Abstract
Intronic polymorphisms in the α-ketoglutarate–dependent dioxygenase gene (FTO) that are highly associated with increased body weight have been implicated in the transcriptional control of a nearby ciliary gene, retinitis pigmentosa GTPase regulator-interacting protein-1 like (RPGRIP1L). Previous studies have shown that congenital Rpgrip1l hypomorphism in murine proopiomelanocortin (Pomc) neurons causes obesity by increasing food intake. Here, we show by congenital and adult-onset Rpgrip1l deletion in Pomc-expressing neurons that the hyperphagia and obesity are likely due to neurodevelopmental effects that are characterized by a reduction in the Pomc/Neuropeptide Y (Npy) neuronal number ratio and marked increases in arcuate hypothalamic–paraventricular hypothalamic (ARH-PVH) axonal projections. Biallelic RPGRIP1L mutations result in fewer cilia-positive human induced pluripotent stem cell–derived (iPSC-derived) neurons and blunted responses to Sonic Hedgehog (SHH). Isogenic human ARH-like embryonic stem cell–derived (ESc-derived) neurons homozygous for the obesity-risk alleles at rs8050136 or rs1421085 have decreased RPGRIP1L expression and have lower numbers of POMC neurons. RPGRIP1L overexpression increases POMC cell number. These findings suggest that apparently functional intronic polymorphisms affect hypothalamic RPGRIP1L expression and impact development of POMC neurons and their derivatives, leading to hyperphagia and increased adiposity.
Keywords: Metabolism, Neuroscience
Keywords: Embryonic development, NPY, Obesity
Retinitis Pigmentosa GTPase Regulator-Interacting Protein-1 Like (RPGRIP1L) hypomorphism in POMC neurons affects POMC- and NPY-expressing neurons, and projections to the paraventricular hypothalamus.
Introduction
Genome-wide association studies have identified common obesity variants that map at over 100 loci across the genome (1). Our aim is to deconvolute the strongest genetic signal for adiposity within an ~47-kb interval in the first intron of α-ketoglutarate–dependent dioxygenase gene (FTO). Specific noncoding alleles within this interval are associated with ~3.5-pound and ~2-pound body weight differences, per risk allele, in adults and children, respectively (2–6). Two of the associated single nucleotide polymorphisms (SNPs), rs8050136 and rs1421085, alter DNA-binding of CUX1 (7, 8), a transcription factor involved in neurodevelopment and highly expressed in the arcuate hypothalamus (ARH) (7, 9, 10).9. In the CNS, CUX1 regulates the expression of FTO and 5′-adjacent RPGRIP1L (7, 8, 11) encoding a protein located at the transition zone of the primary cilium (12). Gene mutations in components of the primary cilium cause syndromic and nonsyndromic forms of obesity, such as Bardet-Biedl Syndrome (BBS), Alström, tubby (13–15), and MC4R/AC3 (16). The subcellular localization of RPGRIP1L at the gate between cytoplasm and cilium suggests it may regulate entry and exit of ciliary signaling components relating to energy balance. In support of this hypothesis, the leptin receptor long isoform (Lepr-b) colocalizes with RPGRIP1L (17). We have also reported that Rpgrip1l expression is downregulated in the hypothalamus of leptin-deficient mice and is upregulated by leptin administration (11), indicating that RPGRIP1L may convey anorectic effects via the canonical leptin signaling pathway. Mice with congenital systemic deletion of 1 Rpgrip1l copy, or 2 Rpgrip1l copies in leptin-receptor isoform b-expressing neurons, are hyperphagic, obese, and display diminished suppression of food intake in response to leptin administration (8, 17). Hence, it is possible that RPGRIP1L modulates food intake in adults through cilia-dependent regulation of Lepr-b trafficking and function.
Embryonic lethality due to Rpgrip1l global deletion in mice is accompanied by severe CNS abnormalities of the cerebellar vermis and midbrain, as well as dilation of ventricles (18, 19). Consistent with phenotypic effects in the CNS, Rpgrip1l hypomorphism in the developing cerebral cortex impairs neuronal migration (20). In humans, RPGRIP1L mutations in Joubert Syndrome (JBST) cause “the molar tooth” characterized by cerebral vermian hypoplasia, accompanied by elongation of the caudal tegmentum and dysplasia of the caudal medulla (19). The severity of developmental defects in individuals who lack functional RPGRIP1L may mask a potential role in energy homeostasis. However, a recent report described a JBST patient with mutations in ciliary adenosine diphosphate (ADP) ribosylation factor–like GTPase 13B (ARL13B) who presented with obesity and structural brain abnormalities similar to those individuals with RPGRIP1L inactivating mutations (21), suggesting that anatomical/functional CNS abnormalities caused by RPGRIP1L haploinsufficiency may cause obesity in humans.
Loss of RPGRIP1L in human primary cell cultures and mice results in increased ciliary axonemal length (17, 22), and systemic overexpression of Cux1 in mice increases cilia length and causes polycystic kidney disease, a typical ciliopathic phenotype (23). Collectively, these reports suggest that RPGRIP1L and CUX1 may be part of the same cilia-related pathway. Moreover, CUX1 loss of function affects aspects of cortical neurocircuit assembly of the corpus callosum (9, 10). Given the observed CNS and ciliary anatomical changes caused by alterations in RPGRIP1L and CUX1 expression, we hypothesized that CUX1-dependent transcriptional control of RPGRIP1L may account for some of the association of FTO intronic SNPs with adiposity by modulating regulation of food intake via cilia-dependent alterations of hypothalamic structure.
Results
Induction of Rpgrip1l hypomorphism in the adult ARH causes obesity
Arcuate stereotaxic injections.
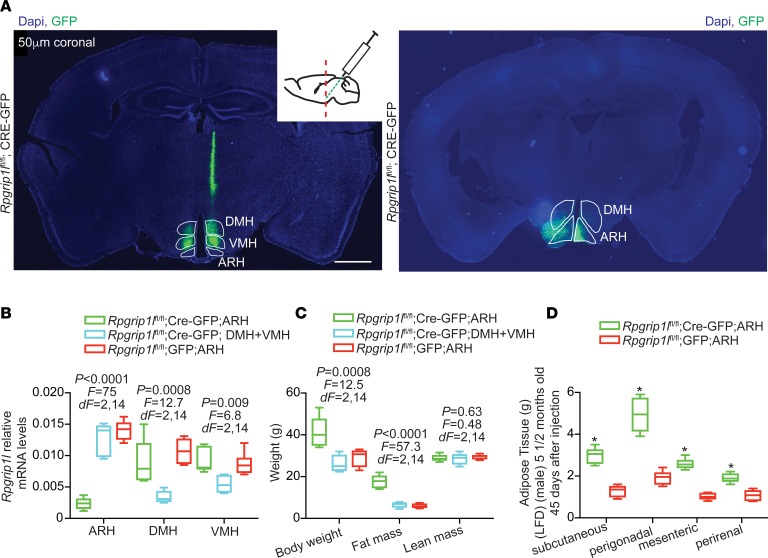
Mice segregating for 1 congenital null Rpgrip1l allele systemically (Rpgrip1l+/–) ingest more regular chow as early as 4 weeks of age and display increased adiposity by 12 weeks of age (17). Just as in congenital Rpgrip1l hypomorphism, Rpgrip1l hypomorphism induced in the adult CNS using tamoxifen-induced Nestin-Cre results in increased body weight and increased fat mass associated with increased food intake (8). To further refine the brain regions responsible for the increased food intake, we stereotaxically injected Cre recombinase–expressing adeno-associated viral particles (Cre-AAV) or GFP-expressing AAV (GFP-AAV) into the ARH of 12-week-old male Rpgrip1lfl/fl mice (Figure 1A). The mouse cohorts injected in the ARH were also compared with littermates that were targeted with Cre-AAV in both the dorsomedial (DMH) and ventromedial (VMH) hypothalami. By 16 weeks of age, ARH expression of Rpgrip1l in Cre-AAV–injected mice was decreased by ~90%; these mice were ~50% heavier than animals in which Cre-AAV had been injected in the DMH + VMH or mice GFP-AAV–injected in the ARH (Figure 1, B and C). The increase in body weight in mice injected with Cre-AAV in the ARH compared with mice injected with GFP-AAV in the ARH was due to an ~3-fold increase in fat mass (Figure 1, C and D). These findings suggest that effects of Rpgrip1l on body weight are conveyed, in part, via the ARH in adult animals and that Rpgrip1l hypomorphism induced in the DMH + VMH of adult animals has no impact on adiposity.
Figure 1. Adult-onset ARH Rpgrip1l deletion increases adiposity.
(A) Stereotaxic injections of Cre-GFP–expressing AAV in the ARH (Rpgrip1lfl/fl;Cre-GFP;ARH) or Cre-expressing AAV particles in the DMH and VMH (Rpgrip1lfl/fl;Cre-GFP;DMH+VMH) of male mice. Scale bars: 1 mm. (B) Decreased Rpgrip1l expression in the ARH of Rpgrip1lfl/fl;Cre-GFP;ARH (n = 6) and in the DMH/VMH of Rpgrip1lfl/fl;Cre-GFP;DMH+VMH mice (n = 5). Rpgrip1l expression is indistinguishable in the ARH, DMH, or VMH of mice stereotaxically injected with GFP-expressing in the ARH (Rpgrip1lfl/fl;GFP;ARH; n = 6). P < 0.05 was statistically significant, by 1-way ANOVA. (C) Increased body weight and fat mass in Rpgrip1lfl/fl;Cre-GFP;ARH mice. P < 0.05 was statistically significant, by 1-way ANOVA. (D) Mass increase of various fat depots of Rpgrip1lfl/fl;Cre-GFP;ARH mice compared with Rpgrip1lfl/fl;GFP;ARH mice. Data in B, C, and D are represented as box-and-whisker plots; boxes are the interquartile range, lines are the median value, and whiskers are minimum and maximum values. *P < 0.01, by 2-tailed Student’s t test.
Cerebellar stereotaxic injections.
