Abstract
Disulfide bonds (SS) are post-translational modifications important for the proper folding and stabilization of many cellular proteins with therapeutic uses, including antibodies and other biologics. With budding advances of biologics and biosimilars, there is a mounting need for a robust method for accurate identification of SS. Even though several mass spectrometry methods have emerged for this task, their practical use rests on the broad effectiveness of both sample preparation methods and bioinformatics tools. Here we present a new protocol tailored toward mapping SS; it uses readily available reagents, instruments, and software. For sample preparation, a 4-h pepsin digestion at pH 1.3 followed by an overnight trypsin digestion at pH 6.5 can maximize the release of SS-containing peptides from non-reduced proteins, while minimizing SS scrambling. For LC/MS/MS analysis, SS-containing peptides can be efficiently fragmented with HCD in a Q Exactive Orbitrap mass spectrometer, preserving SS for subsequent identification. Our bioinformatics protocol describes how we tailored our freely downloadable and easy-to-use software, Spectrum Identification Machine for Cross-Linked Peptides (SIM-XL), to minimize false identification and facilitate manual validation of SS-peptide mass spectra. To substantiate this optimized method, we’ve comprehensively identified 14 out of 17 known SS in BSA.
Keywords: Tandem mass spectrometry, Protein disulfide bond, Pepsin, Trypsin, HCD, SIM-XL
1. Introduction
Disulfide bonds (SS) are formed between the sulfhydryl groups of vicinal cysteines and are among the most common post-translational modifications in proteins. They play important roles in folding proteins and stabilizing functional protein domains [1, 2]. Proper SS arrangements are important for maintaining protein functions and dysregulation of SS formations have been reported in diseases, including neurodegeneration [3], cancer [4], inflammation [5], and heart diseases [6]. Therefore, precise identification of SS is critical for understanding protein functions in cells and assuring accurate productions of therapeutic proteins.
Conventional methods, such as X-ray crystallography [7], Edman sequencing [8], and NMR [9] are widely used to pinpoint SS; however, they require large amounts of purified proteins and thus their applicability is greatly limited. In contrast, high resolution MS has emerged as a frontline method for mapping SS in small quantities of proteins, including in mixtures, thanks to the advancements of soft ionization sources, resolution, sensitivity, and efficient fragmentation methods. A typical workflow for MS identification of SS includes three key steps: (1) Sample preparation: proteins are digested by proteases, usually trypsin, to release peptides that are amenable for LC/MS/MS analysis. An ideal sample preparation method should generate ample amounts of peptides linked by SS, and minimize the formation of artifactual SS, also known as SS scrambling; (2) LC/MS/MS: peptides are separated by HPLC and are fragmented with appropriate MS/MS fragmentation techniques that are sufficiently powerful to dissociate the peptide backbones, yet gentle enough to preserve the SS integrity; and (3) Bioinformatics: MS/MS spectra are analyzed by automated software tools to identify SS-containing peptides and localize the SS sites. A desirable tool can identify SS with high sensitivity and accuracy, and enable easy manual validation to eliminate false SS localization. Despite the tremendous progress of existing tools for this task, routine SS mapping is still challenging, due to low proteolytic digestion efficiencies of non-reduced proteins, SS scrambling, poor fragmentation of large SS-containing peptides, and limitations in bioinformatics tools.
First, in classic proteomics sample preparation steps, the neutral to alkaline (pH 7–9) conditions necessary for optimal tryptic digestion lead to striking SS scrambling [10, 11]. Thus, lowering the pH to acidic conditions can reduce SS scrambling; however, both tryptic digestion specificity and efficiency are vastly reduced, resulting in fewer peptides for SS mapping [11, 12]. To overcome this predicament, some groups have used pepsin at a pH range of 1 to 3 or combined multiple proteases, including trypsin, Lys-C, and Glu-C, at mildly acidic pH to produce sufficient peptides to identify SS [13, 14]. These strategies rely on proteases with efficiencies less than trypsin at optimal pH, thus still produce too few peptides from most proteins for SS mapping. Second, with respect to MS/MS fragmentation, either electron transfer dissociation (ETD) or higher energy collision dissociational (HCD) has been used for successful SS identification [13, 15]. Also, combinations of ETD-MS2 and CID-MS3 or HCD-MS3 approaches have been developed to identify SS in therapeutic proteins [16], capitalizing on the fact that ETD can sometimes cleave SS in peptides, which can then be identified by CID or HCD. Third, the widely adopted database search engines for matching tandem MS spectra with linear peptides, such as Mascot [17] and SEQUEST [18], do not have the capability to identify protein SS. Newer bioinformatic tools such as MassMatrix [19] aim to properly identify SS-linked peptides and localize SS sites; yet, are only recently available commercially and have limited capabilities to support manual validation.
In the present study, we describe a robust and fast SS identification protocol that uses: pepsin followed by trypsin to effectively digest non-reduced proteins in acidic buffers to minimize SS scrambling; HCD mode in a Q Exactive Orbitrap to fragment the peptides, whilst leaving SS largely intact for localization, and SIM-XL [20, 21] that we tailored specifically for identifying and validating SS-containing peptides.
2. Materials and Methods
2.1. Materials
Lysozyme from chicken egg white (SwissProt Accession #: P00698), bovine pancreatic ribonuclease (RNase A, SwissProt Accession #: P61823), BSA (SwissProt Accession #: P02769), N-ethylmaleimide (NEM), formic acid, and trifluoroacetic acid (TFA) were purchased from Sigma (St. Louis, MO). C18 spin columns were purchased from Fisher Scientific (Fair Lawn, NJ). Trypsin (V5113) and pepsin (V195A) were purchased from Promega (Madison, WI). ACN, methanol, acetic acid, and water were purchased from J. T. Baker (Center Valley, PA). SDS, Tris-HCl, polyacrylamide, ammonium persulfate, TEMED, Laemmli buffer, and Coomassie Brilliant Blue were purchased from BioRad (Hercules, CA).
