by initial spot tracking and autoregressive motion tracking with a maximum track length of 30 μm per frame.
Nanostring gene expression array
Gene expression analysis was carried out using a custom 72-plex NanoString assay (NanoString Technologies Inc.). RNA was extracted from explanted mesh tissues using conventional PeqGold (Peqlab) phenolic extraction followed by chloroform-based protein extraction and isopropanol precipitation of total RNA. RNA was hybridized with the probe set according to the manufacturer’s protocols. Data processing was performed using NSolver (NanoString Technologies Inc.) followed by subsequent calculation of differential gene expression between mesh-implanted and sham-operated groups.
Protein mass spectrometry
To isolate proteins for mass spectrometry analysis, 20-μm tissue sections were cut from cryopreserved tissue isolates. For each sample, a minimum of 3 slides with at least 6 sections per slide were prepared. Samples were then stained for 1 minute using a 1% Cresylviolet solution (solved in 50% v/v ethanol), followed by subsequent washing in 70% and 96% ethanol. Mesh tissue was isolated by scraping away the majority of the underlying muscular tissue while leaving intact the extracellular matrix and immune cell infiltrates. For sham-operated animals, abdominal wall sections were used as control tissues and treated accordingly. Tissues were then mechanically harvested from the slides and transferred to Eppendorf tubes and lysed in 30 μl lysis buffer (8 M urea in 50 mM ammonium bicarbonate with 1× phosphatase and protease inhibitors, Serva) for 15 minutes on ice, followed by centrifugation for 20 minutes (4°C, 14,000 g). The lysates (25 μl each) were then separated by SDS-PAGE on 4%–20% gradient gels (Serva). The gels were stained with Coomassie Brilliant Blue. 8–10 gel slices per lane were excised and subjected to in-gel tryptic digestion (51). After digestion, the samples were desalted using homemade C18 tips and dried in a speed vac.
The lyophilized peptides were resuspended in 15 μl 3% FA/5% ACN and then subjected to mass spectrometry analysis. First, the peptides were trapped on a C18 precolumn (Acclaim PepMap100, C18, 5 μm, 100 Å, 300 μm i.d. × 5 mm, Thermo Scientific) using buffer A (0.1% FA) on a nanoLC system (RSLCnano, Thermo Scientific). Subsequently, the peptides were separated on an Easyspray C18 analytical column (Thermo Scientific) coupled with the Easyspray source (2-μm particle size, 75-μm inner diameter, 50 cm length, 40°C column oven temperature, 2 kV; Thermo Scientific) using 80- or 90-minute gradients. The applied gradients were (80 min) as follows: 0–5 minutes, 5% buffer B (80% ACN/0.1% FA); 5–45 minutes, 5%–35% B; 45–52 minutes, 35%–45% B; 52–53 minutes, 45%–99% B; 53–58 minutes, 99% B; 58–59 minutes, 99%–5% B; 59–80 minutes, 5% B. Additional applied gradients were (90 minutes) as follows: 0–10 minutes, 5% buffer B (80% ACN/0.1% FA); 10–45 minutes, 5%–35% B; 45–55 minutes, 35%–50% B; 55–58 minutes, 50%–90% B; 58–63 minutes, 90% B; 63–64 minutes, 99%–5% B; 64–90 minutes 5% B. Mass spectrometry was performed on a Q Exactive Plus instrument (Thermo Scientific) in data-dependent mode. For full MS settings, we used the following parameters: 70,000 resolution; AGC target, 3e6; maximum injection time, 100 milliseconds; scan range, 300–1750 m/z. We used the following dd-MS2 settings: 17,500 resolution; AGC target, 2e5; maximum injection time, 110 milliseconds; top 10 precursor fragmentation; isolation window, 1.8 m/z; collision energy, 27. We used the following dd settings: minimum AGC, 5e2; 10 second dynamic exclusion; only 2+ to 5+ peptides were allowed.
