Abstract
Genome editing reemerged in 2012 with the development of CRISPR/Cas9 technology, which is a genetic manipulation tool derived from the defense system of certain bacteria against viruses and plasmids. This method is easy to apply and has been used in a wide variety of experimental models, including cell lines, laboratory animals, plants, and even in human clinical trials. The CRISPR/Cas9 system consists of directing the Cas9 nuclease to create a site-directed double-strand DNA break using a small RNA molecule as a guide. A process that allows a permanent modification of the genomic target sequence can repair the damage caused to DNA. In the present study, the basic principles of the CRISPR/Cas9 system are reviewed, as well as the strategies and modifications of the enzyme Cas9 to eliminate the off-target cuts, and the different applications of CRISPR/Cas9 as a system for visualization and gene expression activation or suppression. In addition, the review emphasizes on the potential application of this system in the treatment of different diseases, such as pulmonary, gastrointestinal, hematologic, immune system, viral, autoimmune and inflammatory diseases, and cancer.
Keywords: CRISPR/Cas9, genome editing, disease models, pulmonary disease, gastrointestinal disease, hematologic disease, viral disease, cancer, autoimmune disease, inflammatory disease
1. Introduction
In order to determine the function of a gene, the gene can be inactivated by homologous recombination or by blocking its messenger RNA through RNA interference (1). This approach can be applied in cultured cells by transfection or in living organisms by transgenesis (1). Recent advances in genome editing allow the manipulation of any gene at its own locus in a broad variety of species and tissues, including cultured cells and animal organs. Genome editing is a powerful tool for biomedical research and provides hope for correcting some inherited diseases.
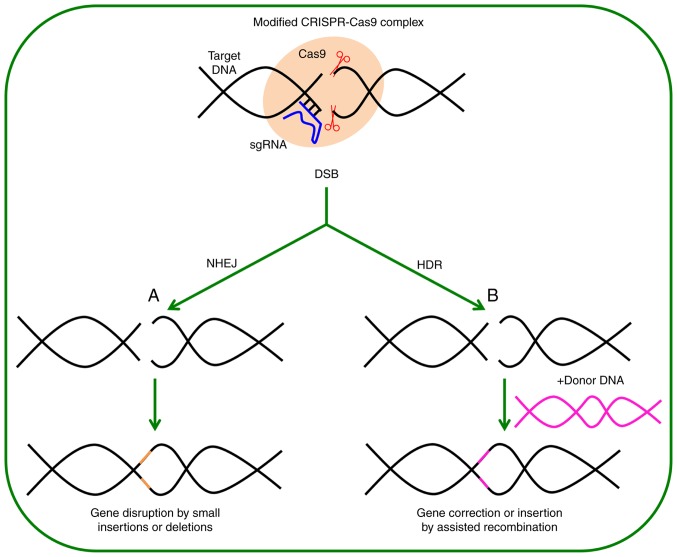
Genome editing is based on the use of highly specific and programmable nucleases, which produce specific changes in regions of interest in the genome by introducing double-strand breaks (DSBs) that are later repaired by cellular mechanisms. These repair mechanisms include the non-homologous end-joining (NHEJ) that is prone to error, and the homology-directed repair (HDR) that is error-free (Fig. 1). Repair permits the generation of insertions, deletions or substitutions in the target area (2-4). These mutations may interrupt, eliminate or correct the defects in genes. The latter possibility affords the ability to correct inherent errors in DNA that cause diseases. The ‘programmable’ nucleases are mainly meganucleases (5), zinc-finger nucleases (6), transcription activator-like effector nucleases (7), and the CRISPR/Cas9 system [involving the Clustered Regularly Interspaced Short Palindromic Repeats and nuclease(s) associated to the CRISPR locus] (8). Usually, the CRISPR/Cas9 system is comprised of a guide RNA (gRNA) that directs the Cas9 nuclease to create a DSB in a specific place of the genome. In the last decade, this system has gained wide acceptance over other systems due to its simplicity, speed and efficiency for modifying endogenous genes in any cell or target tissue, even in the most traditionally difficult-to-treat organisms.
Figure 1.
Genome editing through the CRISPR/Cas9 system. Cas9 and the gRNA create a complex that binds with the DNA close to the PAM site. A DSB is generated in the target site that could be repaired via NHEJ or HDR. (A) The repair by NHEJ usually results in insertions or deletions, or in frameshift that causes the gene knockout by interruption. (B) If a DNA donor with homology in the ends is provided, this DNA can be inserted to the target site to modify the gene, introducing the nucleotides and leading to frameshifts or insertion of cDNA. gRNA, guide RNA; PAM, protospacer adjacent motif; DSB, double-strand break; NHEJ, non-homologous end joining; HDR, homology-directed repair.
2. CRISPR/Cas9 system
Discovery of the CRISPR/Cas9 system
The eubacteria and archaea possess a defense system that adapts, through RNA, to recognize and destroy external DNA and RNA. This provides acquired immunity against invading plasmids and viruses (9). This system is found in approximately 50% of the bacterial genomes that are sequenced and in 87% of the genomes of archaea (9-12). It is also important in a range of additional functions, including replicon divisions (13), high-temperature adaptation (14), chromosomal rearrangements (15) and DNA repair (16).
CRISPR was discovered in the Escherichia coli genome in 1987 as a series of repeated fragments of 29 nucleotides (nt) in length interspaced with variable sequence fragments of 32 nt (17). Interest in the CRISPR system and its associated Cas genes led to the discovery of similar short-repeat palindromic sequences of 24-40 nt in several groups of bacteria and archaea. The repeat sequences are separated by unique variable sequences of 20-58 nt (13,18). The associated genes (Cas) were identified invariably adjacent to a CRISPR locus, suggesting a functional association (19). The initial hypothesis regarding the function of the CRISPR locus proposed roles in cellular DNA repair and replicon partitioning processes; however, in 2005, the first evidence that the CRISPR/Cas system is part of an adaptive prokaryotic immune system was reported through the observation that the majority of the sequences intercalated between the identical repeats were derived from invading phage and plasmid genomes (20-22). In 2007, the incorporation of new spacers was demonstrated in a CRISPR/Cas locus of Streptococcus thermophiles (23), while the CRISPR transcription processing to small mature CRISPR-RNAs (crRNAs) that guide the Cas complex of Escherichia coli was validated experimentally in 2008 (24). In 2010, Cas of Streptococcus thermophilus was demonstrated to create a single DSB at a precise position in target DNA (25), and the following year it was reported that the maturation of crRNA requires trans-encoded small crRNA (tracrRNA), Cas9 and an RNase III in Streptococcus pyogenes (26). Evidence of function in a heterologous system was obtained in 2011 when it was shown that the CRISPR/Cas system of Streptococcus thermophilus on transfer to Escherichia coli provided heterologous immunity against plasmids and phage infection (27). In 2012, the simplification of the CRISPR/Cas9 of Streptococcus pyogenes system was achieved by replacing a tracrRNA and a crRNA with a synthetic single gRNA to direct Cas9 to its target and to perform the cleavage (8). Finally, in 2013, the use of the CRISPR/Cas9 system (type II, Streptococcus pyogenes) was described as a genome editing tool for the induction of site-specific DSBs and subsequent mutagenesis in plant, mouse and human cells, and clinical trials (28-32).
Operation of the CRISPR/Cas9 system as a genome editing tool
The type II CRISPR/Cas system is the most commonly used system for genome editing, using the well-characterized Cas9 endonuclease of Streptococcus pyogenes. In the endogenous system, the mature crRNA joins with a tracrRNA (small RNA that is complementary to the CRISPR sequences) to form a tracrRNA:crRNA complex, which guides the Cas9 to a target site. Thus, CRISPR/Cas9 performs sequence-specific cleavage by simple interaction of crRNA by base pairing at the target site. After joining the target site, the two DNA strands are cleaved by the nuclease domain of Cas9, a HNH domain that cleaves the complementary target strand to the gRNA and a RuvC-like nuclease domain, which cleaves the non-target strand (33-35). The gRNA designed by Jinek et al (8) in 2012 was a chimerical RNA, which contains all the essential components of crRNA and tracRNA to guide Cas9. Since then, multiple variants of CRISPR/Cas9 have been developed, which recognize sequences of 18-24 nt of the gRNA, and 2-4 nt of protospacer adjacent motif (PAM) in target sites (3,36). Therefore, CRISPR/Cas9 can theoretically be directed to a specific sequence of DNA of 22-29 nt, which is unique in most of the genomes, although it has been noted that CRISPR/Cas9 has a high-tolerance for non-specific mating of base pairs between gRNA and its complementary target sequence. This specificity is sensitive to numbers, position and distribution of wrong interactions (3,8,28,29). For instance, the CRISPR/Cas9 of Streptococcus pyogenes (SpCas9) tolerates up to six imbalances of base pairs at target sites (8). The genome editing mediated by CRISPR/Cas9 depends on the generation of the DSB and the subsequent process of DNA repair. The DSB generated by the CRISPR/Cas9 triggers the process of cell repair in DNA, as a NHEJ, which is prone to error and thus can produce mutations involving small insertions and deletions (indels) in target sites, which can interrupt or eliminate the function of the genes or the genomic target elements (such as regulatory regions). Another repair process that can also be triggered is the HDR error-free, which can potentially correct innate disease-causing errors of DNA (genes or regulatory elements) (37).
PAM sequence and off-target cuts
The specificity of CRISPR/Cas9, besides the complementarity of the gRNA/target sequences, requires a PAM sequence that is located immediately after the target sequence. The reliance on the PAM sequence for the cleavage of the DNA restricts the frequency of the cleavage sites in the genomes, thus target sites are found more frequently for small PAM sequences than for longer ones; consequently off-target cut sites are less likely to exist for long PAM sequences than for short ones. The identified PAM sequences vary between different microorganisms, and the following sequences have been reported: 5′-NGG-3′ in Streptococcus pyogenes (SpCas9) (8), 5′-NGGNG-3′ or 5′-NNAGAAW-3′ in Streptococcus thermophiles (St1Cas9) (25,38,39), 5′-NNGRRT-3′ or 5′-NNGRR(N)-3′ in Staphylococcus aureus (SaCas9) (40,41), 5′-NNNRRT-3′ or 5′-NNNNGMTT-3′ in Neisseria meningitidis (NmCas9) (42), and 5′-NGG-3′ in Francisella novivida (FnCpf1) (43,44), where N refers to every nucleotide, R to purines A or G, M to nucleotide A or C, and W to weak bonds A or T (3,11,36,45).
Although the DSB activity of CRISPR/Cas9 is based on the complementarity of target sequences with the gRNAs of ~20 nt in length, the system allows cleavage at genomic locations partially complementary to the gRNA, because the gRNA allows mismatch pairings between the DNA and gRNA (3). These off-target cuts of CRISPR/Cas9 are one of the biggest issues that currently remain unresolved (causing certain undesirable consequences) and differ between different target sites due to the diversity of nucleotide sequence and the genomic context. The CRISPR/Cas9 system has been implemented successfully for gene editing, and for the control of different types of biological systems; however, there is little evidence of the consequences of off-target genome editing Indeed, there are only a few assays of genotoxicity based on cells in culture that allow to quantify, stratify and help prevent biological side effects of gene editing in a given target cell population. Although a number of gene editing studies have reached the phase of clinical trial, clinical evidence showing that gene editing is truly capable of treating disorders is still scarce (30-32).
Several methods have been proposed to optimize gRNA design and minimize off-target cuts to reach the reliability and specificity necessary for safety in therapeutics applications (46). Identification of the optimal gRNA among various candidates for a given target site, and the localization of potential off-target sites can be supported by different bioinformatics tools (47); for example: CRISPRdirect (48), E-CRISPR (49), WU-CRISPR (50), CRISPR gRNA design tool (https://www.atum.bio/eCom-merce/cas9/input), sgRNA Designer (51), sgRNA Scorer 2.0 (52,53), CRISPRscan (54), CRISPR-ERA (55), CCtop (56), CRISPOR (57), Breaking-Cas (46), CHOPCHOP (58), CRISP MultiTarget (59), GT-Scan (60), ge-CRISPR (47), CRISPR Design (61), Cas-Designer (62), Cas-OFFinder (63), COSMID (64), DESKGEN Guide Picker (65), and CRISPR Genome Analyzer (66) (Table I). These software programs have different characteristics and applications. The optimal choice will depend on the type of organism (eukaryotic or prokaryotic) for which the gRNA is designed, the variety of PAM to be used, the size of the DNA target, and the variety of recently-reported Cas-like nucleases (46).
Table I.
