Abstract
Background:
Externalizing disorders are known to be partly heritable, but the biological pathways linking genetic risk to the manifestation of these costly behaviors remain under investigation. This study sought to identify neural phenotypes associated with genomic vulnerability for externalizing disorders.
Methods:
155 White, non-Hispanic veterans were genotyped using a genome-wide array and underwent resting-state functional magnetic resonance imaging. Genetic susceptibility was assessed using an independently-developed polygenic score for externalizing, and functional neural networks were identified using graph theory-based network analysis. Tasks of inhibitory control and psychiatric diagnosis (alcohol/substance use disorders) were used to measure externalizing phenotypes.
Results:
A polygenic externalizing disorder score (PS) predicted connectivity in a brain circuit (10 nodes, 9 links) centered on left amygdala that included several cortical (bilateral IFG pars triangularis, left rostral anterior cingulate cortex) and subcortical (bilateral amygdala, hippocampus, and striatum) regions. Directional analyses revealed that bilateral amygdala influenced left prefrontal cortex (IFG) in participants scoring higher on the externalizing PS, whereas the opposite direction of influence was observed for those scoring lower on the PS. Polygenic variation was also associated with higher Participation Coefficient for bilateral amygdala and left rACC, suggesting that genes related to externalizing modulated the extent to which these nodes functioned as communication hubs.
Conclusions:
Findings suggest that externalizing polygenic risk is associated with disrupted connectivity in a neural network implicated in emotion regulation, impulse control, and reinforcement learning. Results provide evidence that this network represents a genetically-associated neurobiological vulnerability for externalizing disorders.
Keywords: polygenic risk score, neural circuits, substance use, alcohol use, disinhibition
Introduction
The common co-occurrence of behavioral disinhibition, substance use disorders, antisocial behavior, and conduct problems reflects a broad dimension of psychopathology termed the externalizing spectrum (Krueger et al., 2002). Externalizing disorders are associated with poor mental health outcomes, premature death (Eaton et al., 2013; Odgers et al., 2007) and are estimated to cost more than 417 billion dollars annually in the United States alone (National Institute on Drug Abuse, 2015). Twin studies have repeatedly found that externalizing phenotypes are highly heritable across different developmental periods and point to behavioral disinhibition as a common feature uniting externalizing disorders (Tarter et al., 2003; Young et al., 2009; Krueger and Markon, 2006). The goal of this study was to identify neural phenotypes of externalizing by investigating associations between measured genetic risk for externalizing disorders and brain networks. Based on growing evidence that connectivity in resting-state functional networks reflects heritable differences in brain organization and function (Glahn et al., 2010; Smit et al., 2008), we examined patterns of resting-state connectivity as plausible heritable neural phenotypes for externalizing.
Research on the polygenic structure of psychiatric disorders is advancing rapidly through the development of polygenic scores (PSs) derived from genome-wide association studies. PSs are summary measures that weight single nucleotide polymorphism (SNPs) from across the genome to provide a measured index of the genetic propensity for a given disorder. Salvatore and colleagues (Salvatore et al., 2015) recently developed an externalizing polygenic score in adults with alcohol dependence and showed that it explained 6% of the variance in externalizing disorders (alcohol/substance use disorders, antisocial behavior) and 2–7% of the variance in other disinhibited phenotypes (e.g., impulsiveness). Building on these findings, we recently replicated the association between the polygenic score and externalizing symptoms in trauma-exposed veterans and found that the polygenic score predicted impaired performance on inhibitory control tasks (Sadeh et al., 2016). Thus, initial validation studies find reliable polygenic associations with externalizing phenotypes and illustrate the promise of this approach for identifying heritable neural mechanisms associated with genetic predispositions for externalizing. An important next step in this line of research is to examine the neurobiological correlates of the externalizing PS. Research that aims to characterize the influences of genetic variations on brain structures and function has grown exponentially over the past several years, and yielded greater insight into the heritability of neurobiological risk factors for psychopathology, including emerging neurogenetics research on externalizing disorders (e.g., Heitzeg et al., 2014; Karoly, et al., 2013; Shehzad et al., 2012). To our knowledge, no research to date has examined whether the externalizing PS relates to neurobiological function or structure – potentially crucial pathways by which genes create vulnerability for externalizing disorders.
Resting-state functional connectivity provides a measure of neural activation in spatially-distributed brain networks under conditions of rest (i.e., when participants are not engaged in an explicit, goal-directed task; Beckmann et al., 2005). Unlike task-based measures of brain activation, resting-state connectivity is relatively stable over time (Zuo and Xing, 2014), suggesting it indexes more trait-like brain networks and, therefore, acts as a potentially valuable medium for investigating genetic influences on brain networks. Indeed, there is growing data to support the heritability of resting-state neural activity, including aberrant connectivity in these networks in psychiatric populations (Glahn et al., 2010; Smit et al., 2008). Recently developed graph theory tools can be applied to resting-state connectivity data to investigate brain organization and function in psychiatric populations, potentially identifying brain networks that exhibit differential features across psychiatric disorders (Fornito and Bullmore, 2015). Graph theory is an analytic tool involving the calculation of network properties that can be used to examine the organization of network connections and, thus, the functional capabilities of a network and the role that nodes (brain regions) play in global and local circuitry (Stam, 2014). For example, the graph theory property Participation Coefficient measures the extent to which a brain region serves as a “hub” that connects different modules (sub-networks). By assessing distributed functional networks, these methods capture greater complexity in the neural bases of psychiatric disorders than is possible when limiting analysis to single regions or even coupling between pairs of regions, approaches that have traditionally dominated research on the neurobiology of psychiatric disorders. This makes graph theory a potentially powerful tool for uncovering large-scale neural circuits that enable the phenotypic complexity inherent in externalizing disorders.
The goals of this study were to (i) identify genetically-associated neural networks for externalizing by examining polygenic associations with resting-state functional connectivity, and (ii) examine associations between the externalizing PS, neural networks, and behavioral phenotypes of externalizing (e.g., alcohol and substance use disorders). Based on our previous finding that the externalizing PS was associated with poorer performance on tasks of inhibitory control (Sadeh et al., 2016), we expected this PS to predict neural network resting-state connectivity and function in regions of the brain that are central to maintaining such control (e.g., inferior frontal gyrus; dorsolateral prefrontal cortex, anterior cingulate; Aron et al., 2004; Criaud and Boulinguez, 2013; Nee et al., 2007). In light of evidence that externalizing disorders are characterized by dysfunction in mesolimbic reward and emotional-salience systems (Durazzo et al., 2011; Gilman et al., 2014; Glenn and Yang, 2012; Korponay et al., 2017), we hypothesized that the externalizing PS may also moderate network resting-state connectivity and organization in ventral striatum and amygdala. Two graph properties were examined: Participation Coefficient indexes the extent to which a node is a ‘connector hub’ that bridges different functional modules, and Within-Module Degree Z-Score or the extent to which a node is a ‘provincial hub’ that facilitates communication within its own module (Power et al., 2013). We limited our analysis to Participation Coefficient and Within-Module Degree Z-Score, because these centrality metrics that are particularly relevant for identifying hubs that integrate information between and within functionally-distinct subnetworks.
