Abstract
Background
Histone deacetylase 4 (HDAC4) has been proposed as a target for Amyotrophic Lateral Sclerosis (ALS) because it mediates nerve-skeletal muscle interaction and since its expression in skeletal muscle correlates with the severity of the disease. However, our recent studies on the skeletal muscle response upon long-term denervation highlighted the importance of HDAC4 in maintaining muscle integrity.
Methods
To fully identify the yet uncharacterized HDAC4 functions in ALS, we genetically deleted HDAC4 in skeletal muscles of a mouse model of ALS. Body weight, skeletal muscle, innervation and spinal cord were analyzed over time by morphological and molecular analyses. Transcriptome analysis was also performed to delineate the signaling modulated by HDAC4 in skeletal muscle of a mouse model of ALS.
Findings
HDAC4 deletion in skeletal muscle caused earlier ALS onset, characterized by body weight loss, muscle denervation and atrophy, and compromised muscle performance, although the main catabolic pathways were not activated. Transcriptome analysis identified the gene networks modulated by HDAC4 in ALS, revealing UCP1 as a top regulator that may be implicated in worsening ALS features.
Interpretation
HDAC4 plays an important role in preserving innervations and skeletal muscle in ALS, likely by modulating the UCP1 gene network. Our study highlights a possible risk in considering HDAC inhibitors for the treatment of ALS.
Fund
This work was supported by FIRB grant (RBFR12BUMH) from Ministry of Education, Universities and Research, by Fondazione Veronesi, by Sapienza research project 2017 (RM11715C78539BD8) and Polish National Science Center grant (UMO-2016/21/B/NZ3/03638).
Keywords: HDAC4, HDAC inhibitors, SOD1G93A mice, Epigenetics, ALS
Research in context.
Evidence before this study
HDAC4 is a stress-responsive epigenetic factor whose expression is strongly up-regulated in skeletal muscle of Amyotrophic Lateral Sclerosis (ALS) patients and of mice following denervation, where it mediates skeletal muscle response to denervation. Therefore, HDAC4 has been proposed as a therapeutic target for muscular pathologies characterized by neurogenic muscle atrophy. However, pan-HDAC inhibitors failed in a phase-II clinical trial. Moreover, specific inhibition of class II HDACs had no effects on body weight loss and survival in ALS mice. Understanding the molecular mechanism underlying ALS progression and modulated by each HDAC is a prerequisite to ameliorate the therapy presently in use.
Added value of this study
In this study, we dissected the role of HDAC4 in ALS pathogenesis with a genetic approach. Deletion of HDAC4 in skeletal muscle accelerated the pathological features of ALS, preceding and exacerbating skeletal muscle loss and denervation, by modulating several biological processes and an interesting gene network in ALS skeletal muscle.
Implications of all the available evidence
We elucidated a pivotal role of HDAC4 in mediating skeletal muscle homeostasis in ALS. Our study provides new experimental bases for reconsidering the use of pan-HDAC inhibitors as a pharmacological treatment for ALS.
Alt-text: Unlabelled Box
1. Introduction
Amyotrophic Lateral Sclerosis is a neurodegenerative disorder characterized by motor neuron degeneration, skeletal muscle weakness and atrophy, fasciculation, speech and swallowing disabilities, progressive paralysis and, eventually, death by respiratory failure. Prognosis varies from few months to few years, although the majority of patients do not exceed 19 months since diagnosis and 30 months after the onset of the symptoms. About 20% of the familial cases of ALS are caused by one among over 150 mutations in the Cu/Zn superoxide dismutase 1 (SOD1) gene, mostly autosomal dominant [1]. In physiological conditions, SOD1 protects the cell from the accumulation of free radicals by converting the superoxide anion, derived from mitochondrial oxidative phosphorylation, to water or hydrogen peroxide [1]. For years ALS has been considered a pure motor neuron disease exclusively. Coherently with this theory, viral delivery of siRNA for the mutant SOD1 accumulation in skeletal muscle was not able to counteract ALS disease in mice, demonstrating that skeletal muscle is not a significant contributor to ALS [2]. However, more recently, it is emerging that motor neuron degeneration is a non-cell-autonomous phenomenon, involving numerous cell types. For instance, microglial cells actively participate in neuroinflammation by secreting pro-inflammatory cytokines [3], and skeletal muscle activates retrograde signals that influence motor neuron survival and ALS progression [4]. By generating transgenic mice, two independent studies demonstrated that the muscle-specific expression of the mutated SOD1 gene recapitulates some pathological features of ALS, including severe myopathy, neuromuscular junction (NMJ) abnormalities and motor neuron degeneration [5,6]. One of the retrograde signals from the skeletal muscle affects muscle reinnervation and ALS progression is mediated by the microRNAs miR-206 and its downstream target histone deacetylase 4 (HDAC4) [7].
HDAC4 is a class IIa HDAC stress-responsive epigenetic factor that mediates skeletal muscle response to denervation through transcriptional and cytoplasmic actions. Upon denervation, HDAC4 transcription is significantly induced in the skeletal muscle, where it represses the transcription of the Dach2 gene and activates the MAPK-AP1 signaling cascade in the cytoplasm, thereby promoting neurogenic muscle atrophy by two independent pathways converging on the E3 ubiquitin-ligases MuRF1 and atrogin-1 transcription [[8], [9], [10]]. Moreover, by indirectly regulating myogenin expression, HDAC4 is responsible for the transcriptional activation of synaptic acetylcholine receptors, MuSK, and miR-206, thus promoting muscle reinnervation [7,10]. Consistently, mice with a muscle-specific deletion of HDAC4 are resistant to neurogenic atrophy and reinnervate faster than controls mice [7,9].
HDAC4 has been proposed as a therapeutic target for muscular pathologies characterized by neurogenic muscle atrophy [11], because of its crucial role in mediating skeletal muscle response to innervation and since its expression in skeletal muscle correlates with the severity of the ALS progression [12,13]. Accordingly, pan-HDAC inhibitors (HDACi), which unspecifically block the activities of class I and II HDACs, restored muscle mass and function and prolonged the survival of a murine model of ALS [[14], [15], [16]]. However, these compounds failed in a phase-II clinical trial [17,18]. Importantly, no changes in the expression of the different members of the HDAC superfamily were registered in the spinal cords of ALS mice, while a significant increase in the expression of the members of class II HDACs was detected in the skeletal muscle with the progression of the disease [19]. Specific inhibition of class II HDACs in ALS mice induced motor improvement and increased skeletal muscle electrical potentials, although no effects on body weight loss or survival were registered [19]. Low inhibitory efficiency and a lack of selectivity are two of the possible reasons for the low efficacy of the treatment. The inhibition of all the members of class II HDACs, moreover, does not clarify the mechanisms underlying ALS progression. Understanding the molecular mechanism modulated by each HDAC, with a genetic approach, is a prerequisite to ameliorate the therapy presently in use.
In this study, we dissected the role of HDAC4 in ALS pathogenesis, by generating SOD1G93A (SOD) mice with a skeletal muscle-specific deletion of HDAC4 (SOD HDAC4mKO mice). Deletion of HDAC4 in skeletal muscle worsened the pathological features of ALS, preceding and exacerbating skeletal muscle loss and denervation. Moreover, HDAC4 modulated several biological processes and an interesting gene network in ALS skeletal muscle. Considering the pivotal role of HDAC4 in mediating skeletal muscle response to ALS, our study provides new experimental bases for the development of new pharmacological treatments for ALS.
2. Materials and methods
2.1. Mice
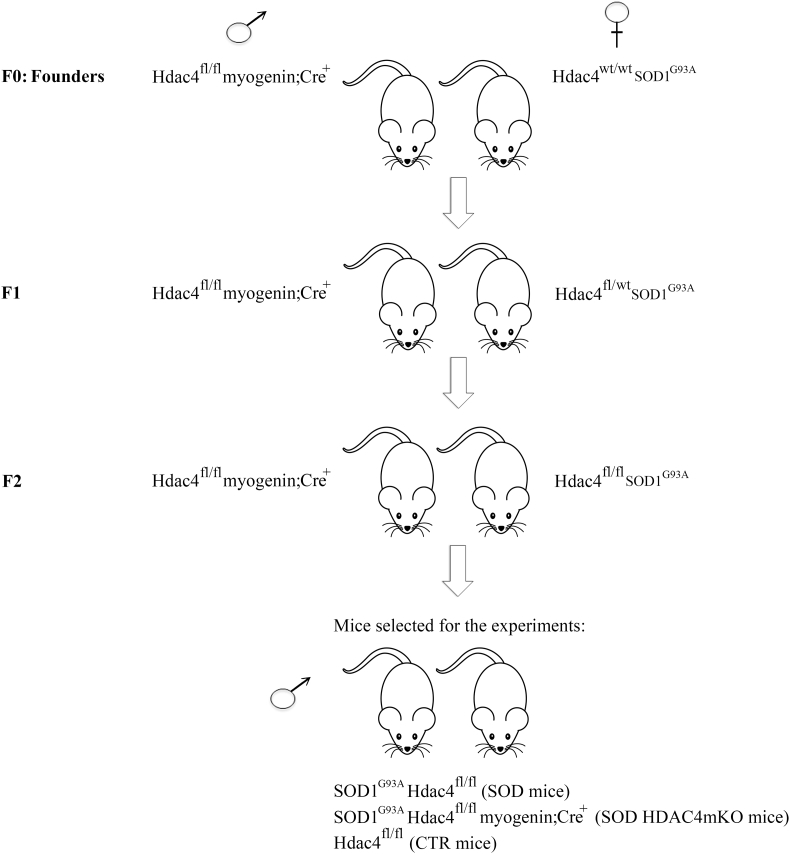
HDAC4 conditional mutant mice were generously provided by Prof. Eric N Olson and Rhonda Bassel-Duby. The reduction of HDAC4 expression in skeletal muscle was confirmed by real-time PCR and western blot analyses in HDAC4mKO mice, compared to controls (CTR) (Supplementary Fig. S1a–c). To study the role of HDAC4 in ALS, Hdac4fl/fl myogenin;Cre mice were crossed with the SOD1G93A transgenic mice (Charles River Laboratories). After several breedings, SOD1G93A Hdac4fl/fl myogenin;Cre mice (mice with ALS disease and HDAC4 deletion in skeletal muscle, called SOD HDAC4mKO mice) and SOD1G93A Hdac4fl/fl mice (mice with ALS disease and homozygous for a floxed Hdac4 allele, referred to as SOD mice) were obtained (Fig. 1). In all the experiments, male SOD HDAC4mKO and SOD mice from the same litter were compared, in order to define HDAC4 function in skeletal muscle in ALS, despite the different genetic backgrounds. In addition, Hdac4fl/fl healthy control (CTR) mice of the same sex and age were used (whenever possible from the same litter, always from the same colony).
Fig. 1.
Breeding strategy. Hdac4fl/fl myogenin;Cre male mice were crossed with Hdac4wt/wt SOD1G93A female mice (F0: Founders). In the F1 breeding, Hdac4fl/fl myogenin;Cre male mice were crossed with Hdac4fl/wt SOD1G93A female mice, to obtain Hdac4fl/fl SOD1G93A mice. Hdac4fl/fl myogenin;Cre males were then crossed with Hdac4fl/fl SOD1G93A females in F2 and, from the litters, only SOD1G93A male mice were used in the experiments: either Hdac4fl/fl (SOD mice) or Hdac4fl/fl myogenin;Cre (SOD HDAC4mKO mice); Hdac4fl/fl as healthy controls (CTR mice).
2.2. Ethics statement
Mice were treated in strict accordance with the guidelines of the Institutional Animal Care and Use Committee, as well as national and European legislation, throughout the experiments. Animal protocols were approved by the Italian Ministry of Health (authorizations # 244/2013 and # 853/2016-PR).
2.3. Replicates
The differences between SOD and SOD HDAC4mKO mice were confirmed in independent experiments, i.e., independent litters, by considering as “n” an independent biological sample, not a technical replicate.
2.4. Functional analyses
Muscle strength analyses were performed by grip test. The grip-strength meter (Ugo Basile) consists of a T-shaped bar connected to a force transducer, in turn connected to the peak amplifier 47,105–001. Mice were held by the tail and allowed to grab the bar. Mice were then pulled backward, exerting a constant force until they lost the grip. The force applied to the bar is recorded as the peak tension. To obtain reproducible data, each animal was subjected to 10 consecutive measurements, and the mean value was used as a single measurement.
2.5. Transcardiac perfusion
For innervations and spinal cord histological analyses, mice were perfused by using “In Vivo Perfusion System 140 ml” (2Biological Instruments). A heparinized (10 units/ml) (Sigma-Aldrich, Saint Louis, Missouri, #84020), in 0.9% NaCl (Sigma-Aldrich, Saint Louis, Missouri, #31434) in water and a 4% paraformaldehyde fixative (Sigma-Aldrich, Saint Louis, Missouri, #1.00496), in PBS: 37 mM NaCl (Sigma-Aldrich, Saint Louis, Missouri, #31434-M); 2.7 mM KCl (Sigma-Aldrich, Saint Louis, Missouri, #P3911); 4.3 mM Na2 HPO4 (Sigma-Aldrich, Saint Louis, Missouri, #S0876); 147 mM KH2PO4 pH 7.4 (Sigma-Aldrich, Saint Louis, Missouri, #P0662) solutions were used. The animals were anesthetized with intraperitoneal (i.p.) injections of 50 mg/Kg Zoletil (Bioz, Los Altos, California) and 10 mg/Kg Xylazine (Sigma-Aldrich, Saint Louis, Missouri, #X1126). To guarantee access to the heart, a thoracotomy was performed, by cutting the ribs and preventing heart and lung damage. The perfusion needle was inserted into the left ventricle, while the right atrium was cut to create a way out for the blood and perfusion fluids, avoiding pressure overloads. The animals were then firstly perfused with the heparinized solution to remove blood from the circulation, and then with the fixative solution. The tail stiffness was used as an index of efficient perfusion.
