Abstract
Stem cells can reside in a state of reversible growth arrest, or quiescence, for prolonged periods of time. Although quiescence has long been viewed as a dormant, low-activity state, increasing evidence suggests that quiescence represents states of poised potential and active restraint, as stem cells “idle” in anticipation of activation, proliferation, and differentiation. Improved understanding of quiescent stem cell dynamics is leading to novel approaches to enhance maintenance and repair of aged or diseased tissues. In this Review, we discuss recent advances in our understanding of stem cell quiescence and techniques enabling more refined analyses of quiescence in vivo.
Throughout the mammalian body, many tissues contain small populations of adult, tissue-specific stem cells whose function is to maintain tissue integrity via ongoing cellular turnover or to serve as source of progenitors to repair or replace damaged tissues after injury. In addition to tissue homeostasis and repair, these cells are also capable of self-renewal in order to maintain the resident stem cell population (Li and Clevers, 2010; Cheung and Rando, 2013). Many tissue-specific stem cells, such as those in blood or brain, are multipotent, but tissues such as the epidermis or skeletal muscle contain stem cells that are unipotent and generate the single differentiated cell type.
Many stem cells experience prolonged periods of quiescence throughout the life of the organism. Cellular quiescence refers to a state of reversible cell cycle arrest in which cells have exited the cell cycle and entered G0 but can re-enter in response to the various stimuli. Terminally differentiated and senescent cells are also in G0 states, but they are irreversibly arrested. Some examples of quiescent stem cell populations are hematopoietic stem cells (HSCs), hair follicle stem cells (HFSCs), muscle stem cells (MuSCs), and neural stem cells (NSCs) (Cheshier et al., 1999; Blanpain et al., 2004; Cheung and Rando, 2013; Codega et al., 2014). The balance between quiescence and proliferation in stem cell populations needs to be regulated carefully as misregulation can lead to excessive cell growth or impaired tissue homeostasis or repair.
While quiescence is a common feature of many stem cell populations, it is not a requisite feature. For example, tissues like the intestinal epithelium and the epidermis rely on a population of continuously cycling stem cells that both self-renew and differentiate into parenchymal cells necessary to maintain the tissue (Li and Clevers, 2010; Hsu et al., 2014). It should be noted that the intestinal crypt also contains quiescent, lineage-restricted stem cells, which have been called “+4 cells.” These cells function as reserve stem cells and can be coaxed to replace the active stem cells in the crypts when there is acute loss of the cycling stem cells (Tetteh et al., 2016; Jadhav et al., 2017). It may be that tissues with continuously cycling stem cells for normal homeostasis also contain a separate “reserve” population of quiescent stem cells that serve as a reservoir to replenish the actively cycling cells under conditions of extreme tissue damage or stress.
In this review, we provide an overview of stem cell quiescence. We start with early studies defining the quiescent state and model systems used to study quiescence in vitro. Next, we discuss methods used to isolate quiescent stem cells to study them ex vivo. Finally, we review how these technologies have been used to characterize the quiescent state of adult stem cells and recent advances in studying the quiescent state in vivo.
In Vitro Studies of Quiescence
In early studies looking at cell proliferation, it was recognized that, within a cell population, some cells are cycling continuously while other cells exist in a non-proliferative, quiescent state with the ability re-enter the cell cycle (Mendelsohn, 1962; Howard and Pelc, 1986). Historically, it was debated whether this quiescent state of cells was a slowing of the cell cycle leading to a prolonged G1 or whether it was a separate phase outside of the cell cycle, termed G0 (Patt and Quastler, 1963). It was soon well accepted that this quiescent state is a distinct phase outside of the cell cycle (Epifanova and Terskikh, 1969; Smith and Martin, 1973). Quiescent cells are similar to cells in the G1 phase, in the sense that both populations are between the M- and S-phases of the cell cycle and contain non-replicated 2n DNA content. However, early studies using 3T3 cells showed that it takes more time for serum-deprived, quiescent cells to transition to S-phase than it does for cells to transition from the end of mitosis to S-phase (Pardee, 1974; Zetterberg and Larsson, 1985). These studies also addressed at which point of the cell cycle cells decide to commit to either quiescence or proliferation. It was noted that cells early in the G1 phase enter the quiescent state in response to serum starvation, but cells in late G1 continue in the cell cycle even when serum was removed (Pardee, 1974; Zetterberg and Larsson, 1985). The transition between these two phases of G1 is defined as a “restriction point” or “R-point” in G1, serving as a threshold to prevent cells from entering the cell cycle if conditions are unfavorable.
Since those early studies, our understanding of the mechanisms that cells use to enter, maintain, and exit quiescence has increased dramatically. This has been accomplished primarily by modeling cellular quiescence in in vitro culture systems using multiple approaches. Quiescence of cells in vitro can be induced by subjecting cells to a variety of quiescence promoting conditions, including mitogen deprivation, nutrient starvation, contact inhibition, and adhesion deprivation (Benecke et al., 1978; Coller et al., 2006). Much of our knowledge about the quiescent state comes from studies performed with the budding yeast Saccharomyces cerevisiae, with the fission yeast Schizosaccharomyces pombe, or with bacteria such as Escherichia coli. Quiescent yeast or bacterial cultures are commonly obtained by growing liquid cultures to saturation in nutrient-rich media or by carbon, nitrogen, or phosphorus deprivation. As the nutrients deplete, cells reach a non-dividing stationary phase and a substantial number of yeast or bacterial cells enter a quiescent state that aids in their long-term survival (Lillie and Pringle, 1980; Werner-Washburne et al., 1993). In contrast to bacteria and yeast, mammalian cells in vivo are more likely to enter quiescence in response to absence of mitogens or spatial cues, rather than absence of nutrients. Not all cells respond the same to various quiescence-inducing cues. For example, both mitogen deprivation and suspension culture induce quiescence in fibroblasts, where myoblast enter the quiescent state by suspension culture in the presence of mitogens while mitogen deprivation induces differentiation (Milasincic et al., 1996; Coller et al., 2006).
Interestingly, in addition to the obvious changes in cell cycle parameters, multiple characteristics of the quiescent state seem to be shared among yeast and mammalian cells, such as condensed chromosomes, decreased cell size, increased rate of autophagy, and resistance to a variety of stressors (Gray et al., 2004; Valcourt et al., 2012). Studies performed in Schizosaccharomyces pombe using deletion mutants indicate the presence of a core set of genes required for both proliferation and quiescence which are conserved among species, including human (Yanagida, 2009). When compared to cells within the cell cycle, quiescent cells display reduced RNA content, reduced rRNA synthesis, and reduced protein synthesis (Fuge et al., 1994; Coller et al., 2006; Lemons et al., 2010; Subramaniam et al., 2013). Studies in Schizosaccharomyces pombe have revealed that as cells transition from a proliferating state to a quiescent state, cell volume decreases by ~ 50% and global mRNA levels decrease by 70%, whereas global protein levels decrease by only 10% (Marguerat et al., 2012). Similar shifts in transcript and protein levels have been seen in quiescent fibroblasts and lymphocytes (Lemons et al., 2010; Lee et al., 2017). Although the total transcript levels diminish when cells transition to the quiescent state, the cells still display a high diversity of transcripts (Marguerat et al., 2012).
Early studies performed in yeast suggest that cells exit the cell cycle and enter an alternate cycle, the “quiescence cycle” (Gray et al., 2004). In this cycle, entry, maintenance, and exit are controlled by specific transcriptional and signaling processes that are distinct from the processes regulating the cell cycle. Later studies performed in fibroblasts and lymphocytes showed that cells kept under quiescence-inducing conditions for long periods of time moved deeper into quiescence and showed reduced sensitivity to mitogenic signals (Coller et al., 2006). These deeply quiescent cells remained viable and metabolically active but take significantly longer to re-enter the cell cycle (Owen et al., 1989; Yanez and O’Farrell, 1989; Lemons et al., 2010). Subsequently, it was shown that quiescence depth can be tuned continuously in fibroblasts via the Retinoblastoma (Rb)-E2F pathway (Yao et al., 2008). In quiescent cells, the CDK substrate Rb is hypophosphorylated, which results in inhibition of the E2F family of transcriptional activators. Upon exposure to mitogenic signals, CDK4/6-dependent phosphorylation of Rb is triggered, in turn initiating activation of E2F. In quiescent cells, the Rb-E2F pathway functions as a bistable switch, converting mitogenic signals into a stop-or-go E2F activation signal underlying the transition from quiescence to proliferation (Yao et al., 2008). More deeply quiescent cells have a higher activation threshold of the Rb-E2F switch, requiring stronger stimulation to re-enter the cell cycle and taking longer to exit quiescence (Kwon et al., 2017). Cyclins and cyclin-dependent kinases play an important role in regulating stem cell quiescence and cycling and these protein families has been extensively reviewed by others (Tesio and Trumpp, 2011; Hao et al., 2016).
Fibroblasts that exit the cell cycle after exposure to quiescence cues induce a unique set of genes specific to the signal inducing quiescence (Coller et al., 2006). For example, quiescence induced by contact inhibition results in differential regulation of genes involved in mechanotransduction, while mitogen deprivation results in downregulation of growth factor signaling cascades (Coller et al., 2006). However, there is also a set of genes whose expression is specific to non-dividing cells and is signal independent, namely the “quiescence program.” Not surprisingly, quiescent cells show decreased transcription of genes associated with cell cycle progression, but also increased expression of genes involved in suppressing differentiation, senescence, and apoptosis (Gos et al., 2005; Coller et al., 2006; Liu et al., 2007). The transcriptional signatures of quiescent fibroblasts have aided in revealing mechanisms that relate to the induction or maintenance of the quiescent state.
Studies using in vitro systems have aided tremendously in increasing our understanding of the quiescent state. They have provided important clues as to the conditions that allow a cell to enter quiescence, insight into the influence of the depth of quiescence on the ability of a quiescent cell to return to the cell cycle, and insight into the nature of a transcriptional program in quiescent cells. Whereas in vitro experiments provide a controlled environment in which cells can be reproducibly manipulated to study quiescence, the in vitro situation does not necessarily reflect the complex in vivo environment in which quiescent cells reside.
