Purpose and Appropriate Sample Types
This 18-color panel was designed to measure vaccine-induced T cell responses in rhesus macaques by intracellular cytokine staining (ICS). It detects seven cytokines, IFN-γ IL-2, IL-4, IL-5, IL-13, IL-17, and IL-21, two memory markers, CD45RA and CCR7, and four follicular helper T cell (Tfh) markers, CXCR3, CXCR5, ICOS, and PD-1. While this panel was optimized for use on cryopreserved rhesus macaque peripheral blood mononuclear cells (PBMC) (Table 1), it can also be used on rhesus macaque tissue samples and human PBMC and tissue samples.
Table 1.
Summary Table for Application of OMIP-XXX
| Purpose | To measure Th1, Th2, Th17, and Tfh responses |
| Species | Rhesus macaque |
| Cell types | PBMC |
| Cross references | OMIP-005: Quality and phenotype of antigen-responsive rhesus macaque T cells |
Background
Experimental vaccines are often tested in nonhuman primates (NHP) for immunogenicity and protection in preclinical trials before proceeding to clinical trials in humans. Historically, vaccine-induced T cell responses have been measured by IFN-γ ELISpot or intracellular cytokine staining (ICS) for IFN-γ, IL-2, and TNF. However, antibody levels, rather than Th1 responses, correlate with protection for many vaccines including the HIV vaccine used in the RV144 clinical trial (1). As such, measuring only Th1 responses may not be adequate for determining T cell correlates of protection or risk in vaccine studies. Therefore, we expanded our 11-color intracellular cytokine staining (ICS) panel, described in OMIP-005, to 18 colors in order to assess a fuller spectrum of T cell responses.
To decrease the variability of the assay due to age-related differences in T cell differentiation, cytokine measurements can be reported as a frequency of memory T cells using the markers CD45RA and CCR7 (2). Similarly, IL-21 can be reported as a frequency of Tfh using a combination of the markers CXCR3, CXCR5, ICOS, and PD-1. This combination of markers allows for the delineation of Tfh cells by all currently accepted methods (3–5).
Similarity to Published OMIPs
This 18-color rhesus macaque panel is an extension of the 11-color panel described in OMIP-005. The cytokines IL-4, IL-5, IL-13, IL-17, and IL-21 were added as well as the Tfh subset markers CXCR3, CXCR5, ICOS, and PD-1. To accommodate these additional markers, CD28 and TNF were removed. It is similar to the following human panels also used to measure antigen-specific responses by intracellular staining: OMIP-001, OMIP-008, OMIP-009, OMIP-14, OMIP-16, OMIP-22, OMIP-25. However, this panel is designed for staining rhesus macaque PBMC (using NHP cross-reactive antibody clones) and includes additional Tfh markers, as well as IL-5 and IL-13. It is also the only panel with this combination of seven cytokines.
Online Material
Developmental Strategy
Our previous panel and staining protocol, described in OMIP-005 (1), was designed to measure Th1 responses from macaques in preclinical HIV vaccine studies. However, the RV144 HIV vaccine clinical trial results suggest that antibody responses, not Th1 responses, correlate with protection from acquisition of HIV and that IL-13 may have a protective role (2). In addition, the antibodies in the RV144 trial waned rapidly, so the identification of a correlate of antibody durability would be useful. Therefore, since Tfh cells producing IL-21 drive the differentiation of B cells and affinity maturation of antibodies (3), we added Tfh markers and IL-21 to our panel. Furthermore, there is evidence that Th2 and Th17 responses may be deleterious in the generation of protective vaccine responses to some pathogens (4, 5). Combined, these data suggest that further characterization of T cells is necessary in vaccine development.
Recent advancements in cytometer instrumentation and the discovery of new dyes enable development of more complex flow cytometry panels which measure more parameters. As such, we developed an 18-color flow cytometry panel that identifies Th1, Th2, Th17, and Tfh responses. The strategy for developing this panel was based generally on our previous panel design, in which the brightest fluorochromes were reserved for cytokines in order to increase sensitivity to detect rare or low responses. Next, priority was given to the dimmer and less widely available memory and Tfh markers. Last, T cell lineage markers were given the lowest priority since the reagents used to identify these populations are more abundant and available on many different fluorochromes (6). All reagents were titrated and the dilutions that gave the brightest signal and the least amount of background staining were chosen (Online Fig. 1).
