Abstract
Amyloid-β (Aβ) peptides can elongate in the fibril axis and associate in the lateral direction. We present detailed atomic Aβ models with different in-register intermolecular β-sheet-β-sheet associations. We probe structural stability, conformational dynamics, and association force of Aβ oligomers with various sizes and structures for both wild-type and mutated sequences using molecular dynamics (MD) simulations. MD simulations show that an Aβ oligomer that is laterally associated through the C-terminal-C-terminal interface is energetically more favorable than other oligomers with the N-terminal-N-terminal and C-terminal-N-terminal interfaces. We further develop a simple numerical model to describe the kinetics of Aβ aggregation process by considering fibril elongation and lateral association using a Monte Carlo algorithm. Kinetic data suggest that fibril elongation and lateral association are mutually competitive. Single-point mutations of Glu22 or Met35 at the interfaces have profound negative effects on intermolecular β-sheet-β-sheet association. These disease-related mutants (E22K, E22Q, and M35O) display more flexible structures, weaker lateral association, and stronger elongation tendencies than the wild type, suggesting that amyloid oligomerization and neurotoxicity might be linked to fibril longitudinal growth.
Keywords: Aβ, amyloid, Peptide Aggregation, Molecular Dynamics
2. INTRODUCTION
Alzheimer’s disease (AD) is a progressive neurodegenerative disease, which is characterized by the deposition of insoluble fibril plaques in the extracelluar space of the brain issue (1). The major component of these plaques is a 40- to 42-residue peptide, called the β-amyloid (Aβ), which is a product of cleavage of the transmembrane amyloid precursor protein (APP). Although the mechanism of fatal neurotoxicity associated with amyloid fibril formation is still unclear, increasing evidence accumulates to suggest that soluble oligomers, rather than insoluble mature fibrils, are the toxic species (2–4). Whereas atomic force microscopy (AFM), electron microscopy (EM) imaging and fiber diffraction are sufficient to identify the overall size, shape, and morphology of different fibrillar species at different time-points during the growth process, they can not provide the detailed 3D structural organization of amyloid oligomers at the atomic level due to their small size and the dynamic, short-lived nature (5, 6). However, molecular modeling (7) has been widely used to characterize the structural and dynamic properties of amyloid oligomers at the molecular level, which is essential for understanding the mechanism of self- assembly in amyloidosis.
Several structural models of Aβ based on different experimental (i.e. x-ray diffraction and solid-state NMR) and computational techniques have been proposed. Experimentally, Tycko and co-workers (8) suggested a structural model of Aβ1–40 with a cross-β structural motif, in which residues 12–24 and 30–40 form two β-strands connected by a loop with the sequence of Gly25-Ser26-Asn27-Lys28-Gly29. Riek and co-workers recently (9) proposed a generally similar structure 3D structure of the Aβ1–42 although differing in the turn conformation, by using side chain contact constraints from pairwise mutagenesis studies and quenched H/D exchange NMR. Riek’s model also consists of a β-strand-loop-β-strand motif at residues 18–42, with a loop occurring at residues Asn27-Lys28- Gly29-Ala30. In parallel, computations by Ma and Nussinov (10) independently led them to propose a parallel β-sheet model for the Aβ16–35 with a β (Lys16-Glu23)-loop (Asp23-Lys28)-β (Gly29-Met35) motif. All three models revealed that the Aβ peptide adopts a β-strand-loop-β- strand motif, with each Aβ peptide stacked in a parallel and staggered manner within β-sheets. All models remarkably agree that the buried salt bridge between residues Asp23 and Lys28 stabilizes the loop, although each model has distinct loop region that could affect the in-register manner of parallel intermolecular β sheets organization. This β- strand-loop-β-strand motif allows multiple β-sheets to stack on top of each other in the lateral direction through different β-sheet interfaces.
In this work, we perform molecular dynamics (MD) simulations of various models to explore the structural stability and dynamics, and the kinetic properties of Aβ9–40 double-layered oligomers for the wild-type and mutated sequences, with particular attention to different sheet-to-sheet associations in the lateral direction. The cross-section of the Aβ models contains a dimer of two hexamers packed in an antiparallel organization, each of which consists of six parallel β-strands (Figure 1). The binding energies between two β-sheets associated through different interfaces are first evaluated and compared to determine which interface is more energetically favorable. Furthermore, a simple numerical model is developed to study the competitive assembly of the protofilament along both fibril elongation and lateral directions using a Metropolis Monte Carlo (MC) algorithm, in which the association probability is determined by binding energies obtained from MD simulations. Finally, computational mutagenesis of residues of Glu22 and Met35 at interfaces is performed to examine the effects of mutation on Aβ sheet-to-sheet association and kinetic assembly behavior.
Figure 1.
(a) Atomic structure of the monomeric Aβ9–40 peptide labeled with residue names and numbers. Atomic models and the corresponding schematic organization of double-layered Aβ peptides comprising two antiparallel packed hexamers with (b) CC interface, (c) NN interface, and (d) CN interface. Color Id: polar residues in green, non-polar residues in orange, positively charged residues in blue, and negatively charged residues in red. In the schematic presentation, a shadowed circle presents the out-forward orientation of the side-chain of the amino acids.
