Abstract
Targeted RNA-sequencing aims to focus coverage on areas of interest that are inadequately sampled in standard RNA-sequencing experiments. Here we present a novel approach for targeted RNA-sequencing that uses complex pools of reverse transcription primers to enable sequencing enrichment at user-selected locations across the genome. We demonstrate this approach by targeting hundreds to thousands of pre-mRNA splice junctions, revealing high-precision detection of splice isoforms, including rare pre-mRNA splicing intermediates.
Identification of the small subset of RNA-sequencing (RNA-seq) reads that span exon-exon junctions within transcripts has enabled the unambiguous detection of vast numbers of novel splice isoforms in scores of organisms1,2. Yet in spite of the power presented by this approach, the sequencing depth necessary to quantitatively detect many splicing events is significantly higher than most experiments generate. While this limitation of whole-transcriptome profiling has been addressed in part by methods that utilize antisense probes3,4 or PCR enrichment5 to target sequencing coverage to genomic regions of interest, a deeper understanding of the basic mechanisms by which splicing is regulated, and the pathological consequences of its mis-regulation, will be facilitated by methods that enable detection of splicing states within cells with higher resolution and greater precision. Towards this end, we have designed a novel targeted sequencing method that enhances splice junction detection and allows for resolution of splicing intermediates. Building upon the historically validated use of primer extension as a tool for assessing splicing status, we demonstrate the ability to multiplex hundreds to thousands of primer extension assays and evaluate the products by deep sequencing, an approach we hereafter refer to as Multiplexed Primer Extension sequencing, or MPE-seq (Fig1A). In this method, user-selected primers are extended to generate complementary DNA (cDNA) during a reverse transcription reaction, enabling targeting of RNA regions of interest. The use of elevated temperatures during reverse transcription minimizes non-specific primer annealing (FigS1), and each primer is appended with a next-generation sequencing adapter as well as a unique molecular identifier (UMI)6. A strand-extension step similar to template-switching7 appends the second sequencing adapter onto the 3′ terminus of the cDNA molecules. Coupling this approach with paired-end sequencing allows for the simultaneous querying of the 5′ and 3′ ends of the cDNAs from targeted regions (see Methods for full details).
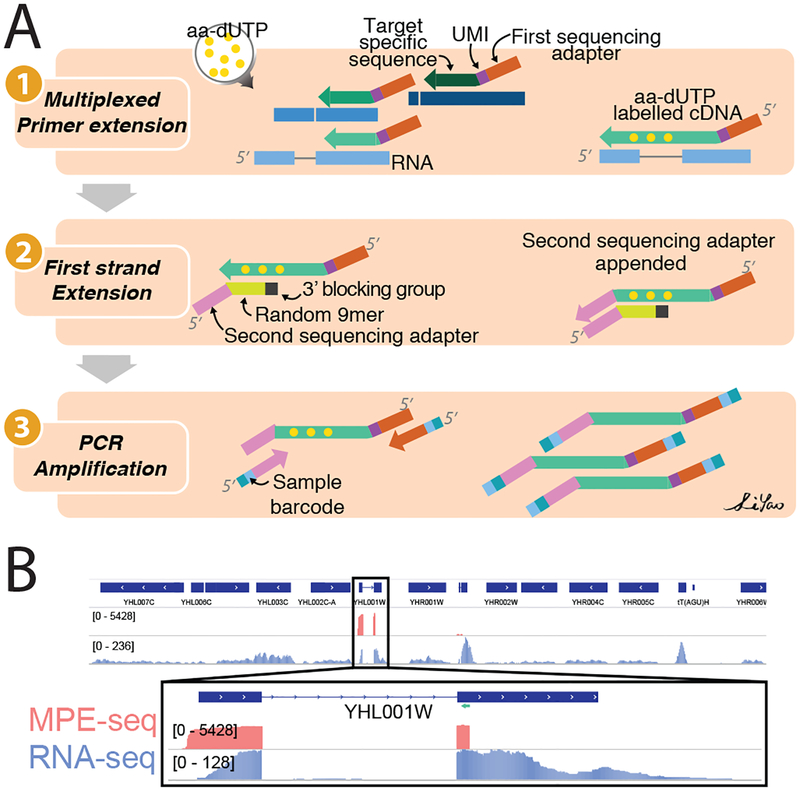
Figure1: MPE-seq uses complex pools of reverse transcription primers to target sequencing to regions of interest.
(A) Outline of MPE-seq protocol. (1) Hundreds to thousands of primers are pooled and extended by reverse transcription at targeted regions of interest. The incorporation of amino-allyl dUTP (aa-dUTP) nucleotides during reverse transcription enables purification of cDNA between subsequent steps. (2) The second sequencing adapter is appended to the 3’ terminus of the cDNA using an oligonucleotide template and Klenow enzyme. (3) Libraries are PCR amplified and sequenced.
(B) Genome browser screenshot of a targeted region in MPE-seq (pink) and conventional RNA-seq (purple). The location of a targeting primer is shown as a green arrow.
