Abstract
Most infections with Mycobacterium tuberculosis (Mtb) manifest as a clinically asymptomatic, contained state, known as latent tuberculosis infection, that affects approximately one-quarter of the global population1. Although fewer than one in ten individuals eventually progress to active disease2, tuberculosis is a leading cause of death from infectious disease worldwide3. Despite intense efforts, immune factors that influence the infection outcomes remain poorly defined. Here we used integrated analyses of multiple cohorts to identify stage-specific host responses to Mtb infection. First, using high-dimensional mass cytometry analyses and functional assays of a cohort of South African adolescents, we show that latent tuberculosis is associated with enhanced cytotoxic responses, which are mostly mediated by CD16 (also known as FcγRIIIa) and natural killer cells, and continuous inflammation coupled with immune deviations in both T and B cell compartments. Next, using cell-type deconvolution of transcriptomic data from several cohorts of different ages, genetic backgrounds, geographical locations and infection stages, we show that although deviations in peripheral B and T cell compartments generally start at latency, they are heterogeneous across cohorts. However, an increase in the abundance of circulating natural killer cells in tuberculosis latency, with a corresponding decrease during active disease and a return to baseline levels upon clinical cure are features that are common to all cohorts. Furthermore, by analysing three longitudinal cohorts, we find that changes in peripheral levels of natural killer cells can inform disease progression and treatment responses, and inversely correlate with the inflammatory state of the lungs of patients with active tuberculosis. Together, our findings offer crucial insights into the underlying pathophysiology of tuberculosis latency, and identify factors that may influence infection outcomes.
Although most Mtb infections do not lead to the manifestation of clinical disease, few studies have focused on delineating the immune factors that are associated with the asymptomatic states that comprise latent tuberculosis infection (LTBI). To broadly characterize this immune state, we used high-dimensional cytometry by time-of-flight (CyTOF), a proteomics technology that assesses the abundance of cell subsets, protein expression and activation of signalling pathways at the single-cell resolution4 (Fig. 1a). We analysed peripheral blood mononuclear cells (PBMCs) from uninfected and latently infected adolescents (aged 13–18 years) from South Africa (Supplementary Table 1). This cohort is from a highly endemic area but has a lower rate of active tuberculosis (TB) than is seen in young children and adults5, indicating a well-controlled Mtb infection.
Fig. 1 |. Schematic representation of the experimental design.
a, Identification of immune features distinguishing uninfected and latently infected individuals from a cohort of South African adolescents. t-SNE, t-distributed stochastic neighbour embedding. viSNE, visualization using t-SNE. b, Analysis of changes in immune cell subset abundance at different stages of infection and end-of-treatment using cell-type deconvolution of transcriptomic data from multiple cohorts, and fluorescence-activated cell sorting (FAC S) analysis of PBMCs from an adult Chinese cohort. c, Evaluation of changes in NK cell frequencies in longitudinal cohorts, for individuals who (1) acquired Mtb infection (QuantiFERON converters); (2) progressed from LTBI to active TB, and (3) patients with active TB who proceeded to treatment completion; and their correlations with pulmonary pathology as measured by PET–CT imaging. ATB, active tuberculosis; EOT, end-of-treatment; UC, uninfected controls.
An initial analysis (Supplementary Table 2) of 14 uninfected controls and 14 individuals with LTBI identified four cell subsets (defined by cell-surface protein expression) with a significantly higher percentage (of total live cells) in individuals with LTBI than uninfected controls (false discovery rate (FDR) of <1%). These four subsets comprised total CD16-expressing cells, natural killer (NK) cells and two closely related populations of CD27−CD8+ αβ T cells that differed in their CD38 expression. By contrast, two other cell subsets, total B cells and naive B cells, were significantly less abundant in individuals with LTBI (Fig. 2a and Extended Data Fig. 1a–c). Similar differences in NK cell and B cell percentages between uninfected controls and individuals with LTBI were also observed in an additional 20 individuals analysed by CyTOF, and another 32 individuals analysed by flow cytometry (Extended Data Fig. 1d). Because latently infected individuals show no significant change in peripheral monocyte or lymphocyte counts compared to uninfected controls6,7, changes in the percentage of a given cell type most likely reflect corresponding alterations in its abundance.
Fig. 2 |. Immune state of TB latency identified in a cohort of South African adolescents.
a,b, Frequencies of cell subsets were defined by surface marker (a) and effector molecule (b) expression that are present in significantly (FDR < 1% by SAM analysis) different abundances between uninfected controls and individuals with LT BI (n = 14 per group) as determined by Citrus analysis of CyTOF results (Extended Data Figs. 1, 2). c, Cytolytic responses of NK cells isolated from PBMCs of uninfected controls and individuals with LT BI (n = 10 per group), quantified by calcein-release from calcein-labelled target (K562) cells upon lysis. d, Percentages of CD16+GZMBhigh cells within each lymphocyte subset in uninfected controls and individuals with LT BI (n = 14 per group) (Extended Data Fig. 2f). e, ADCC response of total PBMCs from uninfected controls and individuals with LTBI (n = 12 per group) as determined by antibody-mediated killing of CFSE-labelled target (P815) cells (Extended Data Fig. 2g). f, Frequencies of phosphorylated ribosomal protein S6 (pS6)+ cells within T cell subsets under different stimulation conditions in uninfected controls and individuals with LTBI (n = 10 per group). g, Volcano plot of plasma protein abundance in uninfected controls and individuals with LTBI (n = 27 per group) (Supplementary Table 3). Throughout, P values were derived using a Mann–Whitney U-test, unless otherwise stated. Mean and error bars representing the 95% confidence intervals are shown for each comparison. See Supplementary Table 1.
Significant differences in the percentage of immune effector cell subsets between samples from uninfected controls and individuals with LTBI were also identified. Granzyme B (GZMB) and perforin (PRF) expressing cells were significantly higher in individuals with LTBI. These mostly consisted of NK cells and GZMB+PRF+IFNγ+TNF+ polyfunctional cells, which largely comprised CD27−CD8+ αβ T cells, but also included NK cells and γδ T cells (Fig. 2b and Extended Data Fig. 2a–c). These cells also expressed significantly higher levels of GZMB (Extended Data Fig. 2d, e), indicating an enhanced cytotoxic potential on a per-cell basis. Indeed, NK cells from individuals with LTBI showed significantly higher target cell lysis than those from uninfected controls (n = 10 per group, P = 0.003; Fig. 2c). Additionally, there were higher percentages of CD16+GZMBhigh cells within the compartments of the NK cells, CD8+ αβ T cells and γδ T cells in PBMCs from individuals with LTBI (Fig. 2d and Extended Data Fig. 2f). PBMCs from individuals with LTBI also mounted significantly higher antibody-dependent cell-mediated cytotoxicity (ADCC) responses than those from uninfected controls (n = 12 per group, P = 0.006; Fig. 2e and Extended Data Fig. 2g). ADCC allows antibodies, in addition to T cells, to contribute to the antigen-specific cytotoxic response. In this context, it was reported that antibodies from individuals with LTBI, compared to those from patients with active TB, have unique Fc functional profiles that promote selective binding to CD16 and effectively drive intracellular Mtb killing8.
