Abstract
Systemic lupus erythematosus (SLE) is a highly variable autoimmune disease that can involve severe organ-threatening symptoms, such as lupus nephritis. Certain drugs, such as mycophenolate mofetil (MMF), are effective at reducing morbidity associated with nephritis; however, the immune pathways associated with disease suppression are poorly defined. Here, we provide evidence that MMF inhibits phosphorylation of STAT3 and other associated immune pathways. Using mass cytometry and bead-based or ELISA assays, the systemic phenotype of SLE patients not taking (MMF–) or taking (MMF+) MMF were studied. MMF+ SLE patients had significant reductions in total numbers of transitional B cells, plasmablasts, and T cells, specifically CD4+ Th17-type and CD4+ Treg-type cells, compared with MMF– patients. Plasma soluble mediators were decreased in MMF+ patients including chemokines (MIG/CXCL9 and SDF-1α/CXCL12) and growth factors (VEGF-A and PDGF-BB). Soluble mediators and cell subsets grouped by functional properties revealed significant modifications associated with STAT3 and B cell pathways. Further, healthy PBMCs treated with IL-6 revealed a reduction in p-STAT3 following the addition of mycophenolic acid (the active metabolite of MMF). In conclusion, the inhibition of STAT3 phosphorylation by MMF may explain the effectiveness of this treatment in SLE patients, since increased levels of p-STAT3 are associated with disease pathology.
Keywords: Autoimmunity, Immunology
Keywords: Drug therapy, Lupus, Signal transduction
Mycophenolate mofetil treatment in systemic lupus erythematosus patients systemically reduces pro-inflammatory pathways associated with the STAT-3 signaling molecule.
Introduction
Systemic lupus erythematosus (SLE) is a chronic, systemic autoimmune disease, characterized by a heterogeneous clinical presentation driven by autoreactive lymphocytes and proinflammatory cytokine production. Lupus inflammation can often lead to renal injury and multiple organ damage even when treated (1). Mycophenolate mofetil (MMF) is an immunosuppressive agent that was first shown to be effective in preventing organ rejection in renal transplant patients, but more recently was found to be efficacious in lupus, especially in patients with lupus nephritis (2, 3). Compared with cyclophosphamide, MMF was found to elicit a similar reduction in disease signs and symptoms in patients with severe lupus, but with a safer side-effect profile (4, 5). MMF is now considered a standard treatment for lupus nephritis, while further investigation into its usefulness for nonrenal manifestations is underway (6). Despite the known safety and utility of MMF in lupus treatment, the systemic impact of MMF treatment on cellular subsets, cell activation, and soluble mediator pathways in SLE remains poorly defined.
MMF is the prodrug of mycophenolic acid (MPA), which reversibly inhibits inosine monophosphate dehydrogenase (IMPDH), and most strongly inhibits the IMPDH II isoform (7). IMPDH is a rate-limiting enzyme in the de novo synthesis of guanine nucleotides and 2-deoxyribonucleotides. The type II isoform is primarily expressed in activated lymphocytes, while the type I isoform is expressed in almost all cell types. While it is recognized that MMF can affect many cell types, the cytostatic effect on lymphocytes is the principal mechanism of action by which immunosuppression is thought to occur (7). In lupus, patients taking MMF were found to have elevated frequencies and numbers of B cells compared with SLE patients not taking immunosuppressants; however, plasmablast numbers and frequencies were decreased in patients taking MMF compared with controls (8, 9). In a separate study, SLE patient samples taken at baseline and 4 weeks after MMF treatment had elevated frequencies of CD4+ T cells and NKT cells and decreased frequencies of CD8+ T cells and NK cells after 4 weeks, but no significant differences in B cells were observed and total numbers by cell type were not reported (10). In addition to phenotypic changes, MMF was found to induce epigenetic modifications in lupus CD4+ T cells by enhancing H4 acetylation and H3K4 trimethylation that increased regulatory miRNA levels and decreased expression of CD40L (11, 12). Variable results, and the absence of data on dendritic cell (DC) and monocyte alterations in SLE patients taking MMF, leave much to be determined for the cellular mechanisms of MMF in SLE pathogenesis.
Although some work has been done to identify phenotypic changes in SLE patients taking MMF, little to no data exist about the effect of MMF on the systemic cytokine milieu. SLE patients are characterized by elevated plasma levels of Th1, Th2, Th17, and myeloid-derived proinflammatory cytokines and chemokines such as interleukin 1α (IL-1α), IL-2, IL-5, IL-10, IL-13, IL-17, interferon-γ (IFN-γ), IP-10, G-CSF, and TNF-α (13). Many SLE patients are also distinguished from healthy controls by elevated blood levels of type I IFN accompanied with upregulation of IFN-inducible genes (13, 14). After 48 hours in vitro, MMF reduced the production of IL-2, IL-3, IL-4, IL-5, IL-6, IL-10, IFN-γ, and TNF-α in human lymphocytes (7). In lupus patients assessed for systemic IFNs, type I IFN was decreased in patients taking MMF compared with patients not taking MMF, but no significant difference was observed in IFN-γ production (15). In CD4+ T cells isolated from lupus patients and treated with either MPA or methanol, IL-17F mRNA levels were downregulated in cells treated with MPA, while no differences were observed in IL-2, IL-17A, or IFN-γ mRNA levels (11).
Using observational data collected from peripheral blood mononuclear cells (PBMCs) of SLE patients taking MMF (MMF+) and control SLE patients matched by age, gender, disease activity, other medication use, and ethnicity that were not taking MMF (MMF–), we assessed differences in cell numbers and activation for all major peripheral cell subsets using a comprehensive mass cytometry staining panel. In addition, 38 plasma soluble mediators were measured to assess the effect of MMF on systemic lupus inflammation and to identify the involvement of particular cellular pathways in MMF-associated immunomodulatory pathways in SLE. To examine STAT3 regulation following MMF treatment, in vitro studies using IL-6–stimulated PBMCs treated with MPA were analyzed by phosphoepitope flow cytometry.
Results
Phenotype evaluation by mass cytometry in MMF+ SLE patients.
