Abstract
Application of the commonly used volatile anesthetic sevoflurane after brain ischemia (sevoflurane postconditioning) attenuates ischemic brain injury. It is not known whether autophagy plays a role in this sevoflurane postconditioning-induced neuroprotection. Human SH-SY5Y cells were induced to become neuron-like cells. These cells were subjected to 1 h oxygen-glucose deprivation (OGD) and then exposed to sevoflurane for 1 h. Chloroquine, an inhibitor of autolysosomes, rapamycin, an autophagy inducer, or 3-methyladenine (3-MA), an autophagy inhibitor, were incubated with cells during OGD and sevoflurane exposure. OGD and the subsequent simulated reperfusion increased lactate dehydrogenase (LDH) release from the cells. This increase was dose-dependent inhibited by sevoflurane postconditioning. OGD increased the ratio of microtubule-associated protein 1 light chain 3 (LC3) II to LC3I and the expression of beclin-1 and p62. These increases were attenuated by sevoflurane. Sevoflurane alone did not have any effects on the expression of p62, beclin-1 and the ratio of LC3II to LC3I. Sevoflurane also enhanced the co-location of autophagosomes and lysosomes. Chloroquine increased the ratio of LC3II to LC3I, p62 and LDH release in cells subjected to OGD. Sevoflurane postconditioning attenuated OGD-induced inactivation of Akt and mechanistic target of rapamycin (mTOR). Inducing autophagosome generation by rapamycin attenuated sevoflurane postconditioning-reduced LDH release. Inhibition of autophagosome generation by 3-MA decreased OGD-induced LDH release. These results suggest that OGD increase autophagosome accumulation via increased formation of autophagosomes and reduced autophagosome clearance and that attenuation of OGD-induced autophagosome accumulation may contribute to sevoflurane postconditioning-induced cell protection.
Keywords: autophagy, autolysosomes, neuroprotection, sevoflurane
1. Introduction
Stroke remains a major global health problem. The Global Burden of Disease (GBD) study ranked stroke as the second most common cause of death (Lozano, et al., 2012) and the third most common cause of disability-adjusted life-years (DALYs) (Murray, et al., 2012) worldwide in 2010. Despite stable incidence rates and declining mortality rates over the past 20 years, the absolute number of people who have a stroke every year, live with the consequences of stroke, and die from their stroke is increasing (Feigin, et al., 2014). The high mortality and disability rates of stroke urge researchers to seek effective treatment. While the majority of clinical trials on exogenous neuroprotective agents for stroke have been disappointing (Ehrenreich, et al., 2009; Ikonomidou, et al., 2002), evoking the endogenous ability for neuroprotection, such as via ischemic conditioning, is a focus of research (Pan, et al., 2016).
Ischemic conditioning refers to a number of related neuroprotective strategies that render neural tissues tolerant to ischemia by conditioning them with brief cycles of ischemia and reperfusion (Murry, et al., 1986), or with some other treatments, such as hypoxia (Zhao, et al., 2005) and volatile anesthetic (Adamczyk, et al., 2010). Although the field of conditioning was started with ischemic preconditioning (brief cycles of ischemia and reperfusion before a long duration of ischemia) (Murry, et al., 1986), its clinical application is limited because the intervention requires accurate prediction of the occurrence of the severe ischemic events. Compared with preconditioning, postconditioning that involves using the conditioning stimulus after the damaging insult has already occurred can also provide neuroprotection (Zhao, et al., 2003). Among the methods that can induce this effect, anesthetic postconditioning has attracted special attention because it is a non-invasive maneuver (Lee, et al., 2008). As a newer volatile anesthetic that is used worldwide in patient care, sevoflurane has been shown to induce a postconditioning effect in the brain (Ren, et al., 2014).
Autophagy is an important physiological process that maintains cellular homeostasis through lysosome-mediated protein degradation and organelle turnover (Mizushima, 2007). Recent studies have shown that autophagy is involved in the cardioprotective effects of ischemic preconditioning and cell protection by the general anesthetic isoflurane (Gidlof, et al., 2016). Anesthetics have effects on autophagy (Ye, et al., 2017). Thus, we hypothesize that the regulation of autophagy is involved in the neuroprotection conferred by sevoflurane postconditioning. To test this hypothesis, we induced human SH-SY5Y cells into neuron-like cells. Oxygen-glucose deprivation (OGD) was used to simulate ischemia in vitro.
2. Materials and methods
2.1. Cell culture
SH-SY5Y cells, a human neuroblastoma cell line, were obtained from the American Type Culture Collection (Manassas, VA) and cultured as we described before (Lin, et al., 2011). Briefly, cells were cultured in a T75 flask containing 13 ml of Eagle’s Minimum Essential Medium (EMEM)/Ham’s F-12 nutrient mixture (1:1) with 10% fetal bovine serum. The cells were kept at 37°C in a humidified incubator gassed with 95% air and 5% CO2. The medium was changed twice per week. When the cells were 70% - 80% confluent, they were exposed to 0.25% trypsin-EDTA solution and sub-cultured in a new flask.
The SH-SY5Y cells were plated at a density of 1 × 105 cells/cm2 in 6-well plates for lactate dehydrogenase (LDH) release assay or Western blotting. One day after plating, cells were incubated in neurobasal medium, supplemented with B-27 supplement (GIBCO, Carlsbad, CA) and L-glutamine (500 μM; Nacalai Tesque Inc., San Diego, CA). Retinoic acid (RA, Sigma, St Louis, MO) at 10 μM was added to the medium for 3 days to induce SH-SY5Y cells to differentiate into a homogenous population of cells with neuronal morphology (Kume, et al., 2008). These cells were used in the experiments.
2.2. Oxygen-glucose deprivation
Cells in control group were washed with phosphate buffered saline (PBS) and incubated in neurobasal medium in a humidified atmosphere of 95% air and 5% CO2 at 37°C. The exposure of cells to OGD was performed as we described before (Wu, et al., 2010). Briefly, neurobasal-A medium that did not contain L-glucose (GIBCO) was bubbled with 100% N2 for 30 min. Cells were washed with PBS once and 2 ml/well of the neurobasal-A medium was added to the cells. These plates were immediately placed in an air-tight chamber (Billups-Rothenberg, Inc., Del Mar, CA) gassed with 100% N2 for 10 min. The oxygen content in the outlet of the chamber was monitored with a DatexTM infrared analyzer (Capnomac, Helsinki, Finland) and reached 2% at 3 – 5 min after the onset of gassing. After closure of the inlet and outlet of the chamber, the chamber was kept at 37°C for 1 h. After confirming that the oxygen content in the chamber was still lower than 2% at the end of the OGD period, the chamber was opened and glucose, B-27 supplement and L-glutamine (500 μM) was added to make the final glucose concentration in the buffer at 4.5 g/l. The plates were kept for 6 h (Western blotting) or 20 h (Western blotting and LDH release assay) in a humidified atmosphere of 95% air and 5% CO2 at 37°C.
2.3. Anesthetic exposure
Cells were placed in an airtight chamber immediately after OGD. The chamber was gassed with 1%, 2%, 3%, 4% or 5% sevoflurane in the carrying gases (95% air and 5% CO2) for 15 min. Volatile anesthetic concentrations in the gases from the outlet of the chamber were monitored with a DatexTM infrared analyzer and reached the target concentrations at ~3 min after the onset of gassing. The chamber was sealed and the incubation was for 1 h at 37°C. At the end of incubation, anesthetic concentrations in the gases from the chamber were confirmed to be at the target concentrations by the infrared analyzer. The plates then were kept for 5 h or 19 h in a humidified atmosphere of 95% air and 5% CO2 at 37 °C.
2.4. Treatment with other reagents
The stock solution of chloroquine diphosphate (CQ) (Sigma, C6628) was prepared in PBS and stored at −20°C until it was used. To block autophagosome clearance, cells in 6-well plates were treated with 50 μM CQ (Pivtoraiko, et al., 2010) at the beginning of OGD or at the corresponding time point for the control group. After the OGD, cells were incubated with CQ at 37°C until they were used for Western blot or LDH assay.
To study the role of autophagy activation, cells were pretreated with rapamycin for 24 h. The final concentration of rapamycin (Sigma, R0395) was 0.2 μM (final concentration of DMSO that was used to dissolve rapamycin was 0.1% v/v) (Xiong, et al., 2011). DMSO was added to the culture medium in other groups. Rapamycin was present until the cells were harvested for assay.
To investigate the effect of autophagy inhibition, 3-methyladenine (3-MA, Sigma, M9281) was dissolved in PBS at 55°C, stored at −20°C and completely dissolved by warming to 55°C just prior to use. The final concentration of 3-MA was 5 mM (Deng, et al., 2013). 3-MA was added 1 h prior to OGD and present until cells were harvested for assy.
2.5. LDH activity assay
LDH activity was determined using an LDH cytotoxicity detection kit as we did before (Kim, et al., 2009a; Kim, et al., 2009b). Briefly, the incubation solution harvested at the end of experiments was centrifuged at 16,060 g for 10 min. One hundred micro-litters of the supernatant were transferred to 96-well plates and incubated with the same volume of reaction mixture from the kit. The samples then were read in a spectrophotometry (Bio-Rad Laboratories, Hercules, CA) with the absorbance wavelength at 492 nm and the reference wavelength at 655 nm. Background absorbance from the cell-free buffer solution was subtracted from all absorbance measurements. After removal of the incubation solution from 6-well plates, 1% triton X-100 lysing solution was applied to each well to dissolve the remaining cells. The percentage of LDH released to incubation buffer in total LDH was calculated as follows: LDH in the buffer × 100 / (LDH in the buffer + intracellular LDH released by triton X-100).
2.6. Western blotting
Human neuron-like cells after various treatments were homogenized in RIPA buffer (Thermo Scientific, 89901) containing protease inhibitor cocktail (Sigma, P2714). The phosphatase inhibitor (Roche, 04906845001) was added when preparing samples for detecting phospho-Akt and phospho-mTOR. Homogenates were centrifuged at 4°C for 20 min at 16,060 g. Protein content in the supernatant was determined using a Bio-Rad Protein Assay Kit. Twenty micrograms of proteins per lane were loaded and electrophoresed in a 12% polyacrylamide gel and then blotted onto a polyvinylidene difluoride membrane. The membrane was blocked with Protein-Free T20 Blocking Buffer (Thermo Scientific, 37573) and then incubated with the following primary antibodies: rabbit anti-LC3B (1:500; Abcam, ab48394) antibody, rabbit anti-beclin-1 (1:1000; Cell Signaling, D40C5) antibody, mouse anti-p62 (1:1000; Abcam, ab56416) antibody, goat anti-cathepsin D (1:200; Santa Cruz, sc-6486) antibody, mouse anti-actin (1:5000; Abcam, ab6276) antibody, rabbit anti-phospho-mTOR (Ser2448) (1:1000, Cell Signaling, 2971) antibody, mouse anti-mTOR (1:1000, Cell Signaling, 4517) antibody, mouse anti-phospho-Akt (Ser473) (1:1000, Cell Signaling, 12694) antibody, or rabbit anti-Akt (1:1000, Cell Signaling, 4685) antibody. Appropriate secondary antibodies were used and protein bands were visualized and quantified using G:Box equipped with gene tools analysis software (Syngene, Frederick, MD, USA). The densities of protein bands were normalized to those of actin to control for errors in protein sample loading and transferring during Western blotting. The results from cells under various experimental conditions then were normalized by those of the corresponding control cells.
2.7. Immunofluorescent staining
Human neuron-like cells plated on Lab-Tek Chamber slides (Thermo Fisher Scientific, 177445) after various treatments at 20 h after the OGD were used for staining. They were washed once with PBS and fixed with 4% paraformaldehyde for 30 min at room temperature. After being washed with PBS for 3 times, cells were incubated with PBS containing 0.1% Triton X-100 for 30 min and washed with PBS for 3 times again. The cells were incubated with blocking solution in PBS containing 5% bovine serum albumin (BSA, MP Biomedicals, 820451) for 1 h at room temperature. Next, they were incubated with appropriate primary antibodies [rabbit anti-LC3B antibody, 1:250, Abcam, ab48394; mouse anti-lysosome-associated membrane glycoprotein 1 precursor −1 (LAMP-1) antibody, 1:200, Abcam, ab25630] diluted in PBS with 5% BSA for 24 h at 4°C. Slides were then rinsed with PBS for 3 times. Cells were subsequently incubated with Alexa Fluor 488 (1:200 dilution, Invitrogen, A21206) or 594 (1:200 dilution, Invitrogen, A21203) secondary antibody in PBS with 5% BSA for 1 h at room temperature in the dark. After being rinsed with PBS for 3 times again, nuclei were stained with DAPI solution (Thermo Fisher Scientific, 62249) for 3 min at room temperature. Cells were washed 3 times with PBS, and covered with coverslips in Aqua-Mount mounting medium (Richard-Allan Scientific, 13800) and left at 4°C overnight.
2.8. Confocal microscopy and co-localization analysis
The slides were examined under an Axio Observer LSM 710 confocal system (Carl Zeiss Microscopy GmbH, Jena, Germany) and analyzed with ZEN Black software (Carl Zeiss Microscopy GmbH). Co-localization analysis of images acquired by confocal laser scanning microscopy was performed using the software FIJI (ImageA 1.45j; Max Planck Society). The plugin “Coloc 2” allows the quantitative determination of co-localizing fluorescence intensities acquired in different channels using methods described previously (Costes, et al., 2004; Manders, et al., 1993). The obtained Mander’s coefficients were used as the fraction of co-localization of both channels, i.e. tM1 represented the fraction of lysosomes that also contain green fluorescent protein LC3 and tM2 represented the fraction of pixels of green signal from LC3 that also had red signal from lysosomes. In addition to the Mander’s coefficients, Pearson correlation coefficients were obtained as another measure for co-localization. One hundred fifty cells in random fields from 3 independent experiments were analyzed.
