Abstract
The gut microbiome is known to play a significant role in human health but its role in aging remains unclear. The objective of this study was to compare the gut microbiome composition between young adult and geriatric nonhuman primates (marmosets) as a model of human health and disease. Stool samples were collected from geriatric (8+ years) and young adult males (2–5 years). Stool 16s rRNA V4 sequences were amplified and sequenced on the Illumina MiSeq platform. Sequences were clustered into operational taxonomic units and classified via mothur’s Bayesian classifier referenced against the Greengenes database. A total of 10 young adult and 10 geriatric marmosets were included. Geriatric marmosets had a lower mean Shannon diversity compared to young marmosets (3.15 vs. 3.46; p=0.0191). Geriatric marmosets had a significantly higher mean abundance of Proteobacteria (0.22 vs. 0.09; p=0.0233) and lower abundance of Firmicutes (0.15 vs. 0.19; p=0.0032) compared to young marmosets. Geriatric marmosets had a significantly higher abundance of Succinivibrionaceae (0.16 vs. 0.01; p=0.0191) and lower abundance of Porphyromonadaceae (0.07 vs. 0.11; p=0.0494). In summary, geriatric marmosets had significantly altered microbiome diversity and composition compared to young adult marmosets. Further studies are needed to test microbiome-targeted therapies to improve healthspan and lifespan.
Keywords: microbiome, gut microbiota, dysbiosis, aging, geriatrics, marmoset
Introduction
An individual’s health and wellbeing were once thought to be solely associated with their own genes and environment; however, it has become clearer that the health of an individual is also influenced by a broader metagenome phenotype as a result of our microbiome. Trillions of microorganisms live commensally within the gut and serve several important physiological functions, including protection against pathogens, immune response regulation, nutrient and energy extraction, and hormone biosynthesis (Belkaid & Hand, 2014; Lynch & Pedersen, 2016). Intestinal microbial diversity in nonhuman primates has been found to be tightly linked to the host phylogeny (Yildirim et al., 2010), to the social group that the individuals are interacting with (Amato et al., 2017), and to the components of their dietary intake (Hicks et al., 2018).
It is not surprising then that dysbiosis (i.e., disturbance or change in the microbiota) has been associated with increased inflammation and altered physiological homeostasis (Cattaneo et al., 2017). Evaluations of nonhuman primates in captive settings have revealed significant loss of microbial diversity for animals maintained in captivity when compared to the same species in wild, or semi-captive settings (Clayton, Gomez, et al., 2018; Clayton et al., 2016). This shift in diversity appears to be associated with changes in dietary components. Red-shanked doucs in captivity have been found to have increased dysbiosis associated with significantly reduced Firmicutes: Bacterioidetes ratios associated with increased risk of wasting syndrome, gastrointestinal disease, and mortality (Clayton, Al-Ghalith, et al., 2018). An evaluation of a macaque model of obesity revealed that the dam’s dietary intake and not her status as obese or lean significantly altered the infant’s developing intestinal microbiome (Ma et al., 2014). In humans, dysbiosis of intestinal microbiota has been associated with obesity, type 2 diabetes, Alzheimer’s disease, cardiovascular disease, and frailty (Barlow, Yu, & Mathur, 2015; Jackson et al., 2016; Komaroff, 2017). In many cases, it remains unclear whether changes in the microbiome are a result of the disease or whether alterations in the microbiome leave the host vulnerable for physiological changes contributing to the disease state.
Human studies have also noted a significant decline in microbiome diversity and composition with age, (Yatsunenko et al., 2012) which might contribute to functional decline and age-related disease status; however, changes in medication exposure, physical activity, and diet with age could also significantly contribute to dysbiosis. A shift in the microbiota toward a Bacteriodetes dominated population is associated with frailer older humans (Jeffery, Lynch, & O’Toole, 2016). Centenarians have been found to have significantly increased populations of pathobionts including Bacteriodetes and Proteobacteria and shifts in Firmicutes diversity (Rampelli et al., 2013). However, these shifts in the gut microbial community are also associated with changes in the diet to low fiber diets with less macronutrient diversity typical of nursing home meals. Therefore, it is unclear whether the microbial shift is a result a dietary change, or whether the microbial shift is associated with the aging process and leads to inflammation and disease. These confounders make the study of the microbiome in aging humans particularly challenging.