The ability of cells to respond to immune challenges is an important factor in maintaining host immune competence. To determine the signalling capacity of immune cells, we used CyTOF (Supplementary Table 2) to investigate their ability to transiently phosphorylate signalling effectors in response to PMA and ionomycin, IFNγ, TNF or combined anti-CD3 and anti-CD28 stimulation. We found that in individuals with LTBI, all T cell subsets exhibited diminished responsiveness through the S6 signalling pathway, irrespective of the stimulation condition, with the exception of γδ T cells after stimulation with both anti-CD3 and anti-CD28, as compared to cells from uninfected controls (Fig. 2f). S6, a ribosomal component, is phosphorylated after mTOR activation. This signalling pathway is critical for ribosome biogenesis, cell growth and proliferation9.
Although alterations in the T cell compartment in individuals with LTBI did not lead to alteration in the levels of T cells in the periphery (Extended Data Fig. 1e), changes in the B cell compartment resulted from a decrease in the abundance of total B cells, which was largely driven by a reduction in circulating naive B cells. Reductions in the levels of peripheral B cells could be due to preferential sequestration of these cells at the site of infection (lungs and associated lymph nodes) and/or altered output of B cells from the bone marrow; inflammation can lead to enhanced myelopoiesis with diminished B cell output10. Analysis of 184 plasma proteins (Supplementary Table 3) showed significantly higher levels of inflammation-associated molecules, such as CXCL8 (also known as IL-8; P = 0.01), adenosine deaminase (ADA; P = 0.035) and NAD kinase (NADK; P = 0.004) in samples from individuals with LTBI relative to uninfected controls (n = 27 per group). By contrast, the plasma levels of CCL23, which has been shown to inhibit myelopoiesis11, was significantly lower in individuals with LTBI (P = 0.02; Fig. 2g and Extended Data Fig. 3), suggesting altered myelopoiesis in LTBI. Taken together, our results identified multiple immune components that operate together in LTBI. Specifically, in the presence of ongoing inflammation coupled with deviations in B and T cell compartments, enhanced cytotoxic responses that are mostly mediated by CD16 and NK cells appeared to be key factors associated with maintaining latency, which we propose represents successful immune control of Mtb infection.
Because alterations in peripheral immune cell distributions have not previously been commonly associated with TB latency, we tested whether such changes were observed in other LTBI cohorts, including those of children and adults (Fig. 1b). Although their PBMCs were not available for analysis, transcriptional profiles of whole-blood or PBMC samples from these cohorts were publicly available. We applied a computational approach to infer leukocyte representations from gene-expression profiles using support vector regression12 with the leukocyte expression signature matrix ‘immunoStates’13. Analysis of gene-expression datasets from 189 uninfected controls and 145 subjects with LTBI from six clinical cohorts (Supplementary Table 4), including children and adults from four continents, showed that NK cells were significantly more abundant (FDR = 0.018%) in cohorts of individuals with LTBI (Fig. 3a). Changes in B cell percentages were heterogeneous across the cohorts. However, analysis from all cohorts combined indicated a significant reduction in the percentages of total B cells (FDR = 1%) and naive B cells (FDR = 0.38%) in samples from individuals with LTBI (Fig. 3a and Extended Data Fig. 4a). No significant differences in total T cell abundance (FDR = 74%) (Fig. 3a) or those of the CD4+ and CD8+ subsets (FDR = 44% for both subsets; Extended Data Fig. 5a) were observed between the uninfected and LTBI cohorts. Thus, the analyses from multiple LTBI cohorts were consistent with our findings obtained from the cohort of South African adolescents.
Fig. 3 |. Peripheral lymphocyte distributions at different infection stages from global cohorts.
Forest plots comparing changes in the levels of NK cells, B cells and T cells were calculated using cell-mixture deconvolution. a, Comparison between uninfected controls (n = 189) and individuals with LTBI (n = 145). b, Comparison between individuals with LT BI (n = 409) and patients with active TB (n = 543). c, Comparison between patients with active TB before (n = 76) and after six months of treatment (end-of-treatment) (n = 97). Cohort GSE identifiers are listed on the left. Boxes represent the standardized mean difference in estimated cellular proportions in a cohort between two comparison groups (effect size). The size of the box is proportional to the sample size of a given cohort. Whiskers represent the 95% confidence interval of the corresponding standardized mean difference in cellular proportions. Diamonds represent the overall difference in cellular proportions between two groups by integrating the standardized mean differences across all individual cohorts-summary effect sizes (Summary). The width of the diamond corresponds to its 95% confidence interval. The q values (FDR) for the summary effect sizes are shown above each plot. d, Percentages of peripheral NK cells, B cells and T cells in a Chinese cohort of uninfected controls (n = 24), individuals with LTBI (n = 17) and patients with active T B (n = 23) assessed by flow cytometry. P values were derived using a one-way ANOVA with Tukey’s multiple comparisons test. Mean and error bars representing the 95% confidence intervals are shown for each comparison. See Supplementary Tables 4, 5.
To evaluate whether and how these immune cell frequencies change during active disease, we analysed transcriptome profiles from PBMC or whole-blood samples from 409 individuals with LTBI and 543 patients with active TB from nine clinical cohorts (Supplementary Table 4), profiling children and adults, with and without HIV comorbidity, across three continents. Deconvolution estimations showed that NK cells were present at significantly (FDR = 0%, P < 2.2 × 10−16) lower frequencies in samples from patients with active TB, with notable consistency across all cohorts (Fig. 3b). Flow cytometry analysis of whole-blood samples from adults collected from 24 uninfected controls, 17 individuals with LTBI and 23 patients with active TB (Supplementary Table 5) from a non-endemic region (Shenzhen) in China also confirmed the stage-specific changes in NK cell frequencies (Fig. 3d).