2.9. Statistical analysis
Results are presented as mean ± S.E.M. (n ≥ 5). Data were analyzed by one-way analysis of variance followed by Tukey test if the data were normally distributed, by one-way analysis of variance on ranks followed by Tukey test if the data were not normally distributed. Differences were considered to be significant at P < 0.05 based on two-tailed hypothesis testing. All statistical analyses were performed with SigmaStat 3.5 (Systat Software, Inc., Point Richmond, CA, USA).
3. Results
3.1. Sevoflurane protected neuron-like cells and CQ enhanced cell injury after OGD-reperfusion
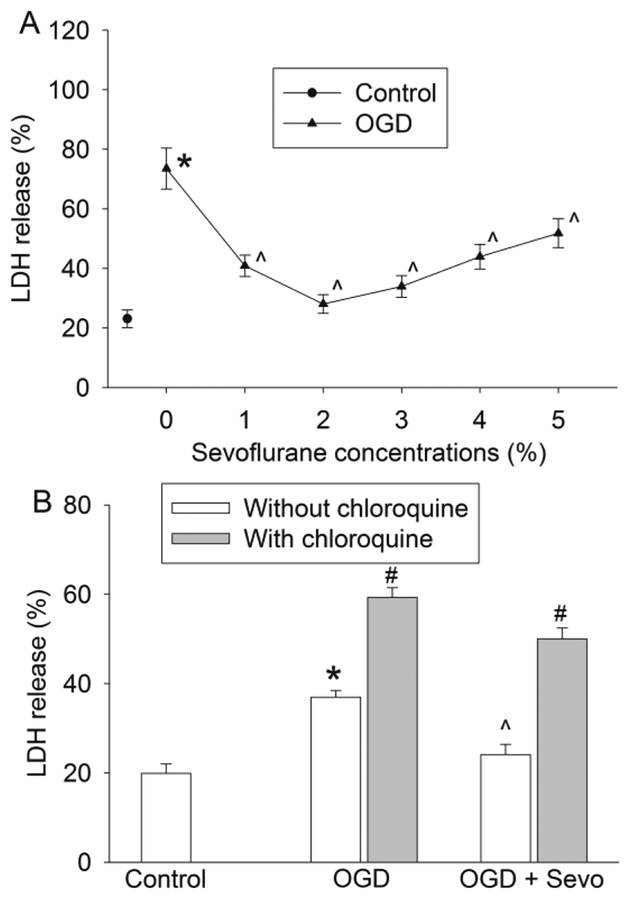
As shown in Fig. 1, the human neuron-like cells differentiated from SH-SY5Y cells responded to OGD and simulated reperfusion with an increase of LDH release into culture medium, suggesting cell injury. This injury was dose-dependently decreased by sevoflurane applied for 1 h immediately after OGD. However, the effects of sevoflurane appeared to be bell-shaped curve with the best protective effect at 2% sevoflurane, a clinically relevant concentration. Based on these results, 2% sevoflurane was used for all subsequent experiments. To determine whether autophagosome-lysosome fusion and autophagosomes play a role in cell injury, we used CQ, a well-known inhibitor of autophagic proteolysis (Kimura, et al., 2013). CQ treatment significantly aggravated the injury of neuron-like cells after OGD-reperfusion injury no matter whether this was in the presence or absence of sevoflurane postconditioning (Fig. 1B). These results suggest the importance of normal autophagy in cell survival.
Fig. 1. Sevoflurane attenuated oxygen-glucose deprivation (OGD)-induced cell injury and chloroquine enhanced the injury.
Neuron-like cells were subjected to 1 h OGD followed with a 20 h simulated reperfusion. A: Cells were exposed to various concentrations of sevoflurane for 1 h immediately after the onset of simulated reperfusion. Results are means ± S.E.M. (n = 18). B: Chloroquine was added just before the OGD and was present till the cells were harvested for LDH release assay. Sevoflurane at 2% was used in this experiment. Results are means ± S.E.M. (n = 9). * P < 0.05 compared to control. ^ P < 0.05 compared to OGD group. # P < 0.05 compared with the corresponding condition without chloroquine. OGD: oxygen-glucose deprivation; Sevo: 2% sevoflurane
3.2. Sevoflurane promoted autophagosome clearance in cells after OGD
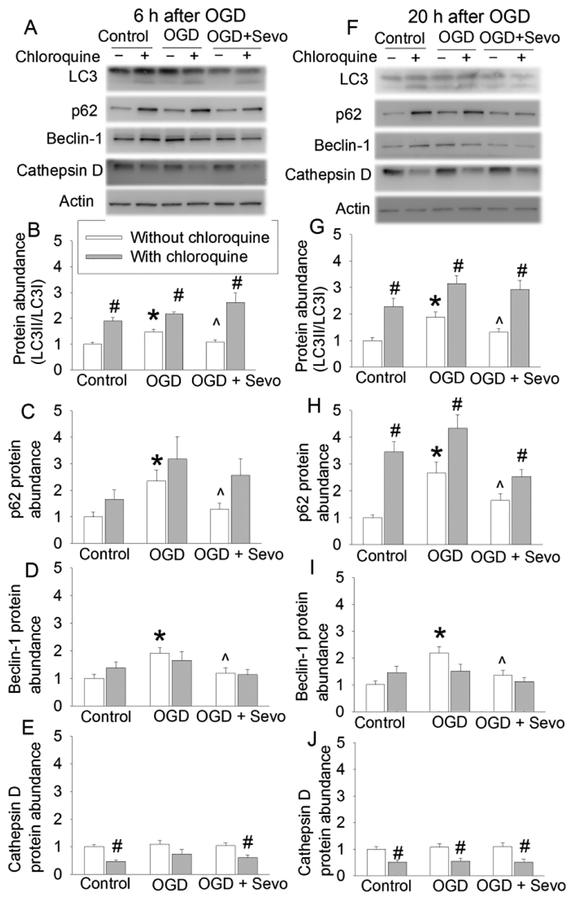
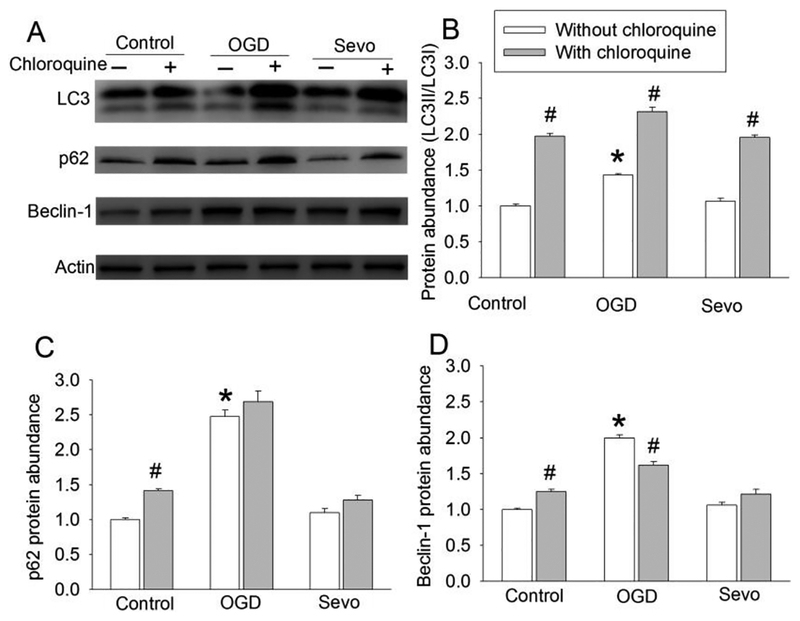
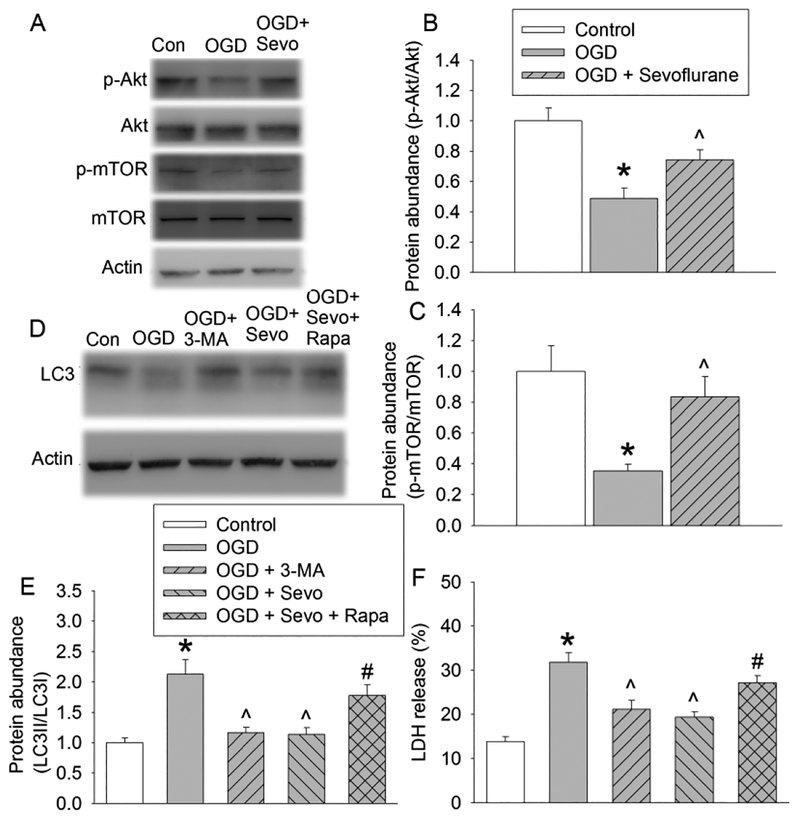
To determine the role of autophagy in the protective effects of sevoflurane, an analysis of the expression of the autophagic markers LC3, beclin-1, p62 and cathepsin D was conducted at 6 h and 20 h after OGD. The ratio of LC3II/LC3I and the expression of beclin-1 and p62 were increased at both time points. This increase was reduced by 2% sevoflurane postconditioning. However, either OGD-reperfusion or 2% sevoflurane postconditioning did not affect the level of cathepsin D (Fig. 2). Because LC3 conversion is a marker for autophagosome formation (Glick, et al., 2010; Mizushima, 2007) and p62 is selectively incorporated into autophagosomes through direct binding to LC3 and then degraded by autophagy (Glick, et al., 2010; Mizushima, 2007; Rusten, et al., 2010), these results suggest that OGD impairs the clearance of autophagosomes and sevoflurane attenuates this impairment. However, the OGD-reperfusion-induced impairment of autophagosome clearance is not likely due to the dysregulation of lysosomal activity because the expression of cathepsin D, an important proteinase contained in lysosomes (Barrett, 1970), was not changed by OGD or sevoflurane compared with control group. We used CQ to clarify the role of sevoflurane-induced autophagosome clearance enhancement in the protection of sevoflurane. Like NH4Cl, CQ neutralizes the lysosomal pH and causes the accumulation of sequestrated materials in either autophagosomes or autolysosomes (Dunmore, et al., 2013; Geng, et al., 2010). Moreover, CQ has been suggested to block autophagosomes fusion with lysosomes (Klionsky, et al., 2016). CQ increased LC3II and p62 (Fig. 2), suggest that the maturation of autophagosomes has been blocked in the presence of CQ. CQ did not change the expression of beclin-1, suggesting that CQ does not affect the induction of autophagosome formation. In addition, CQ reduced the expression of cathepsin D (Fig. 2), consistent with its effects on lysosomes. Interestingly, sevoflurane did not affect the expression of p62, beclin-1 and the ratio of LC3II to LC3I under control condition (Fig. 3), suggesting that sevoflurane does not alter the formation and clearance of autophagosomes under control condition.
Fig. 2. Effects of 2% sevoflurane postconditioning on OGD-induced autophagic markers in human neuron-like cells.
Cells were subjected to 1 h OGD followed with 6 h or 20 h simulated reperfusion. Cells were exposed to 2% sevoflurane for 1 h immediately after the onset of simulated reperfusion. Chloroquine was added before the OGD and was present till the cells were harvested for assay. A: Representative images of Western blotting of samples harvested at 6 h after OGD. B: Quantitative results of the ratio of LC3II to LC3I at 6 h after OGD. C: Quantitative results of p62 at 6 h after OGD. D: Quantitative results of beclin-1 at 6 h after OGD. E: Quantitative results of cathepsin D at 6 h after OGD. F: Representative images of Western blotting of samples harvested at 20 h after OGD. G: Quantitative results of the ratio of LC3II to LC3I at 20 h after OGD. H: Quantitative results of p62 at 20 h after OGD. I: Quantitative results of beclin-1 at 20 h after OGD. J: Quantitative results of cathepsin D at 20 h after OGD. Results are means ± S.E.M. (n = 9). * P < 0.05 compared to control. ^ P < 0.05 compared to OGD group. # P < 0.05 compared with the corresponding condition without chloroquine. OGD: oxygen-glucose deprivation; Sevo: 2% sevoflurane.