Evaluating the interactions between the microbiome and phenotypic aging in a nonhuman primate model offers the advantage of being able to more definitively control environment, diet, and exposure to medication. The marmoset offers a unique nonhuman primate in which to evaluate hypotheses associated with both aging and healthspan. Marmosets have a short lifespan when compared to other primates; they reach sexual maturity between 12–18 months and full adult weight by 24 months (Tardif, Mansfield, Ratnam, Ross, & Ziegler, 2011). They are considered young adults from 2–5 years of age and are considered geriatric over the age of 8 years. Marmosets are relatively inexpensive to house, and they are not zoonotic hosts making them easier and safer to handle for experimentation than many other nonhuman primate biomedical models. Furthermore, the recent expansion of phenotypic assessments of aging have found that marmosets display many aging characteristics that mimic human aging, including cardiovascular changes, inflammatory disease, and cognitive decline (Ross, Davis, Dobek, & Tardif, 2012) (other manuscripts in this special issue). Therefore, our study objective was to compare the gut microbial diversity and composition between young adult and geriatric marmosets. We hypothesized that geriatric marmosets would have significantly less diverse and compositionally different microbiomes than young adult marmosets independent of differences in diet or housing.
Methods
Study Design
This was a cross-sectional study of marmosets housed at the Barshop Institute for Longevity and Aging Studies in San Antonio, TX in August 2017. The study was approved by the Institutional Animal Care and Use Committee at UT Health San Antonio, and follows guidelines set forth by the American Society of Primatologists. Marmosets housed at the Barshop Institute are primarily a breeding colony of animals maintained in barrier (Ross et al., 2017) or semi-barrier conditions (isolated from other nonhuman primates) specifically for aging related research with husbandry protocols following those outlined in (Layne & Power, 2003). The marmosets are maintained on a daily standardized base diet consisting of a mix of commercial products (Harlan Teklad marmoset purified diet, Purina Mazuri, and Zupreem marmoset diet; Table 1). Marmosets in the colony receive limited dietary enrichment items including cheerios, marshmallows and peanuts. Eligibility criteria for the study included animals that were not receiving intervention including probiotics, antibiotics, or rapamycin treatment, and had no recent medical concerns. At the time of the study the Barshop colony consisted of 5 males and 3 females over the age of 10, and 7 males aged 8–9 that met the eligibility criteria. The colony also had 23 males and 23 females that were considered young adult age (2–5) that met eligibility criteria. Given the limitation in number of geriatric female marmosets in our population, this analysis focused on determining whether geriatric male marmoset microbiomes differed from young marmosets. While there is no evidence in the human literature that male and female gut microbiota populations differ significantly, potential sex differences have not been explored in geriatric populations and it remains unclear what effects the changes in sexually specific endocrine cycles might have on the microbiome (Duncan, Colman, & Kramer, 2012; Jašarević, Morrison, & Bale, 2016). In this analysis, we assessed 10 geriatric males (8+ years; mean (± SD) age was 10.2 (±2.03) years) and 10 young adult males (2–5 years; mean 3.1 (±0.7) years) which were randomly selected from the available animals to assess intestinal microbial diversity. None of these animals had a history of disease, diarrhea or dietary shift; all known medical history is outlined in Table 2. A fresh stool sample was collected from each animal, placed in a sterile collection tube and frozen at −80˚ C until shipment for analysis.
Table 1.
Macronutrient composition of marmoset base diets
| Nutrient | Harlan Purified | Mazuri | Zupreem |
|---|---|---|---|
| Protein % | 15.4 | 21.3 | 8.5 |
| Fat % | 13.8 | 7.8 | 2.5 |
| Carbohydrate % | 70.8 | 70.9 | 89 |
| Energy (kcal/g) | 3.6 | 3.4 | 3.4 |
Table 2.