3. MATERIALS AND METHODS
3.1. Systems setup
The starting monomeric structure of a 32-residue Aβ9–40 peptide was extracted from the NMR structures kindly provided by R. Tycko (8). Aβ9–40 consisted of two β- strands, β1 (residues Gly9-Glu22) and β2 (residues Gly29- Val40), connected by a U-bent turn spanning four residues Asp23-Lys28, as shown in Figure 1. Double-layered Aβ oligomers were built from two β-sheets which are at a distance of ~10 Å in an antiparallel organization, each made up of six strands ~4.7 Å apart stacked in parallel to each other. Since Aβ has a U-turn shape, Aβ oligomers can associate through three possible interfaces determined by two stacked β-sheets in the lateral direction perpendicular to the fibril growth axis. The cross-section of the oligomers is made of two “U-shaped” β-sheets, i) with hydrophobic C-terminal–C-terminal (CC) interface through van der Waals contacts from Ile31 to Val40, ii) with N-terminal–N- terminal (NN) interface through partial hydrophobic contacts between Val18 and Phe20 and salt bridges between Hsp14 and Glu22, or iii) with C-terminal–N- terminal (CN) interface through various side-chain contacts (see Figure 1). All interfaces were shielded from the solvent. The wild type sequence was mutated at two positions, at Glu22 and Met35 to examine the sheet-to- sheet association behavior at the interfaces, i.e. Glu22 was mutated to Lys or Gln at the N-terminal, while Met35 was oxidized at the C-terminal. All starting structures of the mutants were built from the wild-type Aβ9–40 by replacing the side-chains of the targeted residues, but without changing the backbone conformations and side-chain orientations. The structure of the designed mutant was first minimized for 500 steps using the steepest decent algorithm with the backbone of the protein restrained before being subjected to the following system setup and production runs.
3.2. MD protocol
All MD simulations were performed using the NAMD program (11) with the CHARMM22 force field (12). The Aβ oligomers and mutants were solvated by a periodic box of TIP3P water molecules that extended 10 Ǻ from any edge of the box to the protein atoms. Counter ions were added to the box by randomly replacing water molecules to neutralize the system, resulting in ionic concentration of ~ 0.15 molar NaCl. We followed a standard protocol for each MD simulation, which consists of initial minimization, heating procedure, equilibrium, and production run. Each system was initially energy minimized to remove bad contacts by using the conjugate gradient method with the peptides constrained and then to relax the atoms without position constrains. The system was then subjected to 1 ns of heating procedure while constraining the backbone atoms of the protein to allow relaxation of water and ions, followed by 5 ns equilibrium run without position constraints on the peptides. The production simulation used the velocity Verlet integrator in the NPT ensemble under periodic boundary conditions with the minimum image convention. Constant pressure (1 atm) and temperature (330 K) on the system were maintained by an isotropic Langevin barostat and a Langevin thermostat. Short-range van der Waals interactions employed a switch function with a twin range cutoff of 10.0 and 12.0 Ǻ. Long-range electrostatic interactions employed particle mesh Ewald summation with a grid size of ~ 1Å. An integration time step of 2 fs was used, with a multiple time-step algorithm employed to compute covalent bonds at every time step, short-range non- bonded interactions every two time steps, and long range electrostatic forces every four time steps. Each system was simulated for 20 ns and trajectories were saved at 2.0 ps intervals for later analysis. The VMD (visual molecular dynamics) program (13) was used for visualization of the trajectories and preparation of figures. The details of the set-up of the simulations were listed in Table 1.
Table 1.
Summary of all simulation of Aβ models with two antiparallel β-sheets, each comprising of six β-strands packed in parallel
| Simulation | Twist Angle (degree) | Sc | |
|---|---|---|---|
| sheet 1 | sheet 2 | ||
| C-C | 15.3 ± 3.0 | 15.5 ± 2.0 | 0.71 ± 0.01 |
| N-N | 11.0 ± 1.2 | 10.7 ± 0.8 | 0.65 ± 0.04 |
| C-N | 21.2 ± 2.2 | 20.9 ± 3.5 | 0.58 ± 0.06 |
| C-C, M35O | 21.1 ± 3.0 | 7.6 ± 1.3 | 0.68 ± 0.03 |
| N-N, E22K | 16.2 ± 3.1 | 14.1 ± 0.9 | 0.57 ± 0.04 |
| N-N, E22Q | 11.9 ± 1.4 | 14.2 ± 1.3 | 0.62 ± 0.05 |
| C-N, M35O | 18.4 ± 3.6 | 12.6 ± 2.4 | 0.57 ± 0.05 |
3.1. Kinetic model
An in-house Monte Carlo (MC) program was developed to study the competitive associations of Aβ oligomerization in both the fibril growth and lateral directions. As shown in Figure 2, an Aβ peptide was modelled by a single pseudo particle that has three binding sites, labeled by N, C, and H where superscript 1 indicates that this binding site is available and 0 when unavailable.
Figure 2.
Schematic illustration of the kinetic model. (a) Pseudo particle with four binding sites labeled by H, N, and C that could be associated through the fibril axis, N- terminal, and C-terminal, respectively. Two monomers assemble into a dimer (b) along the fibril axis, or through (c) CC, (d) NN, and (e) CN association in the lateral direction. Superscript 1 indicates binding site available and 0 unavailable.