The raw data were analyzed using MaxQuant (1.5.1.2, ref. 52) and the built-in Andromeda search engine (53). The spectra were searched against the mouse SwissProt database (version 10/2017) (only reviewed and canonical sequences), including the contaminants function of MaxQuant. MaxQuant default settings (including mass tolerance) were used. Specific settings included protease, trypsin (2 missed cleavages); fixed modification, carbamidomethylation; variable modifications, oxidation (Met) and N-terminal protein acetylation as; false discovery rate, 0.01 for both peptide and protein level; minimum peptide length, 7 amino acids.
FISH
FISH was performed according to standard protocols. Briefly, cryosections were fixed in 4% PFA for 10 minutes, treated with 0.1% Sudan Black B in 70% ethanol for 20 minutes, and afterward were transferred into hybridization buffer (0.6 M NaCl, 0.68 mM sodium citrate, 10% formamide, 0.1% SDS, 1% BSA, pH 7.0) at 65°C for 30 minutes. For denaturation of RNA, the sample was heated to 75°C for 2 minutes and incubated with Nlrp3 specific probe (5 ng/ μl) inside a wet chamber overnight at 65°C (probe sequence: 5′AGATACCATACGGTCCTCCTG-FITC). Consecutive washings were performed in hybridization buffer (at 65°C room temperature) for 30 minutes each. After an additional washing in TBS 0.3% Triton X-100 for 15 minutes, slides were counterstained with Hoechst33258 and covered with mounting medium.
TUNEL apoptosis staining
TUNEL was performed according to standard protocols using the DeadEnd Fluorimetric TUNEL system (Promega). Briefly, 7-μm cryosections were fixed initially using 4% paraformaldehyde and washed with PBS and 0.2% Triton X-100. DNA strands were labeled using fluorescent fluorescein-12-dUTP added to the 3′ end of the DNA by terminal desoxyribonucleotidyl transferase for 60 minutes. Nuclei were stained with Hoechst33258 (1 μg/ml) for 15 minutes.
Quantitative reverse transcription real-time PCR
RNA was purified using conventional PeqGold (Peqlab) phenolic extraction followed by chloroform-based protein extraction and isopropanol precipitation of total RNA. cDNA was synthesized using a Transcriptor First Strand cDNA synthesis kit (Roche). Quantitative SYBR Green–based (Invitrogen) real-time polymerase chain reaction was performed using an ABI7300 real time cycler (Applied Biosystems). Reactions were performed in duplicates, and gene expression was normalized based on β-actin. Primer sequences are provided in the Supplemental Methods Reagents.
Fibroblast coculture
1 × 105 NIH/3T3 fibroblasts were seeded into conventional 6-well cell culture plates and left overnight for adhesion. At the following day, 0.5 cm2 mesh explants were added to the culture plates and were left for 24 hours to respond to the stimuli secreted by the cells within the explanted tissue. The fibroblast response was assessed for the subsequent 24 hours by time-lapse microscopy, Images were taken at an interval of 15 minutes from at least 3 independent fields of view per condition. Imaging was performed using an Axio Observer Z1 (Zeiss) with a climate chamber unit, providing a stable environment (37°C and 5% CO2) over the time course of the experiment. Data analysis was carried out using Fiji version 1.48s; proliferating cells were identified by changes in phase contrast by automated particle analysis following image conversion into binary black and white images and subsequent thresholding to omit nondividing cells from the analysis. Videos were carefully examined to rule out induction of apoptosis on a large scale to rule out false positive signals.
Statistics
Statistical analysis and video microscopy analysis.
Experimental statistics and replicates as well as significance and precision measures (mean ± SEM) are stated in the figure legends. P < 0.05 following 2-tailed unpaired Student’s t test indicates statistically significance. Statistical analysis was performed in PRISM 6 (GraphPad). For intravital microscopy, cells were counted using a spot-tracking algorithm in Imaris 7.7 (Bitplane). For GFP signal enhancement, images were processed with Ilastik 1.2 using a graphical training algorithm prior to spot tracking and quantification analysis.
