Abstract
Wolbachia, an alpha-proteobacterium closely related to Rickettsia, is a maternally transmitted, intracellular symbiont of arthropods and nematodes. Aedes albopictus mosquitoes are naturally infected with Wolbachia strains wAlbA and wAlbB. Cell line Aa23 established from Ae. albopictus embryos retains only wAlbB and is a key model to study host–endosymbiont interactions. We have assembled the complete circular genome of wAlbB from the Aa23 cell line using long-read PacBio sequencing at 500× median coverage. The assembled circular chromosome is 1.48 megabases in size, an increase of more than 300 kb over the published draft wAlbB genome. The annotation of the genome identified 1,205 protein coding genes, 34 tRNA, 3 rRNA, 1 tmRNA, and 3 other ncRNA loci. The long reads enabled sequencing over complex repeat regions which are difficult to resolve with short-read sequencing. Thirteen percent of the genome comprised insertion sequence elements distributed throughout the genome, some of which cause pseudogenization. Prophage WO genes encoding some essential components of phage particle assembly are missing, while the remainder are found in five prophage regions/WO-like islands or scattered around the genome. Orthology analysis identified a core proteome of 535 orthogroups across all completed Wolbachia genomes. The majority of proteins could be annotated using Pfam and eggNOG analyses, including ankyrins and components of the Type IV secretion system. KEGG analysis revealed the absence of five genes in wAlbB which are present in other Wolbachia. The availability of a complete circular chromosome from wAlbB will enable further biochemical, molecular, and genetic analyses on this strain and related Wolbachia.
Keywords: symbiosis, Aa23, mosquito, PacBio, prophage, IS elements
Introduction
Wolbachia are Gram-negative α-proteobacteria of the order Rickettsiales. Maternally transmitted infections are widespread, occurring in an estimated 40–65% of insect species (Hilgenboecker et al. 2008; Werren et al. 2008) including 28% of mosquito species (Kittayapong et al. 2000). Wolbachia infections are not limited to arthropods as nematodes, including several major human pathogens, also harbor the endosymbiont (Fenn et al. 2006; Lefoulon et al. 2016). Currently, Wolbachia strains have been classified as Wolbachia pipientis (Hertig 1936; Lo et al. 2007), which has been divided into 16 major phylogenetic clades termed supergroups, denoted A–Q, mainly based on multilocus sequence typing (MLST) analysis (Baldo et al. 2006). Most supergroups are restricted to arthropods (A, B, E, G, H, I, K, M, N, O, P, and Q) (Lefoulon et al. 2016), whereas supergroups C and D are the major nematode-infecting lineages. Supergroup F is unique as it contains both nematode and arthropod-infecting strains (Lefoulon et al. 2016). The nature of the association between Wolbachia strains and their hosts varies greatly. In nematode species which contain Wolbachia, the prevalence of infection is 100% and the relationship is obligate (Taylor et al. 2005). These attributes have been exploited to enable the use of antibiotics as a novel approach to treat filarial infections (Langworthy et al. 2000; Bazzocchi et al. 2008; Johnston et al. 2014). In contrast, infection is less prevalent in insect hosts and can cause broad effects on insect physiology leading to several phenotypic changes attributed to the ability of Wolbachia to act as manipulators of the host (Werren et al. 2008; Cordaux et al. 2011). Among these manipulations, cytoplasmic incompatibility (CI) is the most common phenotype in mosquitoes (Sinkins 2004) and provides a reproductive advantage to Wolbachia-infected females over uninfected females, resulting in spread and persistence of Wolbachia in populations (Xi et al. 2005). When experimentally transferred to uninfected mosquitoes, Wolbachia can also suppress infection or transmission of viruses (Walker et al. 2011; Aliota et al. 2016; Aliota and Walker 2016; Carrington et al. 2018), Plasmodium parasites (Kambris et al. 2010), and filarial nematodes (Kambris et al. 2009; Andrews et al. 2012) making Wolbachia a particularly attractive agent for control of vector-borne pathogens.
