Abstract
Purpose:
To introduce a quantitative tool that enables rapid forecasting of T1 and T2 parameter map errors due to normal and aliasing noise as a function of the MRF sequence, which can be used in sequence optimization.
Theory and Methods:
The variances of normal noise and aliasing artifacts in the collected signal are related to the variances in T1 and T2 maps through derived quality factors. This analytical result is tested against the results of a Monte-Carlo approach for analyzing MRF sequence encoding capability in the presence of aliasing noise, and verified with phantom experiments at 3T. To further show the utility of our approach, our quality factors are used to find efficient MRF sequences for fewer repetitions.
Results:
Experimental results verify the ability of our quality factors to rapidly assess the efficiency of an MRF sequence in the presence of both normal and aliasing noise. Quality factor assessment of MRF sequences is in agreement with the results of a Monte-Carlo approach. Analysis of MRF parameter map errors from phantom experiments is consistent with the derived quality factors, with T1 (T2) data yielding goodness of fit R2 ≥ 0.92 (0.80). In phantom and in vivo experiments, the efficient pulse sequence, determined through quality factor maximization, led to comparable or improved accuracy and precision relative to a longer sequence, demonstrating quality factor utility in MRF sequence design.
Conclusion:
The here introduced quality factor framework allows for rapid analysis and optimization of MRF sequence design through T1 and T2 error forecasting.
Keywords: Magnetic Resonance Fingerprinting, Pulse Sequence Design, Optimization
Introduction
MRI has long been used as a diagnostic tool, taking advantage of the MR signal’s dependence on underlying tissue properties, such as relaxation times and spin density, to provide natural contrast between tissue types. While many MR experiments are designed to provide qualitative weighting for a particular property, quantitative mapping is a useful diagnostic tool [1], for example, in oncology [2][3], cardiology [4][5], and neurology [6][7].
The desire for quantitative MRI has led to the introduction of Magnetic Resonance Fingerprinting (MRF), which allows for fast, simultaneous quantization of intrinsic tissue properties, including spin density and relaxation times (T1, T2) [8]. Improving upon previously developed quantitative MR techniques, MRF can be performed on a clinically relevant time scale and produces maps of multiple tissue parameters from a single data set. This is achieved by varying imaging parameters such as the flip angle, repetition time (TR), and echo time (TE) over N repetitions (time steps), causing tissues with different underlying characteristics to produce unique signal evolutions. The obtained signals are then matched to a dictionary to generate maps.
It has recently been shown that MRF parameter maps are reliably reproducible given a particular sequence design [9]. However, it is also known that the precision of parameter quantification depends on the chosen imaging sequence (flip angles, TRs, TEs, and N) [10]–[16]. For example, there are known structures that have been implemented in MRF sequence design to enhance T1 or T2 sensitivity, such as inversion pulses to improve T1 sensitivity, time steps with zero degree flip angles to allow for signal recovery, RF phase alternation to build bSSFP signal, and π/2 pulses to improve T2 sensitivity [17][18].
The dependence of T1 and T2 mapping on the sequence design raises the question of optimality. High degrees of freedom in the pulse sequence design add to the challenge of determining an experiment that will result in minimal mapping error. To this end, a number of techniques have been proposed for assessing MRF sequence designs. In Cohen et al. and Sommer et al., techniques are proposed for judging encoding capability, and thus pulse sequence design optimality, based on the correlation between local and global dictionary entries[13][14]. However, Sommer et al. found inconsistency in this method in the presence of undersampling artifacts. With a rich history in statistical estimation theory, several groups have used the Cramer-Rao lower bound to assess MRF parameter estimation in the presence of Gaussian white noise [11][15][16][19]. However, Gaussian white noise does not accurately characterize the undersampling noise that is prevalent in MRF and dependent on signal amplitude.
To address the problem of signal amplitude dependent aliasing noise, Sommer et al. have successfully proposed a trial and error, Monte Carlo technique for assessing MRF sequence performance [13]. While able to accurately predict the efficacy of a given experimental design, the Monte Carlo approach does not provide an analytical tool for quickly understanding errors in MRF maps due to aliasing noise.
In this work we introduce a mathematical framework that relates parameter map errors due to both Gaussian white noise and correlated undersampling artifacts to the MRF signal, and thus to the chosen imaging sequence (N, TR series, flip angle series). As a result of this framework, “quality factors” are found that are inversely related to parameter map variance. These quality factors are compared to the results of the Monte Carlo approach presented by Sommer et al. and experimentally validated. To show further the utility of the approach, our quality factors are used to find an efficient flip angle distribution, requiring fewer acquisitions than the best sequence from the Monte Carlo approach, yet achieving a similar encoding capability. This analytical approach offers increased understandability and usability compared to the previously presented Monte Carlo method.
Theory
In the MRF framework, a dictionary (D) of potential signals (d(θ)) given an experimental design is generated for a range of relevant tissue parameters (θ). Then, the experimentally acquired signal (s) is matched to a dictionary entry to determine tissue properties per voxel. Matching is often performed with maximization of the complex valued dot product between the normalized signal and the dictionary composed of normalized entries ,
| [1] |
To understand errors in MRF parameter mapping, this work analyzes Eq. [1] using appropriate models for ideal and noisy MRF signals. This work specifically concentrates on the case where θ = [T1, T2]; therefore, we focus on FISP-MRF based sequences, which have been shown to be insensitive to off-resonance frequency. [20]
The MRF signal, which behaves according to the Bloch equations, is dependent on a variety of experimental parameters and intrinsic tissue properties. In general, at each RF pulse the magnetization is rotated, then longitudinal magnetization recovers exponentially at a rate determined by R1=1/T1 and transverse magnetization decays exponentially at a rate determined by R2=1/T2 up to the time of the subsequent RF pulse. If the repetition time is short and the signal is not spoiled between RF pulses, the signal contains the magnetization’s history and is dependent on both T1 and T2. In a FISP-MRF experiment, where gradients in the slice select direction are applied at the end of each TR to generate a phase twist greater than 2π, the overall signal from a given voxel is dephased and thus insensitive to off-resonance frequency. [20]
Therefore, the solution to the Bloch equations at each time step in an FISP-MRF experiment is a complicated function of R1, R2, repetition times, echo times, and flip angles. In MRF experiments, typical TE and TR values are short relative to T1 and T2. Therefore, the signal at each time step can be represented by a first-order Taylor Series expansion about chosen values of R1 and R2, R1,0=1/T1,0 and R2,0=1/T2,0. Then, each dictionary entry (corresponding to relaxation rates R1 and R2) is a 1×N dimensional vector that follows the model:
| [2] |
where d0 = d(R1,0, R2,0) is the dictionary entry corresponding to the relaxation rates R1,0 and R2,0, and the 1×N dimensional coefficients c1 and c2 contain the first derivatives of the transverse magnetization with respect to R1 and R2 respectively at each time step.
The acquired signal in an MRF experiment will, in general, contain both Gaussian white noise (η) as the result of thermal contributions, and correlated aliasing noise (α) due to undersampling artifacts. Therefore, a noisy acquired signal from a voxel with R1=R1,0 and R2=R2,0 is expected to be
| [3] |
The thermal noise in the acquired signal is assumed to have zero mean, variance , and to be uncorrelated between real and imaginary channels. Thermal noise is taken to be Gaussian rather than Rician due to the high SNR in fully sampled data [21].