Adequate detection of cytokine responses poses a challenge due to background responses from non-specific cells. Therefore, the use of fluorochromes that will maximize resolution between negative and positive populations is critical. Brilliant Violet (BV) and Brilliant Blue (BB) fluorochromes are amongst the brightest and were used for detecting five of the seven cytokines. IL-2, IL-4, IL-5, IL-13, and IL-17 were paired with BV750, BB700, BB515, BV421, and BV605, respectively. We also wanted to measure the Th2 cytokines IL-4 and IL-5, however, the commercial conjugates available for both reagents were limited. Thus, options for obtaining custom conjugates for both reagents were pursued. Since IL-4 and IL-5 are expressed at low levels, both were conjugated to the brightest fluorophores available BB700 and BB515, respectively. To ensure that the custom conjugates performed as expected, we compared the frequencies of IL-4 and IL-5 positive cells obtained with commercial PE reagents to those obtained with the custom conjugates and observed similar results (Online Fig. 2). While the separation between positive and negative events wasn’t as high with the IL-5 BB515 custom conjugate as with the commercial PE reagent, we chose to use BB515 because frequencies were similar between the two antibody conjugates and we needed PE for CXCR5. Last, we chose IL-21 Ax647 and IFN-γ Ax700 because expression levels of these cytokines are more robust, and we were able to distinguish negative and positive populations with moderately bright fluorochromes. As with OMIP-005, the activation marker CD69 was included in the panel to exclude background responses from CD69− cells (7).
Next, we prioritized memory subset (CD45RA and CCR7) and Tfh markers (PD-1, CXCR5, CXCR3, and ICOS). Relatively bright fluorochromes were also reserved for these markers. CD45RA was placed in the open PE-Cy5 channel and CCR7 BV650 was selected since BV650 is relatively bright. Using a bright fluorochrome for CCR7 is necessary since this receptor is particularly dim in nonhuman primates. In a similar manner, the dim markers CXCR3, CXCR5, ICOS, and PD-1 were put on the moderately bright fluorochromes BV711, PE, PE-Cy7, and BV785, respectively. We initially had CXCR5 in the surface staining portion of the panel; however, we noticed that the frequency of CXCR5+ cells decreased upon stimulation in some animals and that all of the IL-21+ cells were CXCR5− (Online Fig. 3A). Therefore, we tested staining for CXCR5 in the intracellular portion of the assay and discovered that the frequency of CXCR5+ cells increased upon stimulation and approximately 50% of the IL-21+ cells were CXCR5+ (Online Fig. 3B). Thus, although optimal separation was observed during surface staining, we decided to stain for CXCR5 during the intracellular step of the assay since we are interested in measuring IL-21 from Tfh (CXCR5+) cells.
Last, we assigned fluorochromes to the lineage marker antibodies. CD3, CD4, and CD8 were assigned to the open channels APC-Cy7, PE-Cy5.5, and BV570, respectively. As with OMIP-0005, CD3 was stained for during the intracellular step of the assay since the CD3 molecule can be internalized along with the TCR complex upon stimulation (8).
Staining Protocol
Commercial Materials:
PBS (GIBCO, Carlsbad, CA)
RPMI 1640 with phenol red (GIBCO)
RPMI 1640 without phenol red (GIBCO)
Heat-inactivated FBS (GIBCO)
Penicillin/Streptomycin/Glutamine (GIBCO)
BD Cytofix/Cytoperm Fixation/Permeabilization kit (BD Biosciences, San Diego, CA)
Monensin (GolgiStop, BD Biosciences)
96 well tissue-culture treated V-bottom plates (Corning Costar, Corning, NY)
Anti-CD28 BD Biosciences
Anti-CD49d BD Biosciences
BD Horizon Brilliant Stain Buffer BD Biosciences
In-house media:
R10 culture medium:
RPMI 1640 with phenol red
10% FBS
100 IU/ml penicillin
100 μg/ml streptomycin
2mM L-glutamine
Wash buffer:
RPMI 1640 without phenol red
4% FCS
0.02% sodium azide
Note: cells are pelleted in a desktop centrifuge with a 25 cm rotor while reagent master mixes are centrifuged in a microfuge with a 7 cm rotor.