The Asian tiger mosquito, Aedes albopictus, is an aggressive biting mosquito and currently one of the most invasive species in the world. Originally native to Southeast Asia, the species has spread in the past 30–40 years and colonized five continents (Kotsakiozi et al. 2017). It is a significant public health concern as it is a competent vector of several arboviruses that cause severe diseases in humans such as dengue, chikungunya, and Zika (Gratz 2004; Grard et al. 2014; Chouin-Carneiro et al. 2016). Two distinct Wolbachia strains (wAlbA and wAlbB), are present in variable density in Ae. albopictus tissues (Kittayapong et al. 2000; Zouache et al. 2009). wAlbB, belonging to the supergroup B, is capable of altering malaria or viral pathogen loads in a host-dependent manner. Transient somatic infection of Anopheles gambiae with wAlbB inhibits Plasmodium falciparum but enhances Plasmodium berghei parasites (Hughes et al. 2011, 2012). It enhances West Nile virus infection in the mosquito Culex tarsalis (Dodson et al. 2014), whereas it blocks transmission of dengue (Mousson et al. 2012) and chikungunya (Raquin et al. 2015). To potentially reduce the transmission of dengue and Zika viruses to humans, wAlbB has also been introduced into new hosts such as Aedes aegypti mosquitoes (Pan et al. 2018). Investigating the interplay between the host and its new endosymbiont is important when predicting the stability of such associations (Pan et al. 2018).
Cell lines containing Wolbachia represent a simplified model in which to explore the symbiotic relationship and have been used extensively in molecular, biochemical, and genetic studies (O’Neill et al. 1997; Voronin et al. 2012; Saucereau et al. 2017). The Aa23 cell line derived from Wolbachia-infected Ae. albopictus mosquito embryos was the first cell line developed to enable studies on Wolbachia–host cell interactions (Sinkins et al. 1995; O’Neill et al. 1997). While Ae. albopictus mosquitoes are naturally infected with wAlbA and wAlbB, only wAlbB was retained in the Aa23 cell line (Sinkins et al. 1995; O’Neill et al. 1997). The cell line comprises at least two cell types and Wolbachia infection varies, with respect to both the level of infection among individual cells and the overall level of infection in a population (O’Neill et al. 1997). However, high cell density during passaging helps to maintain a relatively stable infection rate, because the duration of exponential growth is affected by cell density (Gerenday and Fallon 1996). wAlbB from Aa23 cells has been used as a source of infection for other insect cell lines (Dobson et al. 2002; Fenollar, Scola, et al. 2003; Xi et al. 2005; Rasgon et al. 2006). Because no nematode-derived cell culture system for Wolbachia exists, the Aa23 insect cell line: wAlbB model system has been used as a proxy to screen for new anti-Wolbachia/filarial compounds (Fenollar, Maurin, et al. 2003).
Due to the importance of the wAlbB-infected Aa23 cell line in studies on symbiosis, pathogen control, and drug screening, a draft genome sequence of this strain was published (Mavingui et al. 2012). For this Wolbachia assembly, Multiple Displacement Amplification of DNA from infected cells was used to construct a mate-paired library containing 6-kb inserts, and sequenced with 454 Titanium pyrosequencing at 76 bp read-length. The resulting genome draft is incomplete with 165 contigs encompassing 49 scaffolds (Mavingui et al. 2012), hampering a comprehensive analysis of the genome.
The short-read technologies, such as 454 and Illumina, cannot easily reconstruct complete microbial chromosomes, and often produce draft assemblies containing gaps. Pacific Biosciences (PacBio) SMRT technology produces long reads, some as long as 100 kb, with average raw read lengths >15 kb, making single and continuous assembly possible (Eid et al. 2008). In addition, the PacBio library preparation process does not include an amplification step, therefore DNA is sequenced as a single molecule in its native form, enabling the detection of covalent base modifications (Flusberg et al. 2010).
In this study, we have assembled the complete circular genome of wAlbB present in the Aa23 cell line, from long-read PacBio sequencing data at 500× median coverage. The long reads enabled sequencing over complex repeat regions which have been difficult to resolve with short-read sequencing. The assembled circular genome is 1,484,007 bp in size, an increase of 321 kb over the published wAlbB draft genome, making it one of the largest sequenced Wolbachia genomes to date. This sequence will serve as important resource for detailed studies of wAlbB and related Wolbachia.
Materials and Methods
Cell Culture
The Aa23 cell line infected with wAlbB was a kind gift from Dr Stephen Dobson. Cells were grown in culture flasks at 28 °C in equal volumes of Mitsuhashi–Maramorosch medium (Sigma M9257) and Schneider’s insect medium (Sigma S0146), supplemented with 10% heat-inactivated fetal bovine serum (O’Neill et al. 1997). The cells retained the morphological heterogeneity originally described (O’Neill et al. 1997) and were routinely subcultured at 7- to 10-day intervals by diluting 1:3 in fresh media to maintain high cell density and contiguous monolayers.