Identification of Quiescent Cells In Vivo
Several methods have been developed to identify quiescent cells. Quiescent cells have been identified by the absence of cell proliferation markers, low RNA content, or label retaining capacity (Kurki et al., 1986; Gerdes et al., 1991; Gothot et al., 1997; Kubota et al., 2008). As quiescent cells are not actively cycling, they do not express endogenous proliferation markers like proliferating cell nuclear antigen (PCNA), Ki67, and phospho-Histone H3 which are extensively used to distinguish quiescent cells from proliferating cells. Cells in G0 as G1 both have unreplicated genomes, so they contain equal DNA content, but G0 cells tend to be less transcriptionally active and thus have a lower total RNA content (Darzynkiewicz et al., 1980; Toba et al., 1995). Therefore, assessment of the ratio of DNA content, measured using DNA binding dyes like Hoechst 33342, propidium iodide, or DAPI, to the RNA content, measured using RNA-binding dyes like pyronin Y or SYTO dyes, within a cell can distinguish G0 cells from G1 cells (Shapiro, 1981; Darzynkiewicz et al., 2004; Kim and Sederstrom, 2015).
The above-mentioned methods can be used only on isolated cells or tissues. To identify slow-cycling cells within their niche in vivo, the ability of such cells to incorporate and then retain DNA or chromatin labels has been analyzed. A pulse of a nucleotide analog, like tritiated thymidine (3H-Tdr), 5-bromo 2′-deoxyuridine (BrdU), or 5-ethynyl-2′-deoxyuridine (EdU), labels actively dividing cells efficiently (Hughes et al., 1958; Gratzner, 1982; Salic and Mitchison, 2008). However, for slow cycling cells to be labeled, a longer exposure to the nucleotide analog is needed. Still, nucleotide labels administered to adult mice might fail to label deeply quiescent cells if they rarely divide. In proliferating cells, the label is diluted after each cell division (Figure 1A). Frequently dividing cells lose the label over time while slow cycling cells retain detectable label for extended periods of time and are thus called “label-retaining cells” (LRCs) (Reznik, 1969; Cotsarelis et al., 1990; Kiel et al., 2007). A limitation of the use of DNA nucleotide analogs is that these labels can only be detected in fixed and permeabilized cells, precluding functional tests of the labeled cells. As an alternative to nucleotide analogs, chromatin labels can be used. For example, the transgenic expression of a form of the histone protein, histone 2B, that is fused to a fluorescent protein (such as GFP) allows for the incorporation of such a reporter (in this case, H2B-GFP) into nucleosome core particles (Tumbar et al., 2004). This approach allows for induction of H2B-GFP expression, and like DNA labels, the H2B-GFP expression will be diluted with each cell division. Cells that retain the H2B-GFP label over time can be easily identified and progeny can be traced through several divisions (Tumbar et al., 2004). One of the advantages is that living, unfixed, H2B-GFP-positive cells can be readily detected, allowing for additional experiments on live cells to assess the properties of quiescent cells.
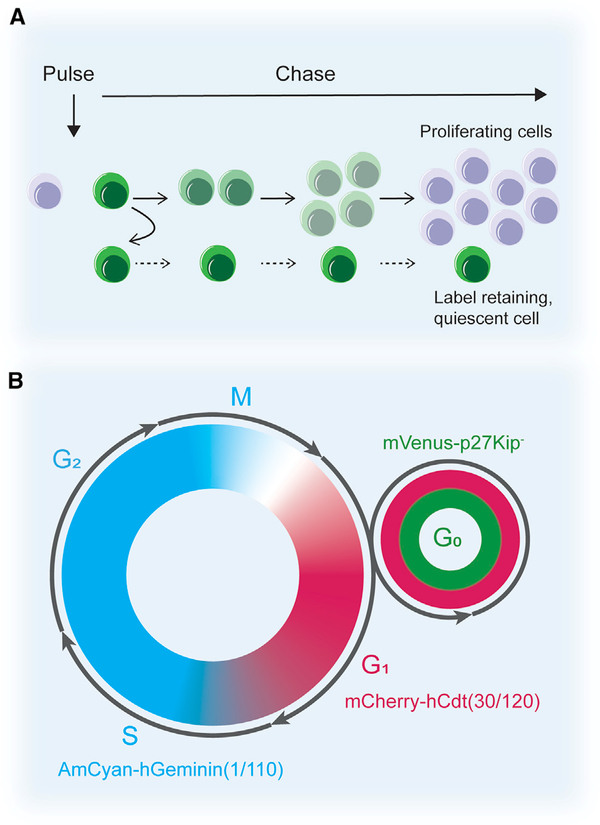
Figure 1. Identification of Quiescent Cells.
(A) Label retention assays. Cells are labeled when they divide during the pulse and, as they further divide, the label is diluted among the daughter cells where it eventually becomes undetectable. Slowly dividing cells (i.e., self-renewed quiescent stem cells) retain the label over time.
(B) The fluorescent ubiquitination-based cell cycle indicator (Fucci) system allows for analysis of the different cell cycle phases. The system uses the fusion of fluorescent proteins to cell-cycle specific proteins, geminin, Cdt1, and p27, to visualize specific cell cycle phases. Based upon the ubiquitination and degradation of these proteins in the various cell cycle phases, nuclei in G1 are labeled red, nuclei in S/G2/M are labeled cyan, and nuclei in G0 are labeled red and green.
Another promising alternative to nucleotide analogs is the use of cell cycle phase sensors that can distinguish between cells in G0 and cells in G1. For example, p27, which is more highly expressed in quiescent cells than in cycling cells, inhibits cell cycle progression at the G0 to G1 and G1 to S transitions (Toyoshima and Hunter, 1994; Coats et al., 1996; Susaki et al., 2007). To visualize cells in G0, a mouse expressing a fusion protein between the fluorescent mVenus protein and a p27 mutant lacking CDK inhibitory activity (p27K−) was developed (Oki et al., 2014). Due to p27 accumulation in quiescent cells, the fluorescent intensity is high in quiescent cells and is rapidly lost as the cells transition into G1 (Figure 1B). In mice expressing the m-Venus-p27K− fusion protein, quiescent cell populations in muscle tissue were identified (Oki et al., 2014). A limitation of these approaches is that it is difficult to discern the exact transition from one phase to another based on the fluorescent signal alone. Other interesting options include the monitoring of p57 and/or p21 expression in G0 and/or G1. However, their expression patterns are dependent on the stem cell type studied (Cheng et al., 2000; Kippin et al., 2005; Matsumoto et al., 2011; Furutachi et al., 2013; Mademtzoglou et al., 2018). Further development of these and other reporter systems will undoubtedly aid in further identification of quiescent cell populations.
Ex vivo Studies of Adult Stem Cell Quiescence
Stem cells are present in almost every tissue in the body and represent an important population of cells for both tissue homeostasis and repair. Most tissue-specific stem cells, with some notable exceptions, persist in a quiescent state and exit quiescence to enter the cell cycle only in response to an increased demand such as occurs in the setting of tissue injury. The main focus of the remainder of this review is the identification and analysis of quiescent stem cells.
In the past, quiescent stem cells have been studied retrospectively, based on functional criteria. Quiescent stem cells from different tissues have been cultured in vitro in conditions that generally do not favor the maintenance of quiescence but rather promote exit from quiescence and entry into the cell cycle. For example, NSCs have been studied in vitro as neurosphere-forming cells. However, quiescent NSCs rarely form neurospheres or give rise to adherent cell colonies (Pastrana et al., 2011; Codega et al., 2014). Sphere-forming assays predominantly allow the expansion of cells that are either poised for proliferation in vivo or are already actively dividing and can, therefore, be rapidly expanded in vitro. Likewise, quiescent MuSCs can be cultured in association with their native fibers allowing the study of the cells as they exit quiescence and begin to divide (Rosenblatt et al., 1995). On the other hand, MuSCs can be maintained in a quiescent state ex vivo for several days under the appropriate culture conditions (Quarta et al., 2016). However, culture conditions have not, to date, allowed for the maintenance of MuSCs, or any other stem cell to our knowledge, in quiescence indefinitely; most current in vitro culture conditions primarily provide a model to study the activation of stem cells from quiescence.
To study the molecular properties and function of quiescent stem cells, it is imperative to be able to prospectively identify and purify the cells. The scarcity of quiescent stem cells in most tissues has made this a difficult task. With the development of marker panels and genetic lineage tracing approaches, it has been possible to prospectively isolate quiescent stem cells. For various quiescent stem cells, panels of molecular markers are available to distinguish the stem cells from other “niche” cells and to distinguish between quiescent and activated stem cells (Kiel et al., 2005; Codega et al., 2014; Liu et al., 2015). These markers allow for purification of quiescent stem cells directly from their niche using techniques such as fluorescent activated cells sorting (FACS) or panning. The characteristics of quiescent stem cells can thus be defined on both a population and single-cell level. This has yielded tremendous insight into the molecular mechanisms regulating stem cells quiescence and activation.
Quiescence in Adult Stem Cells
Adult stem cells, which reside in unique and specialized niches, can exhibit different quiescent profiles. During homeostasis, the majority of HSCs and MuSCs are quiescent but can become activated to proliferate and differentiate in response to various stressors (Schultz et al., 1978; Cheshier et al., 1999; Passegué et al., 2005). HSCs and MuSCs are quiescent for long periods of time and divide infrequently. HSCs are estimated to divide only five times per cell over the lifetime of a mouse (Wilson et al., 2008; Foudi et al., 2009). Unlike the mainly quiescent HSCs and MuSCs, adult NSCs can be found at various stages of activation at any given time and HFSCs undergo bouts of cycling to regenerate the hair follicle (Tumbar et al., 2004; Codega et al., 2014; Llorens-Bobadilla et al., 2015). Hair follicles undergo synchronized cycles of growth (anagen), regression (catagen), and rest (telogen) (Paus and Cotsarelis, 1999; Müller-Röver et al., 2001). During telogen, which can last for months, HFSCs are quiescent within their niche, the bulge (Plikus et al., 2008).
As noted above, dysregulation of the balance between quiescence and proliferation can profoundly affect stem cell functionality. For example, in MuSCs, disruption of Notch signaling via deletion of RBP-jκ or disruption of the microRNA processing pathway by deletion of Dicer results in spontaneous activation of stem cells (Bjornson et al., 2012; Cheung et al., 2012; Mourikis et al., 2012). These spontaneously activated MuSCs fail to self-renew, thereby depleting the stem cell pool and rendering the muscle unable to regenerate in response to injury. Likewise, loss of quiescence in HSCs leads to prolonged expansion, resulting in exhaustion of the HSC population and a decreased adaptive immune function and myeloproliferative disease (Orford and Scadden, 2008; Jacob and Osato, 2009). Studies in NSCs have shown that return to quiescence after activation is required to maintain a pool of adult stem cells (Ziebell etal., 2018). Loss of hippocampal NSC quiescence leads to premature depletion of the stem cell pool, resulting in a reduced supply of new neurons and cognitive deficits (Farioli-Vecchioli et al., 2012; Jones et al., 2015). These studies highlight the importance of stem cell quiescence and show that misregulation of quiescence negatively affects tissue homeostasis and regenerative potential.