Methods and Materials
Participants
Participants were military veterans of Operations Enduring Freedom and Iraqi Freedom. Exclusion criteria were a history of seizures, serious medical illness (including prior cerebrovascular accident or myocardial infarction), acute suicide risk, current psychotic disorder, bipolar disorder, or cognitive disorder due to general medical condition, pregnancy, metal implant, shrapnel, aneurysm clip, or pacemaker, and moderate or severe TBI. To avoid potential genetic confounds related to ancestry, the sample was limited to genetically-confirmed White non-Hispanic individuals. We did not have adequate statistical power in our sample to conduct analyses with other ethnically-homogenous groups. Only participants with viable functional neuroimaging data were analyzed. Study approval was obtained from all relevant Institutional Review Boards and regulatory committees. Participants provided written informed consent and were compensated financially.
The final sample consisted of 155 male and female military veterans ages 19–57. Demographic information and clinical characteristics are presented in Table 1. The predominately male composition of this sample (~92%) is representative of other military veteran samples, but likely limits the generalizability of the findings to men.
Table 1.
Descriptive Characteristics (N = 155)
| Age (M/SD) | 31.2 / 8.1 |
| Male (n, %) | 143 / 92.3 |
| Ethnicity (n, %) | |
| White, Non-Hispanic | 155 / 100.0 |
| Currently Employed (n, %) | 108 / 69.7 |
| Lifetime Mental Health Diagnosis (n, %) | |
| Alcohol/Substance Use Disorder | 101 / 65.6 |
| Unipolar Mood Disorder | 54 / 34.8 |
| Posttraumatic Stress Disorder | 106 / 68.4 |
| Mild Traumatic Brain Injury (n, %) | 103 / 66.5 |
Note. Participants with a diagnosis of current bipolar disorder, schizophrenia or psychotic disorder were ineligible to participate.
Measures
Genotyping.
DNA was extracted from peripheral blood samples and genotyping details are provided in Supplemental Methods.
Polygenic Scores.
To calculate the externalizing polygenic score, we obtained a list of reference alleles and effect sizes for 587,378 SNPs from the investigators of the discovery externalizing disorders GWAS (Salvatore et al., 2015) that reflect polygenic associations with externalizing disorders (e.g., alcohol use disorder, substance use disorder, antisocial personality disorder). We confirmed this list had been pruned of SNPs with ambiguous coding (i.e. A/T and G/C SNPs) in our data. Of the SNPS, 480,856 were genotyped on the Illumina OMNI 2.5–8 array in our sample and available for externalizing risk-score calculations (after removal of SNPs with missing rates >0.01 and Hardy–Weinberg equilibrium p-values <0.000001). Polygenic scores were calculated by PLINK1 (Purcell et al., 2007) using the --score option, which computes a linear function of the additively coded number of reference alleles weighted by the betas from the discovery externalizing GWAS sample.
Externalizing Phenotypes.
Participants completed the color-word interference test (i.e., Stroop) from the Delis-Kaplan Executive Function System (Delis et al., 2001) to measure inhibitory control. The inhibition subtest measures inhibition of an automatic response (word reading) to generate a less salient incongruent response (color naming), and the inhibition/switching subtest measures flexibly switching between these response sets. We used the scaled scores from these subtests adjusted for performance on the color-naming and word-reading component tests [e.g., inhibition - (color naming + word reading)/2]. (See Sadeh et al., 2016 for details).
Lifetime alcohol and substance use disorders was assessed via the Structured Clinical Interview for DSM-IV (First et al., 1994) based on the alcohol, cannabis, cocaine, opioids, amphetamines, polysubstance, and the other substances modules. We did not model the latent externalizing dimension, because we did not have a measure of antisocial behavior or trait constraint.
MRI Acquisition and Preprocessing.
Participants were instructed to remain still with eyes open while 2 EPI runs (voxel size = 3×3×3mm, TR = 3000ms, TE = 30ms, scan time per run = 360s) were acquired on a Siemens 3T TIM Trio. Two MPRAGEs (voxel size = 1×1×1mm, T1 = 1000ms, TR = 2530s, TE = 3.32ms) were acquired and averaged to create a single high contrast-to-noise image.
Individualized cortical parcellations and subcortical segmentations were created via FreeSurfer (Fischl et al., 2002). Cortical surface models were manually checked slice-by-slice and edited for accuracy. The Desikan/Killiany parcellation was used (34 regions per hemisphere), along with subcortical segmentation (7 regions per hemisphere). Frontal and temporal poles were excluded due to frequent susceptibility artifact, and 5 visual regions (the regions of least interest) were excluded to reduce the number of multiple comparisons. The number of brain region connections that were examined, and corrected for, in the network analysis was 2,278.
To ensure that our choice of particular preprocessing stream did not drive the findings, preprocessing was repeated with other options: (i) slice-timing correction, (ii) partialing of the square, autocorrelation, and 1st derivative of motion parameters, (iii) partialing the 1st derivative of global, ventricular, and white matter signals, (iv) determination on a participant-specific basis whether partialling of global signal was necessary (via the Global Negative Index), (v) motion scrubbing. Connectivity matrices remained extremely similar (correlations > .97), indicating that findings were not specific to our preprocessing stream.
Pairwise LiNGAM Analyses.
Given that the Pairwise LiNGAM method requires non-Gaussian information to be retained in the timeseries (Ramsey et al., 2014), preprocessing was repeated substituting in FSL’s non-linear high-pass filter. Only high-pass filtering was performed given evidence that low-pass filtering removes important non-Gaussian information (Ramsey et al., 2014). For each connection, a pairwise LiNGAM coefficient was first estimated for each participant. Significance of analyses was determined via permutation (5000 permutations). In order to ensure that outliers did not drive findings, analyses were repeated after winsorization of LiNGAM coefficients to 2.5SD, after which all findings remained significant.
Graph Property Computation.