Data and software availability
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (54), with the data set identifier PXD009944.
The proteinGroups.txt results derived from the MaxQuant search was further analyzed using Perseus (52). The protein list was filtered for reversed hits, contaminants, and those “only identified by site.” Further requirements for protein inclusion included minimum 1 unique peptide and 2 total peptides (razor+unique). The 3 individual biological replicates were grouped into their corresponding groups (mesh day 7, mesh day 21, sham day 7, sham day 21). Proteins were only included in the final data set if they were identified in all replicates in at least one group. The median of the 3 replicates was taken for the calculation of the individual ratios. Ratios (e.g., mesh day 7 vs. sham day 7) were normalized against the respective median of the ratios (see Supplemental Table 4).
Protein network analysis was performed by performing using STRING (https://string-db.org/). Resulting enrichment process data sets were filtered for the GO pathways associated with stress response, general immunology, humoral and cellular immune activation, inflammation, and wound healing (Supplemental Tables 2 and 3), with a FDR ≤ 0.01 and a cluster of at least 5 proteins. Data were further isolated using Cytoscape 3.6.1. Pathway enrichment analysis was performed using the BiNGO plug-in followed by an enrichment map analysis, using a FDR cutoff of 0.01 and a P value of 0.1 and a Jaccard cutoff of 0.48. String network generation was performed using the StringApp plug-in followed by functional enrichment mapping of the GO pathways.
Custom plug-ins for imaging analysis using Imaris were created using Matlab and implemented into Imaris.
Study approval
Experiments were performed under ethical conditions approved according to German legal requirements (State Agency for Nature, Environment and Consumer Protection in North-Rhine Westphalia). Mesh tissue was collected as approved by the local ethics committee at the University Hospital Aachen.
Author contributions
FH performed the major part of the experimental work. FH, PLJ, and FT were responsible for the conception and writing of the manuscript as well as the design of the individual experiments. KTVT performed additional experiments and participated in the preparation of the manuscript. CP and MG were responsible for the preparation of the protein mass spectrometry data. FH and CP were responsible for the proteomics analysis and interpretation. AAR, KTVT, MB, and UPN were responsible for the human tissue acquisition and human histopathology. LJG and AKF performed additional experiments. TL and CT were involved in the development of the overall study design and aided with the preparation of the manuscript. JC provided additional human mesh explants.
Supplementary Material
Acknowledgments
We thank the Two-Photon Microscopy and the Proteomics Core Facilities of the Interdisciplinary Center for Clinical Research (IZKF) Aachen. This work was supported by the German Research Foundation (DFG; Ta434/3-1 and SFB/TRR57) and IZKF Aachen. The study sponsors had no role in the study design or in the collection, analysis, and interpretation of data.
Version 1. 01/24/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(2):e123862. https://doi.org/10.1172/jci.insight.123862.
Contributor Information
Felix Heymann, Email: fheymann@ukaachen.de.
Christian Preisinger, Email: cpreisinger@ukaachen.de.
Anjali A. Roeth, Email: aroeth@ukaachen.de.
Melanie Geiger, Email: melanie.geiger@rwth-aachen.de.
Lukas Jonathan Geisler, Email: lgeisler@ukaachen.de.
Joachim Conze, Email: jconze@ukaachen.de.
Tom Luedde, Email: tluedde@ukaachen.de.
Christian Trautwein, Email: ctrautwein@ukaachen.de.
Marcel Binnebösel, Email: mbinneboesel@ukaachen.de.
Frank Tacke, Email: frank.tacke@gmx.net.