1.0. Preliminary Operations
1.1 Determine desired 96-well plate layout. One column per animal is recommended. Each sample will likely have at least 2 stimulations each: peptide and negative (DMSO). If more than 3 million cells per sample are to be run, cells should be plated in duplicate or triplicate. Include an extra well of cells for the unstained control for gating on the flow cytometer.
1.2 Prepare the peptide pool(s) by mixing the peptides in DMSO to achieve a final concentration of 400 μg/ml. Peptides are used at a final concentration of 2 μg/ml when added to cells.
1.3 In separate tubes, prepare the appropriate Stimulation Mixes as listed below. The table below is for 100 μl of solution per well for 4 wells with 10% extra (total volume for 5 wells). Adjust the volumes proportionately as needed for individual experiments.
Component uL uL Final conc. Stimulant Peptide-5 DMSO-5 2 μg/ml Anti-CD49d (1 mg/ml) 1 1 1 μg/ml Anti-CD28 (1 mg/ml) 1 1 1 μg/ml Monensin (GolgiStop 3 mM) 1 1 3 μM R10 Medium 492 492 Total Volume 500 500 1.4 Determine the amount of Perm/Wash Buffer needed for the experiment and dilute the 10× Perm/Wash Buffer 1:10 with deionized water. Shake well and filter through a 0.22 micron vacuum flip filter. Store at 4°C for up to 1 week.
1.5 Prepare 25 ml of PBS + Benzonase per plate by diluting 5 μl of Benzonase nuclease stock in 25 ml of PBS.
1.6 Prepare 1% PFA by diluting the 4% stock PFA solution to 1% in 1× PBS. Make enough for 125 μl per sample (plus 10% extra).
1.7 Prepare the live/dead Amine Staining Mix no more than 1 – 2 hours prior to staining cells. Dilute the appropriate amount of amine reactive dye 1:40 in dH20, then 1:20 in 1× PBS at a total volume of 100 μl per sample (plus 10% extra). Store at 4°C in the dark until ready to use.
1.8 Prepare the Surface Staining Mix no more than 1 – 2 hours prior to staining cells. Add the appropriate antibodies to BD Horizon Brilliant Stain Buffer in a microcentrifuge tube, 15 ml conical tube, or a 50 ml conical tube depending on the final volume (be sure to add the Brilliant Staining Buffer to the tube first!). Spin the antibodies in a microcentrifuge at 13,000 rpm (15,700g) for 5 minutes to remove aggregates. For volumes larger that 1.5 ml, split antibody mix evenly between microcentrifuge tubes and spin. Remove the supernatant and adjust the total volume to 100 μl per sample (depending on panel) with Brilliant Staining Buffer.
1.9 Prepare the Intracellular Cytokine Staining Mix no more than 1–2 hours prior to staining cells. Add the appropriate antibodies to BD Horizon Brilliant Staining Buffer in a microcentrifuge tube, 15 ml conical tube, or a 50 ml conical tube depending on the final volume (be sure to add the Brilliant Staining Buffer to the tube first!). Spin the antibodies in a microcentrifuge at 13,000 rpm (15,700g) for 5 minutes to remove aggregates. For volumes larger that 1.5 ml, split antibody mix evenly between microcentrifuge tubes and spin. Remove the supernatant and add the appropriate volume of 10× Perm/Wash Buffer.
2.0. Procedure
2.1 Prepare cells according to the appropriate SOP (cell preparation and/or thawing) using R10 as the final buffer. Cells should be at a final concentration of 10 – 30 million cells/ml.
2.2 Pipette 100 μl of cells into the appropriate wells on the plate (1 to 3 million cells/well) according to plate layout.
2.3 Pipette 100 μl of the peptide and negative control Stimulation Mixes from the table above into the appropriate wells and mix thoroughly with a multi-channel pipette.
2.4 Incubate the plate for 6 hrs (18 hrs if overnight stimulation) in a humidified 5% CO2 incubator at 37oC. At the end of the incubation, the plate may be held at 4oC overnight if desired.
2.5 Centrifuge the plate at 2000 rpm for 3 minutes at RT (18–26°C). All centrifugation steps from this point will be done the same way.
2.6 Flick supernatant into a biohazardous waste container under a laminar flow hood and immediately and quickly blot the plate on a paper towel before returning to an upright position. All flicks and blots from this point will be done the same way.