2.2. Evaluation of pepsin digestion efficiency by gel electrophoresis
Forty micrograms of RNase A, lysozyme, or BSA were mixed with pepsin at a w/w ratio of 50:1 in 1% TFA, pH 1.3, at either room temperature or 37 °C as specified below. At each time point (0 min, 30 min, 1 h, 2 h, 4 h, 8 h and 16 h), two μg of each protein solution was taken and mixed with 5 μl of a 4x Laemmli sample buffer. The proteins were denatured at 95 °C for 5 min, and then separated with 10% SDS-PAGE mini gels. Each 10% SDS-PAGE resolving gel was made from a mixture containing 2.67 ml of 30% acrylamide, 2 ml of 1.5 M Tris-HCl (pH 8.8), 80 μl of 10% SDS, 80 μl of 10% ammonium persulfate, and 8 μl of TEMED in 3.2 ml of H2O. Each 4% stacking gel was made from a mixture containing 0.67 ml of 30 % acrylamide, 1.25 ml of 0.5 M Tris-HCl (pH 6.8), 50 μl of 10% SDS, 50 μl of 10% ammonium persulfate, 5 μl of TEMED in 3 ml of H2O, and was overlaid above each resolving gel. The proteins were separated on 10% SDS-PAGE gels with a constant voltage of 100 V at room temperature for ~ 60 min. The gels were fixed with 50% methanol and 10% acetic acid for 30 min, then stained with the Coomassie Brilliant Blue.
2.3. Comparison of in-solution digestion strategies
For SS analysis of lysozyme and RNase A, two different digestion strategies were compared: 1) proteins were digested with only trypsin at different pH conditions, or 2) proteins were digested first with pepsin at pH 1.3 followed by trypsin at different pH conditions. For the trypsin-only digestion, two μg of each protein was digested at a protein/trypsin w/w ratio of 50:1, at 37 °C overnight, in 100 mM Tris-HCl, at pH of either 6.0, 6.5, 7.0 or 7.5, respectively. For the pepsin/trypsin digestion, two μg of each protein was first digested at a protein/pepsin w/w ratio of 50:1, at 37 °C for 4 h in 1% TFA (pH 1.3). Next, the pH of protein solutions was adjusted with 100 mM Tris-HCl to either 6.0, 6.5, 7.0, or 7.5, respectively. Prior to tryptic digestions in some experiments, peptides produced from the pepsin digestion were alkylated with 2 mM NEM to block the free thiols from SS scrambling. Trypsin was then added to each pepsin digest at a protein/trypsin w/w ratio of 50:1, and incubated at 37 °C overnight. The resulting peptides were desalted with the C18 spin columns and ~1 μg equivalent of each resultant protein digest was injected onto the LC/MS/MS for analysis.
For each BSA SS mapping experiment, two μg of BSA in 50 μl of 1% TFA (pH 1.3) were incubated with 0.04 μg of pepsin at 37 °C for 4 h. The resulting peptide solution was brought to pH 6.0 or 6.5 with 100 mM Tris-HCl. The peptides were then alkylated with 2 mM NEM for 30 min at 37 °C, and incubated with 0.04 μg of trypsin at 37 °C overnight. The resulting peptides were desalted with the C18 spin columns and ~1 μg equivalent of each resultant protein digest was subject to LC/MS/MS analysis.
2.4. LC/MS/MS analysis
A Q Exactive MS coupled with an Ultimate 3000 HPLC System (Thermo Fisher Scientific) was used to analyze the peptides. In brief, the peptides were injected onto a C18 trapping column (Acclaim PepMap 75 μm × 2 cm, 3 μm, 100 Å), and then separated using a nano C18 column (Acclaim PepMap, 75 μm × 50 cm, 2 μm, 100 Å). The mobile phase A consisted of 2% ACN and 0.1% formic acid and mobile phase B consisted of 85% ACN and 0.1% formic acid. An 85-min gradient from 1% to 50% mobile phase B was used at a flow rate of 250 nl/min. After each run, a wash program was engaged to remove the peptide carry-overs on the column. The peptides eluted from the HPLC were electrosprayed into the MS through a Proxeon Flex nanospray source, at a spray voltage of 2.15 kV, and a capillary temperature of 275 °C. All the spectra were acquired in a data-dependent mode. MS spectra were acquired from 400 to 2000 m/z at a resolution of 140,000 FWHM (at 400 m/z). The ten most intense peptide ions with charge states of 3 to 8 were selected for MS2 fragmentation by HCD, with a normalized collision energy (NCE) of 25%. The MS/MS resolution was 17,500 FWHM and AGC value was 50,000.
2.5. Data analysis with SIM-XL
The RAW files were analyzed with SIM-XL (http://patternlabforproteomics.org/sim-xl/). The following parameters were used for the search: the mass tolerance of both precursors and fragments were set at 20 ppm; and disulfide bond was selected as the cross-linker. Both pepsin A and trypsin were chosen as the proteolytic enzymes with up to 4 missed cleavages. The minimal amino acid per chain was set as 2. Intra-link maximum charge was set as 6 and HCD was selected as the fragmentation method. The minimal and maximal [M+H]+ of the linear peptides were set at 600 Da and 4500 Da, respectively. The quality control filter for MS/MS spectra (Xrea) was set at 0.15; so spectra with less than this threshold were not considered; and the number of isotopic possibilities was set as 4. The MS/MS spectra were searched against each protein sequence and its reversed sequence. A False Discovery Rate (FDR) was calculated using the following formulation: , where FP is the number of incorrect assignments above score threshold in reversed sequence data search and TP is the number of correct assignments above the score threshold [22]. Only SS-containing peptides identified at 1% FDR or better and with SIM-XL primary scores of at least 1.5 were manually evaluated in the SIM-XL outputs and discussed in this study.
