Abstract
Purpose:
Fluorescence lifetime imaging microscopy (FLIM) of endogenous fluorescent metabolites permits the measurement of cellular metabolism in cell, tissue and animal models. In parallel, magnetic resonance spectroscopy (MRS) of dynamic nuclear (hyper)polarized (DNP) 13C-pyruvate enables measurement of metabolism at larger in vivo scales. Presented here is the design and initial application of a bioreactor that connects these two metabolic imaging modalities in vitro, using 3D cell cultures.
Theory and Methods:
The model fitting for FLIM data analysis and the theory behind a model for the diffusion of pyruvate into a collagen gel are detailed. The device is MRI-compatible, including an optical window, a temperature control system and an injection port for the introduction of contrast agents. 3D printing, computer numerical control (CNC) machining and laser cutting were used to fabricate custom parts.
Results:
Performance of the bioreactor is demonstrated for 4T1 murine breast cancer cells under glucose deprivation. Mean NADH fluorescence lifetimes were 10% longer and hyperpolarized 13C lactate:pyruvate (Lac:Pyr) ratios were 60% lower for glucose-deprived 4T1 cells compared to 4T1 cells in normal medium. Looking at the individual components of the NADH fluorescent lifetime, τ1 (free NADH) showed no significant change, while τ2 (bound NADH) showed a significant increase, suggesting that the increase in mean lifetime was due to a change in bound NADH.
Conclusion:
A novel bioreactor that is compatible with and can exploit the benefits of both FLIM and 13C MRS, in three-dimensional (3D) cell cultures for studies of cell metabolism has been designed and applied.
Keywords: Magnetic Resonance Spectroscopy (MRS), Optical Imaging, Bioreactor, Multimodal, Fluorescence Lifetime Imaging (FLIM), Metabolism, Lactate Production, NADH
Introduction
Metabolic shifts are known to be a key feature of the cancer disease process. As such, accurate measures of metabolism during cancer progression may guide development of new therapies, new potential drug targets and more accurate prognoses [1]–[3]. In particular, there is growing interest in understanding the evolution of an individual’s metabolic pathway(s) during cancer progression and treatment [4]. Pathways and individual molecules identified as crucial to tumor metabolism become key biomarkers for assessing disease progression [5], [6]. Examples of such biomarkers include lactate dehydrogenase (LDH) activity, which catalyzes the conversion of pyruvate to lactate [7] and partly regulates the redox potential of individual cells [8]. Additionally, a cofactor of LDH activity is nicotinamide adenine dinucleotide (NADH), an intermediate metabolite central to cellular redox potential [9], [10].
In recent years, several techniques to measure these two biomarkers in vitro and in vivo have emerged. Specifically, magnetic resonance spectroscopy (MRS) of hyperpolarized 13C-labeled pyruvate allows for the real-time monitoring of LDH activity [11]–[13], while optical fluorescence lifetime imaging (FLIM) of the intrinsically fluorescent NADH [14], [15] allows for the measurement of its chemical state, whether protein-bound or free in the cytosol [16]. These two metabolic measurement techniques yield complementary information, by probing organ and cellular scales, respectively. Therefore, combined studies that utilize both methods may add value for quantitatively investigating enzyme activity and cofactor status for various metabolic pathways.
Hyperpolarized MRS imaging studies with 13C-pyruvate are moving rapidly to clinical translation [12], principally because of their ability to measure LDH activity and upregulation of glycolysis of cancer in vivo [17], [18]. These recent advances are supported by pre-clinical studies as well as in vitro studies of cell cultures [19] and tumor biopsy tissues [20] using MRS of three dimensional (3D) sample volumes. In contrast, optical imaging experiments are often performed in adherent 2D cell cultures on glass bottom dishes at sub-cellular resolution [21]. Although the cellular resolution is desirable, cells cultured directly on conventional glass bottom dishes lack the 3D microenvironment encountered in vivo [22], [23].
Collagen gels that more closely resemble the native (breast) tumor microenvironment [24] can improve the biological relevance of optical imaging experiments (Supporting Information Figure S1). While in vivo optical experiments using imaging windows implanted above tumors in small animal models enable direct imaging within the tumor microenvironment [25], they have intrinsic limitations including poor depth of field and increased cost and complexity for initial screenings of novel compounds.
Bioreactor systems allowing for a variety of controlled in vitro studies on larger populations of cells have been developed over the past three decades [26]–[30]. Bioreactor designs have been engineered for DNP-MRS studies, most typically for MRS on high field nuclear magnetic resonance (NMR) systems [19], and, independently, for optical imaging [31]. However, as of yet, bioreactors for complementary optical and MRS studies of the same 3D cell culture have not been developed, partly due to engineering challenges. In particular, creating a device that maintains cell cultures, is MR-compatible, has an optical window and is compatible with different hardware is a unique challenge.
The purpose of this work is to design a bioreactor for the study of cellular metabolism in 3D cell culture using both FLIM on an inverted multiphoton laser scanning microscope and 3D volumetric MRS using a horizontal bore preclinical MRI system, at cellular and population scales. Embedding cells in a collagen matrix was the cell culturing technique used for this device, in order to maintain similarity to the native tumor microenvironment. This ability to assess information, such as metabolic status, at different scales and in a model that is similar to in vivo, improves our understanding of the cellular impact on larger scale tissue and organ level changes, thus permitting an improved assessment of disease progression and therapeutic responses.
Theory
Fluorescence Lifetime Measurements
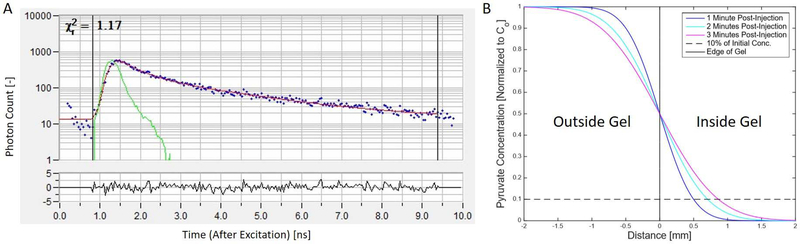
Fluorescence lifetime imaging (FLIM), specifically as a time correlated single photon counting technique, was used to collect the fluorescence lifetime data of NADH in this work. During acquisition, a photon counting detector with timing electronics synced with a pulsed excitation laser is used to collect the fluorescence emission photons resulting from the excitation of any given pixel. In this way, photon fluence can be plotted against time (from excitation) for each pixel in a field of view (Figure 1A). The mean fluorescence lifetime is the average time a molecule spends in the excited state and is calculated by averaging all the measured lifetimes for every photon [14], [15], [32].
Figure 1:
Sample fluorescence lifetime data (A). The two-component decay model fit is shown in red with the collected data shown in a blue scatter plot. The green plot is the instrumentation response function (IRF) which represents the overall time-resolution of the acquisition system. (B) Results of the diffusion model showing concentration profiles of pyruvate as a function of injected concentration at the boundary of the collagen gel. The 10% line (dashed black) and the edge of the gel (solid black) are shown.