For example, two units (unit could be monomer, dimer, trimer, or other high-order oligomers) could associate with each other in either fibril axis (i.e. HH binding) direction or lateral axis normal to fibril axis (i.e. CC, NN, or NC binding). We assumed that oligomers have the equal probability to associate with each other in either fibril or lateral directions. However, in the lateral direction, Aβ peptides can associate with each other through one of three interfaces (i.e. CC, NN, and NC interfaces), which correspond to three different association probabilities. The association probability is proportional to the averaged binding energy (Table 2), while dissociation probability is inversely proportional to the averaged binding energy. For double-layered Aβ models, the binding energies between two adjacent β-sheets forming different interfaces were calculated using the generalized Born method with molecular volume (GBMV) (14) on the basis of all-atom MD trajectories, i.e. 〈∆Ebind〉=〈∆Edoublesheets〉‒〈∆Esheet1〉‒〈∆Esheet2〉. Detailed binding energy calculations can be found in reference (15). The system consisted of 4,000 Aβ monomers at the beginning of the simulations. To initiate a nucleus seed formation, at the beginning of the MC simulations, i.e. when the number of monomers was larger than 3950, only association events occurred. Under other circumstances in the aggregation process, the ratio of association and dissociation was approximately 70%. The probability of an attempt to association/disassociation being accepted was Paccept = min{1,exp[‒β∆E]}, where ∆E is the change in binding energy.
Table 2.
Mean interaction energy and native contacts between two β-sheets by averaging the last 10 ns simulations
| Simulation | Interaction Energy (kcal/mol) | Native Contacts |
|---|---|---|
| C-C | −484.5 ± 16.5 | 145.0 ± 5.9 |
| N-N | −284.7 ± 12.5 | 99.6 ± 4.3 |
| C-N | −210.2 ± 29.5 | 59.4 ± 9.5 |
| C-C, M35O | −373.2 ± 15.7 | 122.4 ± 5.2 |
| N-N, E22K | −174.1 ± 11.7 | 50.9 ± 5.1 |
| N-N, E22Q | −250.4 ± 13.4 | 82.1 ± 5.1 |
| C-N, M35O | −111.2 ± 25.3 | 26.9 ± 7.1 |
4. RESULTS AND DISCUSSION
4.1. Relative structural stability of Aβ oligomers
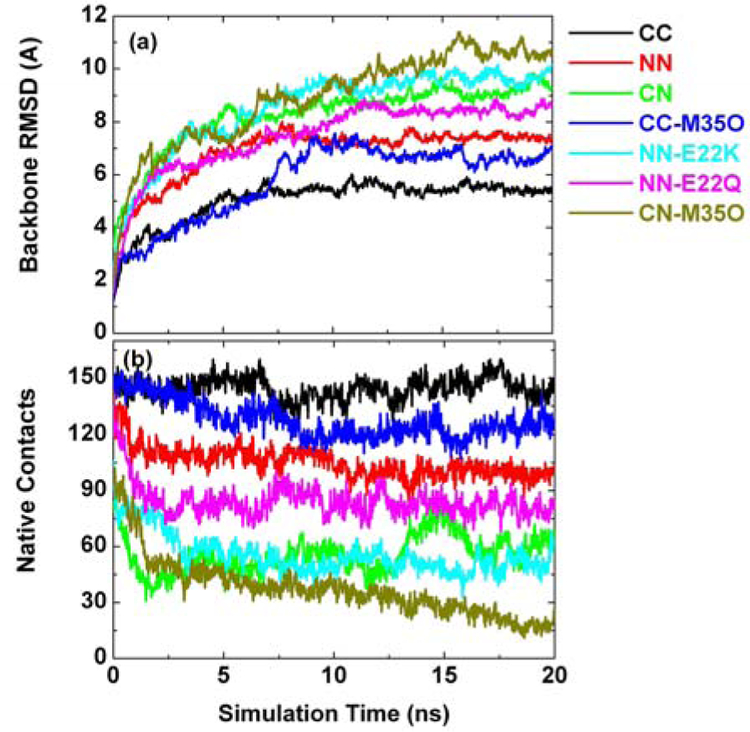
The conformational change and the conservation of the oligomers were monitored by the time evolution of the backbone root mean square deviation (RMSD) through the simulations relative to their initial energy minimized structures, as shown in Figure 3a. It can be seen that the structural deviation of double-layered Aβ 12-mers was strongly dependent on the sheet-to-sheet interfaces. The oligomer with C-terminal-C-terminal (CC) interface experienced the least structural deviation with an averaged RMSD of 5 Å during the 20 ns simulation, while the oligomer with C-terminal-N-terminal (CN) interface displayed dramatic conformational changes with a continuously increasing RMSD up to 12 Å at 20 ns. The oligomer with the N-terminal-N-terminal (NN) interface experienced intermediate structural deviation. As shown in Figure 1, the association force between the β-sheets largely depends on side-chain interactions at the interface. These side chains differ in hydrophobicity, electrostatics, and geometrical characteristics (size and shape), thus introducing different corresponding interactions. Native contacts between two β-sheets were used to roughly estimate those side-chain-side-chain interactions. As shown in Table 2, the average numbers of native contacts, sampled from the last 10 ns of the simulations, were 145, 100, and 60 for the CC, NN, and CN oligomers, respectively. The trends observed in the native contacts (Figure 3b) were in good qualitative agreement with those observed in the RMSD profiles, in which a lower RMSD value is generally correlated with a higher number of native contacts and vice-versa. Additionally, to characterize how two surfaces geometrically fit each other, shape complementarity (Sc) was used to measure the extent of the surface match of two facing β-sheets using the program SC of the CCP4 with default parameters (16). Sc of 1.0 implies perfect interface match, while 0.0 means two unrelated surfaces. As shown in Table 1, as expected, a CC interface has relatively higher Sc of 0.71 than the NN (Sc=0.65) and CN (Sc=0.58) interfaces.