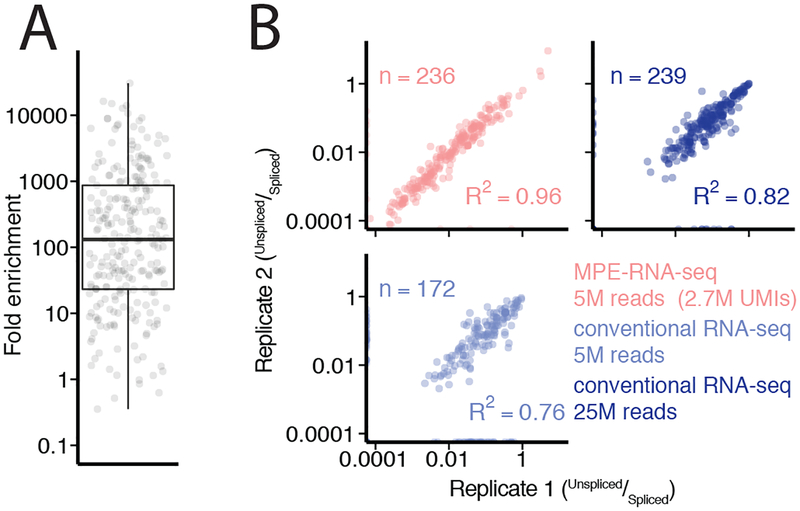
As an initial demonstration of MPE-seq, we examined pre-mRNA splicing in the budding yeast Saccharomyces cerevisiae. For each of the 309 annotated introns in the yeast genome, primers were systematically designed within a 50nt window immediately downstream of the 3′ splice site, ensuring that short extensions would cross splice junctions. Primers were pooled at equimolar concentration and MPE-seq libraries were generated using total cellular RNA from wildtype yeast and sequenced to a depth of only ~5 million reads. As a comparative reference, conventional RNA-seq libraries were generated using poly-A selected RNA and sequenced to ~40 million reads. Whereas the conventional RNA-seq libraries yielded read coverage that comprised full gene bodies across the transcriptome, MPE-seq coverage was focused on the selected genes, precisely targeted to the regions upstream of the designed primers (Fig1B). Just over 75% of sequenced fragments from MPE-seq mapped to targeted regions (FigS2, TableS1), resulting on average in a greater than 100-fold enrichment in sequencing depth at these regions when compared with RNA-seq (Fig2A, FigS3). Although the fold-enrichment varied on a target-by-target basis, it was nevertheless similar across transcripts with a wide range of expression levels (FigS3). From these data we extrapolate that a standard RNA-seq experiment would require ~500 million sequencing reads to achieve a similar level of coverage over the targeted regions as these 5 million MPE-seq reads provided. Given the increased read depth achieved over targeted regions using MPE-seq, we asked how well unspliced isoforms were sampled. Measurements of the fraction of unspliced message from replicate libraries using MPE-seq exhibited superior internal reproducibility compared to the larger, replicate RNA-seq libraries (Fig2B), likely reflecting the sampling noise associated with RNA-seq data with reduced sequencing depth over the targeted regions. Moreover, while MPE-seq is not amenable to de novo discovery of novel splicing events across the entire genome, it did allow for the identification of scores of rare, previously unannotated splicing events at the targeted regions (TableS2). Nevertheless, while MPE-seq provided increased sensitivity and reproducibility of splicing measurements, estimates of the fraction unspliced determined from MPE-seq in a wildtype strain only modestly correlated with those determined by RNA-seq (FigS4A–B). Notably, this correlation improved when comparing how these techniques measured changes in splicing between samples assayed by the same methodology (FigS4C), presumably reflecting inherent technical biases8 present in one or both approaches that are internally well controlled.
Figure2: MPE-seq enrichment enables high-precision measurements of splicing.
(A) Each point represents the fold enrichment of a target region in MPE-seq over conventional RNA-seq. Horizontal lines in boxplots represent the 25th, 50th, and 75th percentiles. Whiskers end at the 0th and 100th percentiles. n=249 target regions that were detected with at least one read in both RNA-seq and MPE-seq libraries for comparison.
(B) Scatter plots depict intron-retention measurements in replicate libraries made from biologically independent samples in MPE-seq and conventional RNA-seq at matched or greater read depth. Pearson correlation coefficients (R2) are indicated. ‘n’ is the number of intron-retention events which were quantified, requiring at least one spliced read and one unspliced read in both experiments.
