Abstract
The live-attenuated measles virus (MV) vaccine based on the Hu191 strain has played a significant role in controlling measles in China. However, it has considerable adverse effects that may cause public health burden. We hypothesize that the safety and efficacy of MV vaccine can be improved by altering the S-adenosylmethionine (SAM) binding site in the conserved region VI of the large polymerase protein. To test this hypothesis, we established an efficient reverse genetics system for the rMV-Hu191 strain and generated two recombinant MV-Hu191 carrying mutations in the SAM binding site. These two mutants grew to high titer in Vero cells, were genetically stable, and were significantly more attenuated in vitro and in vivo compared to the parental rMV-Hu191 vaccine strain. Importantly, both MV-Hu191 mutants triggered a higher neutralizing antibody than rMV-Hu191 vaccine and provided complete protection against MV challenge. These results demonstrate its potential for an improved MV vaccine candidate.
Keywords: Measles virus, Vaccine, Reverse genetics, Methyltransferase, S-adenosylmethionine, Pathogenesis and immunogenicity
Highlights
-
•
An efficient reverse genetics system for Chinese MV-Hu191 strain was developed.
-
•
rMV-Hu191 mutants in SAM binding site are attenuated in vitro and in vivo.
-
•
rMV-Hu191 mutants in SAM binding site enhance the safety and immunogenicity of MV vaccine.
1. Introduction
Measles virus (MV) is an enveloped virus with a non-segmented, negative-sense (NNS) RNA genome in the family Paramyxoviridae, order Mononegavirales (Radecke et al., 1995). In developing countries, measles is still a leading cause of mortality in children (Griffin and Oldstone, 2009, Tangy and Naim, 2005), though vaccination is an effective, economical, and safe way to prevent outbreaks (Bester, 2016, de Vries et al., 2008). In early 1960, a live-attenuated vaccine based on the Hu191 strain of MV was developed and is currently widely used for immunization in all provinces of China (Zhang et al., 2009). While this vaccine is efficacious, it has associated adverse effects. Many vaccinated infants and children in China experienced side effects ranging from skin rashes, itching, swelling, and to high fever (Bester, 2016, Shu et al., 2011). Additionally, outbreaks of measles have been increasing significantly in the past a few years in China, particularly the increasing proportion of adult and infant cases (Ma et al., 2016, Zhang et al., 2016). The infected adults had received measles vaccination during childhood; still remain susceptible to infection with the measles virus, as the population immunity against measles after vaccination gradually reduces with time (Abad and Safdar, 2015, Gao et al., 2017, Ma et al., 2016, Zhang et al., 2016). Thus, there is an increasing urgency to develop a safer, more efficient MV vaccine for eradication of measles in China.
Reverse genetics system has been established for many NNS RNA viruses including the vesiculovirus, morbillivirus, respirovirus, and pneumovirus (Neumann et al., 2002). Similar to other NNS RNA viruses, the minimal machinery for MV transcription and replication is the ribonucleoprotein (RNP) complex, which consists of the nucleocapsid (N)-RNA template tightly associated with the RNA-dependent RNA polymerase, the large (L) protein and the phosphoprotein (P). Assembly of replication-competent RNPs is essential to the rescue of NNS RNA viruses (Bukreyev et al., 1996, Clarke et al., 2000, Garcin et al., 1995, Gassen et al., 2000, Jin et al., 1998, Lawson et al., 1995). This can be achieved by co-transfection of a plasmid encoding a full-length antigenomic cDNA together with plasmids encoding N, P, and L genes. Previously, several groups have already successfully rescued infectious MV from cDNA clones (Duprex et al., 1999, Kovacs et al., 2003, Nakatsu et al., 2006, Parks et al., 1999, Radecke et al., 1995, Sidhu et al., 1995). The reverse genetics system can facilitate the rational design of safer, more efficient measles vaccine candidates.
The L protein of NNS RNA viruses possesses the majority of enzymatic activities for transcription and replication (Ferron et al., 2002, Poch et al., 1990, Whelan et al., 2004). During transcription, NNS RNA viruses synthesize mRNAs that are capped and methylated at the 5’end and polyadenylated at the 3’ end. Recent studies have shown that the entire mRNA capping and methylation machinery of NNS RNA viruses is distinct from their host (Ferron et al., 2002, Furuichi and Shatkin, 2000, Ma et al., 2014, Ogino and Banerjee, 2007, Zhang et al., 2014). Using vesicular stomatitis virus (VSV) as a model, it was found that VSV mRNA capping is catalyzed by an RNA:GDP polyribonucleotidyltransferase (PRNTase) in the L protein that transfers a monophosphate RNA onto a GDP acceptor (Li et al., 2008, Ogino and Banerjee, 2007). The mRNA cap methylation in NNS RNA viruses is also unusual in that a single region in the L protein catalyzes both guanine-N-7 (G-N-7) and ribose 2′-O (2′-O) methylation (Li et al., 2006, Rahmeh et al., 2009). Thus, mRNA cap formation is an excellent target for development of antiviral drugs and live vaccine candidates for NNS RNA viruses. Based on the sequence alignments, the L protein contains six conserved regions (CR) numbered I to VI. Recent studies showed that CR V of the L protein possesses an mRNA capping enzyme whereas CR VI is responsible for mRNA cap methyltransferase (MTase) activity (Li et al., 2008, Ogino et al., 2005). It was shown that mutations to the capping enzyme were lethal to the virus. However, mutations to MTase region yielded recombinant viruses that were attenuated in vitro and in vivo. This suggests mRNA cap MTase is a novel target for rational design of live attenuated vaccines for NNS RNA viruses. This novel concept has recently been tested in several NNS RNA viruses including VSV, avian metapneumovirus (aMPV), human metapneumovirus (hMPV), and rabies virus (RABV) (Li et al., 2006, Ma et al., 2014, Sun et al., 2014, Tian et al., 2015, Zhang et al., 2014). It was shown that recombinant viruses lacking MTase activity are highly attenuated in vitro and in vivo, yet retain optimal immunogenicity.
We hypothesized that engineering mutations to the MTase region of MV L protein would lead to further attenuation of the current live attenuated vaccine strain, enhancing the safety of MV vaccine. To test this hypothesis, we established a robust reverse genetics system based on a Chinese MV vaccine strain MV-Hu-191, allowing us to recover recombinant MV in BHK cells stably expressing T7 RNA polymerase (Xu et al., 2011, Zhang et al., 2014). Subsequently, two recombinant MVs with amino acid (aa) substitutions in the S-adenosylmethionine (SAM) binding site of L protein (rMV-Hu191-G1788A and rMV-Hu191-G1792A) were successfully recovered. These two MTase-defective mutants had delayed replication kinetics, grew to high titers, and were genetically stable through 15 passages in cell culture. Both MV mutants were significantly more attenuated in vitro and in vivo compared to the parental vaccine strain. Interestingly, both mutants induced significantly higher neutralizing antibody titers compared to the parental virus. These results demonstrate that alteration of SAM binding sites in MV L protein enhances both the safety profile and the immunogenicity of the MV vaccine. Thus, mRNA cap MTase can serve a novel approach for rational design of a safer and more efficacious MV vaccine.