3. Results and Discussion
3.1. An optimized pepsin/trypsin digestion procedure to improve the digestion efficiency of non-reduced proteins
The classic tryptic digestion approach that has been effectively used in expression proteomics research is not ideal for SS identification because: 1) under the optimal pH (7.5 to 8.5) of trypsin digestion, SS scrambling frequently occurs; still, 2) under acidic pH to minimize SS scrambling, trypsin digestion efficiency is poor, especially against non-reduced proteins that are necessary for SS identification. Therefore, proteases that are active in acidic pH, e.g. pepsin, have been used for SS identification [14, 23–26]. Among the studies using pepsin, the proteolytic digestion conditions vary extensively: for example, Liu et al. digested the proteins for 2 h at room temperature [14]; Ni et al. performed the digestion for 30 min at 37 °C [26]; and Haniu et al. did a 20-h digestion at 37 °C [24]. However, due to low proteolytic specificity, prolonged pepsin digestion can produce many small peptides that are difficult to be identified with LC/MS/MS [11]. To overcome this dilemma, we hypothesize that a more efficient acidic protein digestion can be achieved with (1) a limited pepsin digestion at pH 1.3 to open the non-reduced proteins, (2) producing peptides that are more accessible for subsequent trypsin to cleave at an acidic pH, and (3) generating partial tryptic peptides that are more amenable for LC/MS/MS analysis, and SS mapping with SIM-XL.
First, to achieve partial pepsin digestion, we carried out a time- and temperature-dependent study of pepsin digestions of the model proteins, lysozyme, RNase A and BSA, all rich in SS. Based on the band densities of intact proteins in the SDS-PAGE, we found that pepsin can efficiently digest lysozyme after a 4-h incubation at 37 °C, but not in the room temperature (Supplemental Fig. S1). In contrast, pepsin can partially digest RNase A and BSA in the room temperature, but more efficiently at 37°C (Supplemental Fig. S1). Hence, in order to minimize non-specific pepsin proteolysis and open up the non-reduced proteins for the subsequent trypsin digestion, we chose to perform 4-h pepsin digestions at 37 °C for all the subsequent experiments.
Next, to determine whether the sequential pepsin/trypsin digestion was superior to the trypsin-alone digestion, we used LC/MS/MS to determine peptide yield from these two approaches, both with trypsin digestions performed at a range of pH, from 6.0 to 7.5. For the pepsin/trypsin digestion, we first digested the model proteins with pepsin at pH 1.3 for 4 h, and then raised the pH of the peptide solutions as described, and incubated them with trypsin, at 37 °C overnight. The combined pepsin/trypsin digestion released drastically more peptides than trypsin alone, at each trypsin digestion pH tested (not shown), especially at pH 6.5 (Fig. 1). The base-peak ion chromatograms show that the combination of pepsin and trypsin produced more peptides from both lysozyme (Fig. 1a) and RNase A (Fig. 1b) than those from the trypsin digestions alone, confirming our hypothesis that at acidic pH, partial pepsin digestion followed by trypsin is more efficient. The effectiveness of the pepsin/trypsin approach could be attributed to the broad protease specificity of pepsin that can cleave non-reduced proteins more readily than trypsin alone, thus opening doors for trypsin to further cut the peptides near the SS-linked peptides.
Fig. 1. Comparison of the digestion efficiency between trypsin and pepsin/trypsin.
Non-reduced lysozyme or RNase A was digested with trypsin at pH 6.5, either alone or after a 4-h pepsin digestion at pH 1.3. The resulting peptides were analyzed by LC/MS/MS. The chromatograms were normalized to the same scale. The base peak ion chromatograms of (a) lysozyme or (b) RNase A, digested with either trypsin alone (upper panels) or pepsin/trypsin (lower panels) are shown. Comparing the peptide peaks in the chromatograms of both proteins, pepsin/trypsin produced more peptides than trypsin alone.
Besides pepsin, chemicals, e.g., oxalic acid has also been successfully used for non-specific cleavages and SS mapping in peptides, especially the ones contain successive cysteines in their sequences [27, 28]. However, care is needed to ensure that the harsh conditions needed for chemical degradation, i.e. heating at 100 °C for oxalic acid, do not incite SS scrambling or chemical modifications, which could complicate data interpretation [29, 30].
3.2. Identification and validation of SS-containing peptides in Lysozyme and RNase A, using SIM-XL
In this study, the acidic pepsin/trypsin digestion strategy generated sufficient SS-containing peptides with lengths that can be efficiently fragmented and identified with HCD in the Q Exactive Orbitrap MS (Fig. 2). To identify SS in the model proteins, we employed SIM-XL. To improve the accuracy of SS identification, we conducted a database search against both forward and reversed protein sequences, and calculated the FDR, using the formula , wherein FP is the number of incorrect assignments above score threshold in the reversed sequence data search and TP is the number of correct assignments above the score threshold [22]. To achieve 1% FDR or better, different SIM-XL primary scores for both inter- and intra-peptide SS links were used as the cutoff score for each LC/MS/MS run, ranging from 1.50 to 2.50. All MS/MS spectra were manually validated by carefully examining the SIM-XL MS2 spectral assignments to remove incorrect identifications. Briefly, in each validated MS2 spectrum: 1) at least 80% of the major peaks (≥ 10% of the base peak intensity) were accounted for; 2) only the mono-isotopic peaks with the correct charge states were considered; 3) each SS-linked peptide set should contain strings of consecutive y- or b- ions, and 4) at least one string of y- or b- ions bracketed the intact disulfide. The peptides identified with the same SS pairs were grouped together and analyzed.