Health history of the young adult and geriatric marmosets
| Animal ID | Age | Housing | Medical History |
|---|---|---|---|
| 14 | 2.0 | paired | Barshop born |
| 2 | 2.0 | paired | SNPRC born, transferred to Barshop 2016 |
| 20 | 2.6 | single | Barshop born |
| 1 | 2.9 | single | Barshop born, given acarbose two weeks 2016 |
| 19 | 3.1 | paired | Barshop born |
| 15 | 3.2 | paired | Barshop born |
| 9 | 3.3 | single | Barshop born, given acarbose two weeks 2016 |
| 7 | 3.7 | paired | Barshop born, given acarbose two weeks 2016 |
| 18 | 3.8 | family | Barshop born, family given probios Feb 2014 |
| 3 | 4.1 | paired | Barshop born |
| 17 | 8.1 | paired | Barshop born, vasectomy July 2016 |
| 4 | 8.2 | single | Barshop born, vasectomy July 2016 |
| 8 | 8.6 | paired | NEPRC born, transferred to SNPRC 2014, transferred to Barshop 2015, vasectomy Oct 2016 |
| 5 | 9.2 | paired | SNPRC born, transferred 2014, vasectomy Oct 2016 |
| 13 | 9.2 | paired | NEPRC born, transferred to SNPRC 2014, vasectomy 2014, transferred to Barshop 2015 |
| 10 | 9.7 | paired | NEPRC born, transferred to SNPRC 2014, vasectomy 2014, transferred to Barshop 2015 |
| 12 | 10.5 | paired | NEPRC born, transferred to SNPRC 2014, vasectomy 2014, transferred to Barshop 2015 |
| 6 | 11.9 | paired | NEPRC born, transferred to SNPRC 2014, vasectomy 2014, transferred to Barshop 2015 |
| 11 | 12.1 | paired | NEPRC born, transferred to SNPRC 2014, vasectomy 2014, transferred to Barshop 2015 |
| 16 | 14.3 | paired | SNPRC born, transferred to Barshop 2013 |
Barshop = UT Health Science Center Barshop Institute Colony, SNPRC = Southwest National Primate Research Center, NEPRC = New England Primate Research Center
Laboratory Procedures
Stool sample sequencing and analysis were completed by Second Genome, Inc., San Francisco, CA. Nucleic acid isolation was performed with the MoBio PowerMag® Microbiome kit (Carlsbad, CA) optimized on the KingFisher Nucleic Acid Purification System (Thermo Fisher, Waltham, MA) for high-throughput processing (Qiagen. PowerMag Microbiome RNA/DNA Isolation Kit). All samples were quantified via the Qubit® Quant-iT dsDNA High Sensitivity Kit (Invitrogen, Life Technologies, Grand Island, NY). To enrich the sample for bacterial 16S V4 rDNA region, DNA was amplified with 515F-806R primers that were tailed with sequences to incorporate Illumina® (San Diego, CA) adapters and indexing barcodes(Caporaso et al., 2011). Each PCR product was quantified using a fluorometric method (Qubit from Invitrogen, Life Technologies, Grand Island, NY), and equimolar amounts were pooled for sequencing. The pooled library was loaded onto the Illumina MiSeq® platform and the amplicons were sequenced using a 250 bp paired-end protocol.
Data and Statistical Analysis
An open reference operational taxonomic unit (OTU) picking strategy was used, such that all sequences could be assigned to a strain OTU. First, sequenced paired-end reads were merged and resulting sequences were compared to a reference database at Second Genome (StrainSelect) using USEARCH (Edgar, 2010). All sequences matching to a unique strain with an identity ≥99% were assigned a strain OTU, which represents ≥99% identical match to a biological sequence within the database. A difference of ≥0.25% between the identity of the best hit and the second best hit was required (e.g., 99.75 versus 99.5). For each strain OTU, one of the matching reads was selected as representative and all sequences were mapped by USEARCH against the strain OTU representatives to calculate strain abundances. The remaining non-strain sequences were quality filtered and de-replicated with USEARCH. Sequences that did not match the reference database then underwent de novo OTU clustering using the UPARSE clustering algorithm, which clusters sequences at 97% similarity (Edgar, 2013). The 97% threshold was derived from a study that found that most strains had 97% 16S rRNA sequence similarity (Konstantinidis & Tiedje, 2005). The UPARSE clustering algorithm comprises a chimera filtering and discards likely chimeric OTUs. All non-strain sequences that passed the quality filtering were mapped to the representative consensus sequences to generate an abundance table for de novo OTUs. All generated OTU sequences were assigned taxonomic classification via Mothur’s bayesian classifier,(Schloss et al., 2009) trained against the Greengenes reference database of 16S rRNA gene sequences clustered at 99% (McDonald et al., 2012). Removal of spurious OTUs was completed by independent filtering, such that OTUs that were seen in at least 10% of the dataset were kept.
Statistical analyses were conducted using R® statistical language and environment. Sample richness was estimated based on the number of OTUs present in a sample. Shannon diversity calculations accounted for OTU richness and relative abundance (Shannon, 1948). The Wilcoxon signed rank test was used to compare alpha diversity between groups. Abundance-weighted sample pair-wise differences were calculated using the Bray-Curtis dissimilarity (Bray & Curtis, 1957). PERMANOVA was used to determine if age significantly contributed to beta diversity of samples. Univariate differential abundance of OTUs was tested using a negative binomial noise model for the overdispersion and Poisson process intrinsic to these data, as implemented in the DESeq2 package (Love, Huber, & Anders, 2014), and described for microbiome applications (McMurdie & Holmes, 2014). DESeq was run under default settings and q-values calculated with the Benjamini-Hochberg procedure to correct p-values, controlling for a false discovery rate of 0.25.
A hierarchical clustering of the samples was used to graphically summarize the relationship between geriatric and young adult marmoset samples. Samples from the distance matrix were clustered hierarchically using the Ward2 method.