In recent years, it has become increasingly clear that the niche in which a stem cell resides has a major impact on the cell cycle status of the cell. Niches are highly specialized, anatomical regions in which stem cells reside together with supportive stromal cells and more differentiated progeny. Stem cells interact with and surrounding cells through cell-cell, cell-extracellular matrix (ECM), and cell-cytokine interactions; such local signals control the balance between quiescence and activation. Single-cell sequencing analysis of NSCs derived from the subventricular zone showed that quiescent NSCs express many receptors involved in transducing external signals and expression of these receptors decreases when the NSCs activate (Shin et al., 2015). The importance of stem cell interactions with other cells in the niche has been shown in the HSC and HFSC compartments where ablation of more differentiated progeny results in activation of the quiescent stem cell population (Hsu et al., 2011; Zhao et al., 2014). In the MuSC and HSC niche compartments, it has been shown that a change in the extracellular matrix components of the niches impacts stem cell quiescence and proliferation (Bentzinger et al., 2013; Urciuolo et al., 2013). There are many components that make up a stem cell niche. Elucidating their roles in regulating quiescence and activation will provide valuable information about the quiescent state and ways to improve in vitro culture conditions to mimic the in vivo environment in order to study quiescence.
Quiescent stem cell populations share a number of characteristics in their quiescent state (Figure 2), such as altered metabolic requirements, reduced protein synthesis, and reduced mRNA levels (Passegué et al., 2005; Simsek et al., 2010; Signer et al., 2014; Llorens-Bobadilla et al., 2015; Shin et al., 2015). Quiescent HSCs and NSCs depend on glycolysis and fatty acid energy metabolism, whereas their activated counterparts use oxidative phosphorylation to meet their energy demands (Simsek et al., 2010; Shin et al., 2015). Detailed, single-cell transcriptome studies of NSCs show that NSC exit from quiescence is marked by downregulation of glycolysis and fatty acid oxidation and by increased ribosome biogenesis, protein synthesis, and mitochondrial oxidative phosphorylation (Llorens-Bobadilla et al., 2015; Shin et al., 2015).
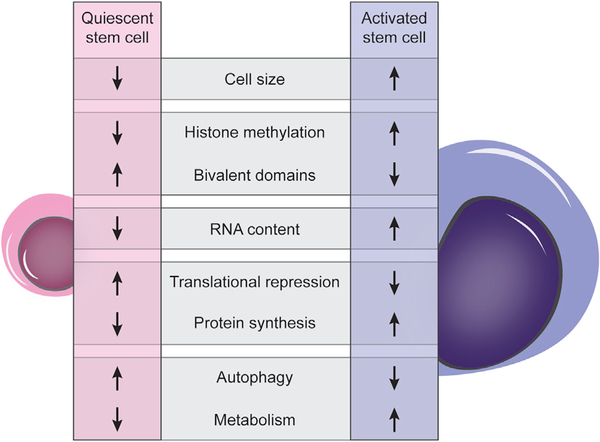
Figure 2. Characteristics of Quiescent and Activated Stem Cells.
Quiescent stem cells are characterized by tight regulation of all cellular processes. Some of the key processes that have been studied in the context of the dynamics of cellular quiescence and the maintenance of the quiescent state are illustrated here, with an indication of whether they are upregulated or downregulated compared to fully activated stem cells.
Both activated NSCs and HSCs have higher rates of protein synthesis than their quiescent counterparts (Signer et al., 2014; Llorens-Bobadilla et al., 2015). As quiescent cells do not proliferate, they have minimal metabolic requirements. Since protein synthesis is the most energy-consuming process in a cell, adult stem cells can avoid intensive energy consumption by keeping protein synthesis at a minimal level (Buttgereit and Brand, 1995). The low rate of protein synthesis in stem cells relates to their low cellular metabolism and increases the fitness and longevity of the cell (Signer et al., 2014; Cai et al., 2015; Blanco et al., 2016). Quiescent HSCs exhibit lower protein synthesis rates than restricted hematopoietic progenitors and similar results have been reported in quiescent NSCs and MuSCs compared to their activated counterparts (Signer et al., 2014; Llorens-Bobadilla et al., 2015; Zismanov et al., 2016). A study using quiescent epidermal stem cells showed that, similar to other quiescent stem cells, these cells synthesize less protein than their immediate progenitors (Blanco et al., 2016). The tight control of protein synthesis rates is necessary for cells either to maintain their quiescent state or to transition into a new state. Genetic perturbations that modestly increase or decrease protein synthesis impair stem cell function. Reduced protein synthesis rates in HSCs impair their ability to exit quiescence and reconstitute the bone marrow of lethally irradiated mice (Signer et al., 2014). On the other hand, in both HSCs and NSCs, increased protein synthesis by deletion of PTEN forces the cells to exit quiescence. The cells start to proliferate and ultimately leads to depletion of the stem cell pool (Bonaguidi et al., 2011; Signer et al., 2014). In quiescent MuSCs, repression of translation is mediated by the phosphorylation of the translation initiation factor eIF2α. MuSCs lacking eIF2α have increased protein synthesis rates, spontaneously activate, and fail to self-renew, leading to depletion of the stem cell pool (Zismanov et al., 2016.
An analysis of transcriptomic profiles of quiescent HSCs, MuSCs, and HFSCs revealed a common quiescent stem cell gene expression signature (Cheung and Rando, 2013). Unsurprisingly, genes involved in RNA replication and cell cycle progression were downregulated in all three quiescent stem cells types compared to their activated counterparts. Conversely, genes that were upregulated in quiescent stem cells included several genes in the autophagy pathway, such as Ulk2, Pink1, and Atg8, and multiple transcriptional regulators with a known function in stem cell fate decisions. Autophagy is particularly critical to non-dividing cells, such as quiescent stem cells, because of the inability of such cells to dilute the effects of dysfunctional components by the process of cell division. Instead, non-dividing cells rely on autophagy to remove unwanted cellular debris, preventing the accumulation of toxic or dysfunctional cellular constituents (Guan et al., 2013). Basal levels of active autophagy have been demonstrated to maintain quiescence in various stem cell populations (García-Prat et al., 2016; Ho et al., 2017). In HSCs, autophagy was shown to regulate the level of reactive oxygen species and to limit oxidative damage mainly through removal of aberrant mitochondria (Mortensen et al., 2011). On the other hand, dysregulation of autophagy in MuSCs results in a delay in their activation or an increase in their propensity to enter a senescent state in aged mice (Tang and Rando, 2014; Garíca-Prat et al., 2016).
Quiescent Stem Cells Are Poised for Activation/Differentiation
Quiescent stem cells can be readily activated to proliferate and differentiate in response to many different stimuli. Even though quiescent stem cells are both transcriptionally and metabolically less active than their cycling counterparts and downstream progenitors, the cells are by no means dormant. Rather, quiescent stem cells have multiple mechanisms in place that allow for rapid cell cycle entry when needed, i.e., they are poised for activation and differentiation.
As a pre-transcriptional regulator, epigenetic DNA modifications can keep genes in a transcriptionally “poised” state. By slightly altering the epigenetic balance, genes can transition from being poised to being either active or repressed. Recent studies have indicated that the quiescent stem cells exhibit unique epigenetic profiles that vary among different lineages. Compared to HFSCs that have exited G0, quiescent HFSCs show a reduction in both tri-methylated histone H3 at lysine 27 (H3K27me3) and tri-methylated histone H3 at lysine 4 (H3K4me3) (Lee et al., 2016). H3K27me3 and H3K4me3 histone marks are associated with inactive and active promotors, respectively (Lee et al., 2016). These chromatin features in quiescent HFSCs are associated with high plasticity. As the cells activate and differentiate, a strong lineage-restricted epigenetic landscape emerges, directing HFSC fate decisions (Lee et al., 2016). In quiescent MuSCs, there is a general lack of the repressive mark H3K27me3, while a high number of genes is marked by H3K4me3, indicating active transcription at these loci and might also be indicative for the cells being poised for activation (Liu et al., 2013). Interestingly, several adult stem cell populations, including MuSCs, HFSCs, and HSCs, show the presence of bivalent chromatin domains displaying both the repressive H3K27me3 mark and permissive H3K4me3 mark (Cui et al., 2009; Lien et al., 2011; Liu et al., 2013). In embryonic stem cells, key developmental genes exist in a bivalent state and are thought to be poised for activation upon initiation of differentiation (Bernstein et al., 2006). During differentiation, the bivalent domains are resolved quickly and poised genes are either activated by removing the repressive H3K27me3 or permanently silenced by loss of the activating H3K4me3 (Bernstein et al., 2006; Mikkelsen et al., 2007). In quiescent HSCs, the resolution of the bivalent domains results in gene expression changes during differentiation and instructs lineage commitment (Cui etal., 2009). The chromatin environment, in quiescent cells, seems permissive for transcription and could allow for active transcription of both key developmental genes and lineage-restricted genes.
In various quiescent stem cell types, the presence of lineage restricted transcripts with the absence of their protein products has been described (Crist et al., 2012; Liu et al., 2013; Llorens-Bobadilla et al., 2015; Grover et al., 2016; de Morrée et al., 2017; Carrelha et al., 2018). This points toward the cells being poised for activation and suggests the presence of post-transcriptional regulatory mechanisms in maintaining quiescence. Multiple post-transcriptional mechanisms have now been identified to play a role in maintenance and exit of quiescence, such as miRNA regulation of mRNA stability and translational repression (Cheung et al., 2012; Crist et al., 2012; Lechman et al., 2012; Hausburg et al., 2015; de Morrée et al., 2017). For example, the microRNA miR-126 can control cell cycle progression in HSCs by targeting the PI3K/AKT/MTOR pathway (Lechman et al., 2012). Overexpression of miR-126 increased the number of quiescent HSCs, whereas knockdown of miR-126 increased HSC proliferation without exhaustion of the stem cell pool. The importance of miRNAs is further highlighted by the fact that disruption of miRNA biogenesis releases stem cells from quiescence (Cheung et al., 2012). In MuSCs, miR-489 maintains the quiescent state by targeting DEK, while miR-31 is sequestered with Myf5 in mRNP particles and targeted for degradation (Cheung et al., 2012; Crist et al., 2012). Upon activation, the mRNP granules dissociate, releasing the Myf5 transcripts, leading to rapid translation initiation. Similarly, it was found that the transcript for another activator of the myogenic program, MyoD, is present in quiescent MuSCs but is translationally repressed through binding of the RNA-binding protein Staufen 1 (de Morrée et al., 2017). As MuSCs activate, Staufen 1 releases MyoD, leading to rapid initiation of translation. This allows the MuSCs to maintain a poised state and to respond rapidly to activating signals without the need for de novo transcription.