Resting data were preprocessed using the Graph Theory GLM (GTG) toolbox version 0.44 (www.nitrc.org/projects/metalab_gtg; RRID: SCR_014075) (Spielberg et al., 2015). Data were motion corrected, detrended (linear and quadratic), bandpass filtered (retaining 0.01–0.10Hz), and the mean global, ventricular, and white matter signals were partialled out, along with estimated motion parameters. Timeseries for FreeSurfer nodes were extracted by calculating mean signal across the node for each time point, for each EPI run. Timeseries for the EPI runs were concatenated after mean-centering each timeseries within run, and a 68×68 Pearson correlation matrix was created for each participant. Before graph properties were computed, each participant’s connectivity matrix was thresholded to include only positive weights and normalized via division by the median positive weight for each matrix (excluding zeros). Normalization was performed to remove bias due to individual differences in overall network weight. Graph theory networks, including modules, were derived from a data-driven approach based on the current sample. Details about module membership are provided in Supplemental Methods.
Statistical Analyses
An externalizing PS computed with a p-value threshold cutoff of <.50 was the primary variable used in all analyses, selected based on prior work (Sadeh et al., 2016). We also report associations between the neural network connectivity measures and externalizing PSs calculated with other p-value thresholds for reference. To identify network connections that varied with polygenic variation, connectivity matrices were entered as dependent variables into the Network Based Statistic (NBS) toolbox (Zalesky et al., 2010), with the PS as the independent variable of interest. An individual connection level threshold of t = 2.9 was used with intensity-based correction for multiple comparisons (5000 permutations) and an overall corrected α < .05. Pairwise LiNGAM 31) was used to gain initial insight into the overall direction of influence of those connections observed in NBS analyses between cortical and subcortical structures. First, we tested the significance of the mean direction (across participants) for each connection via a one-sample t-test. Next, we tested whether the PS moderated the direction of influence for each connection via a regression with the same covariates used in the NBS analyses. False discovery rate was used to correct (across connections) for multiple comparisons.
We then identified graph theory properties that varied with polygenic variation. First, resting-state connectivity matrices were entered into the GTG toolbox, which computes properties for each participant using the Brain Connectivity Toolbox (Rubinov and Sporns, 2010). Two graph properties were examined: Participation Coefficient or the extent to which a node is connected to nodes in different modules, and Within-Module Degree Z-Score or the extent to which a node is connected to other nodes within its own module. Graph properties were examined only for nodes that emerged in the NBS analysis. Properties were entered as dependent variables in robust regressions in the GTG toolbox. Predictor models were the same as NBS analyses. Significance was determined via permutation tests (5000 repetitions). False discovery rate was used to correct (across nodes) for multiple comparisons, and adjusted p-values are in brackets.
We included covariates in our analyses to conduct a rigorous test of our hypotheses and reduce the likelihood that these variables could account for findings. All analyses were adjusted for age, sex, the first two ancestry principal components, deployment-related blast exposure (Fortier et al., 2014), handedness, current employment, and total cholesterol. Covariates were selected based on demonstrated associations with neural connectivity/integrity, genetic effects, and/or externalizing phenotypes in previous research (Han et al., 2014; Price et al., 2006; Spielberg et al., 2017). Given high rates of trauma exposure, PTSD, and mild TBI in this sample, we also examined whether these variables and other comorbidities altered study findings and found that they did not (see Supplemental Results for details). Correlations between the covariates and study variables are reported in the supplemental materials (Supplemental Table 1).
Results
Networks Related to Externalizing Polygenic Score
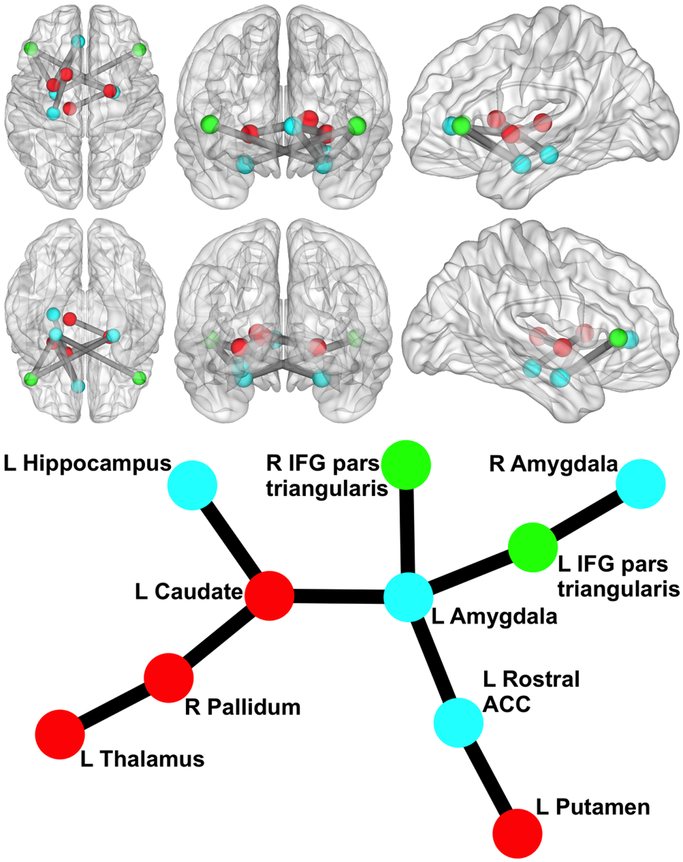
We began by examining whether polygenic variation related to externalizing predicted disturbances in functional network connectivity. The externalizing PS was associated with hyper-connectivity in a network centered on left amygdala (10 nodes, 9 links, corrected p =.046). As displayed in Figure 1, this network encompassed cortical (bilateral inferior frontal gyrus [IFG] pars triangularis and left rostral anterior cingulate cortex [ACC]) and subcortical regions (bilateral amygdala, hippocampus, and striatal areas) that have been implicated in emotion regulation and inhibitory control. The externalizing PS accounted for significance variance (ps<.005) in the strength of connectivity in this resting-state network across multiple significance p-value thresholds (see Table 2).
Figure 1. Network Related to Externalizing Polygenic Score.
Circle/sphere color reflects module. Stick/ball figure created using Kamada-Kawai spring embedder algorithm (SONIA; Bender-deMoll and McFarland, 2006). R = right; L = left; ACC = anterior cingulate cortex; IFG = inferior frontal gyrus. Node color indicates different modules. The six 3d brain images show (clockwise from top left) an axial view from superior to the brain, a coronal view from anterior to the brain, a sagittal view from left of the brain, a sagittal view from right of the brain, a coronal view from posterior to the brain, and an axial view from inferior to the brain (created via BrainNet Viewer; Xia et al., 2013).
Table 2.