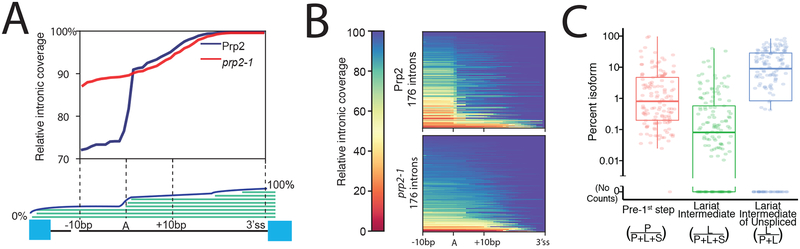
We next sought to determine whether we could detect splicing intermediates using MPE-seq. By identifying the locations of reverse transcription stops, primer extension reactions have historically been used to map a variety of biological features, including transcription start sites (TSSs)9, and the locations of branch sites within the lariat intermediate (LI) species of the pre-mRNA splicing reaction10,11 (FigS5A). Our approach anticipated the possibility of mapping the 3′ ends of the cDNA molecules, and indeed we found in our MPE-seq libraries that the 3′ ends of many cDNAs accumulated at the TSSs as determined by an orthologous method12 (FigS6), indicating that reverse transcription generally proceeded to the 5′ terminus of the RNA. Importantly, we also observed many cDNAs which terminated at or near the annotated branchpoint motifs within introns, with decreased read coverage upstream of the motifs, consistent with the inability of reverse transcriptase to read past the branched adenosine in the LI (Fig3A–B). This drop in read coverage was not apparent in MPE-seq libraries generated from a strain harboring a conditional mutation in Prp2, an RNA helicase required for catalyzing the 1st step of splicing13, corroborating that these cDNAs originate from LIs. We note that these LI-derived cDNAs often contained at their 3′termini a unique signature of mismatches incorporated by reverse transcriptase at the branched adenosine (FigS7) which may serve as a unique tag for de novo identification of branch sites in organisms with less well annotated branch sites14. The ability of MPE-seq to differentiate between unspliced isoforms allowed us to estimate that ~10% of unspliced pre-mRNAs are of the LI form genome-wide under steady state conditions (see FigS5B, TableS3, methods), albeit with significant variation between individual pre-mRNAs (Fig3B–C). Although we identified correlations between transcript- and intron-level features and the abundances of these species (FigS8), none of these correlations held when considering the abundance of pre-1st step RNA relative to LI, a metric that we expect would reflect variation between the relative catalytic rates of the 1st and 2nd steps of splicing. A more complete understanding of the determinants of in vivo splicing efficiency will require kinetic measurements of the individual steps of splicing rather than the steady state levels measured here. The ability of MPE-seq to robustly distinguish these splice isoforms provides an opportunity to do just this.
Figure3: MPE-seq enables genome-wide profiling of lariat intermediates.
(A) Meta-intron coverage plot surrounding predicted branchpoints in a wild-type (Prp2) and step1 splicing mutant strain (prp2–1). The region between the +10 position downstream of the annotated branchpoint and the 3′ splice site (3′ss) was re-scaled for each intron.
(B) Heatmap plots showing the relative coverage at each intron for which lariat intermediate reads were detected.
(C) Estimates of the relative abundance of each isoform for each targeted intron for which reads were detected. Horizontal lines in boxplots represent the 25th, 50th, and 75th percentiles. Whiskers end at the 0th and 100th percentiles. n=141 introns for which we attempted lariat quantification and found at least one spliced read.
Whereas our initial experiments were performed using individually synthesized oligonucleotides as primers, we sought to increase the utility of this approach by examining methods that would facilitate an increase in the number of targeted regions. We developed an approach that used pools of primers derived from array-based syntheses of thousands of oligonucleotides (FigS9A–B). Using this approach, we recreated the 309 previously described S. cerevisiae primers, and an additional 3918 primers that targeted splice junctions in the relatively intron-rich fission yeast Schizosaccharomyces pombe. Importantly, genome-wide splicing efficiencies determined from MPE-seq libraries generated using primers from pooled syntheses were highly correlated with those derived from individually synthesized oligos (FigS9D), validating the utility of this approach. Moreover, MPE-seq libraries generated with primers derived from pooled synthesis also showed strong enrichment for the targeted regions, with levels on par with what was observed using individually synthesized oligonucleotide primers (FigS2 and FigS9C). Not surprisingly, a modest increase in off-target reads was seen when primers from the pooled synthesis were used, consistent with the decreased sequence fidelity of array-based oligo synthesis15 and the increased capacity of these aberrant oligos to prime reverse transcription at undesirable locations. Additionally, as the fraction of the transcriptome that is targeted becomes larger, the fold enrichment over RNA-seq is naturally expected to decrease. Accordingly, when we used the ~4000 targeting primers in fission yeast we achieved a median enrichment at targeted regions of six-fold (TableS4). Nevertheless, this enrichment enabled detection of rare but natural alternative splicing events16 that are poorly sampled using standard RNA-seq library preparation methods (FigS9E). While we see no de facto limitation to the number of unique primer sequences or species that could be used for MPE-seq, with increasing numbers of primers comes an increasing potential for their cross-reactivity with undesirable RNA targets, highlighting the importance of specificity and fidelity in primer design and synthesis.