2.6. Histological analyses
To reduce the numbers of mice in our experiments, considering that a similar phenotype was observed in the Tibialis Anterior (TA) and Gastrocnemius (GA) muscles, TA muscles were consistently used for morphological evaluation of the phenotype. Muscles were dissected, embedded in tissue freezing medium (Leica, Wetzlar, Germany) and frozen in liquid nitrogen pre-cooled isopentane (Sigma-Aldrich, Saint Louis, Missouri, # PHR1661). Cryosections (8 μm or 20 μm) were obtained by using a Leica cryostat.
Ventral spinal cords were isolated from perfused mice, embedded in tissue freezing medium (Leica, Wetzlar, Germany) and frozen in liquid nitrogen pre-cooled isopentane (Sigma-Aldrich, Saint Louis, Missouri, # PHR1661). Cryosections (16 μm) were obtained by using a Leica cryostat.
Hematoxylin and eosin (H&E) (Sigma-Aldrich, Saint Louis, Missouri, # H3136 and # 861006) staining was performed according to the Sigma-Aldrich manufacturer's instructions. Cryosections of spinal cords (16 μm) were also stained with NISSL staining (Sigma-Aldrich, Saint Louis, Missouri, #C5042), performed according to the Sigma-Aldrich manufacturer's instructions.
2.7. Immunofluorescences
Cryosections were fixed in 4% formaldehyde (Sigma-Aldrich, Saint Louis, Missouri, #F8775) for 10 min at room temperature, then washed and blocked with 1% BSA (Sigma-Aldrich, Saint Louis, Missouri, #A9647) and 0.2% Triton (Sigma-Aldrich, Saint Louis, Missouri, # X100) for 1 h. Samples were then incubated with a 1:100 dilution in 1% BSA of rabbit polyclonal anti-Neurofilament antibody (Genetex, Hsinchu City, Taiwan, # GTX110065) or 1:200 anti-p62/SQSTM1 (C terminus) (Progen, #GP62-C) overnight at 4 °C, followed by incubation with a 1:500 dilution in 1% BSA of Alexa Fluor 568 anti-rabbit (ThermoFisher, Waltham, Massachusetts, #A11011) and α-bungarotoxin Alexa fluor-488 conjugate (ThermoFisher, Waltham, Massachusetts, # B13422), or Alexa Fluor 488 anti-guinea pig IgG (ThermoFisher, Waltham, Massachusetts, #A11073) for one hour at room temperature. Nuclei were stained with TO-PRO-3 iodide (ThermoFisher, Waltham, Massachusetts, #T3605).
2.8. Morphometric analyses
Myofiber cross-sectional area and p62 fluorescence were quantified on H&E or immunostained sections, respectively, by using Image J software. The entire muscle section was quantified for each sample.
NMJ images were collected using Leica software (Leica Microsystems) and analyzed using ImageJ software. Endplate and AChR areas were analyzed using “NMJ-morph” ImageJ plugin [20]. Perforation area was calculated by subtraction of AChR area from endplate area.
Innervation was quantified using images acquired with on confocal microscope LEICA-DM-IRE2 (LCSSP2) equipped with LEICA TSCSP2 laser and DC500 camera. About 20 NMJs were quantified per mouse. Innervation was evaluated based on the overlap between neurofilament and postsynaptic (α-488 bungarotoxin) components. About 23 NMJs were quantified per mouse.
2.9. RNA extraction and real-time PCR
To reduce the numbers of mice in our experiments, considering that a similar phenotype was observed in the TA and GA muscles, GA muscles were consistently used for gene expression analyses. Total RNA was isolated and purified from 30 to 50 mg of GA muscles by using Trizol (Invitrogen, Carlsbad, California, #15596018), following the manufacturer's protocol. One microgram of total RNA was converted to cDNA by using the PrimeScript™ RT reagent Kit (Takara, Kyoto, Japan, #RR047A). Real-time PCR was performed with the SDS-ABI Prism 7500 (Applied Biosystem) by using the Sybr Green reaction mix (Applied Biosystem, Foster City, California, #4368577). Gapdh was used as loading control. For micro-RNAs, RT-PCR was performed by using the TaqMan MicroRNA Reverse Transcriptase kit (Applied Biosystems) and the specific primers for miR-206 and U6, as loading control, according to the manufacturer's recommendations. miR-206 expression was analyzed by quantitative real-time PCR, using TaqMan™ MicroRNA Assay (Applied Biosystems).
The primer sequences used are reported below:
Hdac4 for: GTCTTGGGAATGTACGACGC.
Hdac4 rev: GTTGCCAGAGCTGCTATTTG.
Atrogin1 for: GCAAACACTGCCACATTCTCTC.
Atrogin1 rev: CTTGAGGGGAAAGTGAGACG.
MuRF1 for: ACCTGCTGGTGGAAAACATC.
MuRF1 rev: CTTCGTGTTCCTTGCACATC.
LC3b for: CACTGCTCTGTCTTGTGTAGGTG.
LC3b rev: TCGTTGTGCCTTTATTATTAGTGCATC.
p62 for: CCCAGTGTCTTGGCATTCTT.
p62 rev: AGGGAAAGCAGAGGAAGCTC.
Myogenin for: GCACTGGAGTTCGGTCCCAA;
Myogenin rev: TATCCTCCACCGTGATGCTG.
Chrna1 for: CTCTCGACTGTTCTCCTGCTG;
Chrna1 rev: GTAGACCCACGGTGACTTGTA.
Chrng for: GGAGAAGCTAGAGAATGGTCC;
Chrng rev: CCCACTGACAAAGTGACTCTGC.
Dach2 for: ACT GAA AGT GGC TTT GGA TAA.
Dach2 rev: TTC AGA CGC TTT TGC ATT GTA.
UCP1 for: CGACTCAGTCCAAGAGTACTTCTCTTC.
UCP1 rev: GCCGGCTGAGATCTTGTTTC.
SOD1 for: CAC TTC GAG CAG AAG GCA AG.
SOD1 rev: CCC CAT ACT GAT GGA CGT GG.
Gapdh for: ACCCAGAAGACTGTGGATGG.
Gapdh rev: CACATTGGGGGTAGGAACA.
2.10. Protein extraction and western blot analyses
GA muscles or ventral roots of spinal cords were dissected, minced, and homogenized in lysis buffer (50 mM Tris HCl pH 7.4) (Sigma-Aldrich, Saint Louis, Missouri, #RES 3098 T-B7); 1 mM EDTA (VWR, Radnor, Pennsylvania, #A10713.36); 150 Mm NaCl (Sigma-Aldrich, Saint Louis, Missouri #31434-M), 1% Triton (Sigma-Aldrich, Saint Louis, Missouri, #X100) supplemented with protease (Sigma-Aldrich, Saint Louis, Missouri, #P8340) and phosphatase inhibitors (Sigma-Aldrich, Saint Louis, Missouri, #P2850). Proteins (30–50 micrograms) were separated by SDS-PAGE and transferred to nitrocellulose membrane (GE Healthcare Life science Amersham, #10600002) for one hour. For poly-ubiquitinated proteins, proteins were transferred overnight, and membranes were boiled for five minutes. Unspecific binding was blocked with 5% not fat dry milk (Santa Cruz, California, #sc-221188) in TBST buffer: 20 mM Tris HCl ph 7.6 (Sigma-Aldrich, Saint Louis, Missouri, #RES 3098 T-B7); 137 mM NaCl (Sigma-Aldrich, Saint Louis, Missouri, #31434-M); 0.5% Tween 20 (Santa Cruz, California, #SC-29113), and the membranes were incubated overnight, at 4 °C, with primary antibody diluted in 5% BSA (Sigma-Aldrich) in TBST. After washing in TBST, membranes were incubated with secondary antibodies HRP-conjugate (BIO-RAD, Hercules, California, #170–6515 or 170–6516) and signals were detected by using ECL chemistry (Advansta, Menlo Park, California, #K-12045-D20). Images were acquired using films or ChemiDoc MP imaging system (BIO-RAD). Anti-alpha tubulin was used as loading control because its expression resulted unaffected by experimental models. Densitometric analyses were performed by measuring band intensity for each sample using Image J software. For the anti-ubiquitin western blots, the intensity of all the bands in the membrane was quantified. The following primary antibodies were used: HDAC4 (Santa Cruz, California, #sc-11418), Gapdh (Santa Cruz, California, #sc-32233), Alpha-tubulin (Sigma-Aldrich, # T5168), Ubiquitin (Stressgen, San Diego, California, #SPA-203), LC3b (Cell Signaling, Danvers, Massachusetts, #2775), p62 (Sigma-Aldrich, Saint Louis, Missouri, #P0067), Chat (Millipore, Burlington, Massachusetts, #AB144P).
2.11. Proteasome assay
Proteasome activity was assessed as previously described in [21]. Briefly, GA muscles were isolated, minced, and homogenized in western blot lysis buffer. Ten micrograms of proteins were used for measuring proteasome activity, by using the CHEMICON Proteasome Activity Assay Kit (Millipore, Burlington, Massachusetts, #APT280) following manufacturer's instructions.
2.12. Next generation sequence analysis
Total RNA was harvested from five SOD and six SOD HDAC4mKO mice. Samples were pooled together and sequenced as duplicates. Analyses were performed at the IGA Technology Services Srl, Udine, Italy. Libraries were generated from total RNA from GA muscles of 14-week-old mice, using the Stranded mRNA Sample Prep kit” (Illumina, San Diego, CA) and sequenced using HiSeq2500 (Illumina, San Diego, CA). Data were deposited in GEO (reference Series number: GSE121789).
2.13. Bioinformatic analyses
On average 56.4 million reads were produced per sample. Read quality was asserted through FastQC, and quality/adapter trimming was carried out using Cutadapt. Subsequently, reads were pseudoaligned to transcriptome GRCm38 (mm10) using Kallisto version 0.43.1. Subsequently, count tables were generated in R using tximport together with DESeq2 [22]. Differential expression of genes was conducted using DESeq2 with default options. Genes were considered differentially expressed if the p-value was<0.05. Gene ontology analyses and gene networks were obtained by David software (https://david.ncifcrf.gov) or by Ingenuity pathway analysis (QIAGEN Redwood City, www.qiagen.com/ingenuity). Over-representation analysis was carried out using the R packages DOSE and ReactomePA [23,24]. The significance of enrichment is determined through application of the hypergeometric test.
2.14. Statistics and program
Statistical significance was determined by using two-tailed Student's t-test when two conditions were compared, or with one-way or two-way analysis of variance (ANOVA), followed by Tukey's HSD test as a post-hoc test, when more than two conditions needed to be compared. All values were expressed as mean ± standard error of the mean (SEM). VassarStats, a statistical computation website available at http://vassarstats.net/, was used for the statistical analyses. The Kaplan–Meier analysis with Log-rank (Mantel-Cox) test was used for ALS onset, progression and mouse survival. All the artwork was created by using Sigma Plot program.
3. Results
3.1. HDAC4 expression is modulated in a murine model of ALS
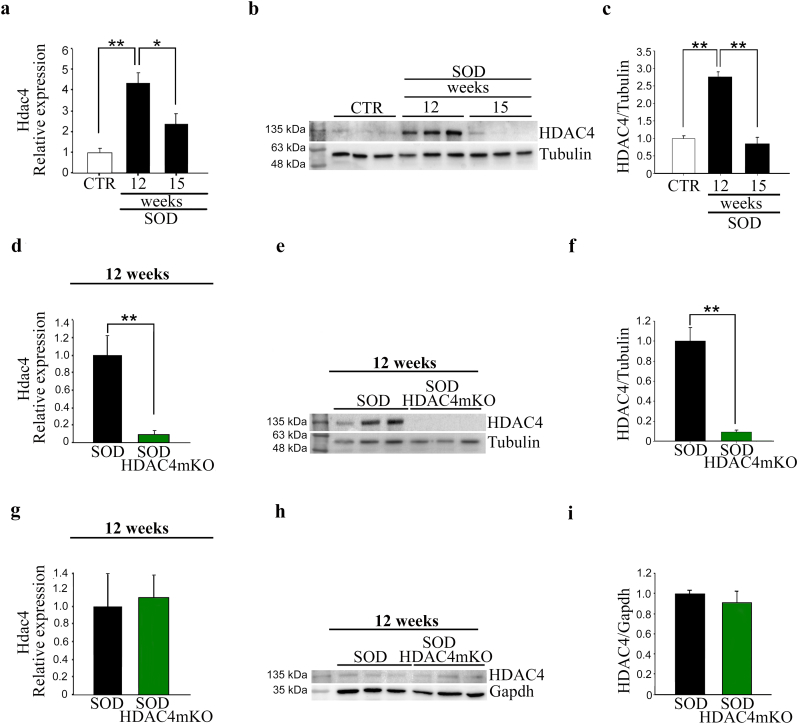
Increased HDAC4 expression in skeletal muscle has been reported to occur in SOD mice or ALS patients [10,12,19]. To evaluate the expression levels of HDAC4 in ALS over time, we analyzed HDAC4 mRNA and protein expression levels in the GA muscles of SOD mice, at a preonset (12 weeks) and at a symptomatic (15 weeks) stage, compared to healthy control (CTR) muscles. HDAC4 mRNA and protein levels were significantly upregulated in skeletal muscle at a preonset stage and decreased at 15 weeks of age (Fig. 2a–c).