Motivated by the work in [13], we model the aliasing noise as proportional to the MRF signal at each time step, with a coefficient of proportionality that is taken to be zero-mean Gaussian noise (ω) with variance . This modeled approximation of aliasing noise is an ansatz, based on the pseudo-randomization of k-space coverage (for example, through golden angle rotation between sparse, variable density spiral acquisitions), and is shown a posteriori to serve as a good approximation of experimentally observed aliasing noise (see Supporting Information Fig. [S1]). Specifically, the nth component of the complex aliasing noise in our model is given by
| [4] |
The components of ω are assumed to be uncorrelated, as are η and ω.
With the presented dictionary and acquired signal models and Eq. [1] expressed as for i=1,2, evaluation of the derivatives with respect to R1 and R2 yields two independent equations with which it is possible to solve for (R1-R1,0) and (R2-R2,0). From the resulting equations, we find expressions for the variance in R1 and R2 [see appendix for a detailed derivation]:
| [5] |
with the quality factors relating to thermal noise given by
| [6] |
and the quality factors relating to aliasing noise given by
| [7] |
where
Our quality factors due to both thermal and aliasing noise contain dependence on R1,0 and R2,0 through d0 and the determination of c1 and c2. Additionally, because the signal components (d0, c1, c2) depend on experimental design parameters (flip angle series, TR series, TE, N), the quality factors are indicative of the ability with which a given experiment can match the noisy acquired signal from a voxel with R1=R1,0 and R2=R2,0 to the appropriate dictionary entry for a given level of noise. Therefore, the comparison of quality factors for different experimental designs allows for the determination of more optimal sequence parameters.
Additionally, noting that c1 and c2 are the derivatives of the MRF signal with respect to R1 and R2, the quality factors for thermal noise (Qi) are determined to be consistent with the Cramer-Rao bounds evaluated for a FISP-MRF experiment. For a complete derivation, please see the Supporting Information. Numerical results can be found in reference [22].
If either thermal noise (i.e. fully sampled data) or aliasing noise (i.e. highly undersampled data) dominates Eq. [5], the variance in parameter quantification becomes approximately proportional to the dominant term. Because MRF is generally performed with large undersampling acceleration factors, this work focuses on the case where aliasing noise is dominant so that
| [8] |
Methods
Verification Experiments
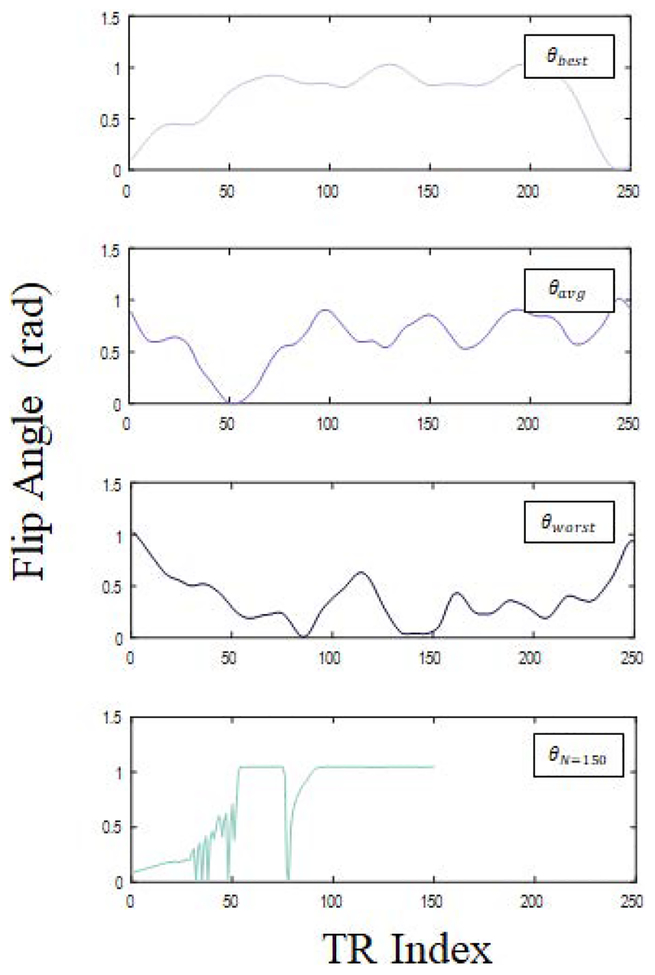
To verify the ability of Eq. [5] to predict the encoding capabilities of a given sequence design, FISP-MRF experiments predicated on the findings of Sommer et al. were performed with a 10-compartment phantom. For these experiments, three FISP-MRF sequences were tested, each containing an inversion pulse (inversion time, TI = 21 ms), constant repetition times (TR = 6.98 ms) and echo times (TE = 2.93 ms), and flip angle patterns with N=250 repetitions. The chosen flip angle distributions (θbest, θaverage, θworst) are based on the three patterns presented in Sommer et al., which were determined to have varying encoding capabilities (best, average, and worst) through their Monte-Carlo approach and with experimental verification (see Fig. [1]) [13].
Figure 1.
Flip angle distributions used in verification and optimization experiments. (a)-(c) Flip angle distributions based on Sommer et al., (θbest, θaverage, θworst), which were determined to have varying encoding capabilities: (a) best, (b) average, and (c) worst. (d) Efficient flip angle distribution with N=150 acquisitions, determined through maximization of aliasing noise quality factors.
Optimization Experiments
As a result of the inverse relationship between and qi, the error in mapping T1 and T2 with MRF in the presence of aliasing noise can be minimized by designing an experiment with maximized q1 and q2, respectively. Optimization of the flip angle sequence through maximization of the herein introduced aliasing noise quality factors was performed in MATLAB (MathWorks, Natick, Massachusetts, USA) with the genetic algorithm. For simplicity, aliasing noise quality factor maximization was performed while allowing the flip angle distribution to vary over the range [0, 60] degrees, fixing TE = 2.93 ms, TR = 6.98 ms, and RF phase = 0°, and executing an initial inversion pulse (TI = 21 ms). Optimization was initially performed with N=250, resulting in a flip angle distribution achieving a small (20%) forecasted improvement in mapping error relative to already optimized θbest as determined by Sommer et al. Therefore, to show the utility of our quality factors, the presented optimization was performed for N=150 to determine a sequence with comparable error and shorter scan time, and therefore improved efficiency, relative to the N=250 step experiment with θbest.
The initial population of the genetic algorithm, containing 900 random entries, was created with a colored noise generator so that 300 entries each had a power spectral density 1/|f|a with a = 20, 50, and 100 to filter out high frequencies [13]. Although the initial population contained relatively smoothly varying flip angle distributions, distributions in future generations could contain rapid variations due to the processes of mutation and crossover employed by the genetic algorithm.
Quality factors are dependent on underlying relaxation time values (T1,0, T2,0) through the calculation of zeroth and first order terms in Eq. [2]; thus, to have an effective formula for use over a large range of relaxation times, optimization was performed by minimizing the weighted sum: with T1,0 = [600, 850, 1300] ms and T2,0 = [45 70 100] ms. Weighting the quality factor terms by T1,0 and T2,0 for this optimization acts to normalize the terms in the sum, preventing any one term from dominating. The T1,0 and T2,0 values chosen for the optimization cover the range of reference relaxation times for white and gray matter at 3T (see Table [1]). The resulting optimized flip angle distribution, θN=150, is shown in Fig. [1]. The results of flip angle optimization for normal noise through maximization of Qi can be found in reference [23].
