Abstract
High macrophage infiltration in cancer is associated with reduced survival in animal models and in patients. This reflects a shift in the macrophage response from a tumor-suppressive to tumor-supportive program governed by transcriptional events regulated by the inflammatory milieu. Although several transcription factors are known to drive a prometastatic program, those that govern an antimetastatic program are less understood. IFN regulatory factor-8 (IRF8) is integral for macrophage responses against infections. Using a genetic loss-of-function approach, we tested the hypothesis that IRF8 expression in macrophages governs their capacity to inhibit metastasis. We found that: (a) metastasis was significantly increased in mice with IRF8-deficient macrophages; (b) IRF8-deficient macrophages displayed a program enriched for genes associated with metastasis; and (c) lower IRF8 expression correlated with reduced survival in human breast and lung cancer, as well as melanoma, with high or low macrophage infiltration. Thus, a macrophagehiIRF8hi signature was more favorable than a macrophagehiIRF8lo signature. The same held true for a macrophageloIRF8hi vs. a macrophageloIRF8lo signature. These data suggest that incorporating IRF8 expression levels within a broader macrophage signature or profile strengthens prognostic merit. Overall, to our knowledge, our findings reveal a previously unrecognized role for IRF8 in macrophage biology to control metastasis or predict outcome.
Keywords: Immunology, Oncology
Keywords: Cancer, Macrophages
IRF8 is a key determinant of macrophage-mediated anti-metastatic activity, whose expression associates with improved survival in patients with solid cancers.
Introduction
Metastasis is the leading cause of most solid cancer–related deaths and, as such, has dominated the field of cancer research for decades (1). Progress in understanding the complexity of the metastatic process was born years ago by Stephen Paget and colleagues who brought forth the “seed” and “soil” hypothesis of metastasis (2). Although progress has been made toward understanding these interactions, the precise mechanisms underlying the prometastatic behavior or, for that matter, the antimetastatic behavior of the soil (i.e., the tumor microenvironment; TME), remain incompletely understood. Numerous studies have demonstrated that metastasis is a complex multistep process (3). In solid cancers, it is well-appreciated that tumor progression to metastasis is governed by both intrinsic and extrinsic factors (4). Intrinsic factors include genetic and epigenetic alterations affecting the expression and function of proto-oncogenes and tumor-suppressor genes (4). However, it is becoming increasingly apparent that such intrinsic alterations alone are insufficient to culminate in successful metastases. Extrinsic factors, which reflect the contribution of stromal elements within the TME, are also important for dissemination of the primary tumor to secondary sites (5).
The TME is heterogeneous, composed of tumor cells with a supporting cast of nonneoplastic cells, including endothelial cells, pericytes, fibroblasts, and diverse immune populations (6). Among the diverse immune populations, macrophages have been identified as one of the most abundant within the TME of many solid cancers (7). Ordinarily, macrophage function is tightly regulated by signaling and transcriptional events that enable them to mediate pro- or antiinflammatory activities, depending upon the type of pathogenic insult (8). When macrophages were first discovered in the TME, they were thought to be immune suppressive and tumor supportive. Macrophages enhanced local tumor invasion, facilitating angiogenesis and tumor cell entry into blood vessels — processes that culminated in metastasis (9). Recent work, however, has underscored the existence of macrophages, which exhibit tumor-suppressive activities (10, 11). This functional dichotomy has shifted the conversation of the macrophage role within the TME from seeking to deplete them to moving to exploit them to boost their tumoricidal properties (12, 13). These antitumor macrophages express higher levels of MHC class I and II, and costimulatory molecules, and they display a cytokine and chemokine signature distinct from their protumor counterparts (12, 14). Regardless, the existence of tumor-supportive vs. tumor-suppressive macrophages within the TME offers important opportunities to reshape the macrophage response during immune surveillance or immunotherapy.
As noted earlier, macrophage function is determined by the activation of transcription factors affected by diverse environmental cues (7). Transcription factors such as HIF-1α, STAT3, and STAT6 are upregulated in protumor macrophages, which in turn activate genes involved in immune suppression, tumor invasion, and angiogenesis (15). Substantial interest has been placed on understanding the regulation of transcriptional events that dictate the tumor-supportive behavior of macrophages (16–18). In contrast, less is known about the diversity of transcription factors that enforce or drive an antitumor/antimetastatic macrophage response. Such knowledge provides an alternative perspective to shift the macrophage response during the metastatic process for potential therapeutic purposes.
IFN regulatory factor-8 (IRF8) is an IFN-γ–inducible transcription factor intimately involved in the differentiation and function of myeloid populations, including macrophages (19). IRF8-null mice, which are globally deficient for IRF8 expression, exhibit a severely compromised myeloid development program. This global IRF8 defect results in a myeloproliferative disorder characterized by a profound accumulation of granulocytes at the expense of monocytes and DCs (19). Consequently, IRF8 has been implicated as a tumor-suppressor gene in certain myeloid leukemias, such as chronic myeloid leukemia (CML) (20). Previous work from our laboratory has also uncovered an unrecognized role for IRF8 in controlling the expansion of myeloid-derived suppressor cells (MDSCs), especially the granulocytic subset, in cancer (21). This observation is consistent with the role of IRF8 in myeloid cell development as a key regulator of the production of granulocytes and monocytes/macrophages (22). However, in addition to the role of IRF8 in myeloid cell development, IRF8 plays important roles in mature myeloid populations at effector or functional levels. In macrophages, IRF8 induction is required for the expression of inducible nitric oxide synthase (iNOS) and IL-12p40, proteins that are instrumental for host defense against infectious disease and adaptive T cell immunity (23, 24). In this study, given the broad importance of IRF8 in host defense, we sought to test the hypothesis that IRF8 expression in macrophages governs their capacity to control tumor metastasis.
Here, we show that IRF8 expression in macrophages is critical for their antimetastatic activity. Using mouse models of mammary cancer and melanoma, we show that lung metastasis was increased when macrophages were rendered deficient in IRF8 expression. IRF8 deficiency in macrophages resulted in a gene expression program enriched for metastasis-associated genes, which expands current knowledge of how IRF8 may regulate host defense in cancer. Lastly, macrophage infiltration in human breast or lung cancer, as well as melanoma stratified according to IRF8 expression levels, was directly associated with overall survival. Thus, we identified a potentially novel role for IRF8 expression in macrophage-mediated antimetastatic activity and clinical outcome, which not only advances our understanding of macrophages in cancer, but also implicates the macrophage-IRF8 axis or profile as a potential prognostic or therapeutic target.
Results
Conditional deletion of IRF8 does not impair myeloid cell development.
To investigate whether IRF8 expression in macrophages is important to tumor growth control, we made use of a conditional IRF8-KO mouse model in which IRF8 was genetically deleted in macrophages via a cell/tissue-specific promoter (25). We bred homozygous IRF8fl/fl mice to mice homozygous for the expression of Cre-recombinase under the control of the macrophage-specific Lyz2 promoter to generate IRF8-deficient progeny (Lyz2Cre-IRF8fl/fl; henceforth referred to as IRF8-cKO) (Supplemental Figure 1, A and B; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.124267DS1). In contrast to the construction of the global KO of Irf8, which impacts BM progenitors (26), this model enables the study of IRF8 function in terminally differentiated myeloid cells (25, 27–31). In agreement with those earlier reports, we observed no impairment in the ability to generate BM-derived macrophages (BMDMs), based on the expression of the macrophage markers CD11b and F4/80 when WT or IRF8-cKO BM cells were cultured with recombinant M-CSF (Figure 1A). We analyzed IRF8 expression in CD11b+F4/80+ macrophages generated from the BM of WT or IRF8-cKO mice treated with vehicle, IFN-γ, or IL-4 for 24 hours (Figure 1B). We used IFN-γ as a positive control, since it is a potent inducer of IRF8 expression, and IL-4 as a negative control, since it signals through an IRF8-independent pathway (32, 33). We observed a significant decrease in IRF8 expression in IRF8-cKO, compared with WT macrophages, either untreated or treated with IFN-γ (Figure 1B and Supplemental Figure 1C). As expected, IL-4 treatment had no effect on IRF8 expression in either genotype (Figure 1B).
Figure 1. Conditional deletion of IRF8 does not impair myelopoiesis.
(A) Flow cytometry plots of cell surface CD11b and F4/80 expression by M-CSF–generated BM-derived macrophages (BMDMs) from C57BL/6-derived WT or IRF8-cKO mice. (B) Intracellular flow cytometric analysis of IRF8 expression by BMDMs from A, incubated with vehicle or IFN-γ (100 U/ml) or IL-4 (20ng/ml) for 24 hours. Data shown as mean fluorescence intensity (MFI) (n = 3 biologic replicates). (C) Flow cytometry plots of CD11c+F4/80+ macrophages from a bronchial alveolar lavage (BAL) of WT or IRF8-cKO mice, as in A. (D) Intracellular flow cytometric analysis of IRF8 expression by BAL-derived macrophages from C, incubated with vehicle or IFN-γ (100 U/ml) for 24 hours. Data shown as MFI (n = 5–6 biologic replicates). (E) iNOS or Arg1 mRNA levels by BMDMs from A incubated with vehicle or IFN-γ (100 U/ml) or IL-4 (1 ng/ml) for 24 hours. (F) Percentages of monocytes in peripheral blood from WT (IRF8fl/fl) or IRF8-cKO mice. (G) Percentages of CD11b+F4/80+ macrophages in spleens from WT (IRF8fl/fl) or IRF8-cKO mice. (H) Percentages of the indicated progenitors of WT or IRF8-cKO mice. n = 6 mice for each group pooled from 2 separate experiments for panels F–H. No significant differences were observed between WT and IRF8-cKO mice for all parameters examined in H. Data represent mean ± SEM, and statistical significance was determined by a 2-tailed Mann-Whitney U test. *P < 0.05.