Variance Accounted for in Neural Parameters Across Externalizing Polygenic Scores Calculated with Different P-Value Thresholds
| P-value threshold | Network Resting-State Connectivity Strength | Left Amygdala Participation Coefficient | Right Amygdala Participation Coefficient | Left Rostral Anterior Cingulate Cortex | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | p-value | R2 | p-value | R2 | p-value | R2 | p-value | |
| <.50 | .21 | <.001 | .07 | .001 | .06 | .003 | .04 | .014 |
| <.40 | .21 | <.001 | .07 | .001 | .06 | .003 | .03 | .029 |
| <.30 | .19 | <.001 | .07 | .001 | .05 | .009 | .02 | .096 |
| <.20 | .17 | <.001 | .07 | .001 | .04 | .018 | .02 | .087 |
| <.10 | .12 | <.001 | .04 | .019 | .02 | .124 | .03 | .047 |
| <.05 | .05 | .004 | .03 | .044 | .01 | .279 | .01 | .165 |
To better understand the direction of influence between nodes (i.e., A→B vs. B→A), we conducted pairwise LiNGAM analyses on links observed in the NBS analyses between cortical and subcortical structures. No links evidenced a significant mean direction of influence. However, the externalizing PS significantly moderated the direction of influence for two links, and these effects survived FDR correction. Specifically, higher polygenic risk was associated with a greater direction of influence from left amygdala→left IFG pars triangularis (p = .007 [corrected p = .033]) and from right amygdala→left IFG pars triangularis (p = .013 [p = .033]). This suggests that bilateral amygdala is influencing left IFG triangularis in participants with stronger polygenic associations with externalizing, whereas the opposite is true in those with weaker polygenic associations with externalizing.
We also examined polygenic effects on the organizational properties of network nodes. Externalizing PS predicted higher Participation Coefficient for left amygdala (p < .001 [corrected p = .004]), right amygdala (p = .002 [p = .012]), and left rostral ACC (p = .012 [p = .039]), suggesting that genetic variation related to externalizing modulates the importance of these nodes for communication between functional modules in the global network. The externalizing PS also nominally predicted higher Within-Module Degree Z-score Coefficient for right pallidum, suggesting externalizing polygenic variation modulates the importance of this node for communication within its functional module, but this did not survive correction for multiple comparisons (p = .046 [corrected p = .460]). These findings suggest that genetic variation related to externalizing modulated the extent to which these nodes functioned as communication hubs between functional modules.
Associations with Cognitive/Psychiatric Phenotypes
Bivariate associations between the study variables are presented in Table 3. We assessed the relevance of the genetic and network metrics for behavioral outcomes by examining relationships between these variables and externalizing phenotypes, specifically inhibitory control and lifetime alcohol/substance use using hierarchical regressions. Covariates were entered in Block 1 of the regression and explanatory variables were entered in Block 2.
Table 3.
Descriptive Statistics and Bivariate Correlations among Study Variables
| M / SD or n / % | 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
|---|---|---|---|---|---|---|---|---|
| 1 Externalizing PS | 0.62/0.12 | -- | ||||||
| Neural Phenotype | ||||||||
| 2 Network Connectivity | 0.05/0.09 | .43* | -- | |||||
| 3 L Amygdala Participation Coefficient | 0.64/0.07 | .28* | .48* | -- | ||||
| 4 R Amygdala Participation Coefficient | 0.63/0.07 | .25* | .52* | .49* | -- | |||
| 5 L rACC Participation Coefficient | 0.65/0.06 | .17* | .18* | −.03 | .03 | -- | ||
| Cognitive Phenotype | ||||||||
| 6 Inhibitory Control | 0.03/1.03 | −.23* | −.20* | −.12 | −.25* | −.13 | -- | |
| Psychiatric Phenotype | ||||||||
| 7 Alcohol/Substance Use Diagnosis | 101/65.6 | .10 | .14 | .03 | .08 | .06 | −.17* | -- |
Note. PS = Polygenic Score. Network Connectivity: average resting-state connectivity in the neural network associated with the externalizing PS (see Figure 1). L= left hemisphere. rACC= Rostral anterior cingulate cortex.
p <.05
At the genomic level, the externalizing PS was not related to a diagnosis of alcohol/substance use disorders (p >.23, ΔR2 = .01). As reported in this sample in Sadeh et al. (2016), the externalizing PS was associated with poorer performance on inhibitory control tasks (p =.01, ΔR2 = .05).
Mean connectivity in the network was negatively associated with performance on inhibitory control tasks (β = −.21, p = .015, ΔR2 = .04) and with a greater likelihood of a lifetime alcohol or substance use diagnosis (Wald X2= 4.9, p = .026, ΔR2 = .03), indicating that hyper-connectivity in the network confers risk for externalizing phenotypes. Examination of the graph properties revealed an inverse association between right amygdala Participation Coefficient and inhibitory control (β = −.24, p = .005, ΔR2 = .05), but no associations emerged for alcohol/substance use diagnosis.
Overall, the externalizing PS explained approximately 5% of the variance in inhibitory control (p = .016), and the neural network parameters (combined) explained approximately 8% of the variance in inhibitory control (p = .023). The combined variance explained across the genomic and neural levels of analysis was also significant (R2 = .10, p = .016). In contrast, the variance explained by the externalizing PS (1%), neural network parameters (4%), and combined genomic and neural levels of analysis (4%) was not significant for alcohol/substance use disorders.
Discussion
The identification of heritable mechanisms that link genes to complex clinical phenotypes is a critical step in mapping the developmental course of psychiatric disorders. We leveraged recent methodological advances in modeling the polygenic architecture of externalizing to uncover novel neural phenotypes for this spectrum of disorders. The externalizing PS predicted hyper-connectivity in a distributed brain network that included prefrontal (PFC) and subcortical regions critical for salience processing, emotion regulation, reinforcement learning, and impulse control. Directional analyses revealed that subcortical regions influenced PFC to a greater extent in participants with stronger polygenic associations with externalizing, whereas the opposite direction of influence was observed for those with weaker polygenic associations. The externalizing PS was associated with the organization of network nodes, specifically higher Participation Coefficient for bilateral amygdala and left rACC. Findings provide preliminary support that disturbed resting-state connectivity in this brain circuit is a genetically-influenced mechanism associated with the manifestation of disinhibited behaviors and alcohol/substance disorders.