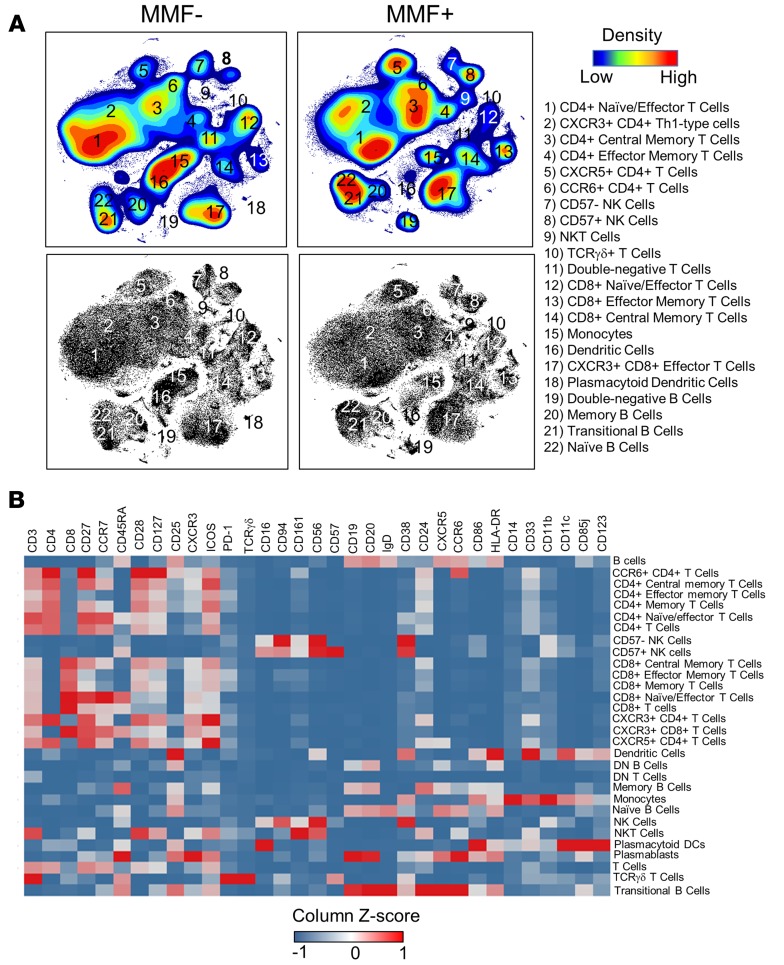
To understand the systemic immune changes that occur in MMF+ SLE patients, we utilized a mass cytometry panel of 33 heavy metal–labeled antibodies to identify major and minor immune cell subsets in patients taking or not taking MMF. The panel included a broad range of markers to allow segregation of T cells (CD4+ and CD8+ memory, naive, and effector cell subsets and TCRγδ+), B cells (naive, transitional, memory, plasmablasts, double-negative, and nonswitched memory), monocytes (classical, nonclassical, and intermediate), DCs, NK cells (CD8+/–, CD57+/–, CD16+/–, CD94+/–), and NKT cells (Supplemental Tables 1 and 2; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124575DS1). Cell activation and chemokine markers were also included to assess activation and functional properties of cell subsets. To evaluate cellular profiles, a high-dimensional method of analysis using t-distribution stochastic neighbor embedding (t-SNE) and a traditional bivariate plot analysis were both utilized. t-SNE creates an unsupervised 2-dimensional map by using the collected cellular data to draw pairwise comparisons and plot similar cells near each other.
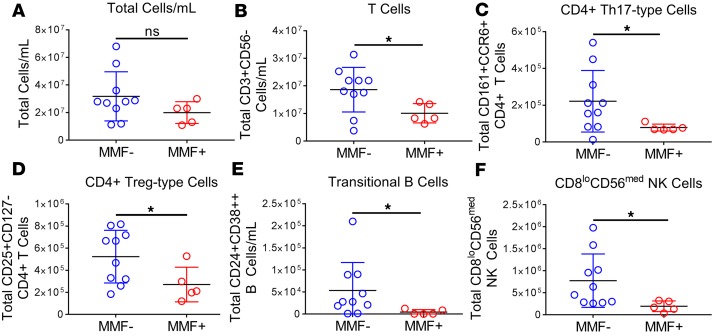
In our analysis, t-SNE reduced 33 parameters into 2 dimensions creating 22 distinct clusters (Figure 1A). The median intensity of each cellular marker within a cluster was assessed by gating and projection into a heatmap to classify cell subsets (Figure 1B). Following gating, cell frequencies were determined and total cell numbers/ml were back-calculated. MMF+ SLE patients trended lower in the total number of circulating cells/ml, but was not statistically significant (Figure 2A). However, 3 populations were found to have significantly fewer numbers in MMF+ patients by t-SNE, namely, T cells (CD3+CD56–), Th17-type cells (CD3+CD4+CCR6+), and plasmablasts (CD19+IgD–CD27+CD38hi) (Supplemental Table 1).
Figure 1. t-SNE analysis pipeline identifies 22 phenotypically distinct populations in PBMCs.
(A) Dimensionality-reduced t-SNE plots (using 33 surface markers) from concatenated PBMC data (110,000 cells) of MMF+ or MMF– SLE patients are shown by density maps and dot plots. Clusters are numbered and described by phenotypic subtype. All plots were derived from 10 individuals in the MMF– group and 5 individuals in the MMF+ group. (B) A heatmap summary of the expression values of all 33 cell surface markers are used to distinguish identified cell subsets. Marker values are displayed on a color scale ranging from blue (levels below the mean) through white (levels equal to the mean) to red (levels greater than the mean) using a column Z score.
Figure 2. Calculated cell numbers reveal decreased lymphocytes in MMF+ SLE patients.
Cell numbers were calculated using frequencies of cell subsets from manual gating and total cell counts. Cell numbers for (A) total cells/ml, (B) T cells, (C) CD4+ Th17-type cells (CCR6+CD161+), (D) CD4+ Treg-type cells (CD25+CD127–), (E) transitional B cells (IgD+CD27–CD24+CD38hi), and (F) CD8loCD56med NK cells are shown. Blue correlates to MMF– patients (n = 10) and red to MMF+ SLE patients (n = 5). *P < 0.05 by Kruskal-Wallis test with Dunn’s multiple comparisons test. Median ± standard deviation shown.
Using a standard biaxial gating strategy, we confirmed t-SNE results and identified additional cell subtypes such as T effector cells (Th1-, Th2-, Th17-, Treg-, and Tfh-type cells), CXCR3+ DCs, and NK cell subsets for a total of 55 cell subsets (Supplemental Figure 1). We identified differences in cell numbers of 5 cell types using bivariate plots, all significantly lower in MMF+ patients. T cells, specifically Th17-type cells (CCR6+CD161+) and Treg-type cells (CD25hiCD127–), transitional B cells (IgD+CD24hiCD38hiCD27–), and CD8loCD56med NK cells (Figure 2, B–F) were significantly reduced in MMF+ patients (Supplemental Table 2). CXCR3+ DCs, plasmacytoid DCs, and CD94– and CD57– NK cells also trended lower in numbers, but did not reach statistical significance (Supplemental Table 2). Based on these observations, MMF targets T cell, B cell, and NK cell subsets in SLE patients, primarily CD4+ T effector cells and CD38hi B cells.
Cell subset activation status and MMF use.