Figure 3.
(a) Backbone root mean square deviations of Aβ oligomers as a function of time. The RMSD is calculated relative to the initial energy minimized structure. (b) Time- dependent native contacts between two β-sheets. The native contacts consist of backbone-backbone hydrogen bonds and side-chain–side-chain contacts.
Taken together, the CC oligomer displayed the highest structural stability, which was mainly attributed to hydrophobic interactions, whereas the NN oligomer folded through weak sidechain–sidechain hydrophobic interactions and unprotected interstrand salt bridges between Hsp14 and Glu22 (17). In parallel, based on experiments, Petkova and co-workers (8) recently proposed a minimal quaternary structure of Aβ1–40 protofibrils using NMR data, in which two intermolecular β-sheets associated through CC sidechain–sidechain hydrophobic interactions in an antiparallel manner. Dolphin and coworkers (18) presented alternative possible quaternary structure of Aβ protofibrils, where two C-terminal strands in each layer were packed in parallel via the central hydrophobic core. All these models suggest that the CC association is favored over other associations, independent of how the β-sheets are packed-in a parallel or antiparallel fashion. Additionally, parallel Aβ strands within the β-sheet provide extra structural stability through backbone/side-chain hydrogen bonds, salt bridges, and in-register π-π interactions (i.e. identical positions along the sequence from two neighboring peptides interact with each other) (19). It should be noted that although all simulations presented here were based on the Tycko’s model of Aβ9–40 (8), our recent Aβ simulations (15) on the basis of the Luhrs’s model of Aβ17–42 (9) also found that the CC interface essentially stabilized by hydrophobic and van der Waals (shape complementarity via M35-M35 contacts) intermolecular interactions was more stable than the NN stabilized by hydrophobic and electrostatic interactions. Although these two Aβ models differ in the length of peptide and detailed turn structure, overall dynamic and association behaviors of Aβ oligomers were consistent with each other.
4.2. Effects of sequence variation on the stability of Aβ oligomers
To examine the effect of sequence variation occurring at the interface on the structural stability of the preformed Aβ oligomers, four mutations were introduced at different interfaces, Met35 oxidation at the CC interface (CC-M35O), Glu22→Lys and Glu22→Gln at the NN interface (NN-E22K and NN-E22Q), and Met35 oxidation at the CN interface (CN-M35O). It is clear that the backbone RMSD difference between the wild type and the corresponding mutants was significant, indicating that the mutations were indeed responsible for changes in the stability. The native contacts between two antiparallel β- sheets revealed the same pattern as the backbone RMSDs. This indicates that the side-chain interactions at the interface are important and affect the stabilities of the Aβ oligomers, in good agreement with previous studies (20). For example, when Met35 residues were oxidized at the the CC interface, neutral hydrophobic Met residues became negatively charged residues, which introduces unfavorable charge-charge repulsion between two β-sheets, as evidenced by the remarkable increase in backbone RMSD and decreased native contacts (Figure 3). A more pronounced electrostatic repulsion was also observed in the E22K mutation at the NN interface. The two β-sheets of the NN-E22K mutant were pushed away from each other (Figure 5e) due to repulsive forces between the K16 and K22 residues. The stabilities of mutants were further compared based on the binding energies between two β- sheets, as shown in Figure 4 and Table 2. The binding affinities of all mutants were larger than the corresponding wild type oligomers, with the following increasing order: CC-M35O < NN-E22Q < NN-E22K < CN-M35O. In general, the NN and CN interfaces were more vulnerable to mutations than the CC interface because they can not attain sufficient packing interactions or the hydrophobic interactions were too weak. On the other hand, the favorable side-chain geometry match between two β-sheets is important for the residue to fit into the interface without disturbing the proper hydrophobic interactions or salt bridges. Consistent with snapshots of mutants at 20 ns in Figure 5, all Sc values of mutants were reduced as compared to the wild type Aβ40. Taken together, although these disease-related mutants (E22K, E22Q, and M35O) experience large overall structural deviations as determined by weak sheet-to-sheet associations, they appear to still maintain each single-layered β-sheet, indicating that their structures are more flexible, and thus they might present increased amyloid oligomerization in the longitudinal direction. Cheng and coworkers (21) recently reported that the “Arctic” mutation (Aβ E22G) accelerated the aggregation of Aβ into protofibrils and fibrils in vitro, but decreased the abundance of nonfibrillar Aβ assemblies as compared with wild-type Aβ, consistent with our simulation observations of higher rates of fibril nucleation and formation.