Bioinformatics programs for the design of gRNA and search of off-target cuts.
| Name | Website | Available species | Use | |
|---|---|---|---|---|
| CRISPR direct | http://crispr.dbcls.jp/ | >200 | Design of gRNAs | Naito et al, 2015 |
| E-CRISPR | http://www.e-crisp.org/E-CRISP/ | 55 | Design of gRNAs | Heigwer et al, 2014 |
| WU-CRISPR | http://crispr.wustl.edu | 2 | Design of gRNAs | Wong et al, 2015 |
| CRISPR gRNA design tool | https://www.atum.bio/eCommerce/cas9/input | 5 | Design of gRNAs for genome editing for Cas9 | https://www.atum.bio/eCommerce/cas9/input |
| sgRNA Designer | https://portals.broadinst-itute.org/gpp/public/analysis-tools/sgrna-design | 2 | Design of gRNAs for S. aureus and D S. pyogenes Cas9 | oench et al, 2014 |
| sgRNA Scorer 2.0 | http://crispr.med.harvard.edu/sgRNAScorerV2 | 2 | Design of gRNAs for Cas9s from S. aureus and S. thermophilus 3 and Cpf1 | Chari et al, 2015 and 2017 |
| CRISPRscan | http://www.crisprscan.org | 19 | Design of gRNAs for Cas9s and Cpf1 of Acidaminococcus and Lachnospiraceae | Moreno-Mateos et al, 2015 |
| CRISPR-ERA | http://CRISPR-ERA.stanford.edu | 9 | Design of gRNAs for genome editing, repression and activation | Liu et al, 2015 |
| CCtop | http://crispr.cos.uni-heidelberg.de | 50 | Design of gRNAs for Cas9s from S. pyogenes and its variants, S. thermophilus, S. aureus, N. Meningitidis, Treponema denticula, Campylobacter jejuni, and Cpf1 of Acidaminococcus and Lachnospiraceae | Stemmer et al, 2015 |
| CRISPOR | http://crispor.tefor.net | 191 | Design of gRNAs for Cas9s from S. pyogenes and its variants, S. thermophilus, S. aureus, N. Meningitidis, Campylobacter jejuni, and Cpf1 of Acidaminococcus and Lachnospiraceae | Haeussler et al, 2016 |
| Breaking-Cas | http://bioinfogp.cnb.csic.es/tools/breakingcas | Any eukaryotic genome available at Ensembl/Ensembl Genomes (>1,000 genomes) | To design gRNAs for Cas9s from S. pyogenes, S. aureus, Cpf1 of Acidaminococcus sp. and Natronobacterium gregoryi (No PAM needed). To evaluate putative undesired off-targets for CRISPR/Cas applications | Oliveros et al, 2016 |
| CHOPCHOP | http://chopchop.cbu.uib.no | 91 | Identifying gRNA targets Cas9 and its variants, and Cpf1 | Montague et al, 2014 |
| CRISP Multi Target | www.multicrispr.net | 12 | Identifying gRNA targets common to several similar sequences or unique to each of these sequences | Prykhozhij et al, 2015 |
| GT-Scan | http://gt-scan.csiro.au | >25 | Identifying unique genomic targets | O’Brien et al, 2014 |
| ge-CRISPR | http://bioinfo.imtech.res.in/manojk/gecrispr/ | 4 | Prediction and analysis of gRNAs genome editing efficiency | Kaur et al, 2016 |
| CRISPR Design | http://crispr.mit.edu | 15 | Selection and validation of gRNAs, as well as prediction of off-target loci for specificity analyses | Hsu et al, 2013 |
| Cas-Designer | http://www.rgenome.net/cas-designer/ | 33 | Design of gRNAs for user-defined PAM sequence with 10 different enzymes in a given DNA sequence. Identifying potential sites off-target using Cas-OF finder | Park et al, 2015 |
| Cas-OFFinder | http://www.rgenome.net/cas-offinder/ | Any given genome or user-provided sequence | Identifying potential off-target sites for given gRNAs | Bae et al, 2014 |
| COSMID | http://crispr.bme.gatech.edu | 7 | Identifying and validating CRISPR/Cas Off-target sites |
Cradick et al, 2014 |
| DESKGEN Guide Picker | https://www.deskgen.com/guide-picker/ | 2 | Meta tool for designing CRISPR experiments | Hough et al, 2017 |
| CRISPR Genome Analyzer | http://crispr-ga.net | Any genomic locus of any organism for which the sequence is available | Assess the quality of gene editing using next gen sequencing data | Güell et al, 2014 |
gRNA, guide RNA; PAM, protospacer adjacent motif.
Strategies and modifications of the enzyme Cas9 to minimize off-target cuts
The design of variations in the traditional SpCas9 system has been examined for controlling its performance, such as using an inducible system for temporary expression of the nuclease (67). Another strategy to decrease activity off-target is to replace the SpCas9 enzyme for mutant nSpCas9 (nCas9), which cuts a single strand through the inactivation of a nuclease domain RuvC or HNH. In this case, in order to perform the DBS, two nCas9 are needed to target opposite strands of DNA in close proximity (separated by no more than 100 bp), with each nCas9 guided by its own sgRNA that is able to cleave only the strand complementary to the sgRNA (45,68). When two nCas9 are used to perform a DSB, off-target activity is reduced by 50 to 1,500 times (40,45,69,70). Furthermore, another strategy consists in increasing the specificity by mitigating the helicase activity (eSpCas9) to disrupt the off-target sites without interfering with specificity and activity (71). Fusing the nuclease Fok1 with dead SpCas9 (dSpCas9) also improves specificity; in this case, dSpCas9 lacks its cleavage activity by inactivation of its two-nuclease HNH and RuvC domains. Consequently, the fusion forms RNA-guided FokI nucleases, thus, the excision activity for the DSB will depend only on the bounds of the two gRNAs to the DNA with a well-defined spacing and orientation, allowing the dimerization of monomers Fok1-dSCas9 to form a catalytically active Fok1 dimer, which reduces the possibility that a suitable target sequence appears again in the genome, and therefore improves specificity (72-74). Another modification is the binary system Split-SpCas9 that uses the expression the nuclease lobe and the α-helical domain independently. These two are naturally in the enzyme Cas9 alone or attached to the DNA through gRNA. The domains do not interact on their own, but the gRNA recruits all of them in a ternary complex that recaps the activity of Cas9 and catalyzes site-specific DNA cleavage. The uses of modified gRNA annul the Cas9 activity dividing the dimer, which allows the development of an inducible and adjustable dimerization system for genome editing applications. This system has been tested in vitro and in vivo in a mouse model (75,76). Among other modifications to date, the SpCas9-cytidine deaminase can be found, which is a fusion of dSpCas9 with the cytidine deaminase enzyme. This action by cytidine deaminases, converts cytosine (C) to uracil (U), working on single-stranded DNA accessible in the ternary complex between Cas9, gRNA and the target DNA for the introduction of point mutations (77).
New variants of system CRISPR
Studies have identified other enzymes of the CRISPR family, including enzymes encoded by Cas genes smaller than SpCas9 (4.2 kb), such as SaCas9, St1Cas9, and NmCas9 (3.2, 3.4, and 3.2 kb, respectively) (41,45). These enzymes would facilitate its packaging into viral vectors (41,45). Another recently identified enzyme called Cpf1 with shorter crRNA sequences can be used instead of SpCas9 (43,78). Also, the most recently discovered C2c2 (Cas13a) and C2c6 (Cas13b), which can cleave RNA (78-82). Cas13b has already been developed as an RNA base editing technology, having been used as a tool for RNA editing using catalytically-inactive Cas13b fused to the adenosine deaminase domain of ADAR2 for programmable adenosine to inosine replacement in transcripts (82).
Other applications of CRISPR/Cas9
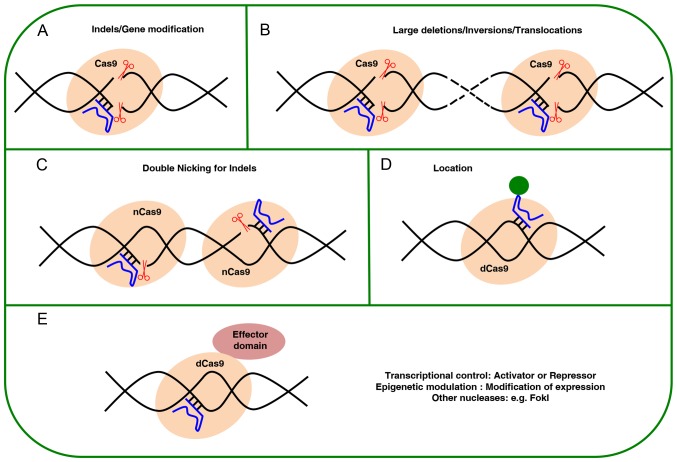
Besides the use of CRISPR/Cas9 to generate indels (Fig. 2), modified versions have been developed for different applications, such as activation or repression through regulation of gene expressions by fusing heterologous domains for modifying epigenetic signatures on histones (i.e. changing methylation patterns) or transcriptional activators/repressors (83-85). Furthermore, CRISPR/Cas9 has been used for the selective labeling of the genome, such as for the monitoring of dynamic chromatin processes (86,87).
Figure 2.
Overview of applications based on Cas9. (A) Nuclease Cas9 directed by a gRNA can induce insertion or deletion mutations; (B) a pair of Cas9 nucleases directed by a gRNA may induce sequence-specific replacement or insertion, large deletions or genomic rearrangements (such as inversions or trans-locations); (C) double cutting by nCas9 to improve the specificity of editing; (D) visualization of specific sites in the genome by dCas9; (E) dCas9 can mediate the regulation of specific endogenous genes by heterologous effector domains or performing histone modifications or DNA methylation. gRNA, guide RNA.
3. CRISPR/Cas9 in disease models
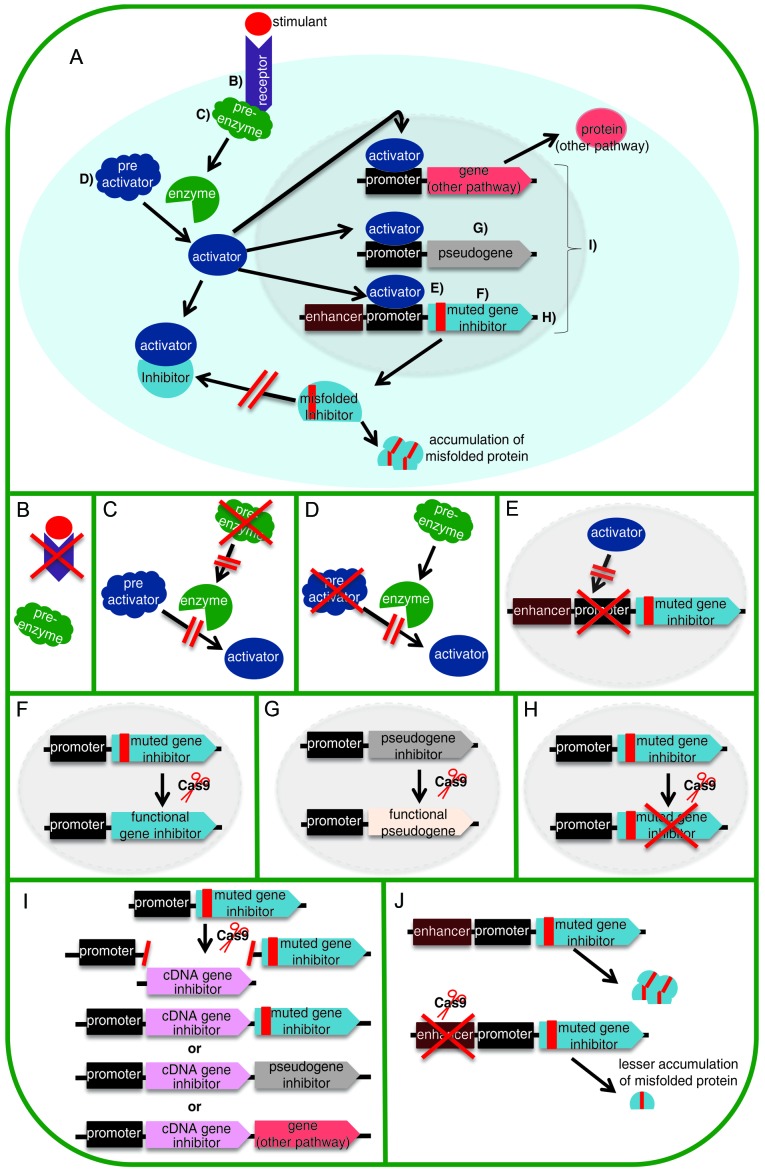
The CRISPR/Cas9 system has an extraordinary therapeutic potential for treating different diseases in which the genetic cause of dysfunction is known, or for the study of these diseases through the creation of cell or animal models. Therapy based on genome editing can lead to the restoration of gene function or compensation of the mutation. Single nucleotide polymorphism (SNP) editing has been approached with different strategies, such as: knocking out the gene that causes the disease (88), introducing a protective mutation (89) or adding a therapeutic transgene (90). When the disease is caused by a virus, cleavage of viral DNA can be performed (70,91-94). Fig. 3 shows certain of the strategies already mentioned and others that could be used to approach different diseases. The purpose of this figure is to expand the panorama of the therapeutic possibilities of CRISPR/Cas9. In the case that a mutation in a gene is difficult to repair due to its genomic context, there may be a pseudogene that could be activated to replace the mutated gene (95). However, if the cause of the disease is a protein that causes damage to the organism by its anomalous characteristics (such as by misfolding and accumulation in a tissue) (e.g. amyloidosis), its expression could be down regulated at several points in its pathway of expression (96).