Fig. 2.
HDAC4 expression is modulated in the skeletal muscle of SOD and SOD HDAC4mKO. (a) Hdac4 expression in GA muscles of SOD mice, at 12 and 15 weeks of age, over healthy control (CTR) muscles, by real-time PCR. Data are expressed as mean +/− SEM. n = 4 mice for each condition. (One-way ANOVA reveals a significant effect: F = 14.34; df 2; p = 0.003; and interaction: *p < 0.05 and **p < 0.01 by Tukey's HSD test). (b) Western blot analyses and (c) densitometry for HDAC4 in GA muscles of CTR and SOD mice, at 12 and 15 weeks of age. Alpha-tubulin was used as a loading control. Data are expressed as mean +/− SEM. n = 3 mice for each condition. (One-way ANOVA reveals a significant effect: F = 57.25; df 2; p < 0.0001; and interaction: **p < 0.01 by Tukey's HSD test). (d) Hdac4 expression in GA muscles of SOD and SOD HDAC4mKO mice, by real-time PCR, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 4 SOD; n = 5 SOD HDAC4mKO. (**p < 0.01 by Student's t-test.) (e) Western blot analyses and (f) densitometry for HDAC4 in GA muscles of SOD and SOD HDAC4mKO mice, at 12 weeks of age. Alpha-tubulin was used as a loading control. Data are expressed as mean +/− SEM. n = 3 mice for each genotype. (**p < 0.01 by Student's t-test). (g) Hdac4 expression in SOD and SOD HDAC4mKO spinal cords, by real-time PCR, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 4 SOD; n = 5 SOD HDAC4mKO. (h) Western blot analyses and (i) densitometry for HDAC4 in SOD and SOD HDAC4mKO spinal cords, at 12 weeks of age. Gapdh was used as a loading control. Data are expressed as mean +/− SEM. n = 3 mice for each condition.
To delineate HDAC4 functions in skeletal muscle in ALS, mice with a muscle-specific deletion of HDAC4 [25] (referred to as HDAC4mKO mice), in which the reduction of HDAC4 expression in skeletal muscle was confirmed by real-time PCR and western blot analyses (Supplementary Fig. S1a-c), were crossed with SOD mice, generating SOD HDAC4mKO mice (Fig. 1). Real-time PCR and western blot analyses confirmed a significant reduction of HDAC4 expression in SOD HDAC4mKO muscles, compared to SOD littermates, at 12 weeks of age (Fig. 2d–f). Instead, no significant changes in HDAC4 expression were registered in spinal cords among healthy CTR, SOD, and SOD HDAC4mKO mice (Fig. 2g–i).
3.2. Deletion of HDAC4 exacerbates ALS phenotype
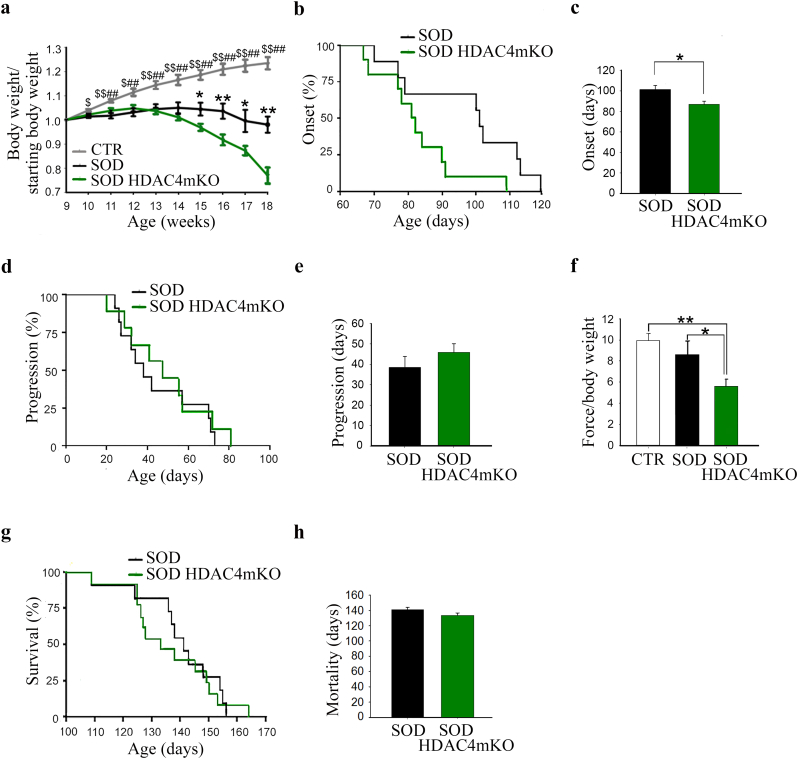
Body weight is a widely used parameter to assess the onset of ALS and to stage it. Therefore, to define the role of HDAC4 in ALS, we monitored the ALS onset and progression in male SOD HDAC4mKO and SOD littermates, by measuring mouse weight over time, compared to male healthy male control (CTR) mice. While CTR mice continued to gain weight over time, SOD mice reached the maximum body weight at 14 weeks of age and deletion of HDAC4 in skeletal muscle induced a precocious and exacerbated body weight loss in SOD HDAC4mKO mice (Fig. 3a). HDAC4 deletion in SOD mice significantly shortens ALS onset (Fig. 3b, c), defined as the time when the body mass starts to decline. Conversely, ALS progression, defined as the time from the onset of the disease to the death of the animal, did not significantly change between SOD HDAC4mKO and SOD mice (Fig. 3d, e). To assess if HDAC4 also affects muscle strength, a grip-test at the preonset stage (i.e., 12 weeks, when HDAC4 expression resulted significantly induced in SOD skeletal muscle) was performed. Importantly, while no significant differences were registered between healthy and SOD mice, SOD HDAC4mKO mice showed a significant reduction in muscle force as soon as 12 weeks of age, compared to SOD littermates and healthy CTR mice (Fig. 3f). Moreover, HDAC4 deletion exacerbated the disease features of SOD mice, including body weight loss, tremor, and kyphosis, although mouse lifespan was not significantly affected (Fig. 3g, h). Of note, HDAC4mKO mice at baseline did not show significant differences in body weight or muscle strength (Supplementary Fig. S1d, e) as previously reported [9,25].
Fig. 3.
Genetic deletion of HDAC4 in skeletal muscle exacerbates the pathological features of ALS in male mice. (a) Body weight of healthy control (CTR), SOD and SOD HDAC4mKO mice, normalized to the body weight at 9 weeks. Data are expressed as mean +/− SEM. n = 8 CTR; n = 14 SOD and n = 14 SOD HDAC4mKO mice. (One-way ANOVA reveals a significant effect at each time point analyzed. 10 weeks: F = 5.02; df 2; p = 0.01249; 11 weeks: F = 10.05; df 2; p = 0.00039; 12 weeks: F = 8.75; df 2; p = 0.000894; 13 weeks: F = 12.29; df 2; p < 0.0001; 14 weeks: F = 17.17; df 2; p < 0.0001; 15 weeks: F = 20.04; df 2; p < 0.0001; 16 weeks: F = 27.56; df 2; p < 0.0001; 17 weeks: F = 21; df 2; p < 0.0001; 18 weeks: F = 31.39; df 2; p < 0.0001 and interactions among variables; *p < 0.05; **p < 0.01 SOD vs SOD HDAC4mKO; $p < 0.05; $$p < 0.01 CTR vs SOD; #p < 0.05; ##p < 0.01 CTR vs SOD HDAC4mKO by Tukey's HSD test.) (b) Age of ALS onset of SOD and SOD HDAC4mKO mice. n = 14 mice for genotype. (*p = 0.039 by log-rank Mantel-Cox test). (c) Average days at the ALS onset of SOD and SOD HDAC4mKO mice. Data are expressed as mean +/− SEM. n = 14 mice for each genotype (*p < 0.05 by Student's t-test.) (d) ALS progression of SOD and SOD HDAC4mKO mice. n = 14 mice for genotype. (e) Average days of ALS progression of SOD and SOD HDAC4mKO mice. n = 14 mice for each genotype. (f) Muscle performance of CTR, SOD and SOD HDAC4mKO mice, by grip test, at 12 weeks of age. Data are expressed as the mean of developed force over the mouse weight, +/− SEM. n = 8 CTR; n = 9 SOD and n = 12 SOD HDAC4mKO mice. (One-way ANOVA reveals a significant effect among genotypes. F = 6.16; df 2; p = 0.009; and interaction: *p < 0.05; **p < 0.01 by Tukey's HSD test.) (g) Survival curve of SOD and SOD HDAC4mKO mice. n = 15 SOD and n = 17 SOD HDAC4 mice. (h) Lifespan of SOD and SOD HDAC4mKO mice. Data are expressed as mean +/− SEM n = 15 SOD and n = 17 SOD HDAC4 mice.
3.3. Muscle atrophy correlates with body weight loss in SOD HDAC4mKO mice
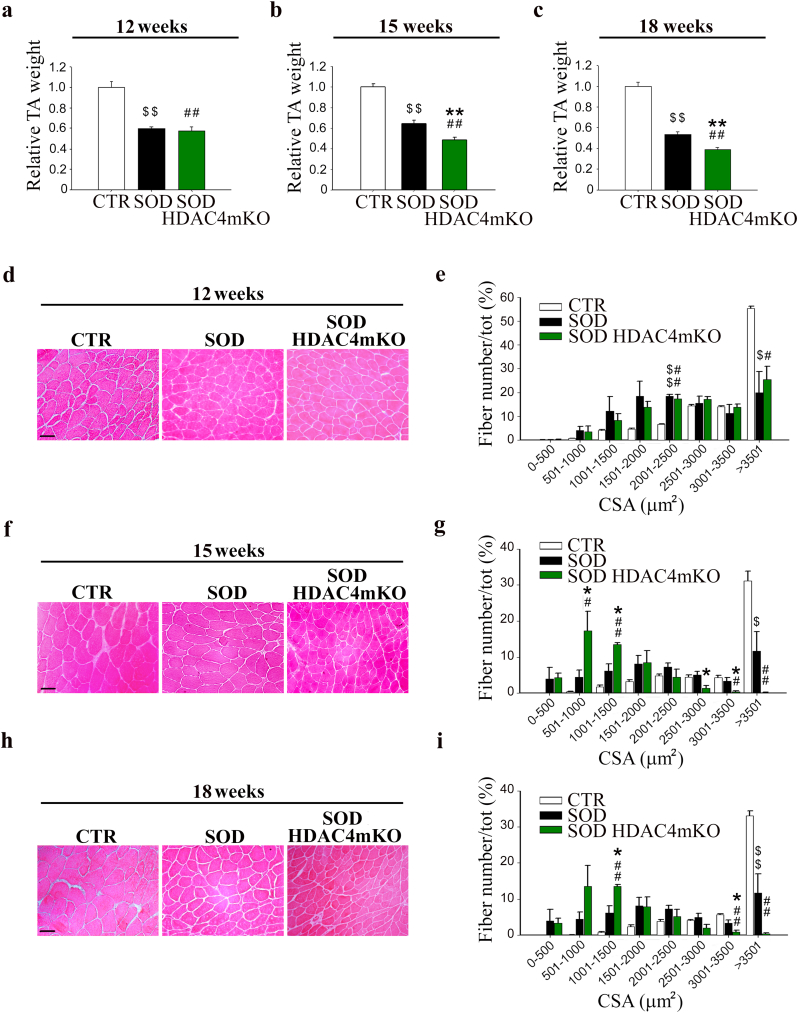
To verify if the body weight loss correlated with muscle mass loss, TA muscles were weighted, and histological analyses were performed at different ages. At 12 weeks (preonset stage), no significant differences in muscle mass or histology were observed between SOD and SOD HDAC4mKO littermates (Fig. 4a, d), despite a significant decrease in muscle mass was registered versus CTR mice. Morphometric analyses of myofiber cross-sectional area (CSA) distribution confirmed these observations, highlighting a significant difference between CTR and either SOD or SOD HDAC4mKO TA myofibers in the 2001–2500 and > 3501 square micron classes (Fig. 4e). At 15 weeks (symptomatic stage), the differences between CTR and SOD or SOD HDAC4mKO mice persisted (Fig. 4b, f, g). Moreover, a significant reduction in muscle mass was apparent in SOD HDAC4mKO mice, compared to SOD littermates (Fig. 4b). Histological and morphometric analyses highlighted a significant increase in the number of small myofibers (501–1500 square micron) and a significant concomitant decrease in the number of big myofibers (2501–3500 square micron) in SOD HDAC4mKO TA muscles compared to SOD littermates, indicative of muscle atrophy (Fig. 4g). At 18 weeks (late stage), the reduction in SOD and SOD HDAC4mKO TA weight persisted, compared to CTR mice (Fig. 4c). Moreover, the differences between SOD and SOD HDAC4mKO TA weight remained (Fig. 4c). Histological and morphometric analyses highlighted that, while SOD mice were characterized by myofibers with heterogeneous size, SOD HDAC4mKO mice showed more pronounced muscle atrophy, characterized by a significantly higher number of smaller myofibers (1001–1500 square micron) and a lower number of bigger myofibers (3001–3500 square micron) (Fig. 4h, i). A similar phenotype was observed in the GA muscles (Supplementary Fig. S2), allowing us to consistently use TA muscles for histological evaluations and GA muscles for molecular biology analyses, to reduce the mouse number in our experiments. Importantly, no significant changes in TA weight, histology or fiber CSA distribution were registered in HDAC4mKO mice at baseline (Supplementary Fig. S1f–h), as previously reported [9,25].