References
- 1.Tang L, Eaton JW. Inflammatory responses to biomaterials. Am J Clin Pathol. 1995;103(4):466–471. doi: 10.1093/ajcp/103.4.466. [DOI] [PubMed] [Google Scholar]
- 2.Rosch R, Junge K, Schachtrupp A, Klinge U, Klosterhalfen B, Schumpelick V. Mesh implants in hernia repair. Inflammatory cell response in a rat model. Eur Surg Res. 2003;35(3):161–166. doi: 10.1159/000070045. [DOI] [PubMed] [Google Scholar]
- 3.Kono H, Rock KL. How dying cells alert the immune system to danger. Nat Rev Immunol. 2008;8(4):279–289. doi: 10.1038/nri2215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fitzgibbons RJ, Forse RA. Clinical practice. Groin hernias in adults. N Engl J Med. 2015;372(8):756–763. doi: 10.1056/NEJMcp1404068. [DOI] [PubMed] [Google Scholar]
- 5.Sheikh Z, Brooks PJ, Barzilay O, Fine N, Glogauer M. Macrophages, foreign body giant cells and their response to implantable biomaterials. Materials (Basel) 2015;8(9):5671–5701. doi: 10.3390/ma8095269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jakubzick CV, Randolph GJ, Henson PM. Monocyte differentiation and antigen-presenting functions. Nat Rev Immunol. 2017;17(6):349–362. doi: 10.1038/nri.2017.28. [DOI] [PubMed] [Google Scholar]
- 7.Heymann F, et al. Hepatic macrophage migration and differentiation critical for liver fibrosis is mediated by the chemokine receptor C-C motif chemokine receptor 8 in mice. Hepatology. 2012;55(3):898–909. doi: 10.1002/hep.24764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Karlmark KR, et al. Hepatic recruitment of the inflammatory Gr1+ monocyte subset upon liver injury promotes hepatic fibrosis. Hepatology. 2009;50(1):261–274. doi: 10.1002/hep.22950. [DOI] [PubMed] [Google Scholar]
- 9.Anderson JM, Rodriguez A, Chang DT. Foreign body reaction to biomaterials. Semin Immunol. 2008;20(2):86–100. doi: 10.1016/j.smim.2007.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krenkel O, Tacke F. Liver macrophages in tissue homeostasis and disease. Nat Rev Immunol. 2017;17(5):306–321. doi: 10.1038/nri.2017.11. [DOI] [PubMed] [Google Scholar]
- 11.Dal-Secco D, et al. A dynamic spectrum of monocytes arising from the in situ reprogramming of CCR2+ monocytes at a site of sterile injury. J Exp Med. 2015;212(4):447–456. doi: 10.1084/jem.20141539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ramachandran P, et al. Differential Ly-6C expression identifies the recruited macrophage phenotype, which orchestrates the regression of murine liver fibrosis. Proc Natl Acad Sci USA. 2012;109(46):E3186–E3195. doi: 10.1073/pnas.1119964109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klinge U, Klosterhalfen B, Birkenhauer V, Junge K, Conze J, Schumpelick V. Impact of polymer pore size on the interface scar formation in a rat model. J Surg Res. 2002;103(2):208–214. doi: 10.1006/jsre.2002.6358. [DOI] [PubMed] [Google Scholar]
- 14.Lake SP, Ray S, Zihni AM, Thompson DM, Gluckstein J, Deeken CR. Pore size and pore shape--but not mesh density--alter the mechanical strength of tissue ingrowth and host tissue response to synthetic mesh materials in a porcine model of ventral hernia repair. J Mech Behav Biomed Mater. 2015;42:186–197. doi: 10.1016/j.jmbbm.2014.11.011. [DOI] [PubMed] [Google Scholar]
- 15.Conze J, et al. Polypropylene in the intra-abdominal position: influence of pore size and surface area. Hernia. 2004;8(4):365–372. doi: 10.1007/s10029-004-0268-8. [DOI] [PubMed] [Google Scholar]
- 16.Leber GE, Garb JL, Alexander AI, Reed WP. Long-term complications associated with prosthetic repair of incisional hernias. Arch Surg. 1998;133(4):378–382. doi: 10.1001/archsurg.133.4.378. [DOI] [PubMed] [Google Scholar]