2. Results
2.1. Recovery of rMV-Hu191 from a full-length cDNA clone
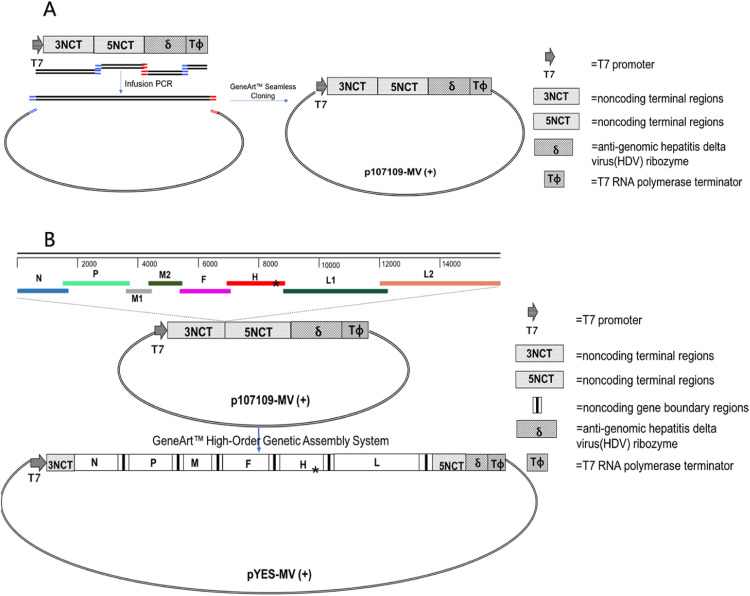
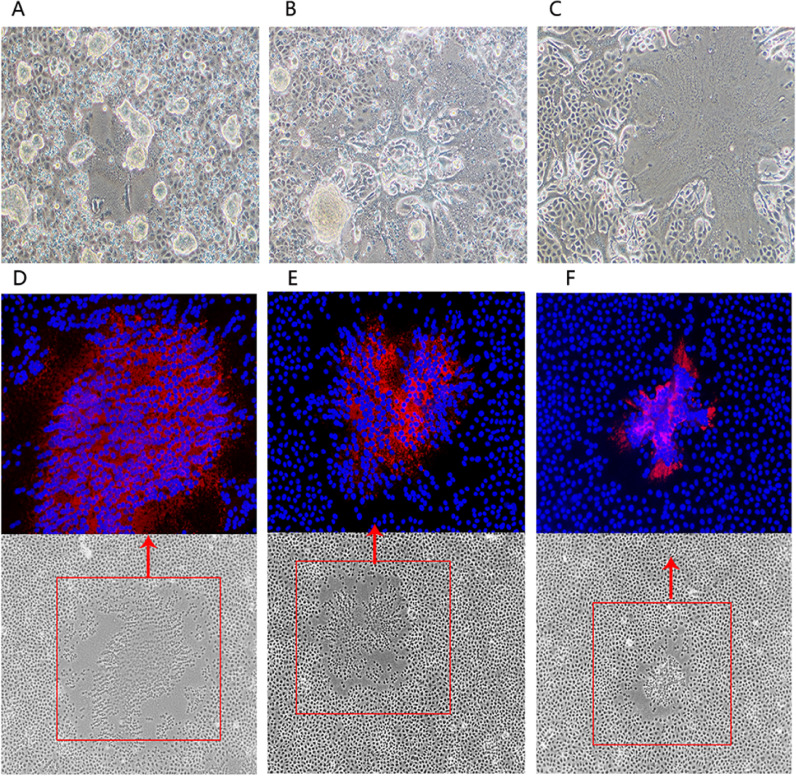
A full-length cDNA clone of MV strain Hu191, pYES-MV(+), was constructed by a novel methodology using the GeneArt™ High-Order Genetic Assembly System. The full-length cDNA clone of MV-Hu191 was successfully assembled by a single step ligation, without the need for restriction endonucleases. The 15,896-nt antigenomic MV cDNA was cloned under the control of a T7 RNA polymerase promoter, a hepatitis delta virus (HDV) ribozyme sequence, and a T7 terminator ( Fig. 1; Table 1). To recover infectious MV, BHK-SR19-T7 cells stably expressing T7 RNA polymerase were co-transfected with full-length cDNA clone pYES-MV(+) and the support plasmids expressing ribonucleoprotein (pT7-Hu191-N, pT7-Hu191-P, and pT7-Hu191-L). Three days post-transfection, cell monolayers were trypsinized and co-cultured with fresh Vero cells. MV-induced syncytia were observed 2–3 days later ( Fig. 2A, B and C). The successful recovery of rMV-Hu191 was further confirmed by detection of N protein expression in Vero cells infected with the rescued rMV-Hu191 by an immunofluorescence assay (Fig. 2D). When extensive syncytia were observed, cells were harvested and the supernatants were used for further passage in Vero cells. After 3–4 passages, recombinant MV was plaque purified, and a large stock of virus was prepared. There is a spontaneous mutation (C to U) at nucleotide (nt) position 8763 within the H gene of the lab-propagated parental virus, which is different from the published Hu191 sequence (GenBank accession no. FJ416067). This site in pYES-MV(+) was mutated back to C by site-directed mutagenesis with PCR, which distinguished the parental virus in the lab but is identical with the published sequence. To confirm the recovered recombinant virus (rMV-Hu191) originated from pYES-MV(+) and not from cross-contamination of the parental MV-Hu191 grown in our laboratory, a region of the rMV H gene was amplified by RT-PCR and sequenced. The result showed that rMV-Hu191 contained the “C” mutation.
Fig. 1.
Construction of a full-length cDNA clone for MV-Hu191. The T7 promoter, 3′ and 5′ non-coding termini (NCT), antigenomic HDV ribozyme and T7 terminator were assembled in several rounds of fusion PCR, and inserted into pYES-2 using a “seamless” cloning strategy, resulted in the construction of p107109-MV(+) (A). Eight overlapping fragments containing the full-length MV genome were assembled into p107109-MV(+), creating pYES-MV(+) (B). A spontaneous mutation (C to U) in the H gene that distinguishes the lab-propagated parental virus and rescued recombinant virus was marked by “*” (B).
Table 1.
Sequences of the oligonucleotides used for PCR.
| Primer | Sequence (5′−3′) |
|---|---|
| MV-5end-F | ACCAAACAAAGTTGGGTAAGGATA |
| MV-N-R | GGTAGGCGGATGTTGTTCTG |
| MV-P-F | CTTCTAGACTAGGTGCGA |
| MV-P-R | GAACTTGGGAGGCAATCACTT |
| MV-M-F | CCAACTAGTACAACCTAAATCCA |
| MV-M-R | GTGGGGAGTTGAGTGTCGTC |
| MV-M-2-F | GGGGGCACCAGTCTTCACATAT |
| MV-M-2-R | CATGAATATGGCAGAGACGT |
| MV-F-F | CCCGACGACACTCAACTCCC |
| MV-F-R | CCTAAGTTTTAATTAACTACCGATA |
| MV-H-F | TCCCTCTGGCCGAACAAT |
| MV-H-R | ACGTTTTTCTTAATTCTGATGTCTAT |
| MV-L1-F | ACATCAGGCATACCCACTA |
| MV-L1-R | CCCACATATGGCTTCTTAG |
| MV-L2-F | GACAAAGAGTCATGTTCAGTG |
| MV-3endR | CAGACAAAGCTGGGAATAG |
| MV-H-M-F | GCCATGCCCCAACATACCTACCTGC |
| MV-H-M-R | GCAGGTAGGTATGTTGGGGCATGGC |
Fig. 2.
Recombinant MV-induced syncytia in Vero monolayers. BHK-SR19-T7 cells were transfected with pYES-MV(+ ) and helper plasmids pT7-Hu191-N, pT7-Hu191-P, and pT7-Hu191-L. At day 3 post-transfection, confluent cell monolayers were trypsinized and transferred onto Vero cell monolayers, and co-cultured for 24 h (A) or 48 h (B). Supernatant (P1) from Vero cells were further passaged onto confluent Vero cell monolayers, and incubated for 36 h (C).
2.2. Recovery of two rMV-Hu191 mutants carrying mutations in the SAM binding site
Having the establishment of robust reverse genetics for rMV-Hu191, we next tested the hypothesis that the rMV-Hu191 vaccine strain can be further attenuated by alteration of the SAM binding site in MV L protein. Previously, this strategy was used in the rational design of live attenuated vaccine candidates for VSV, aMPV, and hMPV (Li et al., 2006, Sun et al., 2014, Zhang et al., 2014). The SAM-dependent MTase superfamily typically contains a conserved G-rich motif for binding the SAM molecule, the methyl donor for RNA methylation (McIlhatton et al., 1997, Schluckebier et al., 1995). Sequence alignment revealed that a GxGxGx motif was conserved in CR VI of the L proteins of all paramyxoviruses and most of the Mononegavirales ( Fig. 3) (Li et al., 2006, McIlhatton et al., 1997, Poch et al., 1990, Zhang et al., 2014). Sequence analysis found that aa residues corresponding to the GxGxGx motif of the MV L protein includes G1788, G1790, and G1792.