Our deconvolution estimations also showed significantly reduced levels of B cells in patients with active TB compared to uninfected individuals (Extended Data Fig. 6). The analysis of B cells in an adult Italian cohort is one such example7. In general, the decrease in the frequencies of B cells and naive B cells was observed in either latency or active disease relative to uninfected individuals (Fig. 3a, b and Extended Data Fig. 4a, b), with the exception of an Indonesian cohort, in which no change in B cell frequencies was observed. Notably, similar to the Indonesian cohort, no change in B cell frequencies was observed in the adult Chinese cohort, assessed by flow cytometry (Fig. 3d). These observations underscore the heterogeneity of immune states in Mtb infection. Although our South African adolescent cohort showed no significant changes in T cell abundance between uninfected controls and individuals with LTBI, both CD4+ and CD8+ αβ T cell populations showed significantly diminished responsiveness through the S6 pathway, which may foretell the eventual drop in peripheral T cell levels observed in patients with active TB (Fig. 3b, Extended Data Fig. 5b). Flow cytometry assessment of the Chinese cohort confirmed the significant decrease in peripheral T cell levels in patients with active TB compared to uninfected controls (Fig. 3d). Whether reduced T cell signalling capacity is a common feature of LTBI requires further analysis. Nonetheless, our results suggest that immune deviations in both T and B cell compartments start in latency and progress further in active disease.
To test whether the disease-induced reductions in peripheral lymphocyte populations recover after successful treatment, we deconvoluted PBMC or whole-blood transcriptome profiles of 76 samples from patients with active TB and 97 samples from patients at the end of treatment from four independent cohorts of HIV-negative adults (Supplementary Table 4) from three continents. NK cell frequencies, together with all major immune cell populations, except CD4+ αβ T cells, were significantly higher in successfully treated individuals relative to patients with active TB, reaching reference levels observed in healthy, Mtb-uninfected individuals (Fig. 3c and Extended Data Fig. 4c, 5c). A longer recovery time for CD4+ αβ T cells might be indicative of a post-treatment inflammatory state due to ongoing subclinical disease as seen by positron emission tomography and computed tomography (PET–CT) analysis14. The trajectories of estimated immune cell frequencies through the different stages of infection and after treatment are summarized in Extended Data Fig. 7.
Because of the apparent correlation between changes in NK cell frequencies and the stages of Mtb infection, we tested whether the levels of peripheral NK cells could inform TB disease progression and response to treatment (Fig. 1c) by analysing three independent longitudinal follow-up studies of South African cohorts, which included individuals who (1) acquired latent Mtb infection (defined as converting from QuantiFERON (QFT)-negative to QFT-positive), (2) progressed from latent infection to active disease (South African adolescent progressor cohort)15,16 and (3) proceeded from active disease to treatment completion (Catalysis-TB cohort)17. Deconvolution estimations showed that relative to pre-infection, levels of NK cells did not change significantly after acquisition of Mtb infection (n =17; Fig. 4a) and decreased during the progression from latent infection to active disease (n = 17; Fig. 4b (left)). Consistent with this latter computational finding, flow cytometry analysis of 32 individuals with LTBI (12 progressors and 20 non-progressors) (Supplementary Table 6) showed that NK cell frequencies decreased in each of the progressors, 0–180 days before TB diagnosis, whereas the non-progressors showed no significant change in peripheral NK cell frequencies during the two-year study period (Fig. 4b (right)). Statistical analysis of the predictive power of NK cell levels for progression to active disease using receiver operating characteristic curves showed an area under the curve of 0.69 (95% confidence interval 0.57–0.82) in the seven months preceding TB diagnosis (Fig. 4c). In addition, deconvolution analysis of gene-expression data from the Catalysis Foundation for Health study of patients with active TB under treatment showed that in individuals who responded to treatment (classified as ‘definite cure’, n = 76), NK cell levels were significantly higher at the end of treatment (week 24) compared to baseline (pre-treatment, P < 0.0001; Fig. 4d). By contrast, treatment non-responders (classified as ‘no cure’, n = 7), showed no significant change in their NK cell percentages between baseline and end of treatment (P = 0.1250; Fig. 4d). Furthermore, we found that the inflammatory burden of the lung indicated by the total glycolytic activity index, as measured by PET–CT14, inversely correlated with peripheral NK cell frequencies at diagnosis (pre-treatment; Fig. 4e) and at week 4 after treatment initiation (Extended Data Fig. 8). Therefore, changes in peripheral NK cell levels reflect changes in the activity level and burden of Mtb in the lung. These observations support the view that circulating NK cells reflect key features of the host immune state, can serve as surrogates of the immune response at the nidus of infection and that longitudinal measurements of peripheral NK cells can inform disease progression and treatment efficacy.
Fig. 4 |. Correlations between peripheral NK cell percentages and disease progression, treatment response and inflammation in the lung.
a, Changes in peripheral NK cell percentages in South African adolescents after acquisition of Mtb infection (Extended Data Fig. 9, n = 17) were determined by cell-mixture deconvolution. Pre-infection (Pre) gene-expression data (180–360 days) were compared to data obtained at the time of infection diagnosis or the nearest time point after diagnosis (0–360 days) (Post). b, Changes in peripheral NK cell percentages during progression from LTBI to active disease at different time points before TB diagnosis and non-progressors over a span of two years (Supplementary Table 6) were determined by cell-mixture deconvolution (17 progressors and 41 non-progressors) (left) and flow cytometry (12 progressors and 20 non-progressors) (right). All P values were derived using a Wilcoxon rank-sum test. Mean and error bars representing the 95% confidence intervals are shown. c, Receiver operating characteristic curves of the potential of estimated NK cell frequencies as a predictor of TB disease progression. d, Estimated NK cell percentages in patients with active TB from the Catalysis-TB cohort at baseline (pre-treatment) and at various time points during treatment. Definite cure indicates sputum culture negative by month 6 after treatment initiation (n = 76); no cure indicates sputum culture positive after six months of treatment initiation (n = 7). P values were derived using a Wilcoxon rank-sum test. Mean and error bars representing the 95% confidence intervals are shown. e, Correlation plot showing the relationship between estimated peripheral NK cell frequencies in patients with active TB at baseline (pre-treatment) and total glycolytic activity index (TGAI) measured by PET–CT imaging of the lungs at baseline. The line represents the best fit and the shaded area the 95% confidence interval. NK cell frequencies were determined by deconvolution.
NK cells have been shown to kill Mtb-infected cells directly or through ADCC18. Moreover, NK cells become activated and expand in the lung during the early response in a mouse model of aerosol exposure with Mtb, but depleting NK cells from these immunocompetent mice does not alter the course of infection19. Nevertheless, in mice with T cell deficiencies, NK cells were found to confer protection against Mtb infection20. Although mice infected with Mtb do not establish latency, these observations are consistent with the proposition that in the immune state that we observed here in TB latency, NK cells could contribute to protective immunity. Along this line, TB is the most common fatal opportunistic infection in HIV/AIDS21, and NK cells are noted for controlling HIV infections22. Progressive impairment of NK cell functions and depletion of NK cells, especially the CD16+ subsets, have been noted in HIV infection23. Additionally, anti-TNF therapy, which is associated with increased incidence of TB disease in autoimmune patients with LTBI24, was shown to impair NK cell function25 and reduce the expression of PRF and granulysin in lymphocytes26.