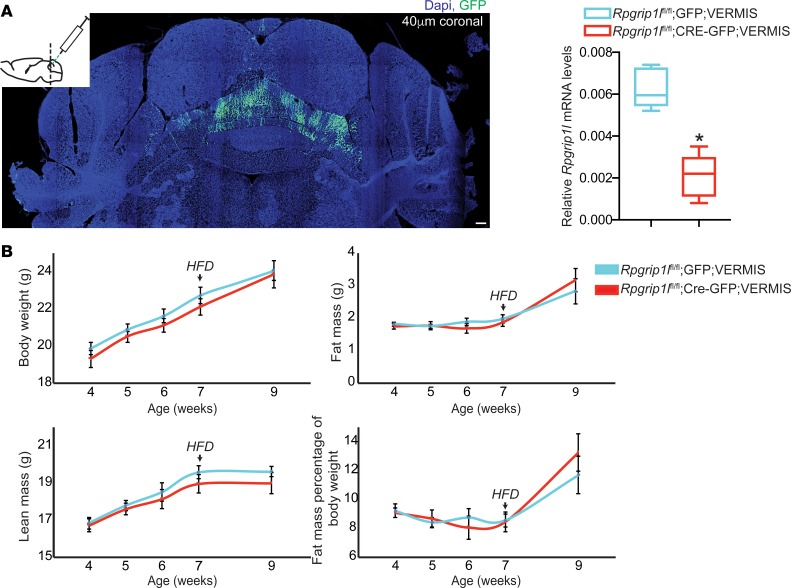
Because RPGRIP1L mutations in JBST patients result in cerebellar vermis hypoplasia/aplasia (24), we stereotaxically injected Cre-AAV into the developing cerebellar vermis of P10 male Rpgrip1lfl/fl mice (Figure 2A). At P10, the cerebellar vermis is still under development (25). Similar to the DMH + VMH, Rpgrip1l hypomorphism in the cerebellar vermis did not increase adiposity in mice fed regular chow or a high-fat diet (Figure 2B). These data indicate that cerebellar Rpgrip1l may not be implicated in the regulation of food intake.
Figure 2. Cerebellar Rpgrip1l hypomorphism has no effect on energy balance.
(A) Stereotaxic injection of P10 male mice at the cerebellar vermis with Cre-GFP–expressing AAV particles (Rpgrip1lfl/fl;Cre-GFP;VERMIS; n = 7) resulted in an ~85% decrease in Rpgrip1l expression compared with mice injected at the cerebellar vermis with GFP-expressing AAV particles (Rpgrip1lfl/fl;GFP;VERMIS; n = 8). Scale bar: 100 μm. (B) No difference in total body weight, fat mass, or lean mass between Rpgrip1lfl/fl;Cre-GFP;VERMIS and Rpgrip1lfl/fl;GFP;VERMIS male mice on ad lib regular chow up to 7 weeks of age or after 2 weeks on high-fat diet. Data in A are represented as box-and-whisker plots; boxes are the interquartile range, lines are the median value, and whiskers are minimum and maximum values. *P < 0.01, 2-tailed Student’s t test.
Adult-onset Rpgrip1l hypomorphism specifically in Pomc neurons does not cause adiposity
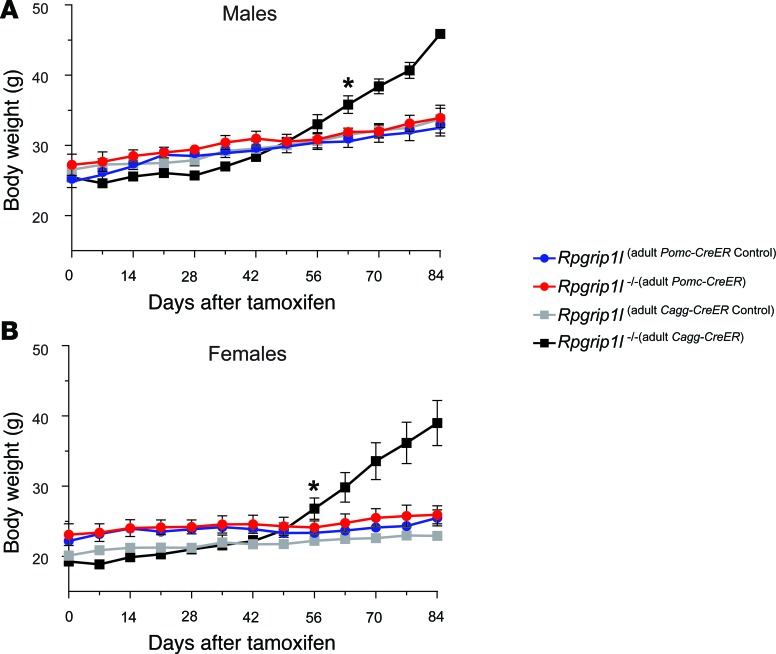
We have reported that 16-week-old mice congenitally hypomorphic for Rpgrip1l in Pomc-expressing neurons (Rpgrip1l–/–[Pomc]) are obese due to increased food intake (8). However, tamoxifen-induced loss of Rpgrip1l in terminally differentiated Pomc neurons in adult animals (Rpgrip1l–/–[adult Pomc–CreER]) at 8 weeks of age did not cause significant increases in body weight (Figure 3). In contrast, systemic Rpgrip1l loss (Rpgrip1l–/–[adult Cagg–CreER]) induced in adults phenocopies the obesity observed in Rpgrip1l–/–(Pomc) mice (Figure 3). These findings suggest that the developmental impact of Rpgrip1l hypomorphism on Pomc neurons is the sole driver for the increased adiposity in Rpgrip1l–/–(Pomc) mice.
Figure 3. Rpgrip1l loss in adult Pomc neurons has no effect on body weight.
(A and B) Loss of Rpgrip1 at 8 weeks of age in both (A) male and (B) female adult POMC neurons (Rpgrip1l–/–[adult POMC–CreER]; n = 4 male [♂] and n = 5 female [♀]) did not result in obesity compared with littermate controls (Rpgrip1l[adult POMC–CreER Control]; n = 9♂ and n = 7♀). In contrast to obese systemic Rpgrip1l loss (Rpgrip1l–/–[adult Cagg–CreER]; n = 5♂ and n = 4♀) compared with littermate controls (Rpgrip1l[adult Cagg–CreER Control]; n = 4♂ and n = 7♀). *P < 0.05 using 2-tailed Student’s t test.
Congenital Rpgrip1l hypomorphism in Pomc neurons alters Pomc and Npy neuron number
Pomc-GFP–positive adult neuron number assessed by IHC.
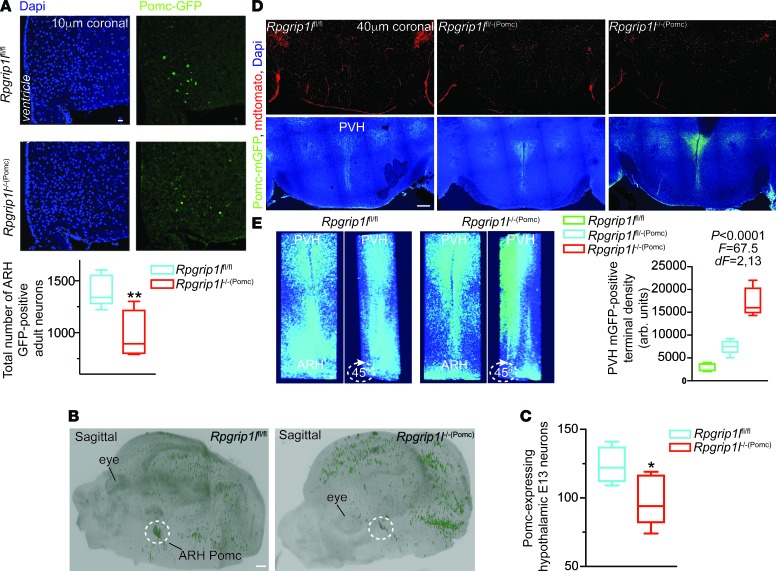
We hypothesized that congenital Rpgrip1l–/–(Pomc) mice may have reduced numbers of POMC ARH neurons. In 10 μm coronal sections spanning the ARH of adult male Rpgrip1l–/–(Pomc) mice segregating for the Pomc-GFP transgene, we found that the number of GFP-positive ARH neurons is reduced by ~50% (Figure 4A).
Figure 4. Neurostructural changes in mice congenitally deleted for Rpgrip1l in Pomc neurons.
(A) Decreased number of GFP-positive ARH neurons in Rpgrip1l–/–(Pomc) male mice (n = 4) compared with Rpgrip1lfl/fl mice (n = 6) segregating for the Pomc-GFP allele. Scale bar: 40 μm. (B) Sagittal view of whole E13 Rpgrip1l–/–(Pomc) and Rpgrip1lfl/fl mouse embryo heads cleared with iDISCO after immunostaining for GFP. Dotted circle indicates ARH Pomc neuroprogenitor localization. Scale bar: 500 μm. (C) Decreased number of Pomc progenitors in Rpgrip1l–/–(Pomc) (n = 5) compared with Rpgrip1lfl/fl embryos (n = 5) assessed by IHC and brain clearing. (D) Coronal sections of the PVH showing allele dose–dependent increase in GFP-positive PVH projections in mice congenitally deleted for 1 (Rpgrip1lfl/–(Pomc)) (n = 5) or 2 (Rpgrip1l–/–(Pomc)) (n = 5) Rpgrip1l alleles compared with Rpgrip1lfl/fl control mice (n = 6). Mice also carried the mT/mG allele. PVH projections are marked with mGFP. mTomato marks all other cells. Scale bar: 500 μm. P < 0.05 was statistically significant, by 1-way ANOVA. (E) Three-dimensional reconstruction from a stack of 40-μm coronal sections covering the whole PVH and part of the ARH in a 6-week Rpgrip1l–/–(Pomc) and Rpgrip1lfl/fl mouse showing increased PVH projections and decreased ARH mGFP-positive cells. Data in A, C, and D are represented as box-and-whisker plots; boxes are the interquartile range, lines are the median value, and whiskers are minimum and maximum values showing increased PVH projections and decreased ARH mGFP-positive cells. Scale bar: 100 μm (E). *P < 0.04, **P < 0.01, by 2-tailed Student’s t test.
Cell death of Pomc neurons assessed by TUNEL.
We examined the possibility that Rpgrip1l deletion in Pomc neurons leads to increased cell death. By TUNEL staining, we were unable to detect increased cell death in the developing hypothalamus of Rpgrip1l–/–(Pomc) E11 embryos (when Pomc expression is initiated) or by induced Rpgrip1l deletion in Nestin-expressing adult hypothalamic neurons (Supplemental Figure 1; supplemental material available online with this article; https://doi.org/10.1172/jci.insight.123337DS1). These findings suggest that congenital Rpgrip1l hypomorphism does not cause cell death.
Pomc progenitor neuron number assessed by iDISCO.
Using the brain-clearing iDISCO system and IHC, we quantified the number of Pomc-expressing ARH neurons of Rpgrip1l–/–(Pomc) mice at E13 when the numbers of Pomc ARH progenitors are in highest (26, 27). We noted an ~20% decrease in Pomc-expressing ARH cells in Rpgrip1l–/–(Pomc) mice (Figure 4, B and C).
Pomc ARH-PVH projections assessed by IHC.
We also utilized the mT/mG allele (28) in which the membrane-bound tdTomato (mT) marker is globally expressed in all tissues except those Pomc-Cre–expressing cells in which recombination results in silencing of mT and expression of membrane-bound GFP (mGFP), allowing visualization of cell bodies, projections, and terminals. We noted a marked increase in the density of anterior medial hypothalamic projections to the paraventricular hypothalamus (PVH) in Rpgrip1l–/–(Pomc) and Rpgrip1fl/–(Pomc) mice (Figure 4, D and E).