Fig. 3.
Sequence conservation of the SAM binding site in the CR VI of L protein in NNS RNA viruses. Partial sequences (amino acid 1745–1841 based on MV) of the L proteins of selected NNS RNA viruses were compared using MegAlign software. The “G” rich motif in the predicted SAM binding site (GxGxGx) is highlighted by gray color. Representative members of the Paramyxiviridae (MV, measles virus; HeV, Hendra virus; NiV, Nipah virus; CDV, canine distemper virus; PDV, porcine distemper virus; PPRV, peste des petits ruminants virus; RPV, Rinderpest virus; BPIV-3, bovine parainfluenza virus 3;HPIV-1, human parainfluenza virus 1; HPIV-3, human parainfluenza virus 3; HPIV-4a, human parainfluenza virus 4a; HPIV-4b, human parainfluenza virus 4b; SeV, Sendai virus; MuV-S79, mumps virus isolate S79 live vaccine; NDV, Newcastle disease virus), Pneumoviridae (PVM, pneumonia virus of mice; BRSV, bovine respiratory syncytial virus; HMPV, human metapneumovirus; HRSVA, human respiratory syncytial virus A; HRSVB, human respiratory syncytial virus B), and Rhabdoviridae (VSVI, vesicular stomatitis virus Indiana serotype) are shown.
Therefore, these amino acids were individually mutated to alanine in an infectious cDNA clone of MV, pYES-MV(+ ). Three recombinant MV clones with a single point mutation (G1788A, G1790A or G1792A) in their SAM binding sites were constructed. Using the reverse genetics system, two recombinant MVs, rMV-Hu191-G1788A and rMV-Hu191-G1792A, were successfully rescued and viral titer gradually increased when they were passaged in Vero cells. The rMV mutants were confirmed by detection of N protein expression in Vero cells infected with the rescued rMV mutants by immunofluorescence (Figs. 2E and 2F). The rMV-Hu191-G1790A mutant was viable, but it grew poorly in Vero cell and further passages of this mutant did not increase viral titer (data not shown). Next, rMV-Hu191-G1788A and rMV-Hu191-G1792A were plaque purified. The sizes of virus-induced plaques differed between the rescued parental and mutant viruses. As demonstrated in Fig. 4, after 6 days of incubation, the parental rMV-Hu191 formed plaques were 1.43 ± 0.22 mm in diameter, whereas the average plaque for rMV-Hu191-G1788A and rMV-Hu191-G1792A was significantly smaller (0.98 ± 0.16 mm and 0.79 ± 0.13 mm, respectively; p < 0.05). This suggests that the two MV mutants likely had impaired growth kinetics that caused the plaque sizes to be reduced. Finally, the entire genome of each MV mutant was amplified by RT-PCR and sequenced. Result showed that each mutant retained the desired mutation. In addition, no other mutations were found in the genome.
Fig. 4.
Plaque morphology of recombinant MVs. (A) Plaque morphology of each recombinant virus carrying mutations in the SAM binding site was shown. Vero cells were infected with each recombinant MV and incubated at 37 °C; all plaques were developed after 6 days of incubation. (B) The average diameter of a total of 15 plaques for each recombinant virus. *** =p < 0.001.
2.3. Recombinant rMV-Hu191 carrying mutations in the SAM binding site were more attenuated in cell culture compared to the parental virus
We next compared the replication kinetics of the rMV-Hu191 mutants and the parental virus in Vero cells in the time course of 120 h after infection ( Fig. 5). Parental rMV-Hu191 reached a peak titer (6.86 ± 0.14 log10PFU/ml) at 72 h post-inoculation (hpi), while peak titers for the two mutants at 96 hpi. Importantly, rMV-Hu191-G1788A was delayed in replication but reached a peak titer of 6.9 ± 0.06 log10PFU/ml at 96 hpi, which was comparable to the parental virus (p > 0.05). The peak titer achieved by rMV-Hu191-G1792A was 6.06 ± 0.20 log10PFU/ml at 96 hpi, which was significantly lower than that of parental rMV-Hu191 at 72 hpi (p < 0.05). Both MV mutants had delayed cytopathic effects (CPE) compared to the parental virus. The parental rMV-Hu191 developed extensive cell-to-cell fusion and large syncytia at 48 hpi and reached maximum CPE at 72 hpi, whereas mutants rMV-Hu191-G1788A and rMV-Hu191-G1792A had a delay in formation of syncytia and reached maximum CPE at 96 hpi ( Fig. 6). These data suggest that rMVs carrying mutations in the SAM binding site were more attenuated in Vero cells than the parental MV vaccine strain.
Fig. 5.
Growth kinetic of recombinant MVs carrying mutations in the SAM binding site. Vero cells were infected with each recombinant MV at an MOI of 0.01; supernatant and cells were harvested after three freeze-thaw cycles, and virus titer was determined by plaque assay in Vero cells. Parental rMV-Hu191 reached a peak titer after 72 h post-infection (hpi) whereas peak titer for the mutants ranged from 84 to 96 hpi. Virus titer for the mutants detected at different time points were different from the parental rMV-Hu191. * =p < 0.05; *** =p < 0.001.
Fig. 6.
Syncytium formation of recombinant MVs. Confluent Vero cells were infected with each recombinant MV at an MOI of 0.01. Syncytia were visualized with a light microscope, and photos taken at 12, 24, 48, 60, 72, 84, and 96 h post-infection.
2.4. Recombinant rMV-Hu191 mutants are genetically stable in cell culture
To investigate whether rMV-Hu191-G1788A and rMV-Hu191-G1792A were genetically stable in vitro, each virus was passaged in Vero cells for 15 times. The mutated region in the L gene was sequenced for each of the first 10 passages. Virus in each passage retained the desired mutation. At passage 15, the entire genome of each mutant was sequenced, showing no additional mutations in the genome.
2.5. rMV-Hu191 carrying mutations in the SAM binding site are more attenuated in vivo than the parental vaccine strain
Four-to-six-week-old specific-pathogen-free (SPF) cotton rats were inoculated intranasally with parental and mutant rMV-Hu191 in order to determine their replication in vivo. No clinical symptoms of respiratory tract infection were found in cotton rats inoculated with any of the rMVs. At day 4 post-inoculation, cotton rats were terminated, and viral titer in the lungs was determined ( Table 2). The parental virus replicated efficiently in lungs with an average titer of 3.75 log10 PFU/g lung tissue. Recombinant rMV-Hu191-G1788A had an average titer of 3.08 log10 PFU/g lung tissue, which was significant lower than rMV-Hu191 (p < 0.05). However, rMV-Hu191-G1792A was the most attenuated mutant. Only 1 out of 5 cotton rats had detectable viral titer in the lung with a titer of 2.56 log10 PFU/g. These results show that the two mutant rMVs were more attenuated in viral replication in vivo compared to the parental vaccine strain.
Table 2.
Replication of rMV-Hu191 mutants in cotton rats.
| Viral replication in lunga |
||
|---|---|---|
| Inoculum | % infected animals | Viral titer (log10 PFU/g)b,c |
| rMV-Hu191 | 100 | 3.75 ± 0.12A |
| rMV-Hu191-G1788A | 100 | 3.08 ± 0.33B |
| rMV-Hu191-G1792A | 20 | 2.56B |
| DMEM | 0 | ND |
ND: not detected.
Each cotton rat was inoculated intranasally with 5.0 × 106 PFU of rMV-Hu191 or rMV-Hu191 mutants in a volume of 100 µl. At day 4 post-infection, the cotton rats were sacrificed, and lungs were collected for both virus titration and RT-PCR.
The viral titer was determined by plaque assay.
Five cotton rats were tested in each group. Values within a column followed by different capital letters (A and B) are significantly different.
To ensure that each mutant was stable in vivo, total RNA was extracted from each lung sample, and the regions harboring mutations were amplified by RT-PCR. The samples from each animal were sequenced, respectively. The result showed that the desired mutation was retained in rMV-Hu191-G1788A or rMV-Hu191-G1792A from each animal. No additional mutations were detected in the sequenced region.