In addition to BMDMs, we examined whether IRF8 deficiency had an impact on tissue-resident bronchial alveolar (BAL) macrophages. Therefore, we analyzed IRF8 expression in BAL macrophages, defined as CD11blo/midF4/80+CD11c+, from WT or IRF8-cKO mice (Figure 1C and Supplemental Figure 1D). Consistent with what we observed with BMDMs, BAL macrophages from IRF8-cKO mice compared with the WT controls expressed little to no IRF8 with or without IFN-γ treatment (Figure 1D and Supplemental Figure 1E).
To determine whether IRF8 deficiency altered the function of BMDMs, we analyzed mRNA levels of the hallmark IFN-γ–inducible IRF8 target gene, iNOS (24). In contrast to WT macrophages, which showed significant iNOS induction after IFN-γ treatment, macrophages from IRF8-cKO mice showed minimal iNOS upregulation under the same conditions (Figure 1E). The expression of the non-IRF8 target gene, Arg1, was similarly induced after IL-4 treatment in both genotypes, demonstrating that macrophages from IRF8-cKO mice are functional (Figure 1E). These data indicate that the loss of IRF8 expression in macrophages in this model did not impair their development, but rather their function, as determined by the lack of induction of iNOS as a prototypical IRF8-regulated target gene.
To further demonstrate that IRF8 deficiency in this Lyz2-Cre model did not impact monocyte/macrophage development under steady-state conditions, we examined peripheral tissues for monocyte and macrophage frequencies. We observed that IRF8 deficiency, compared with the WT controls, did not alter monocyte frequencies in the blood, as well as macrophage percentages in the spleen (Figure 1, F and G). Additionally, we did not observe significant differences in the absolute numbers of RBCs or granulocytes in IRF8-cKO mice compared with the WT controls, as measured by complete blood count analysis (CBC) (Supplemental Figure 2, A and B). Interestingly, we observed a reduction in the absolute numbers of WBCs and lymphocytes in IRF8-cKO mice compared with the WT controls (Supplemental Figure 2, A and C). Despite those differences, the percentages and absolute numbers of splenic B cells, CD3+ T cells, or granulocytes in WT vs. IRF8-cKO mice were not significantly different (Supplemental Figure 2D). Collectively, these data suggested that IRF8 deficiency in this model did not profoundly impact the development of such peripheral leukocyte populations, particularly with respect to an elevation in or expansion of diverse hematopoietic populations typically seen in the IRF8-null mouse model (26).
To further confirm that the loss of IRF8 expression in this model did not globally impact myelopoiesis, relative to that observed in IRF8–global KO mice (26), we quantified progenitors in the BM of WT (IRF8fl/fl) or IRF8-cKO mice by comprehensive flow cytometry. We focused on 3 major progenitor populations: hematopoietic and stem cell progenitors (HSPCs, namely the Lin–Sca-1+Kit– [LSK] fraction), megakaryocyte-erythroid progenitors (MEPs) progenitors, and granulocyte-monocyte progenitors (GMPs, early- or late-stage granulocyte or monocyte progenitors). Although, in some instances, we observed variability among the mice, we observed no significant differences in the frequencies of LSKs, MEPs, or GMPs (early or late stage) in WT vs. IRF8-cKO mice (Figure 1H). Overall, our data demonstrate that conditional deletion of IRF8 in this model did not globally alter hematopoiesis in the BM or the periphery, but it did significantly reduce IRF8 expression in macrophages originating from the BM or a tissue-resident site.
IRF8 deficiency in macrophages leads to increased lung metastasis.
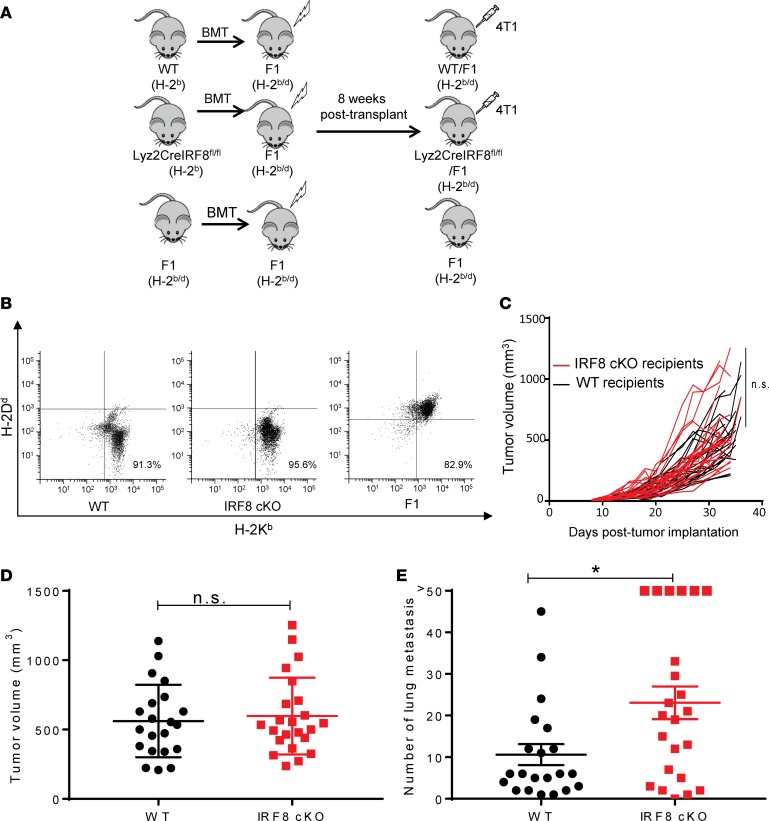
To test our hypothesis that IRF8 expression in macrophages is an important determinant for the control of metastasis, we used the well-characterized orthotopic 4T1 mammary tumor model (34). The 4T1 tumor model is spontaneously metastatic, particularly to the lung, which is a common site of metastasis for breast cancer (35). 4T1 is syngeneic to BALB/c (H-2d) mice, while the IRF8-cKO mouse model is on a C57BL/6 (H-2b) background. Therefore, to make this experiment genetically feasible, we developed a BM chimera approach wherein CB6F1/J mice (H-2b/d), a semisyngeneic strain bearing genetic elements compatible to sustain both the BM transplant and tumor challenge, were first lethally irradiated, followed by hematopoietic reconstitution using T cell–depleted BM cells from either WT or IRF8-cKO donors (Figure 2A).
Figure 2. Reduced IRF8 expression in macrophages leads to increased 4T1 spontaneous lung metastasis.
(A) Experimental design for the BM chimera tumor model. CB6F1 mice (H-2b/d) were lethally irradiated and then transplanted with WT, IRF8-cKO (Lyz2-CreIRF8fl/fl), or F1 T cell–depleted BM cells. Eight weeks after transplant, reconstitution was determined, followed by tumor implantation. (B) Flow cytometry plots of the reconstituted WT, IRF8-cKO, and F1 genotypes, as determined by peripheral blood analysis 8 weeks after transplant. (C) The chimeric recipients were then implanted with 4T1 cells (5 × 104) in the fourth mammary gland, and tumor growth was measured. (D) End-point tumor volumes for WT or IRF8-cKO hosts, which were not significant. (E) Lungs were collected ~30 days after tumor implantation for histopathologic analyses. Whole lung sections were H&E stained and analyzed in a blinded manner for the number of metastatic foci. Metastatic counts beyond 50 were considered too numerous to count accurately and, thus, are recorded as >50. Metastasis in the IRF8-cKO recipients, compared with the WT controls, was statistically significant by a 2-tailed Mann-Whitney U test (mean ± SEM of 21–23 mice per group, *P < 0.05). Data in C–E were compiled from 4 separate experiments.
Flow cytometric analysis of peripheral blood or specific myeloid or lymphoid cell types confirmed efficient hematopoietic repopulation, based on coexpression of donor (H-2b) and host (H-2d) MHC class I alleles (Figure 2B and Supplemental Figure 3). Eight weeks after transplantation, these chimera recipients were implanted with 4T1 tumor cells into the mammary gland, and primary tumor growth was measured over time. No significant difference was observed between the 2 cohorts with respect to primary tumor growth rate (Figure 2, C and D). In contrast, we observed a significant difference in the number of spontaneous lung metastases with the IRF8-cKO recipients exhibiting increased metastatic lesions compared with the WT counterparts at similar endpoint tumor volumes (Figure 2E and Supplemental Figure 4A). While both cohorts displayed demonstrable lesions, it is important to emphasize that the difference in metastasis between the IRF8-cKO cohort and the WT control was significant. It is also important to note that this comparison was performed at endpoint to maximize the contrast between the groups.
Differences in metastatic outcome did not reflect differences in macrophage infiltration into the primary tumor mass, as both WT and IRF8-cKO recipients contained comparable macrophage frequencies (Supplemental Figure 4, B–D). Furthermore, we examined the impact of tumor growth on changes in the frequencies or absolute numbers of several major BM progenitor or peripheral immune populations in WT vs. IRF8-cKO mice. First, we observed no significant differences in the frequencies of LSKs, MEPs, or GMPs (Supplemental Figure 5A). Second, we observed no significant differences in the absolute numbers of lymphocytes, monocytes, or granulocytes, as determined by CBC (Supplemental Figure 5B). Third, we examined macrophages, granulocytes, DCs, and lymphoid cells (CD4+ and CD8+ T cells) in the spleen, primary tumor, and lung microenvironment (Supplemental Figure 5C). Our findings indicated that, other than granulocytes in the spleen, we observed no significant differences in the frequencies of all other populations analyzed. These data suggest that IRF8 deficiency in this model did not profoundly further impact hematopoiesis under tumor-bearing conditions, particularly within the relevant TMEs. We observed no significant difference in angiogenesis, as both cohorts expressed comparable levels of CD31 (Supplemental Figure 6, A and B). Lastly, consistent with what we observed in the 4T1 model regarding the lack of effect of IRF8 deficiency on primary tumor growth, we observed a similar pattern in 2 additional tumor models, the AT-3 mammary carcinoma model (36) and the B16-F10 melanoma (Supplemental Figure 6, C and D).