Npy-positive adult neuron number assessed by IHC.
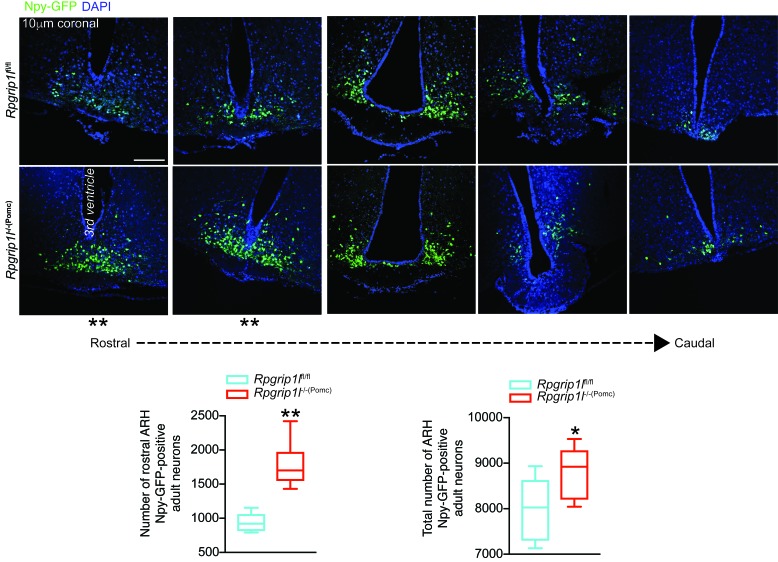
Bbs2- or Bbs6-null obese mice (murine models of the Bardet-Biedl ciliopathy) have an ~20% reduction in mature Pomc ARH neurons, as assessed by an anti-Pomc antibody (29). In contrast, in our congenital Rpgrip1l–/–(Pomc) mice, the Pomc-GFP transgene marks mature Pomc neurons that actively express GFP, as well as ARH neurons derived from Pomc-expressing early developmental progenitors (e.g., Npy neurons; refs. 26, 27). Pomc is expressed by the majority of cells in the developing ventral hypothalamus at E10–E11 and is silenced later in gestation in more than half of these cells, some of which become Npy neurons and some of which are directed to other terminal fates (26, 27). Therefore, the adiposity and hyperphagia of congenital Rpgrip1l–/–(Pomc) mice may be due to developmental changes in both mature Pomc ARH neurons and Pomc-derived ARH neurons. We explored the possibility that Rpgrip1l hypomorphism in Pomc neurons affected the number of Npy-expressing neurons that derive from Pomc-expressing progenitors. The total number of Npy-GFP–positive ARH neurons is increased by ~12% in Rpgrip1l–/–(Pomc) adult mice due to an ~2-fold increase in the number of Npy neurons in the rostral ARH of Rpgrip1l–/–(Pomc) mice (Figure 5). Respective increases and decreases of hypothalamic Pomc and Npy neurons have been implicated in hypophagia (30, 31). The converse changes in the congenital Rpgrip1l–/–(Pomc) mice are consistent with their hyperphagia.
Figure 5. Increased numbers of hypothalamic Npy neurons in male mice congenitally deleted for Rpgrip1l in Pomc neurons.
Increased number of Npy neurons in the rostral and whole ARH of 6-week Rpgrip1l–/–(Pomc) mice (n = 9) compared with Rpgrip1lfl/fl control mice (n = 6) also segregating for the Npy-GFP allele. Scale bar: 100 µm. Data are represented as box-and-whisker plots; boxes are the interquartile range, lines are the median value, and whiskers are minimum and maximum values. *P <0.04, **P < 0.01, by 2-tailed Student’s t test.
Diminished ciliation of iPSC-derived neurons of patients with RPGRIP1L mutations
Generation of iPSCs from JBST patients.
We and others have demonstrated that Sonic Hedgehog (SHH) signaling activation promotes human stem cell differentiation to ARH-type neurons that predominately express POMC (70%–80%) and exhibit other hypothalamus-specific patterns of gene transcription (32–34). We generated iPSCs from three JBST patient primary skin fibroblast lines UW15 (35), UW04-03 (36), and UW04-04 (17) with biallelic mutations in the RPGRIP1L C2 domain that severely disrupt — or are predicted to disrupt — nephrocystin-4 binding that facilitates recruitment of other proteins to the base of the cilium (37). The iPSCs expressed the expected pluripotency markers (Supplemental Figure 2). Pluripotency was further confirmed by injection of the iPSCs into immunocompromised NOD scid γ (NSG) mice resulting in the formation of teratomas that display ectodermal, mesodermal, and endodermal structures (Supplemental Figure 3A). The derived iPSCs had normal karyotypes (Supplemental Figure 3B).
Generation of JBST neurons from iPSCs.
We attempted to generate ARH-like neurons from the JBST iPSCs but were unable to generate NKX2.1-positive neuronal progenitors from which POMC and NPY neurons derive (38, 39), further suggesting the importance of RPGRIP1L in the development of ARH neurons. Subsequently, we utilized a dual-SMAD inhibition protocol (40) to direct differentiation of the JBST iPSCs into neurons positive for neuromarkers Nestin, neuron-specific class III β-tubulin (TUJ1), and Neurofilament (NF) (Supplemental Figure 3C).
Ciliation in iPSC-derived JBST neurons.
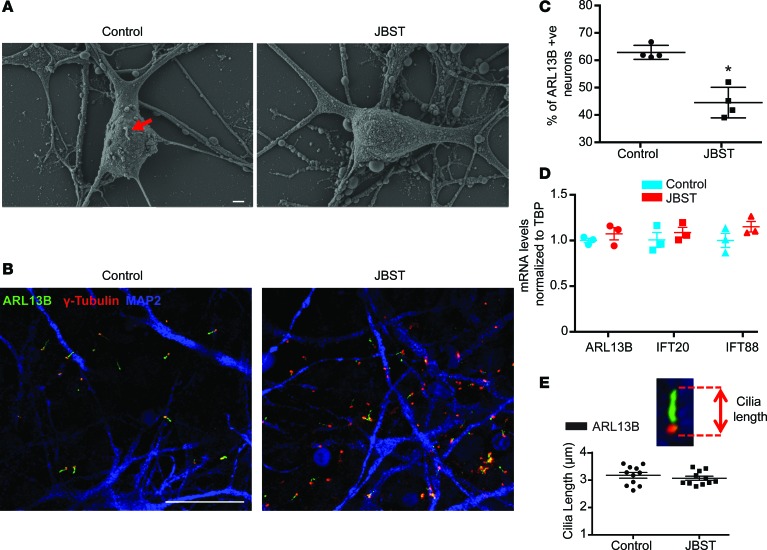
ARL13B is a small GTPase that participates in ciliary formation and function (41). About 60% of control neurons and ~40% of JBST neurons were ARL13B-positive (Figure 6, A–C). ARL13B, IFT20, and IFT88 mRNA levels were indistinguishable between control and JBST neurons (Figure 6D), suggesting that the ~30% decrease in ARL13B-positive JBST neurons is due to lack of cilia. No differences were observed in ciliary length between JBST and control neurons (Figure 6E).
Figure 6. Reduced ciliogenesis in human neurons with hypomorphic mutations of RPGRIP1L.
(A) Scanning microscopy of iPSC-derived Joubert Syndrome patient (JBST) and control neurons. Red arrow indicates a primary cilium. The iPSC-derived JBST neuron lacks a primary cilium. Scale bar: 1 μm. (B) Immunostaining showing decreased number of cilia positive for ARL13B in JBST neurons. MAP2 is a neuronal marker, and γ-tubulin marks the basal body. Scale bar: 20μm. (C) Decreased percentage of ARL13B-positive JBST neurons. Each dot represents 500 neurons. (D) mRNA levels of ARL13B and ciliary structural genes IFT20 and IFT88 are indistinguishable between control and JBST neurons. Each dot represents 1 cell line. Expression normalized to TATA box binding protein (TBP). (E) No difference in ARL13B-positive ciliary axonemal length between control and JBST neurons. Each dot represents the average of 500 neurons. *P < 0.01, by 2-tailed Student’s t test.
RPGRIP1L hypomorphism is associated with perturbed SHH signaling
SHH conveys important hypothalamic neurospecification signals in vitro (32); these are mediated via the primary cilium (39, 42). When SHH binds to its receptor, PATCHED1, the liganded receptor exits the primary cilium, promoting ciliary entry of Smoothened (SMO) and consequential upregulation of downstream effectors, such as GLI1 (43), and negative feedback upregulation of Patched1 (44).
Shh signaling in Pomc progenitors.
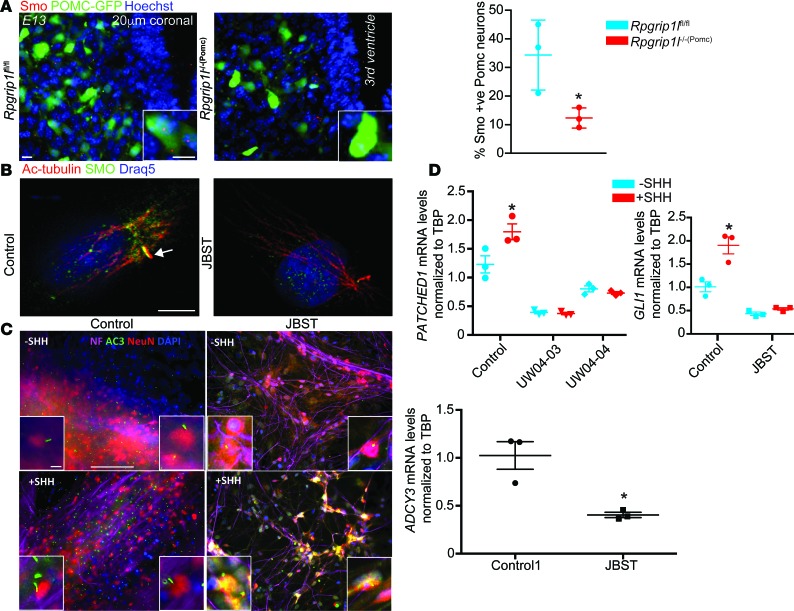
Since Shh signaling is diminished in the neural tube of E10 and E10 embryos deleted for Rpgrip1l (18), we tested the possibility that Shh signaling in the ARH is perturbed by Rpgrip1l hypomorphism. In the developing hypothalamus of Rpgrip1l–/–(Pomc) E13 embryos, ~15% of Pomc neuroprogenitors were Smo-positive compared with ~35% in Rpgrip1lfl/fl control embryos (Figure 7A).
Figure 7. Sonic Hedgehog (SHH) signaling is impaired in JBST patient fibroblasts and neurons.