Table 1.
Relaxation times in white and gray matter at 3T from literature [29] and the results of ROI and nRMSE analysis on in vivo parameter maps obtained with FISP-MRF experiments using θbest, θN=150, and a reference scan based on Jiang et al.
| White Matter | Gray Matter | |||||
|---|---|---|---|---|---|---|
| Sequence | nRMSET1 (ms) | nRMSET2 (ms) | ||||
| Literature | (812.3–1110.0) | (49.5–79.6) | (1275.0–1763.0) | (66.0–110.0) | - | - |
| Reference Scan | 1066 ± 91 | 59 ± 5 | 1610 ± 128 | 78 ± 10 | - | - |
| θbest | 1033 ± 88 | 53 ± 4 | 1579 ± 133 | 71 ± 7 | 0.09 | 0.15 |
| θN=150 | 993 ± 89 | 49 ± 4 | 1629 ± 95 | 73 ± 10 | 0.10 | 0.22 |
Dictionary and Quality Factor Calculation
Dictionaries containing signal time courses for a range of relevant tissue parameters are generated in MATLAB using the Bloch equations. Because these dictionaries are used to fit d0, c1, and c2 for the calculation of quality factors as defined in Eqs. [6] and [7], the array of T1 and T2 values chosen should have small enough increments to perform a linear regression while spanning the typical physiological values of the samples to be imaged. For this work, T1 was chosen to vary over [35, 1000] ms in 5 ms increments and [1010, 2800] ms in 10 ms increments. T2 was chosen to vary over [2, 24] ms in 1 ms increments, [26, 40] ms in 2 ms increments, and [45, 800] ms in 5 ms increments, resulting in 56,659 dictionary entries.
To ensure matching accuracy to experimental data, FISP-MRF dictionaries were calculated with slice profile corrections [24]. This calculation was performed based on work by Hargreaves using RF-pulse and gradient information specific to the performed experiments [25]. Using Bloch simulations over 250 spins per voxel, a slice profile corrected FISP-MRF dictionary with 250 time steps for each entry was generated in three hours with MATLAB on the CWRU High Performance Cluster.
Both normal noise and aliasing noise quality factors were then calculated in MATLAB with Eqs. [6] and [7], using linear regressions over nearest neighbors from the calculated dictionary entries for chosen T1,0 and T2,0 values to determine d0, c1, and c2.
Experimental Design and Image Reconstruction
FISP-MRF experiments were performed on a 3T Siemens Skyra (Siemens Healthcare, Erlangen, Germany) with a 20-channel head-neck array coil. For phantom experiments, an 8 mm slice was excited with an apodized sinc pulse (nz=8, τRF=2 ms) during application of slice select (Gss = 11.74 mT/m, slew rate=117.4 T/m/s) and rephasing (Grephase =−27.4 mT/m, slew rate=119.1 T/m/s) gradients. For in vivo experiments, a 5 mm slice was excited with an apodized sinc pulse (nz=8, τRF=2 ms) during application of slice select (Gss = 11.74 mT/m, slew rate=117.4 T/m/s) and rephasing (Grephase =−27.4 mT/m, slew rate=119.1 T/m/s) gradients. Additionally, a spoiler gradient (Gspoil =27.4 mT/m, slew rate=119.1 T/m/s) was applied at the end of each TR to create a phase twist of 8π in the slice select direction.
Data was collected using a variable density spiral k-space trajectory, which required 48 interleaves to fully cover k-space (192×192, 300 mm2 FOV for phantom experiments; 256×256, 300 mm2 FOV for in vivo experiments) [26]. Fully sampled data sets were acquired for each experiment. Then, data was retrospectively undersampled to achieve acceleration factors ranging from R=5 to R=48, employing golden angle rotation of interleaves within each time step and relative to neighboring time steps [27]. Image reconstruction was performed using the non-uniform fast Fourier transform (NUFFT) [28] and the adaptive method for combining images from multiple coils [29].
Phantom Experiments
Both verification experiments, based on the results of Sommer et al., and optimization experiments, derived through maximization of our aliasing noise quality factors, were performed as described in Experimental Design and Image Reconstruction to image a 10-compartment phantom. Reference T1,0 and T2,0 values for each of the ten phantom compartments were determined with inversion recovery and spin echo sequences, respectively (see Supporting Information Table [1]). For additional comparison to the optimization experiments, reference T1 and T2 maps were obtained using an undersampled (R=48) 1000 step FISP-MRF sequence based on Jiang et al. [20]
Parameter maps (T1 and T2) were generated through complex valued dot product matching with the fully sampled and undersampled data from each experiment. A 5×5 ROI was drawn in each of the ten phantom compartments for calculation of mean and standard deviation values. The obtained mean values were used to calculate qi and Qi for each experiment, and errors in the data were propagated based on the changes in qi and Qi as the result of mean error relative to the reference T1,0 and T2,0 values in Table [1].
In vivo Experiments
Experiments with θbest and θN=150 were performed in vivo as described in Experimental Design and Image Reconstruction to generate T1 and T2 maps from volunteer head scans. Reference T1 and T2 maps were obtained using a 1000 step FISP-MRF sequence based on Jiang et al., which was shown to reliably yield comparable results to standard T1 and T2 mapping techniques [20].
The in vivo parameter maps resulting from experiments with θbest and θN=150 were analyzed through calculations of the relative error in each parameter map compared to the reference map, and the resulting normalized root mean squared errors in T1 and T2 (nRMSET1, nRMSET2), defined as , with n total voxels, the matched value of Ti in the jth voxel given by Ti,j, and the Ti value in the jth voxel of the reference scan given by . Regions of interest were drawn in areas of white and gray matter for calculation of mean , standard deviation values for additional parameter map comparison to reference map and literature values [30].
Results
Verification Experiments: Quality Factors as Functions of T1 and T2
With the generated dictionaries, both aliasing and normal noise quality factors for the three experiments based on Sommer et al. [13] were calculated in MATLAB with T1,0 varying over [350, 1710] ms in increments of 10 ms and T2,0 varying over [30, 110] ms in increments of 2 ms.
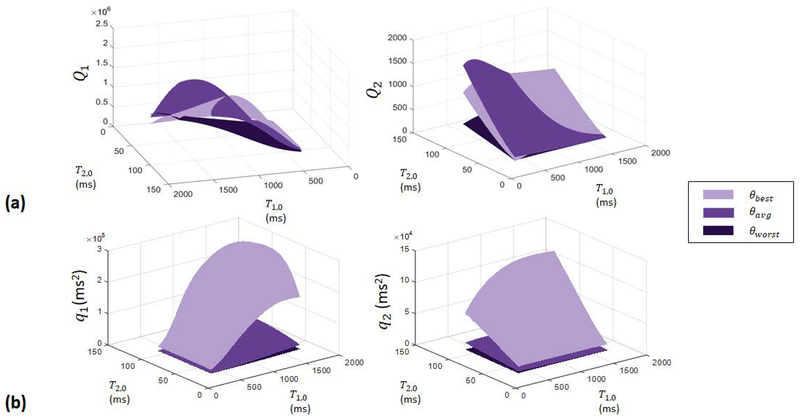
The resulting surface plots of the normal noise quality factor, Qi, versus T1,0 and T2,0 are shown in Fig. [2a], demonstrating that the encoding capability of the three sequences in the presence of normal noise does not necessarily follow the θbest > θaverage > θworst hierarchy predicted for aliasing noise by Sommer et al., but instead the sequences should tend to perform comparably well [13]. Surface plots of qi versus T1,0 and T2,0 shown in Fig. [2b] reveal that θbest results in the highest valued aliasing noise quality factors and θworst results in the lowest valued aliasing noise quality factors over the entire tested range, in agreement with the encoding capability predicted by Sommer et al.