Taken together, our analyses offer a better understanding of the immune state of latent Mtb infection and factors that mediate and/or predict transitions from latent infection to active disease. These findings may be useful for generating hypotheses that could lead to new intervention strategies.
METHODS
Data reporting.
No statistical methods were used to predetermine sample size. The experiments were not randomized and the investigators were not blinded to allocation during experiments and outcome assessment.
Study design and participants.
A South African adolescent cohort (Adolescent Cohort Study (ACS))5, aged 13–18 years, who were either uninfected or latently infected with Mtb (LTBI) (Supplementary Table 1), were analysed to characterize the immune state of TB latency. All adolescents whose parents or legal guardians provided written, informed consent and who provided written, informed assent themselves were enrolled. The study protocols were approved by the relevant human research ethics committees. Individuals were classified as latently infected if diagnosed positive by a QuantiFERON TB Gold In-tube assay (Qiagen; >0.35 IU ml−1). All participants were healthy without signs or symptoms of active disease. Only adolescents who remained disease-free for two years from the time of enrolment were included in the analysis.
Cohorts associated with publicly available datasets27–37 that were used for the analysis of immune cell distributions at different stages of Mtb infection, and end-of-treatment, by cell-mixture deconvolution, are described in Supplementary Table 4.
An adult Chinese cohort (Supplementary Table 5) from Shenzhen, a non-endemic region in China was assessed for immune cell distributions at different stages of Mtb infection using flow cytometry analysis of PBMC samples. Individuals were defined as latently infected if diagnosed positive by the interferon gamma release assay (IGRA+) but showed no symptom or chest X-ray signs suggestive of active disease. Tuberculosis was defined as intrathoracic disease with positive sputum smears and/or cultures for Mtb. The study protocols were approved by the relevant human research ethics committees.
Three independent longitudinal South African progressor cohorts were analysed for the kinetics of the frequency changes in NK cells using deconvolution of gene expression datasets. These cohorts transitioned from: (1) an uninfected state to latency (GSE116014), for which samples were obtained from an Mtb-acquisition sub-cohort, selected from the larger ACS cohort5,16, who were diagnosed as QFT-negative at multiple time points, six months apart, and then converted to QFT-positive, indicating a newly acquired Mtb infection (Extended Data Fig. 9); (2) latency to active disease, individuals enrolled in the ACS cohort described above, were assessed longitudinally every six months during a 2-year follow-up study15. Adolescents who developed active tuberculosis disease during this 2-year follow-up were included as ‘progressors’, and those who did not, were classified as LTBI ‘non-progressors’ (Supplementary Table 6). Active TB was defined as intrathoracic disease, with either two sputum smears that were positive for acid-fast bacilli or one positive sputum culture confirmed to be M. tuberculosis complex (mycobacterial growth indicator tube, BD BioSciences). Participants were excluded if they were known to be HIV-positive; (3) active disease to end-of-treatment, the Catalysis-TB cohort17.
Mass cytometry measurements and analysis.
CyTOF experiments were performed as previously described38. In brief, cryopreserved PBMCs were thawed and rested in complete RPMI with 10% FCS at 37 °C for 2 h at cell densities of approximately 107 cells per ml. Cells from each sample were equally split into two parts and were either left untreated or stimulated for 4 h with 150 ng ml−1 phorbol-12-myristate-13-acetate (PMA) and 1 μM ionomycin in the presence of brefeldin A and monensin (eBioscience). Cells (5 × 106) were then stained (1 h; room temperature) with a mixture of metal-tagged antibodies (a complete list of antibodies and their catalogue numbers is provided in Supplementary Table 2). All antibodies were validated by the manufacturers for mass cytometry applications (as indicated on the manufacturer’s datasheet, available online) and were conjugated using MAXPAR reagents (Fluidigm Inc.). Cisplatin and iridium intercalators were used to identify live and dead cells. We used palladium barcoding (Fluidigm Inc.) according to the manufacturer’s instructions. Cells were washed twice with PBS, fixed in 1.6% paraformaldehyde (PFA) (Sigma-Aldrich; 1 h), washed again in ultrapure water and analysed using CyTOF mass cytometry on a CyTOF 2 instrument (Fluidigm). Intracellular phosphorylated-protein staining was carried out as previously described39. In brief, cryopreserved PBMCs were thawed and rested as described above. Rested cells were incubated with cisplatin for 1 min and immediately quenched with four volumes of complete RPMI with 10% FCS, and rested again for 30 min at 37 °C. Subsequently, cells were split into five tubes; one was left untreated and the others were stimulated with (1) PMA and ionomycin, (2) IFNγ (50 ng ml−1), (3) TNF (50 ng ml−1) or (4) anti-CD3 (500 ng ml−1) and anti-CD28 (2 μg ml−1) for 15 min at 37 °C. The reaction was stopped by adding PBS with 2% PFA (incubated for 10 min; room temperature), followed by palladium barcoding as recommended by the manufacturer (Fluidigm). After barcoding, cell samples were washed and then combined for surface-marker staining (1 h; room temperature). Subsequently, cells were washed and permeabilized in MeOH at −80 °C overnight. The next day, cells were washed and incubated with the cocktail of antibodies to intracellular signalling proteins at room temperature for 1 h, followed by DNA staining as described above.
Cell events were acquired at approximately 500 events s−1. In addition, we spiked each sample with internal metal-isotope bead standards for sample normalization using the CyTOF software (Fluidigm Inc.). Data processing and gating of dead cells and normalization beads was done on the Cytobank website (http://www.cytobank.org). To account for intra-run declines in mean marker intensity over time, we performed a within-sample-over-time normalization step by using a running window to adjust mean marker intensity throughout each individual run, such that the mean expression over time was equal to that measured at the beginning of the run. Data was debarcoded using Fluidigm’s Debarcoder tool. Data was arcsinh-transformed for analysis.
Analysis of CyTOF data.
Citrus (cluster identification, characterization and regression).
Cell subset abundance and functional marker expression in PBMCs from uninfected controls and subjects with LTBI were compared using the Citrus algorithm available from the Cytobank website. Citrus40 uses regularized supervised learning algorithms to identify stratifying clusters (subsets) and cell response features. It is data driven and corrected for multivariate comparisons. In brief, Citrus analysis consists of the following steps. First, FCS files of normalized, ‘live cell/no beads’ samples were randomly sampled for n single-cell events. Second, collected single-cell events were pooled and iteratively hierarchically clustered based on similarity of expression of subsets of the measured channels. This procedure yielded overlapping clusters with the largest cluster encompassing all of the sampled events. Third, the pooled dataset was split back into its constitutive samples, and the relative abundance of cells in each cluster was computed, as well as the median expression of each functional marker in each cluster. Only clusters for which the abundance in one or more of the measured samples was greater than some lower-bound P values were considered for downstream differential analysis. Fourth, to determine differences in cell subset abundances or functional marker medians expression, we used the SAM algorithm in Citrus, which assesses FDR by permutations. For each set of analysis, we set n to 20,000 and the clustering threshold to 1% of total cells and performed the analysis iteratively such that 20,000 events from the entire dataset were chosen randomly for the multiple rounds of analysis. We also analysed the entire dataset in R. These analyses yielded qualitatively similar results.