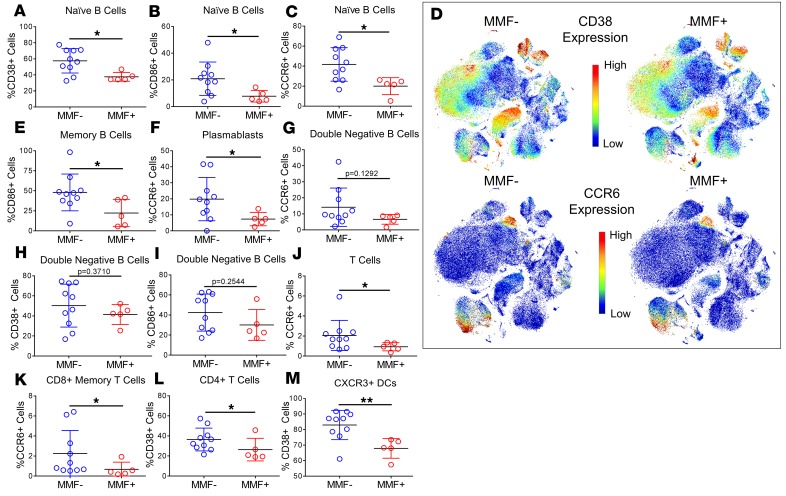
To better define the activation status of our defined cell populations we analyzed the frequencies of activation (CD27, CD28, CD38, CD86, HLA-DR, and ICOS), regulatory (CD85j, CD94, CD33, and PD-1), and chemokine (CCR6, CCR7, CXCR3, and CXCR5) receptor expression on each gated cell subset (Supplemental Table 3). Although MMF+ patients had no observed differences in naive, memory, or double-negative B cell numbers compared with MMF– patients (Supplemental Tables 1 and 2), the expression of CD38, CD86, and CCR6 on naive B cells and CD86 on memory B cells was at significantly lower frequencies on peripheral blood cells of MMF+ patients compared with MMF– patients (Figure 3, A–E). Plasmablasts, which were decreased systemically in MMF+ patients by t-SNE analysis, also had lower expression of CCR6, a chemokine receptor upregulated after activation in B cells and important for the recall response to antigen (16, 17) (Figure 3, D and F). Despite differences in memory B cells, IgD–CD27– double-negative B cells, also known as nonconventional memory B cells, only trended lower in CCR6, CD38, and CD86 expression (Figure 3, G–I).
Figure 3. Cell subset marker expression reveals activated B cell, T cell, and DC subsets, and elevated CCR6 expression in MMF+ SLE patients.
All 55 cell subsets as determined by manual gating were assessed for frequencies of activating, inhibitory, and chemokine receptors. Significant differences in cell surface marker expression are highlighted in MMF– (n = 10) and MMF+ SLE patients’ (n = 5) (A) naive B cells (CD38), (B) naive B cells (CD86), (C) naive B cells (CCR6), (E) memory B cells (CD86), and (F) plasmablasts (CCR6). Nonsignificant differences were found in expression on double-negative B cells of (G) CCR6, (H) CD38, and (I) CD86. Significant differences in expression were also found in (J) T cells (CCR6), (K) CD8+ memory T cells (CCR6), (L) CD4+ T cells (CD38), and (M) CXCR3+ DCs (CD38). Concatenated files of MMF– patients (n = 10) and MMF+ patients (n = 5) were used to create t-SNE dot plots individually colored by channel using arcsinh(5)-transformed expression values for (D) CD38 and CCR6. Cell orientations for the t-SNE plots in D can be found in Figure 1. *P < 0.05, **P < 0.01 by Kruskal-Wallis test with Dunn’s multiple comparisons test. Median ± standard deviation shown.
B cell activation markers and CCR6 expression were significantly lower in MMF+ patients. In addition, T cells also had lower expression of CCR6, specifically on CD8+ memory T cells, in MMF+ patients compared with MMF– patients (Figure 3, D, J, and K). CD4+ T cells and CXCR3+ DCs had lower expression of the activation marker CD38 in MMF-treated SLE patients (Figure 3, L and M), suggesting disease suppression by MMF may occur by hindering both adaptive and innate cell pathways.
Plasma soluble mediator levels negatively correlate with MMF use.
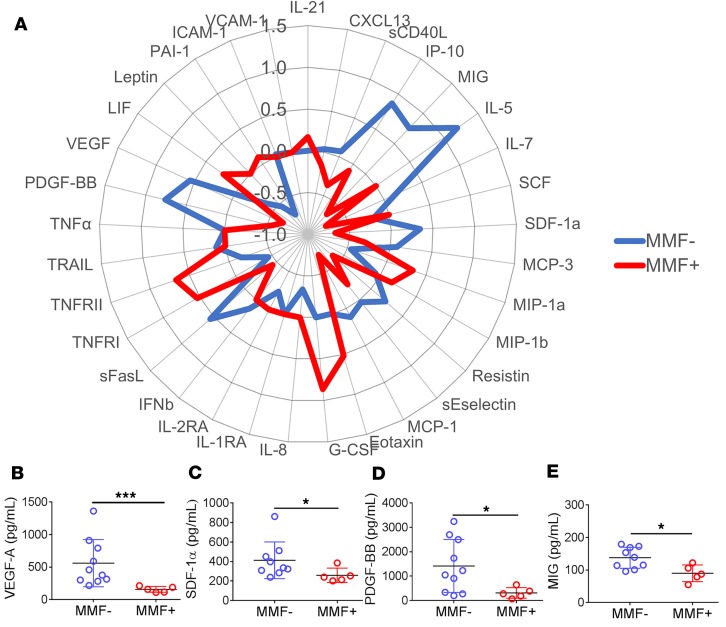
As activation marker frequency was diminished in MMF+ patients, we expected specific proinflammatory cytokines, growth factors, and chemokines to also be downregulated in MMF+ SLE patients. To assess this, 38 soluble mediators were assayed in plasma by multiplex assays and ELISA assays. The majority of soluble mediators were lower in the plasma of MMF+ SLE patients including IL-5, IL-7, IP-10/CXCL10, and MCP-1 compared with MMF– patients (Figure 4A and Supplemental Table 4). In addition, vascular endothelial growth factor (VEGF) and platelet-derived growth factor (PDGF-BB) and the chemokines stromal cell–derived factor 1 (SDF-1α/CXCL12) and IFN-γ–inducible MIG/CXCL9, were all downregulated in MMF+ SLE patients (Figure 4, B–E). The most significantly depressed soluble mediator was VEGF (P = 0.0007), which drives angiogenesis, capillary permeability, and endothelial cell proliferation and chemotaxis.
Figure 4. Growth factor and chemokine downregulation distinguish MMF+ SLE patients from MMF– patients.
Proinflammatory soluble mediators (38 total) were measured by multiplex or ELISA. Overall trends in relative plasma soluble mediator levels are depicted by (A) radar plot using the group median Z scores. MMF– SLE patients (n = 10) are highlighted in blue and MMF+ SLE patients (n = 5) are in red. Soluble mediators are represented by dot plots and include (B) VEGF, (C) PDGF-BB, (D) SDF-1α/CXCL12, and (E) MIG/CXCL9. *P < 0.05, ***P < 0.001 by Kruskal-Wallis test with Dunn’s multiple comparisons test. Median ± standard deviation shown.
STAT3 phosphorylation and associated immune pathways are downregulated by MMF.