To our knowledge, this is the first study to use a polygenic approach to isolate brain circuits related to externalizing. We found that an aggregate measure of genetic risk for externalizing was associated with functional connectivity in a circuit centered on left amygdala that included several cortical (bilateral IFG pars triangularis, left rostral ACC) and subcortical (bilateral amygdala, hippocampus, striatum) regions. These regions are central to psychological processes previously implicated in externalizing, including emotion regulation (amygdala, ACC), impulse control (IFG), and reward learning (caudate, putamen) (Baskin-Sommers et al., 2012; DeVito et al., 2013; Glenn and Yang, 2012; Sadeh et al., 2015). Although connectivity studies on the latent externalizing spectrum are sparse, there is evidence from within-region activation studies of increased amygdala reactivity and heightened engagement of executive control regions in impulsive and externalizing individuals (Foell et al., 2016; Sadeh et al., 2013). Our findings extend this work by showing that externalizing is also associated with increased amygdala connectivity at rest, primarily to PFC (see Figure 1). These findings also converge with neuroimaging studies conducted with specific externalizing disorders, which tend to implicate abnormalities in the structure and function of prefrontal regions important for cognitive and behavioral control (e.g., ACC) and subcortical reward and salience systems (amygdala, ventral striatum) (Cardenas et al., 2011; Dom et al., 2005; Nikolova et al., 2016; Yang and Raine, 2009). Unlike much of the previous research, this study linked resting-state functional connectivity in these key regions to genetic variation for externalizing disorders without relying on the selection of a few brain regions of interest. While this more unbiased approach to identifying an externalizing PS-related neural network is a strength of the current study, its exploratory nature highlights the need for replication in an independent sample.
Although PFC is typically thought to exert top-down control over subcortical structures, our analyses revealed that this was not the case for individuals with higher polygenic externalizing scores. Bilateral amygdala was found to influence left IFG in high polygenic scorers, whereas the opposite direction of influence was observed for low scorers. This connectivity pattern with PFC is in contrast to what has been observed in healthy individuals. For example, Etkin et al. (2006) examined interregional connectivity during an emotional conflict task in a sample of healthy subjects and found a significant inverse relationship between activity in rostral cingulate and right amygdala when emotional conflict resolution demand was high. This finding suggests that rostral cingulate may help regulate emotional reactivity in part by decreasing engagement of the amygdala in response to emotional distractors in healthy individuals. Our findings suggest that this pattern may be reversed in individuals with higher polygenic associations with externalizing disorders, such that amygdala has greater influence on prefrontal cognitive control structures, at least during resting state.
Polygenic burden was also associated with greater bilateral amygdala and left rostral ACC Participation Coefficient, suggesting that these regions are more likely to serve as ‘connector hubs’ as genetic risk increases. Connector hubs provide ‘shortcuts’ to communicate between segregated modules, and such module segregation is necessary for a network to perform multiple types of specialized processes. Thus, our findings suggest that amygdala and rACC influence a wider range of specialized processes as externalizing polygenic scores increase. This may lead to greater influence of amygdala on, not only bilateral IFG as the resting-state connectivity analyses suggested, but on a diverse range of processes. These findings extend previous work on externalizing by suggesting that abnormalities in neural network organization may be a heritable mechanism by which genetic risk for externalizing is conferred.
This study identified genetically-associated circuitry via resting-state coupling in order to examine genomic relations with stable functional networks. Thus, an important next step in this line of research will be to investigate the functional and structural features of externalizing PS-related brain networks. First, examining how connectivity in this network shifts in response to cognitive/emotional challenges, like inhibitory control tasks, will be important to address in future research. Connectivity between PFC executive-control regions (rACC, IFG) and subcortical regions crucial for emotion/salience (amygdala) increases with the demand for inhibitory control (Spielberg et al., 2015). Based on limited research on functional connectivity and impulsivity (Farr et al., 2012; Shannon et al., 2011) we would not expect high externalizing individuals to show this increase in PFC-subcortical connectivity with increasing inhibitory control demands in functional tasks. An alternative hypothesis, based on the present findings, is that bottom-up (e.g., affective) processes would influence inhibitory control to a greater extent in high externalizing individuals, rather than top-down control simply being deficient. Second, it will be important to isolate the anatomical pathways that enable the identified functional network, given that resting-state fMRI coupling does not imply direct anatomic connection and these analyses are not bounded by such connections.
Given that brain circuits, behavioral phenotypes, and environmental mechanisms are likely reciprocally influential (Miller and Rockstroh, 2013), studying how these dynamic relationships evolve across development will be important for establishing developmental pathways to behavioral manifestations of externalizing. The cross-sectional nature of our data prohibits strong conclusions regarding the direction of the proposed effects, and prospective research is needed to ascertain the mechanism(s) by which neural and behavioral phenotypes interact overtime. The observed associations across genetic, neural, and behavioral levels of analysis provides compelling evidence that resting-state connectivity in the identified network is a heritable mechanism that confers risk for externalizing. At the same time, more research is needed to validate this network as an endophenotype, including examining the generalizability of the polygenic associations with network connectivity. Additionally, although this study focused on identifying neurobiological mechanisms related to genetic risk for externalizing, understanding the role environmental factors play in the etiology of externalizing disorders is equally important. There is extensive evidence to suggest that environmental influences exert both direct and interactive effects on externalizing outcomes. For example, a large literature points to exposure to risky contexts (e.g., deviant peers) and stressful events (e.g., trauma) as powerful environments that interact with genes to confer risk for externalizing outcomes (e.g., Enoch, 2012; Hicks et al., 2004), including in studies of polygenic risk for externalizing (Sadeh et al., 2016; Salvatore et al., 2015). Based on these findings, incorporating the environment as a level of analysis in future polygenic-neuroimaging studies will be important for developing comprehensive etiological models of externalizing disorders.
This study has several strengths including analysis of multiple levels of analysis (genes, brain circuits, and behavior), a relatively large neuroimaging-genetics sample, and use of thousands of loci to identify genetically-associated brain circuits. Several limitations should also be considered. First, we were unable to model the latent externalizing spectrum because a measure of antisocial behavior and/or trait constraint was not available, a limitation that may have impacted associations between the externalizing PS and externalizing psychopathology (i.e., alcohol and substance use disorders) in this sample. Given that the externalizing PS was developed to capture genetic risk for comorbidity among externalizing disorders, assessing the covariance among alcohol/substance use disorders and antisocial behavior may be necessary to detect a robust polygenic association with these phenotypes. Second, although our ability to detect small effects was limited by the modest sample size, the sample was comparable to other recent neuroimaging-genetic studies (Little et al., 2015; Pagliaccio et al., 2015). Further, we cannot say definitively that the current results will generalize to the latent externalizing spectrum. Third, we did not have an independent sample to examine the replicability of our findings. Investigating the replicability of our findings in a larger independent sample with more diverse sample characteristics (e.g., greater representation of women, ethnic minorities, and civilians) is needed before strong conclusions can be drawn. Fourth, the cross-sectional nature of our data prohibits conclusions regarding the direction of the proposed effects, and prospective research is needed to ascertain the mechanism(s) by which neural and behavioral phenotypes interact overtime. For example, it is possible that the externalizing PS causes an externalizing phenotype (clinical symptoms) that then leads to variability in the neural phenotypes rather than the genes relating directly to neural network functioning. Only longitudinal studies that assess the interactive effects of multiple levels of analysis over time are well positioned to clarify the direction of these effects. Finally, although use of an independently-derived PS represents a less biased approach to genetic analysis than selecting candidate genes, there are limitations to the polygenic approach. For example, the predictive power of the PS is limited by the robustness of the original GWAS, and the PS assumes an additive genetic model without interactions, which might not optimally reflect the underlying genetic architecture of externalizing. Additionally, the PS does not take into account environmental effects. Given the high level of stress exposure in the current sample and externalizing prone samples in general (e.g., Douglas et al., 2010; Luntz and Widom, 1994), examining the influence of potential epigenetic effects in relation to externalizing disorders will be important to explore in future research, such as whether differentially methylated loci associated with externalizing overlap with the externalizing PS.