Fig. 4.
Deletion of HDAC4 worsens muscle atrophy in ALS. (a, b, c) TA muscle weight of healthy control (CTR), SOD and SOD HDAC4mKO mice at 12, 15 or 18 weeks of age. Data are expressed as mean of TA weight relative to SOD littermates +/− SEM. n = 5 CTR, n = 6 SOD and n = 8 SOD HDAC4mKO mice in a; n = 5 CTR, n = 8 SOD and n = 10 SOD HDAC4mKO mice in b; n = 10 CTR, n = 8 SOD and n = 8 SOD HDAC4mKO mice in c. (One-way ANOVA reveals a significant effect: F = 30.91; df 2; p < 0.0001 in a; F = 48.01; df 2; p < 0.0001 in b; F = 106.6; df 2; p < 0.0001 in c; and interaction: **p < 0.01 SOD vs SOD HDAC4mKO; $$p < 0.01 CTR vs SOD; ##p < 0.01 CTR vs SOD HDAC4mKO by Tukey's HSD test.) (d, f, h) Representative images of TA muscle histology at 12, 15 or 18 weeks of age. Scale bar = 50 μm. (e, g, i) Myofiber CSA distribution of CTR, SOD and SOD HDAC4mKO TA muscles at 12, 15 or 18 weeks of age. Data are expressed as mean +/− SEM. n = 3 mice per each genotype in e; n = 4 mice per each genotype in g and i. (One-way ANOVA reveals a significant effect in the following classes: 2001–2500 μm2 F = 29.28; df 2; p = 0.0008 and > 3500 μm2 F = 9.89; df 2; p = 0.012 in e; in 501–100 μm2 F = 7.86; df 2; p = 0.012; in 1001–1500 μm2 F = 18.97; df 2; p = 0.0009; in 2501–3000 μm2 F = 5.2; df 2; p = 0.035; in 3001–3500 μm2 F = 8.03; df 2; p = 0.012; in >3501 μm2 F = 16.85; df 2; p = 0.001 in g; and in 1001–1500 μm2 F = 22.9; df 2; p = 0.0004; in 3001–3500 μm2 F = 15.08; df 2; p = 0.001 and in >3501 μm2 F = 27.14; df 2; p = 0.0001 in i; and interaction: *p < 0.05 SOD vs SOD HDAC4mKO; $p < 0.05; $$p < 0.01 CTR vs SOD; #p < 0.05; ##p < 0.01 CTR vs SOD HDAC4mKO by Tukey's HSD test.)
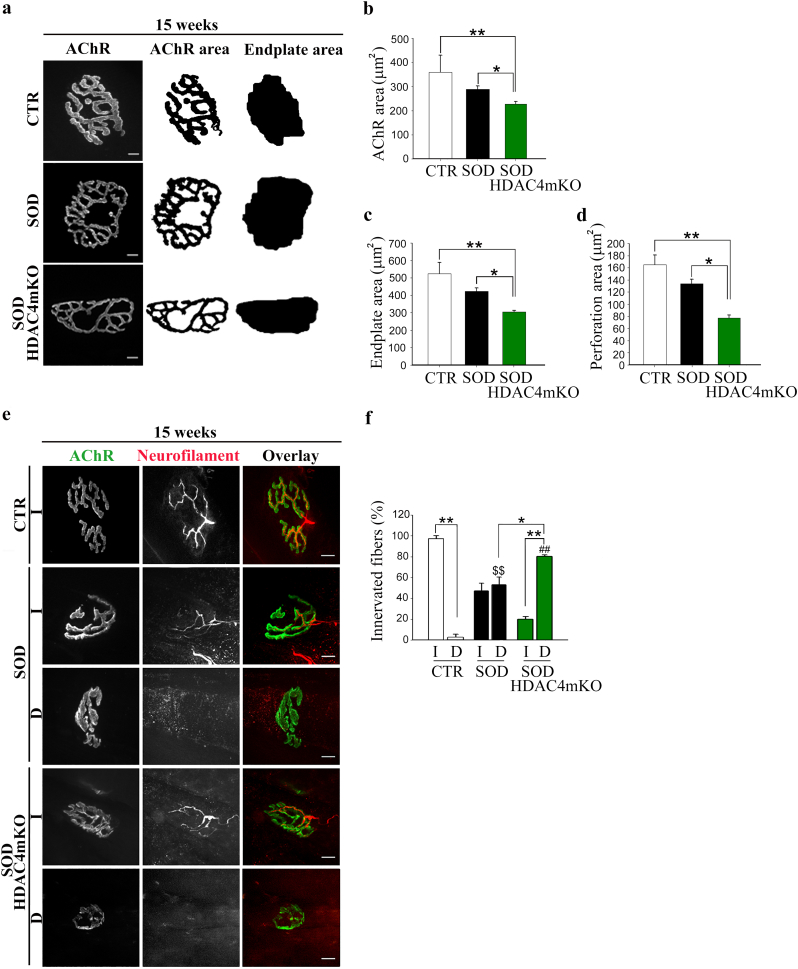
3.4. HDAC4 affects NMJs and innervation in ALS
NMJ degeneration and muscle denervation are the two main features of ALS that precede and contribute to neurogenic muscle atrophy. To evaluate if the HDAC4 deletion affected NMJ shape, acetylcholine receptors (AChRs) were labeled using fluorescently conjugated Bungarotoxin (BTX) in healthy CTR, SOD and SOD HDAC4mKO TA muscles, and the synaptic areas were analyzed at different ages. At a preonset stage, no apparent differences in NMJ morphology were observed among the experimental groups (data not shown). At 15 weeks, however, SOD HDAC4mKO mice had reduced NMJ surface area as compared to SOD littermates or healthy CTR mice, while no major differences were observed between CTR and SOD mice (Fig. 5a, b). This reduction was associated with a decrease of the area occupied by AChRs, as well as of the area of perforations between AChR-rich regions [26] (Fig. 5c, d). To evaluate if HDAC4 deletion affects NMJ innervation in SOD mice, TA muscles were stained with anti-neurofilament antibody and analyzed using confocal microscopy. No differences were detected at a preonset stage (data not shown). At 15 weeks, while almost the totality of the CTR NMJs resulted innervated, as expected, SOD HDAC4mKO mice displayed a significantly higher percentage of denervated NMJs as compared to SOD littermates (Fig. 5e, f). Importantly, NMJs and innervations formed and matured normally in HDAC4mKO mice (Supplementary Fig. S1i), as previously reported [7].
Fig. 5.
HDAC4 alters NMJ structure and innervation in ALS. (a) Representative images of NMJ postsynaptic machinery in healthy control (CTR), SOD and SOD HDAC4mKO TA muscles, at 15 weeks of age. Right images show processing during NMJ analysis for automated quantification of AChR and endplate areas. Scale bar = 10 μm. (b) Quantification of AChR, (c) endplate and (d) perforation areas. Data are expressed as mean +/− SEM. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 9.13; df 2; p = 0.015 in b; F = 9.64; df 2; p = 0.007 in c; F = 18.12; df 2; p = 0.002 in d; and interaction: *p < 0.05; **p < 0.01 by Tukey's HSD test.) (e) Representative images of innervated (I) and denervated (D) NMJs, at 15 weeks of age. Scale bar = 10 μm. (f) Quantification of innervated and denervated NMJs in CTR, SOD and SOD HDAC4mKO mice. Data are expressed as mean +/− SEM. n = 3 CTR; n = 5 SOD or SOD HDAC4mKO mice. (Two-way ANOVA reveals a significant effect of denervation: F = 5.35; df 1; p = 0.033 and interaction between the genotype and denervation: F = 124.64; df 2; p < 0.0001; *p < 0.05; **p < 0.01; $$p < 0.01 D SOD vs D CTR; ##p < 0.01 D SOD HDAC4mKO vs D CTR by Tukey's HSD test.)
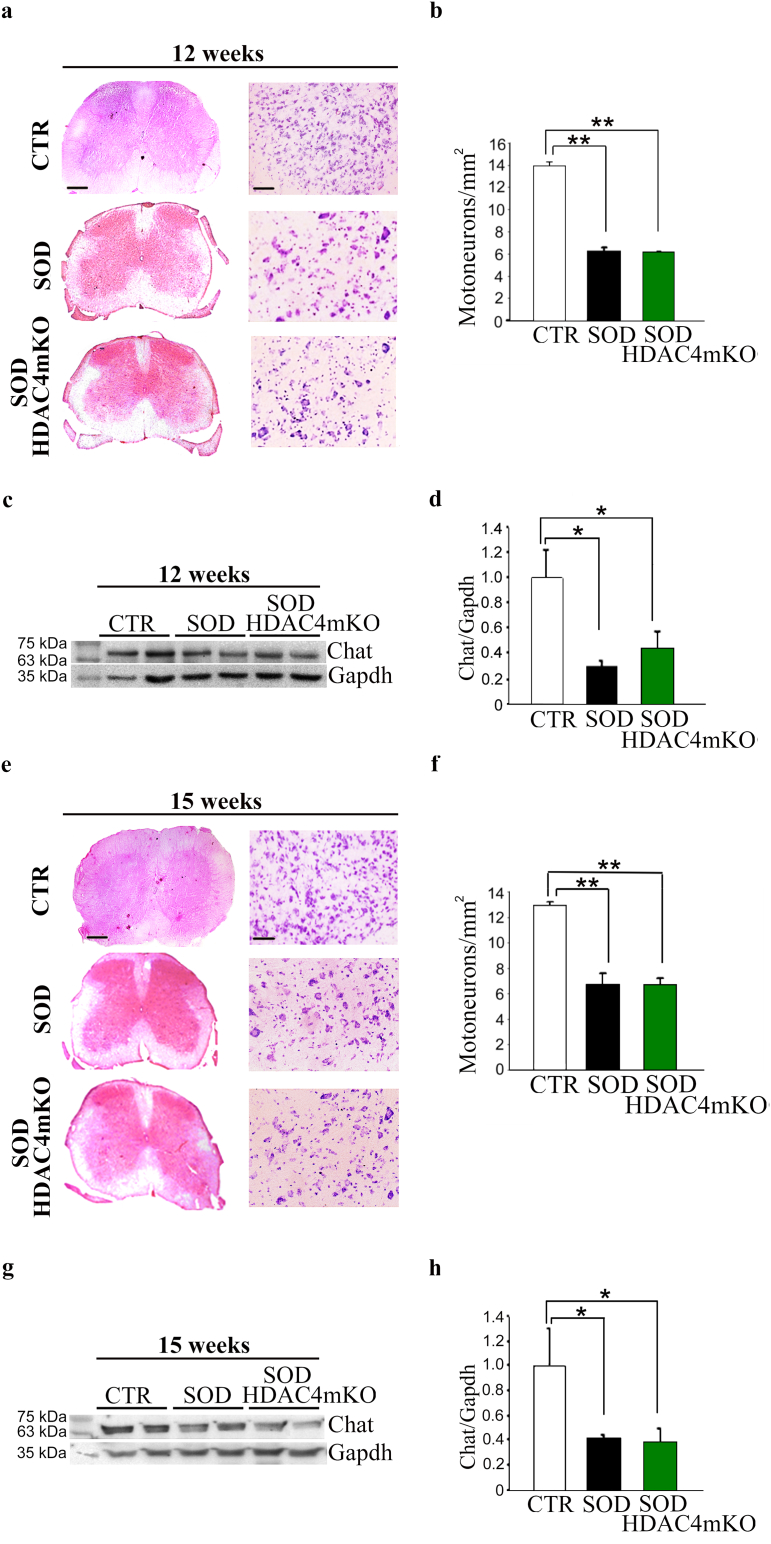
3.5. HDAC4 does not affect motoneuron survival in ALS
To define if HDAC4 from skeletal muscle impacts on the spinal cord, histological and molecular analyses were performed on healthy CTR, SOD, and SOD HDAC4mKO spinal cords over time. Despite the significant decrease in motor neurons between CTR and SOD or SOD HDAC4mKO mice, no differences in the spinal cord ventral horn morphology, or in the number of motor neurons, visualized by NISSL staining, were detected between SOD and SOD HDAC4mKO mice at 12 or 15 weeks of age (Fig. 6a, b, e, f). Western blot analyses from ventral roots for choline acetyltransferase (Chat), a motor neuron marker, confirmed that no significant changes in protein levels occurred between SOD and SOD HDAC4mKO mice, even though SOD mice showed a significant decrease in Chat expression compared to healthy CTR mice (Fig. 6c, d, g, h), as expected. These results indicate that HDAC4 deletion in muscle does not affect motoneuron bodies in the central nervous system in ALS.
Fig. 6.