Results
Sequencing Summary
Prior to filtering, the number of OTUs and sequences generated were 641 and 3,267,331, respectively. A total of 467 OTUs from 3,257,446 sequences passed sample quality check and were used for downstream analyses; 99.84% of all sequences were able to be classified at the phylum, class, and order levels. Only 75.76% and 21.74% of OTUs were classified at the genus and species levels, respectively. Sample library sizes ranged from 137,270 to 198,201 reads in geriatric marmoset samples and 149,348 to 176,782 in the young adult marmoset samples.
Sample Diversity by Age Group
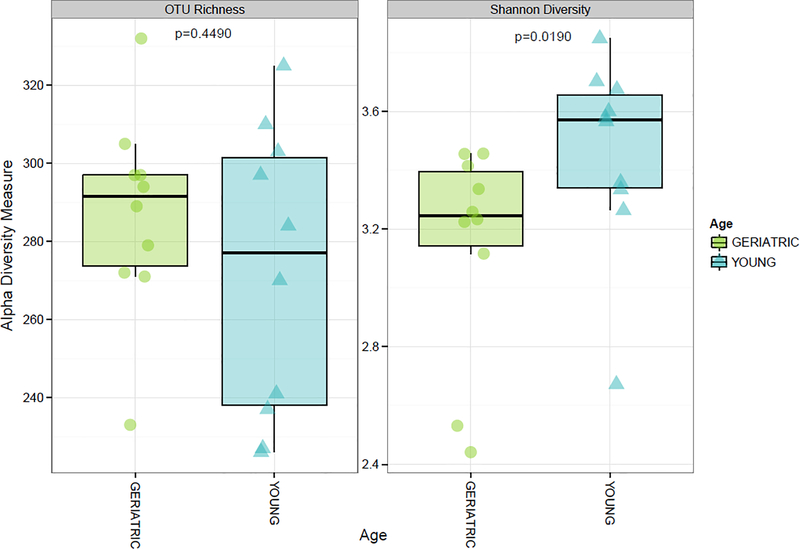
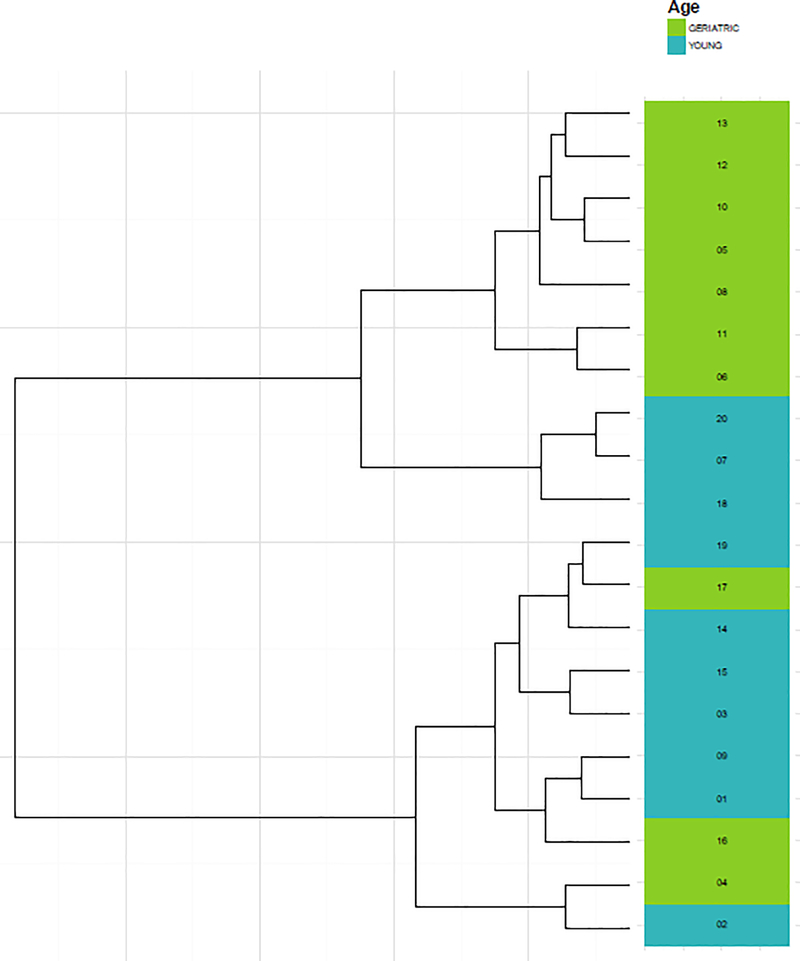
Figure 1 displays alpha diversity measures. Mean (± standard deviation) OTU richness was not statistically different between geriatric (287 ± 24) and young adult (272 ± 37) samples (p=0.4490). However, Shannon diversity was significantly lower in geriatric (3.15 ± 0.37) compared to young adult (3.46 ± 0.33) samples (p=0.0191). Hierarchical clustering largely separated samples by age (Figure 2).