Our work here demonstrates the capacity of MPE-seq to facilitate examinations of pre-mRNA splicing status in a targeted, cost-effective way that improves the precision and sensitivity of splice isoform detection. The improved sensitivity of this approach is perhaps best exemplified by our ability to detect the LI products of the pre-mRNA splicing pathway. Though other recently described methods14,17,18, have reported large-scale detection of upstream-exon splice intermediates and excised lariats, MPE-seq uniquely detects LIs, not excised lariats, from unfractionated cellular RNA. Moreover, these profiling methods that detect RNAs physically associated with the splicoesome, require protein tagging and/or purification steps that necessitate large amounts of starting material, limiting the applications to which they might be applied. Conversely, MPE-seq can be implemented in virtually any system of interest with a need for only microgram quantities of RNA. Additionally, the ability of MPE-seq to query RNA from a wide variety of sources (cytoplasmic/nuclear fractionated RNA, polysome-fractionated RNA, poly-A selected RNA, metabolically labelled RNA) enables an analysis of the cellular location, translational or polyadenylation status and turnover rates of splice isoforms and intermediates. Overall, we expect that the sensitivity, precision and flexibility of this approach will enable a higher-resolution understanding of the splicing pathway. Likewise, primer extension assays have been used to assay RNA secondary structure after in vitro19 or in vivo20 chemical probing, and we expect that MPE-seq could be readily adapted to RNA structure interrogation and other approaches where primer-extension assays or targeted RNA sequencing is applicable.
Online Methods
Strain Maintenance and Growth Conditions.
Unless otherwise indicated, all S. cerevisiae experiments used the wild type (WT) strain BY4741 (MATa, his2Δ1, leu2Δ0, met15Δ0, ura3Δ0). Single colonies were inoculated into liquid YPD media and grown overnight at 30℃. Overnight cultures were then inoculated into fresh liquid YPD, seeding cultures at OD600 ~0.05. Cells were collected by vacuum filtration once cultures reached OD600 ~0.7, immediately followed by flash freezing in liquid nitrogen. Cell pellets were then stored at −80℃. For the temperature sensitive mutant prp2–121 we grew cultures as described above except at 25℃. Once cultures reached OD600 ~0.7, an equal volume of fresh 50℃ YPD media was added to shift cells to the non-permissive temperature of 37℃. The cultures were then maintained at 37℃ for 15 minutes before cell collection as described above. All S. pombe experiments used the wild type strain JP002 (h+, ade6-M210, leu1-32, ura4-D18). Single colonies were inoculated into liquid YES media and grown overnight at 30˚C. Overnight cultures were then inoculated into fresh liquid YES, seeded at OD600 ~0.05. Cells were collected by vacuum filtration upon reaching OD600 ~0.5 and immediately flash frozen in liquid nitrogen. Cell pellets were stored at −80˚C.
MPE-seq Primer and Oligo Design.
Gene specific reverse transcription primer design.
For each of the 309 annotated spliceosomal introns within the S. cerevisiae genome (annotations obtained from UCSC SacCer3) as well as for a subset of introns (3918 in total) within the S. pombe genome (annotations obtained from Ensemble ASM294v2.37), a reverse transcription primer was designed within the first 50 nucleotides downstream of the intron. Targeting to this region ensured that short-read sequencing of the products generated from reverse transcription with these primers would cross the upstream exon-exon or exon-intron boundaries, enabling determination of the splicing status. Primers were designed using OligoWiz, a program initially developed for microarray probe design, but which enables the selection of primer sequences optimized for target specificity relative to a designated genomic background22. We used the standalone version of OligoWiz with default parameters for short (24–26bp) oligo design to obtain optimal sequences within each 50bp window. To the 5′ end of each of these sequences was appended two additional sequence elements: a random 7-nucleotide unique molecular index (UMI) which allows for the detection and removal of amplification artifacts arising from library preparation6; and the P5 region of the Illumina sequencing primer to enable the sequencing of the reverse transcription products. Each of the primers targeting S. cerevisiae junctions was individually synthesized by Integrated DNA Technologies (IDT), the full sequences of which are provided in (TableS5). Array-based oligonucleotide synthesis was performed by LC Sciences using individual OligoMix syntheses for primers from each species (TableS6).