Immunostaining of Wolbachia with Anti-VirB8 Antibody
Cells cultured on glass coverslips within 24-well microtiter plates were fixed in 4% formaldehyde in phosphate-buffered saline (PBS) for 15 min and subsequently permeabilized using chilled 100% methanol (−20 °C) for 1 min. Fixed cells were then incubated in polyclonal rabbit anti-VirB8 antibody (Li and Carlow 2012) diluted 1:2,000 in PBS containing 5% goat serum, followed by Alexa Fluor 488 (green) conjugated goat anti-rabbit secondary antibodies (Molecular Probes; Invitrogen Life Technologies) according to manufacturer’s instructions. Cell nuclei were stained with Hoechst 33342 at 1:10,000 dilution in PBS. Prolong Gold anti-fade reagent (Invitrogen Life Technologies) was used to avoid fading. Images were acquired using an Axiovert 200 M microscope (Carl Zeiss, Oberkochen, Germany) and processed using ZEN software (Carl Zeiss).
DNA Extraction
To harvest host cell-free wAlbB, spent culture media from Aa23 cells (passage #65) was collected and centrifuged at 500 × g to remove cell debris, followed by 5,000 × g to collect the Wolbachia-enriched pellet. Genomic DNA was extracted using a Qiagen MagAttract HMW kit following manufacturer’s instructions. Briefly, 220 μl of buffer ATL and 20 μl of proteinase K were added, and the sample was incubated at 56 °C for 3 h with mixing at 900 rpm (Eppendorf thermomixer C). DNA was eluted with 150 μl of AE buffer and quantified using a NanoDrop and Qubit instruments (Thermo Fisher Scientific). The quality of DNA was assessed using an Agilent 4200 TapeStation System. The DNA obtained was good quality (DIN > 8.2), and high molecular weight, larger than 60 kb in size (supplementary fig. S1, Supplementary Material online).
PacBio and Illumina Library Construction and Sequencing
For library construction, intact genomic DNA was fragmented using a Megaruptor 2 device (Diagenode). Sheared DNA was purified with AMPure PB beads and 2 μg were used to construct a SMRTbell library according to PacBio library construction guidelines with some modifications. Briefly, sheared DNA was repaired using the NEBNext FFPE DNA Repair Mix, followed by end-repair to generate blunt ends. Following purification using AMPure PB beads, PacBio universal hairpin adaptors were ligated to the DNA to produce SMRTbell libraries. After adaptor removal and library clean up, concentration and size of the SMRTbell library were determined using the Qubit HS DNA kit and Agilent TapeStation analysis. To enrich for longer insert sizes, size selection was performed using the BluePippin system (Sage Science), resulting in a library that contained an insert size of approximately 45 kb (supplementary fig. S1, Supplementary Material online). The PacBio sequencing primer was then annealed to the SMRTbell library followed by binding of the polymerase to the primer–library complex. The size-selected library was loaded onto two SMRT cells in the PacBio RSII system using a MagBead binding kit and sequenced with a 300 min collection time. Two additional SMRT cells were loaded with library that did not undergo size selection.
For Illumina library construction, genomic DNA was fragmented to 300 bp average size using a Covaris S2 (Covaris, Inc.) with the following settings: 10% duty cycle, intensity 4,200 cycles per burst and treatment time of 80 s. Libraries were constructed using the NEBNext Ultra II DNA Library Prep Kit for Illumina (New England Biolabs, Inc.). The library quality was assessed using a high sensitivity DNA chip on a Bioanalyzer (Agilent Technologies, Inc.) and was sequenced on an Illumina MiSeq platform (paired-end, 150 nt reads).
Genome Assembly and DNA Modification Analysis
PacBio sequencing reads from all four flow cells were processed and assembled using the HGAP assembler version 3 (Chin et al. 2013) as implemented in the PacBio SMRT® Analysis Server v2.3.0 (https://www.pacb.com/products-and-services/analytical-software/smrt-analysis; last accessed February 20, 2019). The contig corresponding to wAlbB was selected for further polishing and circularization. Overlapping regions at the termini of this contig were identified by BLAST analysis, and were merged to circularize the chromosome. This circular draft assembly sequence was further polished using multiple rounds of the ReSequencing.1 protocol from the PacBio SMRT® Analysis Server v2.3.0. The origin of replication, oriC, was identified by generating a consensus of all 10 Wolbachia oriC sequences obtained from the DoriC database (Gao et al. 2012). The assembled chromosome was verified to be free of any structural errors via the RS.Bridgemapper pipeline available as a part of the PacBio SMRT portal.