Fig. 2. Comparison of the numbers of SS identified at different tryptic digestion pH with alkylation.
After a 4-h pepsin digestion of lysozyme or RNase A at pH 1.3, the resulting peptides were alkylated with NEM and further digested with trypsin, at various pH conditions as indicated. The peptides were analyzed by LC/MS/MS and the SS-containing peptides were identified by SIM-XL. Different peptides containing identical SS were consolidated into a minimal number SS identified for each protein. The number of total (blue bars) and known (orange bars) SS identified from (a) lysozyme or (b) RNase are scaled to the left Y-axis. The % of the known/total SS (yellow line) identified from (a) lysozyme or (b) RNase A is scaled to the right Y-axis. The most known SS and % of the known/total SS from both proteins were identified at pH 6.5. % Known/total: [(known SS)/(total SS)] X100%. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article).
Using SIM-XL, we found that following pepsin digestion, the pH at which trypsin digestion was subsequently performed considerably affected the number of SS-containing peptides identified. Using the known SS reported in the literatures for the model proteins to assess the sensitivity of each pH on SS mapping of the alkylated peptides, we found that trypsin digestion at pH 6.5 outperformed the other 3 pH values evaluated, enabled the identification of 3 known SS in either lysozyme (Fig. 2a & Supplemental Table S1) or RNase A (Fig. 2b &Supplemental Table S2), representing ~75% of the known SS reported for each protein. Interestingly, omitting alkylation prior to trypsin digestion did not noticeably affect the number of known SS identified for either protein (Supplemental Figs. S2a & S2b). Remarkably, trypsin alone digestion was ineffective, led to the identification of only 1 known SS in lysozyme (Supplemental Fig. S2c) and none in RNase A.
To assess the impact of the tryptic digestion pH on the specificity of the known SS identified from the pepsin/trypsin digestions, we compared the % of the known SS over the total SS identified for each protein. We found that tryptic digestion at pH 6.5 was superior to other pH values evaluated, leading to the specific identifications of 60–80% of the SS that are known for both lysozyme and RNase A (Fig. 2). SS scrambling is expected in the peptides prepared from the tryptic digestions performed at 7.0–7.5 [11, 14] (Fig. 2), and thus we prepared our samples with pepsin/trypsin under the acidic conditions, which are intended to impede SS scrambling [11, 31]. Yet, a number of unknown SS were still observed following the acidic digestions (Fig. 2 and Supplemental Tables S1 & S2). Alkylation after pepsin digestion appeared to be effective at lessening SS scrambling; for example, at pH 6.5, unknown SS was slashed from 3–4 (Supplemental Fig. S2, no alkylation) to 1–2 (Fig. 2, with alkylation). Such observations indicate that acidic proteolysis conditions may reduce but not eliminate SS scrambling (Fig. 2, pH 6.0), a phenomenon also reported by others [13, 32]. Likewise, unknown SS may be formed before proteolysis, perhaps during protein extraction and purification [15]. Of course, we cannot eliminate the possibilities of identifying genuinely novel SS in these proteins. From this analysis, we conclude that following a pepsin digestion at pH 1.3, a subsequent trypsin digestion at pH 6.5 is the best approach for preparing protein samples for SS mapping, enabling the identifications of the highest number of native SS, with the lowest % of SS scrambling, among the options we have compared.
For method development, lysozyme and RNase A were chosen as the model proteins because each has 4 well-characterized SS [33–35]. Overall, following the optimized pepsin (pH 1.3)/trypsin (pH 6.5) digestion approach, we identified 3 out of the 4 known SS in each model protein (Supplemental Fig. S3), including Cys30-Cys115, Cys64-Cys80 and Cys76-Cys94 in lysozyme, and Cys52-Cys110, Cys66-Cys121 and Cys84-Cys136 in RNase A (Supplemental Tables S1 & S2).
The development of specialized software tools, including MassMatrix [19], DISULPHIDE [36], SIM-XL and SlinkS [14], have enabled automated SS mapping. However, proper validation of the results obtained from these software tools remain challenging. Because MS/MS spectra from SS-linked peptides tend to contain many and complex fragments, many of spectra can still be falsely assigned to SS-linked peptides [13, 37]. Although the FDR filtering method used in this study has been proven useful [22, 32], manual validation of the spectra is still necessary to assure accurate mapping of SS in peptides. For this purpose, we have designed an interactive Spectrum Viewer in SIM-XL that allows seamless manual validations of isotope peaks in both precursor and fragment ions, and annotations of both spectra and peptides. To the best of our knowledge, SIM-XL is the only software that allows for readily assessment of mass spectra with an integrated 2-D visualization (Fig. 3a) that allows effortlessly clicking and assessing the mass spectra (Fig. 3b) [20], thus, easing manual validation. For example, SIM-XL’s validation tools allowed us to confirm a known disulfide bond between Cys30 and Cys115 (Fig. 4a) between lysozyme peptide22−33 and peptide115−125 and invalidate an unknown disulfide bond between Cys6 and Cys64 between lysozyme peptide6−13 and peptide62−68 (Fig. 4b), due to incomplete fragment ion series. Similarly, we validated a known disulfide bond between Cys66 and Cys121 in RNase A between peptide66−75 and peptide113−124 (Fig. 4c) and invalidated an unknown disulfide bond between Cys52 and Cys121 between peptide37−57 and peptide118−124 (Fig. 4d), due to the presence of several unannotated peaks with high signal-to-noise ratios. Not surprisingly, many of the SS-containing peptides identified in this study have K or R at the C-termini, suggesting tryptic or semi-tryptic peptides from the pepsin/trypsin digestions lend themselves for efficient MS/MS fragmentation, peptide spectral matching and SS localization.