- 17.Wolf MT, et al. Macrophage polarization in response to ECM coated polypropylene mesh. Biomaterials. 2014;35(25):6838–6849. doi: 10.1016/j.biomaterials.2014.04.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faulk DM, et al. ECM hydrogel coating mitigates the chronic inflammatory response to polypropylene mesh. Biomaterials. 2014;35(30):8585–8595. doi: 10.1016/j.biomaterials.2014.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Poppas DP, et al. Hydrogel coated mesh decreases tissue reaction resulting from polypropylene mesh implant: implication in hernia repair. Hernia. 2016;20(4):623–632. doi: 10.1007/s10029-016-1481-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hachim D, LoPresti ST, Yates CC, Brown BN. Shifts in macrophage phenotype at the biomaterial interface via IL-4 eluting coatings are associated with improved implant integration. Biomaterials. 2017;112:95–107. doi: 10.1016/j.biomaterials.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Blázquez R, Sánchez-Margallo FM, Álvarez V, Usón A, Casado JG. Surgical meshes coated with mesenchymal stem cells provide an anti-inflammatory environment by a M2 macrophage polarization. Acta Biomater. 2016;31:221–230. doi: 10.1016/j.actbio.2015.11.057. [DOI] [PubMed] [Google Scholar]
- 22.Narine K, et al. Growth factor modulation of fibroblast proliferation, differentiation, and invasion: implications for tissue valve engineering. Tissue Eng. 2006;12(10):2707–2716. doi: 10.1089/ten.2006.12.2707. [DOI] [PubMed] [Google Scholar]
- 23.Junge K, et al. Mesh biocompatibility: effects of cellular inflammation and tissue remodelling. Langenbecks Arch Surg. 2012;397(2):255–270. doi: 10.1007/s00423-011-0780-0. [DOI] [PubMed] [Google Scholar]
- 24.Klopfleisch R. Macrophage reaction against biomaterials in the mouse model - Phenotypes, functions and markers. Acta Biomater. 2016;43:3–13. doi: 10.1016/j.actbio.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 25.Di Vita G, Patti R, Sparacello M, Balistreri CR, Candore G, Caruso C. Impact of different texture of polypropylene mesh on the inflammatory response. Int J Immunopathol Pharmacol. 2008;21(1):207–214. doi: 10.1177/039463200802100123. [DOI] [PubMed] [Google Scholar]
- 26.García-Moreno F, et al. Comparing the host tissue response and peritoneal behavior of composite meshes used for ventral hernia repair. J Surg Res. 2015;193(1):470–482. doi: 10.1016/j.jss.2014.07.049. [DOI] [PubMed] [Google Scholar]
- 27.Klinge U, Dietz U, Fet N, Klosterhalfen B. Characterisation of the cellular infiltrate in the foreign body granuloma of textile meshes with its impact on collagen deposition. Hernia. 2014;18(4):571–578. doi: 10.1007/s10029-014-1220-1. [DOI] [PubMed] [Google Scholar]
- 28.Wolf MT, et al. Macrophage polarization in response to ECM coated polypropylene mesh. Biomaterials. 2014;35(25):6838–6849. doi: 10.1016/j.biomaterials.2014.04.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Karlmark KR, et al. The fractalkine receptor CX3CR1 protects against liver fibrosis by controlling differentiation and survival of infiltrating hepatic monocytes. Hepatology. 2010;52(5):1769–1782. doi: 10.1002/hep.23894. [DOI] [PubMed] [Google Scholar]
- 30.Brown BN, et al. Macrophage phenotype as a predictor of constructive remodeling following the implantation of biologically derived surgical mesh materials. Acta Biomater. 2012;8(3):978–987. doi: 10.1016/j.actbio.2011.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heymann F, Tacke F. Immunology in the liver--from homeostasis to disease. Nat Rev Gastroenterol Hepatol. 2016;13(2):88–110. doi: 10.1038/nrgastro.2015.200. [DOI] [PubMed] [Google Scholar]