The validity and correctness of chromosome circularization was confirmed by polymerase chain reaction (PCR) and sequencing across the ends of the polished chromosome.
Primers F1 (5′TCCCCTGCCCTACCTGAGTA3′) and R1 (5′GTCATCATCCTGCGCGAGAG3′) were used to amplify a 1,599 bp fragment that spans the junction of circularization; primers F2 (5′TGTTGCTTTCATTGAGGCTGGT3′) and R2 (5′TATTGGACCCACACCGCGAA3′) were used to amplify a 1,081 bp fragment to verify the oriC sequence, using the Q5 HiFi PCR master mix (NEB M0543) following manufacturer’s instructions. Search for potential DNA modifications in the Wolbachia wAlbB genome was carried out using the polished genome as a reference genome in the RS_Modification_and_Motif_Analysis.1 pipeline from the PacBio SMRT® Analysis Server v2.3.0.
To check and correct any potential indel errors typically observed in PacBio-only assemblies (Watson and Warr 2019), Illumina sequencing was performed. After adapter-trimming and filtering of low-quality reads using BBMap package, version 37.17 (https://sourceforge.net/projects/bbmap; last accessed February 20, 2019), the reads were mapped to the PacBio chromosome using bwa version 0.7.15-r1140 (Li and Durbin 2009) in paired-end mode. Pilon software version 1.22 (Walker et al. 2014) was run on the bam file output from bwa, using alignments with mapping quality ≥20 (Pilon flag minmq = 20).
Whole genome alignment of the contigs from the published assembly contigs to the complete circular genome as the reference sequence was performed using the minimap2 software (Li 2018), with the recommended parameter “asm5” for generating whole genome alignments in SAM format. The sam_pileup (v1.1.2) program was used to generate a pileup file, which was then used as an input to bcftools_mpileup (v1.4.0.0) to generate a variant calling format (VCF) file. Single nucleotide variants and indels in the published assembly as compared with the complete circular genome were identified using custom scripts for further analysis of the VCF file. All gene sequences from the published contigs were aligned to the complete genome using BLAST, and the alignments were converted to bed format. Overlaps between the published gene sequences and the new annotations, as well as additional genetic loci found in the genomic regions missing in the published assembly were identified using the “intersect” functionality in the bedtools package (Quinlan and Hall 2010), and by manual inspections of the whole-genome and gene alignments in the IGV genome browser.
Genome Annotation and Analysis
Protein-coding genes, rRNA, tRNA, ncRNA, and pseudogenes were identified using the NCBI prokaryotic annotation pipeline (Angiuoli et al. 2008). Further functional annotation of protein-coding genes was carried out using the eggNOG-Mapper (Huerta-Cepas et al. 2017) web server (http://eggnogdb.embl.de/#/app/emapper; last accessed February 20, 2019) against the eggNOG database (Huerta-Cepas et al. 2016). The completeness of the genome was assessed using the Benchmarking Universal Single-Copy Orthologs (BUSCO) pipeline version 3.0.2 (Simão et al. 2015). Insertion sequence (IS) elements were identified by searching against the ISfinder database (Siguier et al. 2006) via the ISsaga web server (Varani et al. 2011) available at http://issaga.biotoul.fr/issaga_index.php (last accessed February 20, 2019). Pfam domains were annotated using the pfam_scan.pl script version 1.6 from http://ftp.ebi.ac.uk/pub/databases/Pfam/Tools (last accessed February 20, 2019) to search against Pfam database version 31.0 (Finn et al. 2016). Annotation of integrated prophage sequences was carried out using the PHASTER web server (Arndt et al. 2016), available at http://phaster.ca (last accessed February 20, 2019). To search for WO-like islands, which are regions containing genes that are typically found in WO phages but are absent in genomes of phage-free Wolbachia (Bordenstein and Bordenstein 2016; LePage et al. 2017), various BLAST comparisons (BlastN, BlastP, and TBlastN) were made to nucleotide and protein sequences from other Wolbachia prophage sequences. These sequences include WOVitA1 (GenBank: HQ906662.1), WOCauB2 (GenBank: AB478515.1) and WOCauB3 (GenBank: AB478516.1), WOVitB (GenBank: HQ906665.1, HQ906666.1), partial genome sequence of WOCauB1 (GenBank: AB161975.2), and the prophage regions from wMel (GenBank: NC_002978.6), wNo (GenBank: CP003883.1), wPip (Pel strain, Genbank: M999887.1), and the WO prophage region from Wolbachia of the Mediterranean flour moth, Ephestia kuehniella wKue (GenBank: AB036666.1). The various BLAST hits on wAlbB genome were visualized in Integrative Genomics Viewer (IGV) genome browser and clusters of hits were chosen as seed regions for annotation of phage-related regions. Genes and proteins flanking these seed regions were further inspected for any potential hits to WO phage elements to extend the WO-like islands when possible. Circos plots (Krzywinski et al. 2009) for visualizing the distribution of various features across the genome were plotted using the R package circlize, version 0.4.3 (Gu et al. 2014). Search for orthologs across multiple genomes was performed using the OrthoFinder (Emms and Kelly 2015) software version 1.1.4 with default parameters, which include a DIAMOND (Buchfink et al. 2015) e-value cut-off of 0.001. The number of orthogroups common across various Wolbachia was visualized as UpSet plots (Lex et al. 2014) using the R package UpSetR, version 1.3.3 (Conway et al. 2017).