Analyses of abundance from unstimulated and stimulated samples were done separately (see Extended Data Figs. 1, 2) because stimulation changed the expression levels of certain cell-surface markers. Manual inspection of Citrus output was used to identify the closest known gross-cell type. We characterized cell clusters using standard cell subset definitions: B cells (CD19+), CD8+ αβ T cells (CD3+TCRβ+CD8+), CD4+ αβ T cells (CD3+TCRβ+CD4+), γδ T cells (CD3+TCRδ+), monocytes (CD3−CD19−CD33+CD14+HLA-DR+), NK cells (CD3−CD19−CD14−HLA-DR−CD16+CD56bright/dim). In all conditions, we report cluster abundance differences at a FDR < 1%.
viSNE analysis.
Single-cell analysis using the dimensionality reduction technique viSNE reduces the multi-parametric data into two dimensions for visualization of similarity and heterogeneity across individual cells41. To account for different scales between parameters, the data was arcsinh transformed. viSNE analysis was performed on raw CyTOF data from the Cytobank database.
Manual gating.
Manual gating was performed on the Cytobank website on normalized, debarcoded data files. A hierarchical gating strategy was used to identify live, single cells of the main PBMC populations (Extended Data Fig. 1a) and their subsets based on the expression of surface, cytokine or signalling molecules.
NK cell cytotoxicity measured by calcein-release assay.
NK cells were enriched from PBMCs using the NK Cell Isolation Kit from Miltenyi Biotech. The NK cell cytotoxicity assay was carried out as described42 with some modifications. In brief, after cell counting, NK cells were mixed with calcein-acetoxymethyl (calcein-AM) labelled target K562 cells (which are susceptible to NK cell-mediated killing because of the lack of surface MHC class I expression), at an effector to target ratio of 2:1. For staining of the target cells, 2 mM of calcein-AM (Life Technologies) was added to the target cells (2 × 106 per ml) and incubated at 37 °C for 30 min with periodic mixing. The target cells were washed, the enriched NK cells were added, and the mixture was incubated at 37 °C for 4 h. Maximum and spontaneous release controls were set up as three replicates using 1% Triton X-100 (final concentration) and plain medium, respectively. After the 4-h incubation, the cells were gently mixed to evenly distribute the released calcein in the supernatant and the plate was spun at 400g for 2 min to pellet the cells and any debris. For the calcein-release assay, 150 μl of the supernatant was collected and transferred to a flat black-bottom plate. The fluorescence was read using a FlexStation3 microplate reader (excitation/emission: 485/530 nm). The percentage of specific lysis was calculated using the formula: ((test release − spontaneous release)/(maximum release − spontaneous release)) × 100.
FcγR-mediated antibody-dependent cell mediated cytotoxicity.
ADCC was carried out as previously described43 with some modifications. In brief, P815 cells (a mouse leukaemia cell line) were stained with 0.25 μM carboxy-fluorescein succinimidyl ester (CFSE; Molecular Probes) and incubated with the 10 μg ml−1 concentration of the P815-specific monoclonal antibody, 2.4G2. Coated and uncoated P815 cells were then cocultured with previously cryopreserved PBMC samples at an effector:target ratio of 10:1 in 10% FCS, penicillin, glutamine and streptomycin. The percentage of target cells that were killed through ADCC was monitored by flow cytometry staining using 7-amino-actinomycin D (7-AAD) viability staining solution (BioLegend). The percentage of cells killed by FcγR-mediated ADCC was obtained by subtracting the percentage 7-AAD+ CFSE-labelled uncoated target cells from the percentage of 7-AAD+ CFSE-labelled coated target cells. A minimum of 300,000 cells were analysed on a BD LSRII, and analysis was then performed using FlowJo (version 10.2) software.
Plasma protein quantification using a proximity extension assay.
For analysis, 20 μl of frozen (−80 °C) plasma samples were thawed and sent to Olink Proteomics. In proximity extension assays, plasma proteins were dually recognized by pairs of antibodies coupled to a cDNA strand that ligates when brought into proximity to its target, extended by a polymerase and detected using a BioMark HD 96 × 96 dynamic PCR array (Fluidigm). The quantification cycle (Cq) values from a DNA extension control are subtracted from the measured Cq value, an interpolate control is corrected for, and finally a correction factor is subtracted to yield a normalized protein expression value, which is log2-transformed.
Cell-mixture deconvolution and cell percentage meta-analysis.
This was carried out as previously described12,13. In brief, publicly available gene expression datasets were collected, pre-processed using the MetaIntegrator R package44 and annotated45. Each microarray dataset was converted into a gene-expression matrix, whereas for GSE7936215,16, which is an RNA-seq dataset, the total read counts per gene were first computed. A Hedge’s g effect size was then computed to estimate changes in cell subset proportions. Effect sizes from all individual datasets were integrated into a summary effect size and significance was computed as previously described44,46. To delineate cell trajectories over time, we computed a cumulative effect size score for each cell type in uninfected controls, LTBI, active TB disease, and end-of-treatment stages. We started by setting a reference effect size of 0 for healthy, Mtb-uninfected controls and then computed all following scores by adding the summary effect size value corresponding to case class for that specific comparison (LTBI, active TB, treated). We computed cumulative standard errors by assuming summary effect sizes to be normally distributed and independent of each other at each stage. Plots and statistics were generated using the R programming language.
Analysis of cell abundance using flow cytometry.
For flow cytometry experiments, PBMCs were thawed and rested as described in ‘Mass cytometry measurements and analysis’. Cells (3–5 × 106) were incubated with antibodies for 30 min at 4 °C, then washed with FACS staining buffer (PBS containing 1% bovine serum albumin and 0.05% sodium azide). The following monoclonal antibodies were used: Pacific Blue-conjugated anti-CD3 (BioLegend, 300417), PE–Cy7-conjugated anti-CD19 (BioLegend, 302216), APC–Cy7-conjugated anti-CD14 (BioLegend, 325620), APC-conjugated HLA-DR (BioLegend, 307610), PE–Dazzle-conjugated anti-CD16 (BioLegend, 302054) and Brilliant Violet 785-conjugated CD56 (BioLegend, 362550). The live/dead aqua-amine reactive dye was used for gating dead cells. All antibodies were validated by the manufacturers for flow cytometry application, as indicated on the manufacturer’s website. Data were analysed using FlowJo version 10.2.