Levels of Quiescence in Adult Stem Cells
Several recent studies have shown the existence of distinct levels of quiescence in adult stem cells (Figure 3). For example, quiescent MuSCs can cycle between two molecularly distinct states: a canonical G0 quiescent state and a primed but still quiescent state, GAlert (Rodgers et al., 2014). Injury to skeletal muscle induces quiescent MuSCs to activate and move from G0 to G1.However, in response to injury, quiescent MuSCs in non-injured muscle respond to the injury by transitioning into the GAlert state, a state from which the MuSCs are able to contribute to muscle repair more rapidly and more effectively (Rodgers et al., 2014). MuSCs in GAlert show an increase in cell size, increased transcriptional activity, increased mitochondrial activity, and increased levels of cellular ATP when compared to MuSCs in G0. Nevertheless, these metrics in MuSCs in GAlert are still significantly lower than in fully activated MuSCs. Interestingly, other stem cells such as HSCs and and fibro-adipogenic progenitors (FAPs) also enter a GAlert state in response to a distant injury (Rodgers et al., 2014).
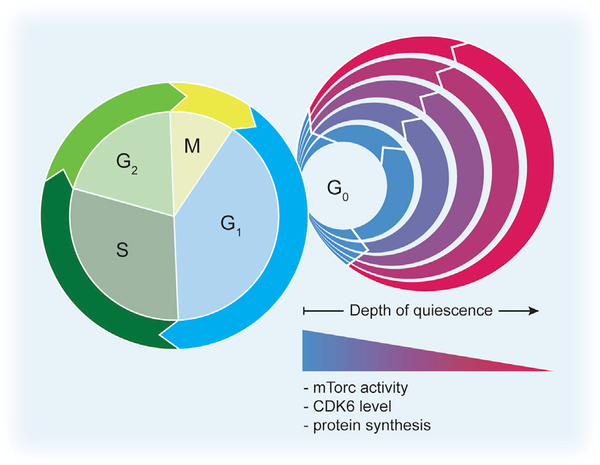
Figure 3. The Cell Cycle and the Quiescence Cycle.
The cell cycle is divided into specific phases, i.e., G1, S, G2, and M. Cells can exit this proliferative cycle in G1 and enter the quiescent state, G0. Recent studies have shown that different levels of quiescence depth exist. Examples of quiescent cells in relatively “shallow quiescent” states, termed “GAlert” or “primed” cells, can enter G1 and the cell cycle faster than more “deeply quiescent” cells. Signaling through mTORC1 has been shown to control the transition from G0 to GAlert in MuSCs, whereas the primed state in HSCs has been shown to be regulated by the level of CDK6 (Rodgers et al., 2014; Laurenti et al., 2015). Undoubtedly, stem cells from different lineages can persist in comparable alternative quiescent states, controlled by other signaling pathways.
The phenomenon of varying depths of quiescence has also been described in HSCs. The timing of exit from quiescence is differentially controlled in subsets of HSCs by CDK6 levels (Laurenti et al., 2015). Long-term (LT)-HSCs lack CDK6 protein and take a longer time to enter the cell cycle than short-term (ST)-HSCs. ST-HSCs are quiescent but contain high levels of CDK6, which allows the cells to enter the cell cycle more rapidly upon mitogenic stimulation than LT-HSCs. This CDK6 primed state in HSCs does not overlap with the GAlert state that has been described in HSCs and MuSCs (Rodgers et al., 2014). CDK6 high and low HSCs display similar levels of mTORC activity, indicating that these cells are not in the GAlert state.
Similarly, a “primed-quiescent” state has also been identified in NSCs (Llorens-Bobadilla et al., 2015). The primed quiescent NSCs were shown to have increased protein synthesis rates when compared to their more deeply quiescent counterparts. This suggests that translational activation is one of the earlier events in the exit from quiescence, which has also been described in MuSCs (Llorens-Bobadilla et al., 2015; Zismanov et al., 2016). Furthermore, primed quiescent NSCs, like MuSCs and HSCs in GAlert and primed HSCs, display accelerated entry into the cell cycle. Studies of the varying depths of quiescence have also highlighted the limited information regarding the G0 to G1 transition. Currently, no G0/G1 checkpoint has been identified and no markers can definitively distinguish G0 from early G1. Overall, the quiescent stem cells in GAlert or the primed quiescent stem cells exhibit enhanced regenerative capacity, which strongly indicates that the position of a stem cell within the quiescence cycle impacts stem cell function.
Stem Cell Quiescence: A State of Cellular “Idling”
The recently uncovered molecular and functional features of quiescent stem cells as described above have revolutionized the view of quiescence. Quiescence was thought to be a state of dormancy; instead, it encompasses features of dynamism, poised potential, and active restraint. Quiescent stem cells are not “off,” but neither are they in gear and charging down a defined pathway. Rather, they are “idling,” with many systems actively engaged, as if keeping one foot on the gas while keeping another foot on the brake. This poises the cells to be able to respond rapidly and have the flexibility to respond to the ever-changing inputs from their environment. Undoubtedly, future studies of quiescent stem cells in their native niche in vivo will further refine this perspective and add additional dimensions to the complexity of the quiescent state. In the following section, we explore in vivo methodologies that are likely to contribute to that growing body of information on the characteristics and functional potential of quiescent stem cells.
New Methodologies to Study Stem Cell Quiescence In Vivo
One of the major downsides of studying stem cells ex vivo is that the cells have to be removed from their niche (Figure 4A). Given that cells undergo rapid alterations in signaling, metabolism, and gene expression in response to changing environmental conditions, removing a stem cell from its niche and purifying it by FACS certainly results in significant molecular changes to the cell. Although freshly isolated stem cells are still quiescent from a cell cycle point of view, activation processes will have been initiated. As such, the isolated cell might not reflect the in vivo state of the stem cell.
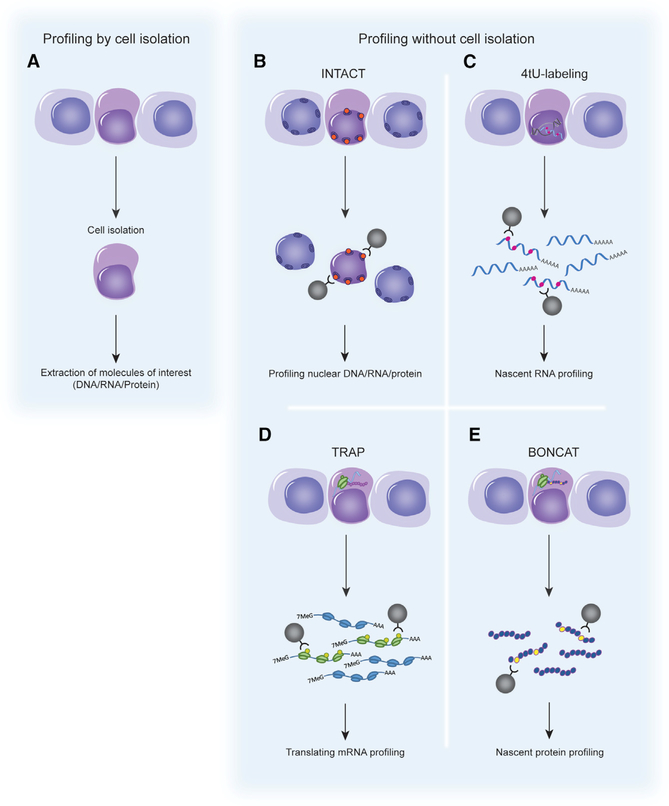
Figure 4. Cell-Specific Profiling with or without Isolation of Cells.
(A) Conventionally, quiescent stem cells are identified and isolated from the tissue by methods such as FACS. Total DNA, RNA, and/or protein is extracted and analyzed.
(B–E) Profiling of quiescent stem cells without cell isolation. In each case, a labeling construct is expressed in a cell-specific manner in vivo. The organelles/molecules of interest are labeled only in these cells. Labeled molecules can be purified from whole tissue homogenates without the need for tissues dissociation and cell purification.
(B) Isolation of nuclei tagged in specific cell types (INTACT). A nuclear targeting fusion (NTF) protein carrying the biotin ligase recognition peptide and the biotin ligase, BirA, are co-expressed in cells of interest. The NTF protein is targeted to the nuclear membrane where it acts as a substrate for BirA. After tissue homogenization, biotinylated nuclei can be recovered.
(C) 4tU tagging to label nascent RNA. The uracil phosphoribosyltransferase (UPRT) enzyme is expressed in a cell-specific manner. 4tU is administered, converted by UPRT to 4-thiouridine, and incorporated into nascent transcripts. The tissue is homogenized and thio-RNAs are extracted.
(D) Translating ribosome affinity purification (TRAP) to isolate actively translated transcripts. Tagged ribosome subunits are expressed in a cell-specific manner. After homogenization of the tissue, mRNAs associated with tagged ribosome/polysome complexes can be extracted.
(E) Bio-orthogonal non-canonical amino acid tagging (BONCAT) to label nascent proteins. A single mutation (L274G) in the amino acid binding site of methionyl-tRNA synthetase (MetRS) enables the loading of the non-canonical amino acid azidonorleucine (ANL) onto methionine tRNA. Nascent proteins can be labeled with ANL in cells that express MetRS L274G.
Two recent studies, one from our group, addressed the impact of isolation of quiescent stem cells from their niche on the transcriptional profile of these cells (van Velthoven et al., 2017; Machado et al., 2017). The transcriptome of MuSCs in vivo was determined by fixing the quiescent stem cells in vivo and compared to the transcriptome of freshly isolated MuSCs. These data showed that the isolation procedure induces major transcriptional changes, indicative of very early activation of quiescent stem cells. Not surprisingly, the isolation procedure induces expression of immediate early genes and genes coding for various heat shock proteins, indicating the induction of stress (van Velthoven et al., 2017; Machado et al., 2017).
To overcome the need for cell isolation prior and eliminate isolation-induced cellular changes, several strategies have been developed by which cell-specific profiling can be performed in vivo (Figure 4B). In Table 1 we summarize the key characteristics of the various methodologies to perform cell-specific profiling in vivo. These include isolation of nuclei tagged in specific cell types (INTACT), biorthogonal labeling of RNA and proteins, and translating ribosome purification (Heiman et al., 2008; Sanz et al., 2009; Deal and Henikoff, 2010; Gay et al., 2014; Mo et al., 2015; Alvarez-Castelao et al., 2017). The major advantage is that tissue disruption is not necessary until after the target molecules have been labeled or captured. This can bypass isolation-induced physiological changes. As such, this kind of profiling without the need for cell isolation better measures the in vivo cell state. These techniques are now being adapted to study specific stem cells in their native environment.