2.6. rMV-Hu191 mutants induce higher neutralizing antibodies than the parental vaccine strain and provide complete protection against MV challenge
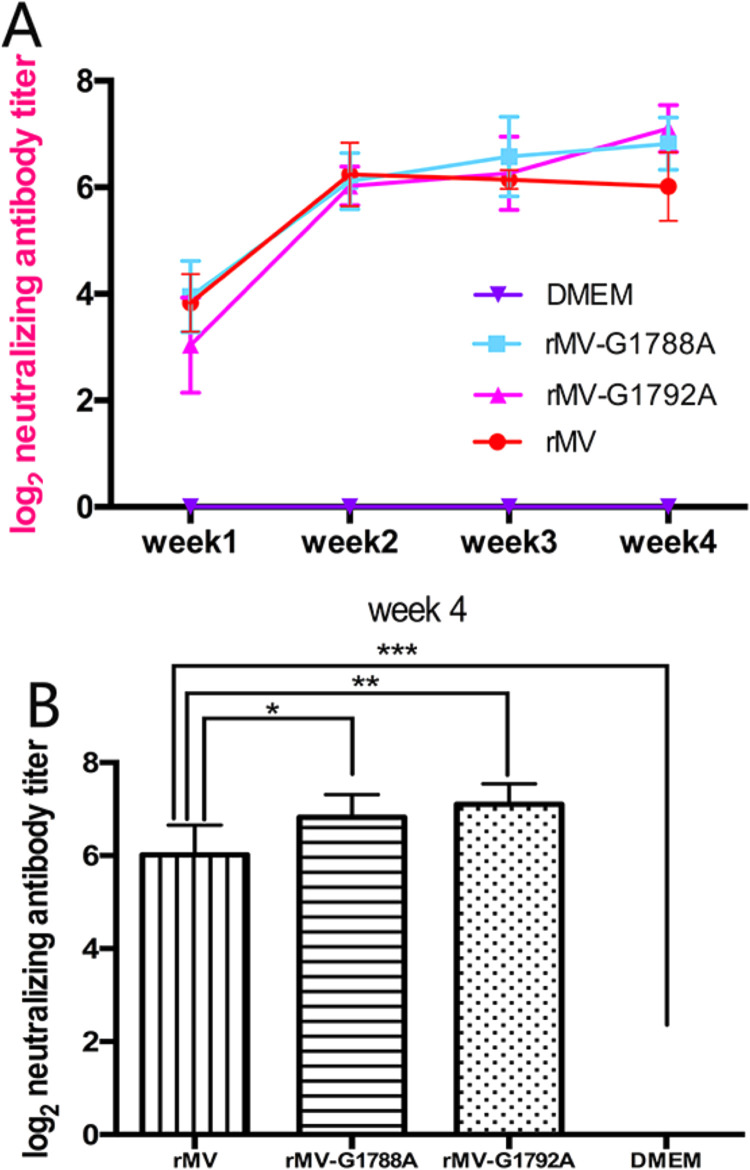
The immunogenicity of rMV-Hu191 mutants was assessed in cotton rats. Briefly, 4–6-week-old SPF cotton rats were intranasally inoculated with 1.0 × 106 PFU of each MV, and were challenged with 1.0 × 107 PFU of rMV-Hu191 at week 4 post-immunization. The two mutant rMVs induced high levels of neutralizing antibodies as early as 1 week after vaccination, and antibodies gradually increased from weeks 2–4. However, antibodies produced by the parental rMV-Hu191 peaked at week 2, and declined during weeks 3 and 4. Overall, the antibodies induced by rMV-Hu191-G1788A and rMV-Hu191-G1792A were comparable to those generated by wild-type rMV-Hu191 at weeks 1–3 (P > 0.05) ( Fig. 7A). However, at week 4, neutralizing antibodies induced by rMV-Hu191 mutants were significantly higher than those from parental rMV-Hu191 (p < 0.05; Fig. 7B). This suggests that rMV-Hu191 mutants were more immunogenic compared to the parental vaccine strain.
Fig. 7.
Neutralizing antibody titers produced by cotton rats after inoculation with recombinant MV. Cotton rats were intranasally inoculated with 1.0 × 106 PFU of rMV-Hu191 or rMV-Hu191 mutants in 0.1 ml of Opti-MEM medium. (A) Weekly blood samples were collected from each cotton rat by facial vein retro-orbital bleeding, and serum was tested for neutralizing antibodies by a plaque-reduction neutralization assay. (B) Recombinant MVs carrying mutations in the SAM binding site elicited significantly higher levels (p < 0.05) of neutralizing antibodies at 4 weeks post-inoculation. * =p < 0.05; * *=p < 0.01; *** =p < 0.001.
At week 4 post-vaccination, cotton rats were challenged with 1.0 × 107 PFU of rMV-Hu191 and all cotton rats were terminated at day 4 post-challenge. No infectious virus was detected in the lung tissue of any of the vaccinated cotton rats ( Table 3). In contrast, an average titer of 4.02 ± 0.37 log10 PFU/g was detected in lung tissue from unvaccinated but challenged controls. These results show that rMV-Hu191-G1788A and rMV-Hu191-G1790A provide complete protection from MV challenge.
Table 3.
Immunogenicity of MTase-defective rMV-Hu191 mutants in cotton ratsa.
| Viral replication in lung |
||
|---|---|---|
| Inoculuma | % infected animals | Viral titer (log10 PFU/g)b |
| rMV-Hu191 | 0 | ND |
| rMV-Hu191-G1788A | 0 | ND |
| rMV-Hu191-G1792A | 0 | ND |
| DMEM | 100 | 4.02± 0.37 |
ND: Not detected.
Cotton rats were intranasally inoculated with 1.0 × 106 PFU of rMV-Hu191 or mutants. At 28 days post-infection, rats were challenged with 1 × 107 PFU of rMV-Hu191. At day 4 post-challenge, cotton rats were sacrificed and lungs were collected for both virus titration and RT-PCR.
The viral titer was determined by plaque assay.
3. Discussion
In this study, we successfully generated two recombinant measles viruses with amino acid substitutions in the SAM binding site of L protein and examined the effects of these mutations on viral replication, safety, and immunogenicity. We found that both rMV-Hu191-G1788A and rMV-Hu191-G1792A were significantly more attenuated compared to parental rMV-Hu191, the widely used vaccine in China. rMVs carrying mutations in the SAM binding site were genetically stable, formed significantly smaller viral plaques, and had delays in CPE and replication kinetics. Recombinant rMV-Hu191-G1788A grew to high titer in Vero cells that was comparable to rMV-Hu191 but exhibited significantly more attenuation in cotton rats. Recombinant rMV-Hu191-G1792A grew to a relatively lower titer in Vero cells but had a greater degree of attenuation in cotton rats compared to rMV-Hu191-G1788A. Both recombinant viruses triggered significantly higher neutralizing antibody compared to rMV-Hu191, and provided complete protection against MV challenge. This indicates that alteration of SAM binding site in MV L protein enhances the safety and immunogenicity of the rMV-Hu191 vaccine strain.
We established a more efficient method to assemble a full-length cDNA clone of MV-Hu191 without using restriction endonucleases. The MV genome was divided into eight overlapping fragments and assembled into a full-length plasmid using the GeneArt™ High-Order Genetic Assembly System. The traditional method for assembly of an infectious cDNA requires multiple cloning steps involved in restriction enzyme digestion and ligation, which are time consuming, labor extensive, and technically challenging. The traditional cloning strategy also often leads to some unexpected deletions, insertions, and mutations in the viral genome, which hamper the subsequent virus rescue. Our assembly strategy was highly efficient, allowing us to obtain full-length cDNA clones in a single step.