(A) IHC showing decreased Smo-positive Pomc-expressing neuroprogenitors (green, Pomc-GFP) in E13 Rpgrip1l–/–(Pomc) (n = 3) compared with Rpgrip1lfl/fl embryos (n = 3). Scale bars: 10 μm. (B) Immunostaining of Smoothened (SMO) and Acetylated-tubulin (Ac-Tub) showing that JBST fibroblasts fail to translocate SMO to the cilium upon SHH stimulation (100 ng/ml). Scale bar: 5μm. Arrow indicates primary cilium. (C) Diminished AC3 expression and localization in the ciliary axoneme of iPSC-derived JBST neurons in the absence of SHH. SHH stimulation resulted in ectopic AC3 localization in the neuron cell body in iPSC-derived JBST neurons. Neuron cell body and projections are also marked with neuro-specific markers Neurofilament (NF) and NeuN. Each column represents the average of 3 cell lines. Scale bar: 100 μm. (D) PATCHED1 and GLI1 expression is not increased in iPSC-derived neurons from JBST patients UW04-03 and UW04-04 upon SHH stimulation. Each column represents the average of 3 cell lines. Expression was normalized to the TATA box binding protein (TBP). *P < 0.01, by 2-tailed Student’s t test.
SHH signaling in JBST fibroblasts and iPSC-derived JBST neurons.
In JBST human fibroblasts, SMO failed to localize to the primary cilium upon SHH exposure (Figure 7B). In iPSC-derived JBST neurons, AC3 expression was decreased by ~60% and was ectopically localized in the neuron cell body rather than the primary cilium upon SHH stimulation (Figure 7C). Finally, upregulation of PATCHED1 and GLI1 expression by ~40% and ~90%, respectively, after SHH treatment in control iPSC-derived neurons, was blunted in iPSC-derived JBST neurons (Figure 7D), further suggesting aberrant SHH signaling.
Alleles at rs1421085/rs8050136 and RPGRIP1L expression correlate with human embryonic stem cell–derived (ESc-derived) POMC-positive ARH neuron number
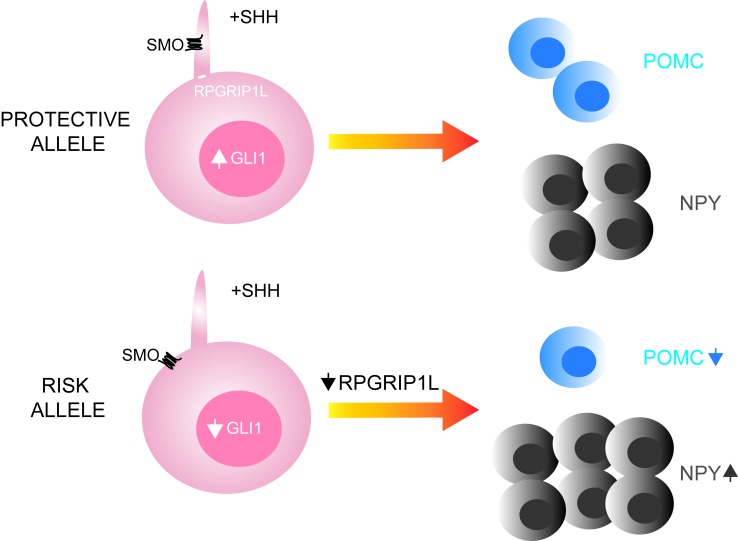
Obesity-associated SNPs in FTO intron 1, rs8050136 and rs1421085, are embedded within 2 binding sites for CUX1 (8). Based on electrophoretic mobility shift assays (7, 11), the rs8050136 A (risk) and rs1421085 C (risk) alleles preferentially bind CUX1 isoform P200, whereas rs8050136 C (protective) and rs1421085 T (protective) alleles preferentially bind CUX1 isoform P110. Luciferase-based promoter probing of the rs8050136 and rs1421085 genomic regions in hypothalamus-derived N2a cells, and systemic overexpression of Cux1 isoforms p200 and p110 in vivo (8, 17), implicate P200 as a repressor of Fto and P110 as an activator of Rpgrip1l expression in the CNS.
Alteration of rs8050136 and rs1421085 by CRISPR.
Using the human H9 ESc line that is heterozygous for rs8050136 and rs1421085 risk alleles, we have used Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated protein-9 (Cas9) technology to create an isogenic allelic series for the adiposity risk (R) or protective-associated (P-associated) alleles at SNP rs8050136 or rs1421085 in human ESc–derived ARH neurons (Supplemental Figure 4A) (45). We have confirmed that these lines are correctly targeted, karyotypically normal, and display no apparent off-target effects (45).
FTO/RPGRIP1L expression per rs8050136/rs1421085 allele in isogenic ARH neurons.
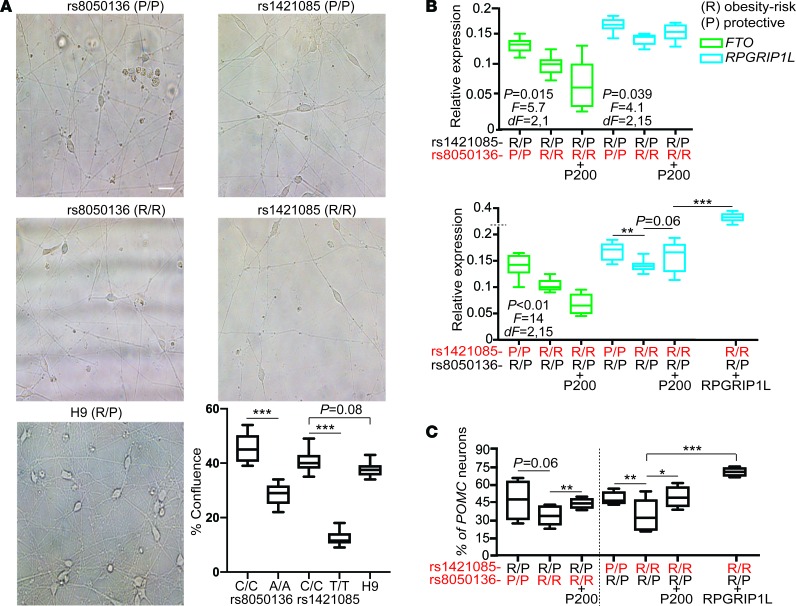
Homozygosity for the risk allele at rs8050136 or rs1421085 resulted in respective ~45% and ~75% decreases in generation of ARH-type neurons compared with neurons homozygous for the protective allele at rs8050136 or rs1421085 (Figure 8A). Homozygosity for the risk allele at rs8050136 or rs1421085 correlated with an ~20%–30% decrease in FTO or RPGRIP1L expression in ARH-type neurons (Figure 8B).
Figure 8. Obesity-risk alleles in FTO intron 1 diminish RPGRIP1L expression and POMC differentiation.
(A) Decreased neuro-confluency of human ESc–derived isogenic ARH neurons (day 30) homozygous for the FTO risk (R/R) allele at rs1421085 or rs8050136 compared with neurons homozygous for the protective (P/P) allele at rs1421085 or rs8050136. Scale bar: 20 μm. (B) Decreased FTO and RPGRIP1L expression in ARH neurons derived from the human H9 ESc line CRISPRed to homozygosity for the risk allele at rs1421085 or rs8050136. P < 0.05 was statistically significant, by 1-way ANOVA. (C) Upregulation of RPGRIP1L expression in response to increased P110 protein levels secondary to P200 overexpression (Supplemental Figure 4B), or in response to virus-induced RPGRIP1L overexpression, correlated with increased POMC neuron number. Red letters signify CRISPRed alleles. Each column represents the average of 6 isogenic cell lines. Data in A, B, and C are represented as box-and-whisker plots; boxes are the interquartile range, lines are the median value, and whiskers are minimum and maximum values. *P < 0.05, **P < 0.04, ***P < 0.01, by 2-tailed Student’s t test.
POMC neuron number per rs8050136/rs1421085 allele in isogenic ARH neurons.
Homozygosity for the risk allele at rs8050136 or rs1421085 was associated with an ~30% decrease in POMC-positive neurons (Figure 8C) compared with neurons homozygous for the protective allele at rs8050136 or rs1421085.
FTO/RPGRIP1L expression per rs8050136/rs1421085 allele in isogenic ARH neurons overexpressing P200.
We overexpressed CUX1 P200 at day 26 of the differentiation protocol, which is 1 day before differentiating cells normally initiate expression of POMC (32, 34) (Claudia Doege, Columbia University, personal communication). P200 overexpression by lentiviral transduction of ARH neurons homozygous for the risk allele (preferentially bound by P200) at rs8050136 or rs1421085 decreased FTO expression by ~60% (Figure 8B). P200 overexpression also resulted in increased P110 protein levels (Supplemental Figure 4, B and C), presumably due to constitutive Cathepsin L processing of P200–P110 (7). Increased P110 protein levels in neurons homozygous for the risk alleles at rs8050136 or rs1421085 were associated with an ~20%–30% increase in RPGRIP1L expression (P = 0.06), presumably due to the lower DNA-binding affinity of P110 for the risk alleles (7).
POMC neuron number per rs8050136/rs1421085 obesity-risk allele in isogenic ARH neurons overexpressing P200.
The increase in RPGRIP1L expression in ARH neurons homozygous for the risk alleles at rs8050136 or rs1421085 overexpressing P200 was associated with restoration of POMC-positive neuron number (Figure 8C). Furthermore, RPGRIP1L overexpression (~2-fold) by lentiviral transduction of neurons homozygous for the obesity-risk allele at rs1421085 was associated with an ~50% increase in the number of POMC-positive neurons (Figure 8C).
Fto does not affect Pomc neuron number. Because (a) RPGRIP1L overexpression alone is sufficient to increase POMC neuron number and (b) the increase in RPGRIP1L expression and decreased FTO expression upon P200 overexpression restores POMC number in neurons homozygous for the risk alleles at rs8050136 or rs1421085, it is implied that RPGRIP1L is important for POMC neuron specification and that FTO protein does not participate in the differentiation of POMC neurons. In support of the latter inference, mice homozygous for the Fto-floxed allele (Ftofl/fl) (8) and also segregating for Pomc-Cre and Pomc-GFP have no apparent change in the number of adult GFP-positive neurons compared with Ftofl/fl littermates (Supplemental Figure 5).
Discussion
The hyperhagia and obesity caused by induced Rpgrip1l deletion in the terminally differentiated CNS (8) (Figure 1) supports the importance of Rpgrip1l in the maintenance and/or neurochemical function of the developed feeding neurocircuitry. It is likely that diminished leptin sensitivity is partly responsible for the increased energy intake. The fact that adult-onset Rpgrip1l deletion in Pomc neurons did not result in increased body weight implies that Rpgrip1l is not essential for the neurochemical function of Pomc adult neurons. Instead, the data presented here support a critical role of RPGRIP1L in the development of Pomc-expressing neuroprogenitors, including Npy neurons that derive from Pomc-expressing progenitors (26, 27). The reciprocal decrease in anorexigenic Pomc neurons and increase in orexigenic Npy neurons may provide a cellular anlage for the hyperphagia of Rpgrip1l–/–(Pomc) mice. Our finding that POMC neuron specification can be altered by modulating RPGRIP1L expression per genotype at obesity risk alleles at rs1421085 or rs8050136 supports the relevance of this mechanistic formulation to the adiposity highly associated with allelic variation at these FTO intronic loci.