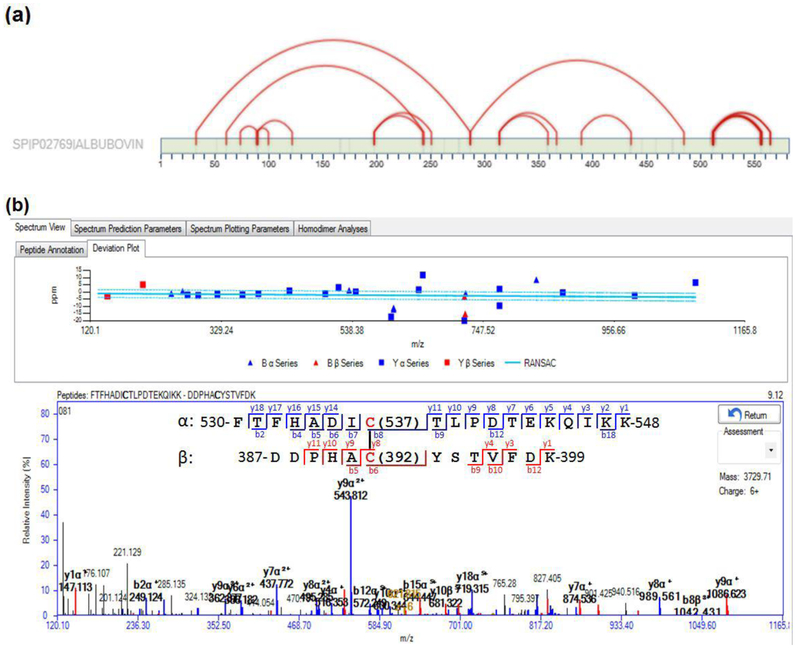
Fig. 3. Using SIM-XL for manual validation of SS.
(a) Through the 2D map, it is possible to assess all identified mass spectra and visualize which protein regions are interacting via SS. (b) An example of a validated MS/MS spectrum of a previously unknown SS (C392-C537) between two peptides in BSA. Similarly, a manual validation can be done for each identification, considering the fragment peaks matched, the identified residues, and the RANSAC curve [43].
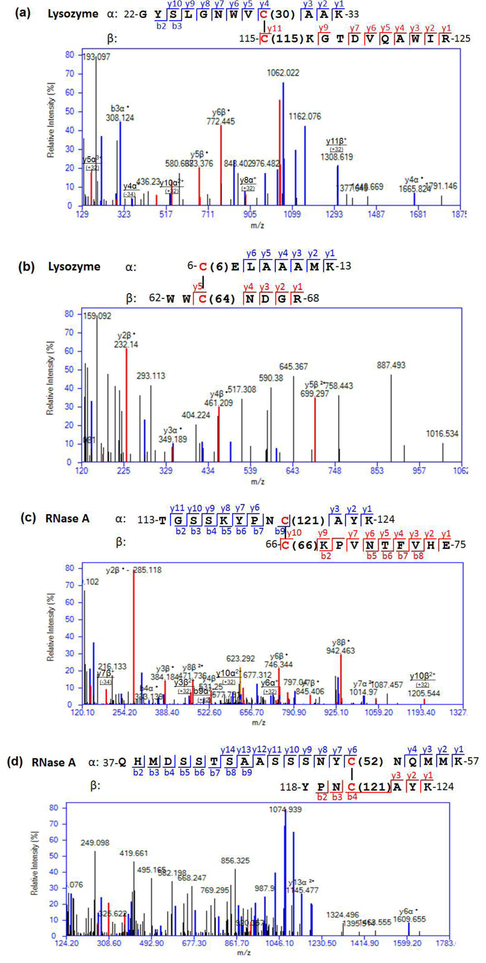
Fig. 4. Examples of manual validation of SS identification from the HCD spectra.
(a) An MS/MS spectrum of a 3H+ ion at m/z 848.08 matched to the lysozyme peptide22–33 linked to peptide115–125, with a SS between Cys30 and Cys115, per a SIM-XL score of 2.47. The strings of b- and y-series ions from lysozyme. The mass difference of 1376.63 amu between the y3 and y4 fragment ions of 22-GYSLGNWVCAAK-33 provided a strong piece of evidence to validate the SS linkage between these two peptides via Cys30 and Cys115. (b) An MS/MS spectrum of a 3H+ ion at m/z 590.59 matched to the lysozyme peptide6–13 linked to peptide62–68, with a SS between Cys6 and Cys64, per a SIM-XL score of 1.32. Since there were not sufficient ions to unambigiously identify either peptide, the SS assignment was not validated. (c) An MS/MS spectrum of a 4H+ ion at m/z 623.04 matched to the RNase A peptide66–75 linked to peptide113–124, with a SS between Cys66 and Cys121, per a SIM-XL score of 2.46. The strings of b- and y-series ions from the spectrum matched to 66-CKPVNTFVHE-75 and 113-TGSSKYPNCAYK-124 in RNase A, and provided a solid piece of inferential evidence to validate the SS linkage between these two peptides via Cys66 and Cys121. (d) An MS/MS spectrum of a 4H+ ion at m/z 791.57 matched to the RNase A peptide37–57 linked to peptide118–124, via a SS between Cys52 and Cys121, with a SIM-XL score of 2.02. Since there were many high intensity ions that are unaccounted for, the SS assignment was not validated. The underlined ions derived from the asymmetric SS cleavages were assigned manually.
Previous studies show that CID fragmentation of SS-containing peptide ions tends to produce proton-induced asymmetric SS cleavages, giving rise to modified cysteines containing either disulfohydryl substituents (+32 amu) or dehydroalanine residues (−34 amu) on the C-S cleavage site [38]. Similarly, we observed asymmetric SS cleavages in HCD spectra (see examples in Figs. 4a & 4c), albeit less prominent than as reported in the CID spectra [38]. Likewise, others also reported that HCD fragmentation of SS-peptides produced only minor asymmetric SS cleavages; still, in HCD spectra, predominant backbone fragmentations containing intact disulfides enabled direct SS mapping [13].