Table 1.
Methodologies to Study Quiescent Cells In Vivo
| Cell Isolation Needed | Molecular Analysis | Key Strength | Transgenic Lines Needed | References | |
|---|---|---|---|---|---|
| Fix-FACS | yes | cell content: DNA, mRNA, rRNA, tRNA, miRNA, non-coding RNA, protein | no transgenic mice or labeling of molecule of interest is needed | none | van Velthoven et al., 2017; Machado et al., 2017 |
| INTACT | no | nuclear content (DNA/RNA/protein) | analysis of DNA, RNA, and protein from same cell | R26-CAG-LSL-Sun1-sfGFP-myc (B6;129-Gt(ROSA)26Sortm1 (CAG- Sun1-sfGFP-Myc)Nat/J; Jax Stock 021039) | Mo et at, 2015 |
| 4tU | no | mRNA, rRNA, tRNA, miRNA, non-coding RNA, “steady-state” RNA with longer labeling times | spatiotemporal control in profiling all RNA species | CAG-L-GFP-SL-UPRT (B6;D2-Tg(CAG-GFP,-Uprt)985Cdoe/J; Jax Stock 021469) | Gay et at, 2013; van Velthoven etal., 2017 |
| TRAP | no | mRNA, subset of non-coding RNAs | identification of translated mRNA serves as a proxy for proteome | Rosa26fsTRAP (R26-CAG-LSL-eGFP/RPL10a-birA; Jax Stock 022367); RiboTag (B6.129-Rpl22tm1.1 Psam/J; Jax Stock 011029) | Heiman et at, 2008; Sanz et at, 2009; Zhou et at, 2013 |
| In vivo BONCAT | no | nascent protein | spatiotemporal control in analyzing proteins | STOPflox R26-MetRS* (Gt(ROSA)26Sortm1 (CAG-GFP, Mars*L274G)Esm; Jax Stock 028071) | Alvarez-Castelao et at, 2017; Liu et at, 2017 |
INTACT: Analysis of Nuclear Content
The different labeling techniques target specific subsets of molecules. Especially when analyzing differential gene expression, it is important to choose the population of mRNA within the total pool that is optimal for addressing one’s specific question. Total mRNA consists of newly transcribed mRNA (unprocessed and processed), steady-state mRNA, actively translated mRNA, and mRNA targeted for degradation. These various populations each give insight into different aspects of molecular regulation within a cell. The use of nuclear labeling strategies such as INTACT have allowed for the distinction between nuclear and cytoplasmic transcripts (Figure 4B). This could aid in understanding not just expression patterns, but also the regulation of specific transcripts. Furthermore, the INTACT strategy allows for the simultaneous analysis of nuclear gene expression, the underlying epigenetic profile, and the nuclear proteome (Deal and Henikoff, 2010; Mo et al., 2015). This technique was used to determine the epigenomic landscape in various neuronal populations in the brain and was able to connect the transcriptional diversity in various types of neurons to their underlying epigenetic diversity (Mo et al., 2015). One caveat of INTACT is the high rate of intron-containing transcripts in nuclear samples. This might lead to a decreased number of RNA-seq reads aligning to exons. Therefore, more RNA-seq reads would be necessary to achieve similar exon coverage as with whole cell RNA samples. However, by combining analysis of the nuclear transcriptome and histone modifications within a cell, INTACT offers the opportunity to identify the transcriptional regulatory networks underlying cell state.
4tU Labeling of Nascent Transcripts In Vivo
Whereas INTACT can be used to isolate nuclear transcripts, which are enriched for nascent transcripts, a more targeted approach to enrich for nascent RNA involves the biosynthetic tagging of RNA using 4-thiouracil (4tU). In order to label nascent RNA in vivo, the Toxoplasma gondii uracil phosphoribosyltransferase (UPRT) is expressed in a cell-specific manner (Gay et al., 2013). UPRT converts 4tU into 4-thiouridine monophosphate which is then incorporated into the RNA that is being transcribed (Figure 4C). We recently used this technique to gain insight into the nascent transcription of quiescent MuSCs in vivo (van Velthoven et al., 2017). Quiescent MuSCs within their niche display widespread, low-level, active transcription. This low but widespread transcription does not necessarily lead to protein expression. However, low-level transcription may be required to maintain a permissive chromatin environment allowing for a rapid response to activation signals (López-Maury et al., 2008). In MuSCs in vivo, we showed that a short 4tU labeling time enriches for transcripts with short half-lives, whereas longer labeling periods enriches for more stable transcripts (van Velthoven et al., 2017). By using various 4tU labeling times, the dynamics of RNA synthesis and turnover can be assessed in vivo. A possible bias of labeling transcripts with 4tU is the excess labeling of rRNA. Since rRNA transcription is always active, a large proportion of nascent transcript labeled with 4tU will be rRNA. It is, therefore, important to remove as much rRNA as possible during sample preparation unless one is specifically interested in studying nascent rRNA synthesis. Another limitation of 4tU labeling is that the ratio of 4tU-labeled RNA to total RNA must lie within a specific range. When cells expressing UPRT are too abundant, the difference between labeled RNA and total RNA will be minimal. Whereas, if very few cells express UPRT or if too short a labeling period is used, the amount of labeled RNA may be too little to show enrichment over total RNA. Therefore, labeling conditions need to be tested and optimized per cell type and/or developmental stage. However, by using various 4tU labeling times, the dynamics of RNA synthesis and turnover can be assessed in a cell-specific manner in vivo.
TRAP: Analysis of Actively Translated Transcripts
Levels of mRNA do not necessarily correlate with the expression level of the proteins they encode (Schwanhäusser et al., 2011; Liu et al., 2016). This lack of correlation between mRNA and protein can be expected given that there are multiple mechanisms regulating translation Moreover, recent studies have shown that translational regulation has a larger impact on the final protein level in cells than was previously assumed (Ingolia et al., 2009). Early responses to a stressor are regulated on a translational level rather than a transcriptional level (Spriggs et al., 2010; Tebaldi et al., 2012). To get a more accurate read on the proteome of a cell than that implied by the total transcriptome, translating ribosome affinity purification (TRAP) can be performed (Figure 4D). Genetic mouse models have been developed that express epitope-tagged ribosomal subunits in specific cell types in vivo (Heiman et al., 2008; Sanz et al., 2009; Zhou et al., 2013). Purification of epitope-tagged ribosomes allows for the isolation of actively translated mRNAs. Analysis of polysome-associated mRNAs can reveal which proteins are potentially important at a certain time in specific cell types and can allow for a more precise prediction of protein abundance within specific cells (Pradet-Balade et al., 2001; Ingolia et al., 2009). The applicability of this approach has been demonstrated in various tissues, like brain, testis, and heart (Heiman et al., 2008; Sanz et al., 2009; Zhou et al., 2013). Comparative analysis of two morphologically indistinguishable, intermixed subclasses of spiny neurons in the brain showed that these subclasses displayed vastly different translational profiles. Moreover, it was sufficiently sensitive to detect the divergent translational changes in the various neuronal subclasses after an experimental manipulation (peripheral cocaine administration) (Heiman et al., 2008). This technique is particularly interesting for studying stem cell quiescence and exit from quiescence. TRAP seems sensitive enough to detect responses to cellular perturbations, and there are indications that the early response to stressors is regulated on a translational rather than a transcriptional level (Heiman et al., 2008; Liu et al., 2016). Therefore, TRAP might facilitate the detection of the transition from G0 to G1. Though TRAP can provide a proxy for protein levels, protein abundance also depends on the rate of translation and protein degradation. Recently, TRAP was combined with ribosome footprinting, which can define the exact location of a ribosome on the transcript and aides in addressing translation rates and provide an even better proxy for protein levels (Sapkota et al., 2018).
BONCAT: Measuring the In Vivo Cellular Proteome
Whereas TRAP methodologies provide a proxy for the cellular proteome in vivo, tools to study the actual proteome in vivo have been notoriously difficult to develop. More recently, a bio-orthogonal non-canonical amino acid tagging (BONCAT) approach to label nascent proteins in vivo was developed (Alvarez-Castelao et al., 2017). The incorporation of amino acids into proteins requires processing through the cell machinery; specific tRNA synthetase, amino acid, tRNA pairs are needed to incorporate an amino acid into a growing peptide, making biorthogonal labeling strategies very suitable for genetic control (Figure 4E). The expression of a methionyl-tRNA synthetase (MetRS) with an expanded amino acid binding site (L274G) enables the methionine tRNA to be loaded with the non-canonical amino acid, azidonorleucine (ANL) (Mahdavi et al., 2016). A gene encoding MetRS L274G can be expressed in a cell-specific manner, allowing for cell-type-specific incorporation of ANL into nascent polypeptides, which can be isolated and analyzed. Using proteomic mass spectrometry, the nascent proteins can be distinguished from the pre-existing proteins in the cell. Enrichment of the nascent proteome enables deeper mass spectrometry analysis as more abundant pre-existing proteins can limit the analysis. Moreover, the nascent proteome will provide insight into the translational control in the cell at a given moment in time. Metabolic labeling of the nascent proteome has been used to compare excitatory hippocampal neurons to inhibitory Purkinje neurons and revealed unique and enriched proteins characteristic of each population (Alvarez-Castelao et al., 2017) Moreover, it was used to detect changes in the hippocampal proteome following housing of mice in an enriched sensory environment. These data indicate that metabolic labeling of the nascent proteome, like TU-tagging and TRAP, might be sensitive enough to characterize the nascent proteome of quiescent stem cells and to assess the very initial changes associated with the transition from G0 to G1.
There are several limitations to the above-mentioned techniques that preclude their ready or general application. First and foremost, there is the need for quiescent stem cell-specific transgenic driver lines to express the labeled molecules in the quiescent stem cell of interest. In addition, the availability of sufficient starting material is a concern as the total amount of labeled molecules of interest is generally a very small subfraction of the total. Moreover, quiescent stem cells generally maintain very low levels of macromolecules such as mRNA and protein compared to proliferating or differentiated cells, and the amount of total starting material is also typically very low. A common technical challenge, related to the relatively low levels of labeled molecules, is the contamination of labeled fractions with low levels of highly abundant unlabeled transcripts or proteins. In the case of incorporation of biorthogonal molecules, a low amount of non-specific incorporation has been observed (Sanz et al., 2009; Gay et al., 2013; Alvarez-Castelao et al., 2017). Awareness of such caveats and limitations allows for correction or compensation during sample preparation and analysis. As such, these and similar methods are and will continue to be invaluable in deciphering the “true” quiescent state of stem cells within their native environment in vivo.