The Pomc-GFP transgene that we used also necessarily marks those neurons that develop from Pomc-expressing neuroprogenitors. We anticipate that specific neuronal subpopulations are affected by Pomc-specific congenital deletion of Rpgrip1l. However, with the exception of Npy neurons — some of which are known to develop from Pomc precursors — we have not identified these neuronal subpopulations. Therefore, the precise neurocellular consequences of the congenital interference with Rpgrip1l expression in Rpgrip1l–/–(Pomc) mice are not known. Future work should be directed toward identifying all the ARH neuronal subpopulations affected in Rpgrip1l–/–(Pomc) mice, as well as assigning the affected ARH-PVH projections to specific ARH neuronal subpopulations.
The increase in PVH projections emanating from the ARH of Rpgrip1l–/–(Pomc) mice may partly reflect the increase in the number of Npy neurons. However, the large magnitude of the increase in projections, and the relatively smaller magnitude in increase of Npy number in Rpgrip1l–/–(Pomc) mice, suggests that other ARH neurons that derive from Pomc-expressing progenitors are also affected. Alternatively, Rpgrip1l hypomorphism may relieve inhibition of axon growth/guidance (46) leading to this apparent change in wiring of the feeding neurocircuitry.
Using a hypothalamic differentiation protocol reported elsewhere (32, 34), the human H9 ESc line and “CRISPERed” isogenic lines produced 70%–80% POMC-positive neurons, whereas less than 1% of ARH neurons were NPY positive. By E18, more than half of POMC neurons convert to NPY-positive neurons during embryogenesis (26). We believe that the differentiation protocol employed here gives rise to early POMC progenitors that may resemble those at E13 in the mouse in vivo. The decreased POMC cell number in the human ESc–derived ARH neurons segregating for the obesity risk alleles at rs1421085 or rs8050136 is in agreement with the reduction in Pomc neuroprogenitors in Rpgrip1l–/–(Pomc) embryos at stage E13.
We were unable to detect any increase in cell death in Rpgrip1l–/–(Pomc) or control embryos at E11 when Pomc expression turns on in the developing hypothalamus; cell death in the ARH is not common at this developmental stage (Lori Zeltser, Columbia University; personal communication). Similarly, adult-onset Rpgrip1l deletion in the CNS using Nestin-Cre did not increase cell death in the adult ARH. It is possible that we have missed a narrow time window at which increased apoptosis may have occurred in Rpgrip1l–/–(Pomc) neurons. Ex vivo live imaging of Pomc progenitors at the developmental stage when POMC expression is normally initiated might provide insights into this issue. However, the fact that Npy neuron number increased in the adult ARH of Rpgrip1l–/–(Pomc) mice suggests a cell specification–related phenotype. Moreover, the human fibroblasts with biallelic mutations in RPGIRP1L used in this study proliferate at normal rates, suggesting that perturbed RPGRIP1L function does not promote apoptosis.
An important question is whether RPGRIP1L hypomorphism causes proliferative and/or cell fate specification changes in POMC-expressing progenitors or neurons that derive from them. RPGRIP1L, a component of the centrosome, and CUX1 are implicated in the control of the cell cycle (47, 48), a cellular process coordinated with ciliary formation arising from the centrosome and ciliary disassembly (49). Nevertheless, it is not clear whether the primary cilium regulates cell division. On the other hand, the importance of the primary cilium in the control of SHH signaling and neuropatterning is well established (42). Shh signaling is disrupted in systemic Rpgrip1l-null embryos in which CNS structural abnormalities are prominent (18, 50). We conclude that RPGRIP1L regulates Pomc neuron specification by a SHH-dependent mechanism.
The primary cilium is also involved in cell migration. The platelet-derived growth factor receptor α (PDGFrα) localizes to the primary cilium and is part of the PDGF signaling pathway that controls cell migration during development and in the adult via AKT signaling and mammalian target of rapamycin complex 1 (mTORC1) pathways (51, 52). RPGRIP1L has been implicated in the mTORC1 pathway (53), and Rpgrip1l hypomorphism results in perturbed neuronal migration of the murine developing cortex (20). The potential effects of Rpgrip1l hypomorphism on Pomc-neuron migration during development, as well as potential effects on neuronal function of Pomc neurons and other neuronal subpopulations affected, are areas of interest.
Pomc is expressed in other sites implicated in feeding behavior, such as the pituitary and brain stem (54, 55), and Pomc neurons in the ARH project to other brain regions, such as the dorso vagal complex (55). Therefore, Rpgrip1l hypomorphism may affect Pomc number and projections in other regions in a manner similar to the ARH.
A recent study indicates that the melanocortin 4 receptor (MC4R) is located on the primary cilium and that hypomorphic Mc4r mutations impair the receptor’s ciliary trafficking in PVH neurons (56). In the same study, inhibition of ciliary Ac3 in Mc4r-expressing neurons was associated with obesity. Shh reduces cAMP levels in the primary cilium by inhibiting adenylyl cyclase activity (57). We observed decreased AC3 (ADCY3) expression and ectopic AC3 localization upon SHH stimulation in JBST iPSC–derived neurons. It is plausible that upregulation of AC3 expression in response to adenylyl cyclase inhibition by Shh in circumstances of RPGRIP1L hypomorphism results in accumulation of AC3 in the cell body due to the deranged AC3 trafficking. However, we find that Rpgrip1l hypomorphism suppresses adiposity in Ac3+/– mice, suggesting that RPGRIP1L is functionally downstream of AC3 and that the AC3-mediated adiposity is not caused by RPGRIP1L hypomorphism (8). Therefore, it is unlikely that decreased AC3 expression and ectopic AC3 localization are driving the developmental defects caused by RPGRIP1L hypomorphism in Pomc neurons. On the other hand, AC3 and cAMP signaling mediate axon outgrowth and guidance in the CNS (58) and may be, at least in part, responsible for the perturbed Pomc neuron axonal outgrowth in circumstances of RPGRIP1L deficiency.
Previous studies have produced conflicting results regarding the correlation of allelic variations at the FTO intronic region with FTO expression or expression of nearby genes. Smemo et al. (59) reported higher expression of iroquois homeobox 3 (IRX3) in human cerebellar and adipose tissue homozygous for FTO obesity risk alleles at rs9930506 compared with tissue from individuals homozygous for the protective FTO alleles. In contrast, partial inhibition of Irx3 in the murine hypothalamus has the opposite effect of what would be predicted by the direction of the association or by Irx3 global deletion (59, 60). Moreover, Claussnitzer et al. (61) failed to see an allele-specific effect on IRX3 expression in a hippocampal neuronal cell line, but they reported effects of allelic variants at rs1421085 on IRX3 and -5 (IRX3/5) in preadipocytes. Claussnitzer et al. proposed a mechanism by which obesity risk alleles of rs1421085 in human mesenchymal adipocyte progenitors increase expression of IRX3/5, resulting in decreased thermogenic potential of new adipocytes. However, a follow-up study presented in vivo and in vitro evidence that Irx3 positively regulates thermogenesis (62). The relevance of these mechanisms in humans is unclear, as the effects of the obesity-risk alleles in the first intron of FTO have been consistently reported to convey effects on food intake and not energy expenditure (6, 63–71). The inconsistencies in correlation of allelic variation to expression of FTO and vicinal genes may be attributed to the possibility that different haplotypes within intron 1 affect different genes and, thereby, different physiological process. This possibility would be consistent with multiple mechanisms (72) at different levels of penetrance and within different race/ethnicities, underlying such a statistically strong effect of noncoding alleles on adiposity. The present study focuses on the action of RPGRIP1L in neurons, in which expression of RPGRIP1L, but not IRX3/IRX5, is correlated in an FTO intronic allele dose-dependent manner (8).
In summary, we present evidence that the very strong association with adiposity of highly prevalent sequence variants in intron 1 of FTO are due, in part, to their indirect impact on the primary cilium. Our data imply that genetic mediation of nonsyndromic, prevalent obesity is conveyed partly by structural alteration of the hypothalamic cells and circuitry that mediate aspects of ingestive behaviors.
Methods
Mouse studies.
Rpgrip1lfl/fl, Pomc-Cre (The Jackson Laboratory; stock number 005965), Pomc-GFP (The Jackson Laboratory; stock number 009593), and Nestin-CreERT2 mice (The Jackson Laboratory; stock number 016261) have been described by us elsewhere (8). The Cagg-CreER allele (Tg[CAG-cre/Esr1*]5Amc/J, The Jackson Laboratory, stock number 004682) used to assess systemic Rpgrip1l loss in adults was induced as previously reported (73). For adult-induced loss of Rpgrip1l specifically in adult terminally differentiated POMC neurons, a POMC-CreER allele that has previously been described (74) was utilized. mT/mG mice were purchased from The Jackson Laboratory (B6.129[Cg]-Gt[ROSA]26Sortm4[ACTB-tdTomato,-EGFP]Luo/J; stock number 007676). Npy-GFP mice were also purchased from The Jackson Laboratory (Npy-GFP [C57BL/6JTgNPY-hrGFP]; stock number 006417). Ftofl/fl mice are described elsewhere (8). Mice where housed in a Barrier facility at room temperature (23°C) on a 12-hour light/12-hour dark cycle (lights off at 7 p.m.), with ad libitum (ad lib) access to food and water.
Human studies.
UW15 (7), UW04-04 (17), and UW04-03 (36) primary skin fibroblast JBST lines were provided by Daniel Doherty (University of Washington, Seattle, Washington, USA). ESc line H9 was purchased by WiCell Research Institute Inc. (catalog WA09).
Diet and dietary treatment.
All mice were fed regular chow (Purina Mills; Picolab, catalog 5058) (16) ad lib unless specified. High-fat diet consists of 65% of calories as fat (catalog D12492; Open Source Diets).
Body mass and composition measurements.
Measurements were taken with a Minispec TD-NMR Analyzer (Bruker Optics) as described elsewhere (8).
Isolation of total RNA and nuclear protein extracts.
Total RNA and protein extraction have been described elsewhere (17). Specific brain regions were micropunched as described in Stratigopoulos et al. (7).
Quantitative PCR (qPCR) and cDNA synthesis.
The method has been described elsewhere (7). The geometric mean of GAPDH and ACTB expression levels was used as loading controls, unless specified. The PCR primers used are shown in Supplemental Table 1.