Statistical analysis.
Analysis of CyTOF data are described in ‘Citrus (cluster identification, characterization and regression)’. In all other experiments, significance levels were determined using Prism version 7 (GraphPad Software). Experiments were analysed using the Mann–Whitney U-test or one-way analysis of variance (ANOVA), as indicated for each experiment. The diagnostic performance of NK cells to discriminate latent TB from active disease cases was evaluated using receiver operating characteristic (ROC) curve analysis, for which the true positive rate (sensitivity) is plotted as a function of the false-positive rate (100 − specificity). The area under the ROC curve is a measure of the probability that a classifier (for example, NK cell frequencies) will rank a randomly chosen positive instance (for example, active TB) higher than a randomly chosen negative one (for example, LTBI). ROC curves were plotted using Prism version 7.
Extended Data
Extended Data Fig. 1 |. Broad alterations of peripheral immune cell distributions in LTBI.
PBMCs from 14 latently infected and 14 uninfected participants of a South African adolescent cohort were characterized using CyTOF with antibody panel 1 (Supplementary Table 2), followed by Citrus analysis and clustering. This unsupervised hierarchical clustering analysis produced a branching structure (dendrogram) that allowed the grouping of total live cells into known immune cell compartments (contoured). Cell clusters are represented as nodes (circles) in this Citrus-derived circular dendrogram, which delineates lineage relationships that were identified from the data. Cluster granularity (that is, cell subset specificity) increases from the centre of the diagram to the periphery. a, Annotation of cluster hierarchy plots based on surface marker expression. The expression intensity of each marker used for cell population characterization is overlaid per cluster on the Citrus circular dendrogram and is indicated, independently for each marker, by the coloured gradient for which the range corresponds to the arcsinh-transformed expression of the median marker expression measured across all Citrus clusters. For each marker, we also provide a dot plot graph demonstrating the marker labelling in the manually gated indicated population. b, Citrus plots showing, based on cell-surface protein expression, clusters (in red, designated A–F) that exhibit significantly different abundances (SAM analysis with FDR < 1%) between the uninfected and latently infected individuals. Individual cell clusters are mapped to well-established, gross-cell types: B cells (CD19+), CD8+ αβ T cells (CD3+TCRβ+CD8+), CD4+ αβ T cells (CD3+TCRβ+CD4+), γδ T cells (CD3+TCRδ+), monocytes (CD3−CD19−CD33+CD14+HLA-DR+), NK cells (CD3−CD19−CD14−HLA-DR−CD16+CD56bright/dim), identifiable by annotated shaded background groupings. c, The phenotype and the composition of cells in each of the stratifying cell subsets (A–F), identified by Citrus analysis. d, Percentages of NK cells and B cells determined by manual gating of 20 additional samples using CyTOF antibody panel 2 (left; Supplementary Table 2) and 32 samples using flow cytometry (right). e, Percentages of CD4+ αβ T cells, CD8+ αβ T cells and γδ T cells in uninfected controls and latently infected individuals, analysed by CyTOF (n = 24 per group; top) and flow cytometry (n = 16 per group; bottom). Throughout, P values were derived using a Mann–Whitney U-test. Mean and error bars representing the 95% confidence intervals are shown for each comparison.
Extended Data Fig. 2 |. Enhanced effector function response in LTBI.
a, Cell subsets, shown as red nodes in a Citrus-derived circular dendrogram and designated as 1–5, were identified as significantly different in abundance (SAM analysis at FDR < 1%) based on CyTOF analysis of effector and cell-surface molecule expression on PBMCs (antibody panel 1, Supplementary Table 2) from uninfected controls and individuals with LT BI (n = 14 per group) after 4-h PMA and ionomycin stimulation. Mapping of individual cell clusters to established, grosscell types are identified by annotated shaded background groupings. b, Expression intensity of selected effector molecules is indicated by the coloured gradient for which the range corresponds to the arcsinh-transformed expression of the median marker expression measured across all C itrus clusters. c, Effector molecule expression and the composition of cells in each of the stratifying cell clusters (1–5), identified by Citrus analysis. d, viSNE analysis of GZMB expression level in immune-cell subsets, representative of 14 uninfected and 14 individuals with LTBI (the colour gradient corresponds to the arcsinh-transformed expression level). e, Quantification of intracellular GZMB expression level in NK cells, CD8+ αβ T cells and γδ T cells in uninfected controls and individuals with LTBI (n = 14 per group). P values were derived using a Mann–Whitney U-test. Mean and error bars representing the 95% confidence intervals are shown for each comparison. f, Dot plots from CyTOF analysis of CD16+GZMBhigh cells within each lymphocyte subset, representative of 14 uninfected controls and 14 individuals with LTBI. g, Gating strategy for ADCC. ADCC was measured using NK-resistant P815 cells, which were either coated with antibody (2.4G2) or left uncoated (control), and labelled with the intracellular dye CFSE, followed by the DNA dye 7AAD. CFSE+7AAD+ cells were defined as dead target cells.
Extended Data Fig. 3 |. Alterations in plasma protein levels in LTBI.
The relative levels of plasma proteins (Supplementary Table 3), shown on a log2 scale, between uninfected controls and individuals with LTBI (n = 27 per group). Plasma proteins that were present at significantly higher levels (a) and significantly lower levels (b) in individuals with LTBI. Plasma protein quantification was performed using the proximity extension assay. P values were derived using a unpaired two-tailed Student’s t-test. Mean and error bars representing the 95% confidence intervals are shown for each comparison.
Extended Data Fig. 4 |. Changes in frequencies of peripheral B cell subsets in LTBI, active TB and after treatment.
Forest plots for estimated frequencies of B cell subsets: naive B cells, memory B cells and plasma cells. a, Comparison between the uninfected state (n = 189) and LTBI (n = 145). b, Comparison between LTBI (n = 409) and active T B (n = 543). c, Comparison between active TB (n = 76) and end-of-treatment (n = 97). Cohort GSE identifiers are listed on the left. In the plots, boxes represent the standardized mean difference in estimated cellular proportions in a cohort between two comparison groups. The size of the box is proportional to the sample size of a given cohort. Lines indicate the 95% confidence interval of the corresponding effect sizes. Diamonds indicate the summary effect size (Summary), obtained by integrating the effect sizes from individual cohorts. The width of the diamond corresponds to its 95% confidence interval. The P values and q values for the summary effect sizes are shown above each plot.
Extended Data Fig. 5 |. Changes in frequencies of peripheral T cell subsets, monocytes and granulocytes in LTBI, active TB and after treatment.