Previously, vaccinia virus vTF-7 or MVA-T7 providing T7 RNA polymerase had often been used to rescue MV in reverse genetics system. However, there was difficulty in separating the rescued MV and the helper viruses. BHK cells stably expressing T7 RNA polymerase, instead, were used to rescue hMPV and bovine respiratory syncytial virus (Buchholz et al., 1999, Zhang et al., 2014). In our study, BHK-SR19-T7 cells were co-transfected with a plasmid expressing antigenomic MV cDNA and support plasmids expressing the MV N, P, and L proteins, allowing for efficient recovery of infectious MV in a vaccinia virus-free cell system. The primary advantage of this system is the elimination of the potential contamination by the vaccinia virus. This rescue system was highly efficient as we were able to recover many mutants in the CR VI of L protein including the two recombinant viruses with aa substitutions in the SAM binding site reported in this study.
A live attenuated vaccine is a very promising vaccine for most human paramyxoviruses, as it does not cause enhanced lung diseases upon re-infection by the same virus. A live-attenuated vaccine based on the Hu191 strain of MV has been developed and is widely used for immunization in Chinese infants and children (Zhang et al., 2009). However, epidemiological study showed that this vaccine still causes some adverse effects that may cause public health burden. In addition to the safety issue, measles outbreaks have been increasing in recent years, likely due to gradual reducing of the population immunity against measles after vaccination with time (Abad and Safdar, 2015, Gao et al., 2017, Ma et al., 2016, Zhang et al., 2016). In this study, we sought to improve the safety and efficacy of current MV vaccine by mutating the SAM binding site in the L protein. Using a robust reverse genetics for rMV-Hu191, rMV-Hu191 carrying mutations in the SAM binding site, rMV-Hu191-G1788A and rMV-Hu191-G1792A, were successfully rescued. These rMV-Hu191 mutants produced smaller plaques, had delayed growth kinetics, and had delayed syncytia formation compared to parental rMV-Hu191. Clearly, both mutants were significantly more attenuated in Vero cells than the parental rMV-Hu191. In cotton rats, rMV-Hu191-G1788A had a significantly lower viral titer in the lungs than rMV-Hu191 (p < 0.05). Recombinant rMV-Hu191-G1792A was even more attenuated, as only 1 out of 5 inoculated cotton rats had detectable viral titer in the lung. Despite the high attenuation phenotype, rMV-Hu191-G1788A grew to high titer compared to parental vaccine virus in Vero cells, the WHO approved cell line for vaccine production. Although rMV-Hu191-G1792A grew to a slightly lower titer (0.5 log less) in Vero cells, it is still economically feasible for vaccine production. Another advantage of using rMV-Hu191-G1792A is that it had a greater degree of attenuation in vitro and in vivo compared to rMV-Hu191-G1788A. Interestingly, both mutants triggered a higher level of neutralizing antibodies than parental rMV-Hu191, suggesting their greater immunogenicity. Finally, cotton rats vaccinated with both mutants were completely protected from the MV challenge. Thus, recombinant rMV-Hu191 carrying mutations in the SAM binding site are potentially improved vaccine candidates for MV.
A novel finding is that recombinant rMV-Hu191 carrying mutations in the SAM binding site triggered a higher neutralizing antibody compared to the parental rMV-Hu191 strain. Although the detailed mechanism is not explored in this study, it is possible that these MV mutants may trigger a higher innate immunity, which in turn triggered a more robust adaptive immunity. In fact, it was shown that coronavirus lacking 2’-O methylation significantly enhanced type I interferon response, which is another advantage of using viral mRNA cap MTase as a target in developing live vaccine candidates. Previously, it was found that two pneumoviruses (aMPV and hMPV) carrying mutations in the SAM binding site in CR VI of L protein were specifically defective in ribose 2′-O methylation but not G-N-7 methylation, and were significantly attenuated but retained wild-type levels of immunogenicity. In addition, it was found that all 2′-O MTase-defective hMPVs were highly sensitive to IFN-α and IFN-β treatment (Sun et al., 2014, Zhang et al., 2014). Given the fact that the MTase domain is highly conserved in L proteins of all NNS RNA viruses, the general mechanism of attenuation of viruses that lack 2′-O methylation may be similarly conserved in all NNS RNA viruses. Future experiments should investigate the mechanisms by which MV mutants enhance immunogenicity.
4. Conclusion
We have established a novel and efficient strategy for assembly of a full-length cDNA clone of MV-Hu191 and established an efficient vaccinia virus-free reverse genetics system for MV-Hu191. We generated two recombinant MV-Hu191 carrying mutations in the SAM binding site, which not only grew to high titer in Vero cells and were genetically stable but also were significantly more attenuated and immunogenic compared to the currently used Chinese MV vaccine strain. These two recombinant viruses may serve as improved vaccine candidates for MV.
5. Materials and methods
5.1. Cells and viruses
Vero cells (African green monkey, ATCC-CCL-81) and BHK-SR19-T7 cells (Zhang et al., 2014) (kindly offered by Apath, LLC, Brooklyn, NY) were grown in Dulbecco's modified Eagle's medium (DMEM; Life Technologies) supplemented with 10% fetal bovine serum (FBS). The Chinese Hu191 vaccine strain of MV (obtained from Dr. Yiyu Lu, Zhejiang CDC) was passaged in Vero cells.
5.2. Cloning and construction of MV-Hu191 genomic plasmids
Viral RNA was extracted from 200 µl of MV-Hu191 using an RNeasy mini-kit (Qiagen), and reverse-transcribed using Super Script® III reverse transcriptase (Invitrogen) and Random Primer Mix (NEB). The genome was amplified in eight overlapping fragments by Q5® High-Fidelity 2 × Master Mix (NEB), using eight pairs of MV-specific primers (Table 1), and cloned into the pEASY-Blunt vector (Transgen) according to the manufacturer's instructions. The resultant eight plasmids containing the full-length MV-Hu191 genome (pEASY-N, pEASY-P, pEASY-M1, pEASY-M2, pEASY-F, pEASY-H, pEASY-L1, pEASY-L2) were sequenced, and found to be identical with the published sequence of MV-Hu191 (GenBank accession no. FJ416067), except for a single point change (C to U) at nt 8763 within the H gene. This mutation in pEASY-H was corrected by site-directed mutagenesis with specific primers (Table 1). The resultant plasmid was named pEASY-H-M.
5.3. Construction of the full-length cDNA clone of MV-Hu191
Several rounds of amplification and “In-fusion” PCR were used to assemble five fragments [the T7 promoter, MV 3′ and 5′ non-coding termini (NCT) (3′−107 nt and 5′−109 nt, respectively) were amplified from pEASY-N and pEASY-L2, respectively, hepatitis delta virus (HDV) ribozyme (84 nt of anti-genomic HDV sequence), and the T7 terminator], and subsequently inserted them into the pYES-2 plasmid using the GeneArt™ Seamless Cloning and Assembly Kit (Invitrogen; Fig. 1A), creating plasmid p107109-MV(+). The primer sequences and approaches used in the PCR assays are available upon request.
The ten fragments were successfully assembled into a full-length cDNA clone using the GeneArt™ High-Order Genetic Assembly System according to the manufacturer's manual (Fig. 1B), creating plasmid pYES-MV(+). Eight MV-Hu191 genomic fragments were amplified with specific primers from pEASY-N, pEASY-P, pEASY-M1, pEASY-M2, pEASY-F, pEASY-H-M, pEASY-L1, and pEASY-L2. The p107109-MV(+) insert (containing the T7 promoter, MV 3′ and 5′ NCTs, HDV ribozyme, and T7 terminator) was divided into two fragments by PCR amplification with specific primers (F:5′-TTCTGCCGCCTGCTTCAAACCG-3’, R:5′-CTCGGATATCCCTAATCC-3’; F:5′-TTGGTTGAACTCCGGAAC-3’, R: 5′-CAGAATGGGCAGACATTACGAATGC-3’).
5.4. Construction of plasmids encoding MV-Hu191 N, P, and L genes
A backbone vector pT7, which contains the T7 RNA polymerase promoter, encephalomyocarditis (EMC) virus internal ribosome entry site (IRES), and the T7 terminator sequences, was used to construct plasmids encoding MV-Hu191 N, P, and L genes. The open reading frames (ORFs) of the MV Hu191 N and P genes were amplified from pEASY-N and pEASY-P using primer pairs MV-CDS-N (+)/(-) and MV-CDS-P (+)/(-), respectively, whereas the ORF of the MV Hu191 L gene was amplified from pEASY-L1 and pEASY-L2 using two primer pairs, MV-CDS-L1 (+)/(-) and MV-CDS-L2 (+)/(-). The MV N, P, and L genes were inserted into the pT7 vector between the IRES and polyA sequences using a “seamless” cloning strategy, resulted in the construction of pT7-Hu191-N, pT7-Hu191-P, and pT7-Hu191-L, respectively. The primer sequences used in the PCR assays are available upon request.