Complex oligo mix amplification method:
The array-based oligos are synthesized at vastly lower quantities than is required for cDNA synthesis in MPE-seq. To generate a quantity of primer pool that is sufficiently large, PCR amplification along with several processing steps were used (FigS9A). This was enabled by addition of 2 key sequence elements appended onto the 3′ end of the individually synthesized oligo primers detailed above. From the 5′ to 3′ direction: (1) a SapI restriction site; and (2) a PCR amplification sequence (TableS5). The oligos were amplified in a standard PCR reaction using Phusion polymerase. This 400 μL PCR reaction contained: 1% of the pooled oligonucleotides from LC Sciences as a template, a forward primer (oHX093) containing a C3 spacer at its 5′ end, and a reverse amplification primer (oHX094) containing a biotin-label at its 5′ end (TableS7). A total of 14 amplification cycles were performed, each consisting of the following conditions: denaturation at 95°C for 10 sec; annealing at 60°C for 20 sec; and extension at 72°C for 30 sec. Upon completion of this initial reaction, the entire reaction was used as a template to seed a larger (40 mL) PCR reaction. For efficient amplification, this large reaction was performed in four 96-well plates with 100μL in each well. Reaction conditions were identical to those described for the first reaction, but a total of 15 cycles were performed for this second amplification. Reactions were purified and concentrated by isopropanol precipitation. To generate single stranded primers for use in MPE-seq, the double-stranded amplicons were first digested using SapI (NEB R0569) in a 150 μL reaction containing 30 μL of enzyme. The reaction was incubated at 37°C overnight, after which the reaction products were concentrated by ethanol precipitation. Next, the 5′ to 3′ lambda exonuclease (NEB M0262) was used to preferentially degrade the two strands containing unmodified 5′ ends. This reaction was performed at 37°C for 2 hours according to the manufacturer’s protocol. The products of this reaction were then purified using Zymo columns using 7× volume binding buffer (2 M guanidinium-HCl, 75% isopropanol). After this step, the remaining DNA consisted of the desired single stranded RT primer, and an undesired single stranded section containing the SapI site plus the amplification primer. Making use of the 5′ biotin tag on the amplification primer, these undesired oligos were removed by affinity capture with streptavidin beads. Specifically, this was accomplished by using 50 μL of Dynabeads MyOne Streptavidin C1 according to the manufacturer’s protocol. The unbound supernatant fraction was retained as it contains the desired products. The recovered material was precipitated and verified using 6% native PAGE stained with SyBr Gold (FigS9B).
1st Strand extension template oligo design.
The oligos were designed with 3 key features from the 5′ to 3′ end of the oligo. First, a portion of the Nextera P7 sequencing adapter. Of the entirety of the P7 adapter, the region 3′ of the i7 barcode was used. This allowed for independent barcoding and amplification of discreet sequencing libraries. Second, a dN9 or dN12 anchor on the 3′ end of the oligo allowed it to randomly anneal to cDNA products. And third, a 3′ carbon block modification (hexanediol, IDT) was added to preclude the ability of Klenow to extend this primer. As such, the oligo may only be used as a template to append the Nextera sequencing adapter onto the end of first strand cDNAs. The full sequence of this primer can be found in TableS7.
MPE-Seq Library Prep
cDNA synthesis.
For S. cerevisiae libraries, RNA was isolated following a hot acid phenol extraction protocol23. Each library was generated using 10 μg of total RNA. From this RNA, cDNA was synthesized by mixing 1 μg of the gene specific primer pool described above with each RNA sample in a 20 μL reaction containing 50 mM Tris-HCl (pH 8.5), 75 mM KCl. The primers were then annealed in a thermocycler with the following cycle: 70°C for 1 minute; 65°C for 5 minutes; hold at 47°C. An equivalent volume of MMLV reverse transcriptase enzyme mix containing 1 mM dATP, 1 mM dGTP, 1 mM dCTP, 0.4 mM aminoallyl-dUTP, 0.6 mM dTTP, 50 mM Tris-HCl (pH 8.5), 150 mM KCl, 6 mM MgCl2, 10 mM DTT was pre-heated to 47°C and added to the primer-annealed RNA mix, resulting in a total reaction volume of 40 μL. Maintaining the samples at 47°C was essential for reducing off-target cDNA synthesis. Reactions were incubated at 47°C for 3 hours, followed by heat inactivation at 85°C for 5 minutes. Remaining RNA was hydrolyzed by addition of ½ volume of 0.3 M NaOH, 0.03 M EDTA and incubation at 65°C for 15 minutes. After neutralization with ½ (original) volume of 0.3 M HCl, the cDNA was purified with a Zymo-5 column using 7× volume of binding buffer (2 M guanidinium HCl, 75% isopropanol). Purified cDNA samples were dried to completion in a SpeedVac. For S. pombe libraries, RNA was isolated as described above. Poly-adenylated RNA was then isolated from 60 μg of total RNA using the NEBNext Poly(A) mRNA Magnetic Isolation Module. RNA was then fragmented to an average size of 200 nucleotides by incubating in 10 mM ZnCl2, 10 mM Tris-HCl pH 7.0 for 10 minutes at 65°C. The reaction was then quenched by addition of EGTA (pH 8.0) to a final concentration of 50 mM. The cDNA synthesis reactions were performed as above with some modifications. For reasons described below, 4 μL of Superscript III (ThermoFisher) reverse transcriptase was used along with the manufacturer supplied 5× buffer. For primer annealing and extension, samples were held at 55°C for 1 hour, followed by heat inactivation at 85°C for 5 minutes.
NHS ester biotin coupling.
Dried cDNA samples were resuspended in 18 μL of fresh 0.1 M Sodium Bicarbonate (pH 9.0), to which 2 μL of 0.1 mg/μL NHS-biotin (ThermoFisher 20217) was added. Reactions were incubated at 65°C for 1 hour followed by purification of biotin coupled cDNA from unreacted NHS-biotin by using Zymo-5 columns using 7× volume of binding buffer (2 M guanidinium HCl, 75% isopropanol).
Streptavidin-biotin purification.