Forest plots for estimated frequencies of CD4+αβ T cells, CD8+αβ T cells, monocytes and granulocytes. a, Comparison between the uninfected state (n = 189) and LTBI (n = 145). b, Comparison between LTBI (n = 409) and active TB (n = 543). c, Comparison between active TB (n = 76) and end-of-treatment (n = 97). Boxes represent the standardized mean difference in estimated cellular proportions in a cohort between two comparison groups. The size of the box is proportional to the sample size of a given cohort. Lines indicate the 95% confidence interval of the corresponding effect sizes. Diamonds indicate the summary effect size (Summary), obtained by integrating the effect sizes from individual cohorts. The width of the diamond corresponds to its 95% confidence interval. The P values and q values for the summary effect sizes are shown above each plot.
Extended Data Fig. 6 |. Comparison of the frequencies of peripheral NK cells, B cells and T cells between uninfected controls and patients with active TB.
Forest plots comparing changes in the levels of NK cells, B cells and T cells between uninfected individuals (n = 191) and patients with active TB (n = 178). Boxes represent the standardized mean difference in estimated cellular proportions in a cohort between two comparison groups. The size of the box is proportional to the sample size of a given cohort. Lines indicate the 95% confidence interval of the corresponding effect sizes. Diamonds indicate the summary effect size (Summary), obtained by integrating the effect sizes from individual cohorts. The width of the diamond corresponds to its 95% confidence interval. The P values and q values for the summary effect sizes are shown above each plot.
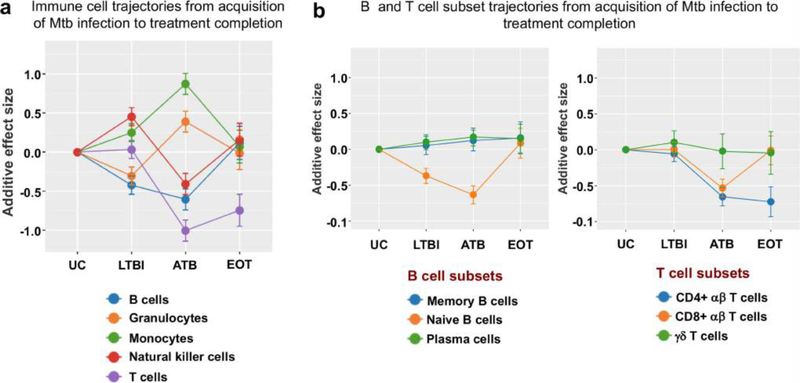
Extended Data Fig. 7 |. Trajectories of different immune cell populations from the acquisition of Mtb infection to end-of-treatment.
Changes in the frequency distribution patterns of different peripheral leukocyte populations (a) and B and T cell subpopulations (b) at the different stages of infection. Lines indicate cumulative effect size scores starting from a healthy baseline level up to treatment of active TB disease. Error bars indicate corresponding standard errors.
Extended Data Fig. 8 |. Correlation between peripheral NK cell percentage and lung inflammation.
Correlation plot showing the relationship between estimated peripheral NK cell frequencies in patients with active TB at week 4 after treatment initiation and total glycolytic activity index (TGAI) of the lung measured by PET–CT imaging at the corresponding time point. The line represents the best fit and the shaded area the 95% confidence interval. NK cell frequencies were determined by deconvolution.
Extended Data Fig. 9 |. Synchronization of the adolescent cohort who underwent QuantiFERON conversion following Mtb acquisition.
To identify changes in peripheral NK cell frequencies after acquisition of Mtb infection by cell-mixture deconvolution analysis, the timescale of the gene expression dataset (GSE116014) was realigned according to the time of first infection diagnosis instead of study enrolment, allowing the identification of gene-expression profiles obtained before infection diagnosis. Each individual is represented by a horizontal bar. The length of the bar represents the number of days between study enrolment and diagnosis with Mtb infection. During follow-up, each individual transitioned from an uninfected state (blue) to infected state (brown), that is, underwent QFT conversion. The black circles represent time points for which gene-expression data were available. Pre-infection (Pre) data (180–360 days) were compared to data obtained at the time of infection diagnosis or the nearest time point after diagnosis (Post) (0–360 days).
Supplementary Material
Acknowledgements
We thank R.-P. S ekaly, A. Filali-Mouhim and K. Ghneim for transcriptional analysis of the Mtb acquisition subcohort of the Adolescent Cohort Study; E. Long, C. Blish for advice and the P815 mouse cell line and K562 human cell line; A. Kasmar for critically reading the manuscript. This work was supported by the Bill and Melinda Gates Foundation (T.J.S., P.K., Y.-h.C.), the National Institutes of Health AI127128 (Y.-h.C.), AI109662, AI057229 and AI125197 (P.K.), K12 5K12HL120001 (F.V.), 5T32AI07290–31 (R.R.C.), VirBio (P.K.), the Natural Science Foundation of China (81525016, 81772145) and the Science and Technology Project of Shenzhen (JSGG20160427104724699) (X.C.).
Reviewer information Nature thanks T. H. M. Ottenhoff and the other anonymous reviewer(s) for their contribution to the peer review of this work.
Footnotes
Competing interests The authors declare no competing interests.
Additional information
Extended data is available for this paper at https://doi.org/10.1038/s41586–018-0439-x.
Supplementary information is available for this paper at https://doi.org/10.1038/s41586–018-0439-x.
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Online content
Any methods, additional references, Nature Research reporting summaries, source data, statements of data availability and associated accession codes are available at https://doi.org/10.1038/s41586–018-0439-x.
Reporting summary. Further information on research design is available in the Nature Research Reporting Summary linked to this paper.
Data availability. The data that support the findings of this study are available from the corresponding authors upon reasonable request.
References
- 1.Houben RM & Dodd PJ The global burden of latent tuberculosis infection: a re-estimation using mathematical modelling. PLoS Med. 13, e1002152 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shea KM, Kammerer JS, Winston CA, Navin TR & Horsburgh CR Jr. Estimated rate of reactivation of latent tuberculosis infection in the United States, overall and by population subgroup. Am. J. Epidemiol 179, 216–225 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.WHO. Global Tuberculosis Report. http://www.who.int/tb/publications/global_report/en/ (2017).