3.3. Comprehensive Identification of SS in BSA
To demonstrate the effectiveness of our pepsin/trypsin sample preparation procedure followed by the SIM-XL bioinformatics analysis for comprehensive SS mapping, we applied this method to identify SS in BSA, which has a complex SS pattern. With this protocol, we identified 14 out of the 17 known (82%) SS in BSA [39] (Fig. 5a & Supplemental Table S3), including an intra-chain disulfide bond between Cys77 and Cys86 (Fig. 5b), and an inter-chain disulfide bond between Cys415 and Cys461 (Fig. 5c). If the results from both pH 6.5 and 6.0 digestions are combined, all 17 known SS in BSA were identified, including 3 SS only identified with tryptic digestion at pH 6.0 (Supplemental Fig. S4). One reason for this observation would be that at pH 6.0 but not pH 6.5, trypsin digestion produced more tryptic and semi-tryptic precursors containing the 3 aforementioned SS, based on the selected MS1 ion signals (not shown). This observation suggests that the slight pH difference could shift either trypsin proteolytic specificities [11] or access to denatured BSA. As such, our results are comparable to the most comprehensive study of BSA SS mapping that we know of, which reported the identification of 15 SS, using an alternating CID/ETD method [15]. As for specificity, we identified only 4 unknown SS in BSA (Supplemental Table S4), demonstrating the high specificity of our method.
Fig. 5. An application of the pepsin/trypsin/SIM-XL method to identify SS in BSA.
(a) Known BSA disulfide bonds identified in this study (red lines). (b) A representative MS/MS spectrum of a BSA peptide66−88, containing an intra-chain SS between Cys77-Cys86, with a SIM-XL score of 2.49. The strings of b- and y-series ions from the spectrum matched to 66-LVNELTEFAKTCVADESHAGCEK-88, and provided a strong piece of inferential evidence to validate the SS linkage between Cys77 and Cys86. (c) A representative MS/MS spectrum of BSA peptide411−420 linked to peptide456−468 via an inter-chain SS between Cys415 and Cys461, with a SIM-XL score of 2.93. The strings of b- and y-series ions from the spectrum matched to 411-IKQNCDQFEK −420 and 456-VGTRCCTKPESER-468, and provided a strong piece of inferential evidence to validate the SS linkage between Cys415 and Cys461. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
In BSA and two other model proteins in this study, we’ve identified previously unknown SS. Confirming whether these SS are novel or de novo SS mapping in non-model proteins will require additional approaches. To minimize SS artifacts and ensure the authenticity of novel and native SS, different MS approaches, or orthogonal methods, e.g. NMR or X-ray crystallography [7, 9], may be utilized for SS validation. For example, a variety of sample preparations for SS identification, e. g. chemical degradation [27, 28], multi-protease digestion [13], partial reduction and differential alkylation [40, 41], or chemical labeling [11, 42], could generate different SS scrambles and artifacts. In contrast, authentic and native SS are likely identified by multiple methods. Thus, the development of multiple SS mapping methods will be valuable for de novo SS mapping in non-model proteins.
4. Conclusion
In this study, we’ve optimized a protocol to effectively digest non-reduced proteins using both pepsin and trypsin in acidic pH, and developed a strategy to accurately identify and validate SS-containing peptides using SIM-XL. We demonstrated that a partial 4-h pepsin digestion at pH 1.3 followed by an overnight trypsin digestion at pH 6.5 could efficiently turn non-reduced proteins into SS-linked peptides. The backbones of these peptides, some with C-terminal K or R, are readily fragmented with HCD, while SS are largely unspoiled. SIM-XL is a free and user-friendly software package, which allows accurate and fast SS identification and convenient spectral validation. Critically, other groups can easily test this method, which uses common reagents, mass spectrometers, and free software. We hope the research community will find this method useful to diverse goals.
Supplementary Material
Significance:
Comprehensive and accurate identification of SS in proteins is critical for elucidating protein structures and functions. Yet, it is far from routine to accomplish this task in many analytical or core laboratories. Numerous published methods require complex sample preparation methods, specialized mass spectrometers and cumbersome or proprietary software tools, thus cannot be easily implemented in unspecialized laboratories. Here, we describe a robust and rapid SS mapping approach that utilizes readily available reagents, instruments, and software; it can be easily implemented in any analytical core laboratories, and tested for its impact on the research community.
The Impact of the Journal of Proteomics.
On the occasion of the 10th anniversary of the Journal of Proteomics, we offer our sincere congratulations to editor Calvete, the editorial board, the reviewers and the staff, for having nurtured this valuable journal for the research community. It is an honor for us to contribute to the 10th anniversary issue of the Journal of Proteomics. Among the coauthors of this study, we have published 22 articles in this journal. This journal has enabled us to share our critical work at different stages of our career and enabled us to build collaborations and contributed to the growth of the proteomics research community, which is crucial for us to receive the critical feedback for continued refinement of our methods and software tools. We wish the continued success of the journal in years to come and will continue to contribute high quality manuscripts that will have broader impact on the research community.
Highlights.
A 4-h pepsin digestion aids the ensuing tryptic digestion of non-reduced proteins at acidic pH.
This digestion method maximizes the release of disulfide-linked peptides for LC/MS/MS analysis.
SIM-XL is a powerful tool for the identification and validation of peptide disulfide bonds.