Figure 2.
Surface plots of (a) normal and (b) aliasing noise quality factors as functions of T1,0 and T2,0 over the range T1,0=[350,1710] ms and T2,0=[30,110] ms, for experiments using θbest, θaverage, and θworst. Figure [2a] demonstrates that the three flip angle distributions, which achieve comparable normal noise quality factors over the tested range in relaxation times, are expected to perform similarly in the presence of normal noise. Figure [2b] shows that experiments with θbest yield the highest aliasing noise quality factors and experiments with θworst yield the lowest aliasing noise quality factors over the tested range, in agreement with the encoding capabilities demonstrated by Sommer et al.
Verification Experiments: Phantom Data
The results of ROI analysis on the T1 and T2 maps obtained with phantom experiments using θbest, θaverage, and θworst based on Sommer et al. are presented. Analysis of parameter maps obtained with fully sampled data show that the three experiments perform comparably well in the absence of aliasing noise. This result is in agreement with the T1 and T2 error forecasted by the normal noise quality factors (see Fig [2a]), which are expected to dominate the relationship presented in Eq. [5] in the absence of aliasing noise. Error in the parameter maps obtained with undersampled data achieving acceleration factors between R=5 and R=10 were determined to be dependent on both normal noise and aliasing noise quality factors, with neither term dominating the relationship between parameter map error and the derived quality factors presented in Eq. [5]. See the attached Supporting Information Figs. [S2] and [S3] for further details regarding parameter map error at low acceleration factors (R≤10).
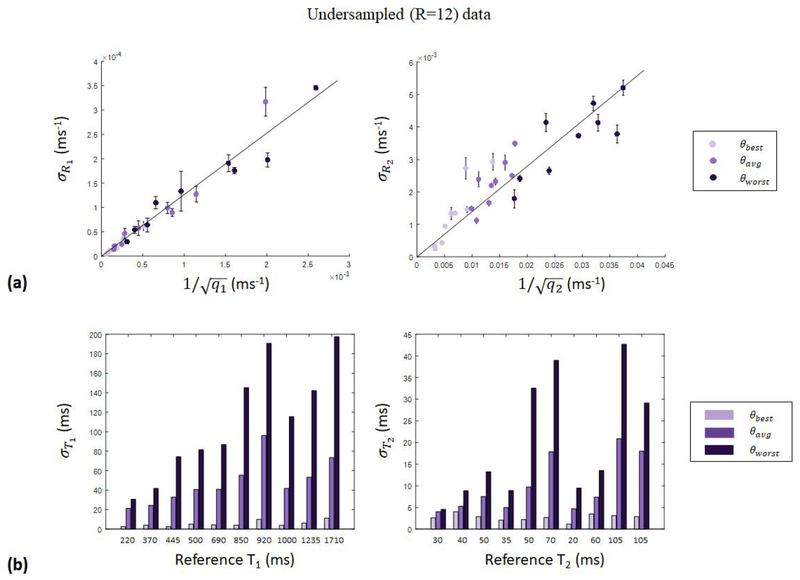
ROI analysis of the T1 and T2 maps obtained with undersampled data achieving acceleration factors R=12 and higher showed that the relationship between parameter map error and quality factors derived in Eq. [5] is dominated by the aliasing noise quality factors, so that the approximation taken in Eq. [8] is appropriate. As a representative example, Fig. [3a] shows the parameter map error per ROI, obtained with undersampled (R=12) data, plotted as a function of the calculated aliasing noise quality factor for the three tested sequences (θbest, θaverage, and θworst). In agreement with Eq. [8], the parameter map error achieves good linearity (R2=0.92 for and R2=0.80 for ) in . The variance of the aliasing noise, corresponding to the slope of the fitted data as a result of Eq. [8], is determined to be σω = 0.12,0.15 from the and data, respectively, agreeing to within 20%. Bar graphs (see Fig. [3b]) of the parameter map error per ROI, obtained with undersampled (R=12) data, show further agreement with the results of Sommer et al. and the surface plots of aliasing noise quality factors as functions of relaxation times presented in Fig. [3a], with θbest achieving the lowest parameter map error and θworst achieving the highest parameter map error in each of the tested ROIs. At higher acceleration factors, parameter map errors continue to show agreement with the quality factor framework in the case where aliasing noise is dominant. For R=16 and R=24, parameter map errors are very well characterized by Eq. [8], with goodness of fit greater than or equal to 0.94 (0.89) for data (see Supporting Information Fig. [S4] for details).
Figure 3.
Results of undersampled (R=12) verification experiments. (a) Parameter map error per ROI, plotted as a function of the calculated aliasing noise quality factor for the three tested sequences (θbest, θaverage, and θworst), achieving goodness of fit R2=0.92 and R2=0.80 in and , respectively. (b) Bar graphs of the parameter map error per ROI, showing further agreement with quality factor analysis, with θbest experiments achieving the lowest error and θworst experiments achieving the highest error, as expected.
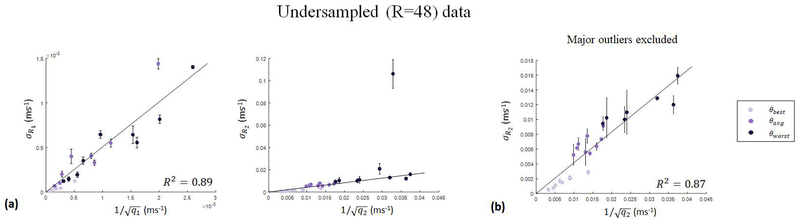
At very high acceleration factors (R=48), while parameter map errors continue to show relative agreement with the presented quality factor framework in the case where aliasing noise is dominant, deviations from the expected results begin to arise. The results of ROI analysis on parameter maps obtained from data at the highest available acceleration factor in this work, R=48, are presented in Fig. [4]. Although the data continues to be well modeled by the aliasing noise quality factors, achieving R2=0.89, the data contains obvious outliers relative to . The divergence from the quality factor framework is, in part, explained by the large mean error in T2 quantification, as verified by the large error bars on the outlying data points. Additional sources of error arise from the first-order approximation of aliasing noise used in the quality factor derivation (see Discussion). Excluding major outliers from the data, the quality factor model is in good agreement with the obtained data, achieving R2=0.87 (see Fig. [4b]).
Figure 4.
Results of undersampled (R=48) verification experiments, plotting parameter map error per ROI as a function of the calculated aliasing noise quality factor for the three tested sequences (θbest, θaverage, and θworst). In (a), all data points are included achieving goodness of fit R2=0.89 and R2=0.20 in and , respectively. In (b), obvious outliers from the are excluded, resulting in goodness of fit R2=0.87.