Concluding Remarks
Advances in identification methods of quiescent adult stem cells have allowed for the isolation and analysis of these cells. It has become clear that the quiescent state is actively maintained and regulated. Quiescence allows stem cells to survive for prolonged periods of time, while being ready to be called upon in times of need. Disruption of the quiescent state is detrimental for tissue homeostasis and the regenerative capacity of a tissue. Uncovering details about the quiescent state of stem cells in their native environment will be essential in understanding the control of cellular quiescence. Recently developed methods allowing for the studies of quiescent adult stem cells in their niche in vivo will provide key information in the maintenance of quiescence during health and disease. Combining global analyses of the quiescent stem cell epigenome, transcriptome, translatome, and proteome will provide a detailed description of the quiescent state. A better understanding of the mechanisms involved in maintaining the quiescent state may result in ways to manipulate this state and ultimately lead to new therapeutic strategies by enhancing stem cell potential.
ACKNOWLEDGMENTS
The authors’ efforts were supported by the Glenn Foundation for Aging Research and by grants from the NIH (P01 AG036695, TR01 AG047820, R01 AR073248, and R37 [MERIT Award] AG023806) and from the Department of Veterans Affairs (BLR&D and RR&D) to T.A.R.
REFERENCES
- Alvarez-Castelao B, Schanzenbächer CT, Hanus C, Glock C, Tom Dieck S, Dörrbaum AR, Bartnik I, Nassim-Assir B, Ciirdaeva E, Mueller A, et al. (2017). Cell-type-specific metabolic labeling of nascent proteomes in vivo. Nat. Biotechnol 35, 1196–1201. [DOI] [PubMed] [Google Scholar]
- Benecke BJ, Ben-Ze’ev A, and Penman S (1978). The control of mRNA production, translation and turnover in suspended and reattached anchorage-dependent fibroblasts. Cell 14, 931–939. [DOI] [PubMed] [Google Scholar]
- Bentzinger CF, Wang YX, von Maltzahn J, Soleimani VD, Yin H, and Rudnicki MA (2013). Fibronectin regulates Wnt7a signaling and satellite cell expansion. Cell Stem Cell 12, 75–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. (2006). A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326. [DOI] [PubMed] [Google Scholar]
- Bjornson CRR, Cheung TH, Liu L, Tripathi PV, Steeper KM, and Rando TA (2012). Notch signaling is necessary to maintain quiescence in adult muscle stem cells. Stem Cells 30, 232–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco S, Bandiera R, Popis M, Hussain S, Lombard P, Aleksic J, Sajini A, Tanna H, Corte´ s-Garrido R, Gkatza N, et al. (2016). Stem cell function and stress response are controlled by protein synthesis. Nature 534, 335–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanpain C, Lowry WE, Geoghegan A, Polak L, and Fuchs E (2004). Self-renewal, multipotency, and the existence of two cell populations within an epithelial stem cell niche. Cell 118, 635–648. [DOI] [PubMed] [Google Scholar]
- Bonaguidi MA, Wheeler MA, Shapiro JS, Stadel RP, Sun GJ, Ming GL, and Song H (2011). In vivo clonal analysis reveals self-renewing and multipotent adult neural stem cell characteristics. Cell 145, 1142–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttgereit F, and Brand MD (1995). A hierarchy of ATP-consuming processes in mammalian cells. Biochem. J 312, 163–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai X, Gao L, Teng L, Ge J, Oo ZM, Kumar AR, Gilliland DG, Mason PJ, Tan K, and Speck NA (2015). Runx1 deficiency decreases ribosome biogenesis and confers stress resistance to hematopoietic stem and progenitor cells. Cell Stem Cell 17, 165–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrelha J, Meng Y, Kettyle LM, Luis TC, Norfo R, Alcolea V, Boukarabila H, Grasso F, Gambardella A, Grover A, et al. (2018). Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature 554, 106–111. [DOI] [PubMed] [Google Scholar]
- Cheng T, Rodrigues N, Shen H, Yang Y, Dombkowski D, Sykes M, and Scadden DT (2000). Hematopoietic stem cell quiescence maintained by p21cip1/waf1. Science 287, 1804–1808. [DOI] [PubMed] [Google Scholar]
- Cheshier SH, Morrison SJ, Liao X, and Weissman IL (1999). In vivo proliferation and cell cycle kinetics of long-term self-renewing hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 96, 3120–3125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, and Rando TA (2013). Molecular regulation of stem cell quiescence. Nat. Rev. Mol. Cell Biol 14, 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung TH, Quach NL, Charville GW, Liu L, Park L, Edalati A, Yoo B, Hoang P, and Rando TA (2012). Maintenance of muscle stem-cell quiescence by microRNA-489. Nature 482, 524–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coats S, Flanagan WM, Nourse J, and Roberts JM (1996). Requirement of p27Kip1 for restriction point control of the fibroblast cell cycle. Science 272, 877–880. [DOI] [PubMed] [Google Scholar]
- Codega P, Silva-Vargas V, Paul A, Maldonado-Soto AR, Deleo AM, Pastrana E, and Doetsch F (2014). Prospective identification and purification of quiescent adult neural stem cells from their in vivo niche. Neuron 82, 545–559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coller HA, Sang L, and Roberts JM (2006). A new description of cellular quiescence. PLoS Biol. 4, e83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotsarelis G, Sun TT, and Lavker RM (1990). Label-retaining cells reside in the bulge area of pilosebaceous unit: implications for follicular stem cells, hair cycle, and skin carcinogenesis. Cell 61, 1329–1337. [DOI] [PubMed] [Google Scholar]
- Crist CG, Montarras D, and Buckingham M (2012). Muscle satellite cells are primed for myogenesis but maintain quiescence with sequestration of Myf5 mRNA targeted by microRNA-31 in mRNP granules. Cell Stem Cell 11, 118–126. [DOI] [PubMed] [Google Scholar]
- Cui K, Zang C, Roh T-Y, Schones DE, Childs RW, Peng W, and Zhao K (2009). Chromatin signatures in multipotent human hematopoietic stem cells indicate the fate of bivalent genes during differentiation. Cell Stem Cell 4, 80–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Sharpless T, Staiano-Coico L, and Melamed MR (1980). Subcompartments of the G1 phase of cell cycle detected by flow cytometry. Proc. Natl. Acad. Sci. USA 77, 6696–6699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darzynkiewicz Z, Juan G, and Srour EF (2004). Differential Staining of DNA and RNA In Current Protocols in Cytometry (NJ, USA: John Wiley & Sons, Inc.). [DOI] [PubMed] [Google Scholar]
- de Morrée A, van Velthoven CTJ, Gan Q, Salvi JS, Klein JDD, Akimenko I, Quarta M, Biressi S, and Rando TA (2017). Staufen1 inhibits MyoD translation to actively maintain muscle stem cell quiescence. Proc. Natl. Acad. Sci. USA 114, E8996–E9005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deal RB, and Henikoff S (2010). A simple method for gene expression and chromatin profiling of individual cell types within a tissue. Dev. Cell 18, 1030–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epifanova OI, and Terskikh VV (1969). On the resting periods in the cell life cycle. Cell Prolif. 2, 75–93. [Google Scholar]
- Farioli-Vecchioli S, Micheli L, Saraulli D, Ceccarelli M, Cannas S, Scardigli R, Leonardi L, Cinà I, Costanzi M, Ciotti MT, et al. (2012). Btg1 is required to maintain the pool of stem and progenitor cells of the dentate gyrus and subventricular zone. Front. Neurosci 6, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foudi A, Hochedlinger K, Van Buren D, Schindler JW, Jaenisch R, Carey V, and Hock H (2009). Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol 27, 84–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuge EK, Braun EL, and Werner-Washburne M (1994). Protein synthesis in long-term stationary-phase cultures of Saccharomyces cerevisiae. J. Bacteriol 176, 5802–5813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutachi S, Matsumoto A, Nakayama KI, and Gotoh Y (2013). p57 controls adult neural stem cell quiescence and modulates the pace of lifelong neurogenesis. EMBO J. 32, 970–981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Prat L, Martínez-Vicente M, Perdiguero E, Ortet L, Rodríguez-Ubreva J, Rebollo E, Ruiz-Bonilla V, Gutarra S, Ballestar E, Serrano AL, et al. (2016). Autophagy maintains stemness by preventing senescence. Nature 529, 37–42. [DOI] [PubMed] [Google Scholar]
- Gay L, Miller MR, Ventura PB, Devasthali V, Vue Z, Thompson HL, Temple S, Zong H, Cleary MD, Stankunas K, and Doe CQ (2013). Mouse TU tagging: a chemical/genetic intersectional method for purifying cell type-specific nascent RNA. Genes Dev. 27, 98–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay L, Karfilis KV, Miller MR, Doe CQ, and Stankunas K (2014). Applying thiouracil tagging to mouse transcriptome analysis. Nat. Protoc 9, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerdes J, Li L, Schlueter C, Duchrow M, Wohlenberg C, Gerlach C, Stahmer I, Kloth S, Brandt E, and Flad HD (1991). Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am. J. Pathol 138, 867–873. [PMC free article] [PubMed] [Google Scholar]
- Gos M, Miloszewska J, Swoboda P, Trembacz H, Skierski J, and Janik P (2005). Cellular quiescence induced by contact inhibition or serum withdrawal in C3H10T1/2 cells. Cell Prolif. 38, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gothot A, Pyatt R, McMahel J, Rice S, and Srour EF (1997). Functional heterogeneity of human CD34(+) cells isolated in subcompartments of the G0 /G1 phase of the cell cycle. Blood 90, 4384–4393. [PubMed] [Google Scholar]
- Gratzner HG (1982). Monoclonal antibody to 5-bromo- and 5-iododeoxyuridine: A new reagent for detection of DNA replication. Science 218, 474–475. [DOI] [PubMed] [Google Scholar]
- Gray JV, Petsko GA, Johnston GC, Ringe D, Singer RA, and WernerWashburne M (2004). “Sleeping beauty”: quiescence in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev 68, 187–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover A, Sanjuan-Pla A, Thongjuea S, Carrelha J, Giustacchini A, Gambardella A, Macaulay I, Mancini E, Luis TC, Mead A, et al. (2016). Single-cell RNA sequencing reveals molecular and functional platelet bias of aged haematopoietic stem cells. Nat. Commun 7, 11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan J-L, Simon AK, Prescott M, Menendez JA, Liu F, Wang F, Wang C, Wolvetang E, Vazquez-Martin A, and Zhang J (2013). Autophagy in stem cells. Autophagy 9, 830–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao S, Chen C, and Cheng T (2016). Cell cycle regulation of hematopoietic stem or progenitor cells. Int. J. Hematol 103, 487–497. [DOI] [PubMed] [Google Scholar]