Every fluorescent molecule has a different fluorescence decay profile as the number of emission photons following excitation decays away to zero. Previous work has shown that the fluorescence of NADH fits a two-component decay model very well, with both a shorter lived and a longer lived component. These components correspond to NADH that is unbound and NADH that is protein-bound, respectively. The fitted decay curve has the form [33], [34]:
In the above equation, τ1 and τ2 are fitted lifetime values that, in the case of NADH lifetime imaging, correspond to the shorter-lived fluorescence of unbound NADH and the longer-lived fluorescence of protein-bound NADH, respectively.
Diffusion Model
The observed lactate signal during the presented MRS experiments depends on the extent to which the injected [1-13C] pyruvate is able to diffuse into the collagen gel containing the target cells. It is possible to create a simple model of diffusion to get a reasonable estimate of the drug penetration into the collagen gel during the time course of the experiments by looking at diffusion theory.
Fick’s Second Law of Diffusion can be solved with the appropriate conditions to produce a concentration profile for many different setups [35]:
Using some simplifying assumptions (Supporting Information Figure S2), we can use the initial conditions where C = Co for x < 0 (outside the gel) and C = 0 for x > 0 (inside the gel) at t = 0. This yields the following equation for concentration [35]:
The variable D in the above equation is the diffusion coefficient of pyruvate for the experimental conditions. Søgaard et al. reported a diffusion coefficient for pyruvate in vivo in 2014 as D = 1.266 um2/ms [36]. This is a conservative number as this experimental setup is in solution, but it was used for the purposes of this model. Solving the concentration equation at any given time therefore yields an estimated concentration profile into the collagen gel (Figure 1B).
Methods
The design requirements for this dual-modality bioreactor included (1) allowing for the maintenance of large 3D cell cultures in a collagen matrix, (2) compatibility with an existing preclinical MR system, (3) integration of an optical window and (4) creating flexibility to allow for extension to other imaging hardware. Additionally, this device must allow for environmental control, such as temperature, and include access for drug and contrast agent delivery. Presented here is the design and initial application of such a device. This bioreactor consists of both custom and commercial parts (Table 1). To demonstrate its utility, the bioreactor was used for a proof of principle study on the effects of glucose deprivation in 3D murine breast cancer cell cultures using complementary 2D optical FLIM (Figure 2C, Supporting Information Figure S3) and 3D volumetric MRS (Figure 2B) measures of metabolism at the cell and population scales (Figure 2).
Table 1:
Bioreactor and supporting equipment parts list. For commercial parts, the part numbers of the ones used here are included, but, in most cases, there are several alternatives to those listed.
| # (Fig. 2) | Description | Manufacturer | Part # | Component Of |
|---|---|---|---|---|
| 1 | Bioreactor body | Custom | N/A | Bioreactor |
| 2 | Optical window compression piece | Custom | N/A | Bioreactor |
| 3 | Bioreactor top | Custom | N/A | Bioreactor |
| 4 | Optical window gasket | Custom | N/A | Bioreactor |
| 5 | Cover glass | Corning Life Sciences | 2850–22 | Bioreactor |
| 6 | Rubber bands | McMaster-Carr | 12205T51 | Bioreactor |
| 7 | 6–32 nylon bolts | McMaster-Carr | 95868A293 | Bioreactor |
| 8 | Luer lock fittings | McMaster-Carr | 51525K222 | Bioreactor |
| 9 | Sealed capillary tube | Pyrex | 9530–3 | Bioreactor |
| 10 | Tube fittings | Variety Used | N/A | Supporting Equip. |
| 11 | MR temperature probe | SA Instruments Inc | N/A | Supporting Equip. |
| 13 | Plastic tubing | Variety Used | N/A | Supporting Equip. |
| 14 | Hot water bath | Thermo Scientific | N/A | Supporting Equip. |
| 15 | 6V DC Diaphragm Pump | UXCell | N/A | Supporting Equip. |
Figure 2:
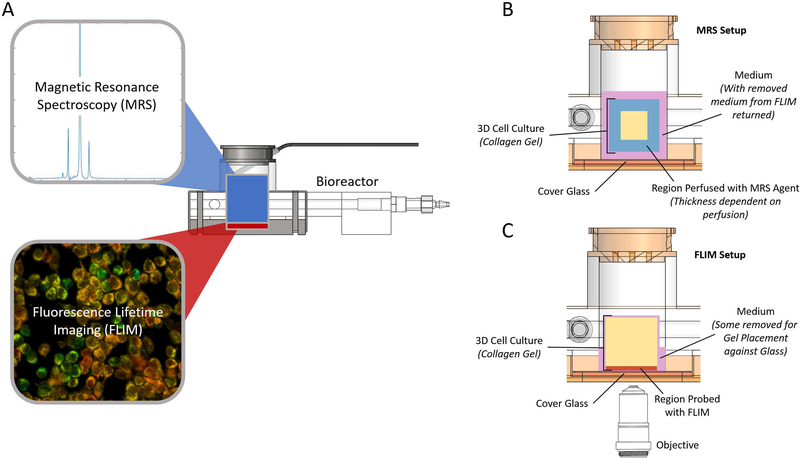
Schematic showing the bioreactor design concept (A). Combining MRS and FLIM allows the collection of complementary metabolic information from the 3D culture volume (blue MR sensitive region) and at cellular scales (red optical port). (B) Cutaway view of the sample area for the MRS experiments. Medium that was removed for optical imaging was returned prior to these experiments. The region of the collagen gel probed by MRS is shown in blue. The thickness of this region depends on perfusion of the drug into the collagen gel. (C) Cutaway view of the sample area for the FLIM experiments. Medium was removed and reserved to ensure that the gel contacted the cover glass for imaging. The region of the collagen gel that can be probed by optical imaging is shown in red.