Fig. 3. Effects of 2% sevoflurane autophagic markers in human neuron-like cells.
Cells were exposed to 2% sevoflurane for 1 h and then harvested 19 h later for Western blotting. A: Representative images of Western blotting. B: Quantitative results of the ratio of LC3II to LC3I. C: Quantitative results of p62. D: Quantitative results of beclin-1. Results are means ± S.E.M. (n = 8). * P < 0.05 compared to control. # P < 0.05 compared with the corresponding condition without chloroquine. OGD: oxygen-glucose deprivation; Sevo: 2% sevoflurane.
3.3. Sevoflurane improved autophagosome-lysosome fusion
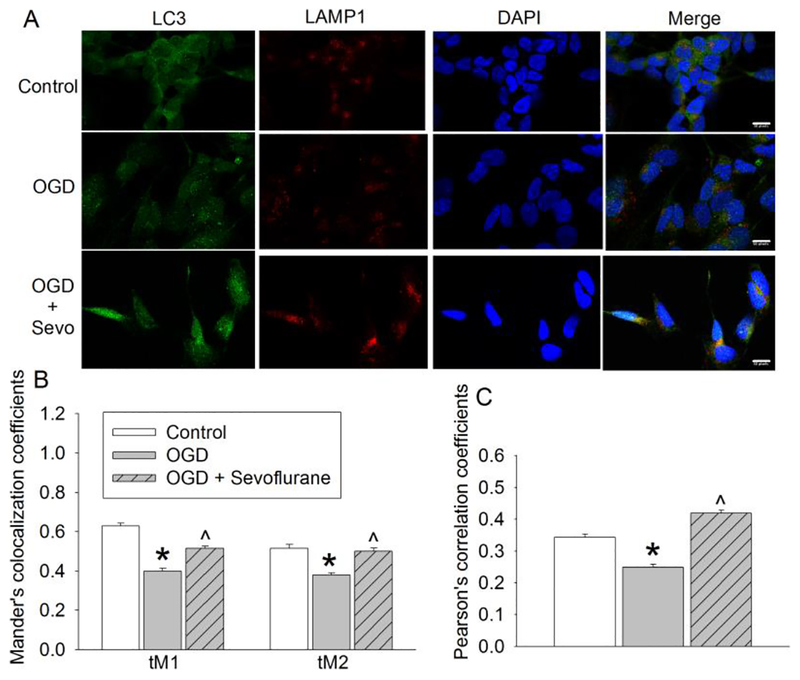
To better understand how sevoflurane improved autophagosome clearance, we examined autolysosome formation. Autophagosomes were visualized via LC3 puncta (green fluorescence), and lysosomes were visualized using LAMP-1 staining (red fluorescence). LAMP-1 is a lysosomal protein (Carlsson, et al., 1989). Quantification data showed that OGD-reperfusion significantly decreased the numbers of autolysosomes (positive staining for both LC3 and LAMP-1) compared with control group. Sevoflurane postconditioning increased the co-localization rate of LC3 with LAMP-1 (Fig. 4). These results indicate that 2% sevoflurane postconditioning improves autophagosome-lysosome fusion.
Fig. 4. Sevoflurane improved autophagosome-lysosome fusion.
Human neuron-like cells were subjected to 1 h OGD followed with 20 h simulated reperfusion. Cells were exposed to 2% sevoflurane for 1 h immediately after the onset of simulated reperfusion. A: Representative confocal microscopic images. Scale bar = 10 μm. B: Mander’s co-localization coefficients. C: Pearson’s correlation coefficients. Results are means ± S.E.M. (n = 150 cells). * P < 0.05 compared to control. ^ P < 0.05 compared to OGD only. OGD: oxygen-glucose deprivation
3.4. Sevoflurane attenuated OGD-induced inactivation of Akt-mTOR pathway to inhibit autophagosome accumulation to protect human neuron-like cells against OGD
The reduction of OGD-reperfusion-induced accumulation of autophagosomes by sevoflurane could be caused by either a decrease in autophagosome generation or increase of autolysosome clearance. Thus, we also examined whether sevoflurane maintained the activation of the Akt-mTOR signaling because activation of this signaling could reduce autophagosome production. Fig. 5 showed that the levels of p-Akt and p-mTOR in the OGD-reperfusion group were lower than that of control group. Sevoflurane postconditioning significantly increased the expression of p-Akt and p-mTOR. In addition, rapamycin, an mTOR inhibitor, attenuated sevoflurane postconditioning-induced decrease of LC3 conversion (Fig. 5). These results suggest that sevoflurane maintains the activation of Akt-mTOR signaling to reduce the generation of autophagosomes under OGD condition.
Fig. 5. Role of Akt-mTOR signaling in sevoflurane postconditioning-induced decrease of autophagy and cell protection.
Human neuron-like cells were subjected to 1 h OGD followed with 20 h simulated reperfusion. Cells were exposed to 2% sevoflurane for 1 h immediately after the onset of simulated reperfusion. 3-MA was added to cells 1 h prior to OGD. Rapamycin was added to cells 24 h prior to OGD. They were present until cells were harvested for assay. A: Representative images of Western blotting. B: Quantitative results of phospho-Akt. C: Quantitative results of phospho-mTOR. D: Representative images of Western blotting. E: Quantitative results of the ratio of LC3II to LC3I. F: LDH release. Results are means ± S.E.M. (n = 8 for panels B and C, = 10 for panel E, = 9 for panel F). * P < 0.05 compared to control. ^ P < 0.05 compared to OGD group. # P < 0.05 compared with OGD plus sevoflurane postconditioning. Con: control; OGD: oxygen-glucose deprivation; 3-MA: 3-methyladenine; Sevo: 2% sevoflurane; Rapa: rapamycin.
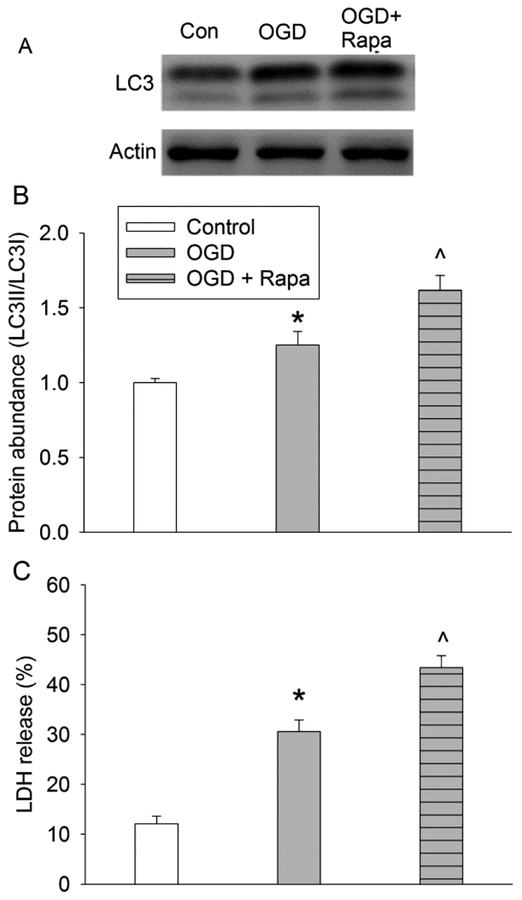
Consistent with its known effects, 3-MA reduced autophagosome production as reflected by a decrease in the LC3 conversion. Interestingly, 3-MA decreased OGD-reperfusion-induced cell injury and rapamycin attenuated the protective effects of sevoflurane (Fig. 5). Also, rapamycin further increased the ratio of LC3II to LC3I and LDH release in cells with OGD (Fig. 6). These results suggest that the reduced autophagosomes generation may be involved in the protective effects of sevoflurane postconditioning.
Fig. 6. Effects of mTOR inhibition on autophagosome formation and cell injury.
Human neuron-like cells were subjected to 1 h OGD followed with 20 h simulated reperfusion. Rapamycin was added to cells 24 h prior to OGD and was present until cells were harvested for assay. A: Representative images of Western blotting. B: Quantitative results of the ratio of LC3II to LC3I. C: LDH release. Results are means ± S.E.M. (n = 8 for panels B, and = 5 for panel C). * P < 0.05 compared to control. ^ P < 0.05 compared to OGD group. Con: control; OGD: oxygen-glucose deprivation; Rapa: rapamycin.
4. DISCUSSION
Postconditioning with sevoflurane has been shown to induce protection in rodent brains (Ren, et al., 2014). Our current study suggests that sevoflurane postconditioning also protect human neuron-like cells. Here, we showed that a concentration as low as 1% sevoflurane for 1 h applied at the onset of simulated reperfusion was sufficient to reduce cell injury. This protection appeared to peak at 2% sevoflurane. Higher concentrations had a reduced protective effect. These results suggest that sevoflurane at clinically relevant concentrations induces postconditioning effects in human neuron-like cells. The reasons for the decreased protective effect with sevoflurane concentrations higher than 2% are not yet known. One possibility for this phenomenon is that sevoflurane at high concentrations may injure cells, which can partially offset sevoflurane-induced protective effects. For example, exposure to 3% sevoflurane for 2 h for 3 consecutive days leads to significant cognitive impairment with increased neuronal apoptosis and overexpression of neuroinflammatory markers in neonatal mice (Xia, et al., 2017).
There was a significant increase in the levels of autophagic markers (LC3 conversion) in the human neuron-like cells after OGD. One of the possible reasons for OGD-induced increase in autophagic markers is the increase of autophagosome generation. The activation of the Akt-mTOR pathway is known to inhibit autophagosome generation in response to various stresses in different cells (Ryou, et al., 2015; Saiki, et al., 2011). Our results showed that OGD led to a significant decrease in the levels of phosphorylated Akt and mTOR, which suggests that OGD increases autophagosome generation. Consistent with this suggestion, OGD also increases beclin-1, a protein that is involved in inducing autophagosome formation (Glick, et al., 2010; Mizushima, 2007). Sevoflurane postconditioning increases the levels of phosphorylated Akt and mTOR in cells after OGD and rapamycin, an mTOR inhibitor (Lamming, et al., 2012), attenuated sevoflurane effects on LC3 conversion. Rapamycin further increased cell injury and autophagosome generation under OGD condition. Also, sevoflurane decreased beclin-1 under OGD condition but did not affect the expression of p62, beclin-1 and the ratio of LC3II to LC3I under control condition. These results suggest that sevoflurane postconditioning decreased OGD-induced increase of autophagosome generation possibly via attenuating inactivation of Akt-mTOR pathway.
The increase of LC3II level by OGD may also be caused by blockage in autophagosomal maturation and degradation (Mizushima, et al., 2010). Therefore, we examined the changes of autophagic flux by tracking the p62 protein level. p62 is selectively incorporated into the autophagosome through direct binding to LC3 and is degraded by autophagy (Saiki, et al., 2011). Here, we showed that OGD increased p62 protein level and that sevoflurane postconditioning prevented this effect. These results indicate that the increased autophagic marker (LC3II) in OGD-treated cells is not only due to enhanced autophagosome generation but also by suppression of the autophagosome maturation and degradation. Sevoflurane postconditioning promoted the degradation of autophagosomes accumulated in the cells after OGD.
To complete the autophagic process, autophagosomes need to be fused with lysosomes to form autophagolysosomes and its components are degraded by lysosomal hydrolases. The metabolites are transported to the cytosol for reuse (Zhou, et al., 2012). In this process, the autophagosome-lysosome fusion, lysosomal proteases and lysosomal pH are critical factors to complete the degradation (Shen, et al., 2014). We examined fluorescence co-localizations of punctate autophagosomal labeling (LC3) and the lysosomal labeling (LAMP1). Our study showed that OGD reduced the co-localization of LC3 with LAMP1 and that sevoflurane improved this co-localization, suggesting that OGD impairs the fusion of autophagosome with lysosome and that sevoflurane postconditioning attenuates this impairment.
Our study does not appear to show a significant effect on the enzymes of lysosomes by current OGD and simulated reperfusion condition because the level of cathepsin D was not changed by OGD. Sevoflurane postconditioning also did not affect cathepsin D expression. In addition, CQ that can increase lysosomal pH to inhibit lysosomal enzymes (Dunmore, et al., 2013; Geng, et al., 2010) decreased cathepsin D and also further increased the ratio of LC3II to LC3I and p62 no matter whether sevoflurane postconditioning was presented. These results suggest that the current conditions of OGD and sevoflurane postconditioning may not have significant effects on lysosomal pH and enzymes to change the autophagosome clearance.
Previous studies show that impaired autophagosome clearance plays a critical role in many diseases. Impaired autophagosome clearance contributes to neuronal death in a piglet model of neonatal hypoxic-ischemic encephalopathy (Cui, et al., 2017). Autophagosome clearance was markedly impaired in the diabetic heart subjected to myocardial ischemia-reperfusion and increasing autophagosome clearance reduced infarct size and improved cardiac function (Wang, et al., 2017b). Also, autophagy dysregulation or autophagosome accumulation occurs in ischemic spinal cord of rats and traumatic brain injury in mice and appears to contribute to the spinal cord and brain injury in these animals (Liu, et al., 2015; Sarkar, et al., 2014). As discussed above, our results suggest that sevoflurane postconditioning facilitates autophagosome-lysosome fusion and reduces the formation of autophagosome and, therefore, attenuates OGD-induced autophagosome accumulation. Thus, these effects shall contribute to sevoflurane postconditioning-induced cell protection. To support this possibility, rapamycin increased autophagosome formation and attenuated sevoflurane postconditioning-induced cell protection. Inhibition of autophagosome formation by 3-MA protected the human neuron-like cells against OGD.