IHC, immunofluorescence, and antibodies.
IHC was performed on cryosections after perfusion fixation, as described elsewhere (17). Teratomas were embedded in paraffin and further cut into 8 μm–thick sections. Sections were stained with H&E using a standard protocol (http://www.ihcworld.com/_protocols/special_stains/h&e_ellis.htm). Cells growing on cultured-cell slides pretreated with Poly–L-ornithine/laminin were fixed with 4% paraformaldehyde for 10 minutes at room temperature. Primary fibroblasts were starved for 36 hours before visualization of primary cilia by immunofluorescence. The following primary antibodies were used: anti-GFP (1:100; Abcam, catalog ab6662), TUJ1 (1:500; MilliporeSigma, catalogs T2200 and T5076), MAP2 (1:10,000; Abcam, catalog AB5392), AC3 (1:100; Santa Cruz Biotechnology Inc., catalog sc-588), ARL13B (1:1,000; gift from Tamara Caspary, Emory University, Atlanta, Georgia, USA), SMO (1:500; gift from Kathryn V. Anderson, Memorial Sloan Kettering Cancer Center, New York, New York, USA), NF (1:10,000; MilliporeSigma, catalog AB5539), γ-tubulin (1:500; MilliporeSigma, catalog T6557), NESTIN (1:500; Stemgent, catalog 09-0045), NeuN (1:100; MilliporeSigma, catalog MAB377), Acetylated tubulin (1:500; MilliporeSigma, catalog T6793), POMC (1:500; Abcam, catalog ab14064), NPY (1:500; Novus, catalog npb1-19808), αMSH (1:500; MilliporeSigma, catalog AB5087), and Gad67 (1:500; MilliporeSigma, catalog MAB5406).
TUNEL.
TUNEL staining was performed using the DeadEnd Fluorometric TUNEL System (Promega, catalog G3250) according to the manufacturer’s instructions.
Scanning electron microscopy.
iPSC-derived neurons were grown on 13 mm glass coverslips and fixed in 2.5% glutaraldehyde in 0.1 M Cacodylate buffer for 30 minutes at room temperature. Sample preparation and imaging for scanning electron microscope was done by the analytical imaging facility at Albert Einstein College of Medicine (New York, New York, USA). Experimental details are described elsewhere (75). Images were obtained at 5000× magnification.
Brain clearing and imaging.
For brain clearing, we followed the protocol described in great detail at https://idisco.info/idisco-protocol/ Briefly, signal was enhanced by treatment with anti-GFP antibody (Aves Laboratories, catalog GFP-1020) and secondary Alexa Fluor 647 (anti-chicken; Thermo Fisher Scientific, catalog A-21449), followed by dehydration performed with tert-Butanol. Delipidization was achieved by treatment with dichloromethane (DCM). Then, samples were stored overnight in Benzyl alcohol-Benzyl benzoate/Diphenyl ether (4:1 vol/vol; BABB-4D) before imaging. Whole E13 embryo heads were recorded with a light sheet fluorescent Ultramicroscope II (Lavision Biotech) utilizing the filters (emission/excitation; Em/Ex) 470/425 nm for tissue autofluorescence and 630/680nm for amplified GFP signal. ARH area was recorded in detail at 4× zoom with a 2-μm plane size. Cell counting was performed with Arrivis Vision4D software (Arrivis) after 3-dimensional (3-D) reconstruction, background removal and cell segmentation.
Viruses.
Ready-to-transduce lentiviral particles carrying human CUX1 P200 (NM_181552) and RPGRIP1L (NM_015272) were purchased by ORIGENE. AAV1 particles used for stereotaxic injections (AAV1.CMV.PI.eGFP.WPRE.bGH, AAV1.CMV.HI.eGFP-Cre.WPRE.SV40) were purchased from the Penn Vector Core (University of Pennsylvania, Philadelphia, Pennsylvania, USA).
Cerebellar stereotaxic injections.
Male C57BL/6J mice aged P11 were anesthetized with a combination of ketamine (80 mg/kg) and xylazine (12 mg/kg) (MilliporeSigma, catalog K113), administered i.p. Mice were then placed in a stereotaxic frame adapted for juveniles, and the scalp was clipped of fur and sterilized. A longitudinal incision was made through the scalp, and the skull was leveled. The target site for injection, defined as 2.6 mm caudal to lambda along the midline, was measured stereotaxically, and the skull above the target was carefully drilled very thin. A glass micropipette loaded in a Drummond Nanoject III device was then filled with AAV1 and positioned over the target site. The pipette was lowered through the soft, thin skull into the cerebellum to a depth of 2.5 mm below the brain surface. Pulses of 20 nl each were injected at 2.5 mm, 2.0 mm, 1.5 mm, 1.0 mm, and 0.5 mm ventral to the brain surface, pausing for 1 minute after each pulse before moving to other sites. Following retraction of the pipette, the incision was closed and mice were permitted to recover from anesthesia prior to being returned to nursing dams.
ARH and DMH/VMH stereotaxic injections.
Injections of 12-week-old male Rpgrip1lfl/fl mice were performed on a stereotaxic frame using a glass micropipette and an air pressure injector system (76, 77). Each animal was injected at 3 sites bilaterally for a total of 6 injections and a total volume of ~150 nl to ensure complete coverage of the ARH (mediolateral [ML], ±0.3 mm; anteroposterior [AP], –1.6 mm; dorsoventricular [DV], –6 mm) or DMH/VMH (ML, ±0.3; AP, –1.6; DV, –5.8mm). The accuracy of injections was ~50%. Injections that missed the targeted regions did not result in body weight changes. These were excluded from the analysis but provide evidence of the cellular specificity of the phenotypic effects. All surgical procedures were performed using an aseptic technique according to the approved protocols at Columbia University.
Generation and characterization of iPSCs.
JBST primary fibroblasts were reprogrammed into iPSCs using retroviruses expressing cMYC, OCT4, SOX2, and KLF4 (78). All iPSCs lines were cultured on mouse embryonic fibroblast (MEF) cells with human ES medium: 500 ml KO DMEM (Thermo Fisher Scientific, catalog 10829018), 90 ml KO Serum (Thermo Fisher Scientific, catalog A3181502), 6.5 ml GlutaMAX (Thermo Fisher Scientific, catalog 35050061), 6.5 ml NEAA (Thermo Fisher Scientific, catalog 11140050), 6.5 ml penicillin/streptomycin (Thermo Fisher Scientific, catalog 15140122), 0.65 ml β-mercaptoethanol (Thermo Fisher Scientific, catalog 21985023), and 10 ng/ml bFGF (Thermo Fisher Scientific, catalog 13256029). Stable iPSC clones (after 8 passages) were characterized by immunostaining with pluripotency markers SOX2 (1:500; Stemgent, catalog 09-0024), OCT4 (1:500; Stemgent, catalog 09-0023), NANOG (1:500; R&D Systems, catalog AF1997), SSEA4 (1:300; R&D Systems, catalog MAB1435), and TRA-1-60 (1:300; MilliporeSigma, catalog MAB4360), as well as by qPCR analysis to confirm retroviral gene silencing and expression of endogenous pluripotency genes, as described elsewhere (Supplemental Figure 2B) (32, 34). Pluripotency was further confirmed by a teratoma assay: JBST iPSCs (5 million/200μl matrigel) were injected s.c. into NSG mice (The Jackson Laboratory, NOD.Cg-Prkdcscid Il2rgtm1Wjl/SzJ, catalog 005557), and after 2–3 months, teratomas were isolated and fixed with 4% paraformaldehyde for >16 hours for immunohistochemical analysis. For karyotyping, control and JBST iPSC lines were cultured in T75 flasks and karyotyped by Cell Line Genetics.
Neuron differentiation protocols.
Human iPSCs were cultured and maintained in human ESc medium on MEF (111,000 cells/well in 6-well plates; Thermo Fisher Scientific, catalog A34180). Upon confluence, iPSC cells were cultured for 4 days in EB medium (human ESc medium without basic fibroblast growth factor (bFGF) plus dual SMAD inhibitors — A83 (1 μM; Stemgent, catalog 04-0014) and LDN (250 nM; Stemgent, catalog 04-0074). From days 5–8, EB medium was replaced with N2 medium in steps from 75% to 50%, to 25%, and to 0%, while maintaining A83 and LDN at constant concentrations. Neuron progenitors were harvested on day 10 and further expanded for 2–3 passages in N2 medium plus A83/LDN on poly–L-ornithine/Laminin–coated plates. For further neuronal differentiation, cells were cultured on Poly–L-ornithine/Laminin–coated plates (Thermo Fisher Scientific, catalog 08-772-66) at 50,000 cells per 24-well or 4-well plate and at 500,000 cells per 6-well plate. After culturing 3 weeks in N2 medium supplemented with B27 (Thermo Fisher Scientific, catalog 17504044), DAPT (10 μM; Stemgent, catalog 04-0041), and BDNF (20 ng/ml; R&D Systems, catalog 248-BD), neurons were fixed/harvested for IHC or RNA analysis.
H9 and H9-derived isogenic lines were differentiated to ARH neurons following the detailed protocol described in a Methods manuscript by Wang et al. (34).
CRISPR.
The H9 ES cell line was propagated and subjected to CRISPR as extensively described elsewhere (45). Six isogenic lines per genotype were used for analysis.
Three-dimensional reconstructions of the PVH/ARH.
We utilized the Zeiss Axio Scan.Z1 (Baymax) automated slide scanner to visualize nuclei by DAPI (Zeiss Filter Set 49), as well as mGFP-positive (FS 38 HE) Pomc neurons and their projections in 60 μm coronal sections spanning the whole PVH and ARH. Images were aligned in Adobe Photoshop; Z-stacks were generated in ImageJ (NIH) and reconstructed in 3-D using Vaa3D (https://github.com/Vaa3D) and CamStudio (www.camstudio.org).
Statistics.
Data are expressed as means ± SEM unless specified. Statistical analysis was performed using 2-tailed Student’s t test (StatView 5.0, SAS Institute Inc.). Statistical significance of the difference between 3 means was determined by 1-way ANOVA using GraphPad Prism 7. Levels of statistical significance were set at P < 0.05.
Study approval.
Animals were maintained according to Columbia University animal welfare guidelines. All procedures were approved by the Columbia University IACUC. Human subjects provided written informed consent, and human subject–related research was reviewed and approved by the Columbia Stem Cell Committee and the Columbia IRB.
Author contributions
Conceptualization was contributed by LW, CAD, DE, NB, RLL, and GS; methodology was contributed by LW, AJDS, YG, KEB, SEE, CAD, HLA, GJM, NB, and GS; investigation was contributed by LW, AJDS, YG, KEB, SEE, RT, JML, XL, RR, MP, JYL, JT, MCDR, and GS; resources were contributed by CAD, HLA, JCB, DE, RC, NB, RLL, and GS; writing the original draft was contributed by GS; writing, reviewing, and editing the draft were contributed by LW, AJDS, YG, KEB, CAD, JYL, NB, RLL, and GS; and supervision was contributed by CAD, HLA, JCB, DE, NB, RLL, and GS.