IRF8 deficiency in macrophages leads to increased spontaneous lung metastasis after surgical resection of the primary tumor.
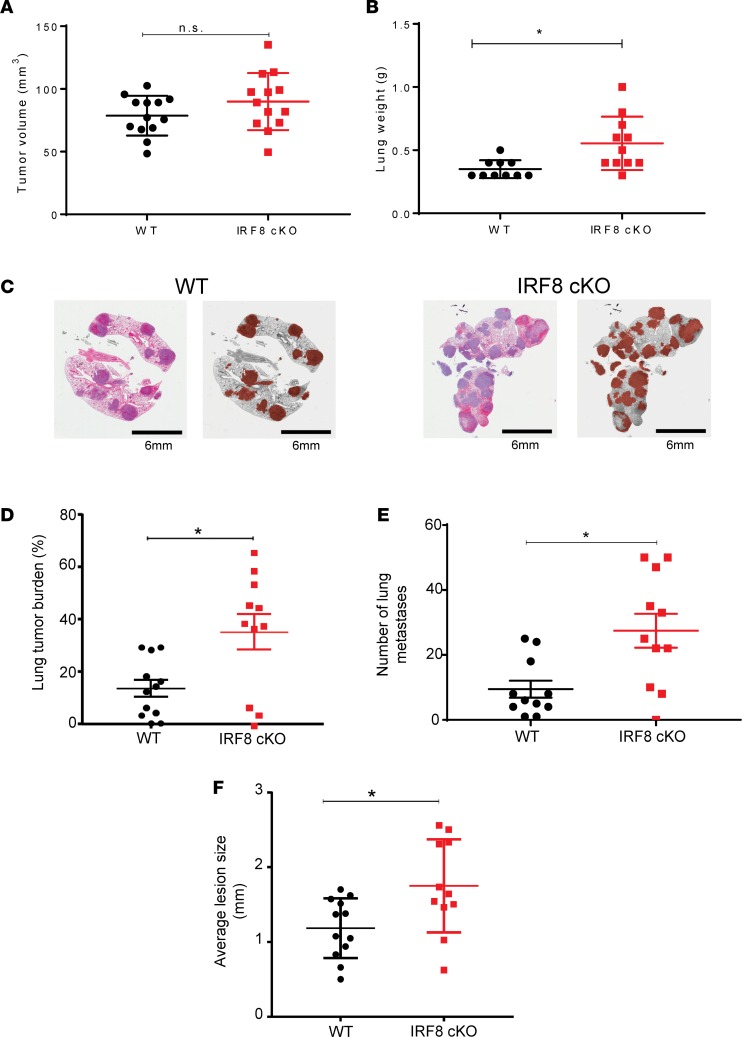
To further explore how the IRF8-macrophage axis governs spontaneous metastasis, we quantified lung metastasis after surgical resection of the primary tumor. 4T1 cells were implanted into both recipient cohorts, and when tumor size reached approximately 100 mm3 (Figure 3A), the tumor mass was surgically removed. Mice were then monitored for signs of morbidity for approximately 31–36 days. Afterward, lungs were removed and recorded for metastatic disease, based on weight, burden (i.e., percent occupied with cancer), and quantification of foci and average size of foci (Figure 3, B–F). Our data revealed that IRF8-cKO recipients displayed significantly enhanced metastatic burden compared with the WT recipients based on all of these quantifiable criteria. Moreover, the average size of tumor foci was significantly larger in IRF8-cKO compared with the WT recipients (Figure 3F), suggesting that IRF8 deficiency affected metastasis at multiple steps, including outgrowth or colonization. Thus, even in the absence of the primary tumor, IRF8 deficiency in macrophages played a significant role in governing spontaneous lung metastasis after surgery.
Figure 3. Reduced IRF8 expression in macrophages leads to increased spontaneous lung metastasis after surgical resection of the primary tumor.
As in Figure 2, recipient mice were implanted with 4T1 tumor cells and, when the primary tumor volume reached ~100 mm3, it was removed. (A) Mean primary tumor volumes on the day of surgery. (B) Thirty-one to 36 days after surgery, lungs were collected and weighed. (C) Representative photomicrographs of H&E-stained whole lung tissue (left), with a corresponding gray scale image with lung metastases shaded in brown (right). Scale bar: 6 mm. (D–F) Lung metastatic burden in WT vs. IRF8-cKO mice was quantified and reported as percent tumor burden in the lungs (D); quantification of discrete foci (E), similar to Figure 2; and average size of lung lesions (F). H&E-stained slides were analyzed in a blinded-manner for data in D–F. n = 10–11/group in panels A, B, and D–F, and statistical significance was determined by a 2-tailed Mann-Whitney U test. *P < 0.05.
IRF8 deficiency in macrophages leads to increased experimental lung metastasis.
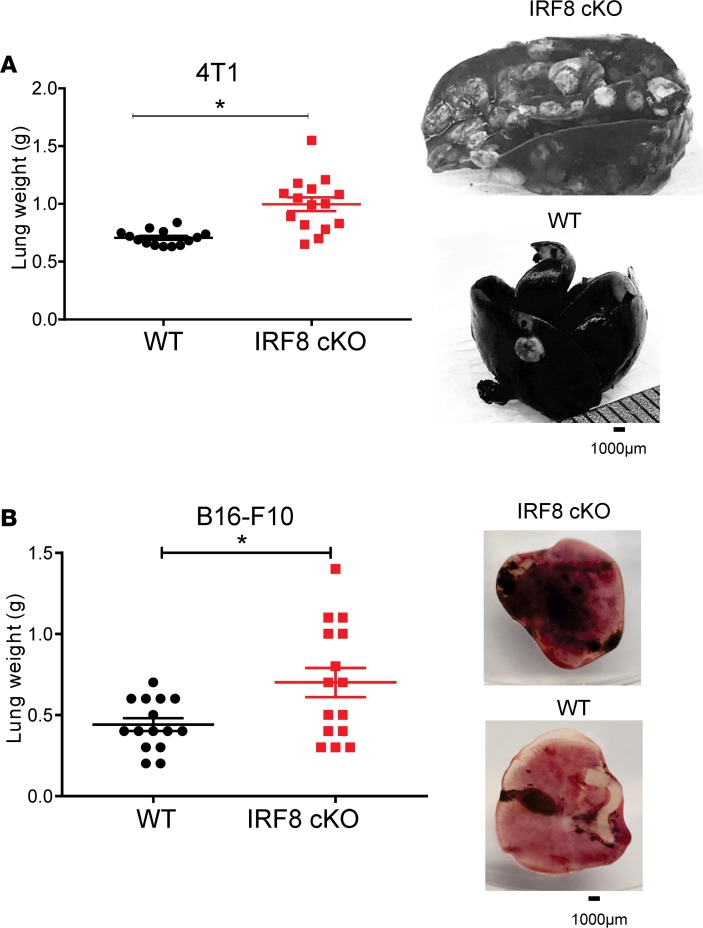
To further elucidate the mechanisms underlying metastasis in the absence of the primary tumor, we utilized an experimental lung metastasis approach, whereby 4T1 tumor cells were injected into the bloodstream. We assessed lung tumor burden approximately 30 days after tumor inoculation when 1 group of mice (i.e., IRF8-cKO) appeared moribund. Lungs were processed for India ink staining, weighed, and photographed to document the extent of tumor burden. Our findings revealed a significant increase in lung weight of IRF8-cKO recipients compared with the WT counterparts (Figure 4A), consistent with enhanced disease burden. Visual inspection supported the interpretation of differences in disease burden between the 2 recipient cohorts. Lungs from IRF8-cKO recipients displayed readily visible nodules compared with the lungs from WT controls (Figure 4A and Supplemental Figure 7).
Figure 4. Reduced IRF8 expression in macrophages leads to increased experimental lung metastasis.
(A) 4T1 cells (5 × 104) were injected i.v. into chimera mice reconstituted with WT (IRF8fl/fl) or IRF8-cKO BM cells, as in Figure 2. (B) B16-F10 melanoma cells (2 × 105) were injected similarly but in syngeneic WT or IRF8-cKO genotypes. Metastatic burden was quantified ~30 days after tumor inoculation by lung weight (Mann-Whitney U test, mean ± SEM of 15 mice per group; *P < 0.05). Right, representative images of whole lung. For the 4T1 model, India ink was used to stain the metastatic nodules, which appeared white against a black tissue background, while in the B16-F10 model, the nodules were readily detectable without stain, which appeared black against a pink tissue background.
These findings were extended to a second tumor model using the B16-F10 melanoma cell line. In this case, we made use of a fully syngeneic C57BL/6 system in which B16-F10 cells were injected i.v. into WT or IRF8-cKO mice. As with the 4T1 studies, the extent of lung tumor burden was determined by weight and visual inspection approximately 30 days after tumor injection. Lungs from IRF8-cKO mice were significantly heavier compared with those of WT mice (Figure 4B). Lung tumor growth was clearly visible without India ink staining, as the nodules appeared black due to the pigmented nature of the cell line (Figure 4B). Thus, our data demonstrate that IRF8 deficiency in macrophages enhanced experimental metastasis or colonization in both mouse models of mammary cancer and melanoma.
IRF8-deficient macrophages associate with an enrichment of metastasis-associated genes.