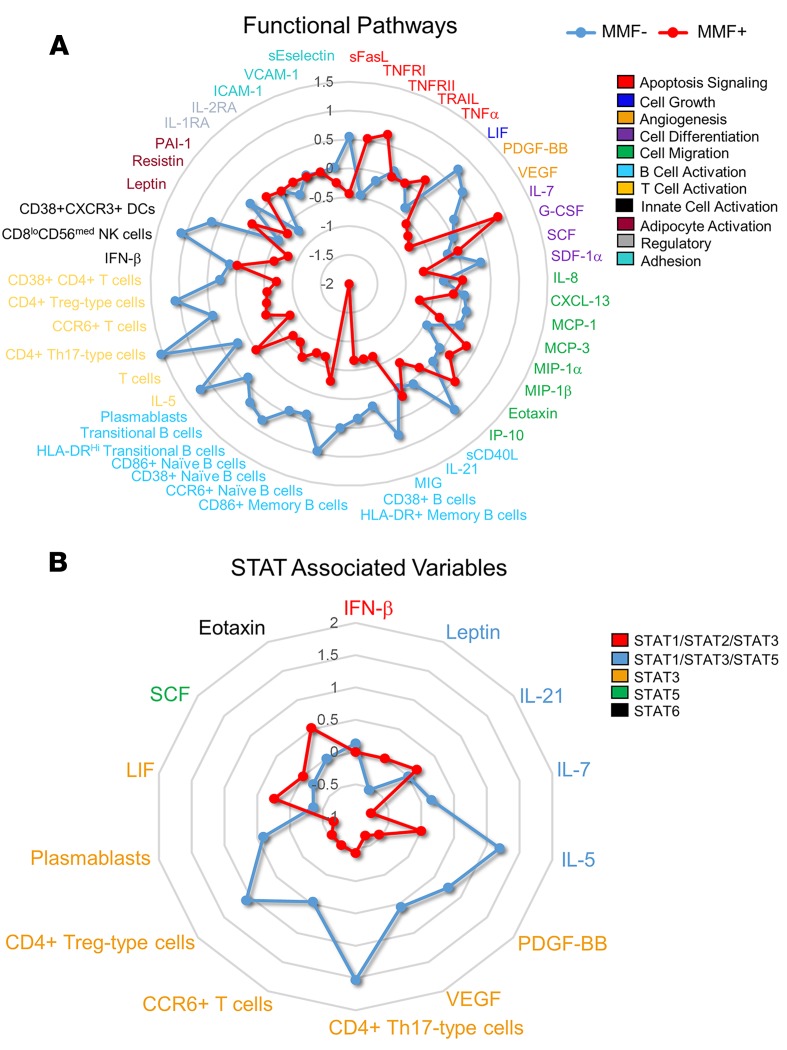
To determine the cellular pathways most affected by MMF treatment in SLE patients, cytokines, chemokines, and significant cell populations were grouped by functional properties (Figure 5A) or biomarkers/variables impacted by STAT signaling (Figure 5B) using the PANTHER classification system, which categorized our variables into the following groups: apoptosis signaling, cell growth, angiogenesis, cell differentiation, cell migration, B cell activation, T cell activation, innate cell activation, adipocyte activation, regulatory and adhesion groups, or STAT1/2/3 associated, STAT1/3/5 associated, STAT3 associated, STAT5 associated, and STAT6 associated. Group medians of each variable were converted to Z scores and plotted using a radar plot (Figure 5, A and B). STAT3-associated variables, namely VEGF, PDGF-BB, CD4+ Th17-type cells, and CD4+ Treg-type cells, were most significantly reduced in MMF+ SLE patients, followed closely by diminished activation in B cell pathways, where STAT3 is also required to form long-lived antibody responses (18).
Figure 5. Downregulation of B and T cell activation and STAT3-associated pathways distinguish MMF+ SLE patients from MMF– SLE patients.
A radar plot using the group median of the variable converted to Z scores show significant differences between MMF– SLE patients (n = 10) (blue) and MMF+ SLE patients (n = 5) (red). Cytokines, chemokines, and significant cell numbers and frequencies were grouped according to functional properties (A) or STAT-associated pathways (B).
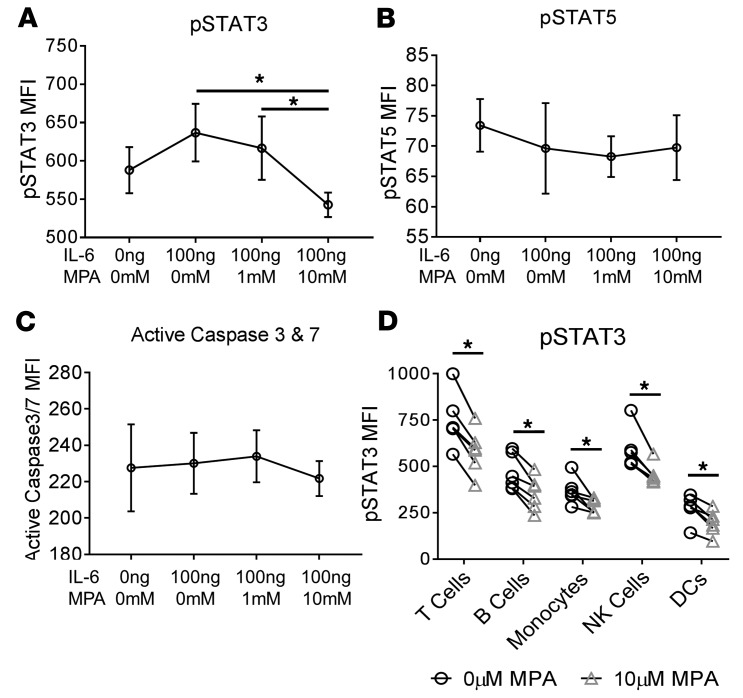
To investigate whether MMF reduced STAT3 phosphorylation, PBMCs from healthy individuals were stimulated with IL-6 and incubated with MPA for 3 hours at varying concentrations. Phosphoepitope flow analysis revealed a significant reduction in phosphorylated STAT3 following incubation with 10 μM MPA, a concentration equivalent to therapeutic levels found in SLE patients (19, 20) (Figure 6A), with no differences observed in phosphorylated STAT5 (Figure 6B) or cell apoptosis (Figure 6C) after 3 hours. Furthermore, reductions in STAT3 phosphorylation were similar across all major cell populations (Figure 6D). Together, these data suggest that p-STAT3 reduction is a principal mechanism of MMF-induced SLE disease suppression.
Figure 6. Mycophenolic acid reduced STAT3 phosphorylation in vitro.
Healthy control PBMCs from 6 individuals were treated with IL-6 (100 ng/ml) for 15 minutes prior to incubation with varying levels of MPA (0 μM, 1 μM, or 10 μM) for 3 hours. Phosphoepitope flow assessment of (A) p-STAT3 MFI, (B), p-STAT5 MFI, and (C) active caspase 3/7 MFI were determined following fixation and staining of cells immediately following the 3-hour incubation. Major cell populations were assessed for differences in (D) p-STAT3 inhibition including T cells, B cells, monocytes, NK cells, and dendritic cells compared between 0 μM MPA and 10 μM MPA. *P < 0.05, ***P < 0.001 by Kruskal-Wallis test with Dunn’s multiple comparisons test. Median ± standard deviation shown.