Acknowledgements
The project described was supported by the National Institute of General Medical Sciences (R01GM112415 to HL and AB, and R01GM067640 to AB), the National Institute of Neurological Disorders and Stroke (P30NS046593 to HL), and the Office of the Director of the National Institutes of Health (1S10OD025047 to HL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- ETD
electron transfer dissociation
- FDR
false discovery rate
- HCD
higher-energy collisional dissociation
- NCE
normalized collision energy
- NEM
N-ethylmaleimide
- RNase A
bovine pancreatic ribonuclease A
- SIM-XL
spectrum identification machine for cross-linked peptides
- SS
disulfide bonds
- TFA
trifluoroacetic acid
Footnotes
Conflict of interest
The authors declare that there is no conflict of interest.
References:
- [1].Hogg PJ, Disulfide bonds as switches for protein function, Trends Biochem Sci 28(4) (2003) 210–4. [DOI] [PubMed] [Google Scholar]
- [2].Thornton JM, Disulphide bridges in globular proteins, J Mol Biol 151(2) (1981) 261–87. [DOI] [PubMed] [Google Scholar]
- [3].Nakamura T, Lipton SA, Cell death: protein misfolding and neurodegenerative diseases, Apoptosis 14(4) (2009) 455–68. [DOI] [PubMed] [Google Scholar]
- [4].Dranoff G, Targets of protective tumor immunity, Ann N Y Acad Sci 1174 (2009) 74–80. [DOI] [PubMed] [Google Scholar]
- [5].Cho J, Protein disulfide isomerase in thrombosis and vascular inflammation, J Thromb Haemost 11(12) (2013) 2084–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J, A redox-dependent pathway for regulating class II HDACs and cardiac hypertrophy, Cell 133(6) (2008) 978–93. [DOI] [PubMed] [Google Scholar]
- [7].Jones TA, Kjeldgaard M, Electron-density map interpretation, Methods Enzymol 277 (1997) 173–208. [DOI] [PubMed] [Google Scholar]
- [8].Haniu M, Acklin C, Kenney WC, Rohde MF, Direct assignment of disulfide bonds by Edman degradation of selected peptide fragments, Int J Pept Protein Res 43(1) (1994) 81–6. [DOI] [PubMed] [Google Scholar]
- [9].Klaus W, Broger C, Gerber P, Senn H, Determination of the disulphide bonding pattern in proteins by local and global analysis of nuclear magnetic resonance data. Application to flavoridin, J Mol Biol 232(3) (1993) 897–906. [DOI] [PubMed] [Google Scholar]
- [10].Ryle AP, Sanger F, Disulphide interchange reactions, Biochem J 60(4) (1955) 535–40. [PMC free article] [PubMed] [Google Scholar]
- [11].Gorman JJ, Wallis TP, Pitt JJ, Protein disulfide bond determination by mass spectrometry, Mass Spectrom Rev 21(3) (2002) 183–216. [DOI] [PubMed] [Google Scholar]
- [12].Yen TY, Joshi RK, Yan H, Seto NO, Palcic MM, Macher BA, Characterization of cysteine residues and disulfide bonds in proteins by liquid chromatography/electrospray ionization tandem mass spectrometry, J Mass Spectrom 35(8) (2000) 990–1002. [DOI] [PubMed] [Google Scholar]
- [13].Lu S, Fan SB, Yang B, Li YX, Meng JM, Wu L, Li P, Zhang K, Zhang MJ, Fu Y, Luo J, Sun RX, He SM, Dong MQ, Mapping native disulfide bonds at a proteome scale, Nat Methods 12(4) (2015) 329–31. [DOI] [PubMed] [Google Scholar]
- [14].Liu F, van Breukelen B, Heck AJ, Facilitating protein disulfide mapping by a combination of pepsin digestion, electron transfer higher energy dissociation (EThcD), and a dedicated search algorithm SlinkS, Mol Cell Proteomics 13(10) (2014) 2776–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Rombouts I, Lagrain B, Scherf KA, Lambrecht MA, Koehler P, Delcour JA, Formation and reshuffling of disulfide bonds in bovine serum albumin demonstrated using tandem mass spectrometry with collision-induced and electron-transfer dissociation, Sci Rep 5 (2015) 12210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu SL, Jiang H, Lu Q, Dai S, Hancock WS, Karger BL, Mass spectrometric determination of disulfide linkages in recombinant therapeutic proteins using online LC-MS with electron-transfer dissociation, Anal Chem 81(1) (2009) 112–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Perkins DN, Pappin DJ, Creasy DM, Cottrell JS, Probability-based protein identification by searching sequence databases using mass spectrometry data, Electrophoresis 20(18) (1999) 3551–67. [DOI] [PubMed] [Google Scholar]
- [18].Eng JK, McCormack AL, Yates JR, An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database, J Am Soc Mass Spectrom 5(11) (1994) 976–89. [DOI] [PubMed] [Google Scholar]
- [19].Xu H, Zhang L, Freitas MA, Identification and characterization of disulfide bonds in proteins and peptides from tandem MS data by use of the MassMatrix MS/MS search engine, J Proteome Res 7(1) (2008) 138–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lima DB, Melchior JT, Morris J, Barbosa VC, Chamot-Rooke J, Fioramonte M, Souza T, Fischer JSG, Gozzo FC, Carvalho PC, Davidson WS, Characterization of homodimer interfaces with cross-linking mass spectrometry and isotopically labeled proteins, Nat Protoc 13(3) (2018) 431–458. [DOI] [PubMed] [Google Scholar]
- [21].Lima DB, de Lima TB, Balbuena TS, Neves-Ferreira AGC, Barbosa VC, Gozzo FC, Carvalho PC, SIM-XL: A powerful and user-friendly tool for peptide cross-linking analysis, J Proteomics 129 (2015) 51–55. [DOI] [PubMed] [Google Scholar]