Figure 3.
Strategies for editing a signaling pathway. (A) Signaling pathway where there is a mutation in the inhibitory gene that prevents the correct folding of encoded protein and therefore its function as an inhibitor. Indents B-E display different strategies that can be approached for preventing that inhibitory protein from being produced. (B) Knockout of the gene that codes for receptor, preventing it from acting on the pre-enzyme and from producing activator protein. (C) Knockout of the gene that codes for pre-enzyme, preventing it from acting on the pre-activator protein and from producing activator protein. (D) Knockout of the gene coding for pre-activator protein, preventing the enzyme acting on it from producing activator protein. (E) Mutation of the binding site of promoter so that the activator protein cannot bind. Indents F-I display different strategies that can be applied for the production of inhibitory protein. (F) Edition of a defective gene to restore production of an inhibitory protein to produce a functional inhibitory protein. (G) In the case that mutations in the inhibitor gene are difficult to repair, the pseudogene inhibitor is repaired to produce a functional inhibitory protein. (H) If a deleterious mutation is difficult to repair and causes the accumulation of a misfolded protein, the gene could be totally inactivated and the pseudogene can be reactivated to produce a functional protein. (I) Another strategy is the addition of the functional cDNA of the inhibitor gene in any of the genes or pseudogene stimulated by the activator protein. (J) Finally, mutation of the enhancer results in reduced production of inhibitory protein.
CRISPR/Cas9 in pulmonary and gastrointestinal diseases
The homozygous ∆508 mutation in the cystic fibrosis trans-membrane conductance regulator (CFTR) gene was corrected using CRISPR/Cas9 in vitro in intestinal stem cells of cystic fibrosis patients (97). Edited stem cells were differentiated into intestinal organoids and exhibited a functional CFTR product (97). CRISPR/Cas9 also shows potential in cases requiring a liver transplant, such as drug therapy-refractory metabolic liver disorders (88,98). CRISPR has been used to suppress genes, thus reprogramming the metabolic pathway, achieving a benign phenotype with treatment of hereditary type I tyrosinemia in mice (88). By eliminating the 4-hydroxyphenylpyruvate dioxygenase (HPD) gene in order to inhibit the second step in tyrosine catabolism, the modified hepato-cytes (FAH−/−/HPD−/−) exhibited a growth advantage over the unedited hepatocytes (FAH−/−/HPD+/+). In a number mice, the replacement was almost complete (92-99%) in 8 weeks. The HPD mutation increased tyrosine catabolism avoiding the pathologic effect of FAH deficiency and the accumulation of tyrosine and toxic metabolites, such as fumarylacetoacetate and succinylacetone in hepatocytes, which in turn results in severe liver damage with increased risk of hepatocarcinoma (88).
The CRISPR/Cas9 system can also be used as therapy in Hirschsprung disease and the megacystis-microcolon-intestinal hypoperistalsis syndrome (MMIHS), in which certain mutations were identified through complete exome sequencing in patients diagnosed with those diseases (99,100). In Hirschsprung disease, four genes (DENND3, NCLN, NUP98 and TBATA) have been linked to the neuronal processes shared by the central nervous system and the enteric nervous system; this function was verified in vivo through a gene knockout by CRISPR/Cas9 (99,100). A zebrafish model was used to observe that the knockout of the afore mentioned genes resulted in the loss of function and interruption of the development of the enteric nervous system (loss of enteric neurons) causing a similar Hirschsprung phenotype in vivo (99). The LMOD1 gene is involved in establishing normal smooth muscle cytoskeletal-contractile coupling, and a mutation causes MMIHS. Mice with knockout of LMOD1 gene exhibited a similar phenotype to this syndrome (protein levels and pathology consistent with MMIHS), displaying beginning of bladder distention at 18.5 days of embryonic development. In certain cases, the developing bladder expanded until it encroached the abdominal cavity, while histological analysis detected early onset thinning of the detrusor muscle of the bladder of Lmod1−/− mice. These results suggest a role for the LMOD1 gene in establishing normal smooth muscle cytoskeletal-contractile coupling (100).
CRISPR/Cas9 in hematologic diseases
β-hemoglobinopathies, including sickle-cell disease (SCD) and β-thalassemia, are caused by mutations in the β-globin (HBB) gene, and affect millions of people worldwide. Several studies have used CRISPR/Cas9 and donor sequences to achieve homologous recombination in the HBB gene in induced pluripotent stem cells (iPSCs) and hematopoietic stem cells (HSCs) (101-104). For instance, efficient correction of the Glu6Val mutation responsible of the SCD was achieved using progenitor cells that were derived from patients and differentiated to erythrocytes, resulting in the expression of the mRNA of adult β-globin (HbA) and confirming the intact transcriptional regulation of modified HBB alleles (101,102). Another genome editing strategy to treat hemoglobinopathies is inserting a mutation to generate a benign genetic condition. In the hereditary persistence of fetal hemoglobin (HPFH), a benign genetic condition, the mutations attenuate the shift of γ-globin to β-globin, causing a high level of fetal hemoglobin expression (HbF) throughout life, which can relieve the clinical signs of β-thalassemia or SCD. In a previous study, CRISPR/Cas9 was used to mimic the HPFH mutation in promoters of the HBG1 and HBG2 genes in human blood progenitor cells. The edited progenitor cells produced red blood cells (RBCs) with increased HbF levels that were sufficient to inhibit the pathological hypoxia of RBCs found in SCD (105). Another strategy applied to increase the HbF levels was CRISPR/Cas9 mutation of BCL11A erythroid enhancer, a validated repressor of HbF and therapeutic target for β-hemoglobin disorders (106). Furthermore, in β-thalassemia caused by the expression-suppression point mutations or deletions in the β-globin gene, effective correction of the HBB mutations by CRISPR/Cas9 was achieved in iPSCs cells derived from a patient, which differentiated in erythroblasts having restored the expression of β-hemoglobin (103,104).
Finally, CRISPR/Cas9 has been applied in the treatment of multiple other hematological diseases, such as alloimmune bleeding disorders, including fetal and neonatal alloimmune thrombocytopenia and post-transfusion purpura, to transform Leu33+ megakaryocyte-like DAMI cells and iPSCs to the Pro33 allotype, which is responsible for generating the human platelet alloantigen 1a and 1b epitopes (107). Furthermore, this technology has been used for treating Fanconi anemia by correcting point mutation in patient-derived fibroblasts (108), as well as in hemophilia for the restoration of factor VIII deficiency in mice (109-112). Notably, CRISPR/Cas9 has been used for the generation of mutant pigs in the vWTF gene to serve as an animal model for von Willebrand disease, or to facilitate bleeding prior to the meat processing in the meat industry (113).
CRISPR/Cas9 in viral diseases
Viruses are obligate intracellular pathogens that infect cells through specific receptors and depend on cellular components of the host for their replication. Upon entering the cell, the viral genome is reproduced, transcribed and translated to complete its life cycle (114). A number of genomes of DNA viruses and retroviruses are integrated into the cellular genome. When a virus infects a human, it can cause severe disease with high mortality, morbidity and/or subsequent transmission to other people. Certain viral infections can be reduced by vaccination immunity, while this is not possible for others. Viral infections that require more attention due to their nature and social impact are those caused by the human papilloma virus (HPV), the human immunodeficiency virus (HIV) and the hepatitis B virus (HBV).
HPV16 expresses variants of the viral oncoproteins E6 and E7, which are tightly linked to the development and maintenance of malignant phenotypes that can result in cervical cancer (93), the second most frequent cause of cancer in women worldwide (92). CRISPR/Cas9 has been used alone or in combination with other treatments in in vitro and in vivo studies to combat HPV16 and HPV18 etiologic factors of cervical cancer (92,93,115). CRISPR-Cas9 targeting HPV16 and HPV18 oncogenes E6 and E7 in cervical carcinoma cell lines, including HeLa and SiHa cells, led to the arrest of the cell cycle and eventual death of the malignant cells (115). In another study, mutation of the HPV16 E6 and E7 viral oncogenes inhibited tumor growth in vivo, demonstrating that treatment with CRISPR/Cas9 works as a therapy with cisplatin, one of the first-line treatments most commonly used as a chemotherapeutic agent (93).
In HIV, the chemokine receptor 5 (CCR5) works as an essential co-receptor to HIV-1; thus, loss of CCR5 receptor function protects against viral infection. CRISPR/Cas9 was used to edit the CCR5 gene in CD4+ cells (116) and in human iPSCs (hiPSCs) cells (89). The mutant hiPSCs differentiated into macrophages that became resistant to trophic CCR5 HIV-1 virus (89).
The persistence of covalently closed circular DNA (cccDNA) of HBV is a major barrier to antiviral therapy for chronic hepatitis B eradication. A cure would require the elimination of persistent cccDNA or removal of the hepatocytic viral load (91). With specific gRNAs against the HBV in multiple studies, the CRISPR/Cas9 system significantly reduced the production of HBV core and surface proteins in Huh-7 (70,91), HepG2 (117), HepG2.2.15 (117,119) and HepG2-H1.3 cell lines (119), which were transfected with a HBV expression vector. Furthermore, in a mouse model, this system cleaved the intrahepatic plasmid containing the HBV genome and facilitated its clearance in vivo, resulting in a reduction of the serum antigen surface levels. This suggests that the CRISPR/Cas9 system was able to disrupt HBV both in vitro and in vivo, indicating its potential in the eradication of persistent infection by this virus (91,118,119). CRISPR/Cas9 has not only been used in the treatment of this virus, but also in the study of cellular mechanisms that lead to carcinogenesis (120).
In the case of hepatitis C virus (HCV) that causes chronic hepatitis, liver cirrhosis and hepatocellular carcinomas, CRISPR/Cas9 has been used in the identification of critical host components for HCV infections (121). In addition, the Epstein-Barr virus (EBV) establishes a persistent infection throughout life in 90-95% of the adults. Although it does not cause disease in healthy carriers, the infection is etiologically associated with lymphoid and epithelial neoplasias, such as Burkitt’s lymphoma, Hodgkin’s disease and nasopharyngeal carcinoma (122). The CRISPR/Cas9 system has been used in vitro against EBV in the Raji cell line, demonstrating a marked reduction in proliferation and viral load, as well as restoring the apoptotic pathway in cells subsequent to treatment (123). Another strategy applied to this virus was the removal of the BART promoter region (558 nt), which is one of two clusters that codes for 22 differentially expressed pre-microRNAs during latent EBV infection and is believed to be involved in epithelial cell transformation (122). This region was deleted in specific human epithelial cell lines with latent EBV infection, including nasopharyngeal carcinoma C666-1 cells. This was tested in order to determine if it is required for infection and transformation of epithelial cells, resulting in the loss of BART miRNA expression and activity. It was identified that EBV performance with pBART deletion was lower in comparison with that of WT virus measured in Raji cells, indicating the importance of miR-BARTs in the viral infection of epithelial cells, and that it will be of great interest to investigate whether they are particularly required for the infection and transformation of epithelial cells (122).
CRISPR/Cas9 in vector diseases
In the case of diseases transmitted by a vector, CRISPR/Cas9 has been used for the study of gene function and for gene drive to eradicate important vector-transmitted diseases, such as dengue, chikungunya, yellow fever and malaria (124,125).