Discussion
One of the more severe manifestations of SLE, lupus nephritis, occurs in up to 60% of patients during the course of the disease (21). MMF is becoming a mainstay of treatment of lupus nephritis (4, 5). In general, the use of MMF in severe SLE patients leads to significant disease suppression, and maintenance therapy with MMF can prevent relapse of nephritis. The therapeutic efficacy of MMF in reducing disease activity is undisputed; however, the cellular alterations and systemic inflammatory pathways responsible for the successful actions of MMF have been unclear. In the current study, in-depth phenotyping of 55 cell subsets and 38 plasma soluble mediators, and in vitro p-STAT3 assessment, provide pivotal insight into the mechanism of MMF in SLE.
STAT3 is a critical regulator of numerous cellular processes including pathways that drive cell proliferation, differentiation, migration, inflammation, and apoptosis (22). The significance of STAT3 in biological actions is revealed in the disruption of the STAT3 gene in mice, which results in early embryonic death (23). STAT3 is the only STAT gene that is developmentally lethal. Conditional deletions of STAT3 have revealed its importance in CD4+ T cell differentiation, and effects on Th17, Treg, and Tfh pathways (24–26). Further, STAT3 is required for memory B cell development and T-dependent antibody responses (27). In SLE, total and phosphorylated levels of STAT3 are elevated and correlate with increased Th17 cells, organ involvement, and SLE disease activity of patients (28–30). Our study is the first to our knowledge to demonstrate that MMF reduces phosphorylation of IL-6–driven STAT3 in healthy PBMCs and may be a primary method for limiting disease activity in SLE.
Further support that MMF hinders the STAT3 pathway was observed in systemic reductions of Th17-type cells (CCR6+CD161+) (31) in MMF+ SLE patients. Th17 cells and IL-17A levels are elevated during active SLE (32, 33). CCR6 and CD161 expression distinguish CD4+ T cell populations that produce IL-17 (32), and reductions were observed in Th17-type cells of SLE patients treated with MMF. In vitro studies with CD4+ lupus T cells found decreased IL-17F mRNA expression in MPA-treated cells compared with methanol-treated cells, but no differences in IL-17A mRNA levels (11). Interestingly, IL-17+ CD4+ T cells have been found to be targeted by MMF compared with IFN-γ+ CD4+ T cells in a mouse diabetes model (34). The decrease in Th17 cells of MMF+ lupus patients was also associated with a reduction in Treg-type cells (CD25hiCD127–). Although this may seem counterintuitive as the number and function of Treg cells negatively correlate with lupus disease activity (33), MMF has been found to decrease CD4+CD25+Foxp3+ Treg cells in transgenic T cell, graft-versus-host, and experimental ischemia-reperfusion mouse models (31, 35, 36). Further, STAT3 is required for differentiation and expression of FoxP3 in Tregs (37). Our data suggest that MMF may have a more profound impact on Th17 and Treg cells in SLE patients that may be important in disease suppression; however, the differential effects on induction and maintenance of Th17 and Treg cells in a longitudinal study looking at IL-17 and FoxP3 by single-cell analysis should be considered.
The results of the current study indicate that B cells are a major target of MMF, which had a significant effect on almost all B cell subsets, except IgD–Ig27– double-negative B cells. Decreases in cell activation were observed in naive B cells, transitional B cells, memory B cells, nonswitched memory B cells, and plasmablasts in MMF+ patients. Furthermore, a decrease in CD38hi B cell subsets (plasmablasts and transitional B cells), as found either by t-SNE or manual gating, were found in patients taking MMF. A reduction in CD38hi B cells in transplant patients and a reduction of plasmablasts in MMF-treated SLE patients has been previously observed; thus, our data confirm the impact of MMF on activated B cell populations in SLE (8, 9, 38, 39).
In addition, MPA inhibited expansion, activation, and immunoglobulin production in primary B cells in vitro from both healthy individuals and rheumatoid arthritis patients; however, MPA had little effect on the expansion of terminally differentiated plasma cells (40). This suggests that reductions in plasmablast numbers in MMF-treated SLE patients may largely be due to blocking of plasma cell differentiation. Decreases in transitional B cells also suggest a possible impact of MMF on B cell maturation out of the bone marrow, and further in vitro investigation into cell subset–specific maturation and expansion in SLE patients is of interest. Despite the known effects of MMF on B cell proliferation (40), double-negative B cells were not reduced in patients taking MMF in this study. The lack of differences may be due to the known exhausted or anergic phenotype of double-negative B cells, also known as nonconventional memory cells, which may result in a reduced ability to expand and respond to stimuli (41). Our data suggest that MMF-induced STAT3 inhibition profoundly effects B cells, most specifically later-stage CD38hi B cells to suppress lupus disease.
Finally, 38 systemic cytokines were assessed in SLE patients on MMF with significant differences found in only 4 key cytokines, VEGF, PDGF-BB, SDF-1α/CXCL12, and MIG/CXCL9, which were all reduced in patients taking MMF. VEGF and PDGF-BB, both of which are angiogenesis factors, were most significantly affected by MMF treatment. SLE is known to be associated with immune-mediated vascular damage and angiogenesis. Elevated VEGF levels correlate with vascular damage in SLE patients, which positively associates with disease activity, suggesting VEGF is a potential driver of SLE endothelial dysfunction (42). VEGF has also been found to be downregulated following treatment with MMF in several murine models (43, 44). The VEGF gene is regulated directly by the STAT3 protein, and decreases in VEGF may directly contribute to decreases in peripheral B cell and T cell numbers (45–48). Further, we also found significant reductions in 2 chemokines, MIG/CXCL9 and SDF-1α/CXCL12, in patients taking MMF. MIG is largely produced by activated B cells, monocytes, and DCs, and reduction in MIG may be due to decreases in B cell activation and numbers seen in MMF-treated patients. In addition, SDF-1α is critical for hematopoiesis including B lymphocyte development within the bone marrow niche (49). Reductions in SDF-1α likely contribute to lower B cell numbers and alterations in the B cell compartment, including decreased transitional B cells, seen in MMF+ patients.
STAT inhibitors are a promising family of therapeutic targets for lupus. Indeed, STAT3 inhibition in MRL/lpr lupus-prone mice inhibited the onset of lupus nephritis, decreased anti-dsDNA antibodies, and reduced inflammatory cytokine production (50). We have identified inhibition of activated STAT3 as a putative mechanism of disease suppression by MMF in patients with SLE. These data hold promising information for drug therapy development utilizing MMF and associated pathway inhibitors. Future studies utilizing a larger cohort assessed longitudinally for STAT3 phosphorylation and upstream proteins, such as the mammalian target of rapamycin pathway and the mitogen-activated protein kinase pathway, following MMF treatment are of particular interest and necessary in determining the involvement of MMF in STAT3 inhibition.
Methods
Study population.
PBMCs and plasma samples from 10 SLE patients not taking MMF and 5 SLE patients taking MMF were studied. Patients were recruited at the Oklahoma Medical Research Foundation (OMRF). SLE patients fulfilled the American College of Rheumatology (ACR) criteria for SLE classification. Subjects were matched by gender, ethnicity, age ± 5 years, medication use, and disease activity by SELENA-SLEDAI (Supplemental Table 5) (51). Similar frequencies of SLE patients with active disease, as defined by SLEDAI ≥ 4, are found in both groups, with 50% of MMF– patients and 60% of MMF+ patients. PBMCs were isolated using Lymphocyte Separation Medium (Mediatech, Inc.) and stored in freezing media (20% human serum and 10% DMSO in RPMI) at –80°C until use. Plasma was also collected and stored at –80°C until use.