20 μL of Dynabeads MyOne Streptavidin C1 (ThermoFisher 65602) per sample were pre-washed twice in 500 μL of 1× bind and wash buffer (5 mM Tris-HCl (pH 7.5), 0.5 mM EDTA and 1 M NaCl) as per manufacturer’s protocol. Washed beads were resuspended in 50 μL of 2× bind and wash buffer per sample and 50 μL was combined with each 50 μL purified cDNA sample. Biotin-streptavidin binding was allowed to proceed for 30 minutes at room temperature with rotation. Bound material was washed twice with 500 μL of 1× bind and wash buffer, followed by an additional wash with 100 μL of 1× SSC. To ensure purification of only single-stranded cDNAs, beads were then incubated with 0.1 M NaOH for two consecutive room temperature washes for 10 minutes and 1 minute, respectively. Finally, the bound material was washed 3 times with 100 μL 1× TE. The cDNA was eluted from the beads by heating samples to 90°C for 2 minutes in the presence of 100 μL of 95% formamide, 10 mM EDTA. The eluate was then purified using Zymo-5 columns as described above, and the cDNA was eluted from columns in 40 μL of water.
First strand extension.
Primers were annealed to purified cDNA by combining: 1uL 1st strand extension oligo (100 μM oJP788 for S. cerevisiae and oJP789 for S. pombe), 5 μL 10× NEB buffer 2, 40 μL purified cDNA sample, and 1 μL of 10 mM (each) dNTP mix. Samples were then incubated at 65°C for 5 minutes, followed by cooling to room temperature on the bench top. To each sample was added 3 μL of Klenow exo-fragment (NEB M0212) and reactions were incubated for 5 minutes at room temperature, after which they were moved to 37°C for 30 minutes. Samples were subsequently purified using streptavidin beads following the protocol described above. Samples were then concentrated using Zymo-5 columns, with elution in 33 μL of water.
PCR amplification.
Amplification of the reaction products was accomplished by using 10 μL of the purified material generated in the 1st strand extension reaction as a template in a PCR reaction. Illumina Nextera (i5) and (i7) indexing primers were used in a standard 50 μL PCR reaction with Phusion polymerase (ThermoFisher F530S). Cycling conditions were as follows: denaturation at 95°C for 10 sec; annealing at 62°C for 20 sec; and extension at 72°C for 30 sec. Libraries typically required between 14 and 20 cycles of amplification, depending upon the efficiency of library preparation. Each PCR reaction was then run on a 6% native poly-acrylamide gel, and the DNA was resolved by staining with SyBr gold. Libraries were size selected from 200bp to 800bp and DNA was extracted from gel fragments via passive diffusion overnight in 0.3 M sodium acetate (pH 5.3). Libraries were then ethanol precipitated and quantified.
cDNA synthesis temperature experiment.
Due to the target specific nature of MPE-seq cDNA synthesis, any reverse transcription (RT) events at non-target sites will reduce the fraction of on-target reads. Indeed, these off-target events contribute significantly to the nonspecific class reads in a typical MPE-seq experiment (Fig2A). One way to reduce off-target RT events is through increasing the specificity of the RT primers. We assessed this by testing the effect of increased temperature during the RT reaction on off-target sequencing reads. MPE-seq libraries were generated using the above described protocol with one primary difference: Increased reaction temperatures required the use of a thermostable enzyme. For this reason, Superscript III (ThermoFisher) was used along with the manufacturer supplied buffer (reaction concentration: 50 mM Tris-HCl pH 8.3, 75 mM KCl, 3 mM MgCl2). Primer annealing and reactions were carried out at 47°C, 51°C, and 55°C in replicate.
MPE-Seq Data Anlysis
Sequencing and alignment.
S. cerevisiae MPE-seq libraries were sequenced on the NextSeq platform by the BRC Genomics Facility at Cornell university using 60bp (P5) + 15bp (P7) paired-end chemistry. PCR duplicates were removed from the dataset by filtering out non-unique reads with respect to all base calls in both reads, including the 7bp UMI. In other words, for each set of identical paired-end reads, a single read-pair was retained for analysis. MPE-seq reads were aligned to the yeast genome (Reference genome assembly R64-1-124) using the STAR aligner25 with the following alignment parameters: {--alignEndsType EndToEnd --alignIntronMin 20 --alignIntronMax 1000 --alignMatesGapMax 400 --alignSplicedMateMapLmin 16 --alignSJDBoverhangMin 1 --outSAMmultNmax 1 --outFilterMismatchNmax 3 --clip3pAdapterSeq CTGTCTCTTATACACATCTCCGAGCCCACGAGAC --clip5pNbases 7 0}. Alignment files were filtered to exclude read mappings deriving from inserts less than 30bases. We believe these short fragments represent unextended reverse transcription primers that were retained in the sequencing libraries. These small fragments can sometimes erroneously map to splice junctions or target introns, even though we believe they are not derived from cellular RNA. S. pombe MPE-seq libraries were sequenced on the MiSeq platform by the BRC Genomics Facility at Cornell university using 100bp (P5) + 50bp (P7) paired-end chemistry. Reads were trimmed to 60bp and 15bp and processed as above for FigS9C while full length reads were processed as above for FigS9E.