Figure 5.
Snapshots of double-layered Aβ models at 20 ns for (a) CC-WT, (b) NN-WT, (c) CN-WT, (d) CC-M35O, (e) NN- E22K, (f) CN-M35O, and (g) NN-E22Q oligomers. Color Id: polar residues in green, non-polar residues in orange, positively charged residues in blue, and negatively charged residues in red.
Figure 4.
Interaction energies for the wild type and mutated sequences between two antiparallel β-sheets associated through different interfaces. The interaction energy is calculated by the GBSW implicit solvent model in the CHARMM program.
4.3. Water channel and twisted β-sheet
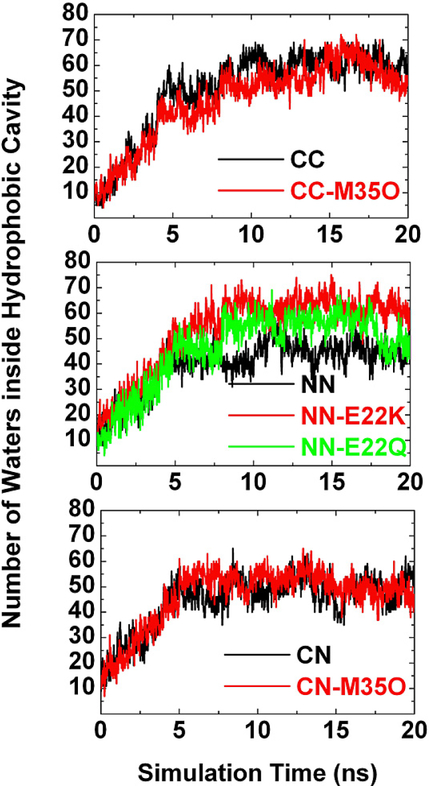
Aβ peptide has a β-strand-loop-β-strand motif. In general, the β-strand–loop–β-strand motif has inherent structural stability due to intrastrand side-chain–side-chain interactions including salt bridges, hydrogen bonds, and hydrophobic interactions. In the simulations, this native motif was well preserved over time in all models. Thus, when Aβ peptides pack in parallel along the fibril axis, they possess internal cavities near the Asp23-Lys28 salt bridges. The sizes of these internal cavities were maintained with a diameter of ~6–8 Å during the 20 ns simulations. It should be noted that there were no water molecules and ions inside the cavity at the beginning of simulations. Figure 6 shows the water occupancy profiles inside the Aβ hydrophobic cavity as a function of time. It can be seen that as the simulation proceeded the number of water molecules gradually increased during the first 6–8 ns and reached up to a relative constant plateau, indicating that water molecules were able to penetrate into these internal cavities, despite the hydrophobic character of the cavities. Due to the limited interior space and restricted side-chain movement, the entering water molecules almost aligned into a continuous spine of hydration along the fibril axis, interacting with the pore-facing side-chains via hydrogen bonding. A typical snapshot of the water inside the hydrophobic cavity of the double-layered CC model is shown in Figure 7. Similar hydrated cavities were also observed in the simulations (22, 23). Entering water molecules can form hydrogen bonds with the oligomer backbones and sidechains to maintain the integrity of the cavities. The hydrated cavity can help maintain and stabilize the oligomeric pore structures composed of the β- strand–loop–β-strand motif. The desolvation of the interior of the Aβ oligomers would impose a large energy and entropic penalty on the structure and stability of oligomers, consistent with the observations of Hummer and co-workers (22, 24). Unlike these interior hydrated cavities made of the internal β-strand–loop–β-strand motif, Aβ peptides can also form annular pores. Jang and co-workers (25) recently modeled Aβ annular structures which were fully embedded in the DOPC lipid bilayer. The average diameter of the Aβ annular structures containing 24 Aβ17–42 monomers is ~20 Å, which is large enough to conduct ions along with water molecules. Most importantly, they found that these Aβ annular structures have strong selective affinity for only Ca2+, supporting experimental observation of calcium-selective β-amyloid ion channel. Lashuel and coworkers (26, 27) reported that many amyloid proteins have channel- or pore-like properties in vitro. In addition, similar to other stable oligomers formed by amyloid peptides, Aβ strands are twisted, with a twist angle between 11 and 21º between consecutive strands (Table 1). The twisted β-strands result from optimizing the side-chain- side-chain interactions, hydrogen bonding, and backbone conformation. Twisting can assist in avoiding electrostatic repulsion by segregating the charged residues between the in-register β-strands.
Figure 6.
Water occupancy profiles inside hydrophobic cavities of double-layered models with (a) CC interface, (b) NN interface, and (c) CN interface. The hydrophobic cavity is defined by the inward pointing side-chains of Phe19, Ala21, Asp23, Lys28, Ala30, and Ile32. Any water molecule within a 3.2 Ǻ cutoff of this cavity is counted as hydration number.
Figure 7.
Representative cross-section of the interior spine of hydration of Aβ CC-WT model, viewed along the protofibrils axis. Water molecules (color in green) are trapped in the interior cavity near the Asp23-Lys28 salt bridges.