Our results may not support that sevoflurane has a direct effect on autophagy under control condition because sevoflurane alone did not affect the expression of p62, beclin-1 and the ratio of LC3II to LC3I. However, our data indicate that sevoflurane postconditioning may reduce OGD-induced autophagosome accumulation to attenuate cell injury because OGD induced autophagosome accumulation and cell injury, sevoflurane postconditioning attenuated this accumulation and provided neuroprotection, and increasing autophagosome formation by rapamycin or deceasing autophagosome clearance by chloroquine attenuated sevoflurane postconditioning-induced reduction of autophagosome accumulation and neuroprotection. Mechanistically, sevoflurane postconditioning may decrease OGD-induced autophagosome accumulation through signaling events that regulate autophagy because sevoflurane attenuated OGD-induced inactivation of mTOR and inhibition of mTOR by rapamycin reduced sevoflurane-induced autophagy and neuroprotection. Nevertheless, attenuating autophagosome accumulation may be a downstream focused event for sevoflurane postconditioning-induced cell protection.
Our results suggest the effects on autophagy as a novel mechanism for sevoflurane postconditioning-induced cell protection. Increased autophagy has been suggested as a mechanism for isoflurane-induced protection in the brain and liver (Rao, et al., 2017; Sheng, et al., 2014). Increased autophagy has also been indicated as a mechanism for electroacupunctureor hyperbaric oxygen-induced neuroprotection (Fang, et al., 2017; Yan, et al., 2011). However, decreased autophagy has been proposed to mediate the neuroprotective effects of electroacupuncture, ketamine and dexmedetomidine (Luo, et al., 2017; Wang, et al., 2017a; Wu, et al., 2015). The reasons for these contradictory conclusions are not known. However, it is known that proper autophagy may be beneficial to save energy and clean damaged organelles and excessive autophagy contributes to cell death caused by ischemia-reperfusion injury (Koike, et al., 2008; Wang, et al., 2016). Thus, achieving appropriate levels of autophagy is needed for cell protection (Ren, et al., 2017). Our results with sevoflurane postconditioning suggest the importance of this delicate balance.
In summary, the present study has shown that OGD and simulated reperfusion increased autophagosome generation and reduced autophagosome clearance to enhance the accumulation of autophagosomes. Sevoflurane postconditioning improves autophagosome clearance and inhibits autophagosome generation via attenuating the inactivation of Akt-mTOR pathway under OGD condition. These sevoflurane effects on autophagy may be responsible for its neuroprotection.
Grant support:
This study was supported by grants (GM098308, HD089999 and AG056995 to Z Zuo) from the National Institutes of Health, Bethesda, MD, the Robert M. Epstein Professorship endowment, University of Virginia, Charlottesville, VA.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosure/conflict of interest: The authors declare no competing financial interests.
References
- Adamczyk S, Robin E, Simerabet M, Kipnis E, Tavernier B, Vallet B, Bordet R Lebuffe G, 2010. Sevoflurane pre- and post-conditioning protect the brain via the mitochondrial K ATP channel. Br J Anaesth 104, 191–200. [DOI] [PubMed] [Google Scholar]
- Barrett AJ, 1970. Cathepsin D. Purification of isoenzymes from human and chicken liver. Biochem J 117, 601–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlsson SR Fukuda M, 1989. Structure of human lysosomal membrane glycoprotein 1. Assignment of disulfide bonds and visualization of its domain arrangement. J Biol Chem 264, 20526–20531. [PubMed] [Google Scholar]
- Costes SV, Daelemans D, Cho EH, Dobbin Z, Pavlakis G Lockett S, 2004. Automatic and quantitative measurement of protein-protein colocalization in live cells. Biophys J 86, 3993–4003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui D, Sun D, Wang X, Yi L, Kulikowicz E, Reyes M, Zhu J, Yang ZJ, Jiang W Koehler RC, 2017. Impaired autophagosome clearance contributes to neuronal death in a piglet model of neonatal hypoxic-ischemic encephalopathy. Cell Death Dis 8, e2919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng YN, Shi J, Liu J Qu QM, 2013. Celastrol protects human neuroblastoma SH-SY5Y cells from rotenone-induced injury through induction of autophagy. Neurochem Int 63, 1–9. [DOI] [PubMed] [Google Scholar]
- Dunmore BJ, Drake KM, Upton PD, Toshner MR, Aldred MA Morrell NW, 2013. The lysosomal inhibitor, chloroquine, increases cell surface BMPR-II levels and restores BMP9 signalling in endothelial cells harbouring BMPR-II mutations. Hum Mol Genet 22, 3667–3679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrenreich H, Weissenborn K, Prange H, Schneider D, Weimar C, Wartenberg K, Schellinger PD, Bohn M, Becker H, Wegrzyn M, Jahnig P, Herrmann M, Knauth M, Bahr M, Heide W, Wagner A, Schwab S, Reichmann H, Schwendemann G, Dengler R, Kastrup A, Bartels C Group, E.P.O.S.T., 2009. Recombinant human erythropoietin in the treatment of acute ischemic stroke. Stroke 40, e647–656. [DOI] [PubMed] [Google Scholar]
- Fang B, Qin M, Li Y, Li X, Tan W, Zhang Y Ma H, 2017. Electroacupuncture preconditioning and postconditioning inhibit apoptosis and neuroinflammation induced by spinal cord ischemia reperfusion injury through enhancing autophagy in rats. Neurosci Lett 642, 136–141. [DOI] [PubMed] [Google Scholar]
- Feigin VL, Forouzanfar MH, Krishnamurthi R, Mensah GA, Connor M, Bennett DA, Moran AE, Sacco RL, Anderson L, Truelsen T, O’Donnell M, Venketasubramanian N, Barker-Collo S, Lawes CM, Wang W, Shinohara Y, Witt E, Ezzati M, Naghavi M, Murray C, Global Burden of Diseases, I., Risk Factors, S. the, G.B.D.S.E.G., 2014. Global and regional burden of stroke during 1990–2010: findings from the Global Burden of Disease Study 2010. Lancet 383, 245–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y, Kohli L, Klocke BJ Roth KA, 2010. Chloroquine-induced autophagic vacuole accumulation and cell death in glioma cells is p53 independent. Neuro-Onc 12, 473–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gidlof O, Johnstone AL, Bader K, Khomtchouk BB, O’Reilly JJ, Celik S, Van Booven DJ, Wahlestedt C, Metzler B Erlinge D, 2016. Ischemic Preconditioning Confers Epigenetic Repression of Mtor and Induction of Autophagy Through G9a-Dependent H3K9 Dimethylation. J Am Heart Assoc 5, e004076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glick D, Barth S Macleod KF, 2010. Autophagy: cellular and molecular mechanisms. J Path 221, 3–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikonomidou C Turski L, 2002. Why did NMDA receptor antagonists fail clinical trials for stroke and traumatic brain injury? The Lancet. Neurol 1, 383–386. [DOI] [PubMed] [Google Scholar]
- Kim J, Li L Zuo Z, 2009a. Delayed treatment with isoflurane attenuates lipopolysaccharide and interferon? -induced activation and injury of mouse microglial cells. Anesthesiology 111, 566–573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JA, Li L Zuo Z, 2009b. Isoflurane induces a postconditioning effect on bovine pulmonary arterial endothelial cells exposed to oxygen-glucose deprivation. Eur J Pharm 615, 144–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura T, Takabatake Y, Takahashi A Isaka Y, 2013. Chloroquine in cancer therapy: a double-edged sword of autophagy. Cancer Res 73, 3–7. [DOI] [PubMed] [Google Scholar]
- Klionsky DJ, Abdelmohsen K, Abe A, Abedin MJ, Abeliovich H, Acevedo Arozena A, Adachi H, Adams CM, Adams PD, Adeli K, Adhihetty PJ, Adler SG, Agam G, Agarwal R, Aghi MK, Agnello M, Agostinis P, Aguilar PV, Aguirre-Ghiso J, Airoldi EM, Ait-Si-Ali S, Akematsu T, Akporiaye ET, Al-Rubeai M, Albaiceta GM, Albanese C, Albani D, Albert ML, Aldudo J, Algul H, Alirezaei M, Alloza I, Almasan A, Almonte-Beceril M, Alnemri ES, Alonso C, Altan-Bonnet N, Altieri DC, Alvarez S, Alvarez-Erviti L, Alves S, Amadoro G, Amano A, Amantini C, Ambrosio S, Amelio I, Amer AO, Amessou M, Amon A, An Z, Anania FA, Andersen SU, Andley UP, Andreadi CK, Andrieu-Abadie N, Anel A, Ann DK, Anoopkumar-Dukie S, Antonioli M, Aoki H, Apostolova N, Aquila S, Aquilano K, Araki K, Arama E, Aranda A, Araya J, Arcaro A, Arias E, Arimoto H, Ariosa AR, Armstrong JL, Arnould T, Arsov I, Asanuma K, Askanas V, Asselin E, Atarashi R, Atherton SS, Atkin JD, Attardi LD, Auberger P, Auburger G, Aurelian L, Autelli R, Avagliano L, Avantaggiati ML, Avrahami L, Awale S, Azad N, Bachetti T, Backer JM, Bae DH, Bae JS, Bae ON, Bae SH, Baehrecke EH, Baek SH, Baghdiguian S, Bagniewska-Zadworna A, Bai H, Bai J, Bai XY, Bailly Y, Balaji KN, Balduini W, Ballabio A, Balzan R, Banerjee R, Banhegyi G, Bao H, Barbeau B, Barrachina MD, Barreiro E, Bartel B, Bartolome A, Bassham DC, Bassi MT, Bast RC Jr., Basu A, Batista MT, Batoko H, Battino M, Bauckman K, Baumgarner BL, Bayer KU, Beale R, Beaulieu JF, Beck GR Jr., Becker C, Beckham JD, Bedard PA, Bednarski PJ., Begley TJ, Behl C, Behrends C, Behrens GM, Behrns KE, Bejarano E, Belaid A, Belleudi F, Benard G, Berchem G, Bergamaschi D, Bergami M, Berkhout B, Berliocchi L, Bernard A, Bernard M, Bernassola F, Bertolotti A, Bess AS, Besteiro S, Bettuzzi S, Bhalla S, Bhattacharyya S, Bhutia SK, Biagosch C, Bianchi MW, Biard-Piechaczyk M, Billes V, Bincoletto C, Bingol B, Bird SW, Bitoun M, Bjedov I, Blackstone C, Blanc L, Blanco GA, Blomhoff HK, Boada-Romero E, Bockler S, Boes M, Boesze-Battaglia K, Boise LH, Bolino A, Boman A, Bonaldo P, Bordi M, Bosch J, Botana LM, Botti J, Bou G, Bouche M, Bouchecareilh M, Boucher MJ, Boulton ME, Bouret SG, Boya P, Boyer-Guittaut M, Bozhkov