Given that IRF8 controls numerous functional properties of macrophages, such as those important for host defense and adaptive immunity (37), we used RNA sequencing (RNA-seq) experiments to identify potentially novel IRF8-dependent pathways or processes to further expand knowledge of macrophages in cancer. We compared gene expression profiles using BMDMs derived from syngeneic WT (IRF8fl/fl) or IRF8-cKO mice, since our intent was to focus on a highly purified population (i.e., void of contaminating or residual tumor or other stromal cell types) reflecting a single variable (i.e., presence or absence of IRF8). We used IFN-γ as a tool to induce IRF8 expression in vitro and to maximize the contrast between WT and IRF8-cKO macrophages in their regulation of IRF8-dependent pathways. All data are deposited under accession no. GSE116904 (https://www.ncbi.nlm.nih.gov/geo/). It is important to emphasize that the absence of IRF8 in macrophages does not globally impair their ability to respond to IFN-γ, since IRF8 is not the only transcription factor utilized for IFN-γ signaling (38, 39).
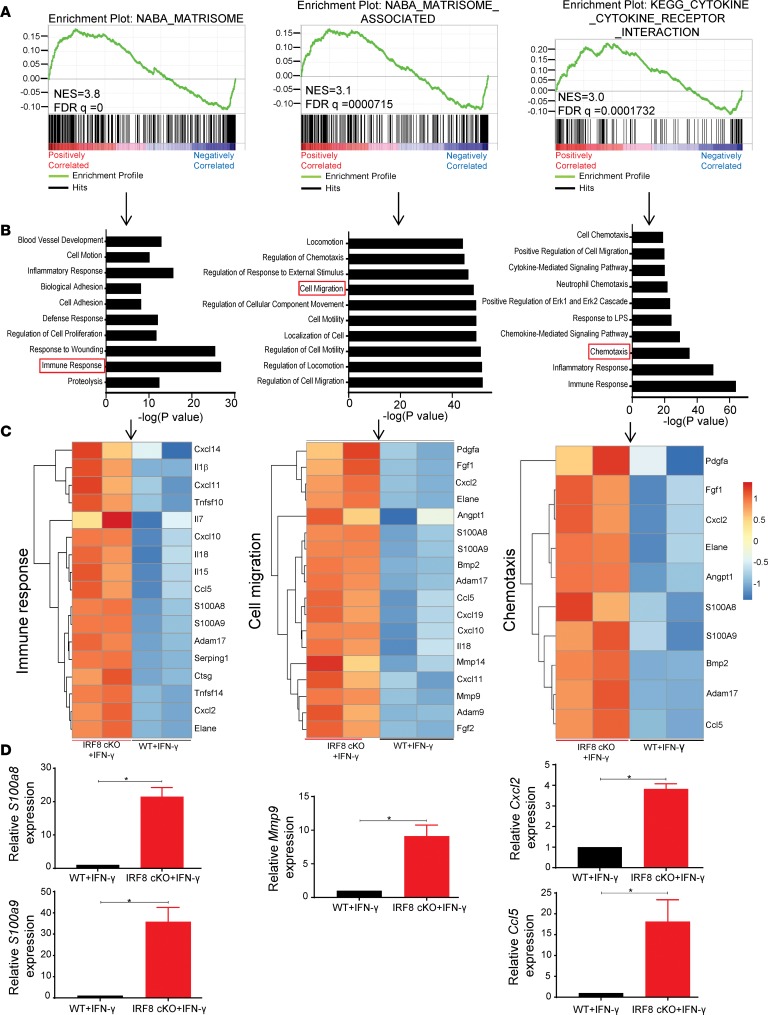
Gene set enrichment analysis (GSEA) was performed on genes up- or downregulated in IRF8-cKO compared with WT macrophages on both sets of macrophages after IFN-γ treatment. Interestingly, genes upregulated in the IRF8-cKO macrophages showed significant enrichment of matrisome, matrisome-associated, and cytokine-cytokine receptor gene sets (Figure 5A). This suggests that IRF8-deficiency in macrophages affects the ensemble of extracellular matrix proteins that are critical in regulating diverse facets of cellular behavior, including survival and migration. Using the genes from the 3 top pathways in the enrichment analysis, we performed a gene ontology analysis using the Database for Annotation, Visualization and Integrated Discovery (DAVID) (40). The top cellular processes included pathways for genes involved in the immune system, cell migration, and locomotion (Figure 5B). The genetic signature of IRF8-deficient macrophages treated with IFN-γ was distinctly different from WT macrophages treated with IFN-γ (Figure 5C and Supplemental Figure 8). Genes such as S100a8, S100a9, Mmp9, Cxcl2, Ccl5, and Mmp14 were upregulated in IFN-γ–treated IRF8-cKO macrophages compared with the controls (Figure 5C). These genes have been previously implicated in the metastatic process (41–46). We validated the expression of several of these highly upregulated genes by quantitative PCR (qPCR) analyses. S100a8, S100a9, Mmp9, Cxcl2, and Ccl5 were all upregulated in IFN-γ–treated IRF8-cKO macrophages compared with the controls (Figure 5D).
Figure 5. Reduced IRF8 expression in macrophages associates with an enrichment of metastasis-associated genes.
Transcriptome analysis of BMDMs from WT (n = 2 biologic replicates) or IRF8-cKO (n = 2 biologic replicates) mice after treatment with IFN-γ (100 U/ml for 24 hours) to maximize IRF8 induction. (A) GSEA for positively enriched genes in BMDMs from the IRF8-cKO model. NES, normalized enrichment score. (B) Overrepresented biologic processes showing genes upregulated in BMDMs from IFN-γ–treated IRF8-cKO mice compared with the IFN-γ–treated WT controls, as determined by gene ontology (GO) analysis. (C) Heatmaps depicting a subset of upregulated genes involved in immune response, cell migration, and chemotaxis (as boxed in B) of BMDMs from the same groups in A and B. Full heatmaps can be found in Supplemental Figure 9. (D) Validation of the indicated genes by qPCR analyses (mean ± SEM of 6 replicates pooled from 2 biologic or independent experiments). Statistical analysis was determined by a 2-tailed Mann-Whitney U test. *P < 0.05.
Lower tumor IRF8 expression levels are associated with decreased survival in human breast and lung cancer, as well as melanoma.
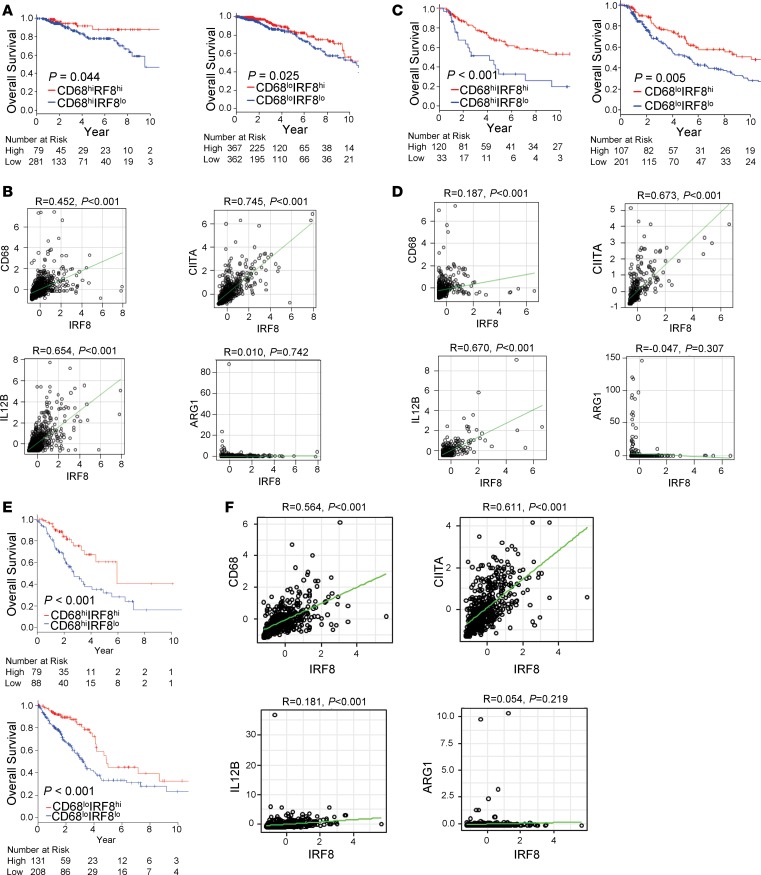
It is known that increased macrophage infiltration has been associated with poor prognosis in many solid cancers, including breast, lung, and melanoma (47–52). In an effort to translate our preclinical findings regarding the macrophage-IRF8 axis to patient outcome, we asked whether IRF8 expression is associated with patient survival in tumors (breast, melanoma, and lung) with higher macrophage infiltration, as well as those with lower macrophage infiltration. We utilized the The Cancer Genome Atlas (TCGA), a publicly available dataset, which has both gene expression data from RNA sequence and patient survival information. CD68, a hallmark pan marker of human macrophages (49), was used to determine macrophage presence in the tumors. Patients were initially stratified into CD68hi or CD68lo based on their expression levels. In the breast cancer dataset, we observed that lower IRF8 expression was associated with worse overall survival in patients with a higher macrophage infiltration (P = 0.044) (Figure 6A, left). Moreover, lower IRF8 expression was associated with worse overall survival in patients with a lower macrophage infiltration (P = 0.025) (Figure 6A, right). These findings imply that, even in patients with a low macrophage infiltration, IRF8 expression associates with breast cancer outcome.
Figure 6. A macrophage signature with higher IRF8 expression compared with lower IRF8 expression is associated with increased survival in patients with breast cancer, melanoma, and lung cancer.