The KEGG automatic annotation server, KAAS (Moriya et al. 2007), available online at https://www.genome.jp/kegg/kaas (last accessed February 20, 2019), was used to find functional annotations of genes in the wAlbB genome. wAlbB protein sequences were used as query sequences and BLAST (bidirectional best hit) searched against a manually curated set of ortholog groups in KEGG to generate KEGG pathways and functional classifications. The KEGG orthology (KO) assignments of wAlbB proteins from KEGG pathway analysis were then compared with the KEGG pathways of Wolbachia wRi from Drosophila simulans and wPip from Culex quinquefasciatus available in the KEGG database, to identify any missing proteins in wAlbB.
Results
High Levels of Wolbachia Infection Enable Production of Host Cell-Free Wolbachia
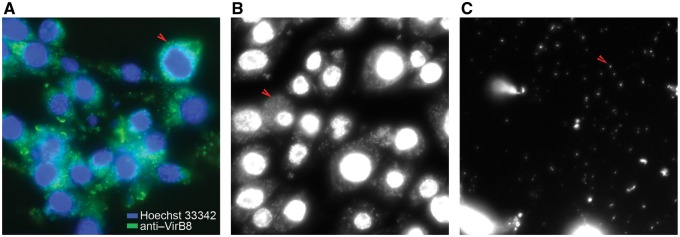
To preserve high levels of Wolbachia infection in Aa23 cells, maintenance of a high-density monolayer of cells was found to be necessary, which was achieved by passaging at high cell densities. Approximately 80% of cells were infected with a high Wolbachia load as verified by staining with an α-virB8 antibody (fig. 1A) or a Hoechst 33342 DNA dye (fig. 1B). The large numbers of host cell-free Wolbachia observed in spent culture media (fig. 1C), obviated the need for further separation of Wolbachia from host cells.
Fig. 1.
—Detection of Wolbachia in Aa23 cells and in culture supernatants. Immunostaining of Wolbachia using an anti-VirB8 antibody (green) and Hoechst staining (blue) of host nuclei (A). Hoechst staining of Wolbachia in cells (B) and in spent media (C). Arrows indicate Wolbachia.
Assembly and Annotation
Processing of the combined PacBio data obtained from four SMRT cells produced 944,546 filtered subreads and a total of 3 billion bases, with the longest subread at 61 kb and median length 3.4 kb. HGAP3 assembly of this data generated 581 contigs. The longest contig was 1,511,710 bp, at ∼500× median coverage (supplementary fig. S1, Supplementary Material online) and 99.9994% consensus concordance. This single contig contained all the contig sequences from the published wAlbB assembly (RefSeq assembly accession GCF_000242415.2), and was selected for further polishing. No other contig matched the wAlbB genome. One contig at 2,400× median coverage corresponded to Ae. albopictus mitochondrial DNA. Of the remaining contigs, megablastn matches against the NCBI nr/nt database were found in sequences from various Aedes species (n = 263), or the mosquito Guato virus (n = 2). The largest number of contigs (n = 302) have no hits in NCBI nr/nt database (supplementary file S1, Supplementary Material online).