In order to investigate the function of specific genes, studies have been conducted on the Plasmodium sp. genome, the parasite that causes malaria (125-127). Since the parasite resides in the RBCs, the transfection efficiency is lower as it has to cross four membranes (the RBC membrane, parasitophorous membrane, parasite cytoplasm membrane and parasite nuclear membrane). However, using CRISPR/Cas9 technology, 100% efficiency of gene deletion, 22-45% efficiency in tagging and 25% efficiency in nucleotide replacement were achieved, which is a significant advance in new studies of this parasite (125). Furthermore, CRISPR/Cas9 can be directed to the primary vector that transmits the disease, such as Aedes aegypti, which is the primary vector of several viruses. In this study, CRISPR/Cas9 was used to research the genetic and neurological basis of innate chemosensory behavior to achieve stable and precise loss-of-function mutations in five genes (124).
CRISPR/Cas9-mediated gene drive involves stimulating biased inheritance of particular alleles, such as gene knockouts, gene replacements and genetic transformations, in order to alter entire populations of organisms. This methodology is important since several species harm human interests, including human health or agriculture (128). CRISPR/Cas9 as part of a gene drive tool can be used to provide a deleterious trait (such as distorted sex ratio, reduced fertility and chemical sensitivity) (129). For instance, in Anopheles gambiae, the main vector for malaria, the CRISPR/Cas9 system was used to confer a recessive female-sterility phenotype upon disruption of three target genes: AGAP005958, AGAP011377 and AGAP007280 (130). This study demonstrated that disrupting AGAP007280 gene alone was sufficient for a successful gene drive targeting female reproduction in an insect population (130).
CRISPR/Cas9 in cancer
Cancer is a group of diseases characterized by multiple genetic and epigenetic alterations in oncogenes and tumor suppressor genes. Experimentation to manipulate normal and cancerous cell genomes is vital for modeling the disease, as well as for the systematic study of genes involved in the process of initiation, progression and therapeutic response of cancer.
The fast modeling of genetic events has taken on a major relevance due to the need to elucidate the importance of genetic alterations present in human tumors, detected by large-scale sequencing of the cancer genome. This is necessary to discern between passenger mutations that are assumed to not directly affect the tumorigenic process, and those that directly or indirectly induce mutations that promote the transformation of normal cells into cancer cells by mutating oncogenes (promoting gain of function) and/or inactivation of tumor suppressor genes (promoting loss of function) (131).
Through the CRISPR/Cas9 technology, it was identified that NANOG and NANOGP8 genes contribute to the high malignant potential of prostate cancer. Knockouts of NANOG and NANOGP8 in human prostate DU145 cells significantly attenuated the malignant potential, including sphere formation, anchorage independent growth, migration ability and drug resistance, as compared with that of parental DU145 cells. Cell proliferation was not inhibited in vitro, but in immune deficient mice the tumorigenic potential decreased significantly in vivo (132). To assess the impact of BCR-ABL fusion on the leukemic processes in the Boff-p210 cell line, a hematopoietic cell line that is independent of interleukin (IL)-3 and expresses BCR-ABL p210, CRISPR/Cas9 was used to eliminate expression of the p210 oncoprotein. This resulted in the loss of ability of Boff-p210 cell line to grow in the absence of IL-3 and showed a significant increase in apoptosis levels (133). In immunosuppressed mice, the edited BCR/ABL cells developed smaller tumors compared with those originating from the parental Boff-p210 cells (133). Furthermore, a single-cell clone of edited BCR/ABL cells with a unique frameshift mutation (averting expression of the p210 oncoprotein) was unable of develop tumors, similar to the results in the Baf/3 parental line (133).
Although important, prior to CRISPR, the generation of animal models to test anti-cancer agents and undetected drug resistance mechanisms was an expensive and slow process (131). To provide a flexible and effective method to investigate the somatic alterations of loss of function, and their influence in tumorigenesis, CRISPR/Cas9 was used for the disruption of somatic genes, by the individual deletion of PTCH1 or the triple deletion of TRP53, PTEN and NF1 genes in the mouse brain, resulting in the development of medulloblastoma and glioblastoma, respectively (134). In some instances, chromosomal rearrangements serve a central role in the pathogenesis of human cancer, such as the oncogenic fusion between the echinoderm microtubule-associated protein like 4 (EML4) gene and anaplastic lymphoma kinase (ALK) gene. The resulting EML4-ALK oncogene is detected in a subset of human non-small cell lung cancer (NSCLC), and this is clinically relevant since it imparts sensitivity to inhibitors of ALK. CRISPR/Cas9 was used to generate a mouse model of lung cancer driven by the EML4-ALK gene rearrangement. The resulting tumors harbored the EML4-ALK inversion, expressed the EML4-ALK fusion gene, presented typical histopathological and molecular characteristics of human ALK+ NSCLC, and responded to treatment with ALK inhibitors (135).
Currently clinical protocols in cancer are underway to assess the application of the CRISPR/Cas9 technology in lung, prostate, renal, esophageal and bladder cancer, as well as in neoplasias associated with HPV and EBV (Table II; https://clinicaltrials.gov/ct2/results?cond=&term=crispr&cntry=&state=&city=&dist=).
Table II.
Clinical gene editing trials with CRISPR/Cas9.
| Disease | Target | Strategy | Edited cells | ClinicalTrials.gov identifier | Status |
|---|---|---|---|---|---|
| HPV-related malignant neoplasm | HPV E6/E7, 16 and 18 | NHEJ | HPV16 and 18 | NCT03057912 | Not yet recruiting |
| HIV-infected subjects with hematological malignances | CCR5 | NHEJ CD | 34+ | NCT0316435 | Recruiting |
| Relapsed or refractory CD19+ on B-cell leukemia and lymphoma | TCR and B2M | NHEJ | UCART19 | NTC03166878 | Recruiting |
| Advanced esophageal cancer | PD-1 | NHEJ | T cell | NCT03081715 | Recruiting |
| Muscle-invasive bladder cancer stage IV | PD-1 | NHEJ | T cell | NCT02863913 | Not yet recruiting |
| Hormone refractory prostate cancer | PD-1 | NHEJ | T cell | NCT02867345 | Not yet recruiting |
| Metastatic renal cell carcinoma | PD-1 | NHEJ | T cell | NCT02867332 | Not yet recruiting |
| Metastatic non-small cell lung cancer | PD-1 | NHEJ | T cell | NCT02793856 | Recruiting |
| Advanced stage EBV associated malignancies (stage IV gastric carcinoma; stage IV nasopharyngeal carcinoma; T-Cell lymphoma stage IV; stage IV adult Hodgkin lymphoma; stage IV diffuse large B-cell lymphoma) | PD-1 | NHEJ | T cell | NCT03044743 | Recruiting |
HPV, human papilloma virus; HIV, human immunodeficiency virus; EBV, Epstein-Barr virus; CCR5, chemokine receptor type 5; TCR, T-cell receptor; B2M, β-2-microglobulin; PD-1, programmed cell death protein 1; NHEJ, non-homologous end joining.
Autoimmune and inflammatory diseases
The wide range of rheumatic diseases extends from rare monogenic auto-inflammatory diseases to complex polygenic autoimmune diseases (136). High levels of IL-1 characterize monogenic autoinflammatory diseases. Genetically, certain of these syndromes result from mutations in the NLRP3 gene, with autosomal dominant inheritance and variable penetrance (136,137). In the complex polygenic autoimmune diseases, such as rheumatoid arthritis (RA), multiple genetic factors and environmental triggers are involved; for instance, the heritability of RA is considered to be ~65% (138). A number of these diseases are potential clinical targets for CRISPR/Cas9, where this technology could be used for in vitro and in vivo functional genomics studies to elucidate the single and combined role of single nucleotide polymorphisms identified by association studies at the genomic level through the rapid creation of cellular and animal models. Additionally, this technology can be applied in individualized therapy as a tool to correct mutations, in strategies adapted for each patient.
In this sense, CRISPR/Cas9 has been used for the creation of a rat chondrosarcoma cell line that stably expresses Cas9 for the study of complex interactions that regulate function, differentiation and chondrocyte homeostasis, and to examine the role of genes associated with cartilage degenerating diseases (139). CRISPR/Cas9 can accelerate the in vitro and in vivo functional elucidation of the role played by genes in the inflammatory and bone components of these diseases. In this regard, CRISPR/Cas9 technology was applied to a murine macrophage cell line to demonstrate that the RAS-GRP3 gene limits the inflammatory response by activating Rap1 (140). In another study, microRNA (miR)-155 was shown to exert pro-inflammatory and pro-osteoclastogenic effects through the CRISPR/Cas9-induced mutation of the miR-155 binding site to the SHIP1 gene, a negative inflammation regulator (141). In addition, CRISPR/Cas9 technology may also incorporate a cell therapy strategy, for instance in RA, a disease characterized by deregulated responses to pro-inflammatory cytokines such as IL-1 and tumor necrosis factor-α (TNF-α). CRISPR/Cas9 was used to program murine induced iPSCs with the ability to respond to an inflammatory stimulus with potent and autonomously regulated production of anti-cytokines. TNF-a and IL-1 are two of the most potent stimulators of CCL2 gene expression. If an antagonistic gene to TNF-α or IL-1 is placed under the control of the CCL2 promoter, it will respond to cytokine levels and cause a self-regulated inflammatory process. This allows the control of the cell expression of biological therapies (90).
CRISPR/Cas9 in primary immunodeficiency
Primary immunodeficiency (PID) is a group of heterogeneous and rare chronic diseases, in which part of the immune system functions inappropriately or does not function at all. They are caused by numerous genetic defects, of which >230 have been identified with very variable clinical manifestations. Among the PID diseases, severe combined immunodeficiency (SCID) results in a blockage of T cell development with an additional primary or secondary defect in B cells, and natural killer (NK) cells may or may not be affected. The most common form of SCID is the X chromosome-linked syndrome X-SCID, which is caused by mutations in the gene that encode the γ receptor of IL-2 (namely the IL2RG gene) (142).
PID, including SCID, can be treated by allogeneic transplantation of healthy HSCs; however, histocompatibility problems may be faced, together with the risk of acquisition of transmitted diseases (143). These problems can be overcome by the correction of the patient’s own HSCs through inserting a copy of the functional gene by a viral vector. Although there are success cases in clinical trials, serious complications may also occur due to viral integration close to oncogenes and hence, their activation (144,145). A promising alternative is the use of CRISPR/Cas9 to induce a targeted homologous repair, having a clear advantage over traditional gene therapy. Alternatively, in certain immune deficiencies, there is loss-of-function or gain-of-function in genes subjected to a strict control of expression. Examples included the X-linked agammaglobulinemia and the signal transducer and activator of transcription 3 (STAT3) loss-of-function, as well as the STAT3 and STAT1 gain-of-function, or activated PI3K-δ syndrome. These diseases ideally require the correction of the existing gene to restore normal function or regulation, which may be achieved by CRISPR/Cas9 (146).
It is important to mention that the therapeutic efficacy of gene editing depends on several factors, including editing efficiency, which varies significantly depending on the cell type, senescence status, and cell cycle status of the target. Other factors that also influence therapeutic efficiency include cell aptitude, which refers to the feasibility of reaching a therapeutic modification threshold, and the efficient delivery of programmable nuclease system to the target tissue, which is only considered to be effective if the programmable nuclease system arrives safely and efficiently to the nucleus of the target cell. Finally, the specificity of the editing is another important factor, which refers to only editing the target DNA without affecting any other genes (4,147). Since hematopoietic cells are the most common target in immunological diseases, autologous hematopoietic stem cell transplantation is a viable grafting method of cells that are already corrected ex vivo by CRISPR/Cas9, replacing all or part of the hematopoietic stem cell compartment (146).
CRISPR/Cas9 technology was recently applied in iPSCs to develop an in vitro model of Janus kinase 3 deficiency. A blockage in T cell development was eliminated after performing a CRISPR/Cas9-mediated editing (148). Chronic granulomatous disease (CGD) is characterized by severe and persistent infections due to the lack of an anti-pathogenic oxidative burst normally performed by phagocytic cells to contain and eliminate bacterial and fungal growth. CRISPR/Cas9 was used in iPSCs derived from a patient with CGD to correct a single mutation in the CYBB gene intron and to restore oxidative function in phagocytes (149). Additionally, CRISPR/Cas9 was used to delete GT dinucleotide in the NCF1 gene, which encodes human p47phox protein, in an acute myeloid leukemia cell line (PLB-985) to generate a CGD cell model that reacts to the genetic background of the disease in order to be used for preclinical vector tests (150).