In summary, findings suggest that disturbed resting-state connectivity in a circuit implicated in emotion regulation, impulse control, and reinforcement learning is a putative biological pathway linking polygenic risk for externalizing with psychiatric outcomes. Preliminary data provide compelling evidence that resting-state connectivity in this network represents an inherited neurobiological vulnerability for externalizing disorders.
Supplementary Material
Acknowledgments
This research was supported in part by NIGMS grant 2P20GM103653-06-6527, NIMH grant R21MH102834, and the Translational Research Center for TBI and Stress Disorders (National Network Research Center B9254-C) and the Cooperative Studies Program, Department of Veterans Affairs. This research is the result of work supported with resources and the use of facilities at the Pharmacogenomics Analysis Laboratory, Research and Development Service, Central Arkansas Veterans Healthcare System, Little Rock, Arkansas. This work was also supported by a Career Development Award to EJW from the United States Department of Veterans Affairs, Clinical Sciences Research and Development Program. The views expressed in this article are those of the authors and do not necessarily reflect the position or policy of the Department of Veterans Affairs or the United States Government.
We would like to thank the Collaborative Study on the Genetics of Alcoholism (COGA) who supplied data for calculating the polygenic risk scores used in this study. COGA Principal Investigators B. Porjesz, V. Hesselbrock, H. Edenberg, L. Bierut, includes eleven different centers: University of Connecticut (V. Hesselbrock); Indiana University (H.J. Edenberg, J. Nurnberger Jr., T. Foroud); University of Iowa (S. Kuperman, J. Kramer); SUNY Downstate (B. Porjesz); Washington University in St. Louis (L. Bierut, J. Rice, K. Bucholz, A. Agrawal); University of California at San Diego (M. Schuckit); Rutgers University (J. Tischfield, A. Brooks); University of Texas Medical Center in San Antonio (L. Almasy), Virginia Commonwealth University (D. Dick), Mount Sinai School of Medicine (A. Goate), and Howard University (R. Taylor). Other COGA collaborators include: L. Bauer (University of Connecticut); D. Koller, S. O’Connor, L. Wetherill, X. Xuei (Indiana University); Grace Chan (University of Iowa); D. Chorlian, N. Manz, K. Chella, A. Pandey (SUNY Downstate); J-C Wang (Mount Sinai School of Medicine) and F. Aliev (Virginia Commonwealth University). A. Parsian and M. Reilly are the NIAAA Staff Collaborators. COGA continues to be inspired by our memories of Henri Begleiter and Theodore Reich, founding PI and Co-PI of COGA, and also owes a debt of gratitude to other past organizers of COGA, including Ting-Kai Li, currently a consultant with COGA, P. Michael Conneally, Raymond Crowe, and Wendy Reich, for their critical contributions. This national collaborative study is supported by NIH Grant U10AA008401 from the National Institute on Alcohol Abuse and Alcoholism (NIAAA) and the National Institute on Drug Abuse (NIDA).
Financial Disclosures
Drs. Sadeh, Spielberg, Logue, Hayes, Wolf, McGlinchey, Milberg, Schichman, Stone, and Miller have no conflicts of interests or financial disclosures to declare.
References
- Aron AR, Robbins TW, Poldrack RA (2004). ‘Inhibition and the right inferior frontal cortex’, Trends in Cognitive Sciences, 8(4), pp. 170–177. [DOI] [PubMed] [Google Scholar]
- Baskin-Sommers AR, Curtin JJ, Larson CL, Stout D, Kiehl KA Newman JP (2012). ‘Characterizing the anomalous cognition–emotion interactions in externalizing’, Biological Psychology, 91, pp. 48–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beckmann CF, DeLuca M, Devlin JT Smith SM (2005). ‘Investigations into resting-state connectivity using independent component analysis’, Philosophical Transactions of the Royal Society of London B: Biological Sciences, 360(1457), pp.1001–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender-deMoll S, McFarland DA (2006). ‘The art and science of dynamic network visualization’, Journal of Social Structure, 7(2), pp. 1–38. [Google Scholar]
- Cardenas VA, Durazzo TC, Gazdzinski S, Mon A, Studholme C Meyerhoff DJ (2011). ‘Brain morphology at entry into treatment for alcohol dependence is related to relapse propensity’, Biological Psychiatry, 70(6), pp. 561–567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardon LR, Palmer LJ (2003). ‘Population stratification and spurious allelic association’, The Lancet, 361(9357), pp.598–604. [DOI] [PubMed] [Google Scholar]
- Criaud M, Boulinguez P (2013). ‘Have we been asking the right questions when assessing response inhibition in go/no-go tasks with fMRI? A meta-analysis and critical review’, Neuroscience and Biobehavioral Reviews, 37(1), pp. 11–23. [DOI] [PubMed] [Google Scholar]
- Delis DC, Kaplan E, Kramer JH (2001). Delis-Kaplan Executive Function System (D-KEFS). The Psychological Corporation, New York. [Google Scholar]
- DeVito EE, Meda SA, Jiantonio R, Potenza MN, Krystal JH Pearlson GD (2013). ‘Neural correlates of impulsivity in healthy males and females with family histories of alcoholism’, Neuropsychopharmacology, 38(10), pp. 1854–1863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dom G, Sabbe BGCC, Hulstijn W Van Den Brink W (2005). ‘Substance use disorders and the orbitofrontal cortex’, The British Journal of Psychiatry, 187(3), pp. 209–220. [DOI] [PubMed] [Google Scholar]