Skeletal muscle-specific HDAC4 deletion does not affect the central nervous system in ALS. (a, e) Representative pictures of spinal cord sections of healthy control (CTR), SOD and SOD HDAC4mKO mice, stained with hematoxylin and eosin (left panel; scale bar = 20 μm) and NISSL staining (right panel; scale bar = 50 μm). (b, f) Quantification of motor neurons of CTR, SOD and SOD HDAC4mKO mice, at 12 or 15 weeks of age. Data are expressed as mean +/− SEM. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 1112.3; df 2; p < 0.0001 in b; F = 54.14; df 2; p = 0.0001 in f; **p < 0.01 by Tukey's HSD test.) (c, g) Western blot analysis and (d, h) densitometry for Chat in SOD and SOD HDAC4mKO ventral roots of spinal nerves, over CTR, at 12 or 15 weeks of age. Gapdh was used as a loading control. Data are presented as mean +/− SEM. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 6.34; df 2; p = 0.0147 in d; F = 9.98; df 2; p = 0.0123 in h; and interaction: *p < 0.05 by Tukey's HSD test.)
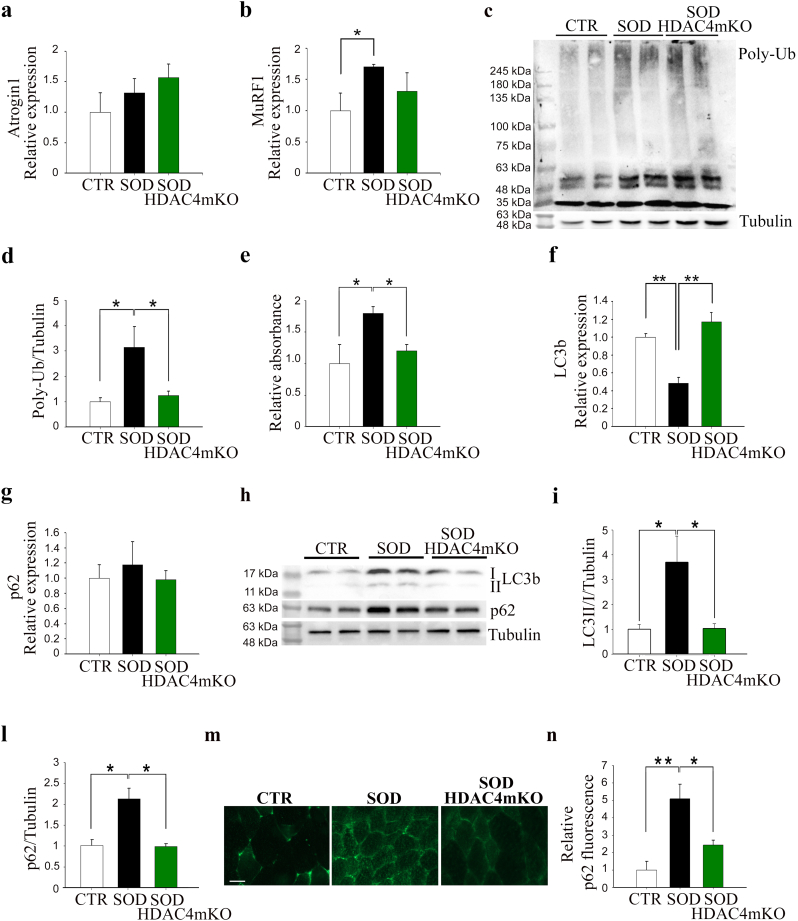
3.6. HDAC4 mediates the activation and progression of the catabolic pathways in ALS
Activation of catabolic pathways, such as the ubiquitin-proteasome system (UPS) and autophagy, is associated with muscle atrophy [27,28]. We wondered if the earlier occurrence of muscle atrophy in SOD HDAC4mKO muscles could depend on a previous activation of the catabolic pathways. Therefore, we monitored the activation of both UPS and autophagic markers in GA muscles, at 12 and 15 weeks of age. No significant differences regarding UPS activation were observed among the experimental groups at 12 weeks of age, as assessed by the expression levels of the E3 ubiquitin ligases Atrogin1 and MuRF1, the amount of poly-ubiquitinated proteins and the proteasome activity (Supplementary Fig. S3a–e). Importantly, no differences in UPS activation were registered at baseline between CTR and HDAC4mKO mice [29]. No changes among genotypes occurred in the expression and protein levels of the two autophagic markers, LC3b and p62, at this time point (Supplementary Fig. S3f–i), as well as in HDAC4mKO mice at baseline [29]. In contrast, at 15 weeks of age, a significant up-regulation of MuRF1 expression was observed in SOD mice, compared to healthy CTR, suggesting an activation of the proteasome pathway, but not in SOD HDAC4mKO (Fig. 7a, b). The evaluation of poly-ubiquitinated proteins and the quantification of the proteasome activity confirmed this result. Indeed, poly-ubiquitinated proteins and proteasome activity were significantly increased in SOD mice, when compared to CTR (Fig. 7c–e). Conversely, SOD HDAC4mKO mice displayed levels of poly-ubiquitinated proteins and proteasome activity similar to those of CTR animals (Fig. 7c–e). Moreover, at 15 weeks of age, SOD mice showed significant down-regulation of the LC3b expression but no changes in p62 mRNA levels, when compared to CTR or SOD HDAC4mKO mice (Fig. 7f, g). However, SOD mice showed significant accumulation of both LC3b II and p62 proteins with respect to CTR mice, suggesting a compromised autophagic flux, while protein accumulation was not present in SOD HDAC4mKO mice (Fig. 7h–l). Significant differences in p62 protein levels among CTR, SOD and SOD HDAC4mKO mice, were also detected and quantified by immunofluorescence analyses, at 15 weeks of age (Fig. 7m, n), confirming the above results.
Fig. 7.
SOD HDAC4mKO mice do not activate the catabolic pathways. (a) Expression levels of Atrogin1 and (b) MuRF1 in healthy control (CTR), SOD and SOD HDAC4mKO GA muscles, at 15 weeks of age, by real-time PCR. n = 4 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 5.45; df 2; p = 0.032; and interaction *p < 0.05 by Tukey's HSD test.) (c) Western blot analyses and (d) quantification of poly-ubiquitinated proteins in CTR, SOD and SOD HDAC4mKO GA muscles, at 15 weeks of age. Alpha-tubulin was used as a loading control. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect among genotypes (F = 5.7; df 2; p = 0.018); *p < 0.05 by Tukey's HSD test.) (e) Proteasome activity in CTR, SOD and SOD HDAC4mKO GA muscles, at 15 weeks of age. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 6.85; df 2; p = .018; and interaction: *p < 0.05 by Tukey's HSD test.) (f) LC3b and (g) p62 expression levels in CTR, SOD and SOD HDAC4mKO GA muscles, at 15 weeks of age, by real-time PCR. n = 4 mice for each genotype. (One-way ANOVA reveals a significant effect in f: F = 17.1; df 2; p = 0.0004; and interaction: **p < 0.01 by Tukey's HSD test.) (h) Western blot analyses and (i, l) densitometry for LC3b and p62 proteins of CTR, SOD and SOD HDAC4mKO GA muscles at 15 weeks of age. Alpha-tubulin was used as a loading control. n = 5 mice for each genotype. Data are shown as mean ± SEM, over CTR mice. (One-way ANOVA reveals a significant effect: F = 6.17; df 2; p = 0.014 in i; F = 16.38; df 2; p = 0.0003 in l; and interaction: *p < 0.05 by Tukey's HSD test.) (m) Representative images and (n) quantification of immunofluorescence for p62 of CTR, SOD and SOD HDAC4mKO muscles at 15 weeks of age. Scale bar = 25 μm. Data are presented as mean +/− SEM. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 12.84; df 2; p = 0.0067; and interaction: *p < 0.05; **p < 0.01 by Tukey's HSD test.)
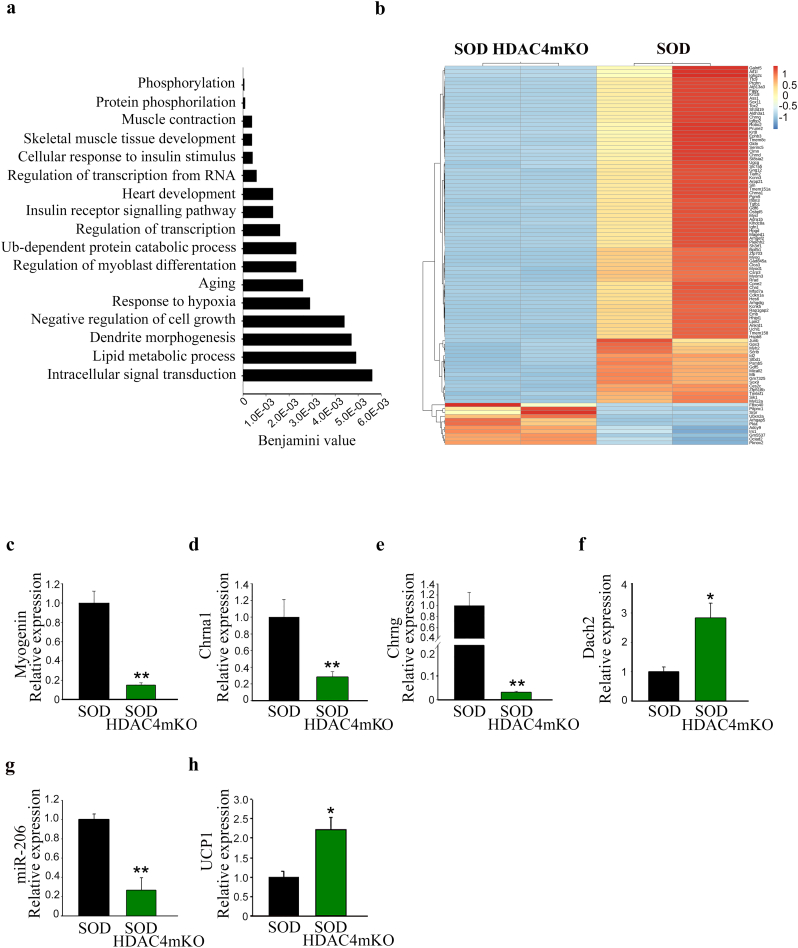
3.7. HDAC4 modulates different cellular pathways in ALS
To define the role of HDAC4 in ALS, we delineated the molecular pathways globally modulated by HDAC4 in skeletal muscle. Transcriptome analysis was performed by comparing SOD and SOD HDAC4mKO GA muscles, at the onset of ALS, i.e. at 14 weeks of age. Total RNA was harvested from five SOD and six SOD HDAC4mKO mice. Samples were pooled together and sequenced as duplicates of SOD and SOD HDAC4mKO muscles. HDAC4 significantly (p < .05) modulated 2508 genes: 49% resulted up-regulated in SOD HDAC4mKO mice, while 51% was down-regulated, as depicted in the MA-plot graph (Supplementary Fig. S4a). Gene ontology analysis revealed that, among biological processes, HDAC4 mainly affected intracellular signal transduction (phosphorylation, protein phosphorylation, positive regulation of transcription from RNA polymerase II promoter, positive regulation of transcription, response to hypoxia, intracellular signal transduction), muscle development and contraction (muscle contraction, skeletal muscle tissue development, heart development, positive regulation of myoblast differentiation, negative regulation of cell growth), metabolism (cellular response to insulin signaling, insulin receptor signaling pathway, lipid metabolic process), ubiquitin-dependent protein catabolic process, aging and the nervous system (dendrite morphogenesis) (Fig. 8a). Top enriched gene sets between SOD HDAC4mKO and SOD mice nicely correlated with the gene ontology results and with the mutant phenotype. Indeed, in the reactome pathways, muscle contraction, transcriptional regulation by Runx2, a known HDAC4 target [30] and metabolism were found (Supplementary Fig. S4b). In disease ontology pathways, muscular disease, muscle tissue disease and myopathy were the top three hits (Supplementary Fig. S4c). A heatmap of the top 100 most significantly modulated genes revealed that HDAC4 modulates genes involved in response to denervation in ALS (Fig. 8b and Supplementary table). Among these, the expression levels of myogenin, Chrna1 and Chrng were confirmed to be down-regulated in SOD HDAC4mKO compared to SOD mice, by real-time PCR on independent samples (Fig. 8c–e). Moreover, Dach2 expression was significantly up-regulated, while miR-206 significantly down-regulated in SOD HDAC4mKO mice, if compared to SOD (Fig. 8f, g), as expected [7,9]. Of note, SOD1 expression did not significantly change between SOD and SOD HDAC4mKO mice, despite the significant increase compared to healthy CTR mice (Supplementary Fig. S4d). An interesting gene network and top upstream modulator, the uncoupling protein 1 (UCP1), were identified by Ingenuity Pathway Analysis (IPA) (Supplementary Fig. S4e). Considering that the over-expression of UCP1 in skeletal muscle in a mouse model of ALS is sufficient to increase NMJ instability and muscle denervation, worsening ALS features [31], we monitored the expression of UCP1 in the skeletal muscle of SOD and SOD HDAC4mKO mice, at 14 weeks of age. UCP1 resulted significantly up-regulated in SOD HDAC4mKO mice, with respect to SOD mice (Fig. 8h), suggesting it may be a key mediator of the SOD HDAC4mKO phenotype. Of note, the expression of none of these molecular markers resulted perturbed in HDAC4mKO mice at baseline (Supplementary Fig. S1 l–o).
Fig. 8.