S. cerevisiae RNA-seq libraries were sequenced on an Illumina HiSeq2500 by the BRC Genomics Facility at Cornell University using 100bp single end reads. S. pombe RNA-seq data26 were downloaded from NCBI (accession code SRS167019) and R2 of read pairs was discarded to make read lengths comparable to our other libraries. Reads were aligned using the STAR aligner with the following alignment parameters: {--alignEndsType EndToEnd --alignIntronMin 20 --alignIntronMax 1000 --alignSJDBoverhangMin 1 --outSAMmultNmax 1 --outFilterMismatchNmax 3 --clip3pAdapterSeq CTGTCTCTTATACACATCTCCGAGCCCACGAGAC}.
When applicable, replicate libraries were combined prior to alignment. However, to assess technical reproducibility of MPE-seq, replicate libraries were subsampled to varying read depths, aligned separately, and compared to RNA-seq libraries also subsampled to varying read depths.
Estimating fraction of on-target reads and MPE-seq enrichment.
Using bedtools27, read 1 alignments extending into a targeted intron or crossing a targeted exon-exon junction were considered on-target. Read 1 alignments which mapped downstream of a targeted intron but did not extend into an intron or cross an exon-exon junction were considered unextended primers. Read 1 alignments that mapped to the genome at non-targeted loci were considered off-target. Unmapped reads were then realigned to the genome with the same parameters as above (except with --clip5pNbases 31 0) and subsequent read 1 alignments to non-targeted loci were also considered off-target. Remaining reads were considered unmapped. Enrichment was calculated by dividing the number of read-count normalized exon-exon junctions found in MPE-seq by the number of read-count normalized exon-exon junctions in RNA-seq datasets for each targeted intron.
Estimating splice isoform abundances from MPE-Seq data.
For each intron, the relative abundance of unspliced and spliced isoforms was determined by counting spliced and unspliced reads. Spliced reads (S) were counted using the SJ.out.tab file created by the aligner. Unspliced reads were counted using bedtools27 to count the number of reads that cover any part of the intron, considering only the first read of the paired-end reads. Unspliced read counts were further categorized as deriving from a lariat intermediate (L) or pre-1st step RNA (P) by considering the mapping location of the second read of the paired end reads, which we observed to often terminate near the TSS, or in the case of a lariat-intermediate-derived cDNA, near the branchpoint-A of the intron. Based on paired end mapping locations, each fragment was categorized into one of six categories (See FigS5) and the counts within those six categories were used to calculate S, P, and L as follows:
Locations of branchpoints (TableS8) were determined by consolidating the most used branchpoint from lariat sequencing data28 and previously described branch locations based on sequence motif searches29.
Heatmaps and meta-gene plots:
To generate metagene plots which illustrate read coverage around features of interest, we used the deepTools ComputeMatrix command30 in conjunction with a BigWig coverage file of the 3′ terminating bases and a bedfile containing TSS-positions as determined by PRO-cap12 or a bedfile containing the annotated branchpoint regions detailed above. Importantly, this bedfile was filtered to only include branchpoint regions that would produce a lariat intermediate that is within the size range captured by library size-selection of MPE-seq libraries (see column “AttemptedLariatQuantification?” in TableS3 of Supplemental Data).
RNA-seq Experiments
Library prep:
For each RNA-seq library, 1 μg of total RNA was input into the “NEBNext Ultra Directional RNA Library Prep Kit for Illumina”. Libraries were prepared following the manufacturer’s protocol.
Estimating splice isoform abundances from RNA-seq data:
Similar to MPE-seq data, spliced reads from target introns were counted using the SJ.out.tab file created by the aligner. Unspliced reads were counted using the bedtools software package27 to count the number of reads which overlapped an intron. Spliced and unspliced read counts for each intron were then length normalized for the feature’s potential mapping space. The potential mapping space for a spliced read is equal to 2 × read length minus the minimum splice junction overhang length. The potential mapping space for an unspliced read is equal to the 2× read length minus minimum splice junction overhang length plus length of the intron. Reads counts assigned to each feature were then divided by the length. Fraction unspliced was calculated for each intron as the quotient of length normalized unspliced reads and spliced reads.
Gene expression normalization
Relative transcript expression was calculated from RNA-seq data via transcripts per million (TPM) normalization31, only considering exonic reads and exonic gene-lengths. For S. cerevisiae MPE-seq data, a similar TPM metric was calculated by summing the reads per gene and dividing by the number of library mapped reads. Given that a single RNA corresponds to a single primer extension event, and because nearly all targeted transcripts have only a single targeting primer, normalization by gene length was not done in this calculation of TPM.
Supplementary Material
Acknowledgements
We thank members of the J.A.P., H. Kwak, and A. Grimson laboratories, as well as the anonymous reviewers for critical feedback on this work. We thank Li Yao for initial drafts of Fig1A and FigS9A. We thank P. Schweitzer, J. Grenier, and the BRC Genomics Facility at Cornell for outstanding technical support with Illumina sequencing. This work was funded by a Research Scholars Grant from the American Cancer Society (to J.A.P.) and NIH grant R01GM098634 (to J.A.P.)