Optimization Experiments: Quality Factors as Functions of T1 and T2
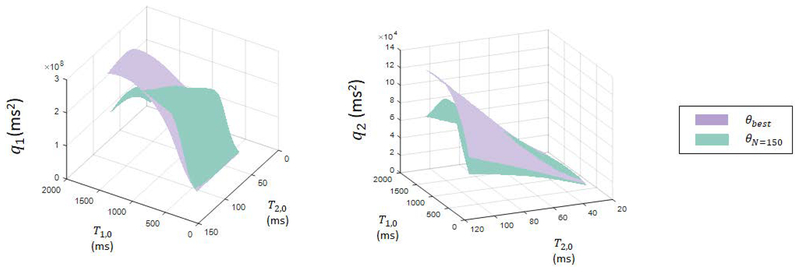
Aliasing noise quality factors for the 250 step experiment with θbest based on Sommer et al. and the efficient experiment with θN=150 derived through maximization of the aliasing noise quality factors were calculated in MATLAB using Eq. [7]. The resulting surface plots of quality factors for aliasing noise as functions of T1,0 and T2,0 (see Fig. [5]) reveal that the θbest and θN=150 experiments yield comparable values of qi, and therefore comparable forecasted errors, over a large range in T1 and T2 values. With comparable error and decreased scan time due to fewer acquisitions, θN=150 is expected to be more efficient in T1 and T2 mapping than θbest. Note, however, the θN=150 experiment is expected to perform poorly at large values of T1 (>1500 ms), which is likely due to the short total scan time relative to T1,0 at high values.
Figure 5.
Surface plots of aliasing noise quality factors as functions of T1,0 and T2,0 over the range T1,0=[350,1710] ms and T2,0=[30,110] ms, comparing experiments using θbest and θN=150. The two experiments yield comparable aliasing noise quality factors, except at large T1, where θN=150 is expected to yield large errors.
Optimization Experiments: Phantom Data
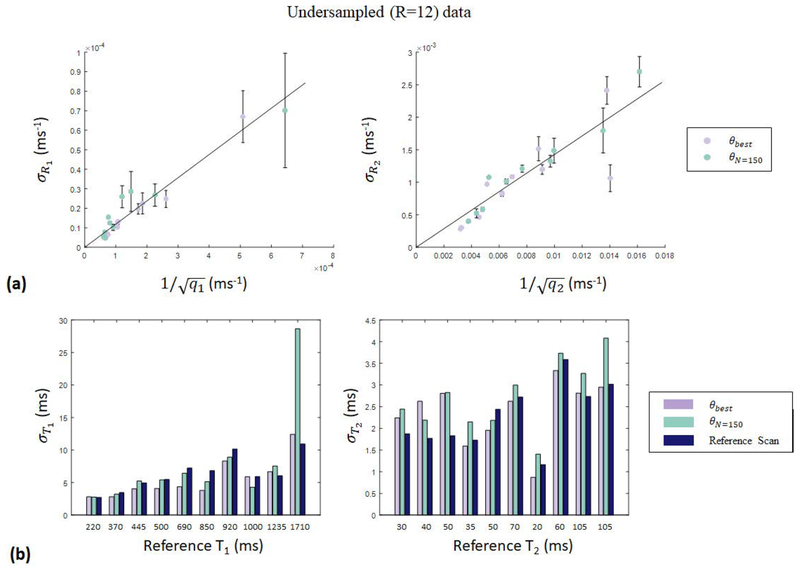
The results of ROI analysis on the T1 and T2 maps obtained with phantom experiments using the 250 step θbest based on Sommer et al. and the 150 step θN=150 derived through quality factor maximization are presented. Because θN=150 was designed to achieve aliasing noise quality factors comparable to θbest, error analysis was designed to specifically probe the relationship between qi and . In agreement with the results of verification experiments, plots of the obtained parameter map error per ROI as functions of aliasing noise quality factors showed that the qi term dominates the relationship in Eq. [5] for undersampled data with acceleration factors at and above R=12. As a representative case, Fig. [6a] shows the parameter map error per ROI, obtained with undersampled (R=12) data, plotted as a function of the calculated aliasing noise quality factor , achieving R2=0.92 and R2=0.70 for and for data, respectively. (We note that without the outlying data point (related to a very low valued T2,0 for which the first order approximation taken in Eq. [2] is less accurate) in the plot comparing our data to , the goodness of fit is increased from 0.70 to 0.92, so that both plots demonstrate excellent agreement with the expected linear relationship in Eq. [8]). Bar graphs of the parameter map error per ROI, obtained with undersampled (R=12) data from experiments using θbest and θN=150 are presented in Fig. [6b]. For additional comparison, Fig. [6b] also shows per ROI, obtained with the reference scan based on the FISP-MRF sequence presented in Jiang et al. and performed at the usual acceleration factor R=48. The graphs in Fig. [6b] continue to show agreement with parameter map error forecasted by aliasing noise quality factors, with generally comparable parameter map error per ROI for θbest and θN=150, except at large T1, where the 150 step experiment is expected to incur high mapping errors. In addition, Fig. [6b] demonstrates that the parameter map error from undersampled data (R=12) obtained with the two short sequences (θbest and θN=150) is comparable to that from the reference sequence.
Figure 6.
Results of undersampled (R=12) phantom optimization experiments. (a) Parameter map error per ROI, plotted as a function of the calculated aliasing noise quality factor for experiments with θbest and θN=150. achieving goodness of fit R2=0.92 and R2=0.70 in and , respectively. (b) Bar graphs of parameter map error per ROI, showing that experiments with θbest and θN=150 performed at R=12, and the standard FISP-MRF experiment based on Jiang et al. yield comparable errors. With fewer acquisitions (shorter scan time) and comparable error, θN=150 is more efficient than θbest.