HDAC4 modulates gene expression in GA skeletal muscle in ALS. (a) Gene ontology of the biological processes most significantly modulated by HDAC4 in ALS. (b) Heatmap of top 100 most significantly differentially expressed genes. Significance is determined by Benjamini-Hochberg corrected p-values between the two genotypes. (c-h) Expression levels of indicated genes in SOD HDAC4mKO mice, relative to SOD littermates, at 14 weeks of age, by real-time PCR. Data are expressed as mean +/− SEM. n = 6 SOD; n = 5 SOD HDAC4mKO mice. (*p < 0.05; **p < 0.01 by Student's t-test.)
4. Discussion
ALS is a progressive, still incurable neurodegenerative disease. Although the primary cause of ALS is motor neuron degeneration, skeletal muscle emerged as a primary target of the disease. As the etymology suggests, “a-myo-trophic” (from Greek) literally means “lack of muscle nourishment”. Without the nerve, skeletal muscle undergoes progressive atrophy, and this contributes to the progression of the disease, coherently with the “dying-back” model of the disease [4]. According to this theory, since muscle weakness and NMJ degeneration precede and contribute to motor neuron loss, treatments aimed at improving skeletal muscles may delay motor impairment [32]. In agreement, treatments to rescue motor neurons have shown only limited success in ALS mice or patients.
HDAC4 is a crucial mediator of skeletal muscle atrophy and reinnervation following denervation [7,9]. The expression of HDAC4 in the skeletal muscle of ALS patients correlates with the severity of the disease progression [12,13]. Our present data complement this information, registering an early up-regulation of HDAC4 in muscle at 12 weeks of age, followed by a decrease at 15 weeks of age. This expression pattern strongly suggests a function of HDAC4 in skeletal muscle at the preonset stage. Class II HDACi administration partially ameliorated ALS features in mice [19], although failing to clarify the specific role of each class II HDAC member in the progression of the disease. Moreover, even though the continuous studies and the advances in pharmacology improved the quality of HDACi and reduced the side effects, long-term use of HDACi is not well-tolerated in humans [33]. Some side effects can be routinely managed (e.g. nausea and vomiting); others are transient and reversible (such as thrombocytopenia, neutropenia, and anemia), but most of them negatively affect the patients' quality of life. The primary toxicities associated with HDACi are nausea, vomiting, anorexia, and fatigue, with body weight loss and severe neurological events, such as neurocognitive impairment and status epilepticus [[34], [35], [36], [37]]. Some side effects and the partial ineffectiveness of the treatments, as well, may depend on the broad action of the pan- or class-specific HDACi. Based on this evidence, understanding the molecular mechanism modulated by each HDAC member is a prerequisite for ameliorating HDACi-based therapies.
In this study, we elucidated the HDAC4 role in skeletal muscle in ALS by using a genetic approach. Muscle-specific mutant mice (SOD HDAC4mKO) were generated and HDAC4 expression in skeletal muscle, but not in spinal cords, was significantly downregulated if compared to SOD littermates and comparable to healthy CTR mice. SOD transgenic mice are a widely used animal model in the study of ALS. In these mice, the transgenic over-expression of mutated human SOD1, a gene mutated in a subset of familial ALS, results in a phenotype closely recapitulating the human disease features [38]. Hindlimb tremors, as first clinical signs of motor neuron disease, occur around 12 weeks of age in SOD mice [39], although reduced rotarod performance was detected earlier, at around 8.5 weeks of age, and correlates with skeletal muscle denervation [40]. Significant body weight loss is registered around 14 weeks of age, while muscle atrophy is detected by MRI imaging around 8.5 weeks of age in SOD mice [39,40]. Deletion of HDAC4 in skeletal muscle drastically worsened the pathological features of ALS by causing a precocious disease onset and by inducing a significant decrease in body weight and muscle atrophy. Interestingly, SOD HDAC4mKO mice showed a significant reduction in muscle strength in as early as 12 weeks. At this age, no differences in body weight, muscle mass or CSA were detected between genotypes, thus indicating that intrinsic defects in myofibers occurred before the decrease in body weight, supporting the “dying-back” model. Besides, a significant increase in muscle atrophy in SOD HDAC4mKO mice was revealed at the late stage of the disease, while the presence of very small and very big myofibers was registered in SOD muscles.
Neuromuscular junction degeneration is one of the main ALS features, causing neurogenic muscle atrophy and, eventually, lethal respiratory failure [41]. Skeletal muscle actively influences the neurodegeneration process in ALS by expressing and releasing signals that influence the maintenance of synaptic connections, axonal growth and neuron survival [42,43]. Aiming to investigate if HDAC4 affected muscle-nerve interface, evaluation of NMJ shape and innervation was performed over time. While no significant differences were detected among genotypes at 12 weeks of age, at 15 weeks, a significant reduction in the synapse area and a higher number of denervated NMJ were found in SOD HDAC4mKO mice. These data demonstrate that skeletal muscle HDAC4 regulates ALS onset, having a protective role for NMJs and muscle innervations, even though NMJ development was not affected by HDAC4 in HDAC4mKO mice, as already reported [7]. In agreement with this phenotype, the expression of an important neurotrophic factor in ALS progression, i.e., glial cell-derived neurotrophic factor (GDNF) [44], was significantly down-regulated in the transcriptome of SOD HDAC4mKO mice (Supplementary Fig. S4f). To investigate if HDAC4 might retrogradely affect the central nervous system in ALS, motor neurons were examined over time, by histological and molecular analyses. Interestingly, no differences were detected between the two experimental groups, indicating that HDAC4 does not affect motor neuron survival in ALS. Of note, all the differences seen between SOD HDAC4mKO and SOD mice, i.e., body and muscle weight, muscle performance and histology, NMJs and innervations, confirmed a specific stress response role of HDAC4 in skeletal muscle in pathological conditions, since no differences were detected at baseline in HDAC4mKO mice [9,25,29].
The decline in body weight correlated with muscle atrophy in SOD HDAC4mKO mice, indicating the importance of preserving skeletal muscle mass in ALS. To search for the molecular mechanisms underlying the enhanced atrophic phenotype, the activation of the two major catabolic pathways in skeletal muscle, i.e., the UPS and autophagy [45], was examined. SOD HDAC4mKO mice did not trigger UPS activation in ALS, coherently with the HDAC4 function following long-term denervation [29]. The lack of UPS activation could contribute to worsening the ALS phenotype in SOD HDAC4mKO mice. Accordingly, treatment with a known activator of the proteasome, i.e., methylene blue, preserves the neuromuscular function of SOD mice [46]. Regarding autophagy, at 15 weeks, SOD mice showed a significant accumulation of LC3b and p62 proteins, without a corresponding increase in gene expression, thus suggesting an impairment of the autophagy flux, in agreement with previous observations [47]. Instead, SOD HDAC4mKO mice showed expression levels of autophagic markers similar to those of healthy CTR mice, thus suggesting that HDAC4 mediates the autophagic flux in skeletal muscle in ALS, as previously observed following long-term denervation [29]. Dysregulation of the autophagic flux occurs in various neurodegenerative diseases [48]. Regarding ALS, even though an increase in autophagy was found in the spinal cords of ALS patients and animal models [[49], [50], [51], [52]], controversial results were also reported. For instance, a study showed that treatment with rapamycin, an autophagy activator, increased degradation of misfolded proteins and decreased the autophagosome accumulation in motor neurons [52]. Others reported that rapamycin administration increased motor neuron degeneration and negatively affected muscle performance in ALS mouse models [49,53], thus rendering it difficult to consider rapamycin as a potential treatment [54]. In skeletal muscle, autophagy was first described as one of the major intracellular mechanisms involved in atrophy in a mouse model of ALS [5]. However, it was recently demonstrated that the autophagic flux is impaired in SOD mice skeletal muscle, and probably abnormal autophagy contributes to muscle degeneration in ALS progression [47].
Muscle regeneration has not been reported as a clinical feature of ALS. Although HDAC4 is crucial for satellite cell proliferation and differentiation [55], satellite cells do express HDAC4 until myogenin expression occurs in SOD HDAC4mKO mice, and HDAC4mKO satellite cells do not show impaired differentiation ability in vitro if compared to controls (data not shown). From all these considerations, we believe HDAC4 functions in ALS are beyond the regulation of muscle regeneration.
Aiming to identify the molecular mechanisms regulated by HDAC4 in ALS, a transcriptome analysis was performed by comparing the skeletal muscle of SOD and SOD HDAC4 at the onset of the disease. Bioinformatic analysis revealed that HDAC4 mostly modulates intracellular signal transduction, muscle development, metabolism and ubiquitin-dependent catabolism in ALS muscles, in line with the phenotype observed. Additional bioinformatic analyses of the top enriched gene sets confirmed that HDAC4 affects muscle contraction and metabolism and that the transcriptome data effectively reflect muscle disease and myopathy. A heatmap of the 100 most significantly modulated genes revealed that HDAC4 mediates skeletal muscle response to denervation in ALS. Indeed, HDAC4 is necessary for the activation of several downstream cellular responses, such as the activation of UPS, autophagy and oxidative stress response [29]. IPA analysis of the differentially expressed genes identified a gene network controlled by UCP1 and UCP1 expression resulted in up-regulated in SOD HDAC4mKO muscles, but not in HDAC4mKO ones. UCP1 is a mitochondrial protein, typically expressed in brown adipose tissue, that induces non-shivering thermogenesis by uncoupling the mitochondrial electron transport chain [56,57]. Overexpression of UCP1 in skeletal muscle is sufficient to induce muscle atrophy, without activating the UPS, to stimulate hypermetabolism and mild motoneuron degeneration [31]. Most importantly, overexpression of UCP1 in the skeletal muscle of SOD mice reduced NMJ stability and contributed to muscle denervation, similarly to deletion of HDAC4 in ALS mice. Accordingly, muscle innervation is also controlled by metabolism. Indeed, hypermetabolism induces a chronic energy deficit in the skeletal muscle of SOD mice, preceding and contributing to muscle denervation and ALS progression [58]. As proof of concept, a high-fat diet aimed to improve the energy deficit is sufficient to improve muscle innervation and prolong the survival of SOD mice, though this strategy has not been suggested for ALS patients due to expected side effect [59]. Moreover, alterations in mitochondrial network and function have been described in skeletal muscle of ALS mice and patients at the preonset stage, being sufficient to cause NMJ degeneration and death of motoneurons in mice, worsening the ALS progression [60,61]. Several pharmacological or gene therapy approaches aimed at resolving mitochondrial dysfunction, such as increasing mitochondrial function and survival or reducing oxidative stress, have been tried. However, despite showing promising results in animal models, ALS remains an incurable and fatal disease.
5. Conclusions
ALS is emerging as a multisystemic disease in which skeletal muscle actively participates with structural and metabolic alterations, contributing to ALS progression in a “dying back” process. This study identified new functions of HDAC4 in skeletal muscle in ALS, whose deletion is sufficient to induce an earlier onset of the disease, characterized by muscle denervation, decrease in NMJ size and functional deficit, accompanied by exacerbated muscle atrophy over time. Given that pan- or class II HDACi indirectly affect HDAC4 targets, our study provides the experimental basis to reconsider the use of such non-specific drugs for the treatment of ALS. More studies are necessary to better delineate the functions of each member of the HDAC superfamily in ALS, in order to propose more specific and efficient therapeutic approaches.
The following are the supplementary data related to this article.
HDAC4mKO mice do not show obvious phenotype. (a) HDAC4 expression in GA muscles of CTR and HDAC4mKO mice, by real-time PCR, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 3 CTR; n = 4 HDAC4mKO (**p < 0.01 by Student's t-test.) (b) Western blot analyses and (c) densitometry for HDAC4 in GA muscles of CTR and HDAC4mKO mice, at 12 weeks of age. Gapdh was used as a loading control. Data are expressed as mean +/− SEM. n = 3 mice for each genotype. (**p < 0.01 by Student's t-test) (d) Body weight of male CTR and HDAC4mKO mice, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 7 CTR; n = 5 HDAC4mKO mice. (e) HDAC4mKO and CTR muscle performance, by grip test, at 12 weeks of age. Data are expressed as the mean of developed force over the mouse weight, +/− SEM. n = 5 CTR and n = 4 HDAC4mKO mice. (f) HDAC4mKO and CTR TA weight, at 12 weeks of age. Data are expressed as mean of TA weight relative to CTR littermates +/− SEM. n = 5 mice for each genotype. (g) Representative pictures of HDAC4mKO and CTR TA histology, at 12 weeks of age. Scale bar = 50 μm. (h) Myofiber CSA distribution of HDAC4mKO and CTR TA muscles, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 3 CTR and n = 4 HDAC4mKO mice. (i) Representative images of NMJ postsynaptic machinery in HDAC4mKO and CTR mice, at 12 weeks of age. n = 3 mice per genotype. Scale bar = 10 μm. (l–o) Expression levels of indicated genes in HDAC4mKO GA muscles, relative to CTR littermates, at 14 weeks of age, by real-time PCR. Data are expressed as mean +/− SEM. n = 5 mice for each genotype.
Deletion of HDAC4 worsens muscle atrophy in GA muscles of ALS mice. (a) Representative images of GA muscle histology at 18 weeks of age. Scale bar = 50 μm. (b) Myofiber CSA distribution of SOD HDAC4mKO and SOD GA muscles at 18 weeks of age. Data are expressed as mean +/− SEM. n = 3 mice per each genotype; *p < 0.05, **p < 0.005 by Student's t-test.