2.7 Add 200 μl of PBS + Benzonase to each of the wells and mix thoroughly by pipetting up and down with a multi-channel pipette. Centrifuge, flick, and blot.
2.8 Add 100 μl of the prepared Amine Staining mix to each of the wells and mix thoroughly. Incubate for 20 min at RT protected from light.
2.9 Wash the cells by adding 120 μl of wash buffer directly to the wells containing the cells in the Amine Staining Mix. Centrifuge, flick, and blot.
2.10 Add 200 μl of wash buffer to the wells and mix thoroughly. Centrifuge, flick, and blot.
2.11 Add 100 μl of Surface Staining Mix to each of the wells, mixing well. Incubate for 20 min at RT protected from light.
2.12 Wash the cells by adding 120 μl of wash buffer directly to the wells containing the cells in Surface Staining Mix. Centrifuge, flick, and blot.
2.13 Add 200 μl of wash buffer to the wells and mix thoroughly. Centrifuge, flick, and blot.
2.14 Add 100 μl of Cytofix/Cytoperm reagent to each of the wells, mixing well. Incubate for 20 min at 4°C protected from light.
2.15 Wash the cells by adding 120 μl of 1× Perm/Wash buffer directly to the wells containing cells in Cytofix/Cytoperm. Centrifuge, flick, and blot.
2.16 Add 200 μl of 1× Perm/Wash buffer to the wells and mix thoroughly. Centrifuge, flick, and blot.
2.17 Add 100 μl of Intracellular Staining Mix to the wells, mixing well. Incubate for 20 min at RT in the dark.
2.18 Wash the cells by adding 120 μl of 1× Perm/Wash buffer directly to the wells containing cells in the Intracellular Staining Mix. Centrifuge, flick, and blot.
2.19 Add 200 μl of 1× Perm/Wash buffer to the cells and mix thoroughly. Centrifuge, flick, and blot.
2.20 Repeat step 2.19 for a third wash.
2.21 Add 125 μl of 1× PBS to the cells and mix.
2.22 Fix by adding 125 μl of 1% formaldehyde to the cells in 1× PBS for a total volume of 250 μl (0.5% formaldehyde) per well and mix. Store covered in foil at 4°C until ready to run on the flow cytometer (within 48 hours).
Supplementary Material
Online Figure 1. Antibody titrations. Frozen PBMC were thawed and stained with each of the antibodies separately using two-fold serial dilutions starting at 1:10. For antibodies that are used in the intracellular staining portion of the assay, cells were stimulated with the super antigen Staphylococcal enterotoxin B (SEB) and treated with BD Perm/Fix prior to staining. The titrations that gave the highest frequency of positive events along with the brightest MFI and minimal background staining were chosen. Optimal antibody concentrations are highlighted in red.
Online Figure 2. Comparison of IL-4 and IL-5 commercial and custom conjugates. The same clones on commercially available IL-4 PE and IL-5 PE were compared to custom IL-4 BB700 (A) and IL-5 BB515 (B), respectively, using the same rhesus macaque PBMC sample within each comparison. Cells were gated first on single cells, followed by lymphocytes, and then CD4+ T cells. Similar frequencies were observed between each commercially available reagent and its custom counterpart.
Online Figure 3. Comparison of surface and intracellular staining of CXCR5 on total CD4 T cells. Three rhesus macaque (RM) PBMC samples were stained with the 18-color panel with CXCR5 either in the surface or intracellular steps of the protocol. CD4 T cells were selected by gating first on single cells, followed by live/SSC low cells, then CD3+/FSC low cells, and finally CD4+ T cells. With surface staining (A), frequencies of CXCR5+ cells decreased with SEB stimulation in comparison to no stimulation (DMSO) and IL-21+ cells were all CXCR5−. However, with intracellular staining of CXCR5 (B), frequencies of CXCR5+ cells increased with SEB stimulation in comparison to no stimulation and ~50% of the IL-21+ cells were CXCR5+.
Figure 1.