Optimization Experiments: In Vivo Data
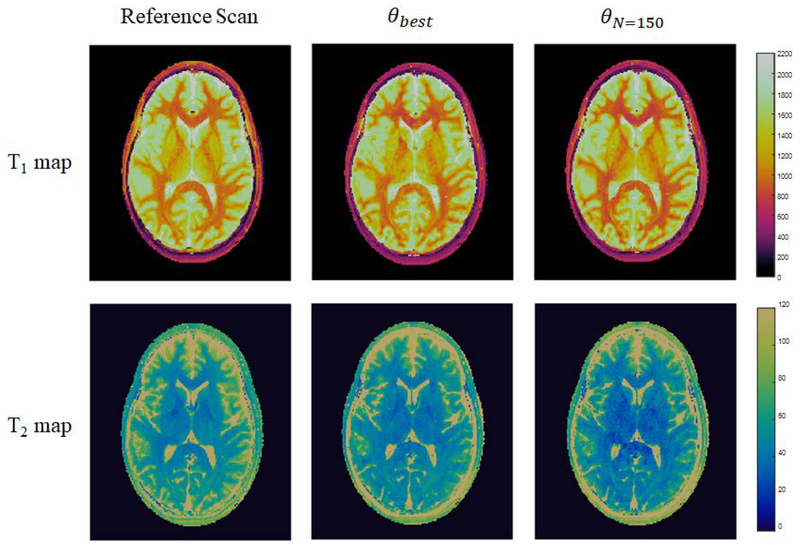
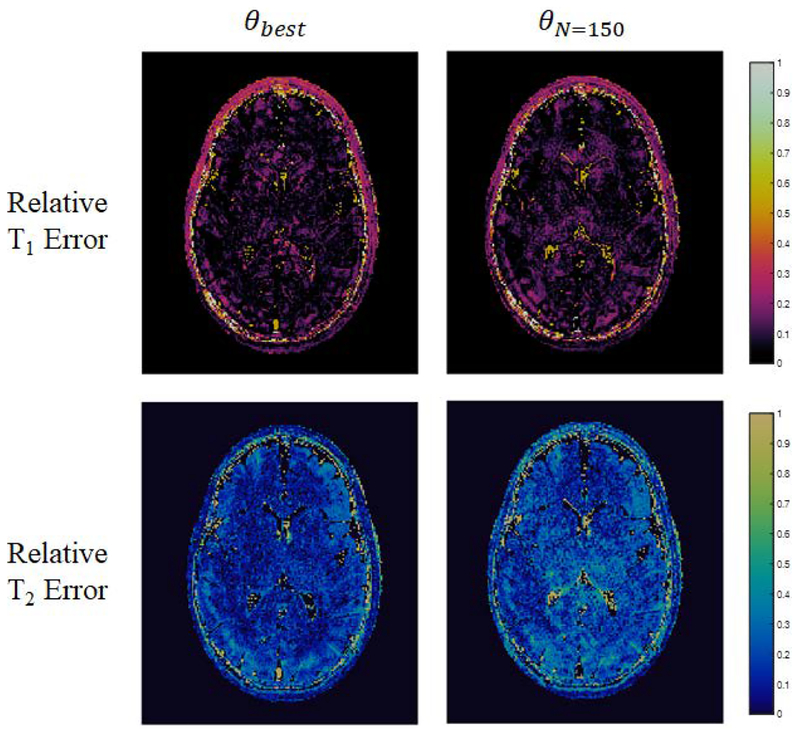
The results of in vivo experiments using the 250 step θbest based on Sommer et al. and the 150 step θN=150 derived through quality factor maximization are presented. As a representative example for the case where the aliasing noise term is expected to dominate Eq. [5] based on the findings of Optimization Experiments: Phantom Data, all presented results were obtained with an acceleration factor R=12. Parameter maps from experiments using θbest and θN=150, as well as reference maps from a FISP-MRF scan based on Jiang et al., are presented in Fig. [7]. Relative error maps from experiments using θbest and θN=150 compared to the reference maps are presented in Fig. [8]. Both the parameter maps and the relative error maps demonstrate the expected comparable mapping capabilities of θbest and θN=150. The results of nRMSE and ROI analysis on maps from the two sequences, detailed in Table [1], further demonstrate the comparable performance of the 150 step experiment to the 250 step experiment in terms of mean and standard deviation relaxation time values. White matter and gray matter ROI analysis on parameter maps from both sequences are in agreement with the reference maps and in agreement with literature values [29]. In addition, θbest and θN=150 are found to have comparable nRMSE in both T1 (0.09, 0.10) and T2 (0.15, 0.22). Due to its comparable performance and decreased scan time, the results of the in vivo experiments with θN=150 continue to demonstrate the utility of the quality factor framework in designing efficient MRF experiments.
Figure 7.
In vivo T1 and T2 maps from experiments using θbest and θN=150, as well as reference maps from a FISP-MRF scan based on Jiang et al., demonstrating the expected comparable mapping capabilities of θbest and θN=150.
Figure 8.
Relative error maps from experiments using θbest and θN=150 compared to a reference scan based on Jiang et al., demonstrating the expected comparable mapping capabilities of θbest and θN=150.
Discussion
In this work, the genetic algorithm was chosen for aliasing noise quality factor optimization, with an initial population of 900 sequences. Optimization was initially performed with N=250, reaching agreement with the already optimized θbest as determined by Sommer et al. within the first iteration (approximately 1 minute). Convergence to a result similar in encoding capability to θbest on this first iteration was anticipated due to the similarity between our initial population and the population of sequences tested by Sommer et al. Allowing further iteration, the optimization produced sequences with incremental improvements over the best sequence at the initial iteration, producing an MRF sequence leading to an approximately 20% improvement in forecasted error relative to θbest. This result suggests two advantages of the quality factor framework in MRF sequence evaluation and optimization. Due to this demonstrated ability to quickly evaluate given MRF sequences, the quality factor framework is expected to provide significant advantages in computation time compared to other, trial and error approaches. In addition, while trial and error approaches are limited to the initial population of MRF sequences, optimizations with our quality factors can produce effective sequences that are dissimilar from the initial population, and therefore do not require a priori knowledge.
To show the efficacy of our approach, optimization was performed for N=150, to determine a sequence with comparable error and therefore improved efficiency relative to the N=250 step experiment with θbest. While still fast (1 minute), the initial iteration of the optimization over 150 steps did not yield a sequence with forecasted error comparable to the 250 step θbest, suggesting that the initial population created with a colored noise generator did not contain the required qualities for an efficient MRF sequence. Therefore, the ability to diverge from the initial population provided by our quality factor framework was required to find an MRF sequence comparable to θbest. As in the optimization over 250 steps, not having a priori information built into our initial population did not affect the capability of the quality factor approach to find an efficient MRF sequence; however, thousands of iterations (approximately 26 hours) were required to find θN=150.
The flip angle distribution optimized for aliasing noise (θN=150) is observed to contain some rapid variation, resulting in a correspondingly varying signal time course (see Fig. [1]). In contrast to the common belief that MRF sequences should be smoothly varying [11][16], this result suggests that while flip angle distributions used in MRF with undersampling should not contain significant and very rapid variations to prevent the unequal weighting of aliasing noise at various time steps, some controlled rapid variations in the flip angle distribution may be beneficial.
The error assessment and optimization in this work has been focused on comparisons with the MRF sequences presented in Sommer et al. For example, we find a 150-step sequence that performs as well as the best, 250-step sequence found with Monte Carlo methods at equal acceleration factors, and, therefore, θN=150 is more efficient than θbest. Short sequences at lower acceleration factors may not be as efficient as longer sequences at higher acceleration factors. This decreased efficiency can be mitigated, for example, in multi-slice imaging where the wait time between k-space acquisitions can be used to acquire data in other slices. In addition, sequences with fewer steps, are expected to be useful in specialized applications, for example in imaging patients for whom breath holds are challenging.
As an example, it is of interest to discuss experiments with the standard 1000-step distribution based on Jiang et al. acquired at R=48. At lower acceleration factors (R=12), the short sequences (θN=150 and θbest) have comparable T1 and T2 mapping error to the standard sequence performed at R=48, as evidenced by both the phantom and the in vivo data presented herein (Fig. [6b], Table 1). Although θN=150 and θbest have fewer time steps than the standard sequence, necessary wait times between the k-space trajectories, required to achieve the reduced acceleration factor, result in a less efficient scan in terms of total sequence duration. However, outside the aforementioned focus of this work, our quality factors can be employed to find new distributions with increased efficiency relative to the standard sequence. In an example, the presented framework has been used to find a flip angle distribution with 500 time steps, achieving higher q1 (reduced T1 mapping error) and comparable q2 (comparable T2 mapping error) relative to the standard sequence in [20]. Other examples employing our quality factors to analyze and optimize MRF sequences with larger number of time steps have been reported in [23][31][32].