4.5. Kinetic of associations
Amyloid-forming peptides not only grow along the fibril axis, but also assemble in the lateral direction leading to the formation of a continuous three-dimensional network, as revealed by AFM and EM experiments (28–30). To obtain insight into the kinetic properties of the Aβ aggregation process, we monitored the numbers of associations and of the oligomers as a function of time using MC. The longitudinal growth was measured by horizonal-horizontal (HH) associations, while the lateral assembly was measured by associations through NN, CC, and CN interfaces. In Figure 8a, all associations in both directions gradually increased, indicating that lateral association of aggregates occurs concurrently with fibril elongation growth, although elongation growth is dominant for all cases. As expected, mutations (E22K, E22Q, and M35O) occurring at the interfaces have a large effect on the corresponding lateral associations, which are determined by low affinity binding at the interfaces. For example, comparison of wild type with the CC-M35O mutant in Figure 8a, indicates that it took ~6,000 MC events for the wild type to consume all monomers, much less than the ~7,000 MC events in the case of CC-M35O mutant due to its unfavorable sheet-to-sheet binding energy induced by M35 oxidation at the CC interface. At 6,000 MC events, the number of HH elongation associations increased to 1,373 for the CC-M35O mutant as compared to 1,098 HH associations for the wild type, while the number of CC lateral associations decreased to 650 as compared to 761 NN lateral associations for the wild type. At the final stage of the aggregation process, the HH and NN associations of the CC-M35O mutant reached 1,619 and 875 at 7,000 MC events, respectively. Other mutants displayed similar association behavior. Thus, as compared to the wild type, the weaker lateral association in mutants led to the enhancement of normal fibril elongation, suggesting that Aβ could aggregate into amyloid fibrils via two competing directions depending on the binding affinity. Experimental evidence (31–33) indicated that E22K and E22Q mutations had significantly higher rate of fibril nucleation and elongation and were more toxic than wild type Aβ40/Aβ42. Thus, comparison between experiments and simulations suggests that fibril elongation, rather than lateral association, might be mainly responsible for neurotoxicity.
Figure 8.
The kinetics of Aβ aggregation process. (a) The number of associations in the fibril axis and lateral direction, and (b) the population of low-order oligomers (dimer, trimer, tetramer, and hexamer), as a function of MC events.
As can be observed in Figure 8b, the monomers decreased monotonically and fused to form low-order oligomers (i.e. dimers, trimers, tetramers, and hexamers), consistent with typical monomer depletion curve (34, 35). The aggregation kinetic of low-order oligomers was nearly identical. The number of these oligomers increased at the early stage of the aggregation process; once reaching a particular size, these aggregates of different sizes were consumed and further assembled into high-order oligomers in a progressive manner, and eventually all converted to the protofibrils, as indicated by the disappearance of these oligomers. The intensity of lower-order oligomer populations had the following deceased order: dimer > trimer > tretramer > hexamer > other high-order oligomers.There was no lag phase observed for dimer and trimer. The absence of lag phase suggests that the dimer and trimer formation is very rapid and unstable, thus neither can serve as minimal nucleus seeds that require highly organized and stable aggregates. But, tetramer, hexamer, and other high- order oligomers exhibited a lag phase. The presence and length of a lag phase is determined by nucleus formation, a thermodynamically unfavorable state that requires a series of monomer association/disassociation (36). The lag time was significantly lengthened with the increasing the size of oligomers (Figure 8b and Figure 9), suggesting that monomers and lower-order oligomers are the dominant species at the beginning of the aggregation, but they finally fuse to give higher-order structures, consistent with experimental observations (37, 38). It should be mentioned that since all aggregates were modeled as spherical particles without structural details, our model did not consider energy barrier sinduced by geometry and collision effects when two aggregates tend to associate into high- order oligomers during the aggregation process.
Figure 9.

The population of high-order oligomers (32- and 64-mer), as a function of MC events.
5. CONCLUSIONS
In this work, we explore the effect of β-sheet-to- β-sheet organization on the structure, dynamics, and association of Aβ40 oligomers in solution using MD and MC simulations. Simulation results show that Aβ oligomers with the CC interface form the most stable and energetically favorable structure as compared to other oligomers with the NN and NC interfaces, in good agreement with experimental data. The stabilizing forces at the CC interface are mainly contributed to hydrophobic interactions between Ile31 and Val40 and steric zippers via Met35-Met35 contacts. All Aβ models having the β-strand- turn-β-strand motif present intramolecular hydrated cavities near the Asp23-Lys28 salt bridges. Designed mutants with substituted residues at the Glu22 and Met35 positions occurring at the interfaces display lower structural stability, weaker lateral association, and stronger elongation than the corresponding wild types, suggesting that fibril elongation and lateral association are competitive to each other. These disease-related mutants (E22K, E22Q, and M35O) have more flexible structures and are more toxic in cell culture, suggesting that these mutants display fast amyloid oligomerization, and that fibril elongation rather than lateral association might be mainly responsible for neurotoxicity.