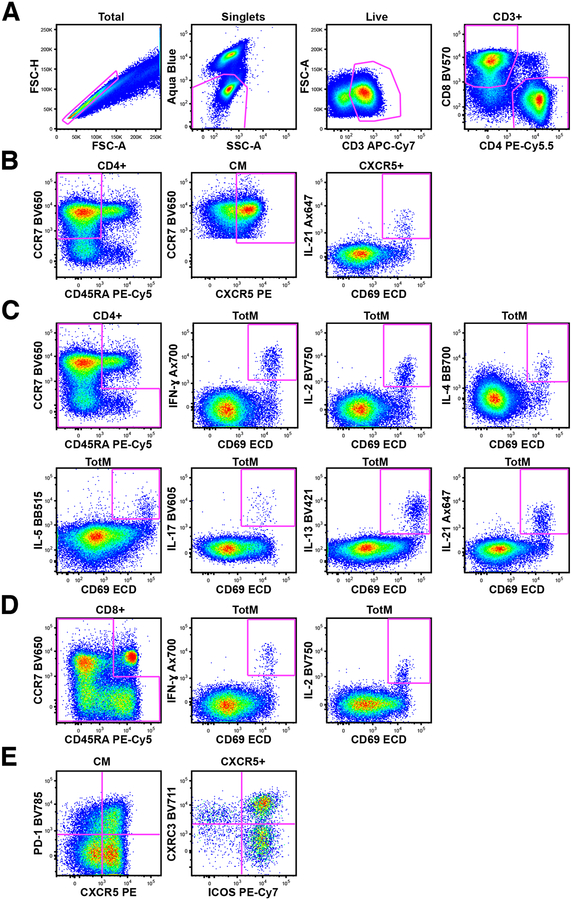
Representative staining and gating. Since this panel covers diverse populations and T cell polarization states, plots from multiple rhesus macaques in the same experiment were chosen in order to best display staining of all populations and production of all cytokines. (A) T cell populations were selected by subsequent gating: first single cells, followed by live/SSC low cells, then CD3+/FSC low cells, and finally CD4+ and CD8+ T cells. (B) To measure IL-21 production from Tfh, CD4 T cells were gated first on central memory (CM) T cells (CCR7+CD45RA−) then CXCR5+ CM cells to identify Tfh. Subsequently, from CXCR5+ Tfh, CD69+IL-21+ were gated. (C and D) Cytokine production from total memory (TotM) CD4 (C) and CD8 (D) T cells was measured by first gating on total memory cells (central memory CCR7+CD45RA− plus effector memory CCR7−CD45RA− plus terminal effector memory CCR7−CD45RA+ T cells) while excluding naïve CCR7+CD45RA+ T cells, then CD69+cytokine+ gates were drawn on total memory T cells. (E) Tfh can be further characterized using CXCR3 and ICOS after gating on CXCR5+ CM CD4 T cells.
Table 2.
Reagents Used for OMIP-XXX
| Specificity | Clone | Fluorochrome | Purpose |
|---|---|---|---|
| Dead cells | Aqua Blue | Exclusion | |
| CD3 | SP34.2 | APC-Cy7 | T cells |
| CD4 | S3.5 | PE-Cy5.5 | |
| CD8 | RPA-T8 | BV570 | |
| CD45RA | 5H9 | PE-Cy5 | Memory markers |
| CCR7 (CD197) | G043H7 | BV650 | |
| CXCR3 (CD183) | 1C6/CXCR3 | BV711 | Tfh markers |
| CXCR5 (CD185) | MU5UBEE | PE | |
| PD-1 (CD279) | EH12.2H7 | BV785 | |
| ICOS (CD278) | C398.4A | PE-Cy7 | |
| CD69 | TP1.55.3 | ECD | Background reduction |
| IFN-γ | B27 | Ax700 | Cytokines |
| IL-2 | MQ1–17H12 | BV750 | |
| IL-4 | MP4–25D2 | BB700 | |
| IL-5 | JES1–39D10 | BB515 | |
| IL-13 | JES10–5A2 | BV421 | |
| IL-17 | BL168 | BV605 | |
| IL-21 | 3A3-N2.1 | Ax647 |
Acknowledgments
We thank Mario Roederer and Pratip K. Chattopadhyay for advice and help in developing and optimizing this panel. This work was funded by 1) the Intramural Research Program of the National Institute of Allergy and Infectious Diseases, NIH and 2) the Foundation for the National Institutes of Health for the Comprehensive Cellular Vaccine Immune Monitoring Consortium award OPP1147555.