(A) Kaplan-Meier survival curves based on TCGA data showing an association of higher IRF8 expression in breast cancer patient tumors with CD68+ macrophages (high [n = 360; left panel] or low [n = 729; right panel] macrophage infiltration) correlates with improved overall survival in breast cancer. (B) Positive correlation between IRF8 and CD68, CIITA, or IL12B and a negative (or no) correlation with ARG1 in breast cancer, as determined by Pearson correlation. (C) Kaplan-Meier survival curves in melanoma patients based on TCGA data, as in A (high [n = 153; left panel] or low [n = 308; right panel] macrophage infiltration). (D) Correlation plots as in B. (E) Kaplan-Meier survival curves in lung cancer patients based on TCGA data, as in A (high [n = 167; top panel] or low [n = 339; bottom panel] macrophage infiltration). (F) Correlation plots as in B. Statistical analysis of the survival curves was determined by the Log-rank test.
We further assessed the correlation between IRF8 and the expression of other genes known to be involved in macrophage function. In the breast cancer patient dataset, we observed that the expression of CD68, which reflects macrophage infiltration, was positively correlated with IRF8 expression (R = 0.452). The expression of class II transactivator (CIITA; R = 0.745) and IL12B (R = 0.654), whose upregulation in macrophages are associated with increased antigen presentation and activation of adaptive immunity (37), showed strong positive correlations with IRF8 expression (Figure 6B). In contrast, Arg1 expression, which is not known to be regulated by IRF8, did not correlate with IRF8 expression (R = 0.01).
Similar patterns were observed in the TCGA melanoma cohort. Patients with lower IRF8 expression showed worse overall survival, not only in patient tumors with a higher macrophage infiltration (P < 0.001), but also in those with a lower macrophage infiltration (P = 0.005) (Figure 6C). CD68 expression correlated with IRF8 expression (R = 0.187), although not as robustly as in the breast cancer cohort (Figure 6D). As with the breast cancer dataset, IRF8 expression positively correlated strongly with CIITA and IL12B expression (R = 0.673 and R = 0.670, respectively), but not Arg1 expression (R = 0.047) (Figure 6D). We extended our TCGA analysis to lung cancer, a cancer type also enriched in macrophages that correlate with a poorer overall survival (50–52). Consistent with what we observed in breast cancer and melanoma, we observed a similar macrophage-IRF8 signature in the lung cancer cohort (Figure 6, E and F). Lastly, since these TCGA datasets did not provide adequate information on metastasis-free survival, we took an alternative approach and analyzed patients in stages 3 and 4, which have metastatic disease. Importantly, we observed similar relationships between CD68 and IRF8 expression levels with survival in breast cancer and melanoma. In lung cancer, however, patient data were insufficient to perform statistical analysis (Supplemental Figure 9). Altogether, these data demonstrate that stratifying macrophages on the basis of varying IRF8 levels imparted clinical utility, suggesting that incorporating IRF8 expression levels as part of a macrophage signature strengthens prognostic merit.
Discussion
It is now well recognized that metastasis arises from the primary tumor via intrinsic and extrinsic mechanisms (4). Extrinsic mechanisms reflect the contribution of stromal and immune cells that infiltrate the primary TME or the metastatic niche, where they may precondition the secondary site for tumor dissemination. One of the most abundant immune populations is the macrophage, and its accumulation has been associated with reduced overall survival (7). Ordinarily, macrophages are critical for host defense against cancer. As tumors progress, however, macrophages may transition from tumor-suppressive to tumor-supportive states in response to the chronic inflammatory conditions (53). In this context, macrophages acquire the ability to suppress antitumor T cell responses and/or facilitate tumor invasion, angiogenesis, and colonization. Given the now well-appreciated role of macrophages in cancer biology, intensive efforts are devoted to understanding mechanisms that dictate such divergent functional states. Such functional states are governed by transcriptional pathways, and in this study, we identified a previously unrecognized role for IRF8 expression in macrophage-metastatic tumor biology.
While STAT1 has been linked to macrophage-mediated mechanisms of host defense (54), it is ubiquitously expressed and may comprise only part of the transcriptional network operational in antitumor macrophage biology. In contrast, IRF8 has long been tied to the regulation of myeloid development and function (37). It is important to note that increased IRF8 expression occurs in response to IFN-γ in a STAT1-dependent manner, and thus, IRF8 in macrophages is a downstream target of STAT1. IRF8, in turn, activates or represses the transcription of numerous downstream target genes that ultimately dictate macrophage developmental or functional fate (19).
In mouse models of mammary cancer and melanoma, we demonstrated that IRF8 expression in macrophages was an important determinant to inhibit lung metastasis, a common site for tumor dissemination (35). This important observation was tested in multiple cancer settings — metastasis in the setting of an intact primary tumor, metastasis after surgery, and metastasis in the complete absence of the primary tumor. Altogether, the use of multiple settings (a) provided additional rigor to support our major findings, (b) suggested that both primary and metastatic microenvironments are potentially relevant sites for macrophage-dependent mechanisms of metastasis, and (c) reflected different clinical scenarios whereby patients may present with metastasis either in the presence or absence of the primary malignancy. Future detailed studies are warranted to interrogate precisely which TME, primary or secondary, these interactions occur to impact metastatic outcome. Moreover, detailed work is needed to expand these findings to additional models across multiple tumor types, as well as to track IRF8 levels in the various IRF8-expressing cell types during disease progression.
The importance of IRF8 in myelopoiesis was elegantly unveiled in an IRF8-null mouse model. In this model, IRF8 expression was compromised through a germline alteration designed to impact hematopoietic fate. Consequently, progeny develop a myeloproliferative phenotype characterized by extensive granulopoiesis and impaired monocyte and DC development (26). In contrast to the role of IRF8 in developmental myeloid cell biology, we now demonstrate a previously unrecognized role for IRF8 in the regulation of antitumor responses by terminally differentiated macrophages. To that end, we made use of a mouse model with myeloid-specific IRF8 deletion after macrophage development (25, 27–31). Our data support this earlier finding, wherein we observed no alterations in BM progenitor and peripheral blood leukocyte subset frequencies under steady-state conditions, which is distinct from the IRF8-null model.
To gain insights into how IRF8 may affect macrophage behavior as it relates to metastasis, we adopted a broad-based unbiased approach using RNA-seq. We show that the loss of IRF8 in macrophages results in a set of genes that aligns with genes previously implicated to metastasis. For these molecular studies, we used in vitro–derived macrophages, which provided us with a highly purified population of cells that lacked potential residual contamination of other cell types generally composed within in vivo tissues. This approach also allowed us to focus on a single variable, the presence or absence of IRF8. As a result, we identified genes, namely S100a8, S100a9, Mmp9, Cxcl2, and Ccl5, which were all upregulated in IRF8-deficient macrophages compared with the controls. S100A8 and S100A9 are produced predominantly by myeloid cells, including macrophages, and act as chemoattractants to recruit suppressive myeloid populations that facilitate the formation of the premetastatic niche (46). The potential relevance of S100A8 and S100A9 in human breast cancer outcome is supported by data revealing that increased expression levels of these factors were associated with a significantly increased risk of death (55). Moreover, increased S100A8 protein levels, as measured by IHC, were strongly correlated with diminished metastasis-free survival in breast cancer patients (55).
CXCL2 and CCL5 may also act as chemoattractants for macrophages and other suppressive myeloid populations and contribute to metastasis. In fact, genetic ablation of CXCR2, the cognate receptor for CXCL2, suppresses metastasis (56). MMP9 was also identified and is involved in the degradation of extracellular matrix proteins, which enables tumor cells to invade basement membrane as part of the angiogenic process (57). MMP9 also plays a key role in the angiogenic switch through the release of VEGF, a major angiogenic growth factor (43). To assess the effects of IRF8 on tumor angiogenesis, we performed immunostaining of tumors for the endothelial marker CD31. Although individual tumors in IRF8-cKO mice showed lower vessel counts compared with controls, there was a considerable degree of heterogeneity between 4T1 tumors in the IRF8-cKO mice. As a result, we did not observe significant differences in microvessel density (MVD) between tumors established in WT and IRF8-cKO mice. This was somewhat surprising, given that we did observe higher levels of MMP9 expression in IRF8-cKO macrophages compared with the controls. However, several possibilities may account for a lack of an effect on angiogenesis, such as the notion that MMP9 alone was insufficient and/or the lung metastatic microenvironment may be a more relevant site. In this regard, noninvasive imaging methods could enable functional assessment of longitudinal changes in the lungs during the premetastatic phase and following establishment of metastases. Future work is required, however, to extend these findings using macrophages isolated from metastatic sites, as well as to validate the causal role of selected genes or gene combinations in gain- and loss-of-function studies.
Importantly, we extended the relevance of IRF8 expression to human breast and lung cancer, as well as melanoma. All 3 cancer types have been shown to be heavily infiltrated with immune cells — especially macrophages, which correlate with a poorer prognosis (47–52). While higher macrophage density has been associated with poorer clinical outcome in such solid cancer types, it is important to note that this relationship with survival is not all-or-none, meaning that not all patients with high macrophage content die and not all patients with low macrophage content survive. Thus, additional features of macrophages may help to explain variable survival outcomes within a given macrophage cohort. Our data support the hypothesis that stratification of a given macrophage cohort on the basis of IRF8 expression levels may be one factor that further teases apart such survival heterogeneity. This is evident by our findings that patients with a macrophagehiIRF8hi profile displayed a better survival outcome than patients with a macrophagehiIRF8lo profile. Similarly, patients with a macrophageloIRF8hi profile displayed a better survival outcome than patients with a macrophageloIRF8lo profile. These data suggest that incorporating IRF8 expression levels within a broader macrophage signature strengthens prognostic merit.