Development of Bioreactor Design
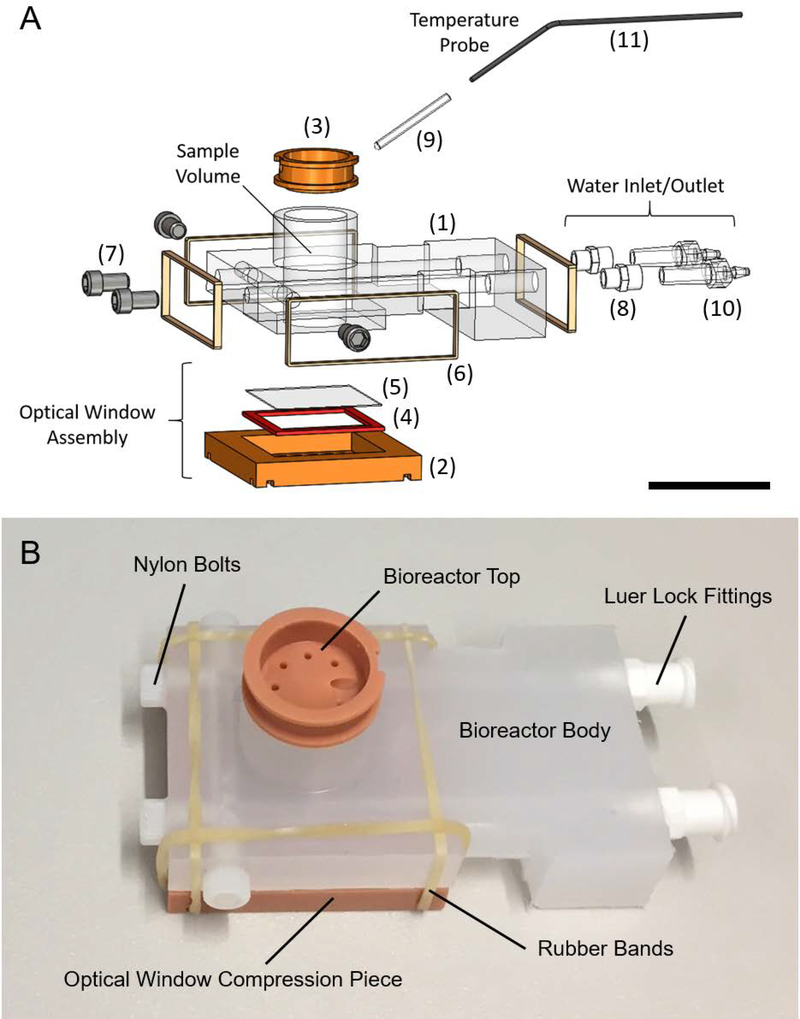
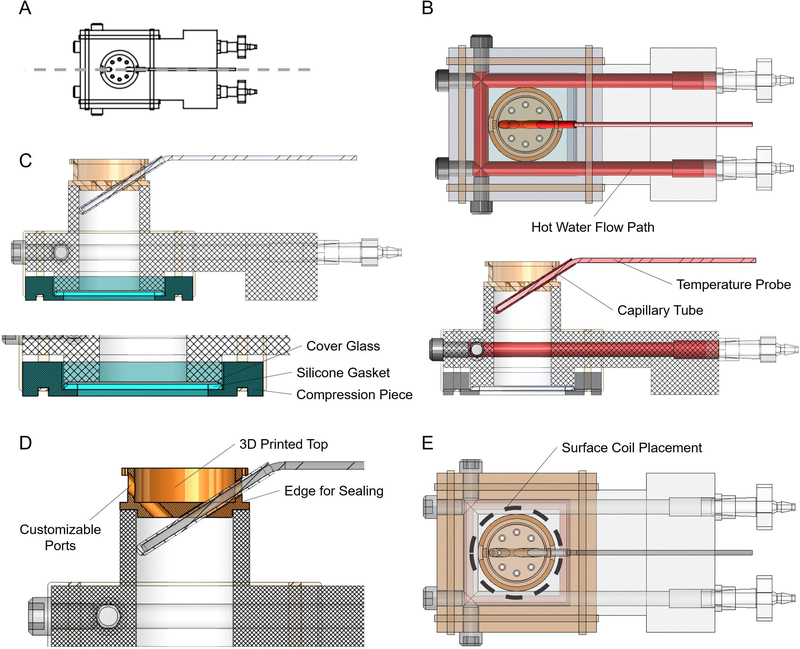
The individual components and the overall assembly of the bioreactor were designed using SolidWorks (Dassault Systemes, Velizy-Villacoublay, France) 3D modeling software. The design went through several iterations. The final design consists of four custom parts and 5 off the shelf parts. An exploded view with individual parts numbered can be seen in Figure 3A. Detailed descriptions of each part are provided in Table 1, along with descriptions of supporting equipment necessary to use the device.
Figure 3:
Bioreactor design and components. (A) Computer aided design (CAD) rendering of the bioreactor assembly in an exploded view. Key components are indicated, including the optical window assembly, water inlet and outlet and the temperature probe. The sample volume is cylindrical and is where 3D cell cultures are prepared for combined MRS and FLIM experiments. Numbers refer to parts described in Table 1. (B) Picture of an assembled bioreactor with visible components labeled. Scale bar in Panel A is approximately 25 mm.
Fabrication of Custom Components
The bioreactor body (Figure 3A, #1) was machined out of a piece of polypropylene stock material (McMaster-Carr, Elmhurst, IL, USA) using a Haas Super Mini Mill 2, a computer numerical control (CNC) machining center (Haas Automation, Inc., Oxnard, CA, USA). Two of the custom components of the bioreactor, the optical window compression piece and the bioreactor top piece (Figure 3A, #2 and Figure 3A, #3, respectively), were 3D-printed using a Perfactory 3 3D Printer (EnvisionTEC Inc., Dearborn, MI, USA) and RC31, a photopolymer (EnvisionTEC). The final custom piece, the optical window silicone gasket (Figure 3A, #4), was laser cut out of a 1/16” thick silicone sheet (McMaster-Carr), using a ULS 6.75 Laser Cutter (Universal Laser Systems, Inc., Scottsdale, AZ, USA). Printing files (.STL) for the two 3D-printed parts, G-code for the CNC-machined part and a.DXF file for the laser cut part are available for download at: https://morgridge.org/designs. Fabricated components on an assembled bioreactor can be seen in Figure 3B.
Before any components of this device were fabricated, a cytotoxicity study was conducted to ensure that any materials used in components that contacted cells were not cytotoxic (Supporting Information Figure S4). Delrin and RC31 both exhibited some level of cytotoxicity and were avoided where possible. The flexibility of 3D printing was necessary for the two bioreactor components that were fabricated with the 3D printing material RC31. However, RC31 was not in direct contact with the cell cultures during these experiments.
4T1 Cell Culture and Collagen Polymerization
4T1 murine breast cancer cells were used for the combined MRS and FLIM experiments shown. Cells were prepared in a 3D collagen matrix as previously described [24]. In brief, stock collagen was neutralized with 2x HEPES buffer, then further diluted to 2mg/ml with cells and medium. A total of 8 million cells were cultured in a total of 1 ml of collagen. The collagen gel was polymerized in the bioreactor for 15 min in a biological safety cabinet then moved to a 37°C cellular incubator for 1 hour. Total experimental volume in the bioreactor was 1.5 mL. The collagen gel initially occupies most of this volume, but contracts over time as the cultured cells interact with the collagen fibers. Medium used was Gibco RPMI 1640 medium (ThermoFisher Scientific, Waltham, MA, USA). For the glucose free state, normal medium was removed and replaced with glucose-free medium roughly an hour before the beginning of imaging. For this proof of concept work, HEPES buffered medium (100mM in 2x PBS) was used so that gas control would not be needed. However, future studies can be done to explore the effect on these metabolic markers of different gas environments, such as in 5% CO2.