6. ACKNOWLEDGEMENT
This work was in part supported by startup funds (J. Zheng) from The University of Akron. We thank the Center for Cancer Research Nanobiology Program (CCRNP) at the National Cancer Institute for providing computational facilities to carry out this work. We thank Dr. Robert Tycko for providing Aβ9–40 atomic structures. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract number NO1-CO- 12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported (in part) by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
7. REFERENCES
- 1.Selkoe DJ: Alzheimer’s disease: genes, proteins, and therapy. Physiol. Rev, 81 (2), 741–766 (2001) [DOI] [PubMed] [Google Scholar]
- 2.Bucciantini M, Giannoni E, Chiti F, Baroni F, Formigli L, Zurdo J, Taddei N, Ramponi G, Dobson CM and Stefani M: Inherent toxicity of aggregates implies a common mechanism for protein misfolding diseases. Nature, 416 (6880), 507–511 (2002) [DOI] [PubMed] [Google Scholar]
- 3.Cleary JP, Walsh DM, Hofmeister JJ, Shankar GM, Kuskowski MA, Selkoe DJ and Ashe KH: Natural oligomers of the amyloid-[beta] protein specifically disrupt cognitive function. Nat Neurosci, 8 (1), 79–84 (2005) [DOI] [PubMed] [Google Scholar]
- 4.Lin H, Bhatia R and Lal R: Amyloid {beta} protein forms ion channels: implications for Alzheimer’s disease pathophysiology. FASEB J, 15 (13), 2433–2444 (2001) [DOI] [PubMed] [Google Scholar]
- 5.Mousseau N and Derreumaux P: Exploring the early steps of amyloid peptide aggregation by computers. Acc. Chem. Res, 38 (11), 885–891 (2005) [DOI] [PubMed] [Google Scholar]
- 6.Makabe K, McElheny D, Tereshko V, Hilyard A, Gawlak G, Yan S, Koide A and Koide S: Atomic structures of peptide self-assembly mimics. Proc. Natl. Acad. Sci, 103 (47), 17753–17758 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zheng J, Ma B, Tsai C-J and Nussinov R: Structural stability and dynamics of an amyloid-forming peptide GNNQQNY from the yeast prion sup-35 Biophysical Journal, 91 (3), 824–833 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Petkova AT, Yau WM and Tycko R: Experimental constraints on quaternary structure in alzheimer’s beta-amyloid fibrils. Biochemistry, 45 (2), 498–512 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Luhrs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Dobeli H, Schubert D and Riek R: 3D structure of Alzheimer’s amyloid-{beta} (1–42) fibrils. Proc. Natl. Acad. Sci, 102 (48), 17342–17347 (2005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ma B and Nussinov R: Stabilities and conformations of Alzheimer’s beta -amyloid peptide oligomers (Abeta 16– 22, Abeta 16–35, and Abeta 10–35): Sequence effects. Proc. Natl. Acad. Sci, 99 (22), 14126–14131 (2002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K and Schulten K: NAMD2: greater scalability for parallel molecular dynamics. Journal of Computational Physics, 151 (1), 283–312 (1999) [Google Scholar]
- 12.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S and Karplus M: CHARMM - a program for macromolecular energy, minimization, and dynamics calculations. J. Computational Chemistry, 4 (2), 187–217 (1983) [Google Scholar]
- 13.Humphrey W, Dalke A and Schulten K: VMD - visual molecular dynamics. J. Molec. Graphics, 14 (1), 33–38 (1996) [DOI] [PubMed] [Google Scholar]
- 14.Lee MS, Feig M, S.F. R.Jr and Brooks Charles L.: New analytic approximation to the standard molecular volume definition and its application to generalized Born calculations. Journal of Computational Chemistry, 24 (11), 1348–1356 (2003) [DOI] [PubMed] [Google Scholar]
- 15.Zheng J, Jang H, Ma B, Tsai C-J and Nussinov R: Modeling the alzheimer a{beta}17–42 fibril architecture: tight intermolecular sheet-sheet association and intramolecular hydrated cavities. Biophys. J, 93 (9), 3046–3057 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lawrence MC and Colman PM: Shape complementarity at protein/protein interfaces. Journal of Molecular Biology, 234 (4), 946–950 (1993) [DOI] [PubMed] [Google Scholar]
- 17.Missimer JH, Steinmetz MO, Baron R, Winkler FK, Kammerer RA, Daura X and van Gunsteren WF: Configurational entropy elucidates the role of salt-bridge networks in protein thermostability. Protein Sci, 16 (7), 1349–1359 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dolphin GT, Dumy P and Garcia J: Control of amyloid [beta]-peptide protofibril formation by a designed template assembly. Angewandte Chemie International Edition, 45 (17), 2699–2702 (2006) [DOI] [PubMed] [Google Scholar]
- 19.Zheng J, Ma B and Nussinov R: Consensus features in amyloid fibrils: sheet-sheet recognition via a (polar or nonpolar) zipper structure. Physical Biology, 3 (3), P1–P4 (2006) [DOI] [PubMed] [Google Scholar]