CRISPR/Cas9 has been used to generate ‘knockout’ models that had not been susceptible to efficient genetic modification for the study of immune system diseases, such as rabbits with knockout of 1-5 genes at the same time (IL2RG, RAG1, RAG2, TIKI1, and ALB) with efficiencies ranging from 100% (for 1 gene) to 33.3% (for 5 genes) by microinjection into pronuclear stage embryos cytoplasm (151). Rabbits with IL2RG and RAG1 gene knockout are an important animal model of immune deficiencies characterized by the absence of mature T, B and NK cells (151). Other model examples involve hamsters carrying knockouts of the STAT2 gene for the study of viral infections, of the KCNQ1 gene for cardiovascular function investigation and of the PPP1R12C gene for transgenic integration (152). In addition, pigs carrying knockout of the gene coding for the JH region of the IgM heavy chain, which is crucial for the development and differentiation of B cells, resulted in piglets lacking antibody-producing B cells. The generation of a B cell-deficient mutant is the first step in producing human antibody repertoires in large animal models (153).
CRISPR/Cas9 in other immune diseases
Regarding the use of CRISPR/Cas9 to investigate allergic diseases, this technology has been used to examine the role of certain genes, such as the MUC18 gene, which is also known as CD146 or melanoma cell adhesion molecule. Through CRISPR/Cas9, the knockout of MUC18 gene was conducted in human nasal airway epithelial cells, which is the first line of defense against environmental factors, such as pathogens and pollution. This led to a reduced response of IL-8 following stimulation of the Toll-like receptor agonist, suggesting a pro-inflammatory role of MUC18 gene in response to bacterial or viral stimulation (154). Another example is the use of this technology in X-linked hyper IgM syndrome, where CRISPR/Cas9 was used to correct mutations in the CD40 ligand (155). Furthermore, CRISPR/Cas9 was used to induce recombination of IgH chain class changes in desired subclasses in murine and human B cells. It was also used to produce Fab fragments instead of the whole IgH molecule in mouse hybridoma cells, with the aim of a more careful scrutiny of Ig subclasses and novel methods for accessible production of Fab fragments for research or therapeutic uses (156).
Monkeys serve as one of the most valuable animal models for the development of therapeutic strategies due to their close similarities with humans (157). CRISPR/Cas9 was successfully used in the generation of monkeys with knockout in the NR0B1, PPAR-γ and RAG1 genes (158), being able to serve as models of X-linked adrenal hypoplasia congenita and hypogonadotropic hypogonadism (159), lipodystrophy metabolism (160), insulin sensitivity, obesity and inflammatory disease (161). Furthermore, monkey knockout models of RAG gene could be used in regenerative medicine, allograft and xenograft transplantation, and reconstitution experiments associated with the immune system (162).
CRISPR/Cas9 in other diseases
The CRISPR/Cas9 system has been applied in cellular and animal models to study and search treatments for different neurological disorders, such as Parkinson’s disease (163). Mutation in PARK2 or PINK1 genes leads to early onset Parkinson’s disease, as an autosomal recessive disease in humans (164). The PARK2 gene encodes a protein called parkin, which is a component of the multiprotein complex E3 ubiquitin ligase, while the PINK1 gene encodes for the PTEN-induced putative kinase 1, a mitochondrial serine/threonine kinase protein. The CRISPR/Cas9 system has also been applied in amyotrophic lateral sclerosis (165,166), Huntington’s disease (167), schizophrenia (168) and autism (168). It has also been applied in movement diseases, such as Duchenne muscular dystrophy (169-172), in metabolic diseases, such as type I diabetes (173) and hypercholesterolemia (174), and in inherited diseases that affect vision, such as retinitis pigmentosa (175), among numerous others.
4. Conclusion
It is expected that CRISPR/Cas9 technology will soon be validated through ex vivo clinical protocols already in course in humans to test against various types of cancer. In the meantime, it is fully applicable to cell therapy experiments and deserves to be vigorously developed as a research tool in an unusually diverse range of biological systems. This will allow the necessary improvements for future biomedical technologies and applications.
Acknowledgments
The authors would like to thank Alec Christopher Escalante Gomez (School of Nutrition of the Universidad Autónoma de Nuevo León, Monterrey) for aid in the translation of this review and Jessica García for valuable suggestions.
Funding
Authors wish to acknowledge Tecnologico de Monterrey and Mission XXI for their financial support regarding the publication charges.
Availability of data and materials
Not applicable.
Authors’ contributions
DRRR reviewed the basic science literature and wrote the manuscript; MAGE reviewed the clinical issues; RRS and MDLGR supported DRRR in revisions of the manuscript; HABS conceived the project and supervised the writing of the manuscript.
Ethics approval and consent to participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Im W, Moon J, Kim M. Applications of CRISPR/Cas9 for gene editing in hereditary movement disorders. J Mov Disord. 2016;9:136–143. doi: 10.14802/jmd.16029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gaj T, Gersbach CA, Barbas CF., III ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31:397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nat Biotechnol. 2014;32:347–355. doi: 10.1038/nbt.2842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cox DBT, Platt RJ, Zhang F. Therapeutic genome editing: Prospects and challenges. Nat Med. 2015;21:121–131. doi: 10.1038/nm.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Epinat JC, Arnould S, Chames P, Rochaix P, Desfontaines D, Puzin C, Patin A, Zanghellini A, Pâques F, Lacroix E. A novel engineered meganuclease induces homologous recombination in yeast and mammalian cells. Nucleic Acids Res. 2003;31:2952–2962. doi: 10.1093/nar/gkg375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Urnov FD, Rebar EJ, Holmes MC, Zhang HS, Gregory PD. Genome editing with engineered zinc finger nucleases. Nat Rev Genet. 2010;11:636–646. doi: 10.1038/nrg2842. [DOI] [PubMed] [Google Scholar]
- 7.Miller JC, Tan S, Qiao G, Barlow KA, Wang J, Xia DF, Meng X, Paschon DE, Leung E, Hinkley SJ, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29:143–148. doi: 10.1038/nbt.1755. [DOI] [PubMed] [Google Scholar]
- 8.Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science. 2012;337:816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327:167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 10.Sorek R, Kunin V, Hugenholtz P. CRISPR-a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6:181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 11.Singh V, Braddick D, Dhar PK. Exploring the potential of genome editing CRISPR-Cas9 technology. Gene. 2017;599:1–18. doi: 10.1016/j.gene.2016.11.008. [DOI] [PubMed] [Google Scholar]
- 12.Makarova KS, Wolf YI, Alkhnbashi OS, Costa F, Shah SA, Saunders SJ, Barrangou R, Brouns SJ, Charpentier E, Haft DH, et al. An updated evolutionary classification of CRISPR-Cas systems. Nat Rev Microbiol. 2015;13:722–736. doi: 10.1038/nrmicro3569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mojica FJ, Ferrer C, Juez G, Rodríguez-Valera F. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17:85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 14.Riehle MM, Bennett AF, Long AD. Genetic architecture of thermal adaptation in Escherichia coli. Proc Natl Acad Sci USA. 2001;98:525–530. doi: 10.1073/pnas.98.2.525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.DeBoy RT, Mongodin EF, Emerson JB, Nelson KE. Chromosome evolution in the Thermotogales: Large-scale inversions and strain diversification of CRISPR sequences. J Bacteriol. 2006;188:2364–2374. doi: 10.1128/JB.188.7.2364-2374.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Makarova KS, Aravind L, Grishin NV, Rogozin IB, Koonin EV. A DNA repair system specific for thermophilic Archaea and bacteria predicted by genomic context analysis. Nucleic Acids Res. 2002;30:482–496. doi: 10.1093/nar/30.2.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ishino Y, Shinagawa H, Makino K, Amemura M, Nakata A. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169:5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mojica FJ, Díez-Villaseñor C, Soria E, Juez G. Biological significance of a family of regularly spaced repeats in the genomes of Archaea, Bacteria and mitochondria. Mol Microbiol. 2000;36:244–246. doi: 10.1046/j.1365-2958.2000.01838.x. [DOI] [PubMed] [Google Scholar]
- 19.Jansen R, Embden JD, Gaastra W, Schouls LM. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43:1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 20.Mojica FJ, Díez-Villaseñor C, García-Martínez J, Soria E. Intervening sequences of regularly spaced prokaryotic repeats derive from foreign genetic elements. J Mol Evol. 2005;60:174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 21.Bolotin A, Quinquis B, Sorokin A, Ehrlich SD. Clustered regularly interspaced short palindrome repeats (CRISPRs) have spacers of extrachromosomal origin. Microbiology. 2005;151:2551–2561. doi: 10.1099/mic.0.28048-0. [DOI] [PubMed] [Google Scholar]
- 22.Pourcel C, Salvignol G, Vergnaud G. CRISPR elements in Yersinia pestis acquire new repeats by preferential uptake of bacteriophage DNA, and provide additional tools for evolutionary studies. Microbiology. 2005;151:653–663. doi: 10.1099/mic.0.27437-0. [DOI] [PubMed] [Google Scholar]
- 23.Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, Romero DA, Horvath P. CRISPR provides acquired resistance against viruses in prokaryotes. Science. 2007;315:1709–1712. doi: 10.1126/science.1138140. [DOI] [PubMed] [Google Scholar]
- 24.Brouns SJJ, Jore MM, Lundgren M, Westra ER, Slijkhuis RJH, Snijders APL, Dickman MJ, Makarova KS, Koonin EV, van der Oost J. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321:960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garneau JE, Dupuis MÈ, Villion M, Romero DA, Barrangou R, Boyaval P, Fremaux C, Horvath P, Magadán AH, Moineau S. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468:67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 26.Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, Eckert MR, Vogel J, Charpentier E. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011;471:602–607. doi: 10.1038/nature09886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sapranauskas R, Gasiunas G, Fremaux C, Barrangou R, Horvath P, Siksnys V. The Streptococcus thermophilus CRISPR/Cas system provides immunity in Escherichia coli. Nucleic Acids Res. 2011;39:9275–9282. doi: 10.1093/nar/gkr606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, Norville JE, Church GM. RNA-guided human genome engineering via Cas9. Science. 2013;339:823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cornu TI, Mussolino C, Cathomen T. Refining strategies to translate genome editing to the clinic. Nat Med. 2017;23:415–423. doi: 10.1038/nm.4313. [DOI] [PubMed] [Google Scholar]
- 31.Cyranoski D. CRISPR gene-editing tested in a person for the first time. Nature. 2016;539:479–479. doi: 10.1038/nature.2016.20988. [DOI] [PubMed] [Google Scholar]
- 32.Cyranoski D. Chinese scientists to pioneer first human CRISPR trial. Nature. 2016;535:476–477. doi: 10.1038/nature.2016.20302. [DOI] [PubMed] [Google Scholar]
- 33.Shub DA, Goodrich-Blair H, Eddy SR. Amino-acid-sequence motif of group I intron endonucleases is conserved in open reading frames of group II introns. Trends Biochem Sci. 1994;19:402–404. doi: 10.1016/0968-0004(94)90086-8. [DOI] [PubMed] [Google Scholar]
- 34.Al-Attar S, Westra ER, van der Oost J, Brouns SJ. Clustered regularly interspaced short palindromic repeats (CRISPRs): The hallmark of an ingenious antiviral defense mechanism in prokaryotes. Biol Chem. 2011;392:277–289. doi: 10.1515/bc.2011.042. [DOI] [PubMed] [Google Scholar]