Supplementary Material
Acknowledgments
We thank Joel Elmquist (UT Southwestern, Dallas, Texas, USA) and Daniel Doherty (University of Washington, Seattle, Washington, USA) for providing the POMC-CreER mice and JBST fibroblast lines, respectively. Also, we thank Lisa Cole Burnett (Columbia University) for microscopy. Work was partially funded by a Russell Berrie Foundation Neuroscience of Obesity Initiative pilot grant, RO1 DK52431-23, and the MBMG Core of the NYNORC (5P30DK026687). the New York Stem Cell Foundation, the Rudin Foundation, the Russell Berrie Foundation Program in Cellular Therapies, and the Diabetes (P30 DK63608-12) and Obesity Research (P30 DK26687-33) Centers of Columbia University. Yossef Goffer was supported by NIDDK F30DK108564. Nicolas F. Berbari was supported by R01 DK114008 and CDMD NIH/NIDDK P30 DK097512. Alain J. De Solis was supported by MSCA-IF-2016 (752319, ARC-Neurohet).
Version 1. 02/07/2019
Electronic publication
Footnotes
Conflict of interest: The authors have declared that no conflict of interest exists.
License: Copyright 2019, American Society for Clinical Investigation.
Reference information: JCI Insight. 2019;4(3):e123337. https://doi.org/10.1172/jci.insight.123337.
Contributor Information
Liheng Wang, Email: lw2381@cumc.columbia.edu.
Alain J. De Solis, Email: alain.desolis@sf.mpg.de.
Yossef Goffer, Email: yg2149@cumc.columbia.edu.
Ross Tanis, Email: Ross.Tanis@uscmed.sc.edu.
Xueting Li, Email: xl2479@cumc.columbia.edu.
Richard Rausch, Email: rr2305@columbia.edu.
Manika Purohit, Email: mp3393@columbia.edu.
Jen-Yi Lee, Email: jenyilee33@gmail.com.
Jerica Tan, Email: jmt2202@barnard.edu.
Maria Caterina De Rosa, Email: md3291@cumc.columbia.edu.
Claudia A. Doege, Email: cad2114@cumc.columbia.edu.
Gabriela J. Martins, Email: gm2743@columbia.edu.
Jens C. Brüning, Email: bruening@nf.mpg.de.
Dieter Egli, Email: de2220@cumc.columbia.edu.
Rudolph L. Leibel, Email: rl232@columbia.edu.
George Stratigopoulos, Email: gs2172@columbia.edu.
References
- 1.Goodarzi MO. Genetics of obesity: what genetic association studies have taught us about the biology of obesity and its complications. Lancet Diabetes Endocrinol. 2018;6(3):223–236. doi: 10.1016/S2213-8587(17)30200-0. [DOI] [PubMed] [Google Scholar]
- 2.Frayling TM, et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316(5826):889–894. doi: 10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scuteri A, et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3(7):e115. doi: 10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meyre D, et al. Genome-wide association study for early-onset and morbid adult obesity identifies three new risk loci in European populations. Nat Genet. 2009;41(2):157–159. doi: 10.1038/ng.301. [DOI] [PubMed] [Google Scholar]
- 5.Thorleifsson G, et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat Genet. 2009;41(1):18–24. doi: 10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 6.Cecil JE, Tavendale R, Watt P, Hetherington MM, Palmer CN. An obesity-associated FTO gene variant and increased energy intake in children. N Engl J Med. 2008;359(24):2558–2566. doi: 10.1056/NEJMoa0803839. [DOI] [PubMed] [Google Scholar]
- 7.Stratigopoulos G, LeDuc CA, Cremona ML, Chung WK, Leibel RL. Cut-like homeobox 1 (CUX1) regulates expression of the fat mass and obesity-associated and retinitis pigmentosa GTPase regulator-interacting protein-1-like (RPGRIP1L) genes and coordinates leptin receptor signaling. J Biol Chem. 2011;286(3):2155–2170. doi: 10.1074/jbc.M110.188482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stratigopoulos G, et al. Hypomorphism of Fto and Rpgrip1l causes obesity in mice. J Clin Invest. 2016;126(5):1897–1910. doi: 10.1172/JCI85526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cubelos B, Briz CG, Esteban-Ortega GM, Nieto M. Cux1 and Cux2 selectively target basal and apical dendritic compartments of layer II-III cortical neurons. Dev Neurobiol. 2015;75(2):163–172. doi: 10.1002/dneu.22215. [DOI] [PubMed] [Google Scholar]
- 10.Rodríguez-Tornos FM, et al. Cux1 Enables Interhemispheric Connections of Layer II/III Neurons by Regulating Kv1-Dependent Firing. Neuron. 2016;89(3):494–506. doi: 10.1016/j.neuron.2015.12.020. [DOI] [PubMed] [Google Scholar]
- 11.Stratigopoulos G, et al. Regulation of Fto/Ftm gene expression in mice and humans. Am J Physiol Regul Integr Comp Physiol. 2008;294(4):R1185–R1196. doi: 10.1152/ajpregu.00839.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Reiter JF, Blacque OE, Leroux MR. The base of the cilium: roles for transition fibres and the transition zone in ciliary formation, maintenance and compartmentalization. EMBO Rep. 2012;13(7):608–618. doi: 10.1038/embor.2012.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chung WK, Goldberg-Berman J, Power-Kehoe L, Leibel RL. Molecular mapping of the tubby (tub) mutation on mouse chromosome 7. Genomics. 1996;32(2):210–217. doi: 10.1006/geno.1996.0107. [DOI] [PubMed] [Google Scholar]
- 14.Borman AD, et al. A homozygous mutation in the TUB gene associated with retinal dystrophy and obesity. Hum Mutat. 2014;35(3):289–293. doi: 10.1002/humu.22482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun X, Haley J, Bulgakov OV, Cai X, McGinnis J, Li T. Tubby is required for trafficking G protein-coupled receptors to neuronal cilia. Cilia. 2012;1(1):21. doi: 10.1186/2046-2530-1-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barroso I. ADCY3, neuronal primary cilia and obesity. Nat Genet. 2018;50(2):166–167. doi: 10.1038/s41588-018-0043-x. [DOI] [PubMed] [Google Scholar]
- 17.Stratigopoulos G, et al. Hypomorphism for RPGRIP1L, a ciliary gene vicinal to the FTO locus, causes increased adiposity in mice. Cell Metab. 2014;19(5):767–779. doi: 10.1016/j.cmet.2014.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vierkotten J, Dildrop R, Peters T, Wang B, Rüther U. Ftm is a novel basal body protein of cilia involved in Shh signalling. Development. 2007;134(14):2569–2577. doi: 10.1242/dev.003715. [DOI] [PubMed] [Google Scholar]
- 19. Parisi M, Glass I. Joubert Syndrome. In: Pagon RA, et al (eds.). GeneReviews. Seattle, WA: University of Washington, Seattle; 2003: https://www.ncbi.nlm.nih.gov/books/NBK1325/ [PubMed]
- 20.Guo J, et al. Developmental disruptions underlying brain abnormalities in ciliopathies. Nat Commun. 2015;6:7857. doi: 10.1038/ncomms8857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Thomas S, et al. Identification of a novel ARL13B variant in a Joubert syndrome-affected patient with retinal impairment and obesity. Eur J Hum Genet. 2015;23(5):621–627. doi: 10.1038/ejhg.2014.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gerhardt C, et al. The transition zone protein Rpgrip1l regulates proteasomal activity at the primary cilium. J Cell Biol. 2015;210(1):115–133. doi: 10.1083/jcb.201408060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cadieux C, et al. Polycystic kidneys caused by sustained expression of Cux1 isoform p75. J Biol Chem. 2008;283(20):13817–13824. doi: 10.1074/jbc.M709332200. [DOI] [PubMed] [Google Scholar]
- 24.Suzuki T, et al. Molecular genetic analysis of 30 families with Joubert syndrome. Clin Genet. 2016;90(6):526–535. doi: 10.1111/cge.12836. [DOI] [PubMed] [Google Scholar]
- 25.Sotelo C. Cellular and genetic regulation of the development of the cerebellar system. Prog Neurobiol. 2004;72(5):295–339. doi: 10.1016/j.pneurobio.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 26.Padilla SL, Carmody JS, Zeltser LM. Pomc-expressing progenitors give rise to antagonistic neuronal populations in hypothalamic feeding circuits. Nat Med. 2010;16(4):403–405. doi: 10.1038/nm.2126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Padilla SL, Reef D, Zeltser LM. Defining POMC neurons using transgenic reagents: impact of transient Pomc expression in diverse immature neuronal populations. Endocrinology. 2012;153(3):1219–1231. doi: 10.1210/en.2011-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muzumdar MD, Tasic B, Miyamichi K, Li L, Luo L. A global double-fluorescent Cre reporter mouse. Genesis. 2007;45(9):593–605. doi: 10.1002/dvg.20335. [DOI] [PubMed] [Google Scholar]
- 29.Seo S, Guo DF, Bugge K, Morgan DA, Rahmouni K, Sheffield VC. Requirement of Bardet-Biedl syndrome proteins for leptin receptor signaling. Hum Mol Genet. 2009;18(7):1323–1331. doi: 10.1093/hmg/ddp031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kokoeva MV, Yin H, Flier JS. Neurogenesis in the hypothalamus of adult mice: potential role in energy balance. Science. 2005;310(5748):679–683. doi: 10.1126/science.1115360. [DOI] [PubMed] [Google Scholar]
- 31.Bewick GA, et al. Post-embryonic ablation of AgRP neurons in mice leads to a lean, hypophagic phenotype. FASEB J. 2005;19(12):1680–1682. doi: 10.1096/fj.04-3434fje. [DOI] [PubMed] [Google Scholar]
- 32.Wang L, et al. Differentiation of hypothalamic-like neurons from human pluripotent stem cells. J Clin Invest. 2015;125(2):796–808. doi: 10.1172/JCI79220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Merkle FT, et al. Generation of neuropeptidergic hypothalamic neurons from human pluripotent stem cells. Development. 2015;142(4):633–643. doi: 10.1242/dev.117978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang L, Egli D, Leibel RL. Efficient Generation of Hypothalamic Neurons from Human Pluripotent Stem Cells. Curr Protoc Hum Genet. 2016;90:21.5.1–21.5.14. doi: 10.1002/cphg.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Arts HH, et al. Mutations in the gene encoding the basal body protein RPGRIP1L, a nephrocystin-4 interactor, cause Joubert syndrome. Nat Genet. 2007;39(7):882–888. doi: 10.1038/ng2069. [DOI] [PubMed] [Google Scholar]