Figure 1.
Comparison of alpha diversity measures between geriatric and young adult marmoset fecal microbiome samples.
Figure 2.
Hierarchical clustering of geriatric and young adult marmoset microbiome samples
Sample Composition by Age Group
Age was associated with significant changes in relative abundance of certain bacterial phyla and families. Table 3 provides the proportional abundance of the 5 most common bacterial phyla and 8 families identified. At the phylum level, geriatric marmosets had a significantly higher mean abundance of Proteobacteria (0.22 vs. 0.09; p=0.0233) and lower abundance of Firmicutes (0.15 vs. 0.19; p=0.0032) compared to young adult marmosets. At the family level, geriatric marmosets had a significantly higher abundance of Succinivibrionaceae (0.16 vs. 0.01; p=0.0191) and lower abundance of Porphyromonadaceae (0.07 vs. 0.11; p=0.0494). Age significantly contributed to beta diversity of the samples (PERMANOVA p- =0.0190).
Table 3.
Comparison of select taxa relative abundance between geriatric and young adult marmoset samples
| Bacteria | Geriatric (n=10) | Young Adult (n=10) | Raw p-value | Adjusted p-valuea |
|---|---|---|---|---|
| Phylum, mean (SD) | ||||
| Bacteroidetes | 0.35 (0.06) | 0.349 (0.1) | 0.7055 | 0.2222 |
| Actinobacteria | 0.21 (0.1) | 0.254 (0.2) | 0.5967 | 0.1667 |
| Firmicutes | 0.15 (0.04) | 0.2 (0.03) | 0.0032 | 0.0278 |
| Proteobacteria | 0.22 (0.11) | 0.09 (0.05) | 0.0233 | 0.0556 |
| Fusobacteria | 0.08 (0.1) | 0.12 (0.15) | 0.7055 | 0.2500 |
| Family, mean (SD) | ||||
| Bifidobacteriaceae | 0.12 (0.05) | 0.15 (0.11) | 0.9397 | 0.2500 |
| Bacteroidaceae | 0.07 (0.05) | 0.12 (0.06) | 0.0696 | 0.1111 |
| Coriobacteriaceae | 0.09 (0.05) | 0.10 (0.05) | 0.4497 | 0.1667 |
| Fusobacteriaceae | 0.08 (0.10) | 0.11 (0.15) | 0.7055 | 0.2222 |
| Porphyromonadaceae | 0.07 (0.04) | 0.11 (0.06) | 0.0494 | 0.0556 |
| Prevotellaceae | 0.13 (0.14) | 0.04 (0.06) | 0.1509 | 0.1389 |
| Succinivibrionaceae | 0.16 (0.11) | 0.01 (0.01) | 0.0191 | 0.0278 |
| Veillonellaceae | 0.07 (0.02) | 0.08 (0.02) | 0.5453 | 0.1944 |
P-values adjusted using the Benjamini-Hochberg procedure and a false discovery rate of 0.25; significance level set at 0.0556 for phyla comparisons and 0.1111 for the family comparisons
Discussion
Variability in microbiome diversity has been associated with differences in nonhuman primate environmental exposure, and dietary preferences, as well as human aging, obesity, and pathogenic disease state. Investigating causality and associations of microbiome change within the human population, or in wild primate populations, is broadly confounded by the inability to control for these environmental and dietary exposures. Further, it is difficult to determine the associations of microbial dysbiosis with human disease due to the inability to control for, or often account for pharmaceutical treatments. In order to determine whether changes in microbial diversity are associated with phenotypic aging, it is necessary to examine microbiome diversity in an animal model, preferably a closely related nonhuman primate species, which can be evaluated in captivity in a somewhat controlled exposure environment. This cross-sectional analysis of young adult and geriatric male marmosets identified significant differences in gut microbiome diversity and composition, with partial clustering of samples by age. Specifically, we found a less diverse microbiota that favored expansion of Proteobacteria and a reduction in the dominant Firmicutes phylum in geriatric marmosets compared to younger adult marmosets. To our knowledge, this is the first study to evaluate the effect of old age on the microbiome in a nonhuman primate.