Results in this work demonstrate that the aliasing noise term dominates Eq. [5] for acceleration factors R=12 and above. Parameter map error is found to be in very good agreement with the relationship in Eq. [8] for acceleration factors between R=12 and 24; however, at the highest available acceleration factor in this work, R=48, deviations from the derived model arise (see Fig. [4]). These deviations are believed to result, in part, from the first-order approximation of the aliasing noise taken in the quality factor derivation, which is not accurate for the large aliasing artifacts (approximately equal to the signal amplitude) present in the undersampled data for R=48. Therefore, at very large acceleration factors, the derived parameter map model in terms of aliasing noise quality factors is an appropriate guide in forecasting the parameter map error of an MRF sequence, serving as a first approximation to the true, more complicated relationship. Further investigation into improving the derived quality factor model for very large acceleration factors will be a topic for future work.
While the quality factors defined by Eqs. [6–7] are specific to T1 and T2 mapping, there are several additional imaging parameters that can be mapped with MRF. In addition to off-resonance frequency mapping, which is necessary in bSSFP-MRF, MRF has expanded into the quantification of properties such as B1+ [24][33][34] and diffusion parameters [35][36]. Therefore, broadening the quality factor framework to include these additional parameters is of great interest.
Because the quality factor framework derived herein is based on the theory of small perturbations to a system, specifically those arising from TR/Ti << 1, the expansion of quality factors to include a third mapping parameter is straightforward if the added parameter of interest can also be modeled as a small perturbation. For example, initial investigations have successfully quantified the error in T1, T2, and B1+ mapping when the flip angle distribution is required to be small (<15°). Alternatively, an expansion can be made assuming a small variation about a central value of B1+. Then quality factors can be calculated at various central B1+ values to analyze mapping capability over a large range in B1+ values, analogous to the procedure for comparing quality factors over a large range of relaxation times used in this work. In addition, initial investigation into the application of quality factors to T1, T2, and ADC mapping is promising. However, more work is required for a robust expansion of the quality factor framework to include added properties such as off-resonance frequency and B1+, which in general do not produce small perturbations to the system.
Conclusion
In this work, quality factors for the assessment of MRF pulse sequence designs in the presence of normal noise and aliasing artifacts were derived, tested, and shown to accurately forecast parameter map error. Due to quality factor dependence on the MRF signal, optimized MRF sequences can be rapidly evaluated and optimized relative to normal and aliasing noise through quality factor maximization, to yield efficient MRF experiments.
Supplementary Material
Acknowledgements
This work made use of the High Performance Computing Resource in the Core Facility for Advanced Research Computing at Case Western Reserve University and was funded by NIH T32EB007509, OTF IPP TECG20140138, NSF/CBET 1553441, NIH R01HL094557, NIH R01DK098503, NIH R01EB016728, and NIH R01BB017219. We are grateful for a useful correpondence with Karsten Sommer on the modeling of aliasing noise. Additionally, we would like to thank Michael Martens and Dan Ma for their assistance and support.
Funding Information: Grant sponsor: NIH, Grant numbers: T32EB007509, R01HL094557, R01DK098503, R01EB016728, R01BB017219; Grant sponsor: OTF; Grant number: IPP TECG20140138; Grant sponsor: NSF/CBET; Grant number: 1553441
Appendix
Quality Factor Derivation
With presented models for the dictionary entries and acquired signal, as detailed in the theory section,
the initial step in our quality factor derivation involves noting that matching through maximization of the complex dot product implies that the derivative of the dot product between the normalized signal and the matched normalized dictionary entry will be zero:
| (A.1) |
In a FISP-MRF experiment where spoiler gradients are applied at the end of each TR to create a phase twist greater than 2π in the slice select direction and excitation pulses are applied along a single axis (ex. along x), the complex signal (d = dx + idy) will be dominated by either the real or the imaginary component (ex. d ≈ idy). Therefore, the imaginary components of Eq. [A.1] vanish, leaving
| (A.2) |
For ease of notation, now let (a⋅b*) indicate the real part of the complex dot product. Using Eq. [A.2] and noting that s is related to the ground truth relaxation rates (R1,0 and R2,0), and thus independent of Ri, to first order in Ri and Rj we find,
| (A.3) |
Using Eq. [2] to write Eq. [A.3] in terms of d0, ci, cj, η, and α, keeping only first order terms in Ri, Rj, η, and α:
| (A.4) |
With
Using the two equations (i=1,2 and j=2,1) to solve for (R1 – R1,0) and (R2 – R2,0):
| (A.5) |
Where
Because ω and α have zero mean and are uncorrelated, averaging Eq. [A.5] over the time-step population yields a vanishing result on the right-hand side of the equation, leading to R1=R1,0. Averaging over the square of both sides leads to the following variance relationships:
| (A.6) |
| (A.7) |
In general, for vectors u = iuy and ν = iνy with vanishing real components and Gaussian white noise η with 〈(η)m(η)n〉 = ση2δm,n:
In the case of aliasing noise with (α)n = |(d0)n|(ω)n and 〈(ω)m(ω)n〉 = σω2δm,n:
Therefore, we find
| (A.8) |
Finally, combining Eqs. [A.6] and [A.8],
With the quality factors relating to thermal noise given by
And the quality factors relating to aliasing noise given by
References
- 1.Cheng HL, Stikov N, Ghugre NR, Wright GA. Practical medical applications of quantitative MR relaxometry. J Magn Reson Imaging 2012;36(4):805–824. doi: 10.1002/jmri.23718. [DOI] [PubMed] [Google Scholar]
- 2.Gao Y, Chen Y, Ma D, Jiang Y, Herrmann KA, Vincent JA, et al. Preclinical MR fingerprinting (MRF) at 7 T: effective quantitative imaging for rodent disease models. NMR Biomed 2015;28(3):384–394. doi: 10.1093/neuonc/nou264.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Langer DL, van der Kwast TH, Evans AJ, Trachtenberg J, Wilson BC, Haider MA. Prostate cancer detection with multi-parametric MRI: logistic regression analysis of quantitative T2, diffusion-weighted imaging, and dynamic contrast enhanced MRI. J Magn Reson Imaging 2009;30(2):327–334. doi: 10.1002/jmri.21824. [DOI] [PubMed] [Google Scholar]
- 4.Roller FC, Harth S, Schneider C, Krombach GA. T1, T2 Mapping and Extracellular Volume Fraction (ECV): Application, Value and Further Perspectives in Myocardial Inflammation and Cardiomyopathies. Rofo 2015;187(9):760–770. doi: 10.1055/s-0034-1399546. [DOI] [PubMed] [Google Scholar]