- 20.Tsai H-H, Zanuy D, Haspel N, Gunasekaran K, Ma B, Tsai C-J and Nussinov R: The stability and dynamics of the human calcitonin amyloid peptide DFNKF. Biophys. J, 87 (1), 146–158 (2004) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheng IH, Scearce-Levie K, Legleiter J, Palop JJ, Gerstein H, Bien-Ly N, Puolivali J, Lesne S, Ashe KH, Muchowski PJ and Mucke L: Accelerating amyloid-beta fibrillization reduces oligomer levels and functional deficits in alzheimer disease mouse models. J. Biol. Chem, 282 (33), 23818–23828 (2007) [DOI] [PubMed] [Google Scholar]
- 22.Buchete N-V and Hummer G: Structure and dynamics of parallel {beta}-sheets, hydrophobic core, and loops in alzheimer’s A{beta} fibrils. Biophys. J, 92 (9), 3032–3039 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han W and Wu Y-D: Molecular dynamics studies of hexamers of amyloid-beta peptide (16–35) and its mutants: Influence of charge states on amyloid formation. Proteins: Structure, Function, and Bioinformatics, 66 (3), 575–587 (2007) [DOI] [PubMed] [Google Scholar]
- 24.Buchete N-V, Tycko R and Hummer G: Molecular dynamics simulations of alzheimer’s [beta]-amyloid protofilaments. Journal of Molecular Biology, 353 (4), 804–821 (2005) [DOI] [PubMed] [Google Scholar]
- 25.Jang H, Zheng J and Nussinov R: Models of {beta}- amyloid ion-channels in the membrane suggest that channel formation in the bilayer is a dynamic process. Biophys. J, 93 (6), 1938–1949 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lashuel HA, Hartley D, Petre BM, Walz T and Lansbury PT: Amyloid pores from pathogenic mutations. Nature, 418 (6895), 291–291 (2002) [DOI] [PubMed] [Google Scholar]
- 27.Lashuel HA and Lansbury PT: Are amyloid diseases caused by protein aggregates that mimic bacterial pore-forming toxins? Quarterly Review of Biophysics, 39 (2), 167–201 (2006) [DOI] [PubMed] [Google Scholar]
- 28.Makarava N, Bocharova OV, Salnikov VV, Breydo L, Anderson M and Baskakov IV: Dichotomous versus palm-type mechanisms of lateral assembly of amyloid fibrils. Protein Sci, 15 (6), 1334–1341 (2006) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shahi P, Sharma R, Sanger S, Kumar I and Jolly RS: Formation of amyloid fibrils via longitudinal growth of oligomers. Biochemistry, 46 (25), 7365–7373 (2007) [DOI] [PubMed] [Google Scholar]
- 30.Raspanti M, Viola M, Sonaggere M, Tira ME and Tenni R: Collagen fibril structure is affected by collagen concentration and decorin. Biomacromolecules, 8 (7), 2087–2091 (2007) [DOI] [PubMed] [Google Scholar]
- 31.Meredith SC: Protein denaturation and aggregation. cellular responses to denatured and aggregated proteins. Annals of the New York Academy of Sciences, 1066 (1), 181–221 (2006) [DOI] [PubMed] [Google Scholar]
- 32.Klein WL, Stine JWB and Teplow DB: Small assemblies of unmodified amyloid β-protein are the proximate neurotoxin in Alzheimer’s diease. Neurobiology of Aging, 25 (5), 569–580 (2004) [DOI] [PubMed] [Google Scholar]
- 33.Hou L, Kang I, Marchant RE and Zagorski MG: Methionine 35 oxidation reduces fibril assembly of the amyloid A[beta] - (1–42) peptide of Alzheimer’s disease. J. Biol. Chem, 277 (43), 40173–40176 (2002) [DOI] [PubMed] [Google Scholar]
- 34.Murphy RM: Kinetics of amyloid formation and membrane interaction with amyloidogenic proteins. Biochimica et Biophysica Acta (BBA) - Biomembranes, 1768 (8), 1923–1934 (2007) [DOI] [PubMed] [Google Scholar]
- 35.Benseny-Cases N, Cocera M and Cladera J: Conversion of non-fibrillar [beta]-sheet oligomers into amyloid fibrils in Alzheimer’s disease amyloid peptide aggregation. Biochemical and Biophysical Research Communications, 361 (4), 916–921 (2007) [DOI] [PubMed] [Google Scholar]
- 36.Yamamoto S, Hasegawa K, Yamaguchi I, Goto Y, Gejyo F and Naiki H: Kinetic analysis of the polymerization and depolymerization of [beta]2-microglobulin-related amyloid fibrils in vitro. Biochimica et Biophysica Acta (BBA) - Proteins & Proteomics, 1753 (1), 34–43 (2005) [DOI] [PubMed] [Google Scholar]
- 37.Lewandowska A, Matuszewska M and Liberek K: Conformational properties of aggregated polypeptides determine ClpB-dependence in the disaggregation process. Journal of Molecular Biology, 371 (3), 800–811 (2007) [DOI] [PubMed] [Google Scholar]
- 38.Meinhardt J, Tartaglia GG, Pawar A, Christopeit T, Hortschansky P, Schroeckh V, Dobson CM, Vendruscolo M and Fandrich M: Similarities in the thermodynamics and kinetics of aggregation of disease-related Abeta (1–40) peptides. Protein Sci, 16 (6), 1214–1222 (2007) [DOI] [PMC free article] [PubMed] [Google Scholar]