- 32.Chen B, et al. S100A9 induced inflammatory responses are mediated by distinct damage associated molecular patterns (DAMP) receptors in vitro and in vivo. PLoS ONE. 2015;10(2):e0115828. doi: 10.1371/journal.pone.0115828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dessing MC, et al. The calcium-binding protein complex S100A8/A9 has a crucial role in controlling macrophage-mediated renal repair following ischemia/reperfusion. Kidney Int. 2015;87(1):85–94. doi: 10.1038/ki.2014.216. [DOI] [PubMed] [Google Scholar]
- 34.Okada K, et al. CD68 on rat macrophages binds tightly to S100A8 and S100A9 and helps to regulate the cells’ immune functions. J Leukoc Biol. 2016;100(5):1093–1104. doi: 10.1189/jlb.2A0415-170RRR. [DOI] [PubMed] [Google Scholar]
- 35.Malik AF, et al. Inflammasome components Asc and caspase-1 mediate biomaterial-induced inflammation and foreign body response. Proc Natl Acad Sci USA. 2011;108(50):20095–20100. doi: 10.1073/pnas.1105152108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nilsson B, Ekdahl KN, Mollnes TE, Lambris JD. The role of complement in biomaterial-induced inflammation. Mol Immunol. 2007;44(1-3):82–94. doi: 10.1016/j.molimm.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 37.Hachim D, LoPresti ST, Yates CC, Brown BN. Shifts in macrophage phenotype at the biomaterial interface via IL-4 eluting coatings are associated with improved implant integration. Biomaterials. 2017;112:95–107. doi: 10.1016/j.biomaterials.2016.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Herrtwich L, et al. DNA damage signaling instructs polyploid macrophage fate in granulomas. Cell. 2016;167(5):1264–1280. doi: 10.1016/j.cell.2016.09.054. [DOI] [PubMed] [Google Scholar]
- 39.Gordon S, Plüddemann A, Martinez Estrada F. Macrophage heterogeneity in tissues: phenotypic diversity and functions. Immunol Rev. 2014;262(1):36–55. doi: 10.1111/imr.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Klopfleisch R, Jung F. The pathology of the foreign body reaction against biomaterials. J Biomed Mater Res A. 2017;105(3):927–940. doi: 10.1002/jbm.a.35958. [DOI] [PubMed] [Google Scholar]
- 41.Jimenez F, et al. CCR2 plays a critical role in dendritic cell maturation: possible role of CCL2 and NF-kappa B. J Immunol. 2010;184(10):5571–5581. doi: 10.4049/jimmunol.0803494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Serbina NV, Pamer EG. Monocyte emigration from bone marrow during bacterial infection requires signals mediated by chemokine receptor CCR2. Nat Immunol. 2006;7(3):311–317. doi: 10.1038/ni1309. [DOI] [PubMed] [Google Scholar]
- 43.Chiu BC, et al. Impaired lung dendritic cell activation in CCR2 knockout mice. Am J Pathol. 2004;165(4):1199–1209. doi: 10.1016/S0002-9440(10)63380-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bank RA, Zandstra J, Room H, Petersen AH, van Putten SM. Biomaterial encapsulation is enhanced in the early stages of the foreign body reaction during conditional macrophage depletion in transgenic macrophage Fas-induced apoptosis mice. Tissue Eng Part A. 2017;23(19-20):1078–1087. doi: 10.1089/ten.tea.2016.0499. [DOI] [PubMed] [Google Scholar]
- 45.Blázquez R, Sánchez-Margallo FM, Álvarez V, Usón A, Casado JG. Surgical meshes coated with mesenchymal stem cells provide an anti-inflammatory environment by a M2 macrophage polarization. Acta Biomater. 2016;31:221–230. doi: 10.1016/j.actbio.2015.11.057. [DOI] [PubMed] [Google Scholar]