5.5. Recovery of recombinant MV-Hu191 (rMV-Hu191) from full-length cDNA clone
To recover recombinant MV, BHK-SR19-T7 cells were grown overnight in six-well plates to approximately 90% confluence, and were transfected with 5 µg of pYES-MV(+), 1.5 µg of pT7-Hu191-N, 1.5 µg of pT7-Hu191-P, and 0.5 µg of pT7-Hu191-L using previously described procedure (Carsillo et al., 2009, Kovacs et al., 2003, Singh and Billeter, 1999). At 72 h post-transfection, cell monolayers were trypsinized and directly transferred onto Vero cell monolayers (P0) at 75% confluence and co-cultured at 37 °C for 3–5 days. Cells were subjected to three freeze-thaw cycles when extensive CPE (MV-induced syncytia) was observed. After a brief centrifugation, supernatants (P1) were harvested and used for further passages on confluent Vero cell monolayers. At P2 or P3, the recovered viruses were plaque purified and sequenced.
5.6. Immunofluorescence assay
Vero cells grown in 24-well tissue culture plates were infected with rMV or rMV-mutant. After 1 h incubation, the cells were washed two times with PBS before cultivating them in DMEM containing 2% FBS. At 24 or 48 h postinfection, the cells were fixed with 4% paraformaldehyde in PBS for 20 min at RT, permeabilized with 0.1% Triton X-100 (Merck Millipore) in PBS for 10 min at RT, and blocked with 1% BSA in PBS containing 0.05% Tween 20 for 1 h at RT. The cells were stained with mouse anti-measles virus N antibody (ab106292, abcam) for 1 h at RT. After washing with PBS, Alexa Fluor ® 594 donkey anti-mouse IgG (H+L) (A21203, Invitrogen) were added and incubated for 1 h at RT. Then, the cells were stained with 4,6-diamidino-2-phenylindole (DAPI) for 10 min at RT. Images were obtained using a Zeiss cLSM780 confocal laser scanning microscope and Zen 2012 software.
5.7. Site-directed mutagenesis
Amino acids (G1788, G1790, and G1792) in the SAM binding site in the Hu191 L protein were mutated to alanine individually (ggt to gct at aa posi tion 1788, gga to gca at aa position 1790, and ggt to gct at aa position 1792; the mutated nucleotides were underlined) in an infectious cDNA clone of MV-Hu191 [pYES-MV(+)] using a Q5® site-directed mutagenesis kit (NEB) according to the manufacturer's instructions. The resultant plasmids were named pYES-MV(+)-G1788A, pYES-MV(+)-G1790A, and pYES-MV(+)-G1792A. Mutations were confirmed by DNA sequencing.
5.8. Virus titration
The titers of rMV-Hu191 viruses were determined by plaque assay in Vero cells. Briefly, Vero cells were seeded in six-well plates at a density of 5 × 105 cells per well, incubated for 18 h, and the medium was removed prior to infection of cell monolayers with serial dilutions of rMV-Hu191. After 1 h of adsorption with constant shaking, the medium was removed and cell monolayers were covered with 2.5 ml of Eagle's minimal essential media (MEM) containing 2% agarose, 0.75% sodium bicarbonate(NaHCO3), 5% FBS, 20 nM HEPES, 2 mM L-Glutamine, and 4 mg/ml of streptomycin. At 6 dpi, cells were fixed in 4% (vol/vol) paraformaldehyde for 2 h, and the plaques were visualized by staining with 0.05% (wt/vol) crystal violet.
5.9. Viral replication kinetics in Vero cells
Confluent Vero cells in six-well plates were infected with rMV-Hu191 viruses at a multiplicity of infection (MOI) of 0.01. After 1 h incubation, the inoculum was removed and the cells were washed three times with PBS. Fresh maintenance media (DMEM supplemented with 2% FBS) was added, and the infected cells were incubated at 37 °C. At different time points post-infection, the cells were harvested by three freeze-thaw cycles, and the supernatant collected by centrifugation at 3000×g in an Allegra 6R centrifuge (Beckman Coulter) for 15 min. Virus titers were determined by plaque assay in Vero cells.
5.10. Genetic stability of rMV-Hu191 mutants in cell culture
Confluent Vero cells in T25 flasks were infected with each rMV-Hu191 mutant at an MOI of 0.01, and cell culture supernatants were collected after appearance of CPE and used to infect new confluent Vero cells in fresh T25 flasks. Each mutant was serially passaged 15 times in Vero cells. Viral RNA was extracted from cell culture supernatant harvested from each passage. The CR VI of the L gene was amplified by RT-PCR and sequenced. Additionally, the entire genome of each recombinant virus was amplified by RT-PCR and sequenced at passage 15.
5.11. RT-PCR and sequencing
All the plasmids, viral stocks, and virus isolates from the lungs of cotton rats were sequenced. Viral RNA was isolated using an RNeasy mini-kit (Qiagen) according to the manufacturer's instructions. Viral RNA was treated with DNase I to eliminate possible contamination from original transfecting plasmid DNA, and no-RT PCR controls were carried out to confirm complete digestion of plasmid DNA.
A 1.2 kb DNA fragment of the H protein gene was amplified by a One-Step RT-PCR kit (Qiagen) using primers rMV-H-8583-Forward (5’-gttcagggatggacctatac-3’) and rMV-L-9793-Reverse (5’-ggtgtgtgtctcctcctat-3’). PCR products were sequenced to ensure that the isolated virus was rescued from pYES-MV(+) and not from the contamination of the wild type MV-Hu191 grown in our laboratory.
A 1.1 kb DNA fragment spanning CR VI of the MV L-protein was amplified by a One-Step RT-PCR kit (Qiagen) using primers rMV-L-14128-Forward (5’-gaccggtagagaaatgtgcag-3’) and rMV-L-15222-Reverse (5’-gcttaatggataggatgtgac-3’). PCR products were sequenced to confirm that each recombinant virus contained the desired mutation.
5.12. Purification of rMV-Hu191
The rMV-Hu191 stocks for use in animal experiments were grown in Vero cells and purified by ultracentrifugation. Twenty T150 flasks with confluent Vero cells were infected with each rMV at MOI of 0.01. After 1 h of adsorption with constant shaking, 15 ml of DMEM (supplemented with 2% FBS) was added to each flask and incubated at 37 °C until extensive CPE was observed. The cells were harvested using a cell-scraper, and suspensions were clarified by centrifugation at 3000×g for 20 min at 4 °C in an Allegra 6 R centrifuge (Beckman Coulter). The cell pellets were resuspended in 2 ml of DMEM and subjected to three freeze-thaw cycles, clarified by low-speed centrifugation, and the supernatants were combined. The virus was pelleted by ultracentrifugation at 30,000×g in a Beckman Ty 50.2 rotor for 2 h, and resuspended in 0.3 ml of DMEM, aliquoted, and stored. Viral titer was determined by plaque assay.
5.13. Replication of rMV-Hu191 in cotton rats
Twenty 4–6 week-old female specific-pathogen-free (SPF) cotton rats (Envigo, Indianapolis, IN) were randomly divided into four groups (5 cotton rats per group), and housed within the ULAR facilities at The Ohio State University according to IACUC policies and guidelines (animal protocol no. 2009A0221). Each inoculated group was separately housed in rodent cages under biosafety level 2 conditions; rats were anesthetized with isoflurane before virus inoculation. Cotton rats in groups 1–3 were inoculated with parental rMV-Hu191, rMV-Hu191-G1788A, and rMV-Hu191-G1792A, respectively. Cotton rats in group 4 were mock-infected with DMEM, and served as uninfected controls. Each cotton rat was inoculated intranasally with 5 × 106 PFU of virus in a volume of 100 µl. At 4 dpi, cotton rats were sacrificed and lungs were collected for virus titration and RT-PCR.