PV, Brady N, Braga VM, Brancolini C, Braus GH, Bravo-San Pedro JM, Brennan LA, Bresnick EH, Brest P, Bridges D, Bringer MA, Brini M, Brito GC, Brodin B, Brookes PS, Brown EJ, Brown K, Broxmeyer HE, Bruhat A, Brum PC, Brumell JH, Brunetti-Pierri N, Bryson-Richardson RJ, Buch S, Buchan AM, Budak H, Bulavin DV, Bultman SJ, Bultynck G, Bumbasirevic V, Burelle Y, Burke RE, Burmeister M, Butikofer P, Caberlotto L, Cadwell K, Cahova M, Cai D, Cai J, Cai Q, Calatayud S, Camougrand N, Campanella M, Campbell GR, Campbell M, Campello S, Candau R, Caniggia I, Cantoni L, Cao L, Caplan AB, Caraglia M, Cardinali C, Cardoso SM, Carew JS, Carleton LA, Carlin CR, Carloni S, Carlsson SR, Carmona-Gutierrez D, Carneiro LA, Carnevali O, Carra S, Carrier A, Carroll B, Casas C, Casas J, Cassinelli G, Castets P, Castro-Obregon S, Cavallini G, Ceccherini I, Cecconi F, Cederbaum AI, Cena V, Cenci S, Cerella C, Cervia D, Cetrullo S, Chaachouay H, Chae HJ, Chagin AS, Chai CY, Chakrabarti G, Chamilos G, Chan EY, Chan MT, Chandra D, Chandra P, Chang CP, Chang RC, Chang TY, Chatham JC, Chatterjee S, Chauhan S, Che Y, Cheetham ME, Cheluvappa R, Chen CJ, Chen G, Chen GC, Chen G, Chen H, Chen JW, Chen JK, Chen M, Chen M, Chen P, Chen Q, Chen Q, Chen SD, Chen S, Chen SS, Chen W, Chen WJ, Chen WQ, Chen W, Chen X, Chen YH, Chen YG, Chen Y, Chen Y, Chen Y, Chen YJ, Chen YQ, Chen Y, Chen Z, Chen Z, Cheng A, Cheng CH, Cheng H, Cheong H, Cherry S, Chesney J, Cheung CH, Chevet E, Chi HC, Chi SG, Chiacchiera F, Chiang HL, Chiarelli R, Chiariello M, Chieppa M, Chin LS, Chiong M, Chiu GN, Cho DH, Cho SG, Cho WC, Cho YY, Cho YS, Choi AM, Choi EJ, Choi EK, Choi J, Choi ME, Choi SI, Chou TF, Chouaib S, Choubey D, Choubey V, Chow KC, Chowdhury K, Chu CT, Chuang TH, Chun T, Chung H, Chung T, Chung YL, Chwae YJ, Cianfanelli V, Ciarcia R, Ciechomska IA, Ciriolo MR, Cirone M, Claerhout S, Clague MJ, Claria J, Clarke PG, Clarke R, Clementi E, Cleyrat C, Cnop M, Coccia EM, Cocco T, Codogno P, Coers J, Cohen EE, Colecchia D, Coletto L, Coll NS, Colucci-Guyon E, Comincini S, Condello M, Cook KL, Coombs GH, Cooper CD, Cooper JM, Coppens I, Corasaniti MT, Corazzari M, Corbalan R, Corcelle-Termeau E, Cordero MD, Corral-Ramos C, Corti O, Cossarizza A, Costelli P, Costes S, Cotman SL, Coto-Montes A, Cottet S, Couve E, Covey LR, Cowart LA, Cox JS, Coxon FP, Coyne CB, Cragg MS, Craven RJ, Crepaldi T, Crespo JL, Criollo A, Crippa V, Cruz MT, Cuervo AM, Cuezva JM, Cui T, Cutillas PR, Czaja MJ, Czyzyk-Krzeska MF, Dagda RK, Dahmen U, Dai C, Dai W, Dai Y, Dalby KN, Dalla Valle L, Dalmasso G, D’Amelio M, Damme M, Darfeuille-Michaud A, Dargemont C, Darley-Usmar VM, Dasarathy S, Dasgupta B, Dash S, Dass CR, Davey HM, Davids LM, Davila D, Davis RJ, Dawson TM, Dawson VL, Daza P, de Belleroche J, de Figueiredo P, de Figueiredo RC, de la Fuente J, De Martino L, De Matteis A, De Meyer GR, De Milito A, De Santi M, de Souza W, De Tata V, De Zio D, Debnath J, Dechant R, Decuypere JP, Deegan S, Dehay B, Del Bello B, Del Re DP, Delage-Mourroux R, Delbridge LM, Deldicque L, Delorme-Axford E, Deng Y, Dengjel J, Denizot M, Dent P, Der CJ, Deretic V, Derrien B, Deutsch E, Devarenne TP, Devenish RJ, Di Bartolomeo S, Di Daniele N, Di Domenico F, Di Nardo A, Di Paola S, Di Pietro A, Di Renzo L, DiAntonio A, Diaz-Araya G, Diaz-Laviada I, Diaz-Meco MT, Diaz-Nido J, Dickey CA, Dickson RC, Diederich M, Digard P, Dikic I, Dinesh-Kumar SP, Ding C, Ding WX, Ding Z, Dini L, Distler JH, Diwan A, Djavaheri-Mergny M, Dmytruk K, Dobson RC, Doetsch V, Dokladny K, Dokudovskaya S, Donadelli M, Dong XC, Dong X, Dong Z, Donohue TM Jr., Doran KS, D’Orazi G, Dorn GW 2nd, Dosenko V, Dridi S, Drucker L, Du J, Du LL, Du L, du Toit A, Dua P, Duan L, Duann P, Dubey VK, Duchen MR, Duchosal MA, Duez H, Dugail I, Dumit VI, Duncan MC, Dunlop EA, Dunn WA Jr., Dupont N, Dupuis L, Duran RV, Durcan TM, Duvezin-Caubet S, Duvvuri U, Eapen V, Ebrahimi-Fakhari D, Echard A, Eckhart L, Edelstein CL, Edinger AL, Eichinger L, Eisenberg T, Eisenberg-Lerner A, Eissa NT, El-Deiry WS, El-Khoury V, Elazar Z, Eldar-Finkelman H, Elliott CJ, Emanuele E, Emmenegger U, Engedal N, Engelbrecht AM, Engelender S, Enserink JM, Erdmann R, Erenpreisa J, Eri R, Eriksen JL, Erman A, Escalante R, Eskelinen EL, Espert L, Esteban-Martinez L, Evans TJ, Fabri M, Fabrias G, Fabrizi C, Facchiano A, Faergeman NJ, Faggioni A, Fairlie WD, Fan C, Fan D, Fan J, Fang S, Fanto M, Fanzani A, Farkas T, Faure M, Favier FB, Fearnhead H, Federici M, Fei E, Felizardo TC, Feng H, Feng Y, Feng Y, Ferguson TA, Fernandez AF, Fernandez-Barrena MG, Fernandez-Checa JC, Fernandez-Lopez A, Fernandez-Zapico ME, Feron O, Ferraro E, Ferreira-Halder CV, Fesus L, Feuer R, Fiesel FC, Filippi-Chiela EC, Filomeni G, Fimia GM, Fingert JH, Finkbeiner S, Finkel T, Fiorito F, Fisher PB, Flajolet M, Flamigni F, Florey O, Florio S, Floto RA, Folini M, Follo C, Fon EA, Fornai F, Fortunato F, Fraldi A, Franco R, Francois A, Francois A, Frankel LB, Fraser ID, Frey N, Freyssenet DG, Frezza C, Friedman SL, Frigo DE, Fu D, Fuentes JM, Fueyo J, Fujitani Y, Fujiwara Y, Fujiya M, Fukuda M, Fulda S, Fusco C, Gabryel B, Gaestel M, Gailly P, Gajewska M, Galadari S, Galili G, Galindo I, Galindo MF, Galliciotti G, Galluzzi L, Galluzzi L, Galy V, Gammoh N, Gandy S, Ganesan AK, Ganesan S, Ganley IG, Gannage M, Gao FB, Gao F, Gao JX, Garcia Nannig L, Garcia Vescovi E, Garcia-Macia M, Garcia-Ruiz C, Garg AD, Garg PK, Gargini R, Gassen NC, Gatica D, Gatti E, Gavard J, Gavathiotis E, Ge L, Ge P, Ge S, Gean PW, Gelmetti V, Genazzani AA, Geng J, Genschik P, Gerner L, Gestwicki JE, Gewirtz DA, Ghavami S, Ghigo E, Ghosh D, Giammarioli AM, Giampieri F, Giampietri C, Giatromanolaki A, Gibbings DJ, Gibellini L, Gibson SB, Ginet V, Giordano A, Giorgini F, Giovannetti E, Girardin SE, Gispert S, Giuliano S, Gladson CL, Glavic A, Gleave M, Godefroy N, Gogal RM Jr., Gokulan K, Goldman GH, Goletti D, Goligorsky MS, Gomes AV, Gomes LC, Gomez H, Gomez-Manzano C, Gomez-Sanchez R, Goncalves DA, Goncu E, Gong Q, Gongora C, Gonzalez CB, Gonzalez-Alegre P, Gonzalez-Cabo P, Gonzalez-Polo RA, Goping IS, Gorbea C, Gorbunov NV, Goring DR, Gorman AM, Gorski SM, Goruppi S, Goto-Yamada S, Gotor C, Gottlieb RA, Gozes I, Gozuacik D, Graba Y, Graef M, Granato GE, Grant GD, Grant S, Gravina GL, Green DR, Greenhough A, Greenwood MT, Grimaldi B, Gros F, Grose C, Groulx JF, Gruber F, Grumati P, Grune T, Guan JL, Guan KL, Guerra B, Guillen C, Gulshan K, Gunst J, Guo C, Guo L, Guo M, Guo W, Guo XG, Gust AA, Gustafsson AB, Gutierrez E, Gutierrez MG, Gwak HS, Haas A, Haber JE, Hadano S, Hagedorn M, Hahn DR, Halayko AJ, Hamacher-Brady A, Hamada K, Hamai A, Hamann A, Hamasaki M, Hamer I, Hamid Q, Hammond EM, Han F, Han W, Handa JT, Hanover JA, Hansen M, Harada M, Harhaji-Trajkovic L, Harper JW, Harrath AH, Harris AL, Harris J, Hasler U, Hasselblatt P, Hasui K, Hawley RG, Hawley TS, He C, He CY, He F, He G, He RR, He XH, He YW, He YY, Heath JK, Hebert MJ, Heinzen RA, Helgason GV, Hensel M, Henske EP, Her C, Herman PK, Hernandez A, Hernandez C, Hernandez-Tiedra S, Hetz C, Hiesinger PR, Higaki K, Hilfiker S, Hill BG, Hill JA, Hill WD, Hino K, Hofius D, Hofman P, Hoglinger GU, Hohfeld J, Holz MK, Hong Y, Hood DA, Hoozemans JJ, Hoppe T, Hsu C, Hsu CY, Hsu LC, Hu D, Hu G, Hu HM, Hu H, Hu MC, Hu YC, Hu ZW, Hua F, Hua Y, Huang C, Huang HL, Huang KH, Huang KY, Huang S, Huang S, Huang WP, Huang YR, Huang Y, Huang Y, Huber TB, Huebbe P, Huh WK, Hulmi JJ, Hur GM, Hurley JH, Husak Z, Hussain SN, Hussain S, Hwang JJ, Hwang S, Hwang TI, Ichihara A, Imai Y, Imbriano C, Inomata M, Into T, Iovane V, Iovanna JL, Iozzo RV, Ip NY, Irazoqui JE, Iribarren P, Isaka Y, Isakovic AJ, Ischiropoulos H, Isenberg JS, Ishaq M, Ishida H, Ishii I, Ishmael JE, Isidoro C, Isobe K, Isono E, Issazadeh-Navikas S, Itahana K, Itakura E, Ivanov AI, Iyer AK, Izquierdo JM, Izumi Y, Izzo V, Jaattela M, Jaber N, Jackson DJ, Jackson WT, Jacob TG, Jacques TS, Jagannath C, Jain A, Jana NR, Jang BK, Jani A, Janji B, Jannig PR, Jansson PJ, Jean S, Jendrach M, Jeon JH, Jessen N, Jeung EB, Jia K, Jia L, Jiang H, Jiang H, Jiang L, Jiang T, Jiang X, Jiang X, Jiang X, Jiang Y, Jiang Y, Jimenez A, Jin C, Jin H, Jin L, Jin M, Jin S, Jinwal UK, Jo EK, Johansen T, Johnson DE, Johnson GV, Johnson JD, Jonasch E, Jones C, Joosten LA, Jordan J, Joseph AM, Joseph B, Joubert AM, Ju D, Ju J, Juan HF, Juenemann K, Juhasz G, Jung HS, Jung JU, Jung YK, Jungbluth H, Justice MJ, Jutten B, Kaakoush NO, Kaarniranta K, Kaasik A, Kabuta T, Kaeffer B, Kagedal K, Kahana A, Kajimura S, Kakhlon O, Kalia M, Kalvakolanu DV, Kamada Y, Kambas K, Kaminskyy VO, Kampinga HH, Kandouz M, Kang C, Kang R, Kang TC, Kanki T, Kanneganti TD, Kanno H, Kanthasamy AG, Kantorow M, Kaparakis-Liaskos M, Kapuy O, Karantza V, Karim MR, Karmakar P, Kaser A, Kaushik S, Kawula T, Kaynar AM, Ke PY, Ke ZJ, Kehrl JH, Keller KE, Kemper JK, Kenworthy AK, Kepp O, Kern A, Kesari S, Kessel D, Ketteler R, Kettelhut Ido C, Khambu B, Khan MM, Khandelwal VK, Khare S, Kiang JG, Kiger AA, Kihara A, Kim AL, Kim CH, Kim DR, Kim DH, Kim EK, Kim HY, Kim HR, Kim JS, Kim JH, Kim JC, Kim JH, Kim KW, Kim MD, Kim MM, Kim PK, Kim SW, Kim SY, Kim YS, Kim Y, Kimchi A, Kimmelman AC, Kimura T, King JS, Kirkegaard K, Kirkin V, Kirshenbaum LA, Kishi S, Kitajima Y, Kitamoto K, Kitaoka Y, Kitazato K, Kley RA, Klimecki WT, Klinkenberg M, Klucken J, Knaevelsrud H, Knecht E, Knuppertz L, Ko JL, Kobayashi S, Koch JC, Koechlin-Ramonatxo C, Koenig U, Koh YH, Kohler K, Kohlwein SD, Koike M, Komatsu M, Kominami E, Kong D, Kong HJ, Konstantakou EG, Kopp BT, Korcsmaros T, Korhonen L, Korolchuk VI, Koshkina NV, Kou Y, Koukourakis MI, Koumenis C, Kovacs AL, Kovacs T, Kovacs WJ, Koya D, Kraft C, Krainc D, Kramer H, Kravic-Stevovic