CyTOF immunophenotyping.
Assays were performed in the Human Immune Monitoring Center at Stanford University. Briefly, PBMCs were stained with a surface antibody cocktail specific for CCR6, CCR7, CD11b, CD11c, CD123, CD127, CD14, CD16, CD161, CD19, CD161, CD19, CD20, CD24, CD25, CD27, CD28, CD3, CD33, CD38, CD4, CD45RA, CD56, CD57, CD8, CD85j, CD86, CD94, CXCR3, CXCR5, HLA-DR, ICOS, IgD, PD-1, and TCRγδ (see Supplemental Table 6 for clone list, supplier, and metal isotope utilized) for 60 minutes on ice. PBMCs were then stained with DOTA-maleimide (Macrocyclics) containing natural-abundance indium (viability marker). The cells were resuspended in iridium-containing DNA intercalator and incubated at room temperature for 20 minutes. The cells were washed twice and diluted before injection into the CyTOF (DVS Sciences). Data analysis was performed using Cytobank by gating on intact cells based on the iridium isotopes of the intercalator, and then on singlets by Ir-191 versus cell length, and then on live cells (indium-Live/Dead–negative population), followed by cell subset–specific gating or viSNE analysis.
Soluble mediator measurement.
Plasma of SLE patients was analyzed for the concentration of soluble mediators using a multiplex bead-based assay (eBioscience/Affymetrix) to measure sCD40L, CXCL13, eotaxin, G-CSF, GROα, ICAM-1, IFN-α, IFN-β, IFN-γ, IL-1α, IL-1β, IL-1RA, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IL-2, IL-21, IL-23, IL-2RA, IL-4, IL-5, IL-6, IL-7, IL-8, IP-10, leptin, LIF, MCP-1, MCP-3, MIG, MIP-1β, MIP-1α, NGF-β, PAI-1, PDGF-BB, RANTES, resistin, SCF/c-kit ligand, SDF-1α/CXCL12, sEselectin, sFasL, TGF-β, TNF-α, TNFRI, TNFRII, TRAIL, VCAM-1, and VEGF and run on a Bio-Plex 200 suspension array reader (Bio-Rad). A standard control serum was included on each plate (Cellgro human AB serum, Mediatech). Soluble mediator concentrations (pg/ml) were calculated using Bio-Plex manager software using 5-parameter logistic curve fitting. Soluble mediators with >60% of concentration values reported as out of range in both treatment groups were dropped from the analysis to minimize data loss from row-based deletion of missing values. These mediators included IFN-α, IFN-γ, IL-1α, IL-1β, IL-10, IL-12p70, IL-13, IL-15, IL-17A, IL-23, IL-4, NGF-β, and TGF-β. The 38 soluble mediators that passed quality control were normalized and values were used for subsequent analyses. Plasma levels of BLyS were assessed using enzyme-linked immunosorbent assay (ELISA) per the manufacturer’s protocol (R&D Systems).
Pathway analyses.
Functional properties and STAT pathways for all cytokines and relevant cell receptors from significant cell populations were assessed using the PANTHER classification system (version 13.1) (52), which is used to classify protein family/subfamilies, biological process, molecular function, and signaling pathways. All proteins were assessed for pathways and functional properties within PANTHER, and are reported in Supplemental Table 7. Cytokines and cell subsets were grouped according to function or STAT-associated pathway outputs, and radar plots were created as mentioned in the data analysis section below.
STAT3 phosphorylation by phosphoepitope flow.
Previously frozen PBMCs were thawed, washed, and resuspended in supplemented RPMI 1640 at 1 × 106 cells/well in a 24-well plate. Cells were rested 1 hour, and then stimulated with IL-6 (100 ng/ml) for 15 minutes prior to addition of MPA, the active metabolite of MMF, at varying concentrations (0 μM, 1 μM, or 10 μM). Cells were incubated with MPA for either 0 or 3 hours, after which cells were washed with cold PBS and stained with near-IR Live/Dead stain (Thermo Fisher Scientific). PBMCs were fixed in paraformaldehyde, washed, and Fc receptor blocked. Cells were then permeabilized with saponin and stained for lineage markers (CD3, CD4, CD8, CD20, CD14, CD11c, and CD56) followed by staining for intracellular markers for p-STAT3 and p-STAT5 (BD Biosciences). FLICA caspase 3/7 stain (ImmunoChemistry Technologies, LLC) was added 1 hour prior to the end of MPA incubation. Cells were resuspended in FACS buffer and immediately run on a FACSCelesta (BD Biosciences).
Data analysis.
t-SNE analysis were performed using Cytobank’s viSNE (53). t-SNE was performed after gating off live, intact singlet cells. For frequency analysis, 24,000 events were used per sample. Cell clusters were gated and frequencies exported for cell number calculations and analysis. Concatenated files of 110,000 cells per group were used to generate representative t-SNE images. Manual gating was also performed on individual subject files for B cells, monocytes, NK cells, T cells, and DCs in Cytobank (Supplemental Figure 1, A–D).
Statistics.
Dot plots, heatmaps, statistical analyses, and multiple testing corrections were performed using GraphPad Prism 6.0 for Windows or TIBCO Spotfire 6.0.1 (TIBCO Software Inc.). Radar plots were generated by using the group median of modified Z scores in Microsoft Excel. P values less than 0.05 were considered significant (*P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001). All error bars indicate the standard deviation.
Study approval.
All work contained in this publication was approved by the Institutional Review Board at the Oklahoma Medical Research Foundation. Blood donors gave written informed consent prior to study inclusion in accordance with the ethics principles stated in the Helsinki Declaration.
Author contributions
SSW analyzed data, wrote the manuscript, and designed and conducted experiments. JMG and JAJ designed research studies and experiments, and wrote the manuscript. EFC designed research studies. HC, RL, and KB analyzed data. SM designed and conducted experiments and acquired data. HTM and PJU designed and conducted experiments.
Supplementary Material
Acknowledgments
We would like to thank Virginia Roberts and Wade DeJager for technical assistance. In addition, we would like to thank the Stanford Human Immune Monitoring Center (HIMC) for performing the CyTOF phenotyping assays. Research reported was supported by NIAID, NIAMS, and NIGMS of the NIH (U19AI082714, U19AI082719, U54GM104938, RC1AR058554, P30GM103510, U01AI101934, P30AR053483, T32AI007633 [to SSW], and S10RR026735) and was conceived through interactions of the NIAID Autoimmunity Center of Excellence. The contents are solely the responsibility of the authors and do not necessarily represent the official views of the NIH or its institutes.
Version 1. 01/24/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(2):e124575. https://doi.org/10.1172/jci.insight.124575.
Contributor Information
Samantha Slight-Webb, Email: samantha-webb@omrf.org.
Hua Chen, Email: Hua-Chen@omrf.org.
Rufei Lu, Email: rufei-lu@ouhsc.edu.