- 35.Wright AV, Nuñez JK, Doudna JA. Biology and applications of CRISPR systems: Harnessing nature’s toolbox for genome engineering. Cell. 2016;164:29–44. doi: 10.1016/j.cell.2015.12.035. [DOI] [PubMed] [Google Scholar]
- 36.Zhang F, Wen Y, Guo X. CRISPR/Cas9 for genome editing: Progress implications and challenges. Hum Mol Genet. 2014;23:R40–R46. doi: 10.1093/hmg/ddu125. [DOI] [PubMed] [Google Scholar]
- 37.Canver MC, Bauer DE, Orkin SH. Functional interrogation of non-coding DNA through CRISPR genome editing. Methods. 2017:121–122. 118–129. doi: 10.1016/j.ymeth.2017.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Karvelis T, Gasiunas G, Miksys A, Barrangou R, Horvath P, Siksnys V. crRNA and tracrRNA guide Cas9-mediated DNA interference in Streptococcus thermophilus. RNA Biol. 2013;10:841–851. doi: 10.4161/rna.24203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gasiunas G, Barrangou R, Horvath P, Siksnys V. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109:E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Friedland AE, Baral R, Singhal P, Loveluck K, Shen S, Sanchez M, Marco E, Gotta GM, Maeder ML, Kennedy EM, et al. Characterization of Staphylococcus aureus Cas9: A smaller Cas9 for all-in-one adeno-associated virus delivery and paired nickase applications. Genome Biol. 2015;16:257. doi: 10.1186/s13059-015-0817-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, Zetsche B, Shalem O, Wu X, Makarova KS, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520:186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, Thomson JA. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci USA. 2013;110:15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen F, Ding X, Feng Y, Seebeck T, Jiang Y, Davis GD. Targeted activation of diverse CRISPR-Cas systems for mammalian genome editing via proximal CRISPR targeting. Nat Commun. 2017;8:14958. doi: 10.1038/ncomms14958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Price AA, Sampson TR, Ratner HK, Grakoui A, Weiss DS. Cas9-mediated targeting of viral RNA in eukaryotic cells. Proc Natl Acad Sci USA. 2015;112:6164–6169. doi: 10.1073/pnas.1422340112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Murovec J, Pirc Ž, Yang B. New variants of CRISPR RNA-guided genome editing enzymes. Plant Biotechnol J. 2017;15:917–926. doi: 10.1111/pbi.12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oliveros JC, Franch M, Tabas-Madrid D, San-León D, Montoliu L, Cubas P, Pazos F. Breaking-Cas-interactive design of guide RNAs for CRISPR-Cas experiments for ENSEMBL genomes. Nucleic Acids Res. 2016;44:W267–W271. doi: 10.1093/nar/gkw407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kaur K, Gupta AK, Rajput A, Kumar M. ge-CRISPR-an integrated pipeline for the prediction and analysis of sgRNAs genome editing efficiency for CRISPR/Cas system. Sci Rep. 2016;6:30870. doi: 10.1038/srep30870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Naito Y, Hino K, Bono H, Ui-Tei K. CRISPRdirect: Software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics. 2015;31:1120–1123. doi: 10.1093/bioinformatics/btu743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Heigwer F, Kerr G, Boutros M. E-CRISP: Fast CRISPR target site identification. Nat Methods. 2014;11:122–123. doi: 10.1038/nmeth.2812. [DOI] [PubMed] [Google Scholar]
- 50.Wong N, Liu W, Wang X. WU-CRISPR: Characteristics of functional guide RNAs for the CRISPR/Cas9 system. Genome Biol. 2015;16:218. doi: 10.1186/s13059-015-0784-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Doench JG, Hartenian E, Graham DB, Tothova Z, Hegde M, Smith I, Sullender M, Ebert BL, Xavier RJ, Root DE. Rational design of highly active sgRNAs for CRISPR-Cas9-mediated gene inactivation. Nat Biotechnol. 2014;32:1262–1267. doi: 10.1038/nbt.3026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chari R, Mali P, Moosburner M, Church GM. Unraveling CRISPR-Cas9 genome engineering parameters via a library-on-library approach. Nat Methods. 2015;12:823–826. doi: 10.1038/nmeth.3473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chari R, Yeo NC, Chavez A, Church GM. sgRNA scorer 2.0: A species-independent model to predict CRISPR/Cas9 activity. ACS Synth Biol. 2017;6:902–904. doi: 10.1021/acssynbio.6b00343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Moreno-Mateos MA, Vejnar CE, Beaudoin JD, Fernandez JP, Mis EK, Khokha MK, Giraldez AJ. CRISPRscan: Designing highly efficient sgRNAs for CRISPR-Cas9 targeting in vivo. Nat Methods. 2015;12:982–988. doi: 10.1038/nmeth.3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu H, Wei Z, Dominguez A, Li Y, Wang X, Qi LS. CRISPR-ERA: A comprehensive design tool for CRISPR-mediated gene editing, repression and activation. Bioinformatics. 2015;31:3676–3678. doi: 10.1093/bioinformatics/btv423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stemmer M, Thumberger T, Del Sol, Keyer M, Wittbrodt J, Mateo JL. CCTop: An intuitive, flexible and reliable CRISPR/Cas9 target prediction tool. PLoS One. 2015;10:e0124633. doi: 10.1371/journal.pone.0124633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Haeussler M, Schönig K, Eckert H, Eschstruth A, Mianné J, Renaud JB, Schneider-Maunoury S, Shkumatava A, Teboul L, Kent J, et al. Evaluation of off-target and on-target scoring algorithms and integration into the guide RNA selection tool CRISPOR. Genome Biol. 2016;17:148. doi: 10.1186/s13059-016-1012-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E. CHOPCHOP: A CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res. 2014;42:W401–W407. doi: 10.1093/nar/gku410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Prykhozhij SV, Rajan V, Gaston D, Berman JN. CRISPR multitargeter: A web tool to find common and unique CRISPR single guide RNA targets in a set of similar sequences. PLoS One. 2015;10:e0119372. doi: 10.1371/journal.pone.0119372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.O’Brien A, Bailey TL. GT-Scan: Identifying unique genomic targets. Bioinformatics. 2014;30:2673–2675. doi: 10.1093/bioinformatics/btu354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, Li Y, Fine EJ, Wu X, Shalem O, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31:827–832. doi: 10.1038/nbt.2647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Park J, Bae S, Kim JS. Cas-Designer: A web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics. 2015;31:4014–4016. doi: 10.1093/bioinformatics/btv537. [DOI] [PubMed] [Google Scholar]
- 63.Bae S, Park J, Kim JS. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics. 2014;30:1473–1475. doi: 10.1093/bioinformatics/btu048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cradick TJ, Qiu P, Lee CM, Fine EJ, Bao G. COSMID: A web-based tool for identifying and validating CRISPR/Cas off-target sites. Mol Ther Nucleic Acids. 2014;3:e214. doi: 10.1038/mtna.2014.64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hough SH, Kancleris K, Brody L, Humphryes-Kirilov N, Wolanski J, Dunaway K, Ajetunmobi A, Dillard V. Guide Picker is a comprehensive design tool for visualizing and selecting guides for CRISPR experiments. BMC Bioinformatics. 2017;18:167. doi: 10.1186/s12859-017-1581-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Güell M, Yang L, Church GM. Genome editing assessment using CRISPR genome analyzer (CRISPR-GA) Bioinformatics. 2014;30:2968–2970. doi: 10.1093/bioinformatics/btu427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cao J, Wu L, Zhang SM, Lu M, Cheung WK, Cai W, Gale M, Xu Q, Yan Q. An easy and efficient inducible CRISPR/Cas9 platform with improved specificity for multiple gene targeting. Nucleic Acids Res. 2016;44:e149. doi: 10.1093/nar/gkw660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, Scott DA, Inoue A, Matoba S, Zhang Y, Zhang F. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154:1380–1389. doi: 10.1016/j.cell.2013.08.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sato T, Sakuma T, Yokonishi T, Katagiri K, Kamimura S, Ogonuki N, Ogura A, Yamamoto T, Ogawa T. Genome editing in mouse spermatogonial stem cell lines Using TALEN and double-nicking CRISPR/Cas9. Stem Cell Reports. 2015;5:75–82. doi: 10.1016/j.stemcr.2015.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sakuma T, Masaki K, Abe-Chayama H, Mochida K, Yamamoto T, Chayama K. Highly multiplexed CRISPR-Cas9-nuclease and Cas9-nickase vectors for inactivation of hepatitis B virus. Genes Cells. 2016;21:1253–1262. doi: 10.1111/gtc.12437. [DOI] [PubMed] [Google Scholar]
- 71.Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Tsai SQ, Wyvekens N, Khayter C, Foden JA, Thapar V, Reyon D, Goodwin MJ, Aryee MJ, Joung JK. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32:569–576. doi: 10.1038/nbt.2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zhang H, Yan Z, Li M, Peabody M, He TC. CRISPR clear? Dimeric Cas9-Fok1 nucleases improve genome-editing specificity. Genes Dis. 2014;1:6–7. doi: 10.1016/j.gendis.2014.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32:577–582. doi: 10.1038/nbt.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wright AV, Sternberg SH, Taylor DW, Staahl BT, Bardales JA, Kornfeld JE, Doudna JA. Rational design of a split-Cas9 enzyme complex. Proc Natl Acad Sci USA. 2015;112:2984–2989. doi: 10.1073/pnas.1501698112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chew WL, Tabebordbar M, Cheng JK, Mali P, Wu EY, Ng AH, Zhu K, Wagers AJ, Church GM. A multifunctional AAV-CRISPR-Cas9 and its host response. Nat Methods. 2016;13:868–874. doi: 10.1038/nmeth.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420–424. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Murugan K, Babu K, Sundaresan R, Rajan R, Sashital DG. The revolution continues: Newly discovered systems expand the CRISPR-Cas toolkit. Mol Cell. 2017;68:15–25. doi: 10.1016/j.molcel.2017.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shmakov S, Abudayyeh OO, Makarova KS, Wolf YI, Gootenberg JS, Semenova E, Minakhin L, Joung J, Konermann S, Severinov K, et al. Discovery and functional characterization of diverse class 2 CRISPR-Cas systems. Mol Cell. 2015;60:385–397. doi: 10.1016/j.molcel.2015.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Abudayyeh OO, Gootenberg JS, Konermann S, Joung J, Slaymaker IM, Cox DB, Shmakov S, Makarova KS, Semenova E, Minakhin L, et al. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. Science. 2016;353:aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Abudayyeh OO, Gootenberg JS, Essletzbichler P, Han S, Joung J, Belanto JJ, Verdine V, Cox DBT, Kellner MJ, Regev A, et al. RNA targeting with CRISPR-Cas13. Nature. 2017;550:280–284. doi: 10.1038/nature24049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Cox DBT, Gootenberg JS, Abudayyeh OO, Franklin B, Kellner MJ, Joung J, Zhang F. RNA editing with CRISPR-Cas13. Science. 2017;358:1019–1027. doi: 10.1126/science.aaq0180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Balboa D, Weltner J, Eurola S, Trokovic R, Wartiovaara K, Otonkoski T. Conditionally stabilized dCas9 activator for controlling gene expression in human cell reprogramming and differentiation. Stem Cell Reports. 2015;5:448–459. doi: 10.1016/j.stemcr.2015.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, Hsu PD, Habib N, Gootenberg JS, Nishimasu H, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517:583–588. doi: 10.1038/nature14136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Piatek A, Ali Z, Baazim H, Li L, Abulfaraj A, Al-Shareef S, Aouida M, Mahfouz MM. RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol J. 2015;13:578–589. doi: 10.1111/pbi.12284. [DOI] [PubMed] [Google Scholar]