T, Krek W, Kretz-Remy C, Krick R, Krishnamurthy M, Kriston-Vizi J, Kroemer G, Kruer MC, Kruger R, Ktistakis NT, Kuchitsu K, Kuhn C, Kumar AP, Kumar A, Kumar A, Kumar D, Kumar D, Kumar R, Kumar S, Kundu M, Kung HJ, Kuno A, Kuo SH, Kuret J, Kurz T, Kwok T, Kwon TK, Kwon YT, Kyrmizi I, La Spada AR, Lafont F, Lahm T, Lakkaraju A, Lam T, Lamark T, Lancel S, Landowski TH, Lane DJ, Lane JD, Lanzi C, Lapaquette P, Lapierre LR, Laporte J, Laukkarinen J, Laurie GW, Lavandero S, Lavie L, LaVoie MJ, Law BY, Law HK, Law KB, Layfield R, Lazo PA, Le Cam L, Le Roch KG, Le Stunff H, Leardkamolkarn V, Lecuit M, Lee BH, Lee CH, Lee EF, Lee GM, Lee HJ, Lee H, Lee JK, Lee J, Lee JH, Lee JH, Lee M, Lee MS, Lee PJ, Lee SW, Lee SJ, Lee SJ, Lee SY, Lee SH, Lee SS, Lee SJ, Lee S, Lee YR, Lee YJ, Lee YH, Leeuwenburgh C, Lefort S, Legouis R, Lei J, Lei QY, Leib DA, Leibowitz G, Lekli I, Lemaire SD, Lemasters JJ, Lemberg MK, Lemoine A, Leng S, Lenz G, Lenzi P, Lerman LO, Lettieri Barbato D, Leu JI, Leung HY, Levine B, Lewis PA, Lezoualc’h F, Li C, Li F, Li FJ, Li J, Li K, Li L, Li M, Li M, Li Q, Li R, Li S, Li W, Li W, Li X, Li Y, Lian J, Liang C, Liang Q, Liao Y, Liberal J, Liberski PP, Lie P, Lieberman AP, Lim HJ, Lim KL, Lim K, Lima RT, Lin CS, Lin CF, Lin F, Lin F, Lin FC, Lin K, Lin KH, Lin PH, Lin T, Lin WW, Lin YS, Lin Y, Linden R, Lindholm D, Lindqvist LM, Lingor P, Linkermann A, Liotta LA, Lipinski MM, Lira VA, Lisanti MP, Liton PB, Liu B, Liu C, Liu CF, Liu F, Liu HJ, Liu J, Liu JJ, Liu JL, Liu K, Liu L, Liu L, Liu Q, Liu RY, Liu S, Liu S, Liu W, Liu XD, Liu X, Liu XH, Liu X, Liu X, Liu X, Liu Y, Liu Y, Liu Z, Liu Z, Liuzzi JP, Lizard G, Ljujic M, Lodhi IJ, Logue SE, Lokeshwar BL, Long YC, Lonial S, Loos B, Lopez-Otin C, Lopez-Vicario C, Lorente M, Lorenzi PL, Lorincz P, Los M, Lotze MT, Lovat PE, Lu B, Lu B, Lu J, Lu Q, Lu SM, Lu S, Lu Y, Luciano F, Luckhart S, Lucocq JM, Ludovico P, Lugea A, Lukacs NW, Lum JJ, Lund AH, Luo H, Luo J, Luo S, Luparello C, Lyons T, Ma J, Ma Y, Ma Y, Ma Z, Machado J, Machado-Santelli GM, Macian F, MacIntosh GC, MacKeigan JP, Macleod KF, MacMicking JD, MacMillan-Crow LA, Madeo F, Madesh M, Madrigal-Matute J, Maeda A, Maeda T, Maegawa G, Maellaro E, Maes H, Magarinos M, Maiese K, Maiti TK, Maiuri L, Maiuri MC, Maki CG, Malli R, Malorni W, Maloyan A, Mami-Chouaib F, Man N, Mancias JD, Mandelkow EM, Mandell MA, Manfredi AA, Manie SN, Manzoni C, Mao K, Mao Z, Mao ZW, Marambaud P, Marconi AM, Marelja Z, Marfe G, Margeta M, Margittai E, Mari M, Mariani FV, Marin C, Marinelli S, Marino G, Markovic I, Marquez R, Martelli AM, Martens S, Martin KR, Martin SJ, Martin S, Martin-Acebes MA, Martin-Sanz P, Martinand-Mari C, Martinet W, Martinez J, Martinez-Lopez N, Martinez-Outschoorn U, Martinez-Velazquez M, Martinez-Vicente M, Martins WK, Mashima H, Mastrianni JA, Matarese G, Matarrese P, Mateo R, Matoba S, Matsumoto N, Matsushita T, Matsuura A, Matsuzawa T, Mattson MP, Matus S, Maugeri N, Mauvezin C, Mayer A, Maysinger D, Mazzolini GD, McBrayer MK, McCall K, McCormick C, McInerney GM, McIver SC, McKenna S, McMahon JJ, McNeish IA, Mechta-Grigoriou F, Medema JP, Medina DL, Megyeri K, Mehrpour M, Mehta JL, Mei Y, Meier UC, Meijer AJ, Melendez A, Melino G, Melino S, de Melo EJ, Mena MA, Meneghini MD, Menendez JA, Menezes R, Meng L, Meng LH, Meng S, Menghini R, Menko AS, Menna-Barreto RF, Menon MB, Meraz-Rios MA, Merla G, Merlini L, Merlot AM, Meryk A, Meschini S, Meyer JN, Mi MT, Miao CY, Micale L, Michaeli S, Michiels C, Migliaccio AR, Mihailidou AS, Mijaljica D, Mikoshiba K, Milan E, Miller-Fleming L, Mills GB, Mills IG, Minakaki G, Minassian BA, Ming XF, Minibayeva F, Minina EA, Mintern JD, Minucci S, Miranda-Vizuete A, Mitchell CH, Miyamoto S, Miyazawa K, Mizushima N, Mnich K, Mograbi B, Mohseni S, Moita LF, Molinari M, Molinari M, Moller AB, Mollereau B, Mollinedo F, Mongillo M, Monick MM, Montagnaro S, Montell C, Moore DJ, Moore MN, Mora-Rodriguez R, Moreira PI, Morel E, Morelli MB, Moreno S, Morgan MJ, Moris A, Moriyasu Y, Morrison JL, Morrison LA, Morselli E, Moscat J, Moseley PL, Mostowy S, Motori E, Mottet D, Mottram JC, Moussa CE, Mpakou VE, Mukhtar H, Mulcahy Levy JM, Muller S, Munoz-Moreno R, Munoz-Pinedo C, Munz C, Murphy ME, Murray JT, Murthy A, Mysorekar IU, Nabi IR, Nabissi M, Nader GA, Nagahara Y, Nagai Y, Nagata K, Nagelkerke A, Nagy P, Naidu SR, Nair S, Nakano H, Nakatogawa H, Nanjundan M, Napolitano G, Naqvi NI, Nardacci R, Narendra DP, Narita M, Nascimbeni AC, Natarajan R, Navegantes LC, Nawrocki ST, Nazarko TY, Nazarko VY, Neill T, Neri LM, Netea MG, Netea-Maier RT, Neves BM, Ney PA, Nezis IP, Nguyen HT, Nguyen HP, Nicot AS, Nilsen H, Nilsson P, Nishimura M, Nishino I, Niso-Santano M, Niu H, Nixon RA, Njar VC, Noda T, Noegel AA, Nolte EM, Norberg E, Norga KK, Noureini SK, Notomi S, Notterpek L, Nowikovsky K, Nukina N, Nurnberger T, O’Donnell VB, O’Donovan T, O’Dwyer PJ, Oehme I, Oeste CL, Ogawa M, Ogretmen B, Ogura Y, Oh YJ, Ohmuraya M, Ohshima T, Ojha R, Okamoto K, Okazaki T, Oliver FJ, Ollinger K, Olsson S, Orban DP, Ordonez P, Orhon I, Orosz L, O’Rourke EJ, Orozco H, Ortega AL, Ortona E, Osellame LD, Oshima J, Oshima S, Osiewacz HD, Otomo T, Otsu K, Ou JH, Outeiro TF, Ouyang DY, Ouyang H, Overholtzer M, Ozbun MA, Ozdinler PH, Ozpolat B, Pacelli C, Paganetti P, Page G, Pages G, Pagnini U, Pajak B, Pak SC, Pakos-Zebrucka K, Pakpour N, Palkova Z, Palladino F, Pallauf K, Pallet N, Palmieri M, Paludan SR, Palumbo C, Palumbo S, Pampliega O, Pan H, Pan W, Panaretakis T, Pandey A, Pantazopoulou A, Papackova Z, Papademetrio DL, Papassideri I, Papini A, Parajuli N, Pardo J, Parekh VV, Parenti G, Park JI, Park J, Park OK, Parker R, Parlato R, Parys JB, Parzych KR, Pasquet JM, Pasquier B, Pasumarthi KB, Patschan D, Patterson C, Pattingre S, Pattison S, Pause A, Pavenstadt H, Pavone F, Pedrozo Z, Pena FJ, Penalva MA, Pende M, Peng J, Penna F, Penninger JM, Pensalfini A, Pepe S, Pereira GJ, Pereira PC, Perez-de la Cruz V, Perez-Perez ME, Perez-Rodriguez D, Perez-Sala D, Perier C, Perl A, Perlmutter DH, Perrotta I, Pervaiz S, Pesonen M, Pessin JE, Peters GJ, Petersen M, Petrache I, Petrof BJ, Petrovski G, Phang JM, Piacentini M, Pierdominici M, Pierre P, Pierrefite-Carle V, Pietrocola F, Pimentel-Muinos FX, Pinar M, Pineda B, Pinkas-Kramarski R, Pinti M, Pinton P, Piperdi B, Piret JM, Platanias LC, Platta HW, Plowey ED, Poggeler S, Poirot M, Polcic P, Poletti A, Poon AH, Popelka H, Popova B, Poprawa I, Poulose SM, Poulton J, Powers SK, Powers T, Pozuelo-Rubio M, Prak K, Prange R, Prescott M, Priault M, Prince S, Proia RL, Proikas-Cezanne T, Prokisch H, Promponas VJ, Przyklenk K, Puertollano R, Pugazhenthi S, Puglielli L, Pujol A, Puyal J, Pyeon D, Qi X, Qian WB, Qin ZH, Qiu Y, Qu Z, Quadrilatero J, Quinn F, Raben N, Rabinowich H, Radogna F, Ragusa MJ, Rahmani M, Raina K, Ramanadham S, Ramesh R, Rami A, Randall-Demllo S, Randow F, Rao H, Rao VA, Rasmussen BB, Rasse TM, Ratovitski EA, Rautou PE, Ray SK, Razani B, Reed BH, Reggiori F, Rehm M, Reichert AS, Rein T, Reiner DJ, Reits E, Ren J, Ren X, Renna M, Reusch JE, Revuelta JL, Reyes L, Rezaie AR, Richards RI, Richardson DR, Richetta C, Riehle MA, Rihn BH, Rikihisa Y, Riley BE, Rimbach G, Rippo MR, Ritis K, Rizzi F, Rizzo E, Roach PJ, Robbins J, Roberge M, Roca G, Roccheri MC, Rocha S, Rodrigues CM, Rodriguez CI, de Cordoba SR, Rodriguez-Muela N, Roelofs J, Rogov VV, Rohn TT, Rohrer B, Romanelli D, Romani L, Romano PS, Roncero MI, Rosa JL, Rosello A, Rosen KV, Rosenstiel P, Rost-Roszkowska M, Roth KA, Roue G, Rouis M, Rouschop KM, Ruan DT, Ruano D, Rubinsztein DC, Rucker EB 3rd, Rudich A, Rudolf E, Rudolf R, Ruegg MA, Ruiz-Roldan C, Ruparelia AA, Rusmini P, Russ DW, Russo GL, Russo G, Russo R, Rusten TE, Ryabovol V, Ryan KM, Ryter SW, Sabatini DM, Sacher M, Sachse C, Sack MN, Sadoshima J, Saftig P, Sagi-Eisenberg R, Sahni S, Saikumar P, Saito T, Saitoh T, Sakakura K, Sakoh-Nakatogawa M, Sakuraba Y, Salazar-Roa M, Salomoni P, Saluja AK, Salvaterra PM, Salvioli R, Samali A, Sanchez AM, Sanchez-Alcazar JA, Sanchez-Prieto R, Sandri M, Sanjuan MA, Santaguida S, Santambrogio L, Santoni G, Dos Santos CN, Saran S, Sardiello M, Sargent G, Sarkar P, Sarkar S, Sarrias MR, Sarwal MM, Sasakawa C, Sasaki M, Sass M, Sato K, Sato M, Satriano J, Savaraj N, Saveljeva S, Schaefer L, Schaible UE, Scharl M, Schatzl HM, Schekman R, Scheper W, Schiavi A, Schipper HM, Schmeisser H, Schmidt J, Schmitz I, Schneider BE, Schneider EM, Schneider JL, Schon EA, Schonenberger MJ, Schonthal AH, Schorderet DF, Schroder B, Schuck S, Schulze RJ, Schwarten M, Schwarz TL, Sciarretta S, Scotto K, Scovassi AI, Screaton RA, Screen M, Seca H, Sedej S, Segatori L, Segev N, Seglen PO, Segui-Simarro JM, Segura-Aguilar J, Seki E, Sell C, Seiliez I, Semenkovich CF, Semenza GL, Sen U, Serra AL, Serrano-Puebla A, Sesaki H, Setoguchi T, Settembre C, Shacka JJ, Shajahan-Haq AN, Shapiro IM, Sharma S, She H, Shen CK, Shen CC, Shen HM, Shen S, Shen W, Sheng R, Sheng X, Sheng ZH, Shepherd TG, Shi J, Shi Q, Shi Q, Shi Y, Shibutani S, Shibuya K, Shidoji Y, Shieh JJ, Shih CM, Shimada Y, Shimizu S, Shin DW, Shinohara ML, Shintani M, Shintani T, Shioi T, Shirabe K, Shiri-Sverdlov R, Shirihai O, Shore GC, Shu CW, Shukla D, Sibirny AA, Sica V, Sigurdson CJ, Sigurdsson EM, Sijwali PS, Sikorska B, Silveira WA, Silvente-Poirot S, Silverman GA, Simak J, Simmet T, Simon AK, Simon HU, Simone C, Simons M, Simonsen A, Singh R, Singh SV, Singh SK, Sinha D, Sinha S, Sinicrope FA, Sirko A, Sirohi K, Sishi BJ, Sittler A, Siu PM, Sivridis E, Skwarska A, Slack R, Slaninova I, Slavov N, Smaili SS, Smalley KS, Smith DR, Soenen SJ, Soleimanpour SA, Solhaug A, Somasundaram K, Son JH, Sonawane A, Song C, Song F, Song HK, Song JX, Song W, Soo KY, Sood