Experimental Setups
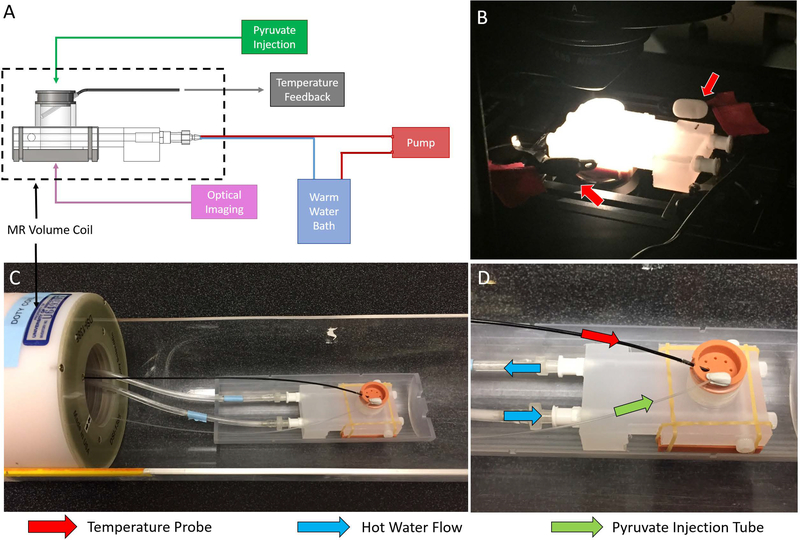
A schematic of the bioreactor and supporting systems can be seen in Figure 4A with key features labeled. Images of bioreactors being used for optical imaging and for MRS experiments can be seen in Figure 4B and Figure 4C-D, respectively. In order to ensure contact with the cover glass for optical imaging, some medium was removed from the chamber and reserved. This medium was returned prior to the MRS studies (Figure 2B-C). The volume coil used for the MRS experiments can be seen in Figure 4C. A heated chamber around the microscope or a warm water flow system through the bioreactor base maintained the sample at biological temperatures (36 ± 1°C) during optical imaging and MRS experiments, respectively. Temperature feedback was achieved with temperature sensors and their positions for each setup are indicated (Figure 4B and Figure 4D).
Figure 4:
Schematic of the bioreactor layout with respect to supporting equipment (A). Optical setup (B) and MRS setup (C, D) of the bioreactor. Temperature feedback for optical experiments was achieved with two independent temperature probes placed near the sample (B, red arrows). The volume coil used in the MRS experiments is shown (C). In MRS experiments, temperature feedback was achieved through the use of an MR-compatible temperature probe placed into a thermal well in the sample (D, red arrow). During MRS experiments, pyruvate was injected into the sample through an injection tube (D, green arrow) and hot water was pumped through channels around the sample to maintain it at 36 ± 1°C (D, blue arrows).
Data Collection
Both NADH FLIM and MRS were performed on each 3D cell culture in a single imaging day. 2D FLIM was performed with a 20x air objective (NA =0.75) (Nikon) on a custom multiphoton laser scanning system built around an inverted Nikon TE300 microscope [37]–[39]. NADH was excited with a multiphoton excitation of 740 nm and a 450/70 bandpass filter (Semrock, Rochester, NY, USA) was used to collect emitted light. Collection times for each field of view were 150 seconds.
MRS experiments were performed in a 4.7T preclinical horizontal bore MRI scanner (Agilent, Palo Alto, CA). Shimming was performed on the proton channel to ensure a uniform field over the sample volume before switching to the carbon channel. A thermally polarized 13C urea phantom was placed adjacent to the sample for calibration in the carbon channel. For MRS experiments, 30 μL aliquots of [1-13C] pyruvic acid (PA) (Cambridge Isotope Laboratories Inc., Tewksbury, MA) and 15 mM trityl radical (Ox063, GE Healthcare, Waukesha, WI) were polarized in a Hypersense polarizer (Oxford Instruments, Abingdon, United Kingdom) for ~1 hour [40]. Samples were dissoluted with 4 mL of solvent containing 1.2 mL 426 mM NaOH, 1.4 mL 400 mM Tris buffer and 1.4 mL 250 mg/L EDTA. [1-13C] PA, with a final polarization fraction of approximately ~18%, was drawn off and 150 μL were rapidly injected into the sample volume of the bioreactor.
Dynamic global MR spectra (flip angle (FA)=10º, repetition time (TR)=3000 ms) were acquired with a dual-tuned 1H/13C volume coil (Doty Scientific, Columbia, SC). Overall diameter and length of the coil were 10.5 cm and 13 cm, respectively, and the entire bioreactor fit inside the 5 cm coil bore. N=9 FLIM (3 fields of view each from 3 bioreactor preparations) and N=5 MRS (2 bioreactors for MRS only in addition to the 3 used for combined FLIM and MRS studies) measurements were acquired for each state (glucose-starved and non-starved).
For the presented data, FLIM was performed first, followed by MRS, although the order could be reversed for future experiments. FLIM took between 1 and 2 hours including setup and MRS took between 1.5 and 2.5 hours including setup and hyperpolarization of pyruvate. Working in pairs minimized setup times for each modality. Additionally, the bioreactor was placed in a live-cell incubator during downtime for setups when possible.
Data Analysis
Mean lifetimes for cellular images were calculated using SPCImage (Becker & Hickl GmbH, Berlin, Germany) for each 2D field of view (FOV). Intensity thresholds were selected for each image prior to processing to dismiss pixels with minimal signal. For pixels above threshold, the time from excitation of each photon was averaged to yield the mean fluorescence lifetime for that pixel. This value was then either represented as a colormap image or averaged across the image to generate a single value for the FOV. To examine the free and protein-bound components of NADH fluorescence lifetime, a two-component decay curve was fitted to the photon counting data for each pixel. The amplitude of the fitted time constants, τ1 and τ2, corresponded to the individual components of the fluorescence lifetimes. All Χ2 values were minimized to ensure the best fit. The mean fluorescence lifetimes and individual components of each pixel were averaged to derive mean NADH fluorescence lifetime and individual component values for each field of view. These values were used to generate the boxplots in the final results. Instrument response functions were collected with urea crystals during each imaging day to account for variability in detection equipment between experiments.
Raw MRS data was reconstructed using an in-house script for MATLAB (Mathworks, Natick, MA, USA). Lac:Pyr ratios were calculated from temporally-summed MR spectra to improve signal-to-noise after background subtraction. Spectra were summed over the same range of 65 time points within the dynamic spectra collected. Lactate and pyruvate signals were integrated from summed spectra over a span of 0.5 ppm centered on the peak in each case. Box plots were generated using R (https://www.r-project.org/), an open source statistical framework. Statistical significance was determined using the Wilcoxon rank-sum test within the R framework.
Results
Meeting the original design criteria of this device resulted in several novel design elements (Figure 5). Maintaining biological temperatures during imaging was important for these studies, as changes in temperature can cause changes in cellular metabolism. To accomplish this with MR-compatible materials, hot water flow was used to warm samples to 36 ± 1°C (Figure 6B). Temperature feedback was achieved with an MR-compatible temperature probe inserted into a sealed capillary tube inside the sample volume of the bioreactor (Figure 5B)
Figure 5:
Key design elements of the bioreactor. (A) Orientation for the cutaway views in panels B, C and D. (B) Top view (top) and side view cutaway (bottom) with the components used for temperature control highlighted in shades of red, including the hot water flow path, the temperature probe and the capillary tube. (C) Side view cutaway with the three components of the optical window highlighted in shades of blue, including the cover glass, silicone gasket and compression piece. (D) Side view cutaway with the Bioreactor Top highlighted in orange with key features labeled, including the customizable ports and the edge for easy sealing with Parafilm. (E) Top view of the bioreactor assembly with the location of a potential surface coil highlighted with a dashed grey line.