Literature Cited
- 1.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R and others. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012;366:1275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature 1999;401:708–12. [DOI] [PubMed] [Google Scholar]
- 3.Morita R, Schmitt N, Bentebibel SE, Ranganathan R, Bourdery L, Zurawski G, Foucat E, Dullaers M, Oh S, Sabzghabaei N and et al. Human blood CXCR5(+)CD4(+) T cells are counterparts of T follicular cells and contain specific subsets that differentially support antibody secretion. Immunity 2011;34:108–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Locci M, Havenar-Daughton C, Landais E, Wu J, Kroenke MA, Arlehamn CL, Su LF, Cubas R, Davis MM, Sette A and others. Human circulating PD-1+CXCR3-CXCR5+ memory Tfh cells are highly functional and correlate with broadly neutralizing HIV antibody responses. Immunity 2013;39:758–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boswell KL, Paris R, Boritz E, Ambrozak D, Yamamoto T, Darko S, Wloka K, Wheatley A, Narpala S, McDermott A and others. Loss of circulating CD4 T cells with B cell helper function during chronic HIV infection. PLoS Pathog 2014;10:e1003853. [DOI] [PMC free article] [PubMed] [Google Scholar]
Online Literature Cited
- 1.Foulds KE, Donaldson M, Roederer M. OMIP-005: Quality and phenotype of antigen-responsive rhesus macaque T cells. Cytometry A 2012;81:360–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R and others. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 2012;366:1275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Spolski R, Leonard WJ. Interleukin-21: basic biology and implications for cancer and autoimmunity. Annu Rev Immunol 2008;26:57–79. [DOI] [PubMed] [Google Scholar]
- 4.Knudson CJ, Hartwig SM, Meyerholz DK, Varga SM. RSV vaccine-enhanced disease is orchestrated by the combined actions of distinct CD4 T cell subsets. PLoS Pathog 2015;11:e1004757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mangodt TC, Van Herck MA, Nullens S, Ramet J, De Dooy JJ, Jorens PG, De Winter BY. The role of Th17 and Treg responses in the pathogenesis of RSV infection. Pediatr Res 2015;78:483–91. [DOI] [PubMed] [Google Scholar]
- 6.Mahnke YD, Roederer M. Optimizing a multicolor immunophenotyping assay. Clin Lab Med 2007;27:469–85, v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Picker LJ, Singh MK, Zdraveski Z, Treer JR, Waldrop SL, Bergstresser PR, Maino VC. Direct demonstration of cytokine synthesis heterogeneity among human memory/effector T cells by flow cytometry. Blood 1995;86:1408–19. [PubMed] [Google Scholar]
- 8.Alcover A, Alarcon B. Internalization and intracellular fate of TCR-CD3 complexes. Crit Rev Immunol 2000;20:325–46. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Online Figure 1. Antibody titrations. Frozen PBMC were thawed and stained with each of the antibodies separately using two-fold serial dilutions starting at 1:10. For antibodies that are used in the intracellular staining portion of the assay, cells were stimulated with the super antigen Staphylococcal enterotoxin B (SEB) and treated with BD Perm/Fix prior to staining. The titrations that gave the highest frequency of positive events along with the brightest MFI and minimal background staining were chosen. Optimal antibody concentrations are highlighted in red.
Online Figure 2. Comparison of IL-4 and IL-5 commercial and custom conjugates. The same clones on commercially available IL-4 PE and IL-5 PE were compared to custom IL-4 BB700 (A) and IL-5 BB515 (B), respectively, using the same rhesus macaque PBMC sample within each comparison. Cells were gated first on single cells, followed by lymphocytes, and then CD4+ T cells. Similar frequencies were observed between each commercially available reagent and its custom counterpart.
Online Figure 3. Comparison of surface and intracellular staining of CXCR5 on total CD4 T cells. Three rhesus macaque (RM) PBMC samples were stained with the 18-color panel with CXCR5 either in the surface or intracellular steps of the protocol. CD4 T cells were selected by gating first on single cells, followed by live/SSC low cells, then CD3+/FSC low cells, and finally CD4+ T cells. With surface staining (A), frequencies of CXCR5+ cells decreased with SEB stimulation in comparison to no stimulation (DMSO) and IL-21+ cells were all CXCR5−. However, with intracellular staining of CXCR5 (B), frequencies of CXCR5+ cells increased with SEB stimulation in comparison to no stimulation and ~50% of the IL-21+ cells were CXCR5+.