- Hausburg MA, Doles JD, Clement SL, Cadwallader AB, Hall MN, Blackshear PJ, Lykke-Andersen J, and Olwin BB (2015). Post-transcriptional regulation of satellite cell quiescence by TTP-mediated mRNA decay. eLife 4, e03390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heiman M, Schaefer A, Gong S, Peterson JD, Day M, Ramsey KE, Suárez-Fariñas M, Schwarz C, Stephan DA, Surmeier DJ, et al. (2008). A translational profiling approach for the molecular characterization of CNS cell types. Cell 135, 738–748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho TT, Warr MR, Adelman ER, Lansinger OM, Flach J, Verovskaya EV, Figueroa ME, and Passegué E (2017). Autophagy maintains the metabolism and function of young and old stem cells. Nature 543, 205–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard A, and Pelc SR (1986). Synthesis of desoxyribonucleic acid in normal and irradiated cells and its relation to chromosome breakage. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med 49, 207–218. [Google Scholar]
- Hsu Y-C, Pasolli HA, and Fuchs E (2011). Dynamics between stem cells, niche, and progeny in the hair follicle. Cell 144, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu Y-C, Li L, and Fuchs E (2014). Transit-amplifying cells orchestrate stem cell activity and tissue regeneration. Cell 157, 935–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes WL, Bond VP, Brecher G, Cronkite EP, Painter RB, Quastler H, and Sherman FG (1958). Cellular proliferation in the mouse as revealed by autoradiography with tritiated thymidine. Proc. Natl. Acad. Sci. USA 44, 476–483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ingolia NT, Ghaemmaghami S, Newman JRS, and Weissman JS (2009). Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science 324, 218–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob B, and Osato M (2009). Stem cell exhaustion and leukemogenesis. J. Cell. Biochem 107, 393–399. [DOI] [PubMed] [Google Scholar]
- Jadhav U, Saxena M, O’Neill NK, Saadatpour A, Yuan G-C, Herbert Z, Murata K, and Shivdasani RA (2017). Dynamic reorganization of chromatin accessibility signatures during dedifferentiation of secretory precursors into Lgr5+ intestinal stem cells. Cell Stem Cell 21, 65–77.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones KM, Sarić N, Russell JP, Andoniadou CL, Scambler PJ, and Basson MA (2015). CHD7 maintains neural stem cell quiescence and prevents premature stem cell depletion in the adult hippocampus. Stem Cells 33, 196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiel MJ, Yilmaz ÖH, Iwashita T, Yilmaz OH, Terhorst C, and Morrison SJ (2005). SLAM family receptors distinguish hematopoietic stem and progenitor cells and reveal endothelial niches for stem cells. Cell 121, 1109–1121. [DOI] [PubMed] [Google Scholar]
- Kiel MJ, He S, Ashkenazi R, Gentry SN, Teta M, Kushner JA, Jackson TL, and Morrison SJ (2007). Haematopoietic stem cells do not asymmetrically segregate chromosomes or retain BrdU. Nature 449, 238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim KH, and Sederstrom JM (2015). Assaying Cell Cycle Status Using Flow Cytometry In Current Protocols in Molecular Biology (NJ, USA: John Wiley & Sons, Inc; ), pp. 28.6.1–28.6.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kippin TE, Martens DJ, and van der Kooy D (2005). p21 loss compromises the relative quiescence of forebrain stem cell proliferation leading to exhaustion of their proliferation capacity. Genes Dev. 19, 756–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubota Y, Takubo K, and Suda T (2008). Bone marrow long label-retaining cells reside in the sinusoidal hypoxic niche. Biochem. Biophys. Res. Commun 366, 335–339. [DOI] [PubMed] [Google Scholar]
- Kurki P, Vanderlaan M, Dolbeare F, Gray J, and Tan EM (1986). Expression of proliferating cell nuclear antigen (PCNA)/cyclin during the cell cycle. Exp. Cell Res 166, 209–219. [DOI] [PubMed] [Google Scholar]
- Kwon JS, Everetts NJ, Wang X, Wang W, Della Croce K, Xing J, and Yao G (2017). Controlling depth of cellular quiescence by an Rb-E2F network switch. Cell Rep. 20, 3223–3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurenti E, Frelin C, Xie S, Ferrari R, Dunant CF, Zandi S, Neumann A, Plumb I, Doulatov S, Chen J, et al. (2015). CDK6 levels regulate quiescence exit in human hematopoietic stem cells. Cell Stem Cell 16, 302–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lechman ER, Gentner B, van Galen P, Giustacchini A, Saini M, Boccalatte FE, Hiramatsu H, Restuccia U, Bachi A, Voisin V, et al. (2012). Attenuation of miR-126 activity expands HSC in vivo without exhaustion. Cell Stem Cell 11, 799–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Kang S, Lilja KC, Colletier KJ, Scheitz CJF, Zhang YV, and Tumbar T (2016). Signalling couples hair follicle stem cell quiescence with reduced histone H3 K4/K9/K27me3 for proper tissue homeostasis. Nat. Commun 7, 11278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H-J, Jedrychowski MP, Vinayagam A, Wu N, Shyh-Chang N, Hu Y, Min-Wen C, Moore JK, Asara JM, Lyssiotis CA, et al. (2017). Proteomic and metabolomic characterization of a mammalian cellular transition from quiescence to proliferation. Cell Rep. 20, 721–736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons JMS, Feng X-J, Bennett BD, Legesse-Miller A, Johnson EL, Raitman I, Pollina EA, Rabitz HA, Rabinowitz JD, and Coller HA (2010). Quiescent fibroblasts exhibit high metabolic activity. PLoS Biol. 8, e1000514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, and Clevers H (2010). Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien W-H, Guo X, Polak L, Lawton LN, Young RA, Zheng D, and Fuchs E (2011). Genome-wide maps of histone modifications unwind in vivo chromatin states of the hair follicle lineage. Cell Stem Cell 9, 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lillie SH, and Pringle JR (1980). Reserve carbohydrate metabolism in Saccharomyces cerevisiae: responses to nutrient limitation. J. Bacteriol 143, 1384–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Adler AS, Segal E, and Chang HY (2007). A transcriptional program mediating entry into cellular quiescence. PLoS Genet. 3, e91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, Hurgo BMC, Leavitt T, Shih J, Brunet A, and Rando TA (2013). Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Cheung TH, Charville GW, and Rando TA (2015). Isolation of skeletal muscle stem cells by fluorescence-activated cell sorting. Nat. Protoc 10, 1612–1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Beyer A, and Aebersold R (2016). On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550. [DOI] [PubMed] [Google Scholar]
- Liu Y, Conboy MJ, Mehdipour M, Liu Y, Tran TP, Blotnick A, Rajan P, Santos TC, and Conboy IM (2017). Application of bio-orthogonal proteome labeling to cell transplantation and heterochronic parabiosis. Nat. Commun 8, 643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llorens-Bobadilla E, Zhao S, Baser A, Saiz-Castro G, Zwadlo K, and Martin-Villalba A (2015). Single-cell transcriptomics reveals a population of dormant neural stem cells that become activated upon brain injury. Cell Stem Cell 17, 329–340. [DOI] [PubMed] [Google Scholar]
- López-Maury L, Marguerat S, and Bähler J (2008). Tuning gene expression to changing environments: from rapid responses to evolutionary adaptation. Nat. Rev. Genet 9, 583–593. [DOI] [PubMed] [Google Scholar]
- Machado L, Esteves de Lima J, Fabre O, Proux C, Legendre R, Szegedi A, Varet H, Ingerslev LR, Barrés R, Relaix F, and Mourikis P (2017). In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993. [DOI] [PubMed] [Google Scholar]
- Mademtzoglou D, Asakura Y, Borok MJ, Alonso-Martin S, Mourikis P, Kodaka Y, Mohan A, Asakura A, and Relaix F (2018). Cellular localization of the cell cycle inhibitor Cdkn1c controls growth arrest of adult skeletal muscle stem cells. eLife 7, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahdavi A, Hamblin GD, Jindal GA, Bagert JD, Dong C, Sweredoski MJ, Hess S, Schuman EM, and Tirrell DA (2016). Engineered aminoacyl-tRNA synthetase for cell-selective analysis of mammalian protein synthesis. J. Am. Chem. Soc 138, 4278–4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marguerat S, Schmidt A, Codlin S, Chen W, Aebersold R, and Bähler J (2012). Quantitative analysis of fission yeast transcriptomes and proteomes in proliferating and quiescent cells. Cell 151, 671–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto A, Takeishi S, Kanie T, Susaki E, Onoyama I, Tateishi Y, Nakayama K, and Nakayama KI (2011). p57 is required for quiescence and maintenance of adult hematopoietic stem cells. Cell Stem Cell 9, 262–271. [DOI] [PubMed] [Google Scholar]
- Mendelsohn ML (1962). Autoradiographic analysis of cell proliferation in spontaneous breast cancer of C3H mouse. III. The growth fraction. J. Natl. Cancer Inst 28, 1015–1029. [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim T-K, Koche RP, et al. (2007). Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448, 553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milasincic DJ, Dhawan J, and Farmer SR (1996). Anchorage-dependent control of muscle-specific gene expression in C2C12 mouse myoblasts. In Vitro Cell. Dev. Biol. Anim 32, 90–99. [DOI] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic signatures of neuronal diversity in the mammalian brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortensen M, Soilleux EJ, Djordjevic G, Tripp R, Lutteropp M, SadighiAkha E, Stranks AJ, Glanville J, Knight S, Jacobsen SE, et al. (2011). The autophagy protein Atg7 is essential for hematopoietic stem cell maintenance. J. Exp. Med 208, 455–467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourikis P, Sambasivan R, Castel D, Rocheteau P, Bizzarro V, and Tajbakhsh S (2012). A critical requirement for notch signaling in maintenance of the quiescent skeletal muscle stem cell state. Stem Cells 30, 243–252. [DOI] [PubMed] [Google Scholar]