AK, Soong TW, Soontornniyomkij V, Sorice M, Sotgia F, Soto-Pantoja DR, Sotthibundhu A, Sousa MJ, Spaink HP, Span PN, Spang A, Sparks JD, Speck PG, Spector SA, Spies CD, Springer W, Clair DS, Stacchiotti A, Staels B, Stang MT, Starczynowski DT, Starokadomskyy P, Steegborn C, Steele JW, Stefanis L, Steffan J, Stellrecht CM, Stenmark H, Stepkowski TM, Stern ST, Stevens C, Stockwell BR, Stoka V, Storchova Z, Stork B, Stratoulias V, Stravopodis DJ, Strnad P, Strohecker AM, Strom AL, Stromhaug P, Stulik J, Su YX, Su Z, Subauste CS, Subramaniam S, Sue CM, Suh SW, Sui X, Sukseree S, Sulzer D, Sun FL, Sun J, Sun J, Sun SY, Sun Y, Sun Y, Sun Y, Sundaramoorthy V, Sung J, Suzuki H, Suzuki K, Suzuki N, Suzuki T, Suzuki YJ, Swanson MS, Swanton C, Sward K, Swarup G, Sweeney ST, Sylvester PW, Szatmari Z, Szegezdi E, Szlosarek PW, Taegtmeyer H, Tafani M, Taillebourg E, Tait SW, Takacs-Vellai K, Takahashi Y, Takats S, Takemura G, Takigawa N, Talbot NJ, Tamagno E, Tamburini J, Tan CP, Tan L, Tan ML, Tan M, Tan YJ, Tanaka K, Tanaka M, Tang D, Tang D, Tang G, Tanida I, Tanji K, Tannous BA, Tapia JA, Tasset-Cuevas I, Tatar M, Tavassoly I, Tavernarakis N, Taylor A, Taylor GS, Taylor GA, Taylor JP, Taylor MJ, Tchetina EV, Tee AR, Teixeira-Clerc F, Telang S, Tencomnao T, Teng BB, Teng RJ, Terro F, Tettamanti G, Theiss AL, Theron AE, Thomas KJ, Thome MP, Thomes PG, Thorburn A, Thorner J, Thum T, Thumm M, Thurston TL, Tian L, Till A, Ting JP, Titorenko VI, Toker L, Toldo S, Tooze SA, Topisirovic I, Torgersen ML, Torosantucci L, Torriglia A, Torrisi MR, Tournier C, Towns R, Trajkovic V, Travassos LH, Triola G, Tripathi DN, Trisciuoglio D, Troncoso R, Trougakos IP, Truttmann AC, Tsai KJ, Tschan MP, Tseng YH, Tsukuba T, Tsung A, Tsvetkov AS, Tu S, Tuan HY, Tucci M, Tumbarello DA, Turk B, Turk V, Turner RF, Tveita AA, Tyagi SC, Ubukata M, Uchiyama Y, Udelnow A, Ueno T, Umekawa M, Umemiya-Shirafuji R, Underwood BR, Ungermann C, Ureshino RP, Ushioda R, Uversky VN, Uzcategui NL, Vaccari T, Vaccaro MI, Vachova L, Vakifahmetoglu-Norberg H, Valdor R, Valente EM, Vallette F, Valverde AM, Van den Berghe G, Van Den Bosch L, van den Brink GR, van der Goot FG, van der Klei IJ, van der Laan LJ, van Doorn WG, van Egmond M, van Golen KL, Van Kaer L, van Lookeren Campagne M, Vandenabeele P, Vandenberghe W, Vanhorebeek I, Varela-Nieto I, Vasconcelos MH, Vasko R, Vavvas DG, Vega-Naredo I, Velasco G, Velentzas AD, Velentzas PD, Vellai T, Vellenga E, Vendelbo MH, Venkatachalam K, Ventura N, Ventura S, Veras PS, Verdier M, Vertessy BG, Viale A, Vidal M, Vieira HL, Vierstra RD, Vigneswaran N, Vij N, Vila M, Villar M, Villar VH, Villarroya J, Vindis C, Viola G, Viscomi MT, Vitale G, Vogl DT, Voitsekhovskaja OV, von Haefen C, von Schwarzenberg K, Voth DE, Vouret-Craviari V, Vuori K, Vyas JM, Waeber C, Walker CL, Walker MJ, Walter J, Wan L, Wan X, Wang B, Wang C, Wang CY, Wang C, Wang C, Wang C, Wang D, Wang F, Wang F, Wang G, Wang HJ, Wang H, Wang HG, Wang H, Wang HD, Wang J, Wang J, Wang M, Wang MQ, Wang PY, Wang P, Wang RC, Wang S, Wang TF, Wang X, Wang XJ, Wang XW, Wang X, Wang X, Wang Y, Wang Y, Wang Y, Wang YJ, Wang Y, Wang Y, Wang YT, Wang Y, Wang ZN, Wappner P, Ward C, Ward DM, Warnes G, Watada H, Watanabe Y, Watase K, Weaver TE, Weekes CD, Wei J, Weide T, Weihl CC, Weindl G, Weis SN, Wen L, Wen X, Wen Y, Westermann B, Weyand CM, White AR, White E, Whitton JL, Whitworth AJ, Wiels J, Wild F, Wildenberg ME, Wileman T, Wilkinson DS, Wilkinson S, Willbold D, Williams C, Williams K, Williamson PR, Winklhofer KF, Witkin SS, Wohlgemuth SE, Wollert T, Wolvetang EJ, Wong E, Wong GW, Wong RW, Wong VK, Woodcock EA, Wright KL, Wu C, Wu D, Wu GS, Wu J, Wu J, Wu M, Wu M, Wu S, Wu WK, Wu Y, Wu Z, Xavier CP, Xavier RJ, Xia GX, Xia T, Xia W, Xia Y, Xiao H, Xiao J, Xiao S, Xiao W, Xie CM, Xie Z, Xie Z, Xilouri M, Xiong Y, Xu C, Xu C, Xu F, Xu H, Xu H, Xu J, Xu J, Xu J, Xu L, Xu X, Xu Y, Xu Y, Xu ZX, Xu Z, Xue Y, Yamada T, Yamamoto A, Yamanaka K, Yamashina S, Yamashiro S, Yan B, Yan B, Yan X, Yan Z, Yanagi Y, Yang DS, Yang JM, Yang L, Yang M, Yang PM, Yang P, Yang Q, Yang W, Yang WY, Yang X, Yang Y, Yang Y, Yang Z, Yang Z, Yao MC, Yao PJ, Yao X, Yao Z, Yao Z, Yasui LS, Ye M, Yedvobnick B, Yeganeh B, Yeh ES, Yeyati PL, Yi F, Yi L, Yin XM, Yip CK, Yoo YM, Yoo YH, Yoon SY, Yoshida K, Yoshimori T, Young KH, Yu H, Yu JJ, Yu JT, Yu J, Yu L, Yu WH, Yu XF, Yu Z, Yuan J, Yuan ZM, Yue BY, Yue J, Yue Z, Zacks DN, Zacksenhaus E, Zaffaroni N, Zaglia T, Zakeri Z, Zecchini V, Zeng J, Zeng M, Zeng Q, Zervos AS, Zhang DD, Zhang F, Zhang G, Zhang GC, Zhang H, Zhang H, Zhang H, Zhang H, Zhang J, Zhang J, Zhang J, Zhang J, Zhang JP, Zhang L, Zhang L, Zhang L, Zhang L, Zhang MY, Zhang X, Zhang XD, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhang Y, Zhao M, Zhao WL, Zhao X, Zhao YG, Zhao Y, Zhao Y, Zhao YX, Zhao Z, Zhao ZJ, Zheng D, Zheng XL, Zheng X, Zhivotovsky B, Zhong Q, Zhou GZ, Zhou G, Zhou H, Zhou SF, Zhou XJ, Zhu H, Zhu H, Zhu WG, Zhu W, Zhu XF, Zhu Y, Zhuang SM, Zhuang X, Ziparo E, Zois CE, Zoladek T, Zong WX, Zorzano A Zughaier SM, 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 12, 1–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koike M, Shibata M, Tadakoshi M, Gotoh K, Komatsu M, Waguri S, Kawahara N, Kuida K, Nagata S, Kominami E, Tanaka K Uchiyama Y, 2008. Inhibition of autophagy prevents hippocampal pyramidal neuron death after hypoxic-ischemic injury. Am J of Path 172, 454–469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kume T, Kawato Y, Osakada F, Izumi Y, Katsuki H, Nakagawa T, Kaneko S, Niidome T, Takada-Takatori Y Akaike A, 2008. Dibutyryl cyclic AMP induces differentiation of human neuroblastoma SH-SY5Y cells into a noradrenergic phenotype. Neurosci Lett 443, 199–203. [DOI] [PubMed] [Google Scholar]
- Lamming DW, Ye L, Katajisto P, Goncalves MD, Saitoh M, Stevens DM, Davis JG, Salmon AB, Richardson A, Ahima RS, Guertin DA, Sabatini DM Baur JA, 2012. Rapamycin-induced insulin resistance is mediated by mTORC2 loss and uncoupled from longevity. Science 335, 1638–1643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Li L, Jung H-H Zuo Z, 2008. Postconditioning with isoflurane reduced ischemia-induced brain injury in rats. Anesthesiology 108, 1055–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin D, Feng C, Cao M Zuo Z, 2011. Volatile anesthetics may not induce significant toxicity to human neuron-like cells. Anesth Analg 112, 1194–1198. [DOI] [PubMed] [Google Scholar]
- Liu S, Sarkar C, Dinizo M, Faden AI, Koh EY, Lipinski MM Wu J, 2015. Disrupted autophagy after spinal cord injury is associated with ER stress and neuronal cell death. Cell Death Dis 6, e1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano R, Naghavi M, Foreman K, Lim S, Shibuya K, Aboyans V, Abraham J, Adair T, Aggarwal R, Ahn SY, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Barker-Collo S, Bartels DH, Bell ML, Benjamin EJ, Bennett D, Bhalla K, Bikbov B, Bin Abdulhak A, Birbeck G, Blyth F, Bolliger I, Boufous S, Bucello C, Burch M, Burney P, Carapetis J, Chen H, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahodwala N, De Leo D, Degenhardt L, Delossantos A, Denenberg J, Des Jarlais DC, Dharmaratne SD, Dorsey ER, Driscoll T, Duber H, Ebel B, Erwin PJ, Espindola P, Ezzati M, Feigin V, Flaxman AD, Forouzanfar MH, Fowkes FG, Franklin R, Fransen M, Freeman MK, Gabriel SE, Gakidou E, Gaspari F, Gillum RF, Gonzalez-Medina D, Halasa YA, Haring D, Harrison JE, Havmoeller R, Hay RJ, Hoen B, Hotez PJ, Hoy D, Jacobsen KH, James SL, Jasrasaria R, Jayaraman S, Johns N, Karthikeyan G, Kassebaum N, Keren A, Khoo JP, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Lipnick M, Lipshultz SE, Ohno SL, Mabweijano J, MacIntyre MF, Mallinger L, March L, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGrath J, Mensah GA, Merriman TR, Michaud C, Miller M, Miller TR, Mock C, Mocumbi AO, Mokdad AA, Moran A, Mulholland K, Nair MN, Naldi L, Narayan KM, Nasseri K, Norman P, O’Donnell M, Omer SB, Ortblad K, Osborne R, Ozgediz D, Pahari B, Pandian JD, Rivero AP, Padilla RP, Perez-Ruiz F, Perico N, Phillips D, Pierce K, Pope CA 3rd, Porrini E, Pourmalek F, Raju M, Ranganathan D, Rehm JT, Rein DB, Remuzzi G, Rivara FP, Roberts T, De Leon FR, Rosenfeld LC, Rushton L, Sacco RL, Salomon JA, Sampson U, Sanman E, Schwebel DC, Segui-Gomez M, Shepard DS, Singh D, Singleton J, Sliwa K, Smith E, Steer A, Taylor JA, Thomas B, Tleyjeh IM, Towbin JA, Truelsen T, Undurraga EA, Venketasubramanian N, Vijayakumar L, Vos T, Wagner GR, Wang M, Wang W, Watt K, Weinstock MA, Weintraub R, Wilkinson JD, Woolf AD, Wulf S, Yeh PH, Yip P, Zabetian A, Zheng ZJ, Lopez AD, Murray CJ, AlMazroa MA Memish ZA, 2012. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2095–2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo C, Ouyang MW, Fang YY, Li SJ, Zhou Q, Fan J, Qin ZS Tao T, 2017. Dexmedetomidine Protects Mouse Brain from Ischemia-Reperfusion Injury via Inhibiting Neuronal Autophagy through Up-Regulating HIF-1alpha. Front Cell Neurosci 11, 197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manders E, Vanderbeek F Aten J, 1993. Measurement of colocalization of objects in dual-color confocal images. J Microoscopy 169 Part 3, 375–382. [DOI] [PubMed] [Google Scholar]