Footnotes
Competing interests statement
The authors have no competing interests to declare.
Code Availability
Code for basic analysis steps are available at https://github.com/bfairkun/ScerSpliceSeq.
Data Availability
All sequencing data are available through NCBI’s Sequence Read Archive (SRA) at accession number SRP148810.
References
- 1.Merkin J, Russell C, Chen P & Burge CB Evolutionary dynamics of gene and isoform regulation in Mammalian tissues. Science 338, 1593–9 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbosa-Morais NL et al. The evolutionary landscape of alternative splicing in vertebrate species. Science (80-.) 338, 1587–1593 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Mercer TR et al. Targeted RNA sequencing reveals the deep complexity of the human transcriptome. Nat. Biotechnol 30, 99–104 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mercer TR et al. Targeted sequencing for gene discovery and quantification using RNA CaptureSeq. Nat. Protoc 9, 989–1009 (2014). [DOI] [PubMed] [Google Scholar]
- 5.Blomquist TM et al. Targeted RNA-Sequencing with Competitive Multiplex-PCR Amplicon Libraries. PLoS One 8, e79120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kivioja T et al. Counting absolute numbers of molecules using unique molecular identifiers. Nat. Methods 9, 72–74 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Zhu YY, Machleder EM, Chenchik A, Li R & Siebert PD Reverse transcriptase template switching: A SMART™ approach for full-length cDNA library construction. BioTechniques 30, 892–897 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Zheng W, Chung LM & Zhao H Bias detection and correction in RNA-Sequencing data. BMC Bioinformatics 12, (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carey MF, Peterson CL & Smale ST The primer extension assay. Cold Spring Harb. Protoc 8, 164–173 (2013). [DOI] [PubMed] [Google Scholar]
- 10.Coombes CE & Boeke JD An evaluation of detection methods for large lariat RNAs. RNA 11, 323–31 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Padgett RA et al. Nonconsensus branch-site sequences in the in vitro splicing of transcripts of mutant rabbit beta-globin genes. Proc. Natl. Acad. Sci. U. S. A 82, 8349–8353 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Booth GT, Wang IX, Cheung VG & Lis JT Divergence of a conserved elongation factor and transcription regulation in budding and fission yeast. Genome Res 26, 799–811 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim SH & Lin RJ Spliceosome activation by PRP2 ATPase prior to the first transesterification reaction of pre-mRNA splicing. Mol. Cell. Biol 16, 6810–6819 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen W et al. Transcriptome-wide Interrogation of the Functional Intronome by Spliceosome Profiling. Cell 173, 1031–1044.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wan W, Lu M, Wang D, Gao X & Hong J High-fidelity de novo synthesis of pathways using microchip-synthesized oligonucleotides and general molecular biology equipment. Sci. Rep 7, 6119 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stepankiw N, Raghavan M, Fogarty EA, Grimson A & Pleiss J a. Widespread alternative and aberrant splicing revealed by lariat sequencing. Nucleic Acids Res 43, 8488–501 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nojima T et al. Mammalian NET-Seq Reveals Genome-wide Nascent Transcription Coupled to RNA Processing. Cell 161, 526–540 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burke J et al. Spliceosome profiling visualizes the operations of a dynamic RNP in vivo at nucleotide resolution. Cell 173, 1014–1030 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lucks JB et al. Multiplexed RNA structure characterization with selective 2’-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq). Proc. Natl. Acad. Sci 108, 11063–11068 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rouskin S, Zubradt M, Washietl S, Kellis M & Weissman JS Genome-wide probing of RNA structure reveals active unfolding of mRNA structures in vivo. Nature 505, 701–5 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hartwell LH, McLaughlin CS & Warner JR Identification of ten genes that control ribosome formation in yeast. MGG Mol. Gen. Genet 109, 42–56 (1970). [DOI] [PubMed] [Google Scholar]
- 22.Wernersson R & Nielsen HB OligoWiz 2.0 - Integrating sequence feature annotation into the design of microarray probes. Nucleic Acids Res 33, (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Collart MA & Oliviero S in Current Protocols in Molecular Biology (2001). doi: 10.1002/0471142727.mb1312s23 [DOI] [PubMed] [Google Scholar]
- 24.Engel SR et al. The reference genome sequence of Saccharomyces cerevisiae: then and now. G3 (Bethesda) 4, 389–98 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dobin A et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rhind N et al. Comparative functional genomics of the fission yeasts. Science 332, 930–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Quinlan AR & Hall IM BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayerle M et al. Structural toggle in the RNaseH domain of Prp8 helps balance splicing fidelity and catalytic efficiency. Proc. Natl. Acad. Sci 114, 4739–4744 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grate L & Ares M Searching yeast intron data at Ares lab web site. Methods in Enzymology 350, 380–392 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Ramírez F et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res 44, W160–W165 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Conesa A et al. A survey of best practices for RNA-seq data analysis. Genome Biology 17, (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.