Susan Macwana, Email: Susan-Macwana@omrf.org.
Krista Bean, Email: kristaklingler.byu@gmail.com.
Paul J. Utz, Email: pjutz@stanford.edu.
Judith A. James, Email: Judith-James@omrf.org.
References
- 1.Tsokos GC. Systemic lupus erythematosus. N Engl J Med. 2011;365(22):2110–2121. doi: 10.1056/NEJMra1100359. [DOI] [PubMed] [Google Scholar]
- 2.Ginzler EM, Aranow C. Mycophenolate mofetil in lupus nephritis. Lupus. 2005;14(1):59–64. doi: 10.1191/0961203305lu2061oa. [DOI] [PubMed] [Google Scholar]
- 3.Sollinger HW. Mycophenolate mofetil for the prevention of acute rejection in primary cadaveric renal allograft recipients. U.S. Renal Transplant Mycophenolate Mofetil Study Group. Transplantation. 1995;60(3):225–232. doi: 10.1097/00007890-199508000-00003. [DOI] [PubMed] [Google Scholar]
- 4.Appel GB, et al. Mycophenolate mofetil versus cyclophosphamide for induction treatment of lupus nephritis. J Am Soc Nephrol. 2009;20(5):1103–1112. doi: 10.1681/ASN.2008101028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ginzler EM, et al. Mycophenolate mofetil or intravenous cyclophosphamide for lupus nephritis. N Engl J Med. 2005;353(21):2219–2228. doi: 10.1056/NEJMoa043731. [DOI] [PubMed] [Google Scholar]
- 6.Sahin A. Mycophenolate mofetil in the treatment of systemic lupus erythematosus. Eurasian J Med. 2009;41(3):180–185. [PMC free article] [PubMed] [Google Scholar]
- 7.Allison AC, Eugui EM. Mycophenolate mofetil and its mechanisms of action. Immunopharmacology. 2000;47(2-3):85–118. doi: 10.1016/S0162-3109(00)00188-0. [DOI] [PubMed] [Google Scholar]
- 8.Eickenberg S, Mickholz E, Jung E, Nofer JR, Pavenstadt HJ, Jacobi AM. Mycophenolic acid counteracts B cell proliferation and plasmablast formation in patients with systemic lupus erythematosus. Arthritis Res Ther. 2012;14(3):R110. doi: 10.1186/ar3835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fassbinder T, et al. Differential effects of cyclophosphamide and mycophenolate mofetil on cellular and serological parameters in patients with systemic lupus erythematosus. Arthritis Res Ther. 2015;17:92. doi: 10.1186/s13075-015-0603-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhao L, Jiang Z, Jiang Y, Ma N, Wang K, Zhang Y. Changes in immune cell frequencies after cyclophosphamide or mycophenolate mofetil treatments in patients with systemic lupus erythematosus. Clin Rheumatol. 2012;31(6):951–959. doi: 10.1007/s10067-012-1958-8. [DOI] [PubMed] [Google Scholar]
- 11.Yang Y, et al. The effect of mycophenolic acid on epigenetic modifications in lupus CD4+T cells. Clin Immunol. 2015;158(1):67–76. doi: 10.1016/j.clim.2015.03.005. [DOI] [PubMed] [Google Scholar]
- 12.Tang Q, et al. Mycophenolic acid upregulates miR-142-3P/5P and miR-146a in lupus CD4+T cells. Lupus. 2015;24(9):935–942. doi: 10.1177/0961203315570685. [DOI] [PubMed] [Google Scholar]
- 13.Slight-Webb S, et al. Autoantibody-positive healthy individuals display unique immune profiles that may regulate autoimmunity. Arthritis Rheumatol. 2016;68(10):2492–2502. doi: 10.1002/art.39706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baechler EC, et al. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proc Natl Acad Sci USA. 2003;100(5):2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fürnrohr BG, Rhodes B, Munoz LE, Weiß K, Vyse TJ, Schett G. Osteoclast differentiation is impaired in a subgroup of SLE patients and correlates inversely with mycophenolate mofetil treatment. Int J Mol Sci. 2015;16(8):18825–18835. doi: 10.3390/ijms160818825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wiede F, et al. CCR6 is transiently upregulated on B cells after activation and modulates the germinal center reaction in the mouse. Immunol Cell Biol. 2013;91(5):335–339. doi: 10.1038/icb.2013.14. [DOI] [PubMed] [Google Scholar]
- 17.Elgueta R, et al. CCR6-dependent positioning of memory B cells is essential for their ability to mount a recall response to antigen. J Immunol. 2015;194(2):505–513. doi: 10.4049/jimmunol.1401553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Avery DT, et al. B cell-intrinsic signaling through IL-21 receptor and STAT3 is required for establishing long-lived antibody responses in humans. J Exp Med. 2010;207(1):155–171. doi: 10.1084/jem.20091706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mino Y, Naito T, Shimoyama K, Ogawa N, Kawakami J. Effective plasma concentrations of mycophenolic acid and its glucuronide in systemic lupus erythematosus patients in the remission-maintenance phase. J Clin Pharm Ther. 2012;37(2):217–220. doi: 10.1111/j.1365-2710.2011.01269.x. [DOI] [PubMed] [Google Scholar]
- 20.Pou L, et al. Mycophenolic acid plasma concentrations: influence of comedication. Ther Drug Monit. 2001;23(1):35–38. doi: 10.1097/00007691-200102000-00007. [DOI] [PubMed] [Google Scholar]
- 21.Cameron JS. Lupus nephritis. J Am Soc Nephrol. 1999;10(2):413–424. doi: 10.1681/ASN.V102413. [DOI] [PubMed] [Google Scholar]
- 22.Camporeale A, Poli V. IL-6, IL-17 and STAT3: a holy trinity in auto-immunity? Front Biosci (Landmark Ed) 2012;17:2306–2326. doi: 10.2741/4054. [DOI] [PubMed] [Google Scholar]
- 23.Takeda K, et al. Targeted disruption of the mouse Stat3 gene leads to early embryonic lethality. Proc Natl Acad Sci USA. 1997;94(8):3801–3804. doi: 10.1073/pnas.94.8.3801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tangye SG, Cook MC, Fulcher DA. Insights into the role of STAT3 in human lymphocyte differentiation as revealed by the hyper-IgE syndrome. J Immunol. 2009;182(1):21–28. doi: 10.4049/jimmunol.182.1.21. [DOI] [PubMed] [Google Scholar]
- 25.Kane A, Deenick EK, Ma CS, Cook MC, Uzel G, Tangye SG. STAT3 is a central regulator of lymphocyte differentiation and function. Curr Opin Immunol. 2014;28:49–57. doi: 10.1016/j.coi.2014.01.015. [DOI] [PubMed] [Google Scholar]
- 26.Hsu P, et al. IL-10 potentiates differentiation of human induced regulatory T cells via STAT3 and Foxo1. J Immunol. 2015;195(8):3665–3674. doi: 10.4049/jimmunol.1402898. [DOI] [PubMed] [Google Scholar]