- 46.Holt DJ, Chamberlain LM, Grainger DW. Cell-cell signaling in co-cultures of macrophages and fibroblasts. Biomaterials. 2010;31(36):9382–9394. doi: 10.1016/j.biomaterials.2010.07.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jannasch M, et al. In vitro chemotaxis and tissue remodeling assays quantitatively characterize foreign body reaction. ALTEX. 2017;34(2):253–266. doi: 10.14573/altex.1610071. [DOI] [PubMed] [Google Scholar]
- 48.Serbina NV, Hohl TM, Cherny M, Pamer EG. Selective expansion of the monocytic lineage directed by bacterial infection. J Immunol. 2009;183(3):1900–1910. doi: 10.4049/jimmunol.0900612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Boring L, et al. Impaired monocyte migration and reduced type 1 (Th1) cytokine responses in C-C chemokine receptor 2 knockout mice. J Clin Invest. 1997;100(10):2552–2561. doi: 10.1172/JCI119798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Heymann F, et al. Long term intravital multiphoton microscopy imaging of immune cells in healthy and diseased liver using CXCR6.Gfp reporter mice. J Vis Exp. 2015;(97):52607. doi: 10.3791/52607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.von Kriegsheim A, Preisinger C, Kolch W. Mapping of signaling pathways by functional interaction proteomics. Methods Mol Biol. 2008;484:177–192. doi: 10.1007/978-1-59745-398-1_12. [DOI] [PubMed] [Google Scholar]
- 52.Tyanova S, Temu T, Cox J. The MaxQuant computational platform for mass spectrometry-based shotgun proteomics. Nat Protoc. 2016;11(12):2301–2319. doi: 10.1038/nprot.2016.136. [DOI] [PubMed] [Google Scholar]
- 53.Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M. Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res. 2011;10(4):1794–1805. doi: 10.1021/pr101065j. [DOI] [PubMed] [Google Scholar]
- 54.Vizcaíno JA, et al. The PRoteomics IDEntifications (PRIDE) database and associated tools: status in 2013. Nucleic Acids Res. 2013;41(Database issue):D1063–D1069. doi: 10.1093/nar/gks1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21(16):3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry proteomics data have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (54), with the data set identifier PXD009944.
The proteinGroups.txt results derived from the MaxQuant search was further analyzed using Perseus (52). The protein list was filtered for reversed hits, contaminants, and those “only identified by site.” Further requirements for protein inclusion included minimum 1 unique peptide and 2 total peptides (razor+unique). The 3 individual biological replicates were grouped into their corresponding groups (mesh day 7, mesh day 21, sham day 7, sham day 21). Proteins were only included in the final data set if they were identified in all replicates in at least one group. The median of the 3 replicates was taken for the calculation of the individual ratios. Ratios (e.g., mesh day 7 vs. sham day 7) were normalized against the respective median of the ratios (see Supplemental Table 4).
Protein network analysis was performed by performing using STRING (https://string-db.org/). Resulting enrichment process data sets were filtered for the GO pathways associated with stress response, general immunology, humoral and cellular immune activation, inflammation, and wound healing (Supplemental Tables 2 and 3), with a FDR ≤ 0.01 and a cluster of at least 5 proteins. Data were further isolated using Cytoscape 3.6.1. Pathway enrichment analysis was performed using the BiNGO plug-in followed by an enrichment map analysis, using a FDR cutoff of 0.01 and a P value of 0.1 and a Jaccard cutoff of 0.48. String network generation was performed using the StringApp plug-in followed by functional enrichment mapping of the GO pathways.
Custom plug-ins for imaging analysis using Imaris were created using Matlab and implemented into Imaris.