- Dosenbach NU, Fair DA, Miezin FM, Cohen AL, Wenger KK, Dosenbach RA, Fox MD, Snyder AZ, Vincent JL, Raichle ME Schlaggar BL (2007). ‘Distinct brain networks for adaptive and stable task control in humans’, Proceedings of the National Academy of Sciences, 104(26), pp.11073–11078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas KR, Chan G, Gelernter J, Arias AJ, Anton RF, Weiss RD, Brady K, Poling J, Farrer L Kranzler HR (2010). ‘Adverse childhood events as risk factors for substance dependence: Partial mediation by mood and anxiety disorders’, Addictive Behaviors, 35, pp. 7–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durazzo TC, Tosun D, Buckley S, Gazdzinski S, Mon A, Fryer SL Meyerhoff DJ (2011). ‘Cortical thickness, surface area, and volume of the brain reward system in alcohol dependence: relationships to relapse and extended abstinence’, Alcoholism: Clinical and Experimental Research, 35(6), pp. 1187–1200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eaton WW, Roth KB, Bruce M, Cottler L, Wu L, Nestadt G, Ford D, Bienvenu OJ, Crum RM, Rebok G, Anthony JC (2013). ‘The relationship of mental and behavioral disorders to all-cause mortality in a 27-year follow-up of 4 epidemiologic catchment area samples’, American Journal of Epidemiology, 178, pp. 1366–1377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA (2012). ‘The influence of gene–environment interactions on the development of alcoholism and drug dependence’, Current Psychiatry Reports, 14(2), pp. 150–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etkin A, Egner T, Peraza DM, Kandel ER, Hirsch J (2006). ‘Resolving emotional conflict: a role for the rostral anterior cingulate cortex in modulating activity in the amygdala’, Neuron, 51(6), pp. 871–882. [DOI] [PubMed] [Google Scholar]
- Farr OM, Hu S, Zhang S, Chiang-shan RL (2012). ‘Decreased saliency processing as a neural measure of Barratt impulsivity in healthy adults’, Neuroimage, 63(3), pp. 1070–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- First MB, Spitzer RL, Gibbon M, Williams JB (1994). Structured Clinical Interview for Axis I DSM-IV Disorders. Biometrics Research, New York. [Google Scholar]
- Fischl B, Salat DH, Busa E, Albert M, Dieterich M, Haselgrove C, Van Der Kouwe A, Killiany R, Kennedy D, Klaveness S, Montillo A (2002). ‘Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain’, Neuron, 33(3), pp. 341–355. [DOI] [PubMed] [Google Scholar]
- Foell J, Brislin SJ, Strickland CM, Seo D, Sabatinelli D Patrick CJ (2015). ‘Externalizing proneness and brain response during pre-cuing and viewing of emotional pictures’, Social Cognitive and Affective Neuroscience,11(7), pp.1102–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fornito A, Bullmore ET (2015). ‘Connectomics: A new paradigm for understanding brain disease’, European Neuropsychopharmacology, 25(5), pp. 733–748. [DOI] [PubMed] [Google Scholar]
- Fortier CB, Amick MM, Grande L, McGlynn S, Kenna A, Morra L, Clark A, Milberg WP, McGlinchey RE (2014). ‘The Boston assessment of traumatic brain injury–lifetime (BAT-L) semistructured interview: Evidence of research utility and validity’, Journal of Head Trauma Rehabilitation, 29(1), pp. 89–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke B, Stein JL, Ripke S, Anttila V, Hibar DP, Van Hulzen KJ, Arias-Vasquez A, Smoller JW, Nichols TE, Neale MC, McIntosh AM (2016). ‘Genetic influences on schizophrenia and subcortical brain volumes: large-scale proof of concept’, Nature Neuroscience, 19, pp. 420–431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman JM, Kuster JK, Lee S, Lee MJ, Kim BW, Makris N, van der Kouwe A, Blood AJ, Breiter HC (2014). ‘Cannabis use is quantitatively associated with nucleus accumbens and amygdala abnormalities in young adult recreational users’, The Journal of Neuroscience, 34(16), pp. 5529–5538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glahn DC, Winkler AM, Kochunov P, Almasy L, Duggirala R, Carless MA, Curran JC, Olvera RL, Laird AR, Smith SM, Beckmann CF (2010). ‘Genetic control over the resting brain’, Proceedings of the National Academy of Sciences, 107(3), pp. 1223–1228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glenn AL,Yang Y (2012). ‘The potential role of the striatum in antisocial behavior and psychopathy’, Biological Psychiatry, 72(10), pp. 817–822. [DOI] [PubMed] [Google Scholar]
- Han K, Mac Donald CL, Johnson AM, Barnes Y, Wierzechowski L, Zonies D, Oh J, Flaherty S, Fang R, Raichle ME, Brody DL (2014). ‘Disrupted modular organization of resting-state cortical functional connectivity in US military personnel following concussive ‘mild’blast-related traumatic brain injury’, Neuroimage, 84, pp. 76–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzeg MM, Villafuerte S, Weiland BJ, Enoch MA, Burmeister M, Zubieta JK, Zucker RA (2014). ‘Effect of GABRA2 genotype on development of incentive-motivation circuitry in a sample enriched for alcoholism risk’, Neuropsychopharmacology, 39(13), pp. 3077–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hicks BM, Krueger RF, Iacono WG, McGue M, Patrick CJ (2004). ‘Family transmission and heritability of externalizing disorders: A twin-family study’, Archives of General Psychiatry, 61(9), pp. 922–928. [DOI] [PubMed] [Google Scholar]
- Karoly HC, Harlaar N, Hutchison KE (2013). ‘Substance use disorders: A theory-driven approach to the integration of genetics and neuroimaging’, Annals of the New York Academy of Sciences, 1282(1), pp.71–91. [DOI] [PubMed] [Google Scholar]
- Korponay C, Pujara M, Deming P, Philippi C, Decety J, Kosson DS, Kiehl KA, Koenigs M (2017). ‘Impulsive-antisocial dimension of psychopathy linked to enlargement and abnormal functional connectivity of the striatum’, Biological Psychiatry: Cognitive Neuroscience and Neuroimaging, 2(2), pp.149–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger RF, Hicks BM, Patrick CJ, Carlson SR, Iacono WG, McGue M (2002). ‘Etiologic connections among substance dependence, antisocial behavior and personality: Modeling the externalizing spectrum’, Journal of Abnormal Psychology, 111, pp. 411–424. [PubMed] [Google Scholar]
- Krueger RF, Markon KE (2006). ‘Reinterpreting comorbidity: A model-based approach to understanding and classifying psychopathology’, Annual Review of Clinical Psychology, 2, pp. 111–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luntz BK, Widom CS (1994). ‘Antisocial personality disorder in abused and neglected children grown up’, American Journal of Psychiatry, 151, pp. 670–674. [DOI] [PubMed] [Google Scholar]
- Miller GA, Rockstroh B (2013). ‘Endophenotypes in psychopathology research: Where do we stand?’, Annual Review Clinical Psychology, 9, pp. 177–213. [DOI] [PubMed] [Google Scholar]
- Nee DE, Wager TD, Jonides J (2007). ‘Interference resolution: insights from a meta-analysis of neuroimaging tasks’, Cognitive, Affective, and Behavioral Neuroscience, 7, pp. 1–17. [DOI] [PubMed] [Google Scholar]