Figure 6:
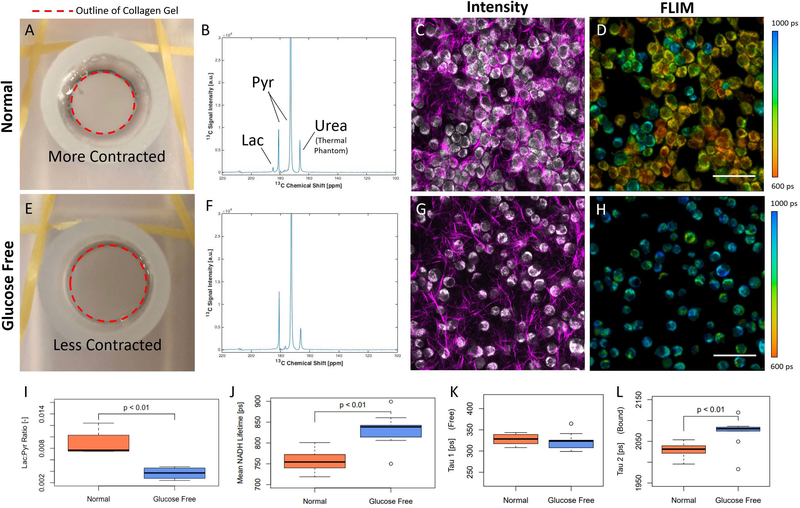
Results of the glucose deprivation study. Example results from a sample cultured in normal medium (A-D). Example results from a sample cultured with glucose-free medium (E-H). Top view picture of a collagen gel in medium (A), example time integrated 13C MR spectra (B), intensity image showing NADH intensity (gray) and collagen from SHG (magenta) (C) and FLIM color-mapped image (D) for cells in normal medium. Top view picture of a collagen gel in medium (E), example time integrated 13C MR spectra (F), intensity image showing NADH intensity (gray) and collagen from SHG (magenta) (G) and FLIM color-mapped image (H) for cells in glucose-free medium. Cell density in glucose-free medium was observed to decrease compared to normal medium, likely because of less gel contraction. Quantitation of hyperpolarized 13C Lac:Pyr ratios comparing normal and glucose-free states for all MRS measurements (I). Mean NADH lifetime measurements for normal and glucose-free conditions for all FLIM measurements (J). τ1, the shorter component of fluorescent lifetime indicative of free NADH, for normal and glucose-free conditions for all FLIM measurements (K). τ2, the longer component of fluorescent lifetime indicative of protein-bound NADH, for normal and glucose-free conditions for all FLIM measurements (L). A thermally polarized 13C urea phantom was included for calibration in MRS studies. Scale bars are 100 microns.
The creation of an optical window that is water-tight, re-usable, easy to assemble and accessible to objectives with low working distances represented a unique engineering challenge. Here, it is accomplished with an assembly of three components (Figure 5C). High resolution grade microcopy cover glass (#1.5) is placed on the bottom of the bioreactor, forming the bottom to the sample chamber, held in place with vacuum grease (Dow-Corning). The cover glass is secured by a rubber gasket and a compression piece (Figure 5C). Easy to apply rubber bands exerted uniform pressure, effectively sealing the device from leaks.
Another engineering challenge addressed with design elements of this bioreactor is the flexibility to translate this device to different hardware, including other horizontal bore MR systems, different acquisition coils, additional sample measurement tools and other inverted optical imaging systems. The Bioreactor Top is 3D printed and can be customized (Figure 5D) to include an array of different ports and sizes of ports to interact with the sample volume in any number of desired ways. The overall shape of the bioreactor can fit on a number of adjustable stages for inverted optical imaging systems. Finally, the cylindrical sample volume allows for the use of a surface coil for MR acquisition, if desired (Figure 5E).
To assess how well the bioreactor serves as a device for determining metabolic status across imaging platforms with different spatial resolution, a proof of principle study was conducted. The goal of this study was to determine whether the two modalities connected with this device could detect similar changes in metabolism with a known perturbation. Thus, the impact of glucose deprivation on 4T1 cell metabolism was tested using the bioreactor to assess feasibility for detecting changes in Lac:Pyr and mean NADH lifetime using MRS and FLIM, respectively.
The results of this study are shown in Figure 6. Data from a sample cultured in normal medium and from a sample with glucose-free medium can be seen in Figure 6A-D and Figure 6E-H, respectively. A qualitative difference between the two states that is apparent in the optical images (Figure 6C-D, G-H), is a decrease in cellular density from the normal medium samples to the glucose-free samples. A contributing factor to this reduction is the contractedness of the collagen gels themselves. Collagen gels in normal medium were visibly more contracted than those in glucose-free medium (Figure 6A, E). This change in local collagen density is also apparent by looking at the collagen signal (magenta) in the intensity images (Figure 6C, G).
MRS experiments with [1-13C] PA in vitro allow an overall measurement of the lactate production from an entire population of cells. Example summed 13C MR spectra from 4T1 cells cultured in normal medium and from 4T1 cells cultured in glucose-free medium can be seen in Figure 6B and Figure 6F, respectively. Lac:Pyr ratio of 4T1 cells cultured in glucose-free medium decreased by 60% (P<0.01) compared to those cultured in normal medium (Figure 6I). This decrease in the Lac:Pyr suggests a reduction in glycolysis. MRS signal strengths are largely dependent on the diffusion of pyruvate into the collagen gel. Diffusion into the gel was estimated and pyruvate concentration profiles were calculated at different time points during the experiments (Figure 1B). According to this model, small amounts of pyruvate will reach 1.5 mm into the gel by t = 3 minutes after injection. Furthermore, the depth at which there is 10% of the initial concentration of pyruvate is 0.5 mm, 0.75 mm and 0.8 mm for t = 1, 2 and 3 minutes, respectively.
NADH FLIM allows for the probing of the free and bound state of NADH and also an overall assessment of cell morphology. In addition to the cell density changes seen between the two states (Figure 6D, H), morphological changes were visually pronounced as well - 4T1 cells cultured in normal medium are densely clumped with multiple protrusions in many cases, while the cells that are glucose deprived are rounder in shape and less aggregated. Measurements of mean NADH lifetime of cells cultured in glucose-free media increased by 10% (P<0.01) compared to those cells cultured in normal medium (Figure 6J).
Looking at the individual components of the fluorescent lifetime data from the fitted model reveals more information about the changes occurring in these cells. The short-lived component of the fitted two-component decay model, τ1, did not exhibit a significant change (p=0.224) between the two states (Figure 6K), while the long-lived component of the fitted decay model, τ2, increased significantly (p<0.01) (Figure 6L). This suggests that the increase in mean lifetime was caused by an increase in the fraction of bound NADH.
Discussion
The novel bioreactor system described here can be used to perform both MRS measurements on a 3D cell culture using a horizontal bore preclinical MRI system and FLIM measurements on individual cells within that culture using an inverted multiphoton laser-scanning microscope. This enables the examination of the heterogeneity of a sample on the cellular scale in the context of a single, sample-wide MRS result. Additionally, it features several novel elements that accomplish the original design requirements of this device. This includes (1) maintaining large 3D cell cultures, (2) containing only MR-compatible materials, (3) integrating a versatile optical window and (4) retaining flexibility of design to allow for hardware changes. This device represents an important bridge between the whole sample measurements of MRS and the cellular scale of optical imaging.