Although our TCGA analysis does not necessarily show IRF8 expression in macrophages, it is reasonable to posit that IRF8 expression in macrophages could be used to improve cell-based approaches for prognosis. Consistent with that notion, it is conceivable that the IRF8 signal is also emanating from other immune cell types, such as DCs, broadening the potential merit of IRF8 as a transcriptional biomarker for immune-based prognostication in tumor biology. Indeed, IRF8 expression correlated with macrophage presence, as well as IL-12B and CIITA expression, which are features necessary for adaptive immunity (58). In conclusion, our study revealed a potentially novel role for IRF8 in the mechanism by which macrophages control metastatic outcome in multiple tumor settings. These findings not only advance our understanding of macrophage-tumor biology, but also implicate the macrophage-IRF8 axis as a potential biomarker of disease status in certain types of solid malignancies.
Methods
Mice.
Mice with a loxP-flanked IRF8 gene [B6(Cg)-Irf8tm1.1Hm/J] were crossed to Lyz2-cre mice (B6.129P2-Lyz2tm1(cre)Ifo/J] [both strains from The Jackson Laboratory]) to generate B6 Lyz2CreIRF8fl/fl (IRF8-cKO). WT C57BL/6 (B6) or IRF8fl/fl B6 mice were used as WT controls. All experiments were performed using female mice 8–12 weeks of age. Female CB6F1 mice were obtained from The Jackson Laboratory.
Genotyping.
Half a centimeter of a tail from each mouse was incubated with DIRECTPCR Lysis Reagent (Viagen Biotech) and Proteinase K solution (Thermo Fisher Scientific) overnight at 55°C in a rotating Hybaid oven (Phoenix Equipment). Proteinase K enzyme was heat-inactivated by placing samples in a water bath at 85°C. DNA was used directly to perform reverse transcription PCR (RT-PCR) to specific primer sets provided by The Jackson Laboratory. Mouse primer sequences are listed in Supplemental Table 1. RT-PCR was conducted on a PTC-200 thermal cycler (MJ Research). To detect Lyz2-Cre in the DNA, the following conditions were used: 95°C for 5 minutes, 35 cycles (64°C for 30 seconds, 72°C for 45 seconds, 95°C for 30 seconds, 64°C for 30 seconds, 72°C for 45 seconds) and 72°C for 10 minutes. To detect the deleted IRF8 band, the following conditions were used: 94°C for 2 minutes, 30 cycles (94°C for 30 seconds, 60°C for 30 seconds, 72°C for 1 minute), and 72°C for 10 minutes. PCR products were then run on a 2% agarose gel stained with Red Safe (FroggaBio) and visualized under UV light using a Bio-Rad Gel-doc system.
Peripheral blood analysis.
Retro-orbital blood draws were performed on mice under anesthesia, and the blood was collected using heparinized capillary tubes (Thermo Fisher Scientific) or Microvette 100/200Z capillary blood tubes (Sarstedt). For CBC, blood was analyzed using IDEXX ProCyte Dx Hematology Analyzer.
In vitro differentiation of BMDMs.
Whole BM was isolated from the femurs and tibias of C57BL/6 WT or IRF8-cKO mice. RBC lysis was achieved using ACK Lysis Buffer (Thermo Fisher Scientific). Macrophages were differentiated and plated at 2 × 105/ml cells with recombinant mouse M-CSF at 30 ng/ml for 5 days. Macrophages were treated with IFN-γ or IL-4 (see Figure 1, Figure 5, Supplemental Figure 1, and Supplemental Figure 8 for details) and were analyzed for IRF8 expression 24 hours after treatment via flow cytometry. All cytokines were obtained from PeproTech.
Isolation of BAL macrophages.
Alveolar macrophages were isolated via bronchioalveolar lavage in 700 μl 3% BSA (Calbiochem) in PBS after mice were euthanized with Avertin (MilliporeSigma). Cells were then washed with RPMI media containing 10% FBS (Corning).
BM transplants.
Semisyngeneic transplants were performed using C57BL/6 WT or IRF8-cKO mice as donors and CB6F1 mice as the recipient hosts. BM from femurs and tibias were collected from the donors and depleted of total T cells using anti-CD90.2 microbeads (Miltenyi Biotec). Host female CB6F1 mice were lethally irradiated with 2 consecutive doses of 5.5 Gy of radiation from a 137Cs source. After 24 hours, recipient mice were injected i.v. with 2 × 106 T cell–depleted BM cells from either C57BL/6 WT or C57BL/6 IRF8-cKO donors. BM reconstitution was confirmed by flow cytometry analysis on peripheral blood samples approximately 8 weeks after transplant.
Cell lines and tumor growth experiments.
The 4T1 and B16-F10 tumor cell lines were purchased from the American Type Culture Collection (ATCC), authenticated at ATCC and maintained as described in ref. 36. The AT-3 tumor cell line was established from a primary mammary gland carcinoma of the PyMT-MMTV transgenic mouse model on a B6 strain and was maintained as described in ref. 36. Cell lines were confirmed to be mycoplasma negative using the Mycoplasma PCR Primer Set (Agilent). 4T1 (5 × 104) or AT-3 (1 × 105) cells were implanted orthotopically into the fourth mammary gland of female mice. Tumor growth was measured 3 times a week, and the volumes were calculated using the formula (w2 × l)/2, where w represents width and l represents length. B16-F10 (2 × 105) cells were injected i.v. into male WT, IRF8fl/fl or IRF8-cKO mice. Mice were euthanized after 28 days, and the lungs were isolated and weighed to determine metastatic burden.
Surgery.
CB6F1 hosts were orthotopically implanted with 4T1 cells in mammary gland no. 4. Tumor masses were surgically resected at approximately 100 mm3. Mice were monitored until ethical/humane endpoints were achieved, and lungs were recovered and quantified in a blinded-fashion for metastatic foci after H&E staining (as described below).
Tumor dissociation.
Primary tumor masses were surgically removed after euthanasia and placed into gentleMACS C-tubes (Miltenyi Biotec). Tumors were dissociated into a single cell suspension using a collagenase/hyaluronidase cocktail (Stemcell Technologies), as per manufacturer instructions, and the gentleMACS dissociator (Miltenyi Biotec). Tumors were then incubated in a Hybaid incubator (Phoenix Equipment) for 1 hour. The resulting mixture was strained through 100-μm SureStrain cell strainers (MTC Bio). The single cell suspension was then used for flow cytometry.
CD31 immunostaining and quantification.
Zinc-fixed paraffin sections were cut at 4 μm, placed on charged slides, and dried at 60°C for 1 hour. Slides were cooled to room temperature and added to the Dako Omnis Autostainer (Agilent Technologies), where they were deparaffinized with Clearify (American Mastertech) and rinsed in water. Slides were incubated with anti-CD31 antibody (clone MEC 13.3; BD Biosciences) at a 1/10 dilution for 50 minutes. Biotin goat anti–rat Ig (BD Biosciences; Supplemental Table 1) was applied for 30 minutes, followed by ELITE ABC (Vector Laboratories) for 30 minutes. DAB (Diaminobenzidine; Dako; Agilent Technologies) was applied for 5 minutes for visualization. Slides were counterstained with Hematoxylin for 8 minutes then placed into water. After removing slides from the autostainer, they were dehydrated, cleared, and cover slipped. Microvessels were manually counted in 5 independent 20× fields for each tumor (n = 5–6 tumors) using ImageJ image analysis software (Version 1.47, NIH) and reported as MVD.
Flow cytometry.
Cells were stained for 20 minutes at room temperature in FACS buffer (1× PBS with 5% FBS) and washed once with FACS buffer. All samples were analyzed on a LSRFortessa cytometer (BD Biosciences) running FACSDiva version 6.1.3, and data files were analyzed using FlowJo version 10 and WinList version 8. Directly conjugated antibodies and isotype controls included those reactive against B220 (RA3-6B2; BioLegend), CD11b (M1/70; BD Biosciences), CD11c (N418; BioLegend), CD16/32 (2.4G2; BD Biosciences), CD45.2 (104; BD Biosciences), F4/80 (BM8; BioLegend), Fixable Viability Stain 780 (BD Biosciences; Supplemental Table 1), H-2Dd (34-2-12; BD Biosciences), H-2Kb (AF6-88.5; BD Biosciences), and IRF8 (REA516; Miltenyi Biotec). Intracellular staining for IRF8 was achieved using the Miltenyi Biotec FoxP3 staining buffer set, per manufacturer recommendations. Directly conjugated antibodies used for cell surface staining for the BM progenitor analysis are listed in Supplemental Table 1.
Histology or India ink staining.
Sections (4 μm) were cut from formalin-fixed, paraffin-embedded lung tissues of tumor-bearing WT or IRF8-cKO mice. Histologic analyses were performed after specimens were stained with H&E. Lungs that received i.v. injections of tumor cells were inflated with 15% India ink solution and preserved in Fekete’s solution (70% EtOH, 37% formaldehyde, glacial acetic acid). Metastatic burden was determined by lung weight or enumeration of discrete metastatic lesions of H&E-stained slides in a blinded fashion. Metastatic counts beyond 50 were considered too numerous to count accurately and, thus, are reported as >50. For additional analyses of metastatic burden, whole tumor sections were captured and digitized using the ScanScope XT system (Aperio Technologies). The extent of metastatic disease in the lung was quantified on digitized images of H&E-stained sections and reported as (a) percent tumor burden (i.e., area of lung metastases expressed as a percentage of the entire lung) and (b) average lesion size in the lungs of WT and IRF-8 KO mice using Imagescope (Version 12.2.1; Aperio Technologies).
Molecular studies.
Total RNA was isolated using RNeasy Mini Kit (Qiagen). cDNA was synthesized using the iSCript cDNA synthesis kit (Bio-Rad). The cDNA was used for qPCR and used for PCR amplification using specific primer sets. qPCR was performed using CFX93 Real Time system (Bio-Rad). SYBR-Green (Invitrogen) was used as the dye for quantification. Data were quantified using the formula-fold change =2-ΔΔCt. All results were reported as ratio of the specific mRNA signal normalized to the indicated housekeeping gene. Mouse primer sequences are listed in Supplemental Table 2.