- 86.Fu Y, Rocha PP, Luo VM, Raviram R, Deng Y, Mazzoni EO, Skok JA. CRISPR-dCas9 and sgRNA scaffolds enable dual-colour live imaging of satellite sequences and repeat-enriched individual loci. Nat Commun. 2016;7:11707. doi: 10.1038/ncomms11707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Qin P, Parlak M, Kuscu C, Bandaria J, Mir M, Szlachta K, Singh R, Darzacq X, Yildiz A, Adli M. Live cell imaging of low- and non-repetitive chromosome loci using CRISPR-Cas9. Nat Commun. 2017;8:14725. doi: 10.1038/ncomms14725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pankowicz FP, Barzi M, Legras X, Hubert L, Mi T, Tomolonis JA, Ravishankar M, Sun Q, Yang D, Borowiak M, et al. Reprogramming metabolic pathways in vivo with CRISPR/Cas9 genome editing to treat hereditary tyrosinaemia. Nat Commun. 2016;7:12642. doi: 10.1038/ncomms12642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kang H, Minder P, Park MA, Mesquitta WT, Torbett BE, Slukvin II. CCR5 disruption in induced pluripotent stem cells using CRISPR/Cas9 provides selective resistance of immune cells to CCR5-tropic HIV-1 virus. Mol Ther Nucleic Acids. 2015;4:e268. doi: 10.1038/mtna.2015.42. [DOI] [PubMed] [Google Scholar]
- 90.Brunger JM, Zutshi A, Willard VP, Gersbach CA, Guilak F. Genome engineering of stem cells for autonomously regulated, closed-loop delivery of biologic drugs. Stem Cell Reports. 2017;8:1202–1213. doi: 10.1016/j.stemcr.2017.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Lin SR, Yang HC, Kuo YT, Liu CJ, Yang TY, Sung KC, Lin YY, Wang HY, Wang CC, Shen YC, et al. The CRISPR/Cas9 system facilitates clearance of the intrahepatic HBV templates in vivo. Mol Ther Nucleic Acids. 2014;3:e186. doi: 10.1038/mtna.2014.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zhen S, Hua L, Takahashi Y, Narita S, Liu YH, Li Y. In vitro and in vivo growth suppression of human papillomavirus 16-positive cervical cancer cells by CRISPR/Cas9. Biochem Biophys Res Commun. 2014;450:1422–1426. doi: 10.1016/j.bbrc.2014.07.014. [DOI] [PubMed] [Google Scholar]
- 93.Zhen S, Lu JJ, Wang LJ, Sun XM, Zhang JQ, Li X, Luo WJ, Zhao L. In vitro and in vivo synergistic therapeutic effect of cisplatin with human papillomavirus16 E6/E7 CRISPR/Cas9 on cervical cancer cell line. Transl Oncol. 2016;9:498–504. doi: 10.1016/j.tranon.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Das D, Smith N, Wang X, Morgan IM. The deacetylase SIRT1 regulates the replication properties of human papilloma-virus 16 E1 and E2. J Virol. 2017;91:e00102-e00117. doi: 10.1128/JVI.00102-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Merling R, Kuhns D, Sweeney C, Wu X, Burkett S, Chu J, Lee J, Koontz S, Di Pasquale G, Afione S, et al. Gene-edited pseudogene resurrection corrects p47 phox-deficient chronic granulomatous disease. Blood Adv. 2016;1:270–278. doi: 10.1182/bloodadvances.2016001214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M. Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature. 2016;533:125–129. doi: 10.1038/nature17664. [DOI] [PubMed] [Google Scholar]
- 97.Schwank G, Koo BK, Sasselli V, Dekkers JF, Heo I, Demircan T, Sasaki N, Boymans S, Cuppen E, van der Ent CK, et al. Functional repair of CFTR by CRISPR/Cas9 in intestinal stem cell organoids of cystic fibrosis patients. Cell Stem Cell. 2013;13:653–658. doi: 10.1016/j.stem.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 98.Yin H, Xue W, Chen S, Bogorad RL, Benedetti E, Grompe M, Koteliansky V, Sharp PA, Jacks T, Anderson DG. Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol. 2014;32:551–553. doi: 10.1038/nbt.2884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gui H, Schriemer D, Cheng WW, Chauhan RK, Antiňolo G, Berrios C, Bleda M, Brooks AS, Brouwer RW, Burns AJ, et al. Whole exome sequencing coupled with unbiased functional analysis reveals new Hirschsprung disease genes. Genome Biol. 2017;18:48. doi: 10.1186/s13059-017-1174-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Halim D, Wilson MP, Oliver D, Brosens E, Verheij JB, Han Y, Nanda V, Lyu Q, Doukas M, Stoop H, et al. Loss of LMOD1 impairs smooth muscle cytocontractility and causes megacystis microcolon intestinal hypoperistalsis syndrome in humans and mice. Proc Natl Acad Sci USA. 2017;114:E2739–E2747. doi: 10.1073/pnas.1620507114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Dever DP, Bak RO, Reinisch A, Camarena J, Washington G, Nicolas CE, Pavel-Dinu M, Saxena N, Wilkens AB, Mantri S, et al. CRISPR/Cas9 β-globin gene targeting in human haematopoietic stem cells. Nature. 2016;539:384–389. doi: 10.1038/nature20134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ou Z, Niu X, He W, Chen Y, Song B, Xian Y, Fan D, Tang D, Sun X. The combination of CRISPR/Cas9 and iPSC technologies in the gene therapy of human β-thalassemia in mice. Sci Re. 2016;6:32463. doi: 10.1038/srep32463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xie F, Ye L, Chang JC, Beyer AI, Wang J, Muench MO, Kan YW. Seamless gene correction of β-thalassemia mutations in patient-specific iPSCs using CRISPR/Cas9 and piggyBac. Genome Res. 2014;24:1526–1533. doi: 10.1101/gr.173427.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Xu P, Tong Y, Liu XZ, Wang TT, Cheng L, Wang BY, Lv X, Huang Y, Liu DP. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2 654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci Rep. 2015;5:12065. doi: 10.1038/srep12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Traxler EA, Yao Y, Wang YD, Woodard KJ, Kurita R, Nakamura Y, Hughes JR, Hardison RC, Blobel GA, Li C, Weiss MJ. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med. 2016;22:987–990. doi: 10.1038/nm.4170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Canver MC, Smith EC, Sher F, Pinello L, Sanjana NE, Shalem O, Chen DD, Schupp PG, Vinjamur DS, Garcia SP, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192–197. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Zhang N, Zhi H, Curtis BR, Rao S, Jobaliya C, Poncz M, French DL, Newman PJ. CRISPR/Cas9-mediated conversion of human platelet alloantigen allotypes. Blood. 2016;127:675–680. doi: 10.1182/blood-2015-10-675751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Osborn MJ, Gabriel R, Webber BR, DeFeo AP, McElroy AN, Jarjour J, Starker CG, Wagner JE, Joung JK, Voytas DF, et al. Fanconi anemia gene editing by the CRISPR/Cas9 system. Hum Gene Ther. 2015;26:114–126. doi: 10.1089/hum.2014.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Park CY, Kim DH, Son JS, Sung JJ, Lee J, Bae S, Kim JH, Kim DW, Kim JS. Functional correction of large factor VIII gene chromosomal inversions in hemophilia a patient-derived iPSCs using CRISPR-Cas9. Cell Stem Cell. 2015;17:213–220. doi: 10.1016/j.stem.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 110.Zhang H, McCarty N. CRISPR-Cas9 technology and its application in haematological disorders. Br J Haematol. 2016;175:208–225. doi: 10.1111/bjh.14297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Chen J, Jiang N, Wang T, Xie G, Zhang Z, Li H, Yuan J, Sun Z, Chen J. DNA shuffling of uricase gene leads to a more ‘human like’ chimeric uricase with increased uricolytic activity. Int J Biol Macromol. 2016;82:522–529. doi: 10.1016/j.ijbiomac.2015.10.053. [DOI] [PubMed] [Google Scholar]
- 112.Guan Y, Ma Y, Li Q, Sun Z, Ma L, Wu L, Wang L, Zeng L, Shao Y, Chen Y, et al. CRISPR/Cas9-mediated somatic correction of a novel coagulator factor IX gene mutation ameliorates hemophilia in mouse. EMBO Mol Med. 2016;8:477–488. doi: 10.15252/emmm.201506039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Hai T, Teng F, Guo R, Li W, Zhou Q. One-step generation of knockout pigs by zygote injection of CRISPR/Cas system. Cell Res. 2014;24:372–375. doi: 10.1038/cr.2014.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Puschnik AS, Majzoub K, Ooi YS, Carette JE. A CRISPR toolbox to study virus-host interactions. Nat Rev Microbiol. 2017;15:351–364. doi: 10.1038/nrmicro.2017.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Kennedy EM, Kornepati AVR, Goldstein M, Bogerd HP, Poling BC, Whisnant AW, Kastan MB, Cullen BR. Inactivation of the human papillomavirus E6 or E7 gene in cervical carcinoma cells by using a bacterial CRISPR/Cas RNA-guided endonuclease. J Virol. 2014;88:11965–11972. doi: 10.1128/JVI.01879-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Wang W, Ye C, Liu J, Zhang D, Kimata JT, Zhou P. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9:e115987. doi: 10.1371/journal.pone.0115987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Zhen S, Hua L, Liu YH, Gao LC, Fu J, Wan DY, Dong LH, Song HF, Gao X. Harnessing the clustered regularly inter-spaced short palindromic repeat (CRISPR)/CRISPR-associated Cas9 system to disrupt the hepatitis B virus. Gene Ther. 2015;22:404–412. doi: 10.1038/gt.2015.2. [DOI] [PubMed] [Google Scholar]
- 118.Ramanan V, Shlomai A, Cox DBT, Schwartz RE, Michailidis E, Bhatta A, Scott DA, Zhang F, Rice CM, Bhatia SN. CRISPR/Cas9 cleavage of viral DNA efficiently suppresses hepatitis B virus. Sci Rep. 2015;5:10833. doi: 10.1038/srep10833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Karimova M, Beschorner N, Dammermann W, Chemnitz J, Indenbirken D, Bockmann JH, Grundhoff A, Lüth S, Buchholz F, Schulze zur Wiesch J, Hauber J. CRISPR/Cas9 nickase-mediated disruption of hepatitis B virus open reading frame S and X. Sci Rep. 2015;5:13734. doi: 10.1038/srep13734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Qi Xu Y, Luo Y, Yang J, Xie J, Deng Q, Su C, Wei N, Shi W, Xu DF, et al. Hepatitis B virus X protein stimulates proliferation, wound closure and inhibits apoptosis of HuH-7 cells via CDC42. Int J Mol Sci. 2017;18:E586. doi: 10.3390/ijms18030586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ren Q, Li C, Yuan P, Cai C, Zhang L, Luo GG, Wei W. A dual-reporter system for real-time monitoring and high-throughput CRISPR/Cas9 library screening of the hepatitis C virus. Sci Rep. 2015;5:8865. doi: 10.1038/srep08865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yuen KS, Chan CP, Wong N-HM, Ho CH, Ho TH, Lei T, Deng W, Tsao SW, Chen H, Kok KH, Jin DY. CRISPR/Cas9-mediated genome editing of Epstein-Barr virus in human cells. J Gen Virol. 2015;96:626–636. doi: 10.1099/jgv.0.000012. [DOI] [PubMed] [Google Scholar]
- 123.Wang J, Quake SR. RNA-guided endonuclease provides a therapeutic strategy to cure latent herpesviridae infection. Proc Natl Acad Sci USA. 2014;111:13157–13162. doi: 10.1073/pnas.1410785111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kistler KE, Vosshall LB, Matthews BJ. Genome engineering with CRISPR-Cas9 in the mosquito Aedes aegypti. Cell Rep. 2015;11:51–60. doi: 10.1016/j.celrep.2015.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Zhang C, Xiao B, Jiang Y, Zhao Y, Li Z, Gao H, Ling Y, Wei J, Li S, Lu M, et al. Efficient editing of malaria parasite genome using the CRISPR/Cas9 system. MBio. 2014;5:e01414-14. doi: 10.1128/mBio.01414-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wagner JC, Platt RJ, Goldfless SJ, Zhang F, Niles JC. Efficient CRISPR/Cas9-mediated genome editing in P. falciparum. Nat Methods. 2014;11:915–918. doi: 10.1038/nmeth.3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ. Genome editing in the human malaria parasite Plasmodium falciparum using the crisPr-cas9 system. Nat Biotechnol. 2014;32:819–821. doi: 10.1038/nbt.2925. [DOI] [PubMed] [Google Scholar]