- 36.Doherty D, et al. Mutations in 3 genes (MKS3, CC2D2A and RPGRIP1L) cause COACH syndrome (Joubert syndrome with congenital hepatic fibrosis) J Med Genet. 2010;47(1):8–21. doi: 10.1136/jmg.2009.067249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Williams CL, et al. MKS and NPHP modules cooperate to establish basal body/transition zone membrane associations and ciliary gate function during ciliogenesis. J Cell Biol. 2011;192(6):1023–1041. doi: 10.1083/jcb.201012116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yee CL, Wang Y, Anderson S, Ekker M, Rubenstein JL. Arcuate nucleus expression of NKX2.1 and DLX and lineages expressing these transcription factors in neuropeptide Y(+), proopiomelanocortin(+), and tyrosine hydroxylase(+) neurons in neonatal and adult mice. J Comp Neurol. 2009;517(1):37–50. doi: 10.1002/cne.22132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Blaess S, Szabó N, Haddad-Tóvolli R, Zhou X, Álvarez-Bolado G. Sonic hedgehog signaling in the development of the mouse hypothalamus. Front Neuroanat. 2014;8:156. doi: 10.3389/fnana.2014.00156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chambers SM, Fasano CA, Papapetrou EP, Tomishima M, Sadelain M, Studer L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat Biotechnol. 2009;27(3):275–280. doi: 10.1038/nbt.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lim YS, Chua CE, Tang BL. Rabs and other small GTPases in ciliary transport. Biol Cell. 2011;103(5):209–221. doi: 10.1042/BC20100150. [DOI] [PubMed] [Google Scholar]
- 42.Lepanto P, Badano JL, Zolessi FR. Neuron’s little helper: The role of primary cilia in neurogenesis. Neurogenesis (Austin) 2016;3(1):e1253363. doi: 10.1080/23262133.2016.1253363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rohatgi R, Milenkovic L, Scott MP. Patched1 regulates hedgehog signaling at the primary cilium. Science. 2007;317(5836):372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 44.Goodrich LV, Johnson RL, Milenkovic L, McMahon JA, Scott MP. Conservation of the hedgehog/patched signaling pathway from flies to mice: induction of a mouse patched gene by Hedgehog. Genes Dev. 1996;10(3):301–312. doi: 10.1101/gad.10.3.301. [DOI] [PubMed] [Google Scholar]
- 45.Stratigopoulos G, De Rosa MC, LeDuc CA, Leibel RL, Doege CA. DMSO increases efficiency of genome editing at two non-coding loci. PLoS ONE. 2018;13(6):e0198637. doi: 10.1371/journal.pone.0198637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juric-Sekhar G, Adkins J, Doherty D, Hevner RF. Joubert syndrome: brain and spinal cord malformations in genotyped cases and implications for neurodevelopmental functions of primary cilia. Acta Neuropathol. 2012;123(5):695–709. doi: 10.1007/s00401-012-0951-2. [DOI] [PubMed] [Google Scholar]
- 47.Lin YW, Yan MD, Shih YL, Hsieh CB. The basal body gene, RPGRIP1L, is a candidate tumour suppressor gene in human hepatocellular carcinoma. Eur J Cancer. 2009;45(11):2041–2049. doi: 10.1016/j.ejca.2009.04.012. [DOI] [PubMed] [Google Scholar]
- 48.Porath B, et al. Cux1 promotes cell proliferation and polycystic kidney disease progression in an ADPKD mouse model. Am J Physiol Renal Physiol. 2017;313(4):F1050–F1059. doi: 10.1152/ajprenal.00380.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Werner S, Pimenta-Marques A, Bettencourt-Dias M. Maintaining centrosomes and cilia. J Cell Sci. 2017;130(22):3789–3800. doi: 10.1242/jcs.203505. [DOI] [PubMed] [Google Scholar]
- 50.Mahuzier A, et al. Dishevelled stabilization by the ciliopathy protein Rpgrip1l is essential for planar cell polarity. J Cell Biol. 2012;198(5):927–940. doi: 10.1083/jcb.201111009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Christensen ST, Pedersen SF, Satir P, Veland IR, Schneider L. The primary cilium coordinates signaling pathways in cell cycle control and migration during development and tissue repair. Curr Top Dev Biol. 2008;85:261–301. doi: 10.1016/S0070-2153(08)00810-7. [DOI] [PubMed] [Google Scholar]
- 52.Umberger NL, Caspary T. Ciliary transport regulates PDGF-AA/αα signaling via elevated mammalian target of rapamycin signaling and diminished PP2A activity. Mol Biol Cell. 2015;26(2):350–358. doi: 10.1091/mbc.E14-05-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Struchtrup A, Wiegering A, Stork B, Rüther U, Gerhardt C. The ciliary protein RPGRIP1L governs autophagy independently of its proteasome-regulating function at the ciliary base in mouse embryonic fibroblasts. Autophagy. 2018;14(4):567–583. doi: 10.1080/15548627.2018.1429874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Clark AJ. 60 YEARS OF POMC: The proopiomelanocortin gene: discovery, deletion and disease. J Mol Endocrinol. 2016;56(4):T27–T37. doi: 10.1530/JME-15-0268. [DOI] [PubMed] [Google Scholar]
- 55.Zheng H, Patterson LM, Phifer CB, Berthoud HR. Brain stem melanocortinergic modulation of meal size and identification of hypothalamic POMC projections. Am J Physiol Regul Integr Comp Physiol. 2005;289(1):R247–R258. doi: 10.1152/ajpregu.00869.2004. [DOI] [PubMed] [Google Scholar]
- 56.Siljee JE, et al. Subcellular localization of MC4R with ADCY3 at neuronal primary cilia underlies a common pathway for genetic predisposition to obesity. Nat Genet. 2018;50(2):180–185. doi: 10.1038/s41588-017-0020-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moore BS, et al. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci USA. 2016;113(46):13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nicol X, Gaspar P. Routes to cAMP: shaping neuronal connectivity with distinct adenylate cyclases. Eur J Neurosci. 2014;39(11):1742–1751. doi: 10.1111/ejn.12543. [DOI] [PubMed] [Google Scholar]
- 59.Smemo S, et al. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature. 2014;507(7492):371–375. doi: 10.1038/nature13138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.de Araujo TM, et al. The partial inhibition of hypothalamic IRX3 exacerbates obesity. EBioMedicine. doi: 10.1016/j.ebiom.2018.11.048. doi: 10.1016/j.ebiom.2018.11.048. [published online ahead of print December 3, 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Claussnitzer M, et al. FTO Obesity Variant Circuitry and Adipocyte Browning in Humans. N Engl J Med. 2015;373(10):895–907. doi: 10.1056/NEJMoa1502214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zou Y, et al. IRX3 Promotes the Browning of White Adipocytes and Its Rare Variants are Associated with Human Obesity Risk. EBioMedicine. 2017;24:64–75. doi: 10.1016/j.ebiom.2017.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speakman JR, Rance KA, Johnstone AM. Polymorphisms of the FTO gene are associated with variation in energy intake, but not energy expenditure. Obesity (Silver Spring) 2008;16(8):1961–1965. doi: 10.1038/oby.2008.318. [DOI] [PubMed] [Google Scholar]
- 64.Wardle J, Llewellyn C, Sanderson S, Plomin R. The FTO gene and measured food intake in children. Int J Obes (Lond) 2009;33(1):42–45. doi: 10.1038/ijo.2008.174. [DOI] [PubMed] [Google Scholar]
- 65.Wardle J, Carnell S, Haworth CM, Farooqi IS, O’Rahilly S, Plomin R. Obesity associated genetic variation in FTO is associated with diminished satiety. J Clin Endocrinol Metab. 2008;93(9):3640–3643. doi: 10.1210/jc.2008-0472. [DOI] [PubMed] [Google Scholar]
- 66.Haupt A, et al. Variation in the FTO gene influences food intake but not energy expenditure. Exp Clin Endocrinol Diabetes. 2009;117(4):194–197. doi: 10.1055/s-0028-1087176. [DOI] [PubMed] [Google Scholar]
- 67.Tanofsky-Kraff M, et al. The FTO gene rs9939609 obesity-risk allele and loss of control over eating. Am J Clin Nutr. 2009;90(6):1483–1488. doi: 10.3945/ajcn.2009.28439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.den Hoed M, Westerterp-Plantenga MS, Bouwman FG, Mariman EC, Westerterp KR. Postprandial responses in hunger and satiety are associated with the rs9939609 single nucleotide polymorphism in FTO. Am J Clin Nutr. 2009;90(5):1426–1432. doi: 10.3945/ajcn.2009.28053. [DOI] [PubMed] [Google Scholar]
- 69.Rutters F, et al. Genetic associations with acute stress-related changes in eating in the absence of hunger. Patient Educ Couns. 2010;79(3):367–371. doi: 10.1016/j.pec.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 70.Qi Q, et al. Dietary Intake, FTO Genetic Variants, and Adiposity: A Combined Analysis of Over 16,000 Children and Adolescents. Diabetes. 2015;64(7):2467–2476. doi: 10.2337/db14-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Obregón Rivas AM, Santos JL, Valladares MA, Cameron J, Goldfield G. Association of the FTO fat mass and obesity-associated gene rs9939609 polymorphism with rewarding value of food and eating behavior in Chilean children. Nutrition. 2018;54:105–110. doi: 10.1016/j.nut.2018.03.001. [DOI] [PubMed] [Google Scholar]
- 72.Carli JFM, LeDuc CA, Zhang Y, Stratigopoulos G, Leibel RL. The role of Rpgrip1l, a component of the primary cilium, in adipocyte development and function. FASEB J. 2018;32(7):3946–3956. doi: 10.1096/fj.201701216R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244(2):305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- 74.Berglund ED, et al. Serotonin 2C receptors in pro-opiomelanocortin neurons regulate energy and glucose homeostasis. J Clin Invest. 2013;123(12):5061–5070. doi: 10.1172/JCI70338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Macaluso FP, Perumal GS, Kolstrup J, Satir P. CLEM Methods for Studying Primary Cilia. Methods Mol Biol. 2016;1454:193–202. doi: 10.1007/978-1-4939-3789-9_12. [DOI] [PubMed] [Google Scholar]
- 76.Cetin A, Komai S, Eliava M, Seeburg PH, Osten P. Stereotaxic gene delivery in the rodent brain. Nat Protoc. 2006;1(6):3166–3173. doi: 10.1038/nprot.2006.450. [DOI] [PubMed] [Google Scholar]
- 77.Peng Y, Gillis-Smith S, Jin H, Tränkner D, Ryba NJ, Zuker CS. Sweet and bitter taste in the brain of awake behaving animals. Nature. 2015;527(7579):512–515. doi: 10.1038/nature15763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Dimos JT, et al. Induced pluripotent stem cells generated from patients with ALS can be differentiated into motor neurons. Science. 2008;321(5893):1218–1221. doi: 10.1126/science.1158799. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.