Microbiome changes noted in this study mimic trends commonly seen in human studies. In healthy human adults, the gut microbiota measured at the bacterial phyla level is relatively stable, though bacterial species can greatly differ between individuals (Yatsunenko et al., 2012). Despite this variation, the overall functional capacity of the microbiome is similar across healthy persons (Qin et al., 2010). In contrast, the microbiome becomes less diverse in the elderly, with microbiota composition significantly correlated with measures of frailty, comorbidities, and inflammation (Claesson et al., 2012). Increasing populations of pathobionts have been found in the gut microbiota of centenarians with increased prevalence of Bacteriodetes and Proteobacteria in comparison to young adults (Rampelli et al., 2013) (Biagi et al., 2010).
The etiology of age-related microbiome changes is not well-defined; however, associations seen between dysbiosis and inflammatory conditions suggest a possible explanation. First, commensal bacteria are a critical regulator of the inflammatory response to acute injury and infection (Belkaid & Hand, 2014). Numerous animal studies and some human studies have noted increased inflammatory markers, as well as inflammatory conditions due to dysbiosis (Buford, 2017). Inflammatory conditions, like inflammatory bowel diseases, are associated with a reduction in species diversity and change in the abundance of bacterial taxa; obligate anaerobes from Bacteroidetes and Firmicutes decrease in abundance while facultative anaerobes from the Proteobacteria phyla are enriched (Buford, 2017). These changes could potentiate the inflammatory response. On the other hand, age-related changes, ranging from gut motility and cellular function to broad shifts in environmental exposure, could contribute to dysbiosis. Exposure to medications, like antibiotics, can also severely disrupt the microbiota and it may never fully recover to its original composition (Dethlefsen, Huse, Sogin, & Relman, 2008; Dethlefsen & Relman, 2011). Broad-spectrum antibiotic use has increased among older adults in recent years, which could contribute to alterations in the microbiome seen in this group (Lee GC, 2014). Physical activity and diet, including dietary preference and nutrient availability, may also change with age, both of which can impact microbiome composition (Blaser & Falkow, 2009). These environmental confounds make it particularly difficult to determine the causal links between microbiome health and healthy aging in human aging populations. Evaluating microbial health in a nonhuman primate model in association with aging is the first step in determining whether evaluations of microbial health may serve as a biomarker for increased risk of unhealthy aging.
While it is impossible to control for all environmental factors in a study examining nonhuman primates, this study examined animals that had lived in the same stable environment for several years, had the same diet that did not change with age, and had known medical exposure over the course of their lifetime. The males in this study were housed singly or with female pair-mate, none of the lived with each other, and none were closely related to each other. The males in this study had no previous exposure to oral antibiotics throughout their lifetime. While the geriatric males had almost all undergone vasectomy several years prior to this study, antibiotics are not standardly given during this procedure. The males examined originated from different colonies but had been in the Barshop colony for at least one year prior to sampling. Only one male had a medical history that included exposure to probiotic treatment, which was provided to the entire family group that he was housed with at the time. Three young males had participated in a study that included dosing with acarbose for two weeks one year prior to this microbiome study. Given the medical histories for the animals that were sampled and the fact that we found a significant alteration in the microbiome associated with age it suggests that changes in microbial diversity in aging subjects may be independent of changes associated with environmental and lifestyle shifts. There are no known husbandry, housing or dietary components that can be used to explain why three geriatric animals clustered more closely with the younger animals. As a short-term cross sectional study that only examined male marmosets, it is possible that the differences found between these young adult and geriatric marmosets do not reflect longitudinal microbiome changes, and it might not be generalizable to female subjects, which will need to be evaluated in the future as geriatric female marmosets become available. However, these findings strongly suggest that further work is needed to evaluate microbiome diversity in geriatric populations of nonhuman primate models in order to determine the causes of declining microbial diversity in aging individuals and to determine how shifts in diversity relate to measures of healthspan. Of particular importance for marmoset aging will be the ability to evaluate the associations between changes in microbial diversity and gut functionality associated with digestibility. Microbiome shifts with age may be a precursor biomarker for age related health decline, and the ability to identify specific targets for changes in diversity may help us to identify individuals at risk for more rapid decline. In summary, geriatric marmosets had significantly altered microbiome diversity and composition compared to young marmosets. These findings indicate a potential role of the gut microbiome in aging nonhuman primate and human health. Further studies are needed to test microbiome-targeted therapies to improve healthspan and lifespan. This work broadens our knowledge of microbial diversity and its association with aging, aside from environmental confounds. This is particularly important for the development of biomarkers for healthy aging as well as potentially offering therapeutic targets for healthy aging interventions.
Acknowledgements
The investigators would like to thank Aubrey Sills and Joselyn Artavia for their dedicated care of the marmosets, the staff at the Barshop Colony for Longevity and Aging Studies, UT Health San Antonio and at Second Genome, Inc. for their assistance with study procedures.