- 27.Fornek JL, Tygrett LT, Waldschmidt TJ, Poli V, Rickert RC, Kansas GS. Critical role for Stat3 in T-dependent terminal differentiation of IgG B cells. Blood. 2006;107(3):1085–1091. doi: 10.1182/blood-2005-07-2871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wen Z, Xu L, Xu W, Xiong S. Detection of dynamic frequencies of Th17 cells and their associations with clinical parameters in patients with systemic lupus erythematosus receiving standard therapy. Clin Rheumatol. 2014;33(10):1451–1458. doi: 10.1007/s10067-014-2656-5. [DOI] [PubMed] [Google Scholar]
- 29.Crispín JC, et al. Expanded double negative T cells in patients with systemic lupus erythematosus produce IL-17 and infiltrate the kidneys. J Immunol. 2008;181(12):8761–8766. doi: 10.4049/jimmunol.181.12.8761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Harada T, Kyttaris V, Li Y, Juang YT, Wang Y, Tsokos GC. Increased expression of STAT3 in SLE T cells contributes to enhanced chemokine-mediated cell migration. Autoimmunity. 2007;40(1):1–8. doi: 10.1080/08916930601095148. [DOI] [PubMed] [Google Scholar]
- 31.Lim DG, et al. Impact of immunosuppressants on the therapeutic efficacy of in vitro-expanded CD4+CD25+Foxp3+ regulatory T cells in allotransplantation. Transplantation. 2010;89(8):928–936. doi: 10.1097/TP.0b013e3181d3c9d4. [DOI] [PubMed] [Google Scholar]
- 32.Yao Y, et al. Balance between inflammatory and regulatory cytokines in systemic lupus erythematosus. Genet Mol Res. 2016;15(2):gmr7626. doi: 10.4238/gmr.15027626. [DOI] [PubMed] [Google Scholar]
- 33.Yang J, et al. Th17 and natural Treg cell population dynamics in systemic lupus erythematosus. Arthritis Rheum. 2009;60(5):1472–1483. doi: 10.1002/art.24499. [DOI] [PubMed] [Google Scholar]
- 34.Kim SM, et al. Targeting T helper 17 by mycophenolate mofetil attenuates diabetic nephropathy progression. Transl Res. 2015;166(4):375–383. doi: 10.1016/j.trsl.2015.04.013. [DOI] [PubMed] [Google Scholar]
- 35.Gandolfo MT, et al. Mycophenolate mofetil modifies kidney tubular injury and Foxp3+ regulatory T cell trafficking during recovery from experimental ischemia-reperfusion. Transpl Immunol. 2010;23(1–2):45–52. doi: 10.1016/j.trim.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wu T, et al. Immunosuppressive drugs on inducing Ag-specific CD4(+)CD25(+)Foxp3(+) Treg cells during immune response in vivo. Transpl Immunol. 2012;27(1):30–38. doi: 10.1016/j.trim.2012.05.001. [DOI] [PubMed] [Google Scholar]
- 37.Pallandre JR, et al. Role of STAT3 in CD4+CD25+FOXP3+ regulatory lymphocyte generation: implications in graft-versus-host disease and antitumor immunity. J Immunol. 2007;179(11):7593–7604. doi: 10.4049/jimmunol.179.11.7593. [DOI] [PubMed] [Google Scholar]
- 38.Bijl M, Horst G, Bootsma H, Limburg PC, Kallenberg CG. Mycophenolate mofetil prevents a clinical relapse in patients with systemic lupus erythematosus at risk. Ann Rheum Dis. 2003;62(6):534–539. doi: 10.1136/ard.62.6.534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weigel G, Griesmacher A, Karimi A, Zuckermann AO, Grimm M, Mueller MM. Effect of mycophenolate mofetil therapy on lymphocyte activation in heart transplant recipients. J Heart Lung Transplant. 2002;21(10):1074–1079. doi: 10.1016/S1053-2498(02)00440-0. [DOI] [PubMed] [Google Scholar]
- 40.Karnell JL, et al. Mycophenolic acid differentially impacts B cell function depending on the stage of differentiation. J Immunol. 2011;187(7):3603–3612. doi: 10.4049/jimmunol.1003319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colonna-Romano G, et al. A double-negative (IgD-CD27-) B cell population is increased in the peripheral blood of elderly people. Mech Ageing Dev. 2009;130(10):681–690. doi: 10.1016/j.mad.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 42.Bărbulescu AL, et al. Vascular endothelial growth factor in systemic lupus erythematosus - correlations with disease activity and nailfold capillaroscopy changes. Rom J Morphol Embryol. 2015;56(3):1011–1016. [PubMed] [Google Scholar]
- 43.Monguilhott Dalmarco E, Mendes de Córdova CM, Fröde TS. Evidence of an anti-inflammatory effect of mycophenolate mofetil in a murine model of pleurisy. Exp Lung Res. 2011;37(7):399–407. doi: 10.3109/01902148.2011.570416. [DOI] [PubMed] [Google Scholar]
- 44.Dalmarco EM, Astolfi G, de Liz R, de Córdova CM, Fröde TS. Modulatory effect of mycophenolate mofetil on carrageenan-induced inflammation in the mouse air pouch model. Int Immunopharmacol. 2012;13(4):476–482. doi: 10.1016/j.intimp.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 45.Chen SH, Murphy DA, Lassoued W, Thurston G, Feldman MD, Lee WM. Activated STAT3 is a mediator and biomarker of VEGF endothelial activation. Cancer Biol Ther. 2008;7(12):1994–2003. doi: 10.4161/cbt.7.12.6967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Niu G, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. 2002;21(13):2000–2008. doi: 10.1038/sj.onc.1205260. [DOI] [PubMed] [Google Scholar]
- 47.Ohm JE, et al. VEGF inhibits T-cell development and may contribute to tumor-induced immune suppression. Blood. 2003;101(12):4878–4886. doi: 10.1182/blood-2002-07-1956. [DOI] [PubMed] [Google Scholar]
- 48.Shrestha B, et al. B cell-derived vascular endothelial growth factor A promotes lymphangiogenesis and high endothelial venule expansion in lymph nodes. J Immunol. 2010;184(9):4819–4826. doi: 10.4049/jimmunol.0903063. [DOI] [PubMed] [Google Scholar]
- 49.Nagasawa T. The chemokine CXCL12 and regulation of HSC and B lymphocyte development in the bone marrow niche. Adv Exp Med Biol. 2007;602:69–75. doi: 10.1007/978-0-387-72009-8_9. [DOI] [PubMed] [Google Scholar]
- 50.Edwards LJ, Mizui M, Kyttaris V. Signal transducer and activator of transcription (STAT) 3 inhibition delays the onset of lupus nephritis in MRL/lpr mice. Clin Immunol. 2015;158(2):221–230. doi: 10.1016/j.clim.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hochberg MC. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 1997;40(9):1725. doi: 10.1002/art.1780400928. [DOI] [PubMed] [Google Scholar]
- 52.Thomas PD, et al. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kotecha N, Krutzik PO, Irish JM. Web-based analysis and publication of flow cytometry experiments. Curr Protoc Cytom. 2010;Chapter 10:Unit 10.17. doi: 10.1002/0471142956.cy1017s53. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.