The glucose deprivation of 4T1 murine breast cancer cells was used as a test application to assess feasibility of the bioreactor for metabolic studies in 3D cell culture. 4T1 cells are extremely glycolytic and sensitive to glucose deprivation [41], and therefore provide a useful benchmark for testing the ability of this device to collect both types of metabolic data under different conditions. The mean NADH lifetimes increased significantly while lactate production decreased significantly between 4T1 cells in normal medium and glucose-deprived 4T1 cells. Furthermore, the τ1 values did not change between the two states, while the τ2 values increased significantly from normal medium to glucose-free medium, suggesting that the increase in mean lifetime observed was primarily caused by a change in bound NADH. Cell density in glucose-free medium was also observed to decrease; the cells without glucose contracted the collagen gels much less than cells in normal medium and this likely contributed to observed changes in cell signaling and overall metabolic rate.
These results are consistent with a shift away from glycolysis, likely due to a combination of a reduction in glycolysis relative to other metabolic processes and a reduction in glycolysis from an overall decrease in total cellular metabolism. The mechanisms behind these metabolic shifts are extremely complex, and this complexity highlights the need for devices like this that allow for the interrogation of samples at multiple spatial scales under controlled interventions within the same 3D cell preparation. Investigations into the metabolic impact of cell culture methods, different cell lines and varied treatment regimens are part of future work. Possible future areas of research also include the use of other hyperpolarized MRS agents, analysis of the time-dependence of MRS signals and different optical agents, where subtle changes in metabolism might be teased out in more detail. Beyond metabolism, this bioreactor design may also be suitable for other applications where MRI and optical imaging offer complementary measures.
In particular, future studies that examine parameters common to both modalities would exploit the usefulness of this device. While the two parameters studied here are related, they are not the same. There are many exogenous fluorescent labels to proteins that also act on hyperpolarized MRS substrates that could be used for a direct comparison of a single parameter. However, this would require an additional treatment of the cell cultures with the desired fluorescent label rather than the more accessible intrinsic fluorescence of NADH used in the current work, demonstrating the basic utility of the device and approach.
The bioreactor system was designed to be easily replicated by others through minimizing the number of custom parts. There were four custom parts in the design and their fabrication was described in detail. Open-source design files are provided, as well as any files required for fabrication, including .STL files for printed pieces and .NC files for CNC machining. These files can be used with machinery and equipment that is common to many fabrication facilities to replicate and iterate on this device.
There are several limitations to the current design of the device that will be addressed in future work. Potential hardware improvements to the system include adding a convenient media flow solution for longitudinal studies and methods to increase drug perfusion to the gel. Perfusion of the collagen gel is a potential source of heterogeneity of substrate distribution for the MRS studies. A lack of perfusion of the drug into the collagen matrix within the observable lifetime of the hyperpolarization would also cause reduced signal within the sample. Therefore, improved lactate signal-to-noise could potentially be achieved by modified methods for casting gel samples with higher surface areas to increase perfusion.
To get a sense of the depth of penetration of pyruvate into the gel during the time course of the experiments presented, a simple model of diffusion was constructed. The results of this model (Supporting Information Figure S2) indicate that small amounts of pyruvate should diffuse as far as 1.5 mm into the gel by the end of an MRS experiment, while the depth at which 10% of the initial concentration of pyruvate is present is about 0.8 mm at the same time point. This indicates a measurement depth that is slightly deeper, but still on the same order to that of FLIM. Several simplifying assumptions were made to enable this model. Notably, the value used for the diffusion coefficient of pyruvate was measured in vivo [36], providing a conservative estimate. Nonetheless, this model provides a useful frame of reference and an increased understanding of the collected data in the bioreactor.
The use of a horizontal bore magnet made the incorporation of an optical window and the use of a large 3D collagen matrix more feasible, which would be much more challenging within the small space of an NMR tube designed for a vertical magnet. While the horizontal bore design enables in vitro studies to be performed on the same system as preclinical in vivo studies, it does limit the use of the device to lower field strengths and limits coil sensitivity. The novelty of using 3D collagen gels provides a more realistic cellular environment compared to other cell culturing techniques, and this attribute ultimately outweighed the limitations that the design introduced.
Furthermore, collagen gels provided both a microenvironment for the cells in this study that is more similar to in vivo than 2D cell culturing methods and a 3D volume to increase overall signal for the MRS experiments. The properties of the gels themselves may affect the metabolism of cells within them. In particular, collagen density and cellular confluence are known to have an effect on the metabolic signature of cells. While the initial number of cells and collagen density were held constant in the presented work, the influence of collagen density on metabolism could be the subject of interesting follow-up studies.
Finally, long-term cell viability with this device will need further investigation for some applications. While all materials were tested for cytotoxicity, the viability of these 3D cell cultures during the duration of long studies will need to be evaluated on the benchtop for given study conditions. Additionally, some experiments require the control of the oxygen environment of cell cultures. Future work will explore the effect of the gas environment of these cultures on cell viability, using a modified top piece to the bioreactor that allows for gas exchange.
This bioreactor constitutes an important step forward in multiscale imaging by bridging optical imaging and MRS onto the same platform. In particular, the ability to examine the heterogeneity of a sample on the cellular scale with optical imaging in the context of a single sample-wide MRS result provides the basis for compelling continuing work. To that end, the device designs have been made open-source, so that it can be iterated upon for additional preclinical research applications.
Supplementary Material
Supporting Information Figure S1: Key differences between common cell culturing techniques. Schematics representing common cell culturing techniques are shown in increasing similarity to in vivo, from left to right. Attributes of each that are important for similarity to in vivo are listed below each with an indicator as to whether each technique does or does not have a given attribute.
Supporting Information Figure S2: (A) Schematic of the collagen gel leading up to and after a pyruvate injection. Immediately following injection, there is a bolus of pyruvate at the top of the sample volume. The pyruvate then diffuses through the medium and then into the gel. (B) Simplified case considered in this diffusion model, with an initial concentration in the medium and a zero concentration within the gel at t ~ 0. (C) Pyruvate concentration profiles at the edge of the gel (solid black line) for t = 1, 2 and 3 minutes post injection. The 10% line (dashed black line) can be used to estimate the depth at which there is 10% of the initial pyruvate concentration at any given time point.
Supporting Information Figure S3: The 3D nature of optical sectioning. Cutaway view of the sample area for the FLIM experiments (left). Medium was removed and reserved to ensure that the gel contacted the cover glass for imaging. The region of the collagen gel that can be probed by optical imaging is shown in red. An orthogonal view of a z-stack of images taken through a collagen gel (inlay, right). Each image was a taken at a different depth into the sample. The signal is from NADH intensity to show the cells inside the collagen gel.