- 5.Dall’Armellina E, Piechnik SK, Ferreira VM, Si QL, Robson MD, Francis JM, et al. Cardiovascular magnetic resonance by non contrast T1-mapping allows assessment of severity of injury in acute myocardial infarction. J Cardiovasc Magn Reson 2012;14:15. doi: 10.1186/1532-429X-14-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bernarding J, Braun J, Hohmann J, Mansmann U, Hoehn-Berlage M, Stapf C, et al. Histogram-based characterization of healthy and ischemic brain tissues using multiparametric MR imaging including apparent diffusion coefficient maps and relaxometry. Magn Reson Med 2000;43(1):52–61. doi:. [DOI] [PubMed] [Google Scholar]
- 7.Gracien RM, Jurcoane A, Wagner M, Reitz SC, Mayer C, Volz S, et al. Multimodal quantitative MRI assessment of cortical damage in relapsing-remitting multiple sclerosis. J Magn Reson Imaging 2016;44(6):1600–1607. doi: 10.1002/jmri.25297. [DOI] [PubMed] [Google Scholar]
- 8.Ma D, Gulani V, Seiberlich N, Liu K, Sunshine JL, Duerk JL, et al. Magnetic resonance fingerprinting. Nature 2013;495(7440):187–192. doi: 10.1038/nature11971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jiang Y, Ma D, Keenan KE, Stupic KF, Gulani V, Griswold MA. Repeatability of magnetic resonance fingerprinting T1 and T2 estimates assessed using the ISMRM/NIST MRI system phantom. Magn Reson Med 2016;doi: 10.1002/mrm.26509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hamilton JI, Wright KL, Jiang Y, Garcia LH, Ma D, Griswold M, et al. Pulse Sequence Optimization for Improved MRF Scan Efficiency. In Proceedings of the 23rd Annual Meeting of the ISMRM, Toronto, Ontario, Canada; 2015. 3386. [Google Scholar]
- 11.Assländer J, Sodickson D, Lattanzi R, Cloos M. Relaxation in Polar Coordinates: Analysis and Optimization of MR Fingerprinting. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 0127. [Google Scholar]
- 12.Sommer K, Amthor T, Koken P, Meineke J, Doneva M. Determination of the Optimum Pattern Length of MRF Sequences. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 1491. [Google Scholar]
- 13.Sommer K, Amthor T, Doneva M, Koken P, Meineke J, Börnert P. Towards predicting the encoding capability of MR fingerprinting sequences. Magn Reson Imaging. 2017;41:7–14. doi: 10.1016/j.mri.2017.06.015. [DOI] [PubMed] [Google Scholar]
- 14.Cohen O, Rosen MS. Algorithm comparison for schedule optimization in MR fingerprinting. Magn Reson Imaging. 2017;41:15–21. doi: 10.1016/j.mri.2017.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao B, Haldar JP, Liao C, Ma D, Griswold MA, Setsompop K, Wald L. Optimal experiment design for magnetic resonance fingerprinting. In Proceedings of the IEEE Eng. Med. Bio. Conf; 2016:453–456. doi: 10.1109/EMBC.2016/7590737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao B, Haldar JP, Setsompop K, Wald L. Optimal Experiment Design for Magnetic Resonance Fingerprinting: Cramer-Rao Bound Meets Spin Dynamics. arXiv:1710.08062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Breithaupt M, Flassbeck S, Ladd M. On Optimizations of MRF Patterns Based on Generalized MRI Sequence Schemes. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 0936. [Google Scholar]
- 18.Hamilton JI, Jiang Y, Chen Y, Ma D, Lo WC, Griswold M, et al. MR fingerprinting for rapid quantification of myocardial T1, T2, and proton spin density. Magn Reson Med 2017;77(4):1446–1458. doi: 10.1002/mrm.26216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.van Valenberg W, Vos FM, Klein S, van Vliet LJ, Poot DHJ. Determining the Time Efficiency of Quantitative MRI Methods Using Bloch Simulations. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 1470. [Google Scholar]
- 20.Jiang Y, Ma D, Seiberlich N, Gulani V, Griswold MA. MR fingerprinting using fast imaging with steady state precession (FISP) with spiral readout. Magn Reson Med 2015;74(6):1621–1631. doi: 10.1002/mrm.25559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gudbjartsson H, Patz S. The Rician Distribution of Noisy MRI Data. Magn Reson Med 1995;34(6):910–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kara D, Fan M, Hamilton J, Griswold MA, Seiberlich N, Brown R. Comparison of dot product quality factors to Cramer-Rao bounds for error analysis in MRF. In Proceedings of the ISMRM Workshop on Magnetic Resonance Fingerprinting, Cleveland, Ohio, USA; 2017. 13. [Google Scholar]
- 23.Kara D, Fan M, Hamilton J, Griswold MA, Seiberlich N, Brown R. Quality Factors for Efficient and Precise MRF Imaging. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 1350. [Google Scholar]
- 24.Ma D, Coppo S, Chen Y, McGivney DF, Jiang Y, Pahwa S, et al. Slice profile and B1 corrections in 2D magnetic resonance fingerprinting. Magn Reson Med 2017. doi: 10.1002/mrm.26580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hargreaves BA. Bloch Equation Simulation;http://mrsrl.stanford.edu/brian/bloch/. Accessed: January 2017.
- 26.Hargreaves BA. Bloch Equation Simulation;http://mrsrl.stanford.edu/brian/vdspiral/. Accessed: January 2017.
- 27.Tolouee A, Alirezaie J, Babyn P. Compressed sensing reconstruction of cardiac cine MRI using golden angle spiral trajectories. J Magn Reson 2015;260:10–19. doi: 10.1016/j.jmr.2015.09.003. [DOI] [PubMed] [Google Scholar]
- 28.Fessler JA. On NUFFT-based gridding for non-Cartesian MRI. J Magn Reson 2007;188(2):191–195. doi: 10.1016/j.jmr.2007.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walsh DO, Gmitro AF, Marcellin MW. Adaptive reconstruction of phased array MR imagery. Magn Reson Med 2000;43(5):682–690. doi:. [DOI] [PubMed] [Google Scholar]
- 30.Hasgall PA, Di Gennaro F, Baumgartner C, Neufeld E, Gosselin MC, Payne D, Klingenböck A, Kuster N. IT’IS Database for thermal and electromagnetic parameters of biological tissues. Version 3.0, September 1, 2015, doi: 10.13099/VIP21000-03-0. [DOI] [Google Scholar]
- 31.Kara D, Fan M, Hamilton J, Griswold MA, Seiberlich N, Brown R. Error minimization with complex valued dot product quality factors. In Proceedings of the ISMRM Workshop on Magnetic Resonance Fingerprinting, Cleveland, Ohio, USA; 2017. 12. [Google Scholar]
- 32.Kara D, Hamilton J, Fan M, Griswold MA, Seiberlich N, Brown R. The statistical error in FISP-MRF experiments. In Proceedings of the 26th Annual Meeting of the ISMRM, Paris, France; 2018. 2780. [Google Scholar]
- 33.Buonincontri G, Sawiak SJ. MR fingerprinting with simultaneous B1 estimation. Magn ResonMed 2016. October;76(4):1127–1135. doi: 10.1002/mrm.26009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cloos MA, Knoll F, Zhao T, Block KT, Bruno M, Wiggins GC, et al. Multiparametric imaging with heterogeneous radiofrequency fields. Nat Commun 2016. August;7:12445. doi: 10.1038/ncomms12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jiang Y, Hamilton JI, Lo WC, Wright KL, Ma D, Coristine AJ, et al. Simultaneous T1, T2, and Diffusion Quantification using Multiple Contrast Prepared Magnetic Resonance Fingerprinting. In Proceedings of the 25th Annual Meeting of the ISMRM, Honolulu, Hawaii, USA; 2017. 1171. [Google Scholar]
- 36.Jiang Y, Ma D, Wright K, Seiberlich N, Gulani V, Griswold MA. Simultaneous T1, T2, Diffusion and Proton Density Quantification with MR Fingerprinting. In Proceedings of the 22th Annual Meeting of the ISMRM, Milan, Italy; 2014. 0028. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.