- 128.Burt A. Site-specific selfish genes as tools for the control and genetic engineering of natural populations. Proc Biol Sci. 2003;270:921–928. doi: 10.1098/rspb.2002.2319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Webber BL, Raghu S, Edwards OR. Opinion: Is CRISPR-based gene drive a biocontrol silver bullet or global conservation threat? Proc Natl Acad Sci USA. 2015;112:10565–10567. doi: 10.1073/pnas.1514258112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Hammond A, Galizi R, Kyrou K, Simoni A, Siniscalchi C, Katsanos D, Gribble M, Baker D, Marois E, Russell S, et al. A CRISPR-Cas9 gene drive system targeting female reproduction in the malaria mosquito vector Anopheles gambiae. Nat Biotechnol. 2016;34:78–83. doi: 10.1038/nbt.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Sánchez-Rivera FJ, Jacks T. Applications of the CRISPR-Cas9 system in cancer biology. Nat Rev Cancer. 2015;15:387–395. doi: 10.1038/nrc3950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Kawamura N, Nimura K, Nagano H, Yamaguchi S, Nonomura N, Kaneda Y. CRISPR/Cas9-mediated gene knockout of NANOG and NANOGP8 decreases the malignant potential of prostate cancer cells. Oncotarget. 2015;6:22361–22374. doi: 10.18632/oncotarget.4293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.García-Tuñón I, Hernández-Sánchez M, Ordoñez JL, Alonso-Pérez V, Álamo-Quijada M, Benito R, Guerrero C, Hernández-Rivas JM, Sánchez-Martín M. The CRISPR/Cas9 system efficiently reverts the tumorigenic ability of BCR/ABL in vitro and in a xenograft model of chronic myeloid leukemia. Oncotarget. 2017;8:26027–26040. doi: 10.18632/oncotarget.15215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zuckermann M, Hovestadt V, Knobbe-Thomsen CB, Zapatka M, Northcott PA, Schramm K, Belic J, Jones DT, Tschida B, Moriarity B, et al. Somatic CRISPR/Cas9-mediated tumour suppressor disruption enables versatile brain tumour modelling. Nat Commun. 2015;6:7391. doi: 10.1038/ncomms8391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Maddalo D, Manchado E, Concepcion CP, Bonetti C, Vidigal JA, Han YC, Ogrodowski P, Crippa A, Rekhtman N, de Stanchina E, et al. In vivo engineering of oncogenic chromosomal rearrangements with the CRISPR/Cas9 system. Nature. 2014;516:423–427. doi: 10.1038/nature13902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Pathak S, McDermott MF, Savic S. Autoinflammatory diseases: Update on classification diagnosis and management. J Clin Pathol. 2017;70:1–8. doi: 10.1136/jclinpath-2016-203810. [DOI] [PubMed] [Google Scholar]
- 137.Sá DC. Inflammasomes and dermatology. An Bras Dermatol. 2016;91:566–578. doi: 10.1590/abd1806-4841.20165577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Kim K, Bang SY, Lee HS, Bae SC. Update on the genetic architecture of rheumatoid arthritis. Nat Rev Rheumatol. 2017;13:13–24. doi: 10.1038/nrrheum.2016.176. [DOI] [PubMed] [Google Scholar]
- 139.Yang M, Zhang L, Stevens J, Gibson G. CRISPR/Cas9 mediated generation of stable chondrocyte cell lines with targeted gene knockouts; analysis of an aggrecan knockout cell line. Bone. 2014;69:118–125. doi: 10.1016/j.bone.2014.09.005. [DOI] [PubMed] [Google Scholar]
- 140.Tang S, Chen T, Yu Z, Zhu X, Yang M, Xie B, Li N, Cao X, Wang J. RasGRP3 limits toll-like receptor-triggered inflammatory response in macrophages by activating Rap1 small GTPase. Nat Commun. 2014;5:4657. doi: 10.1038/ncomms5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Jing W, Zhang X, Sun W, Hou X, Yao Z, Zhu Y. CRISPR/CAS9-mediated genome editing of miRNA-155 inhibits proinflammatory cytokine production by RAW264.7 cells. Biomed Res Int. 2015;2015;326042 doi: 10.1155/2015/326042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Ott de Bruin LM, Volpi S, Musunuru K. Novel genome-editing tools to model and correct primary immunodeficiencies. Front Immunol. 2015;6:250. doi: 10.3389/fimmu.2015.00250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Cowan MJ, Neven B, Cavazanna-Calvo M, Fischer A, Puck J. Hematopoietic stem cell transplantation for severe combined immunodeficiency diseases. Biol Blood Marrow Transplant. 2008;14(1 Suppl 1):S73–S75. doi: 10.1016/j.bbmt.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 144.Deakin CT, Alexander IE, Kerridge I. Accepting risk in clinical research: Is the gene therapy field becoming too risk-averse. Mol Ther. 2009;17:1842–1848. doi: 10.1038/mt.2009.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Kohn DB, Kuo CY. New frontiers in the therapy of primary immunodeficiency: From gene addition to gene editing. J Allergy Clin Immunol. 2017;139:726–732. doi: 10.1016/j.jaci.2017.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Goodman MA, Moradi Manesh D, Malik P, Rothenberg ME. CRISPR/Cas9 in allergic and immunologic diseases. Expert Rev Clin Immunol. 2017;13:5–9. doi: 10.1080/1744666X.2017.1241711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Wang M, Glass ZA, Xu Q. Non-viral delivery of genome-editing nucleases for gene therapy. Gene Ther. 2017;24:144–150. doi: 10.1038/gt.2016.72. [DOI] [PubMed] [Google Scholar]
- 148.Chang CW, Lai YS, Westin E, Khodadadi-Jamayran A, Pawlik KM, Lamb LS, Jr, Goldman FD, Townes TM. Modeling human severe combined immunodeficiency and correction by CRISPR/Cas9-enhanced gene targeting. Cell Rep. 2015;12:1668–1677. doi: 10.1016/j.celrep.2015.08.013. [DOI] [PubMed] [Google Scholar]
- 149.Flynn R, Grundmann A, Renz P, Hänseler W, James WS, Cowley SA, Moore MD. CRISPR-mediated genotypic and phenotypic correction of a chronic granulomatous disease mutation in human iPS cells. Exp Hematol. 2015;43:838–848. e3. doi: 10.1016/j.exphem.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wrona D, Siler U, Reichenbach J. CRISPR/Cas9-generated p47(phox)-deficient cell line for chronic granulomatous disease gene therapy vector development. Sci Rep. 2017;7:44187. doi: 10.1038/srep44187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Yan Q, Zhang Q, Yang H, Zou Q, Tang C, Fan N, Lai L. Generation of multi-gene knockout rabbits using the Cas9/gRNA system. Cell Regen (Lond) 2014;3:12. doi: 10.1186/2045-9769-3-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Fan Z, Li W, Lee SR, Meng Q, Shi B, Bunch TD, White KL, Kong IK, Wang Z. Efficient gene targeting in golden Syrian hamsters by the CRISPR/Cas9 system. PLoS One. 2014;9:e109755. doi: 10.1371/journal.pone.0109755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Chen F, Wang Y, Yuan Y, Zhang W, Ren Z, Jin Y, Liu X, Xiong Q, Chen Q, Zhang M, et al. Generation of B cell-deficient pigs by highly efficient CRISPR/Cas9-mediated gene targeting. J Genet Genomics. 2015;42:437–444. doi: 10.1016/j.jgg.2015.05.002. [DOI] [PubMed] [Google Scholar]
- 154.Chu HW, Rios C, Huang C, Wesolowska-Andersen A, Burchard EG, O’Connor BP, Fingerlin TE, Nichols D, Reynolds SD, Seibold MA. CRISPR-Cas9-mediated gene knockout in primary human airway epithelial cells reveals a proinflammatory role for MUC18. Gene Ther. 2015;22:822–829. doi: 10.1038/gt.2015.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Kuo CY, Hoban MD, Joglekar AV, Kohn DB. Site specific gene correction of defects in CD40 ligand using the Crispr/Cas9 genome editing platform. J Allergy Clin Immunol. 2015;135:AB17. doi: 10.1016/j.jaci.2014.12.987. [DOI] [Google Scholar]
- 156.Cheong TC, Compagno M, Chiarle R. Editing of mouse and human immunoglobulin genes by CRISPR-Cas9 system. Nat Commun. 2016;7:10934. doi: 10.1038/ncomms10934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Rodriguez-Sanchez IP, Guindon J, Ruiz M, Tejero ME, Hubbard G, Martinez-de-Villarreal LE, Barrera-Saldaña HA, Dick EJ, Jr, Comuzzie AG, Schlabritz-Loutsevitch NE. The endocannabinoid system in the baboon (Papio spp.) as a complex framework for developmental pharmacology. Neurotoxicol Teratol. 2016;58:23–30. doi: 10.1016/j.ntt.2016.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Niu Y, Shen B, Cui Y, Chen Y, Wang J, Wang L, Kang Y, Zhao X, Si W, Li W, et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell. 2014;156:836–843. doi: 10.1016/j.cell.2014.01.027. [DOI] [PubMed] [Google Scholar]
- 159.Kang Y, Zheng B, Shen B, Chen Y, Wang L, Wang J, Niu Y, Cui Y, Zhou J, Wang H, et al. CRISPR/Cas9-mediated Dax1 knockout in the monkey recapitulates human AHC-HH. Hum Mol Genet. 2015;24:7255–7264. doi: 10.1093/hmg/ddv425. [DOI] [PubMed] [Google Scholar]
- 160.Kim S, Huang LW, Snow KJ, Ablamunits V, Hasham MG, Young TH, Paulk AC, Richardson JE, Affourtit JP, Shalom-Barak T, et al. A mouse model of conditional lipodystrophy. Proc Natl Acad Sci USA. 2007;104:16627–16632. doi: 10.1073/pnas.0707797104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Croasdell A, Duffney PF, Kim N, Lacy SH, Sime PJ, Phipps RP. PPARγ and the innate immune system mediate the resolution of inflammation. PPAR Res. 2015;2015;549691 doi: 10.1155/2015/549691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Huang J, Guo X, Fan N, Song J, Zhao B, Ouyang Z, Liu Z, Zhao Y, Yan Q, Yi X, et al. RAG1/2 knockout pigs with severe combined immunodeficiency. J Immunol. 2014;193:1496–1503. doi: 10.4049/jimmunol.1400915. [DOI] [PubMed] [Google Scholar]
- 163.Chen Y, Xiong M, Dong Y, Haberman A, Cao J, Liu H, Zhou W, Zhang SC. Chemical control of grafted human PSC-derived neurons in a mouse model of Parkinson’s disease. Cell Stem Cell. 2016;18:817–826. doi: 10.1016/j.stem.2016.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Zhou X, Xin J, Fan N, Zou Q, Huang J, Ouyang Z, Zhao Y, Zhao B, Liu Z, Lai S, et al. Generation of CRISPR/Cas9-mediated gene-targeted pigs via somatic cell nuclear transfer. Cell Mol Life Sci. 2015;72:1175–1184. doi: 10.1007/s00018-014-1744-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Wang L, Yi F, Fu L, Yang J, Wang S, Wang Z, Suzuki K, Sun L, Xu X, Yu Y, et al. CRISPR/Cas9-mediated targeted gene correction in amyotrophic lateral sclerosis patient iPSCs. Protein Cell. 2017;8:365–378. doi: 10.1007/s13238-017-0397-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Bhinge A, Namboori SC, Zhang X, VanDongen AMJ, Stanton LW. Genetic correction of SOD1 mutant iPSCs reveals ERK and JNK activated AP1 as a driver of neurodegeneration in amyotrophic lateral sclerosis. Stem Cell Reports. 2017;8:856–869. doi: 10.1016/j.stemcr.2017.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Merienne N, Vachey G, de Longprez L, Meunier C, Zimmer V, Perriard G, Canales M, Mathias A, Herrgott L, Beltraminelli T, et al. The self-inactivating KamiCas9 system for the editing of CNS disease genes. Cell Rep. 2017;20:2980–2991. doi: 10.1016/j.celrep.2017.08.075. [DOI] [PubMed] [Google Scholar]
- 168.Page SC, Hamersky GR, Gallo RA, Rannals MD, Calcaterra NE, Campbell MN, Mayfield B, Briley A, Phan BN, Jaffe AE, Maher BJ. The schizophrenia- and autism-associated gene, transcription factor 4 regulates the columnar distribution of layer 2/3 prefrontal pyramidal neurons in an activity-dependent manner. Mol Psychiatry. 2018;23:304–315. doi: 10.1038/mp.2017.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Long C, Amoasii L, Mireault AA, McAnally JR, Li H, Sanchez-Ortiz E, Bhattacharyya S, Shelton JM, Bassel-Duby R, Olson EN. Postnatal genome editing partially restores dystrophin expression in a mouse model of muscular dystrophy. Science. 2016;351:400–403. doi: 10.1126/science.aad5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Long C, McAnally JR, Shelton JM, Mireault AA, Bassel-Duby R, Olson EN. Prevention of muscular dystrophy in mice by CRISPR/Cas9-mediated editing of germline DNA. Science. 2014;345:1184–1188. doi: 10.1126/science.1254445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Nelson CE, Hakim CH, Ousterout DG, Thakore PI, Moreb EA, Castellanos Rivera RM, Madhavan S, Pan X, Ran FA, Yan WX, et al. In vivo genome editing improves muscle function in a mouse model of Duchenne muscular dystrophy. Science. 2016;351:403–407. doi: 10.1126/science.aad5143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Tabebordbar M, Zhu K, Cheng JKW, Chew WL, Widrick JJ, Yan WX, Maesner C, Wu EY, Xiao R, Ran FA, et al. In vivo gene editing in dystrophic mouse muscle and muscle stem cells. Science. 2016;351:407–411. doi: 10.1126/science.aad5177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Ackermann AM, Zhang J, Heller A, Briker A, Kaestner KH. High-fidelity Glucagon-CreER mouse line generated by CRISPR-Cas9 assisted gene targeting. Mol Metab. 2017;6:236–244. doi: 10.1016/j.molmet.2017.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Jarrett KE, Lee CM, Yeh YH, Hsu RH, Gupta R, Zhang M, Rodriguez PJ, Lee CS, Gillard BK, Bissig KD, et al. Somatic genome editing with CRISPR/Cas9 generates and corrects a metabolic disease. Sci Rep. 2017;7:44624. doi: 10.1038/srep44624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Mookherjee Yu W, Chaitankar S, Hiriyanna V, Kim S, Brooks JW, Ataeijannati M, Sun Y, Dong X, Li LT, et al. Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nat Commun. 2017;8:14716. doi: 10.1038/ncomms14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Not applicable.