RNA-seq studies.
The sequencing libraries were prepared with the TruSeq Stranded Total RNA kit (Illumina) from 1 μg total RNA. Following the manufacturer’s instructions, the first step depletes rRNA from total RNA. After ribosomal depletion, the remaining RNA was purified, fragmented, and primed for cDNA synthesis. Fragmented RNA was then reverse transcribed into first strand cDNA using random primers. The next step removed the RNA template and synthesized a replacement strand, incorporating dUTP in place of deoxythymidine triphosphate (dTTP) to generate double-stranded (ds) cDNA. AMPure XP beads (Beckman Coulter) were used to separate the ds cDNA from the second strand reaction mix, resulting in blunt-ended cDNA. A single A nucleotide was then added to the 3′ ends of the blunt fragments. Multiple indexing adapters, containing a single T nucleotide on the 3′ end of the adapter, were ligated to the ends of the ds cDNA, preparing them for hybridization onto a flow cell. Adapter ligated libraries were amplified by PCR, purified using Ampure XP beads, and validated for appropriate size on a 4200 TapeStation D1000 Screentape (Agilent Technologies). The DNA libraries were quantitated using KAPA Biosystems qPCR kit and were pooled together in an equimolar fashion, following experimental design criteria. Each pool was denatured and diluted to 2.4 pM with 1% PhiX control library added (Illumina). The resulting pool was then loaded into a 75 cycle NextSeq v2 HO Reagent cartridge and sequenced on a NextSeq500 following the manufacturer-recommended protocol (Illumina).
RNA-sequencing analyses.
Twelve single-end biological samples were obtained. A total of approximately 40 million reads were reported for each sample. First pass base pairs quality control (QC) was performed using fastqc (v0.10.1). Spliced alignment was done using Bowtie (v1.0.1) and TopHat (v2.0.13), allowing a maximum of 1 mismatch per read. Alignment and downstream analyses were performed using GRCm38 (vM10) GENCODE reference and annotation of the mouse genome. Separate lane replicates were merged into a single sample alignment file using MergeSamFiles from Picard (v1.97). Second pass QC was done using alignment output with RSeQC (v2.6.3) in order to check genomic features’ abundances, junction saturation, and gene-body coverage. Read counts were estimated with HTSeq (v0.9.1) using intersection-strict option. Differential expression analyses were performed using DESeq2 (v1.18.1). Downstream and visualization plots are done using regularized log2 transformation implemented by DESeq2. Heatmaps were generated using the R-3.4.1 library pheatmap (v1.0.8). All data are deposited under accession no. GSE116904.
Data acquisition and preprocessing from TCGA.
There were 1093 breast cancer patients with clinical and primary tumor mRNA expression data from RNA sequence available through TCGA. Of those, there are 1090 patients with available overall survival (OS) data. There were 472 melanoma patients with clinical and mRNA expression data from RNA sequence. Among them, there are 432 patients with available OS data. There were 525 lung cancer patients with 506 patients having mRNA expression and OS data. The clinical and the gene expression quantification data (mRNA expression Z-score from RNA sequence) were downloaded through the cBioportal (http://cbioportal.org) (59, 60) and processed, as previously described (61). Patients were classified as either CD68hi or CD68lo, based on expression level using upper tertile cutoff (top 33% high, remaining 67% low), further stratified either IRF8hi or IRF8lo, based on expression level by testing cutoffs using the percentile whose association with OS of the whole cohort had the lowest P value (62). The prognostic differences were analyzed using Kaplan-Meier methods with Log-rank test. Pearson correlation between IRF8 and other genes of interest were analyzed, based on the expression levels. All TCGA statistical analyses were performed using R software (http:///www.r-project.org/) and Bioconductor (http://bioconductor.org/). All tests were 2-sided, and P < 0.05 were considered statistically significant.
Statistics.
Data were analyzed using GraphPad Prism (v7.04). Data were recorded as the mean ± SEM of the indicated number of mice, or biologic or experimental replicates. Differences in primary tumor growth over time between genotypes were determined by a 2-tailed t test at individual time points. Differences in all measurements of metastatic outcome between genotypes were determined by a 2-tailed Mann-Whitney U test. Similarly, differences in cell populations or IRF8 expression levels as assessed by flow cytometry, or mRNA expression levels for the indicated genes as assessed by qPCR, were determined by a 2-tailed Mann-Whitney U test. OS was determined by the log-rank test. Correlation proficiency between 2 variables was determined by Pearson correlation. In all cases, P < 0.05 was considered statistically significant.
Study approval.
All studies in mice were performed under protocol no. 1117M approved by the IACUC of the Roswell Park Comprehensive Cancer Center.
Author contributions
DYFT, MJN, and SIA conceived and designed the study. DYFT, SHC, MS, MJN, MLH, and SIA provided reagents and acquired data. DYFT, SHC, MLH, EK, NCH, ECG, MS, MJN, and SIA analyzed and interpreted data. DYFT, EK, MS, MJN, and SIA wrote the manuscript. SIA supervised the study.
Supplementary Material
Acknowledgments
This work was supported by R01CA172105 from the National Cancer Institute/NIH (to SIA), an NIH training grant T32CA085183 (to both DYFT and SHC), and a predoctoral fellowship award F31CA228396 (to SHC). All Roswell Park core resources were supported through an NCI Cancer Center support grant P30CA016056.
Version 1. 02/07/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(3):e124267. https://doi.org/10.1172/jci.insight.124267.
Contributor Information
Eriko Katsuta, Email: eriko.katsuta@roswellpark.org.
Eduardo Cortes Gomez, Email: Eduardo.CortesGomez@RoswellPark.org.
Mukund Seshadri, Email: mukund.seshadri@roswellpark.org.
References
- 1.Fidler IJ, Kripke ML. The challenge of targeting metastasis. Cancer Metastasis Rev. 2015;34(4):635–641. doi: 10.1007/s10555-015-9586-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Paget S. The distribution of secondary growths in cancer of the breast. 1889. Cancer Metastasis Rev. 1989;8(2):98–101. [PubMed] [Google Scholar]
- 3.Gupta GP, Massagué J. Cancer metastasis: building a framework. Cell. 2006;127(4):679–695. doi: 10.1016/j.cell.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 4.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell. 2017;168(4):670–691. doi: 10.1016/j.cell.2016.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang L, Lin PC. Mechanisms that drive inflammatory tumor microenvironment, tumor heterogeneity, and metastatic progression. Semin Cancer Biol. 2017;47:185–195. doi: 10.1016/j.semcancer.2017.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Binnewies M, et al. Understanding the tumor immune microenvironment (TIME) for effective therapy. Nat Med. 2018;24(5):541–550. doi: 10.1038/s41591-018-0014-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Twum DYF, Burkard-Mandel L, Abrams SI. The Dr. Jekyll and Mr. Hyde complexity of the macrophage response in disease. J Leukoc Biol. 2017;102(2):307–315. doi: 10.1189/jlb.4MR1116-479R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Martinez FO, Gordon S. The M1 and M2 paradigm of macrophage activation: time for reassessment. F1000Prime Rep. 2014;6:13. doi: 10.12703/P6-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condeelis J, Pollard JW. Macrophages: obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 10.Movahedi K, et al. Different tumor microenvironments contain functionally distinct subsets of macrophages derived from Ly6C(high) monocytes. Cancer Res. 2010;70(14):5728–5739. doi: 10.1158/0008-5472.CAN-09-4672. [DOI] [PubMed] [Google Scholar]
- 11.Grugan KD, et al. Tumor-associated macrophages promote invasion while retaining Fc-dependent anti-tumor function. J Immunol. 2012;189(11):5457–5466. doi: 10.4049/jimmunol.1201889. [DOI] [PubMed] [Google Scholar]
- 12.Beatty GL, et al. CD40 agonists alter tumor stroma and show efficacy against pancreatic carcinoma in mice and humans. Science. 2011;331(6024):1612–1616. doi: 10.1126/science.1198443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shime H, et al. Toll-like receptor 3 signaling converts tumor-supporting myeloid cells to tumoricidal effectors. Proc Natl Acad Sci USA. 2012;109(6):2066–2071. doi: 10.1073/pnas.1113099109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guiducci C, Vicari AP, Sangaletti S, Trinchieri G, Colombo MP. Redirecting in vivo elicited tumor infiltrating macrophages and dendritic cells towards tumor rejection. Cancer Res. 2005;65(8):3437–3446. doi: 10.1158/0008-5472.CAN-04-4262. [DOI] [PubMed] [Google Scholar]
- 15.Williams CB, Yeh ES, Soloff AC. Tumor-associated macrophages: unwitting accomplices in breast cancer malignancy. NPJ Breast Cancer. 2016:15025. doi: 10.1038/npjbcancer.2015.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Franklin RA, et al. The cellular and molecular origin of tumor-associated macrophages. Science. 2014;344(6186):921–925. doi: 10.1126/science.1252510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hussain SF, et al. A novel small molecule inhibitor of signal transducers and activators of transcription 3 reverses immune tolerance in malignant glioma patients. Cancer Res. 2007;67(20):9630–9636. doi: 10.1158/0008-5472.CAN-07-1243. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, et al. PD-L1 induced by IFN-γ from tumor-associated macrophages via the JAK/STAT3 and PI3K/AKT signaling pathways promoted progression of lung cancer. Int J Clin Oncol. 2017;22(6):1026–1033. doi: 10.1007/s10147-017-1161-7. [DOI] [PubMed] [Google Scholar]