Supporting Information Figure S4: Assessment of MDA-231 cell growth on various materials. A) Brightfield images of cells grown for 3 days in wells either with no material or in the presence of materials potentially utilized for the bioreactor, including polystyrene (PS) (cell culture plastic control), polypropylene (PP), silicone rubber (SR), Delrin (del) or RC31 (RC31). (B) Graph showing the change, over 3 days, in the density of cells grown in the presence of various materials, normalized to the cell density of that treatment on day 1. (P=0.0113 for materials comparison, two-way ANOVA; * P<0.05, **<0.01, Dunnett’s multiple comparison test vs. no material control, day 3 only). C) Graph showing the cell density on day 3 relative to PS control, which takes into account mechanical disruption of cell contacts resulting from physical presence of the material wafer in the well. (P=0.008, one-way ANOVA; Dunnett’s multiple comparisons test indicate no significant differences when compared to control PS). Scale bar is 100 microns.
Acknowledgments
The authors would like to acknowledge Brett Morris and Patricia Keely for helpful discussions on, and help with, preliminary experiments with collagen polymerization and cell culture techniques. The authors would also to thank George Petry for his help with machining considerations during the construction of the various iterations of the bioreactor. Funding for this project was provided by The Morgridge Institute for Research, the Departments of Medical Physics and Radiology at UW-Madison, UL1TR000427 and TL1TR000429 to UW ICTR from NIH/NCATS, T32CA00926 to Department of Medical Physics from NIH/NCI, NIH R01 CA142833, CA185251 and CA179556 to SMP, and the AAPM 2014 Graduate Fellowship.
Abbreviations
- CNC
Computer Numerical Control
- DNP
Dynamic Nuclear Polarization
- FLIM
Fluorescence Lifetime Imaging
- LDH
Lactate Dehydrogenase
- MR
Magnetic Resonance
- MRS
Magnetic Resonance Spectroscopy
- NADH
Nicotinamide Adenine Dinucleotide
- NMR
Nuclear Magnetic Resonance
- PA
Pyruvic Acid
- 3D
Three-Dimensional
- 2D
Two-Dimensional
References
- [1].Hanahan D and Weinberg RA, “The hallmarks of cancer,” Cell, vol. 100, no. 1, pp. 57–70, January 2000. [DOI] [PubMed] [Google Scholar]
- [2].Hanahan D and Weinberg RA, “Hallmarks of cancer: the next generation,” Cell, vol. 144, no. 5, pp. 646–674, March 2011. [DOI] [PubMed] [Google Scholar]
- [3].Hsu C-C, Tseng L-M, and Lee H-C, “Role of mitochondrial dysfunction in cancer progression,” Exp. Biol. Med. Maywood NJ, vol. 241, no. 12, pp. 1281–1295, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Phan LM, Yeung S-CJ, and Lee M-H, “Cancer metabolic reprogramming: importance, main features, and potentials for precise targeted anti-cancer therapies,” Cancer Biol. Med, vol. 11, no. 1, pp. 1–19, March 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Kelly RS, Vander Heiden MG, Giovannucci E, and Mucci LA, “Metabolomic Biomarkers of Prostate Cancer: Prediction, Diagnosis, Progression, Prognosis, and Recurrence,” Cancer Epidemiol. Biomark. Prev. Publ. Am. Assoc. Cancer Res. Cosponsored Am. Soc. Prev. Oncol, vol. 25, no. 6, pp. 887–906, June 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Chang C-H et al. , “Metabolic Competition in the Tumor Microenvironment Is a Driver of Cancer Progression,” Cell, vol. 162, no. 6, pp. 1229–1241, September 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Le A et al. , “Inhibition of lactate dehydrogenase A induces oxidative stress and inhibits tumor progression,” Proc. Natl. Acad. Sci. U. S. A, vol. 107, no. 5, pp. 2037–2042, February 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Belenky P, Bogan KL, and Brenner C, “NAD+ metabolism in health and disease,” Trends Biochem. Sci, vol. 32, no. 1, pp. 12–19, January 2007. [DOI] [PubMed] [Google Scholar]
- [9].Warburg O and Christian W, “Pyridin, der wasserstoffübertragende Bestandteil von Gärungsfermenten,” Helv. Chim. Acta, vol. 19, no. 1, pp. E79–E88, January 1936. [Google Scholar]
- [10].Harden A and Young WJ, “The Alcoholic Ferment of Yeast-Juice. Part II.--The Conferment of Yeast-Juice,” Proc. R. Soc. Lond. B Biol. Sci, vol. 78, no. 526, pp. 369–375, October 1906. [Google Scholar]
- [11].Brindle K, “Watching tumours gasp and die with MRI: the promise of hyperpolarised 13C MR spectroscopic imaging,” Br. J. Radiol, vol. 85, no. 1014, pp. 697–708, June 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Gallagher FA, Bohndiek SE, Kettunen MI, Lewis DY, Soloviev D, and Brindle KM, “Hyperpolarized 13C MRI and PET: in vivo tumor biochemistry,” J. Nucl. Med. Off. Publ. Soc. Nucl. Med, vol. 52, no. 9, pp. 1333–1336, September 2011. [DOI] [PubMed] [Google Scholar]
- [13].Kennedy BWC, Kettunen MI, Hu D-E, and Brindle KM, “Probing lactate dehydrogenase activity in tumors by measuring hydrogen/deuterium exchange in hyperpolarized l-[1-(13)C,U-(2)H]lactate,” J. Am. Chem. Soc, vol. 134, no. 10, pp. 4969–4977, March 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Chang C-W, Sud D, and Mycek M-A, “Fluorescence lifetime imaging microscopy,” Methods Cell Biol, vol. 81, pp. 495–524, 2007. [DOI] [PubMed] [Google Scholar]
- [15].Elson DS et al. , “Real-time time-domain fluorescence lifetime imaging including single-shot acquisition with a segmented optical image intensifier,” New J. Phys, vol. 6, no. 1, p. 180, 2004. [Google Scholar]
- [16].Drozdowicz-Tomsia K et al. , “Multiphoton fluorescence lifetime imaging microscopy reveals free-to-bound NADH ratio changes associated with metabolic inhibition,” J. Biomed. Opt, vol. 19, no. 8, p. 86016, August 2014. [DOI] [PubMed] [Google Scholar]
- [17].Golman K, in ‘t Zandt R, and Thaning M, “Real-time metabolic imaging,” Proc. Natl. Acad. Sci, vol. 103, no. 30, pp. 11270–11275, July 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Serrao EM and Brindle KM, “Potential Clinical Roles for Metabolic Imaging with Hyperpolarized [1–13C]Pyruvate,” Front. Oncol, vol. 6, March 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Keshari KR et al. , “Hyperpolarized (13)C spectroscopy and an NMR-compatible bioreactor system for the investigation of real-time cellular metabolism,” Magn. Reson. Med, vol. 63, no. 2, pp. 322–329, February 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Tessem M-B et al. , “Evaluation of lactate and alanine as metabolic biomarkers of prostate cancer using 1H HR-MAS spectroscopy of biopsy tissues,” Magn. Reson. Med, vol. 60, no. 3, pp. 510–516, September 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Campos D et al. , “Radiation Promptly Alters Cancer Live Cell Metabolic Fluxes: An In Vitro Demonstration,” Radiat. Res, vol. 185, no. 5, pp. 496–504, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Provenzano PP, Eliceiri KW, Campbell JM, Inman DR, White JG, and Keely PJ, “Collagen reorganization at the tumor-stromal interface facilitates local invasion,” BMC Med, vol. 4, no. 1, p. 38, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Even-Ram S and Yamada KM, “Cell migration in 3D matrix,” Curr. Opin. Cell Biol, vol. 17, no. 5, pp. 524–532, October 2005. [DOI] [PubMed] [Google Scholar]