- Müller-Röver S, Handjiski B, van der Veen C, Eichmüller S, Foitzik K, McKay IA, Stenn KS, and Paus R (2001). A comprehensive guide for the accurate classification of murine hair follicles in distinct hair cycle stages. J. Invest. Dermatol 117, 3–15. [DOI] [PubMed] [Google Scholar]
- Oki T, Nishimura K, Kitaura J, Togami K, Maehara A, Izawa K, Sakaue-Sawano A, Niida A, Miyano S, Aburatani H, et al. (2014). A novel cell-cycle-indicator, mVenus-p27K-, identifies quiescent cells and visualizes G0-G1 transition. Sci. Rep 4, 4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orford KW, and Scadden DT (2008). Deconstructing stem cell self-renewal: genetic insights into cell-cycle regulation. Nat. Rev. Genet 9, 115–128. [DOI] [PubMed] [Google Scholar]
- Owen TA, Soprano DR, and Soprano KJ (1989). Analysis of the growth factor requirements for stimulation of WI-38 cells after extended periods of density-dependent growth arrest. J. Cell. Physiol 139, 424–431. [DOI] [PubMed] [Google Scholar]
- Pardee AB (1974). A restriction point for control of normal animal cell proliferation. Proc. Natl. Acad. Sci. USA 71, 1286–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passegué E, Wagers AJ, Giuriato S, Anderson WC, and Weissman IL (2005). Global analysis of proliferation and cell cycle gene expression in the regulation of hematopoietic stem and progenitor cell fates. J. Exp. Med 202, 1599–1611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastrana E, Silva-Vargas V, and Doetsch F (2011). Eyes wide open: a critical review of sphere-formation as an assay for stem cells. Cell Stem Cell 8, 486–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patt HM, and Quastler H (1963). Radiation effects on cell renewal and related systems. Physiol. Rev 43, 357–396. [DOI] [PubMed] [Google Scholar]
- Paus R, and Cotsarelis G (1999). The biology of hair follicles. N. Engl. J. Med 341, 491–497. [DOI] [PubMed] [Google Scholar]
- Plikus MV, Mayer JA, de la Cruz D, Baker RE, Maini PK, Maxson R, and Chuong C-M (2008). Cyclic dermal BMP signalling regulates stem cell activation during hair regeneration. Nature 451, 340–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pradet-Balade B, Boulmé F, Beug H, Müllner EW, and Garcia-Sanz JA (2001). Translation control: bridging the gap between genomics and proteomics? Trends Biochem. Sci 26, 225–229. [DOI] [PubMed] [Google Scholar]
- Quarta M, Brett JO, DiMarco R, De Morree A, Boutet SC, Chacon R, Gibbons MC, Garcia VA, Su J, Shrager JB, et al. (2016). An artificial niche preserves the quiescence of muscle stem cells and enhances their therapeutic efficacy. Nat. Biotechnol 34, 752–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reznik M (1969). Thymidine-3H uptake by satellite cells of regenerating skeletal muscle. J. Cell Biol 40, 568–571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodgers JT, King KY, Brett JO, Cromie MJ, Charville GW, Maguire KK, Brunson C, Mastey N, Liu L, Tsai C-R, et al. (2014). mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(Alert). Nature 510, 393–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblatt JD, Lunt AI, Parry DJ, and Partridge TA (1995). Culturing satellite cells from living single muscle fiber explants. In Vitro Cell. Dev. Biol. Anim 31, 773–779. [DOI] [PubMed] [Google Scholar]
- Salic A, and Mitchison TJ (2008). A chemical method for fast and sensitive detection of DNA synthesis in vivo. Proc. Natl. Acad. Sci. USA 105, 2415–2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanz E, Yang L, Su T, Morris DR, McKnight GS, and Amieux PS (2009). Cell-type-specific isolation of ribosome-associated mRNA from complex tissues. Proc. Natl. Acad. Sci. USA 106, 13939–13944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sapkota D, Lake AM, Yang W, Yang C, Wesseling H, Guise A, Uncu C, Dalal JS, Kraft A, Lee J-M, et al. (2018). Cell type specific profiling of alternative translation identifies novel protein isoforms in the mouse brain. bioRxiv. 10.1101/324236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz E, Gibson MC, and Champion T (1978). Satellite cells are mitotically quiescent in mature mouse muscle: an EM and radioautographic study. J. Exp. Zool 206, 451–456. [DOI] [PubMed] [Google Scholar]
- Schwanhäusser B, Busse D, Li N, Dittmar G, Schuchhardt J, Wolf J, Chen W, and Selbach M (2011). Global quantification of mammalian gene expression control. Nature 473, 337–342. [DOI] [PubMed] [Google Scholar]
- Shapiro HM (1981). Flow cytometric estimation of DNA and RNA content in intact cells stained with Hoechst 33342 and pyronin Y. Cytometry 2, 143–150. [DOI] [PubMed] [Google Scholar]
- Shin J, Berg DA, Zhu Y, Shin JY, Song J, Bonaguidi MA, Enikolopov G, Nauen DW, Christian KM, Ming GL, and Song H (2015). Single-cell RNA-seq with waterfall reveals molecular cascades underlying adult neurogenesis. Cell Stem Cell 17, 360–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signer RAJ, Magee JA, Salic A, and Morrison SJ (2014). Haematopoietic stem cells require a highly regulated protein synthesis rate. Nature 509, 49–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, and Sadek HA (2010). The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell 7, 380–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JA, and Martin L (1973). Do cells cycle? Proc. Natl. Acad. Sci. USA 70, 1263–1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spriggs KA, Bushell M, and Willis AE (2010). Translational regulation of gene expression during conditions of cell stress. Mol. Cell 40, 228–237. [DOI] [PubMed] [Google Scholar]
- Subramaniam S, Sreenivas P, Cheedipudi S, Reddy VR, Shashidhara LS, Chilukoti RK, Mylavarapu M, and Dhawan J (2013). Distinct transcriptional networks in quiescent myoblasts: a role for Wnt signaling in reversible vs. irreversible arrest. PLoS ONE 8, e65097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Susaki E, Nakayama K, and Nakayama KI (2007). Cyclin D2 translocates p27 out of the nucleus and promotes its degradation at the G0-G1 transition. Mol. Cell. Biol. 27, 4626–4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang AH, and Rando TA (2014). Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 33, 2782–2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tebaldi T, Re A, Viero G, Pegoretti I, Passerini A, Blanzieri E, and Quattrone A (2012). Widespread uncoupling between transcriptome and translatome variations after a stimulus in mammalian cells. BMC Genomics 13, 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tesio M, and Trumpp A (2011). Breaking the cell cycle of HSCs by p57 and friends. Cell Stem Cell 9, 187–192. [DOI] [PubMed] [Google Scholar]
- Tetteh PW, Basak O, Farin HF, Wiebrands K, Kretzschmar K, Begthel H, van den Born M, Korving J, de Sauvage F, van Es JH, et al. (2016). Replacement of lost Lgr5-positive stem cells through plasticity of their enterocyte-lineage daughters. Cell Stem Cell 18, 203–213. [DOI] [PubMed] [Google Scholar]
- Toba K, Winton EF, Koike T, and Shibata A (1995). Simultaneous threecolor analysis of the surface phenotype and DNA-RNA quantitation using 7-amino-actinomycin D and pyronin Y. J. Immunol. Methods 182, 193–207. [DOI] [PubMed] [Google Scholar]
- Toyoshima H, and Hunter T (1994). p27, a novel inhibitor of G1 cyclin-Cdk protein kinase activity, is related to p21. Cell 78, 67–74. [DOI] [PubMed] [Google Scholar]
- Tumbar T, Guasch G, Greco V, Blanpain C, Lowry WE, Rendl M, and Fuchs E (2004). Defining the epithelial stem cell niche in skin. Science 303, 359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urciuolo A, Quarta M, Morbidoni V, Gattazzo F, Molon S, Grumati P, Montemurro F, Tedesco FS, Blaauw B, Cossu G, et al. (2013). Collagen VI regulates satellite cell self-renewal and muscle regeneration. Nat. Commun 4, 1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valcourt JR, Lemons JMS, Haley EM, Kojima M, Demuren OO, and Coller HA (2012). Staying alive: metabolic adaptations to quiescence. Cell Cycle 11, 1680–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Velthoven CTJ, de Morree A, Egner IM, Brett JO, and Rando TA (2017). Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner-Washburne M, Braun E, Johnston GC, and Singer RA (1993). Stationary phase in the yeast Saccharomyces cerevisiae. Microbiol. Rev 57, 383–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson A, Laurenti E, Oser G, van der Wath RC, Blanco-Bose W, Jaworski M, Offner S, Dunant CF, Eshkind L, Bockamp E, et al. (2008). Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell 135, 1118–1129. [DOI] [PubMed] [Google Scholar]
- Yanagida M (2009). Cellular quiescence: are controlling genes conserved? Trends Cell Biol. 19, 705–715. [DOI] [PubMed] [Google Scholar]
- Yanez I, and O’Farrell M (1989). Variation in the length of the lag phase following serum restimulation of mouse 3T3 cells. Cell Biol. Int.Rep 13,453–462. [DOI] [PubMed] [Google Scholar]
- Yao G, Lee TJ, Mori S, Nevins JR, and You L (2008). A bistable Rb-E2F switch underlies the restriction point. Nat. Cell Biol 10, 476–482. [DOI] [PubMed] [Google Scholar]
- Zetterberg A, and Larsson O (1985). Kinetic analysis of regulatory events in G1 leading to proliferation or quiescence of Swiss 3T3 cells. Proc. Natl. Acad. Sci. USA 82, 5365–5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Perry JM, Marshall H, Venkatraman A, Qian P, He XC, Ahamed J, and Li L (2014). Megakaryocytes maintain homeostatic quiescence and promote post-injury regeneration of hematopoietic stem cells. Nat. Med 20, 1321–1326. [DOI] [PubMed] [Google Scholar]
- Zhou P, Zhang Y, Ma Q, Gu F, Day DS, He A, Zhou B, Li J, Stevens SM, Romo D, and Pu WT (2013). Interrogating translational efficiency and lineage-specific transcriptomes using ribosome affinity purification. Proc. Natl. Acad. Sci. USA 110, 15395–15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziebell F, Dehler S, Martin-Villalba A, and Marciniak-Czochra A (2018). Revealing age-related changes of adult hippocampal neurogenesis using mathematical models. Development 145, dev153544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zismanov V, Chichkov V, Colangelo V, Jamet S, Wang S, Syme A, Koromilas AEE, and Crist C (2016). Phosphorylation of eIF2a is a translational control mechanism regulating muscle stem cell quiescence and self-renewal. Cell Stem Cell 18, 79–90. [DOI] [PubMed] [Google Scholar]