- 19.Tamura T, Kurotaki D, Koizumi S. Regulation of myelopoiesis by the transcription factor IRF8. Int J Hematol. 2015;101(4):342–351. doi: 10.1007/s12185-015-1761-9. [DOI] [PubMed] [Google Scholar]
- 20.Abrams SI, Netherby CS, Twum DY, Messmer MN. Relevance of Interferon Regulatory Factor-8 Expression in Myeloid-Tumor Interactions. J Interferon Cytokine Res. 2016;36(7):442–453. doi: 10.1089/jir.2015.0174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Waight JD, et al. Myeloid-derived suppressor cell development is regulated by a STAT/IRF-8 axis. J Clin Invest. 2013;123(10):4464–4478. doi: 10.1172/JCI68189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tamura T, Nagamura-Inoue T, Shmeltzer Z, Kuwata T, Ozato K. ICSBP directs bipotential myeloid progenitor cells to differentiate into mature macrophages. Immunity. 2000;13(2):155–165. doi: 10.1016/S1074-7613(00)00016-9. [DOI] [PubMed] [Google Scholar]
- 23.Giese NA, et al. Interferon (IFN) consensus sequence-binding protein, a transcription factor of the IFN regulatory factor family, regulates immune responses in vivo through control of interleukin 12 expression. J Exp Med. 1997;186(9):1535–1546. doi: 10.1084/jem.186.9.1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simon PS, et al. The NF-κB p65 and p50 homodimer cooperate with IRF8 to activate iNOS transcription. BMC Cancer. 2015;15:770. doi: 10.1186/s12885-015-1808-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Clausen BE, Burkhardt C, Reith W, Renkawitz R, Förster I. Conditional gene targeting in macrophages and granulocytes using LysMcre mice. Transgenic Res. 1999;8(4):265–277. doi: 10.1023/A:1008942828960. [DOI] [PubMed] [Google Scholar]
- 26.Holtschke T, et al. Immunodeficiency and chronic myelogenous leukemia-like syndrome in mice with a targeted mutation of the ICSBP gene. Cell. 1996;87(2):307–317. doi: 10.1016/S0092-8674(00)81348-3. [DOI] [PubMed] [Google Scholar]
- 27.Cosin-Roger J, et al. Gp96 deficiency affects TLR4 functionality and impairs ERK and p38 phosphorylation. PLoS One. 2018;13(2):e0193003. doi: 10.1371/journal.pone.0193003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Faust N, Varas F, Kelly LM, Heck S, Graf T. Insertion of enhanced green fluorescent protein into the lysozyme gene creates mice with green fluorescent granulocytes and macrophages. Blood. 2000;96(2):719–726. [PubMed] [Google Scholar]
- 29.Forget MA, et al. Macrophage colony-stimulating factor augments Tie2-expressing monocyte differentiation, angiogenic function, and recruitment in a mouse model of breast cancer. PLoS One. 2014;9(6):e98623. doi: 10.1371/journal.pone.0098623. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 30.Lu N, et al. Activation of the epidermal growth factor receptor in macrophages regulates cytokine production and experimental colitis. J Immunol. 2014;192(3):1013–1023. doi: 10.4049/jimmunol.1300133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang W, et al. Ptpn11 deletion in a novel progenitor causes metachondromatosis by inducing hedgehog signalling. Nature. 2013;499(7459):491–495. doi: 10.1038/nature12396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kantakamalakul W, et al. Regulation of IFN consensus sequence binding protein expression in murine macrophages. J Immunol. 1999;162(12):7417–7425. [PubMed] [Google Scholar]
- 33.Humblin E, et al. IRF8-dependent molecular complexes control the Th9 transcriptional program. Nat Commun. 2017;8(1):2085. doi: 10.1038/s41467-017-01070-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aslakson CJ, Miller FR. Selective events in the metastatic process defined by analysis of the sequential dissemination of subpopulations of a mouse mammary tumor. Cancer Res. 1992;52(6):1399–1405. [PubMed] [Google Scholar]
- 35.Jin L, Han B, Siegel E, Cui Y, Giuliano A, Cui X. Breast cancer lung metastasis: Molecular biology and therapeutic implications. Cancer Biol Ther. 2018;19(10):858–868. doi: 10.1080/15384047.2018.1456599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stewart TJ, Liewehr DJ, Steinberg SM, Greeneltch KM, Abrams SI. Modulating the expression of IFN regulatory factor 8 alters the protumorigenic behavior of CD11b+Gr-1+ myeloid cells. J Immunol. 2009;183(1):117–128. doi: 10.4049/jimmunol.0804132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tamura T, Ozato K. ICSBP/IRF-8: its regulatory roles in the development of myeloid cells. J Interferon Cytokine Res. 2002;22(1):145–152. doi: 10.1089/107999002753452755. [DOI] [PubMed] [Google Scholar]
- 38.Blanco JC, Contursi C, Salkowski CA, DeWitt DL, Ozato K, Vogel SN. Interferon regulatory factor (IRF)-1 and IRF-2 regulate interferon gamma-dependent cyclooxygenase 2 expression. J Exp Med. 2000;191(12):2131–2144. doi: 10.1084/jem.191.12.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Langlais D, Barreiro LB, Gros P. The macrophage IRF8/IRF1 regulome is required for protection against infections and is associated with chronic inflammation. J Exp Med. 2016;213(4):585–603. doi: 10.1084/jem.20151764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dennis G, et al. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4(5):P3. doi: 10.1186/gb-2003-4-5-p3. [DOI] [PubMed] [Google Scholar]
- 41.Ager EI, et al. Blockade of MMP14 activity in murine breast carcinomas: implications for macrophages, vessels, and radiotherapy. J Natl Cancer Inst. 2015;107(4):djv017. doi: 10.1093/jnci/djv017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Halama N, et al. Tumoral Immune Cell Exploitation in Colorectal Cancer Metastases Can Be Targeted Effectively by Anti-CCR5 Therapy in Cancer Patients. Cancer Cell. 2016;29(4):587–601. doi: 10.1016/j.ccell.2016.03.005. [DOI] [PubMed] [Google Scholar]
- 43.Hiratsuka S, et al. MMP9 induction by vascular endothelial growth factor receptor-1 is involved in lung-specific metastasis. Cancer Cell. 2002;2(4):289–300. doi: 10.1016/S1535-6108(02)00153-8. [DOI] [PubMed] [Google Scholar]
- 44.Kessenbrock K, Plaks V, Werb Z. Matrix metalloproteinases: regulators of the tumor microenvironment. Cell. 2010;141(1):52–67. doi: 10.1016/j.cell.2010.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lim SY, Yuzhalin AE, Gordon-Weeks AN, Muschel RJ. Targeting the CCL2-CCR2 signaling axis in cancer metastasis. Oncotarget. 2016;7(19):28697–28710. doi: 10.18632/oncotarget.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Srikrishna G. S100A8 and S100A9: new insights into their roles in malignancy. J Innate Immun. 2012;4(1):31–40. doi: 10.1159/000330095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen P, et al. Tumor-associated macrophages promote angiogenesis and melanoma growth via adrenomedullin in a paracrine and autocrine manner. Clin Cancer Res. 2011;17(23):7230–7239. doi: 10.1158/1078-0432.CCR-11-1354. [DOI] [PubMed] [Google Scholar]
- 48.Zhao X, et al. Prognostic significance of tumor-associated macrophages in breast cancer: a meta-analysis of the literature. Oncotarget. 2017;8(18):30576–30586. doi: 10.18632/oncotarget.15736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Conway EM, et al. Macrophages, Inflammation, and Lung Cancer. Am J Respir Crit Care Med. 2016;193(2):116–130. doi: 10.1164/rccm.201508-1545CI. [DOI] [PubMed] [Google Scholar]
- 51.Hiraoka K, et al. Inhibition of bone and muscle metastases of lung cancer cells by a decrease in the number of monocytes/macrophages. Cancer Sci. 2008;99(8):1595–1602. doi: 10.1111/j.1349-7006.2008.00880.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mei J, et al. Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: A systemic review and meta-analysis. Oncotarget. 2016;7(23):34217–34228. doi: 10.18632/oncotarget.9079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pollard JW. Tumour-educated macrophages promote tumour progression and metastasis. Nat Rev Cancer. 2004;4(1):71–78. doi: 10.1038/nrc1256. [DOI] [PubMed] [Google Scholar]
- 54.Genard G, Lucas S, Michiels C. Reprogramming of Tumor-Associated Macrophages with Anticancer Therapies: Radiotherapy versus Chemo- and Immunotherapies. Front Immunol. 2017;8:828. doi: 10.3389/fimmu.2017.00828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Drews-Elger K, et al. Infiltrating S100A8+ myeloid cells promote metastatic spread of human breast cancer and predict poor clinical outcome. Breast Cancer Res Treat. 2014;148(1):41–59. doi: 10.1007/s10549-014-3122-4. [DOI] [PubMed] [Google Scholar]
- 56.Steele CW, et al. CXCR2 Inhibition Profoundly Suppresses Metastases and Augments Immunotherapy in Pancreatic Ductal Adenocarcinoma. Cancer Cell. 2016;29(6):832–845. doi: 10.1016/j.ccell.2016.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jablonska J, Lang S, Sionov RV, Granot Z. The regulation of pre-metastatic niche formation by neutrophils. Oncotarget. 2017;8(67):112132–112144. doi: 10.18632/oncotarget.22792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Marquis JF, et al. Interferon regulatory factor 8 regulates pathways for antigen presentation in myeloid cells and during tuberculosis. PLoS Genet. 2011;7(6):e1002097. doi: 10.1371/journal.pgen.1002097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gao J, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Cerami E, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Terakawa T, et al. High expression of SLCO2B1 is associated with prostate cancer recurrence after radical prostatectomy. Oncotarget. 2018;9(18):14207–14218. doi: 10.18632/oncotarget.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Budczies J, et al. Cutoff Finder: a comprehensive and straightforward Web application enabling rapid biomarker cutoff optimization. PLoS One. 2012;7(12):e51862. doi: 10.1371/journal.pone.0051862. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.