5.14. Immunogenicity of rMV-Hu191 in cotton rats
For the immunogenicity study, twenty five 4–6 week-old cotton rats (Envigo) were randomly divided into five groups (5 cotton rats per group). Cotton rats in groups 1 were mock-infected with DMEM and served as uninfected unchallenged control. Cotton rats in groups 2, 3 and 4 were intranasally inoculated with 1.0 × 106 PFU of rMV-Hu191, rMV-Hu191-G1788A, and rMV-Hu191-G1792A, respectively. Cotton rats in groups 5 were mock-infected with DMEM and served as uninfected challenged control. After immunization, the cotton rats were evaluated daily for mortality, and blood samples were collected from each cotton rat weekly by facial vein retro-orbital bleeding, and the serum was used for detection of neutralizing antibodies. At 4 weeks post-immunization, the cotton rats in groups 2–5 were challenged with 1.0 × 107 PFU of parental rMV-Hu191 via intranasal route, and evaluated twice daily for the presence of any clinical symptoms. At 4 days post-challenge, all cotton rats were euthanized by CO2 asphyxiation, and their lungs were collected for virus titration. The immunogenicity of rMV-Hu191 mutants was assessed based on their ability to trigger neutralizing antibodies and the ability to protect MV replication in lungs. Serum neutralization of virus was performed using an endpoint dilution plaque reduction assay; and (ii) quantification of lung viral titers was done by plaque assay.
5.15. Virus-serum neutralization assay
MV-specific neutralizing antibody was determined using an endpoint dilution plaque reduction assay. Briefly, cotton rat sera were collected weekly until challenge. The serum samples were heat inactivated at 56 °C for 30 min. Two-fold dilutions of the serum samples were mixed with an equal volume of DMEM containing approximately 100 PFU/well rMV-Hu191 in a 96-well plate, and the plate was incubated at room temperature for 1 h with constant rotation. The mixtures were then transferred to confluent Vero cells in a 6-well plate in triplicate. After 1 h of incubation at 37 °C, the virus-serum mixtures were removed and the cell monolayers were covered with 2.5 ml of Eagle's minimal essential media (MEM) containing 2% agarose, 0.75% sodium bicarbonate(NaHCO3), 5% FBS, 20 nM HEPES, 2 mM L-Glutamine, and 4 mg/ml of streptomycin. Then, the cells were incubated for another 6 days before virus plaque titration as described above. The plaques were counted, and 50% plaque reduction titers were calculated as the MV-specific neutralizing antibody titers.
5.16. Statistical analysis
Statistical analysis was performed by one-way multiple comparisons; two-way multiple comparisons (ANOVA) using Prism statistical analysis software (version 8.0). P value of < 0.05 was considered statistically significant.
Acknowledgments
This work was supported partly by the National Key Research and Development Program of China (2017YFD0500104), the Zhejiang Provincial Science Foundation for Distinguished Young Scholars (LR14C180001), the National Natural Science Foundation of China (31572518), and the Thousand Young Talent Program of China.
Acknowledgments
Author contributions
Conceived and designed the experiments: Zhengyan Zhao, Yao-Wei Huang, and Jianrong Li. Performed the experiments: Yilong Wang, Rongxian Liu, Mijia Lu, Xiaoqiang Hao, Yingzhi Yang, and Bin Wang. Analyzed the data: Duo Zhou, Dongming Zhou, Bin Wang, Zhengyan Zhao, Yao-Wei Huang, and Jianrong Li. Wrote the manuscript: Yilong Wang, Zhengyan Zhao, Yao-Wei Huang, Jianrong Li, and all other co-authors edited the manuscript.
Conflicts of interest
The authors declare that they have no competing interests.
Contributor Information
Jianrong Li, Email: Li.926@osu.edu.
Yao-Wei Huang, Email: yhuang@zju.edu.cn.
Zhengyan Zhao, Email: Zhaozy@zju.edu.cn.
References
- Abad C.L., Safdar N. The reemergence of measles. Curr. Infect. Dis. Rep. 2015;17:51. doi: 10.1007/s11908-015-0506-5. [DOI] [PubMed] [Google Scholar]
- Bester J.C. Measles and measles vaccination: a review. JAMA Pediatr. 2016;170:1209–1215. doi: 10.1001/jamapediatrics.2016.1787. [DOI] [PubMed] [Google Scholar]
- Buchholz U.J., Finke S., Conzelmann K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999;73:251–259. doi: 10.1128/jvi.73.1.251-259.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bukreyev A., Camargo E., Collins P.L. Recovery of infectious respiratory syncytial virus expressing an additional, foreign gene. J. Virol. 1996;70:6634–6641. doi: 10.1128/jvi.70.10.6634-6641.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carsillo M., Klapproth K., Niewiesk S. Cytokine imbalance after measles virus infection has no correlation with immune suppression. J. Virol. 2009;83:7244–7251. doi: 10.1128/JVI.00148-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke D.K., Sidhu M.S., Johnson J.E., Udem S.A. Rescue of mumps virus from cDNA. J. Virol. 2000;74:4831–4838. doi: 10.1128/jvi.74.10.4831-4838.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries R.D., Stittelaar K.J., Osterhaus A.D., de Swart R.L. Measles vaccination: new strategies and formulations. Expert Rev. Vaccin. 2008;7:1215–1223. doi: 10.1586/14760584.7.8.1215. [DOI] [PubMed] [Google Scholar]
- Duprex W.P., McQuaid S., Hangartner L., Billeter M.A., Rima B.K. Observation of measles virus cell-to-cell spread in astrocytoma cells by using a green fluorescent protein-expressing recombinant virus. J. Virol. 1999;73:9568–9575. doi: 10.1128/jvi.73.11.9568-9575.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferron F., Longhi S., Henrissat B., Canard B. Viral RNA-polymerases -- a predicted 2'-O-ribose methyltransferase domain shared by all Mononegavirales. Trends Biochem. Sci. 2002;27:222–224. doi: 10.1016/s0968-0004(02)02091-1. [DOI] [PubMed] [Google Scholar]
- Furuichi Y., Shatkin A.J. Viral and cellular mRNA capping: past and prospects. Adv. Virus Res. 2000;55:135–184. doi: 10.1016/S0065-3527(00)55003-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao J., Shen B., Xiong J., Lu Y., Jiang Q. The measles epidemic trend over the past 30 years in a central district in Shanghai, China. PLoS One. 2017;12:e0179470. doi: 10.1371/journal.pone.0179470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcin D., Pelet T., Calain P., Roux L., Curran J., Kolakofsky D. A highly recombinogenic system for the recovery of infectious Sendai paramyxovirus from cDNA: generation of a novel copy-back nondefective interfering virus. Embo J. 1995;14:6087–6094. doi: 10.1002/j.1460-2075.1995.tb00299.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassen U., Collins F.M., Duprex W.P., Rima B.K. Establishment of a rescue system for canine distemper virus. J. Virol. 2000;74:10737–10744. doi: 10.1128/jvi.74.22.10737-10744.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffin D.E., Oldstone M.B. Measles. History and basic biology. Introduction. Curr. Top. Microbiol. Immunol. 2009;329:1. [PubMed] [Google Scholar]
- Jin H., Clarke D., Zhou H.Z.Y., Cheng X., Coelingh K., Bryant M., Li S.Q. Recombinant human respiratory syncytial virus (RSV) from cDNA and construction of subgroup A and B chimeric RSV. Virology. 1998;251:206–214. doi: 10.1006/viro.1998.9414. [DOI] [PubMed] [Google Scholar]