- 4.Spitzer MH & Nolan GP Mass cytometry: single cells, many features. Cell 165, 780–791 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahomed H et al. Predictive factors for latent tuberculosis infection among adolescents in a high-burden area in South Africa. Int. J. Tuberc. Lung Dis 15, 331–336 (2011). [PubMed] [Google Scholar]
- 6.Takenami I et al. Blood cells and interferon-gamma levels correlation in latent tuberculosis infection. ISRN Pulmonol. 2013, 256148 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Joosten SA et al. Patients with tuberculosis have a dysfunctional circulating B-cell compartment, which normalizes following successful treatment. PLoS Pathog. 12, e1005687 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lu LL, et al. A functional role for antibodies in tuberculosis. Cell 167, 433–443 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ruvinsky I & Meyuhas O Ribosomal protein S6 phosphorylation: from protein synthesis to cell size. Trends Biochem. Sci 31, 342–348 (2006). [DOI] [PubMed] [Google Scholar]
- 10.Ueda Y, Kondo M & Kelsoe G Inflammation and the reciprocal production of granulocytes and lymphocytes in bone marrow. J. Exp. Med 201, 1771–1780 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shih CH, van Eeden SF, Goto Y & Hogg JC CCL23/myeloid progenitor inhibitory factor-1 inhibits production and release of polymorphonuclear leukocytes and monocytes from the bone marrow. Exp. Hematol 33, 1101–1108 (2005). [DOI] [PubMed] [Google Scholar]
- 12.Schölkopf B, Smola AJ, Williamson RC & Bartlett PL New support vector algorithms. Neural Comput. 12, 1207–1245 (2000). [DOI] [PubMed] [Google Scholar]
- 13.Vallania F et al. Leveraging heterogeneity across multiple data sets increases accuracy of cell-mixture deconvolution and reduces biological and technical biases. Preprint at https://biorxiv.org/content/early/2017/10/20/206466 (2017). [DOI] [PMC free article] [PubMed]
- 14.Malherbe ST et al. Persisting positron emission tomography lesion activity and Mycobacterium tuberculosis mRNA after tuberculosis cure. Nat. Med 22, 1094–1100 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zak DE et al. A blood RNA signature for tuberculosis disease risk: a prospective cohort study. Lancet 387, 2312–2322 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Scriba TJ et al. Sequential inflammatory processes define human progression from M. tuberculosis infection to tuberculosis disease. PLoS Pathog. 13, e1006687 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thompson EG et al. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis 107, 48–58 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esin S & Batoni G Natural killer cells: a coherent model for their functional role in Mycobacterium tuberculosis infection. J. Innate Immun 7, 11–24 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Junqueira-Kipnis AP et al. NK cells respond to pulmonary infection with Mycobacterium tuberculosis, but play a minimal role in protection. J. Immunol 171, 6039–6045 (2003). [DOI] [PubMed] [Google Scholar]
- 20.Feng CG et al. NK cell-derived IFN-γ differentially regulates innate resistance and neutrophil response in T cell-deficient hosts infected with Mycobacterium tuberculosis. J. Immunol 177, 7086–7093 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Kwan CK & Ernst JD HIV and tuberculosis: a deadly human syndemic. Clin. Microbiol. Rev 24, 351–376 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scully E & Alter G NK cells in HIV disease. Curr. HIV/AIDS Rep. 13, 85–94 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mansour I, Doinel C & Rouger P CD16+ NK cells decrease in all stages of HIV infection through a selective depletion of the CD16+CD8+CD3− subset. AIDS Res. Hum. Retroviruses 6, 1451–1457 (1990). [DOI] [PubMed] [Google Scholar]
- 24.Xie X, Li F, Chen JW & Wang J Risk of tuberculosis infection in anti-TNF-α biological therapy: from bench to bedside. J. Microbiol. Immunol. Infect 47, 268–274 (2014). [DOI] [PubMed] [Google Scholar]
- 25.Nocturne G et al. Impact of anti-TNF therapy on NK cells function and on immunosurveillance against B-cell lymphomas. J. Autoimmun 80, 56–64 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Bruns H et al. Anti-TNF immunotherapy reduces CD8+ T cell-mediated antimicrobial activity against Mycobacterium tuberculosis in humans. J. Clin. Invest 119, 1167–1177 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berry MP et al. An interferon-inducible neutrophil-driven blood transcriptional signature in human tuberculosis. Nature 466, 973–977 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maertzdorf J et al. Human gene expression profiles of susceptibility and resistance in tuberculosis. Genes Immun. 12, 15–22 (2011). [DOI] [PubMed] [Google Scholar]
- 29.Maertzdorf J et al. Functional correlations of pathogenesis-driven gene expression signatures in tuberculosis. PLoS ONE 6, e26938 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaforou M et al. Detection of tuberculosis in HIV-infected and -uninfected African adults using whole blood RNA expression signatures: a case–control study. PLoS Med. 10, e1001538 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson ST et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med 370, 1712–1723 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bloom CI et al. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS ONE 7, e46191 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Verhagen LM et al. A predictive signature gene set for discriminating active from latent tuberculosis in Warao Amerindian children. BMC Genomics 14, 74 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cai Y et al. Increased complement C1q level marks active disease in human tuberculosis. PLoS ONE 9, e92340 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ottenhoff TH et al. Genome-wide expression profiling identifies type 1 interferon response pathways in active tuberculosis. PLoS ONE 7, e45839 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tientcheu LD et al. Differential transcriptomic and metabolic profiles of M. africanum- and M. tuberculosis-infected patients after, but not before, drug treatment. Genes Immun. 16, 347–355 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee SW et al. Gene expression profiling identifies candidate biomarkers for active and latent tuberculosis. BMC Bioinformatics 17, S3 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Leipold MD & Maecker HT Phenotyping of live human PBMC using CyTOF™mass cytometry. Bio Protoc. 5, e1382 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fernandez R & Maecker H Cytokine-stimulated phosphoflow of PBMC using CyTOF mass cytometry. Bio Protoc. 5, e1496 (2015). [PMC free article] [PubMed] [Google Scholar]
- 40.Bruggner RV, Bodenmiller B, Dill DL, Tibshirani RJ & Nolan GP Automated identification of stratifying signatures in cellular subpopulations. Proc. Natl Acad. Sci. USA 111, E2770–E2777 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Amir ED et al. viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat. Biotechnol 31, 545–552 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Somanchi SS, McCulley KJ, Somanchi A, Chan LL & Lee DA A novel method for assessment of natural killer cell cytotoxicity using image cytometry. PLoS ONE 10, e0141074 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Salinas-Jazmín N, Hisaki-Itaya E & Velasco-Velázquez MA A flow cytometry-based assay for the evaluation of antibody-dependent cell-mediated cytotoxicity (ADCC) in cancer cells. Methods Mol. Biol 1165, 241–252 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Haynes WA et al. Empowering multi-cohort gene expression analysis to increase reproducibility. Pac. Symp. Biocomput 22, 144–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sweeney TE, Braviak L, Tato CM & Khatri P Genome-wide expression for diagnosis of pulmonary tuberculosis: a multicohort analysis. Lancet Respir. Med 4, 213–224 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Khatri P et al. A common rejection module (CRM) for acute rejection across multiple organs identifies novel therapeutics for organ transplantation. J. Exp. Med 210, 2205–2221 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.