Catabolic pathways are not activated at 12 weeks of age. (a) Atrogin1 and (b) MuRF1 expression levels in healthy control (CTR), SOD and SOD HDAC4mKO GA muscles, at 12 weeks of age, by real-time PCR. n = 4 mice for each genotype. (c) Western blot analyses and (d) quantification of poly-ubiquitinated proteins in CTR, SOD and SOD HDAC4mKO muscles, at 12 weeks of age. Alpha-tubulin was used as a loading control. n = 3 mice for each genotype. (e) Proteasome activity in CTR, SOD and SOD HDAC4mKO GA muscles, at 12 weeks of age. n = 3 mice for each genotype. (f) LC3b and (g) p62 expression levels in CTR, SOD and SOD HDAC4mKO GA muscles, by real time PCR. n = 4 mice for each genotype. (h) LC3b and p62 protein levels of CTR, SOD and SOD HDAC4mKO, at 12 weeks of age, and (i) p62 relative quantification. Alpha-tubulin was used as a loading control. n = 5 mice for each genotype. Data are shown as mean ± SEM, over CTR mice.
Bioinformatic analysis of the genes modulated by HDAC4 in ALS. (a) MA-plot graph of the RNA-sequencing data. Red dots represent counts significantly modulated between SOD HDAC4mKO and SOD samples. (b, c) Top enriched gene sets among the complete set of significantly differentially expressed genes between the genotypes. GeneRatio indicates quotient of differentially expressed genes within the given node. Nodes are derived from Reactome or Disease Ontology. (d) SOD1 expression levels in healthy control (CTR), SOD and SOD HDAC4mKO GA muscles, at 14 weeks of age, by real-time PCR. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 5.42; df 2; p = 0.018; and interaction: *p < 0.05 by Tukey's HSD test.) (e) Gene network regulated by UCP1. Node (gene) and arrows (gene relationship) symbols were pictured whit distinct colors based on their modulation. The intensity of the node color indicated the degree of up-regulation (red) and down-regulation (green), respectively. The arrow color indicated if induce activation (orange), inhibition (blue), or is inconsistent with state of downstream molecule (yellow). (f) Plot showing Gdnf counts between SOD and SOD HDAC4mKO mice. Significance testing was performed as for the rest of the RNA-seq dataset.
Supplementary material
Declarations
Availability of data and material
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Acknowledgments
Acknowledgements
We would like to thank Carla Ramina, Emanuela Greco and Fabrizio Biuso for technical assistance; Eric N Olson and Rhonda Bassel-Duby for providing HDAC4 conditional mutant mice.
Funding sources
This work was supported by FIRB grant (RBFR12BUMH) from Ministry of Education, Universities and Research, Fondazione Veronesi post-doctoral fellowship, partially supported by Sapienza research project 2017 (RM11715C78539BD8) and Polish National Science Center grant (UMO-2016/21/B/NZ3/03638). The funders had no role in study design, data collection, data analysis, interpretation or writing of the report.
Declarations of interests
Dr. Pigna reports grants from Ministry of Education, University and Research, grants from “Sapienza” University of Rome, during the conduct of the study.
Dr. Simonazzi reports grants from Ministry of Education, University and Research, grants from “Sapienza” University of Rome, during the conduct of the study.
Dr. Sanna reports grants from Ministry of Education, University and Research, grants from “Sapienza” University of Rome, during the conduct of the study.
Dr. Bernadzki reports grants from Polish National Science Center, during the conduct of the study.
Dr. Prószyński reports grants from Polish National Science Center, during the conduct of the study.
Dr. Heil has nothing to disclose.
Dr. Palacios has nothing to disclose.
Dr. Adamo reports grants from Ministry of Education, University and Research, grants from “Sapienza” University of Rome, during the conduct of the study.
Dr. Moresi reports grants from Ministry of Education, University and Research, grants from “Sapienza” University of Rome, during the conduct of the study.
Authors' contributions
EP, ES, KS performed the experiments and analyzed the data; KMB, TP performed the NMJ and innervation analyses; CH and DP contributed with the bioinformatics analyses of the RNA-sequencing data; SA and VM conceived the project and critically analyzed the data. EP, TP, DP, SA and VM wrote the manuscript. All authors read and approved the final manuscript.
References
- 1.Musarò A. Understanding ALS: new therapeutic approaches. FEBS J. 2013;280:4315–4322. doi: 10.1111/febs.12087. [DOI] [PubMed] [Google Scholar]
- 2.Miller T.M., Kim S.H., Yamanaka K., Hester M., Umapathi P., Arnson H. Gene transfer demonstrates that muscle is not a primary target for non-cell-autonomous toxicity in familial amyotrophic lateral sclerosis. Proc Natl Acad Sci. 2006;103(51):19546–19551. doi: 10.1073/pnas.0609411103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellezza I., Grottelli S., Costanzi E., Scarpelli P., Pigna E., Morozzi G. Peroxynitrite activates the NLRP3 inflammasome cascade in SOD1(G93A) mouse model of amyotrophic lateral sclerosis. Mol Neurobiol. 2018;55:2350–2361. doi: 10.1007/s12035-017-0502-x. [DOI] [PubMed] [Google Scholar]
- 4.Dadon-Nachum M., Melamed E., Offen D. The “dying-back” phenomenon of motor neurons in ALS. J Mol Neurosci. 2011;43:470–477. doi: 10.1007/s12031-010-9467-1. [DOI] [PubMed] [Google Scholar]
- 5.Dobrowolny G., Aucello M., Rizzuto E., Beccafico S., Mammucari C., Boncompagni S. Skeletal muscle is a primary target of SOD1G93A-mediated toxicity. Cell Metab. 2008;8:425–436. doi: 10.1016/j.cmet.2008.09.002. [DOI] [PubMed] [Google Scholar]
- 6.Wong M., Martin L.J. Skeletal muscle-restricted expression of human SOD1 causes motor neuron degeneration in transgenic mice. Hum Mol Genet. 2010;19(11):2284–2302. doi: 10.1093/hmg/ddq106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Williams A.H., Valdez G., Moresi V., Qi X., McAnally J., Elliott J.L. MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science. 2009;80:326. doi: 10.1126/science.1181046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi M.C., Cohen T.J., Barrientos T., Wang B., Li M., Simmons B.J. A direct HDAC4-MAP kinase crosstalk activates muscle atrophy program. Mol Cell. 2012;47:122–132. doi: 10.1016/j.molcel.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moresi V., Williams A.H., Meadows E., Flynn J.M., Potthoff M.J., McAnally J. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell. 2010;143:35–45. doi: 10.1016/j.cell.2010.09.004. http://www.ncbi.nlm.nih.gov/pubmed/20887891 Internet. cited 2018 Jun 27. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen T.J., Waddell D.S., Barrientos T., Lu Z., Feng G., Cox G.A. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J Biol Chem. 2007;282:33752–33759. doi: 10.1074/jbc.M706268200. [DOI] [PubMed] [Google Scholar]
- 11.Echaniz-Laguna A., Bousiges O., Loeffler J.-P., Boutillier A.-L. Histone deacetylase inhibitors: therapeutic agents and research tools for deciphering motor neuron diseases. Curr Med Chem. 2008;15:1263–1273. doi: 10.2174/092986708784534974. [DOI] [PubMed] [Google Scholar]
- 12.Bruneteau G., Simonet T., Bauché S., Mandjee N., Malfatti E., Girard E. Muscle histone deacetylase 4 upregulation in amyotrophic lateral sclerosis: potential role in reinnervation ability and disease progression. Brain. 2013;136:2359–2368. doi: 10.1093/brain/awt164. http://www.ncbi.nlm.nih.gov/pubmed/23824486 Internet. cited 2018 Jul 2. Available from. [DOI] [PubMed] [Google Scholar]
- 13.DI Pietro L., Baranzini M., Berardinelli M.G., Lattanzi W., Monforte M., Tasca G. Potential therapeutic targets for ALS: MIR206, MIR208b and MIR499 are modulated during disease progression in the skeletal muscle of patients. Sci Rep. 2017;7 doi: 10.1038/s41598-017-10161-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rouaux C., Panteleeva I., Rene F., Gonzalez de Aguilar J.-L., Echaniz-Laguna A., Dupuis L. Sodium valproate exerts neuroprotective effects in vivo through CREB-binding protein-dependent mechanisms but does not improve survival in an amyotrophic lateral sclerosis mouse model. J Neurosci. 2007;27:5535–5545. doi: 10.1523/JNEUROSCI.1139-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ryu H., Smith K., Camelo S.I., Carreras I., Lee J., Iglesias A.H. Sodium phenylbutyrate prolongs survival and regulates expression of anti-apoptotic genes in transgenic amyotrophic lateral sclerosis mice. J Neurochem. 2005;93:1087–1098. doi: 10.1111/j.1471-4159.2005.03077.x. [DOI] [PubMed] [Google Scholar]
- 16.Yoo Y.E., Ko C.P. Treatment with trichostatin a initiated after disease onset delays disease progression and increases survival in a mouse model of amyotrophic lateral sclerosis. Exp Neurol. 2011;231:147–159. doi: 10.1016/j.expneurol.2011.06.003. [DOI] [PubMed] [Google Scholar]
- 17.Piepers S., Veldink J.H., De Jong S.W., Van Der Tweel I., Van Der Pol W.L., Uijtendaal E.V. Randomized sequential trial of valproic acid in amyotrophic lateral sclerosis. Ann Neurol. 2009;66:227–234. doi: 10.1002/ana.21620. [DOI] [PubMed] [Google Scholar]
- 18.Cudkowicz M.E., Andres P.L., Macdonald S.A., Bedlack R.S., Choudry R., Brown R.H. Phase 2 study of sodium phenylbutyrate in ALS. Amyotroph Lateral Scler. 2009;10:99–106. doi: 10.1080/17482960802320487. [DOI] [PubMed] [Google Scholar]
- 19.Buonvicino D., Felici R., Ranieri G., Caramelli R., Lapucci A., Cavone L. Effects of class II-selective histone deacetylase inhibitor on neuromuscular function and disease progression in SOD1-ALS mice. Neuroscience. 2018;379:228–238. doi: 10.1016/j.neuroscience.2018.03.022. [DOI] [PubMed] [Google Scholar]
- 20.Jones R.A., Reich C.D., Dissanayake K.N., Kristmundsdottir F., Findlater G.S., Ribchester R.R. NMJ-morph reveals principal components of synaptic morphology influencing structure-function relationships at the neuromuscular junction. Open Biol. 2016;6 doi: 10.1098/rsob.160240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pigna E., Renzini A., Greco E., Simonazzi E., Fulle S., Mancinelli R. HDAC4 preserves skeletal muscle structure following long-term denervation by mediating distinct cellular responses. Skelet Muscle. 2018;8 doi: 10.1186/s13395-018-0153-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu G., He Q.Y. ReactomePA: an R/Bioconductor package for reactome pathway analysis and visualization. Mol Biosyst. 2016;12:477–479. doi: 10.1039/c5mb00663e. [DOI] [PubMed] [Google Scholar]
- 24.Yu G., Wang L.G., Yan G.R., He Q.Y. DOSE: an R/Bioconductor package for disease ontology semantic and enrichment analysis. Bioinformatics. 2015;31:608–609. doi: 10.1093/bioinformatics/btu684. [DOI] [PubMed] [Google Scholar]
- 25.Potthoff M.J., Wu H., Arnold M.A., Shelton J.M., Backs J., McAnally J. Histone deacetylase degradation and MEF2 activation promote the formation of slow-twitch myofibers. J Clin Invest. 2007;117:2459–2467. doi: 10.1172/JCI31960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Proszynski T.J., Sanes J.R. Amotl2 interacts with LL5, localizes to podosomes and regulates postsynaptic differentiation in muscle. J Cell Sci. 2013;126:2225–2235. doi: 10.1242/jcs.121327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sandri M., Sandri C., Gilbert A., Skurk C., Calabria E., Picard A. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mammucari C., Milan G., Romanello V., Masiero E., Rudolf R., Del Piccolo P. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–471. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- 29.Pigna E., Renzini A., Greco E., Simonazzi E., Fulle S., Mancinelli R. HDAC4 preserves skeletal muscle structure following long-term denervation by mediating distinct cellular responses. Skelet Muscle. 2018;8:6. doi: 10.1186/s13395-018-0153-2. http://www.ncbi.nlm.nih.gov/pubmed/29477142 Internet. cited 2018 Sep 27. Available from. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vega R.B., Matsuda K., Oh J., Barbosa A.C., Yang X., Meadows E. Histone deacetylase 4 controls chondrocyte hypertrophy during skeletogenesis. Cell. 2004;119:555–566. doi: 10.1016/j.cell.2004.10.024. [DOI] [PubMed] [Google Scholar]