- Kovacs G.R., Parks C.L., Vasilakis N., Udem S.A. Enhanced genetic rescue of negative-strand RNA viruses: use of an MVA-T7 RNA polyrnerase vector and DNA replication inhibitors. J. Virol. Methods. 2003;111:29–36. doi: 10.1016/s0166-0934(03)00132-0. [DOI] [PubMed] [Google Scholar]
- Lawson N.D., Stillman E.A., Whitt M.A., Rose J.K. Recombinant vesicular stomatitis viruses from DNA. Proc. Natl. Acad. Sci. USA. 1995;92:4477–4481. doi: 10.1073/pnas.92.10.4477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Rahmeh A., Morelli M., Whelan S.P. A conserved motif in region v of the large polymerase proteins of nonsegmented negative-sense RNA viruses that is essential for mRNA capping. J. Virol. 2008;82:775–784. doi: 10.1128/JVI.02107-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Wang J.T., Whelan S.P. A unique strategy for mRNA cap methylation used by vesicular stomatitis virus. Proc. Natl. Acad. Sci. USA. 2006;103:8493–8498. doi: 10.1073/pnas.0509821103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma C., Yan S., Su Q., Hao L., Tang S., An Z., He Y., Fan G., Rodewald L., Wang H. Measles transmission among adults with spread to children during an outbreak: implications for measles elimination in China, 2014. Vaccine. 2016;34:6539–6544. doi: 10.1016/j.vaccine.2016.02.051. [DOI] [PubMed] [Google Scholar]
- Ma Y., Wei Y., Zhang X., Zhang Y., Cai H., Zhu Y., Shilo K., Oglesbee M., Krakowka S., Whelan S.P., Li J. mRNA cap methylation influences pathogenesis of vesicular stomatitis virus in vivo. J. Virol. 2014;88:2913–2926. doi: 10.1128/JVI.03420-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIlhatton M.A., Curran M.D., Rima B.K. Nucleotide sequence analysis of the large (L) genes of phocine distemper virus and canine distemper virus (corrected sequence) J. Gen. Virol. 1997;78(Pt 3):571–576. doi: 10.1099/0022-1317-78-3-571. [DOI] [PubMed] [Google Scholar]
- Nakatsu Y., Takeda M., Kidokoro M., Kohara M., Yanagi Y. Rescue system for measles virus from cloned cDNA driven by vaccinia virus Lister vaccine strain. J. Virol. Methods. 2006;137:152–155. doi: 10.1016/j.jviromet.2006.05.029. [DOI] [PubMed] [Google Scholar]
- Neumann G., Whitt M.A., Kawaoka Y. A decade after the generation of a negative-sense RNA virus from cloned cDNA - what have we learned? J. Gen. Virol. 2002;83:2635–2662. doi: 10.1099/0022-1317-83-11-2635. [DOI] [PubMed] [Google Scholar]
- Ogino T., Banerjee A.K. Unconventional mechanism of mRNA capping by the RNA-dependent RNA polymerase of vesicular stomatitis virus. Mol. Cell. 2007;25:85–97. doi: 10.1016/j.molcel.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Ogino T., Kobayashi M., Iwama M., Mizumoto K. Sendai virus RNA-dependent RNA polymerase L protein catalyzes cap methylation of virus-specific mRNA. J. Biol. Chem. 2005;280:4429–4435. doi: 10.1074/jbc.M411167200. [DOI] [PubMed] [Google Scholar]
- Parks C.L., Lerch R.A., Walpita P., Sidhu M.S., Udem S.A. Enhanced measles virus cDNA rescue and gene expression after heat shock. J. Virol. 1999;73:3560–3566. doi: 10.1128/jvi.73.5.3560-3566.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poch O., Blumberg B.M., Bougueleret L., Tordo N. Sequence comparison of five polymerases (L proteins) of unsegmented negative-strand RNA viruses: theoretical assignment of functional domains. J. Gen. Virol. 1990;71(Pt 5):1153–1162. doi: 10.1099/0022-1317-71-5-1153. [DOI] [PubMed] [Google Scholar]
- Radecke F., Spielhofer P., Schneider H., Kaelin K., Huber M., Dotsch C., Christiansen G., Billeter M.A. Rescue of measles viruses from cloned DNA. EMBO J. 1995;14:5773–5784. doi: 10.1002/j.1460-2075.1995.tb00266.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahmeh A.A., Li J., Kranzusch P.J., Whelan S.P. Ribose 2'-O methylation of the vesicular stomatitis virus mRNA cap precedes and facilitates subsequent guanine-N-7 methylation by the large polymerase protein. J. Virol. 2009;83:11043–11050. doi: 10.1128/JVI.01426-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schluckebier G., O'Gara M., Saenger W., Cheng X. Universal catalytic domain structure of AdoMet-dependent methyltransferases. J. Mol. Biol. 1995;247:16–20. doi: 10.1006/jmbi.1994.0117. [DOI] [PubMed] [Google Scholar]
- Shu M., Liu Q., Wang J., Ao R., Yang C., Fang G., Wan C., Guo W. Measles vaccine adverse events reported in the mass vaccination campaign of Sichuan province, China from 2007 to 2008. Vaccine. 2011;29:3507–3510. doi: 10.1016/j.vaccine.2009.10.106. [DOI] [PubMed] [Google Scholar]
- Sidhu M.S., Chan J., Kaelin K., Spielhofer P., Radecke F., Schneider H., Masurekar M., Dowling P.C., Billeter M.A., Udem S.A. Rescue of synthetic measles-virus minireplicons - measles genomic termini direct efficient expression and propagation of a reporter gene. Virology. 1995;208:800–807. doi: 10.1006/viro.1995.1215. [DOI] [PubMed] [Google Scholar]
- Singh M., Billeter M.A. A recombinant measles virus expressing biologically active human interleukin-12. J. Gen. Virol. 1999;80(Pt 1):101–106. doi: 10.1099/0022-1317-80-1-101. [DOI] [PubMed] [Google Scholar]
- Sun J., Wei Y., Rauf A., Zhang Y., Ma Y., Zhang X., Shilo K., Yu Q., Saif Y.M., Lu X., Yu L., Li J. Methyltransferase-defective avian metapneumovirus vaccines provide complete protection against challenge with the homologous Colorado strain and the heterologous Minnesota strain. J. Virol. 2014;88:12348–12363. doi: 10.1128/JVI.01095-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tangy F., Naim H.Y. Live attenuated measles vaccine as a potential multivalent pediatric vaccination vector. Viral Immunol. 2005;18:317–326. doi: 10.1089/vim.2005.18.317. [DOI] [PubMed] [Google Scholar]
- Tian D., Luo Z., Zhou M., Li M., Yu L., Wang C., Yuan J., Li F., Tian B., Sui B., Chen H., Fu Z.F., Zhao L. Critical role of K1685 and K1829 in the large protein of rabies virus in viral pathogenicity and immune evasion. J. Virol. 2015;90:232–244. doi: 10.1128/JVI.02050-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan S.P., Barr J.N., Wertz G.W. Transcription and replication of nonsegmented negative-strand RNA viruses. Curr. Top. Microbiol. Immunol. 2004;283:61–119. doi: 10.1007/978-3-662-06099-5_3. [DOI] [PubMed] [Google Scholar]
- Xu P., Li Z., Sun D., Lin Y., Wu J., Rota P.A., He B. Rescue of wild-type mumps virus from a strain associated with recent outbreaks helps to define the role of the SH ORF in the pathogenesis of mumps virus. Virology. 2011;417:126–136. doi: 10.1016/j.virol.2011.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Wei Y., Zhang X., Cai H., Niewiesk S., Li J. Rational design of human metapneumovirus live attenuated vaccine candidates by inhibiting viral mRNA cap methyltransferase. J. Virol. 2014;88:11411–11429. doi: 10.1128/JVI.00876-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Zhou J., Bellini W.J., Xu W., Rota P.A. Genetic characterization of Chinese measles vaccines by analysis of complete genomic sequences. J. Med. Virol. 2009;81:1477–1483. doi: 10.1002/jmv.21535. [DOI] [PubMed] [Google Scholar]
- Zhang Z., Zhao Y., Yang L., Lu C., Meng Y., Guan X., An H., Zhang M., Guo W., Shang B., Yu J. Measles outbreak among previously immunized adult healthcare workers, China, 2015. Can. J. Infect. Dis. Med. Microbiol. 2016;2016:1742530. doi: 10.1155/2016/1742530. [DOI] [PMC free article] [PubMed] [Google Scholar]