- Nikolova YS, Knodt AR, Radtke SR, Hariri AR (2016). ‘Divergent responses of the amygdala and ventral striatum predict stress-related problem drinking in young adults: possible differential markers of affective and impulsive pathways of risk for alcohol use disorder’, Molecular Psychiatry, 21(3), pp. 348–356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odgers CL, Caspi A, Broadbent JM, Dickson N, Hancox RJ, Harrington HL, Poulton R, Sears MR, Thomson WM, Moffitt TE (2007). ‘Conduct problem subtypes in males predict differential adult health burden’, Archives of General Psychiatry, 64(4), pp. 476–484. [DOI] [PubMed] [Google Scholar]
- Power JD, Schlaggar BL, Lessov-Schlaggar CN, Petersen SE (2013). ‘Evidence for hubs in human functional brain networks’, Neuron, 79(4), pp. 798–813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006). ‘Principal components analysis corrects for stratification in genome-wide association studies’, Nature Genetics, 38(8), pp. 904–909. [DOI] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC (2007). ‘PLINK: A tool set for whole-genome association and population-based linkage analyses’, The American Journal of Human Genetics, 81, pp. 559–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JD, Sanchez-Romero R, Glymour C (2014). ‘Non-Gaussian methods and high-pass filters in the estimation of effective connections’, Neuroimage, 84, pp. 986–1006. [DOI] [PubMed] [Google Scholar]
- Rubinov M, Sporns O (2010). ‘Complex network measures of brain connectivity: Uses and interpretations’, Neuroimage, 52(3), pp. 1059–1069. [DOI] [PubMed] [Google Scholar]
- Sadeh N, Spielberg JM, Heller W, Herrington JD, Engels AS, Warren SL, Crocker LD, Sutton BP, Miller GA (2013). ‘Emotion disrupts neural activity during selective attention in psychopathy’, Social Cognitive and Affective Neuroscience, 8(3), pp. 235–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh N, Spielberg JM, Miller MW, Milberg WP, Salat DH, Amick MM, Fortier CB, McGlinchey RE (2015). ‘Neurobiological indicators of disinhibition in posttraumatic stress disorder’, Human Brain Mapping, 36(8), pp. 3076–3086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadeh N, Wolf EJ, Logue MW, Lusk J, Hayes JP, McGlinchey RE, Milberg WP, Stone A, Schichman SA, Miller MW (2016). ‘Polygenic risk for externalizing disorders and executive dysfunction in trauma-exposed veterans’, Clinical Psychological Science, 4(3), pp. 545–558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvatore JE, Aliev F, Bucholz K, Agrawal A, Hesselbrock V, Hesselbrock M, Bauer L, Kuperman S, Schuckit MA, Kramer JR, Edenberg HJ (2015). ‘Polygenic risk for externalizing disorders gene-by-development and gene-by-environment effects in adolescents and young adults’, Clinical Psychological Science, 3(2), pp. 189–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon BJ, Raichle ME, Snyder AZ, Fair DA, Mills KL, Zhang D, Bache K, Calhoun VD, Nigg JT, Nagel BJ, Stevens AA (2011). ‘Premotor functional connectivity predicts impulsivity in juvenile offenders’, Proceedings of the National Academy of Sciences, 108(27), pp. 11241–11245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehzad Z, DeYoung CG, Kang Y, Grigorenko EL, Gray JR (2012). ‘Interaction of COMT val 158 met and externalizing behavior: Relation to prefrontal brain activity and behavioral performance’, Neuroimage, 60(4), pp. 2158–2168. [DOI] [PubMed] [Google Scholar]
- Smit DJ, Stam CJ, Posthuma D, Boomsma DI, De Geus EJ (2008). ‘Heritability of “small-world” networks in the brain: A graph theoretical analysis of resting-state EEG functional connectivity’, Human Brain Mapping, 29(12), pp. 1368–1378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberg JM, McGlinchey RE, Milberg WP, Salat DH (2015). ‘Brain network disturbance related to posttraumatic stress and traumatic brain injury in veterans’, Biological Psychiatry, 78(3), pp. 210–216. [DOI] [PubMed] [Google Scholar]
- Spielberg JM, Miller GA, Heller W, Banich MT (2015). ‘Flexible brain network reconfiguration supporting inhibitory control’, Proceedings of the National Academy of Sciences, 112(32), pp. 10020–10025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spielberg JM, Sadeh N, Leritz EC, McGlinchey RE, Milberg WP, Hayes JP, Salat DH (2017). ‘Higher serum cholesterol is associated with intensified age - related neural network decoupling and cognitive decline in early - to mid - life.’, Human Brain Mapping, 38, pp. 3249–3261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stam CJ (2014). ‘Modern network science of neurological disorders’, Nature Reviews Neuroscience, 15(10), pp. 683–695. [DOI] [PubMed] [Google Scholar]
- Tarter RE, Kirisci L, Mezzich A, Cornelius JR, Pajer K, Vanyukov M, Gardner W, Blackson T, Clark D (2003). ‘Neurobehavioral disinhibition in childhood predicts early age at onset of substance use disorder’, American Journal of Psychiatry, 160(6), pp. 1078–1085. [DOI] [PubMed] [Google Scholar]
- National Institute on Drug Abuse. Trends and statistics. http://www.drugabuse.gov/related-topics/trends-statistics. Updated August, 2015. Accessed February 17, 2016.
- Xia M, Wang J, He Y (2013). ‘Viewer: A network visualization tool for human brain connectomics’, PLoS ONE, 8, e68910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y, Raine A (2009). ‘Prefrontal structural and functional brain imaging findings in antisocial, violent, and psychopathic individuals: a meta-analysis’, Psychiatry Research: Neuroimaging, 174(2), pp. 81–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young SE, Friedman NP, Miyake A, Willcutt EG, Corley RP, Haberstick BC, Hewitt JK (2009). ‘Behavioral disinhibition: liability for externalizing spectrum disorders and its genetic and environmental relation to response inhibition across adolescence’, Journal of Abnormal Psychology, 118(1), pp. 117–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalesky A, Fornito A, Bullmore ET (2010). ‘Network-based statistic: Identifying differences in brain networks’, Neuroimage, 53, pp. 1197–1207. [DOI] [PubMed] [Google Scholar]
- Zuo XN, Xing XX (2014). ‘Test-retest reliabilities of resting-state FMRI measurements in human brain functional connectomics: a systems neuroscience perspective’, Neuroscience and Biobehavioral Reviews, 45, pp. 100–118. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.