- [24].Burkel B, Morris BA, Ponik SM, Riching KM, Eliceiri KW, and Keely PJ, “Preparation of 3D Collagen Gels and Microchannels for the Study of 3D Interactions In Vivo,” J. Vis. Exp. JoVE, no. 111, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Szulczewski JM et al. , “In Vivo Visualization of Stromal Macrophages via label-free FLIM-based metabolite imaging,” Sci. Rep, vol. 6, May 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Gaspar DA, Gomide V, and Monteiro FJ, “The role of perfusion bioreactors in bone tissue engineering,” Biomatter, vol. 2, no. 4, pp. 167–175, October 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Sailon AM, Allori AC, Davidson EH, Reformat DD, Allen RJ, and Warren SM, “A Novel Flow-Perfusion Bioreactor Supports 3D Dynamic Cell Culture,” J. Biomed. Biotechnol, vol. 2009, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tandon N, Taubman A, Cimetta E, Saccenti L, and Vunjak-Novakovic G, “Portable bioreactor for perfusion and electrical stimulation of engineered cardiac tissue,” Conf. Proc. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc. IEEE Eng. Med. Biol. Soc. Annu. Conf, vol. 2013, pp. 6219–6223, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Lauritsen S et al. , “Bioreactor for quantification of cell metabolism by MR-hyperpolarization,” Biomed. Phys. Eng. Express, vol. 1, no. 4, p. 47003, 2015. [Google Scholar]
- [30].Lee J et al. , “High-Throughput Hyperpolarized 13C Metabolic Investigations using a Multi-Channel Acquisition System,” J. Magn. Reson. San Diego Calif 1997, vol. 260, pp. 20–27, November 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Sud D, Mehta G, Mehta K, Linderman J, Takayama S, and Mycek M-A, “Optical imaging in microfluidic bioreactors enables oxygen monitoring for continuous cell culture,” J. Biomed. Opt, vol. 11, no. 5, p. 50504, October 2006. [DOI] [PubMed] [Google Scholar]
- [32].Lakowicz JR, Szmacinski H, Nowaczyk K, and Johnson ML, “Fluorescence lifetime imaging of free and protein-bound NADH,” Proc. Natl. Acad. Sci. U. S. A, vol. 89, no. 4, pp. 1271–1275, February 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lakowicz JR, Szmacinski H, Nowaczyk K, and Johnson ML, “Fluorescence lifetime imaging of free and protein-bound NADH,” Proc. Natl. Acad. Sci. U. S. A, vol. 89, no. 4, pp. 1271–1275, February 1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Walsh AJ, Sharick JT, Skala MC, and Beier HT, “Temporal binning of time-correlated single photon counting data improves exponential decay fits and imaging speed,” Biomed. Opt. Express, vol. 7, no. 4, pp. 1385–1399, April 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Crank J, The Mathematics of Diffusion, 2nd ed. Oxford: Oxford University Press, 1980. [Google Scholar]
- [36].Søgaard LV, Schilling F, Janich MA, Menzel MI, and Ardenkjaer-Larsen JH, “In vivo measurement of apparent diffusion coefficients of hyperpolarized 13C-labeled metabolites,” NMR Biomed, vol. 27, no. 5, pp. 561–569, May 2014. [DOI] [PubMed] [Google Scholar]
- [37].Wokosin DL, Squirrell JM, Eliceiri KW, and White JG, “Optical workstation with concurrent, independent multiphoton imaging and experimental laser microbeam capabilities,” Rev. Sci. Instrum, vol. 74, no. 1, pp. 193–201, January 2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Yan L, Rueden CT, White JG, and Eliceiri KW, “Applications of combined spectral lifetime microscopy for biology,” BioTechniques, vol. 41, no. 3, p. 249, 251, 253 passim, September 2006. [DOI] [PubMed] [Google Scholar]
- [39].Conklin MW, Provenzano PP, Eliceiri KW, Sullivan R, and Keely PJ, “Fluorescence Lifetime Imaging of Endogenous Fluorophores in Histopathology Sections Reveals Differences Between Normal and Tumor Epithelium in Carcinoma In Situ of the Breast,” Cell Biochem. Biophys, vol. 53, no. 3, pp. 145–157, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Gordon JW, Niles DJ, Adamson EB, Johnson KM, and Fain SB, “Application of flow sensitive gradients for improved measures of metabolism using hyperpolarized (13) c MRI,” Magn. Reson. Med, vol. 75, no. 3, pp. 1242–1248, March 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Morris BA et al. , “Collagen Matrix Density Drives the Metabolic Shift in Breast Cancer Cells,” EBioMedicine, vol. 13, pp. 146–156, October 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information Figure S1: Key differences between common cell culturing techniques. Schematics representing common cell culturing techniques are shown in increasing similarity to in vivo, from left to right. Attributes of each that are important for similarity to in vivo are listed below each with an indicator as to whether each technique does or does not have a given attribute.
Supporting Information Figure S2: (A) Schematic of the collagen gel leading up to and after a pyruvate injection. Immediately following injection, there is a bolus of pyruvate at the top of the sample volume. The pyruvate then diffuses through the medium and then into the gel. (B) Simplified case considered in this diffusion model, with an initial concentration in the medium and a zero concentration within the gel at t ~ 0. (C) Pyruvate concentration profiles at the edge of the gel (solid black line) for t = 1, 2 and 3 minutes post injection. The 10% line (dashed black line) can be used to estimate the depth at which there is 10% of the initial pyruvate concentration at any given time point.
Supporting Information Figure S3: The 3D nature of optical sectioning. Cutaway view of the sample area for the FLIM experiments (left). Medium was removed and reserved to ensure that the gel contacted the cover glass for imaging. The region of the collagen gel that can be probed by optical imaging is shown in red. An orthogonal view of a z-stack of images taken through a collagen gel (inlay, right). Each image was a taken at a different depth into the sample. The signal is from NADH intensity to show the cells inside the collagen gel.
Supporting Information Figure S4: Assessment of MDA-231 cell growth on various materials. A) Brightfield images of cells grown for 3 days in wells either with no material or in the presence of materials potentially utilized for the bioreactor, including polystyrene (PS) (cell culture plastic control), polypropylene (PP), silicone rubber (SR), Delrin (del) or RC31 (RC31). (B) Graph showing the change, over 3 days, in the density of cells grown in the presence of various materials, normalized to the cell density of that treatment on day 1. (P=0.0113 for materials comparison, two-way ANOVA; * P<0.05, **<0.01, Dunnett’s multiple comparison test vs. no material control, day 3 only). C) Graph showing the cell density on day 3 relative to PS control, which takes into account mechanical disruption of cell contacts resulting from physical presence of the material wafer in the well. (P=0.008, one-way ANOVA; Dunnett’s multiple comparisons test indicate no significant differences when compared to control PS). Scale bar is 100 microns.