Funding
This work was supported by the National Institutes of Health/National Institute on Aging San Antonio Claude D. Pepper Older Americans Independence Center (1P30AG044271-01A1).
Footnotes
Conflicts of Interest
The authors report no competing interests related to the content of this manuscript.
References
- Amato KR, Van Belle S, Di Fiore A, Estrada A, Stumpf R, White B, . . . Leigh SR (2017). Patterns in Gut Microbiota Similarity Associated with Degree of Sociality among Sex Classes of a Neotropical Primate. Microb Ecol, 74(1), 250–258. doi: 10.1007/s00248-017-0938-6 [DOI] [PubMed] [Google Scholar]
- Barlow GM, Yu A, & Mathur R (2015). Role of the Gut Microbiome in Obesity and Diabetes Mellitus. Nutr Clin Pract, 30(6), 787–797. doi: 10.1177/0884533615609896 [DOI] [PubMed] [Google Scholar]
- Belkaid Y, & Hand TW (2014). Role of the microbiota in immunity and inflammation. Cell, 157(1), 121–141. doi: 10.1016/j.cell.2014.03.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biagi E, Nylund L, Candela M, Ostan R, Bucci L, Pini E, . . . De Vos W (2010). Through ageing, and beyond: gut microbiota and inflammatory status in seniors and centenarians. PLoS One, 5(5), e10667. doi: 10.1371/journal.pone.0010667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blaser MJ, & Falkow S (2009). What are the consequences of the disappearing human microbiota? Nat Rev Microbiol, 7(12), 887–894. doi: 10.1038/nrmicro2245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray JR, & Curtis JT (1957). An Ordination of the Upland Forest Communities of Southern Wisconsin. Ecological Monographs, 27(4), 326–349. [Google Scholar]
- Buford TW (2017). (Dis)Trust your gut: the gut microbiome in age-related inflammation, health, and disease. Microbiome, 5(1), 80. doi: 10.1186/s40168-017-0296-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Lozupone CA, Turnbaugh PJ, . . . Knight R (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc Natl Acad Sci U S A, 108 Suppl 1, 4516–4522. doi: 10.1073/pnas.1000080107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattaneo A, Cattane N, Galluzzi S, Provasi S, Lopizzo N, Festari C, . . . Group I-F (2017). Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol Aging, 49, 60–68. doi: 10.1016/j.neurobiolaging.2016.08.019 [DOI] [PubMed] [Google Scholar]
- Claesson MJ, Jeffery IB, Conde S, Power SE, O’Connor EM, Cusack S, . . . O’Toole PW (2012). Gut microbiota composition correlates with diet and health in the elderly. Nature, 488(7410), 178–184. doi: 10.1038/nature11319 [DOI] [PubMed] [Google Scholar]
- Clayton JB, Al-Ghalith GA, Long HT, Tuan BV, Cabana F, Huang H, . . . Johnson TJ (2018). Associations Between Nutrition, Gut Microbiome, and Health in A Novel Nonhuman Primate Model. Sci Rep, 8(1), 11159. doi: 10.1038/s41598-018-29277-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton JB, Gomez A, Amato K, Knights D, Travis DA, Blekhman R, . . . Johnson TJ (2018). The gut microbiome of nonhuman primates: Lessons in ecology and evolution. Am J Primatol, 80(6), e22867. doi: 10.1002/ajp.22867 [DOI] [PubMed] [Google Scholar]
- Clayton JB, Vangay P, Huang H, Ward T, Hillmann BM, Al-Ghalith GA, . . . Knights D (2016). Captivity humanizes the primate microbiome. Proc Natl Acad Sci U S A, 113(37), 10376–10381. doi: 10.1073/pnas.1521835113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, Huse S, Sogin ML, & Relman DA (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol, 6(11), e280. doi: 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dethlefsen L, & Relman DA (2011). Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc Natl Acad Sci U S A, 108 Suppl 1, 4554–4561. doi: 10.1073/pnas.1000087107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan AE, Colman RJ, & Kramer PA (2012). Sex differences in spinal osteoarthritis in humans and rhesus monkeys (Macaca mulatta). Spine (Phila Pa 1976), 37(11), 915–922. doi: 10.1097/BRS.0b013e31823ab7fc [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics, 26(19), 2460–2461. doi: 10.1093/bioinformatics/btq461 [DOI] [PubMed] [Google Scholar]
- Edgar RC (2013). UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods, 10(10), 996–998. doi: 10.1038/nmeth.2604 [DOI] [PubMed] [Google Scholar]
