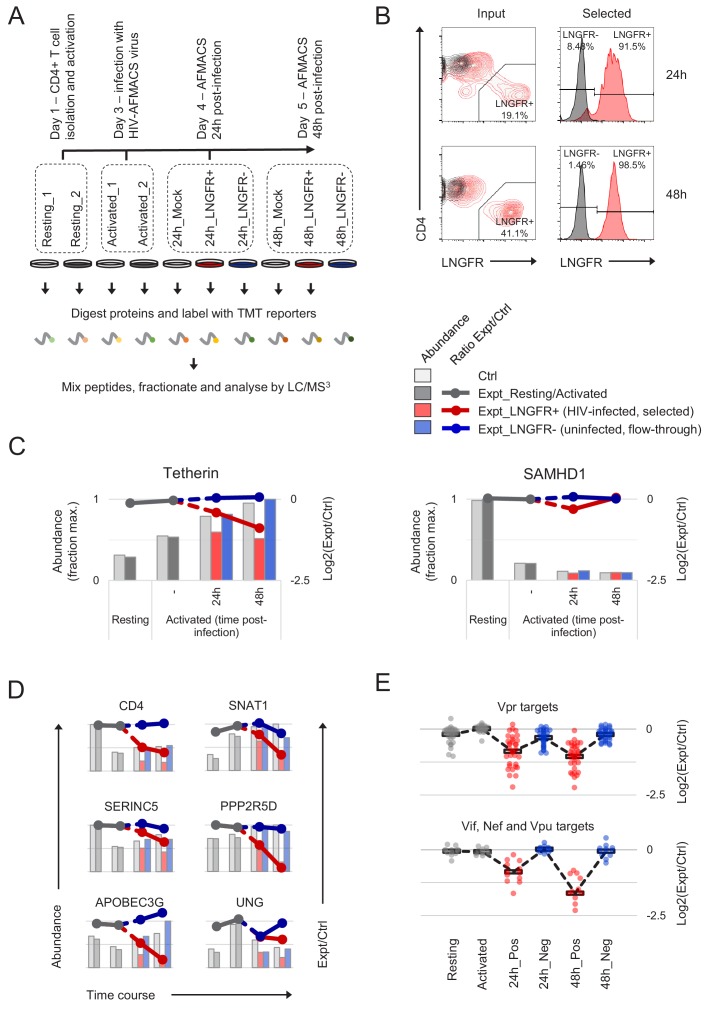
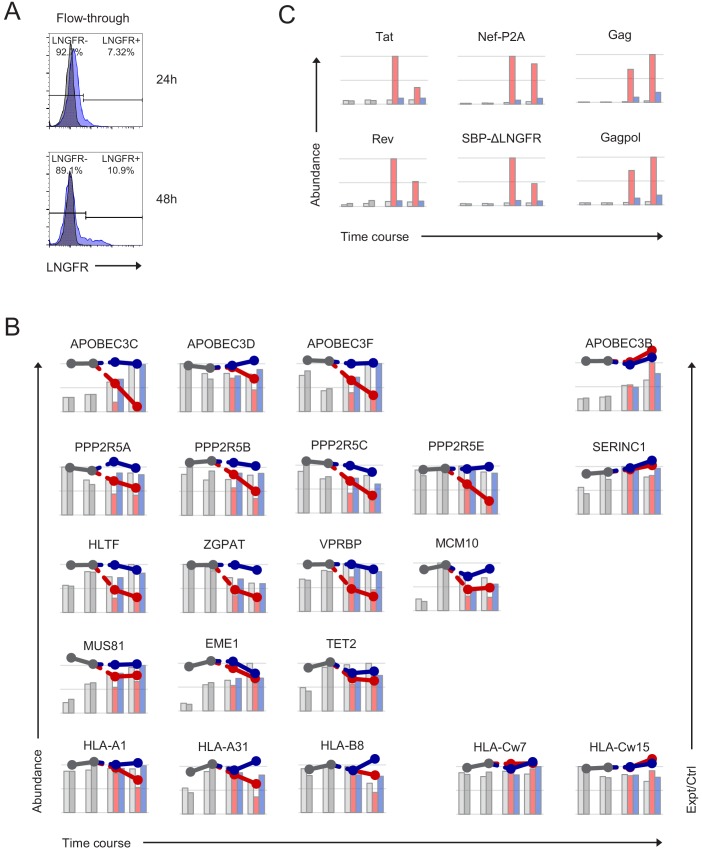
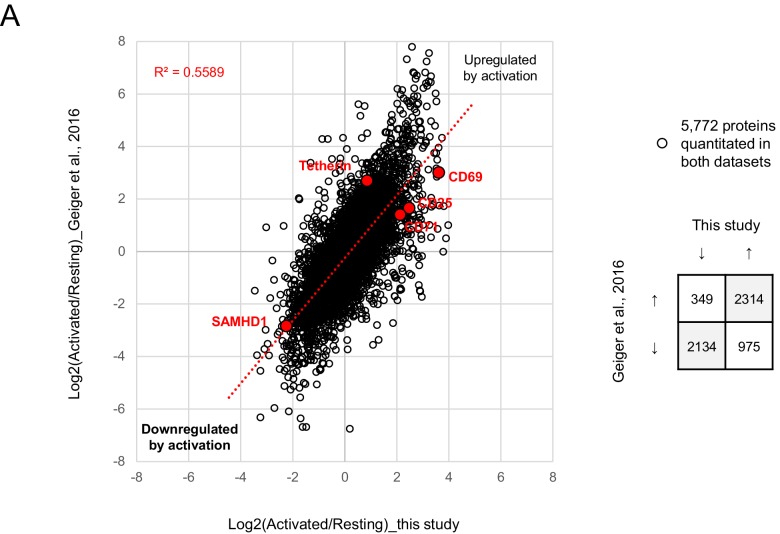
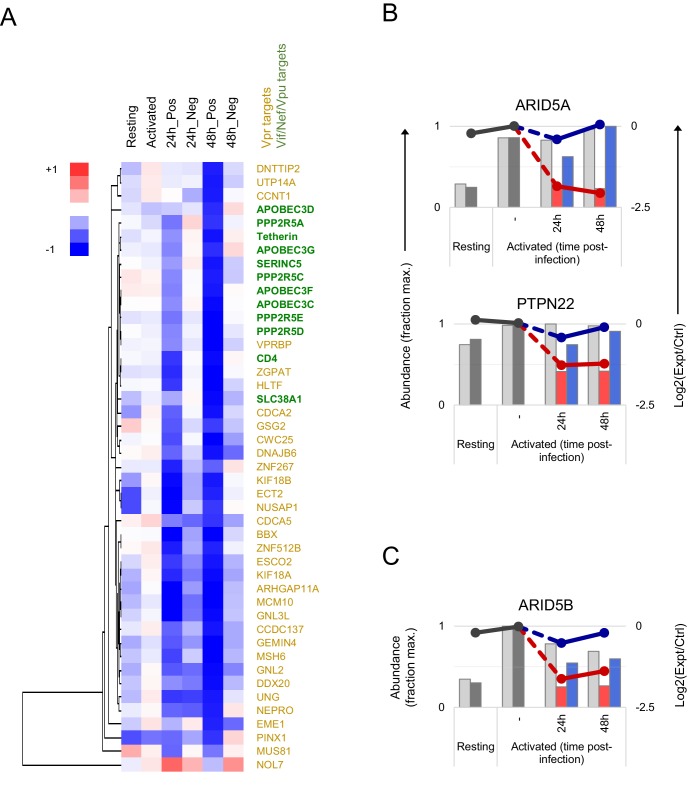
Figure 2. Temporal proteomic analysis of HIV infection in primary T cells.
(A) Overview of time course proteomic experiment. Control (pale grey, resting/activated/mock) and experimental (dark grey, resting/activated; red, LNGFR+, HIV-infected, selected; blue, LNGFR-, uninfected, flow-through) cells are indicated for each condition/time point. (B) Magnetic sorting of HIV-infected (red, LNGFR+, selected) cells used for (A). Corresponding uninfected (LNGFR-, flow-through) cells are shown in Figure 2—figure supplement 1A. Cells were separated using AFMACS at the indicated time points post-infection with HIV-AFMACS, stained with anti-LNGFR and anti-CD4 antibodies and analysed by flow cytometry. Mock-infected cells are shown in grey. (C) Expression profiles of illustrative restriction factors regulated by T cell activation and HIV infection (tetherin) or T cell activation alone (SAMHD1) in cells from (A–B). Relative abundances (bars, fraction of maximum) and log2(ratio)s of abundances (lines) in experimental (Expt):control (Ctrl) cells are shown for each condition/time point and coloured as in (A) (summarised in the key). (D) Expression profiles of illustrative accessory protein targets (CD4, Nef/Vpu; SERINC5, Nef; SNAT1, Vpu; APOBEC3G, Vif; PPP2R5D, Vif; UNG, Vpr) in cells from (A–B). Axes, scales and colours are as in (C). Expression profiles of other accessory protein targets are shown in Figure 2—figure supplement 1B. (E) Patterns of temporal regulation of Vpr vs other accessory protein (Vif/Nef/Vpu) targets in cells from (A–B). Log2(ratio)s of abundances in experimental (Expt):control (Ctrl) cells are shown for 45 accessory protein targets (as in Figure 2—figure supplement 3A). Colours are as in (C), and average profiles (mean, black lozenges/dotted lines) are highlighted for each group of targets.