- Mizushima N, 2007. Autophagy: process and function. Genes Dev 21, 2861–2873. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Yoshimori T Levine B, 2010. Methods in mammalian autophagy research. Cell 140, 313–326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray CJ, Vos T, Lozano R, Naghavi M, Flaxman AD, Michaud C, Ezzati M, Shibuya K, Salomon JA, Abdalla S, Aboyans V, Abraham J, Ackerman I, Aggarwal R, Ahn SY, Ali MK, Alvarado M, Anderson HR, Anderson LM, Andrews KG, Atkinson C, Baddour LM, Bahalim AN, Barker-Collo S, Barrero LH, Bartels DH, Basanez MG, Baxter A, Bell ML, Benjamin EJ, Bennett D, Bernabe E, Bhalla K, Bhandari B, Bikbov B, Bin Abdulhak A, Birbeck G, Black JA, Blencowe H, Blore JD, Blyth F, Bolliger I, Bonaventure A, Boufous S, Bourne R, Boussinesq M, Braithwaite T, Brayne C, Bridgett L, Brooker S, Brooks P, Brugha TS, Bryan-Hancock C, Bucello C, Buchbinder R, Buckle G, Budke CM, Burch M, Burney P, Burstein R, Calabria B, Campbell B, Canter CE, Carabin H, Carapetis J, Carmona L, Cella C, Charlson F, Chen H, Cheng AT, Chou D, Chugh SS, Coffeng LE, Colan SD, Colquhoun S, Colson KE, Condon J, Connor MD, Cooper LT, Corriere M, Cortinovis M, de Vaccaro KC, Couser W, Cowie BC, Criqui MH, Cross M, Dabhadkar KC, Dahiya M, Dahodwala N, Damsere-Derry J, Danaei G, Davis A, De Leo D, Degenhardt L, Dellavalle R, Delossantos A, Denenberg J, Derrett S, Des Jarlais DC, Dharmaratne SD, Dherani M, Diaz-Torne C, Dolk H, Dorsey ER, Driscoll T, Duber H, Ebel B, Edmond K, Elbaz A, Ali SE, Erskine H, Erwin PJ, Espindola P, Ewoigbokhan SE, Farzadfar F, Feigin V, Felson DT, Ferrari A, Ferri CP, Fevre EM, Finucane MM, Flaxman S, Flood L, Foreman K, Forouzanfar MH, Fowkes FG, Fransen M, Freeman MK, Gabbe BJ, Gabriel SE, Gakidou E, Ganatra HA, Garcia B, Gaspari F, Gillum RF, Gmel G, Gonzalez-Medina D, Gosselin R, Grainger R, Grant B, Groeger J, Guillemin F, Gunnell D, Gupta R, Haagsma J, Hagan H, Halasa YA, Hall W, Haring D, Haro JM, Harrison JE, Havmoeller R, Hay RJ, Higashi H, Hill C, Hoen B, Hoffman H, Hotez PJ, Hoy D, Huang JJ, Ibeanusi SE, Jacobsen KH, James SL, Jarvis D, Jasrasaria R, Jayaraman S, Johns N, Jonas JB, Karthikeyan G, Kassebaum N, Kawakami N, Keren A, Khoo JP, King CH, Knowlton LM, Kobusingye O, Koranteng A, Krishnamurthi R, Laden F, Lalloo R, Laslett LL, Lathlean T, Leasher JL, Lee YY, Leigh J, Levinson D, Lim SS, Limb E, Lin JK, Lipnick M, Lipshultz SE, Liu W, Loane M, Ohno SL, Lyons R, Mabweijano J, MacIntyre MF, Malekzadeh R, Mallinger L, Manivannan S, Marcenes W, March L, Margolis DJ, Marks GB, Marks R, Matsumori A, Matzopoulos R, Mayosi BM, McAnulty JH, McDermott MM, McGill N, McGrath J, Medina-Mora ME, Meltzer M, Mensah GA, Merriman TR, Meyer AC, Miglioli V, Miller M, Miller TR, Mitchell PB, Mock C, Mocumbi AO, Moffitt TE, Mokdad AA, Monasta L, Montico M, Moradi-Lakeh M, Moran A, Morawska L, Mori R, Murdoch ME, Mwaniki MK, Naidoo K, Nair MN, Naldi L, Narayan KM, Nelson PK, Nelson RG, Nevitt MC, Newton CR, Nolte S, Norman P, Norman R, O’Donnell M, O’Hanlon S, Olives C, Omer SB, Ortblad K, Osborne R, Ozgediz D, Page A, Pahari B, Pandian JD, Rivero AP, Patten SB, Pearce N, Padilla RP, Perez-Ruiz F, Perico N, Pesudovs K, Phillips D, Phillips MR, Pierce K, Pion S, Polanczyk GV, Polinder S, Pope CA 3rd, Popova S, Porrini E, Pourmalek F, Prince M, Pullan RL, Ramaiah KD, Ranganathan D, Razavi H, Regan M, Rehm JT, Rein DB, Remuzzi G, Richardson K, Rivara FP, Roberts T, Robinson C, De Leon FR, Ronfani L, Room R, Rosenfeld LC, Rushton L, Sacco RL, Saha S, Sampson U, Sanchez-Riera L, Sanman E, Schwebel DC, Scott JG, Segui-Gomez M, Shahraz S, Shepard DS, Shin H, Shivakoti R, Singh D, Singh GM, Singh JA, Singleton J, Sleet DA, Sliwa K, Smith E, Smith JL, Stapelberg NJ, Steer A, Steiner T, Stolk WA, Stovner LJ, Sudfeld C, Syed S, Tamburlini G, Tavakkoli M, Taylor HR, Taylor JA, Taylor WJ, Thomas B, Thomson WM, Thurston GD, Tleyjeh IM, Tonelli M, Towbin JA, Truelsen T, Tsilimbaris MK, Ubeda C, Undurraga EA, van der Werf MJ, van Os J, Vavilala MS, Venketasubramanian N, Wang M, Wang W, Watt K, Weatherall DJ, Weinstock MA, Weintraub R, Weisskopf MG, Weissman MM, White RA, Whiteford H, Wiebe N, Wiersma ST, Wilkinson JD, Williams HC, Williams SR, Witt E, Wolfe F, Woolf AD, Wulf S, Yeh PH, Zaidi AK, Zheng ZJ, Zonies D, Lopez AD, AlMazroa MA Memish ZA, 2012. Disability-adjusted life years (DALYs) for 291 diseases and injuries in 21 regions, 1990–2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet 380, 2197–2223. [DOI] [PubMed] [Google Scholar]
- Murry CE, Jennings RB Reimer KA, 1986. Preconditioning with ischemia: a delay of lethal cell injury in ischemic myocardium. Circulation 74, 1124–1136. [DOI] [PubMed] [Google Scholar]
- Pan J, Li X Peng Y, 2016. Remote ischemic conditioning for acute ischemic stroke: dawn in the darkness. Rev Neurosci 27, 501–510. [DOI] [PubMed] [Google Scholar]
- Pivtoraiko VN, Harrington AJ, Mader BJ, Luker AM, Caldwell GA, Caldwell KA, Roth KA Shacka JJ, 2010. Low-dose bafilomycin attenuates neuronal cell death associated with autophagy-lysosome pathway dysfunction. J Neurochem 114, 1193–1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao Z, Pan X, Zhang H, Sun J, Li J, Lu T, Gao M, Liu S, Yu D Ding Z, 2017. Isoflurane Preconditioning Alleviated Murine Liver Ischemia and Reperfusion Injury by Restoring AMPK/mTOR-Mediated Autophagy. Anesth Analg 125, 1355–1363. [DOI] [PubMed] [Google Scholar]
- Ren G, Zhou Y, Liang G, Yang B, Yang M, King A Wei H, 2017. General Anesthetics Regulate Autophagy via Modulating the Inositol 1,4,5-Trisphosphate Receptor: Implications for Dual Effects of Cytoprotection and Cytotoxicity. Sci Rep 7, 12378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, Wang Z, Ma H Zuo Z, 2014. Sevoflurane postconditioning provides neuroprotection against brain hypoxia-ischemia in neonatal rats. Neurol Sci 35, 1401–1404. [DOI] [PubMed] [Google Scholar]
- Rusten TE Stenmark H, 2010. p62, an autophagy hero or culprit? Nat Cell Biol 12, 207–209. [DOI] [PubMed] [Google Scholar]
- Ryou MG, Choudhury GR, Li W, Winters A, Yuan F, Liu R Yang SH, 2015. Methylene blue-induced neuronal protective mechanism against hypoxia-reoxygenation stress. Neurosci 301, 193–203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saiki S, Sasazawa Y, Imamichi Y, Kawajiri S, Fujimaki T, Tanida I, Kobayashi H, Sato F, Sato S, Ishikawa K, Imoto M Hattori N, 2011. Caffeine induces apoptosis by enhancement of autophagy via PI3K/Akt/mTOR/p70S6K inhibition. Autophagy 7, 176–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar C, Zhao Z, Aungst S, Sabirzhanov B, Faden AI Lipinski MM, 2014. Impaired autophagy flux is associated with neuronal cell death after traumatic brain injury. Autophagy 10, 2208–2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen HM Mizushima N, 2014. At the end of the autophagic road: an emerging understanding of lysosomal functions in autophagy. Trends Bioch Sci 39, 61–71. [DOI] [PubMed] [Google Scholar]
- Sheng R, Zhang TT, Felice VD, Qin T, Qin ZH, Smith CD, Sapp E, Difiglia M Waeber C, 2014. Preconditioning stimuli induce autophagy via sphingosine kinase 2 in mouse cortical neurons. J Biol Chem 289, 20845–20857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang CQ, Ye Y, Chen F, Han WC, Sun JM, Lu X, Guo R, Cao K, Zheng MJ Liao LC, 2017a. Posttraumatic administration of a sub-anesthetic dose of ketamine exerts neuroprotection via attenuating inflammation and autophagy. Neurosci 343, 30–38. [DOI] [PubMed] [Google Scholar]
- Wang M, Li YJ, Ding Y, Zhang HN, Sun T, Zhang K, Yang L, Guo YY, Liu SB, Zhao MG Wu YM, 2016. Silibinin Prevents Autophagic Cell Death upon Oxidative Stress in Cortical Neurons and Cerebral Ischemia-Reperfusion Injury. Mol Neurobiol 53, 932–943. [DOI] [PubMed] [Google Scholar]
- Wang Y, Liang B, Lau WB, Du Y, Guo R, Yan Z, Gan L, Yan W, Zhao J, Gao E, Koch W Ma XL, 2017b. Restoring diabetes-induced autophagic flux arrest in ischemic/reperfused heart by ADIPOR (adiponectin receptor) activation involves both AMPK-dependent and AMPK-independent signaling. Autophagy 13, 1855–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Lin D, Li G Zuo Z, 2010. Statin post-treatment provides protection against simulated ischemia in bovine pulmonary arterial endothelial cells. Eur J Pharmacol 636, 114–120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Zou Z, Zou R, Zhou X Cui S, 2015. Electroacupuncture pretreatment induces tolerance against cerebral ischemia/reperfusion injury through inhibition of the autophagy pathway. Mol Med Rep 11, 4438–4446. [DOI] [PubMed] [Google Scholar]
- Xia Y, Xu H, Jia C, Hu X, Kang Y, Yang X, Xue Q, Tao G Yu B, 2017. Tanshinone IIA Attenuates Sevoflurane Neurotoxicity in Neonatal Mice. Anesth Analg 124, 1244–1252. [DOI] [PubMed] [Google Scholar]
- Xiong N, Jia M, Chen C, Xiong J, Zhang Z, Huang J, Hou L, Yang H, Cao X, Liang Z, Sun S, Lin Z Wang T, 2011. Potential autophagy enhancers attenuate rotenone-induced toxicity in SH-SY5Y. Neurosci 199, 292–302. [DOI] [PubMed] [Google Scholar]
- Yan W, Zhang H, Bai X, Lu Y, Dong H Xiong L, 2011. Autophagy activation is involved in neuroprotection induced by hyperbaric oxygen preconditioning against focal cerebral ischemia in rats. Brain Res 1402, 109–121. [DOI] [PubMed] [Google Scholar]
- Ye F Zuo ZY, 2017. Anesthetic effects on autophagy. Med Gas Res 7, 204–211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao P Zuo Z, 2005. Prenatal hypoxia-induced adaptation and neuroprotection that is inducible nitric oxide synthase-dependent. Neurobiol Dis 20, 871–880. [DOI] [PubMed] [Google Scholar]
- Zhao ZQ, Corvera JS, Halkos ME, Kerendi F, Wang NP, Guyton RA Vinten-Johansen J, 2003. Inhibition of myocardial injury by ischemic postconditioning during reperfusion: comparison with ischemic preconditioning. Am J Physiol Heart Circ Physiol 285, H579–588. [DOI] [PubMed] [Google Scholar]
- Zhou J, Hu SE, Tan SH, Cao R, Chen Y, Xia D, Zhu X, Yang XF, Ong CN Shen HM, 2012. Andrographolide sensitizes cisplatin-induced apoptosis via suppression of autophagosome-lysosome fusion in human cancer cells. Autophagy 8, 338–349. [DOI] [PubMed] [Google Scholar]