- [22].Elias JE, Gygi SP, Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry, Nat Methods 4(3) (2007) 207–14. [DOI] [PubMed] [Google Scholar]
- [23].Wallis TP, Pitt JJ, Gorman JJ, Identification of disulfide-linked peptides by isotope profiles produced by peptic digestion of proteins in 50% (18)O water, Protein Sci 10(11) (2001) 2251–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Haniu M, Horan T, Arakawa T, Le J, Katta V, Hara S, Rohde MF, Disulfide structure and N-glycosylation sites of an extracellular domain of granulocyte-colony stimulating factor receptor, Biochemistry 35(40) (1996) 13040–6. [DOI] [PubMed] [Google Scholar]
- [25].Haniu M, Arakawa T, Bures EJ, Young Y, Hui JO, Rohde MF, Welcher AA, Horan T, Human leptin receptor. Determination of disulfide structure and N-glycosylation sites of the extracellular domain, J Biol Chem 273(44) (1998) 28691–9. [DOI] [PubMed] [Google Scholar]
- [26].Ni W, Lin M, Salinas P, Savickas P, Wu SL, Karger BL, Complete mapping of a cystine knot and nested disulfides of recombinant human arylsulfatase A by multi-enzyme digestion and LC-MS analysis using CID and ETD, J Am Soc Mass Spectrom 24(1) (2013) 125–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Calvete JJ, Jurgens M, Marcinkiewicz C, Romero A, Schrader M, Niewiarowski S, Disulphide-bond pattern and molecular modelling of the dimeric disintegrin EMF-10, a potent and selective integrin alpha5beta1 antagonist from Eristocophis macmahoni venom, Biochem J 345 Pt 3 (2000) 573–81. [PMC free article] [PubMed] [Google Scholar]
- [28].Bauer M, Sun Y, Degenhardt C, Kozikowski B, Assignment of all four disulfide bridges in echistatin, J Protein Chem 12(6) (1993) 759–64. [DOI] [PubMed] [Google Scholar]
- [29].Glocker MO, Arbogast B, Deinzer ML, Characterization of disulfide linkages and disulfide bond scrambling in recombinant human macrophage colony stimulating factor by fast-atom bombardment mass spectrometry of enzymatic digests, J Am Soc Mass Spectrom 6(8) (1995) 638–43. [DOI] [PubMed] [Google Scholar]
- [30].Wang Y, Lu Q, Wu SL, Karger BL, Hancock WS, Characterization and comparison of disulfide linkages and scrambling patterns in therapeutic monoclonal antibodies: using LC-MS with electron transfer dissociation, Anal Chem 83(8) (2011) 3133–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Mo J, Tymiak AA, Chen G, Characterization of disulfide linkages in recombinant human granulocyte-colony stimulating factor, Rapid Commun Mass Spectrom 27(9) (2013) 940–6. [DOI] [PubMed] [Google Scholar]
- [32].Na S, Paek E, Choi JS, Kim D, Lee SJ, Kwon J, Characterization of disulfide bonds by planned digestion and tandem mass spectrometry, Mol Biosyst 11(4) (2015) 1156–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Jaureguiadell J, Jolles J, Jolles P, The disulfide bridges of hen’s egg-white lysozyme, Biochimica et Biophysica Acta 107(1) (1965) 97–111. [PubMed] [Google Scholar]
- [34].Canfield RE, Liu AK, The Disulfide Bonds of Egg White Lysozyme (Muramidase), The J Biol Chem 240 (1965) 1997–2002. [PubMed] [Google Scholar]
- [35].Smyth DG, Stein WH, Moore S, The sequence of amino acid residues in bovine pancreatic ribonuclease: revisions and confirmations, J Biol Chem 238 (1963) 227–34. [PubMed] [Google Scholar]
- [36].Caporale C, Sepe C, Caruso C, Pucci P, Buonocore V, Assignment of protein disulphides by a computer method using mass spectrometric data, FEBS Lett 393(2–3) (1996) 241–7. [DOI] [PubMed] [Google Scholar]
- [37].Lakbub JC, Shipman JT, Desaire H, Recent mass spectrometry-based techniques and considerations for disulfide bond characterization in proteins, Anal Bioanal Chem 410(10) (2018) 2467–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Mormann M, Eble J, Schwoppe C, Mesters RM, Berdel WE, Peter-Katalinic J, Pohlentz G, Fragmentation of intra-peptide and inter-peptide disulfide bonds of proteolytic peptides by nanoESI collision-induced dissociation, Anal Bioanal Chem 392(5) (2008) 831–8. [DOI] [PubMed] [Google Scholar]
- [39].Brown JR, Structure of serum albumin: disulfide bridges, Fed. Proc 33 (1974) 1389–1389. [Google Scholar]
- [40].Foley SF, Sun Y, Zheng TS, Wen D, Picomole-level mapping of protein disulfides by mass spectrometry following partial reduction and alkylation, Anal Biochem 377(1) (2008) 95–104. [DOI] [PubMed] [Google Scholar]
- [41].Jones MD, Hunt J, Liu JL, Patterson SD, Kohno T, Lu HS, Determination of tumor necrosis factor binding protein disulfide structure: deviation of the fourth domain structure from the TNFR/NGFR family cysteine-rich region signature, Biochemistry 36(48) (1997) 14914–23. [DOI] [PubMed] [Google Scholar]
- [42].Huang SY, Hsieh YT, Chen CH, Chen CC, Sung WC, Chou MY, Chen SF, Automatic disulfide bond assignment using a1 ion screening by mass spectrometry for structural characterization of protein pharmaceuticals, Anal Chem 84(11) (2012) 4900–6. [DOI] [PubMed] [Google Scholar]
- [43].Fischler MA, Bolles RC, Random sample consensus: a paradigm for model fitting with applications to image analysis and automated cartography, Communication of the ACM 24(6) (1981) 381–395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.