- 31.Dupuis L., Gonzalez de Aguilar J.L., Echaniz-Laguna A., Eschbach J., Rene F., Oudart H. Muscle mitochondrial uncoupling dismantles neuromuscular junction and triggers distal degeneration of motor neurons. PLoS One. 2009;4 doi: 10.1371/journal.pone.0005390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsitkanou S., Della Gatta P.A., Russell A.P. Skeletal muscle satellite cells, mitochondria, and MicroRNAs: their involvement in the pathogenesis of ALS. Front Physiol. 2016;7:1–9. doi: 10.3389/fphys.2016.00403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bruserud Ø., Stapnes C., Ersvaer E., Gjertsen B.T., Ryningen A. Histone deacetylase inhibitors in cancer treatment: a review of the clinical toxicity and the modulation of gene expression in cancer cell. Curr Pharm Biotechnol. 2007;8:388–400. doi: 10.2174/138920107783018417. http://www.ncbi.nlm.nih.gov/pubmed/18289048 Internet. cited 2018 Jun 27. Available from. [DOI] [PubMed] [Google Scholar]
- 34.Galanis E., Jaeckle K.A., Maurer M.J., Reid J.M., Ames M.M., Hardwick J.S. Phase II trial of vorinostat in recurrent glioblastoma multiforme: a north central cancer treatment group study. J Clin Oncol. 2009;27:2052–2058. doi: 10.1200/JCO.2008.19.0694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gimsing P., Hansen M., Knudsen L.M., Knoblauch P., Christensen I.J., Ooi C.E. A phase I clinical trial of the histone deacetylase inhibitor belinostat in patients with advanced hematological neoplasia. Eur J Haematol. 2008;81:170–176. doi: 10.1111/j.1600-0609.2008.01102.x. Internet. cited 2018 Jun 27. Available from: [DOI] [PubMed] [Google Scholar]
- 36.Gojo I., Jiemjit A., Trepel J.B., Sparreboom A., Figg W.D., Rollins S. Phase 1 and pharmacologic study of MS-275, a histone deacetylase inhibitor, in adults with refractory and relapsed acute leukemias. Blood. 2007;109:2781–2790. doi: 10.1182/blood-2006-05-021873. Internet. cited 2018 Jun 27. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pilatrino C., Cilloni D., Messa E., Morotti A., Giugliano E., Pautasso M. Increase in platelet count in older, poor-risk patients with acute myeloid leukemia or myelodysplastic syndrome treated with valproic acid and all-trans retinoic acid. Cancer. 2005;104:101–109. doi: 10.1002/cncr.21132. [DOI] [PubMed] [Google Scholar]
- 38.Hayworth C.R., Gonzalez-Lima F. Pre-symptomatic detection of chronic motor deficits and genotype prediction in congenic B6.SOD1G93A ALS mouse model. Neuroscience. 2009;164:975–985. doi: 10.1016/j.neuroscience.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weydt P., Hong S.Y., Kliot M., Möller T. Assessing disease onset and progression in the SOD1 mouse model of ALS. Neuroreport. 2003;14:1051–1054. doi: 10.1097/01.wnr.0000073685.00308.89. [DOI] [PubMed] [Google Scholar]
- 40.Mead R.J., Bennett E.J., Kennerley A.J., Sharp P., Sunyach C., Kasher P. Optimised and rapid pre-clinical screening in the SOD1G93A transgenic mouse model of amyotrophic lateral sclerosis (ALS) PLoS One. 2011;6 doi: 10.1371/journal.pone.0023244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wijesekera L.C., Leigh P.N., Brain W., Walton J., Leigh P., Anderton B. Amyotrophic lateral sclerosis. Orphanet J Rare Dis. 2009;4:3. doi: 10.1186/1750-1172-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Funakoshi H., Belluardo N., Arenas E., Yamamoto Y., Casabona A., Persson H. Muscle-derived neurotrophin-4 as an activity-dependent trophic signal for adult motor neurons. Science. 1995;268(80):1495–1499. doi: 10.1126/science.7770776. [DOI] [PubMed] [Google Scholar]
- 43.Lu B., Je H.-S. Neurotrophic regulation of the development and function of the neuromuscular synapses. J Neurocytol. 2003;32:931–941. doi: 10.1023/B:NEUR.0000020633.93430.db. [DOI] [PubMed] [Google Scholar]
- 44.Henderson C.E., Phillips H.S., Pollock R.A., Davies A.M., Lemeulle C., Armanini M. GDNF: a potent survival factor for motoneurons present in peripheral nerve and muscle. Science. 1994;266(80):1062–1064. doi: 10.1126/science.7973664. [DOI] [PubMed] [Google Scholar]
- 45.Glass D.J. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–1984. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 46.Talbot J.D., Barrett J.N., Nonner D., Zhang Z., Wicomb K., Barrett E.F. Preservation of neuromuscular function in symptomatic SOD1-G93A mice by peripheral infusion of methylene blue. Exp Neurol. 2016;285:96–107. doi: 10.1016/j.expneurol.2016.08.013. [DOI] [PubMed] [Google Scholar]
- 47.Xiao Y., Ma C., Yi J., Wu S., Luo G., Xu X. Suppressed autophagy flux in skeletal muscle of an amyotrophic lateral sclerosis mouse model during disease progression. Physiol Rep. 2015;3 doi: 10.14814/phy2.12271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banerjee R., Beal M.F., Thomas B. Autophagy in neurodegenerative disorders: pathogenic roles and therapeutic implications. Trends Neurosci. 2010;33:541–549. doi: 10.1016/j.tins.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li A., Zhang X., Le W. Altered macroautophagy in the spinal cord of SOD1 mutant mice. Autophagy. 2008;4:290–293. doi: 10.4161/auto.5524. [DOI] [PubMed] [Google Scholar]
- 50.Morimoto N., Nagai M., Ohta Y., Miyazaki K., Kurata T., Morimoto M. Increased autophagy in transgenic mice with a G93A mutant SOD1 gene. Brain Res. 2007;1167:112–117. doi: 10.1016/j.brainres.2007.06.045. [DOI] [PubMed] [Google Scholar]
- 51.Sasaki S. Autophagy in spinal cord motor neurons in sporadic amyotrophic lateral sclerosis. J Neuropathol Exp Neurol. 2011;70:349–359. doi: 10.1097/NEN.0b013e3182160690. [DOI] [PubMed] [Google Scholar]
- 52.Zhang X., Li L., Chen S., Yang D., Wang Y., Zhang X. Rapamycin treatment augments motor neuron degeneration in SOD1 G93A mouse model of amyotrophic lateral sclerosis. Autophagy. 2011;7:412–425. doi: 10.4161/auto.7.4.14541. [DOI] [PubMed] [Google Scholar]
- 53.Bhattacharya A., Bokov A., Muller F.L., Jernigan A.L., Maslin K., Diaz V. Dietary restriction but not rapamycin extends disease onset and survival of the H46R/H48Q mouse model of ALS. Neurobiol Aging. 2012;33:1829–1832. doi: 10.1016/j.neurobiolaging.2011.06.002. [DOI] [PubMed] [Google Scholar]
- 54.Nassif M., Hetz C. Targeting autophagy in ALS: a complex mission. Autophagy. 2011;7:450–453. doi: 10.4161/auto.7.4.14700. [DOI] [PubMed] [Google Scholar]
- 55.Marroncelli N., Bianchi M., Bertin M., Consalvi S., Saccone V., De Bardi M. HDAC4 regulates satellite cell proliferation and differentiation by targeting P21 and Sharp1 genes. Sci Rep. 2018;8:3448. doi: 10.1038/s41598-018-21835-7. http://www.nature.com/articles/s41598-018-21835-7 Internet. cited 2018 Jun 27. Available from: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Couplan E., Gelly C., Goubern M., Fleury C., Quesson B., Silberberg M. High level of uncoupling protein 1 expression in muscle of transgenic mice selectively affects muscles at rest and decreases their IIb fiber content. J Biol Chem. 2002;277:43079–43088. doi: 10.1074/jbc.M206726200. [DOI] [PubMed] [Google Scholar]
- 57.Hoerter J., Gonzalez-Barroso M.D.M., Couplan E., Mateo P., Gelly C., Cassard-Doulcier A.M. Mitochondrial uncoupling protein 1 expressed in the heart of transgenic mice protects against ischemic-reperfusion damage. Circulation. 2004;110:528–533. doi: 10.1161/01.CIR.0000137824.30476.0E. [DOI] [PubMed] [Google Scholar]
- 58.Krakora D., MacRander C., Suzuki M. Neuromuscular junction protection for the potential treatment of amyotrophic lateral sclerosis. Neurol Res Int. 2012;2012 doi: 10.1155/2012/379657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dupuis L., Oudart H., Rene F., de Aguilar J.-L.G., Loeffler J.-P. Evidence for defective energy homeostasis in amyotrophic lateral sclerosis: benefit of a high-energy diet in a transgenic mouse model. Proc Natl Acad Sci. 2004;101:11159–11164. doi: 10.1073/pnas.0402026101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Boyer J.G., Ferrier A., Kothary R. More than a bystander: The contributions of intrinsic skeletal muscle defects in motor neuron diseases. Front Physiol. 2013;(4 DEC) doi: 10.3389/fphys.2013.00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dobrowolny G., Martini M., Scicchitano B.M., Romanello V., Boncompagni S., Nicoletti C. Muscle expression of SOD1G93A triggers the dismantlement of neuromuscular junction via PKC-Theta. Antioxidants Redox Signal. 2018:28. doi: 10.1089/ars.2017.7054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
HDAC4mKO mice do not show obvious phenotype. (a) HDAC4 expression in GA muscles of CTR and HDAC4mKO mice, by real-time PCR, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 3 CTR; n = 4 HDAC4mKO (**p < 0.01 by Student's t-test.) (b) Western blot analyses and (c) densitometry for HDAC4 in GA muscles of CTR and HDAC4mKO mice, at 12 weeks of age. Gapdh was used as a loading control. Data are expressed as mean +/− SEM. n = 3 mice for each genotype. (**p < 0.01 by Student's t-test) (d) Body weight of male CTR and HDAC4mKO mice, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 7 CTR; n = 5 HDAC4mKO mice. (e) HDAC4mKO and CTR muscle performance, by grip test, at 12 weeks of age. Data are expressed as the mean of developed force over the mouse weight, +/− SEM. n = 5 CTR and n = 4 HDAC4mKO mice. (f) HDAC4mKO and CTR TA weight, at 12 weeks of age. Data are expressed as mean of TA weight relative to CTR littermates +/− SEM. n = 5 mice for each genotype. (g) Representative pictures of HDAC4mKO and CTR TA histology, at 12 weeks of age. Scale bar = 50 μm. (h) Myofiber CSA distribution of HDAC4mKO and CTR TA muscles, at 12 weeks of age. Data are expressed as mean +/− SEM. n = 3 CTR and n = 4 HDAC4mKO mice. (i) Representative images of NMJ postsynaptic machinery in HDAC4mKO and CTR mice, at 12 weeks of age. n = 3 mice per genotype. Scale bar = 10 μm. (l–o) Expression levels of indicated genes in HDAC4mKO GA muscles, relative to CTR littermates, at 14 weeks of age, by real-time PCR. Data are expressed as mean +/− SEM. n = 5 mice for each genotype.
Deletion of HDAC4 worsens muscle atrophy in GA muscles of ALS mice. (a) Representative images of GA muscle histology at 18 weeks of age. Scale bar = 50 μm. (b) Myofiber CSA distribution of SOD HDAC4mKO and SOD GA muscles at 18 weeks of age. Data are expressed as mean +/− SEM. n = 3 mice per each genotype; *p < 0.05, **p < 0.005 by Student's t-test.
Catabolic pathways are not activated at 12 weeks of age. (a) Atrogin1 and (b) MuRF1 expression levels in healthy control (CTR), SOD and SOD HDAC4mKO GA muscles, at 12 weeks of age, by real-time PCR. n = 4 mice for each genotype. (c) Western blot analyses and (d) quantification of poly-ubiquitinated proteins in CTR, SOD and SOD HDAC4mKO muscles, at 12 weeks of age. Alpha-tubulin was used as a loading control. n = 3 mice for each genotype. (e) Proteasome activity in CTR, SOD and SOD HDAC4mKO GA muscles, at 12 weeks of age. n = 3 mice for each genotype. (f) LC3b and (g) p62 expression levels in CTR, SOD and SOD HDAC4mKO GA muscles, by real time PCR. n = 4 mice for each genotype. (h) LC3b and p62 protein levels of CTR, SOD and SOD HDAC4mKO, at 12 weeks of age, and (i) p62 relative quantification. Alpha-tubulin was used as a loading control. n = 5 mice for each genotype. Data are shown as mean ± SEM, over CTR mice.
Bioinformatic analysis of the genes modulated by HDAC4 in ALS. (a) MA-plot graph of the RNA-sequencing data. Red dots represent counts significantly modulated between SOD HDAC4mKO and SOD samples. (b, c) Top enriched gene sets among the complete set of significantly differentially expressed genes between the genotypes. GeneRatio indicates quotient of differentially expressed genes within the given node. Nodes are derived from Reactome or Disease Ontology. (d) SOD1 expression levels in healthy control (CTR), SOD and SOD HDAC4mKO GA muscles, at 14 weeks of age, by real-time PCR. n = 3 mice for each genotype. (One-way ANOVA reveals a significant effect: F = 5.42; df 2; p = 0.018; and interaction: *p < 0.05 by Tukey's HSD test.) (e) Gene network regulated by UCP1. Node (gene) and arrows (gene relationship) symbols were pictured whit distinct colors based on their modulation. The intensity of the node color indicated the degree of up-regulation (red) and down-regulation (green), respectively. The arrow color indicated if induce activation (orange), inhibition (blue), or is inconsistent with state of downstream molecule (yellow). (f) Plot showing Gdnf counts between SOD and SOD HDAC4mKO mice. Significance testing was performed as for the rest of the RNA-seq dataset.
Supplementary material
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.