- Hicks AL, Lee KJ, Couto-Rodriguez M, Patel J, Sinha R, Guo C, . . . Williams BL (2018). Gut microbiomes of wild great apes fluctuate seasonally in response to diet. Nat Commun, 9(1), 1786. doi: 10.1038/s41467-018-04204-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson MA, Jeffery IB, Beaumont M, Bell JT, Clark AG, Ley RE, . . . Steves CJ (2016). Signatures of early frailty in the gut microbiota. Genome Med, 8(1), 8. doi: 10.1186/s13073-016-0262-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jašarević E, Morrison KE, & Bale TL (2016). Sex differences in the gut microbiome-brain axis across the lifespan. Philosophical Transactions of the Royal Society B: Biological Sciences, 371(1688), 20150122. doi:doi: 10.1098/rstb.2015.0122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery IB, Lynch DB, & O’Toole PW (2016). Composition and temporal stability of the gut microbiota in older persons. ISME J, 10(1), 170–182. doi: 10.1038/ismej.2015.88 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komaroff AL (2017). The Microbiome and Risk for Obesity and Diabetes. JAMA, 317(4), 355–356. doi: 10.1001/jama.2016.20099 [DOI] [PubMed] [Google Scholar]
- Konstantinidis KT, & Tiedje JM (2005). Genomic insights that advance the species definition for prokaryotes. Proc Natl Acad Sci U S A, 102(7), 2567–2572. doi: 10.1073/pnas.0409727102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Layne DG, & Power RA (2003). Husbandry, handling, and nutrition for marmosets. Comp Med, 53(4), 351–359. [PubMed] [Google Scholar]
- Lee GC, R. K., Lewis J, Frei CR. (2014). National trends in outpatient antibiotic prescribing, broad-spectrum prescribing, and antibiotic prescribing for acute respiratory tract infections BMC Medicine. [Google Scholar]
- Love MI, Huber W, & Anders S (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol, 15(12), 550. doi: 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch SV, & Pedersen O (2016). The Human Intestinal Microbiome in Health and Disease. N Engl J Med, 375(24), 2369–2379. doi: 10.1056/NEJMra1600266 [DOI] [PubMed] [Google Scholar]
- Ma J, Prince AL, Bader D, Hu M, Ganu R, Baquero K, . . . Aagaard KM (2014). High-fat maternal diet during pregnancy persistently alters the offspring microbiome in a primate model. Nat Commun, 5, 3889. doi: 10.1038/ncomms4889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D, Price MN, Goodrich J, Nawrocki EP, DeSantis TZ, Probst A, . . . Hugenholtz P (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J, 6(3), 610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMurdie PJ, & Holmes S (2014). Waste not, want not: why rarefying microbiome data is inadmissible. PLoS Comput Biol, 10(4), e1003531. doi: 10.1371/journal.pcbi.1003531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, . . . Wang J (2010). A human gut microbial gene catalogue established by metagenomic sequencing. Nature, 464(7285), 59–65. doi: 10.1038/nature08821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rampelli S, Candela M, Turroni S, Biagi E, Collino S, Franceschi C, . . . Brigidi P (2013). Functional metagenomic profiling of intestinal microbiome in extreme ageing. Aging (Albany NY), 5(12), 902–912. doi: 10.18632/aging.100623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN, Austad S, Brasky K, Brown CJ, Forney LJ, Gelfond JA, . . . Tardif SD (2017). The development of a specific pathogen free (SPF) barrier colony of marmosets (Callithrix jacchus) for aging research. Aging (Albany NY), 9(12), 2544–2558. doi: 10.18632/aging.101340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross CN, Davis K, Dobek G, & Tardif SD (2012). Aging Phenotypes of Common Marmosets (Callithrix jacchus). J Aging Res, 2012, 567143. doi: 10.1155/2012/567143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, . . . Weber CF (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol, 75(23), 7537–7541. doi: 10.1128/AEM.01541-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon CE (1948). A Mathematical Theory of Communication. Bell System Technical Journal, 27(4), 623–656. [Google Scholar]
- Tardif SD, Mansfield KG, Ratnam R, Ross CN, & Ziegler TE (2011). The marmoset as a model of aging and age-related disease. Institute for Laboratory Animal Research Journal, 52(1), 54–65. doi: 10.1093/ilar.52.1.54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yatsunenko T, Rey FE, Manary MJ, Trehan I, Dominguez-Bello MG, Contreras M, . . . Gordon JI (2012). Human gut microbiome viewed across age and geography. Nature, 486(7402), 222–227. doi: 10.1038/nature11053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildirim S, Yeoman CJ, Sipos M, Torralba M, Wilson BA, Goldberg TL, . . . Nelson KE (2010). Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One, 5(11), e13963. doi: 10.1371/journal.pone.0013963 [DOI] [PMC free article] [PubMed] [Google Scholar]