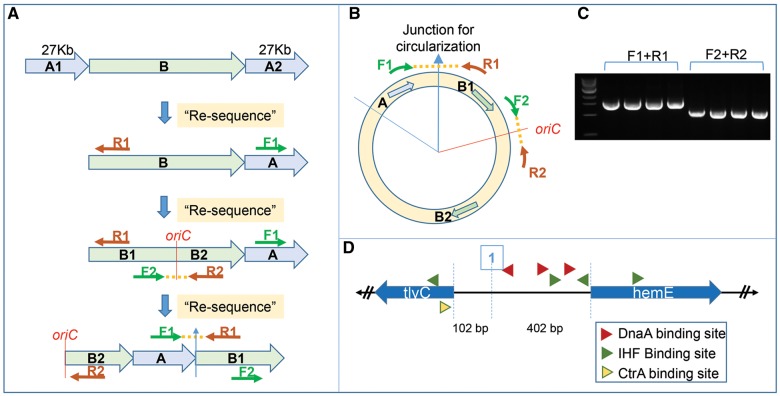
A BLAST analysis of the wAlbB contig to itself identified a highly similar 27 kb region repeated at the beginning and end of the contig, indicating that this contig represents the complete circular chromosome. To validate that these regions (marked A1 and A2 in fig. 2A) represent overlapping ends of the circular chromosome, we collapsed them into a single consensus region (marked A in fig. 2A and B). Using the PacBio ReSequencing.1 pipeline, all sequencing reads were remapped to this corrected chromosome sequence, which generated a polished single contig representing a circular chromosome with non-repeating ends. Primers F1 and R1 (fig. 2A and B) designed to span this candidate junction of circularization produced a PCR product of expected size of 1.5 kb (fig. 2C) and sequence, confirming the correctness of the circularization. For the next round of polishing, the first base of the chromosome sequence was reset to the start of oriC and this permuted sequence was again used as a reference for remapping all the reads using the PacBio ReSequencing.1 pipeline. The sequence of the oriC region was also verified by sequencing the PCR product generated using primers F2 and R2 (fig. 2A–C). The final polished circular genome produced as the output had a median coverage of ∼500× and was used in all subsequent analysis. The identified wAlbB oriC has all the hallmark features typical of Wolbachia oriC regions (Ioannidis et al. 2007). It is flanked by hemE and tlyC genes, with the intergenic regions having binding sites for DnaA, IHF, and CtrA (fig. 2D).
Fig. 2.
—Circularization and polishing of the wAlbB genome assembly and determination of oriC. De novo assembly of PacBio data produced a single contig representing the wAlbB genome, with terminal regions marked A1 and A2 in (A), showing high sequence identity over a 27 kb region, suggesting overlapping ends of a circular chromosome. They were therefore collapsed into a single consensus region “A” to represent the circular the chromosome, with the junction of circularization between regions A and B indicated by a blue arrow (B). This draft circular genome was polished using the PacBio ReSequencing.1 pipeline, and the junction of circularization was validated by Sanger sequencing of a 1.5 kb amplicon (C) produced by primers F1 and R1. The origin of the circular chromosome was reset to the beginning of the oriC locus. The permuted chromosome sequence was again polished using ReSequencing.1 pipeline in SMRT analysis software. The oriC sequence and the new junction of circularization at oriC was verified by primers F2 and R2 (A and B) that successfully produced an amplicon of correct size 1.5 kb (C) and correct sequence as confirmed by Sanger sequencing. The oriC locus in wAlbB has all the hallmarks observed in other Wolbachia oriC sequences, such as flanking genes tlyC and hemE, three DnaA binding sites, four IHF binding sites and one CtrA binding site (D). All PCR were performed on four independent DNA samples.
Indel errors are sometimes observed in assemblies produced from only long-read technologies such as PacBio or Nanopore (Watson and Warr 2019). Therefore, the polished, circular wAlbB genome was checked for any potential errors using Illumina reads. About 4.8 million paired-end reads mapped to the assembly (mapping quality ≥10) providing a ∼630× median coverage of the genome and were used as an input to the Pilon error-detection and correction tool (Walker et al. 2014). This analysis did not identify any indels, but 65 potential single nucleotide polymorphisms (SNPs). However, these SNPs have very low-quality scores and low coverage (∼2× coverage as opposed to a median coverage of ∼630× at all other positions) and therefore did not pass the filter for making changes. In summary, no errors were detected in the PacBio assembly based on the Pilon analysis using high-quality and high-coverage Illumina read alignments. This indicates that the median PacBio coverage of ∼500× and multiple rounds of polishing using the PacBio ReSequencing.1 pipeline was sufficient to produce an accurate assembly.
The final circular chromosome is 1,484,007 bp in size (table 1), an increase of 321 kb over the published wAlbB draft genome. Using the minimap2 tool for genome alignments (Li 2018), all the contigs from the published genome (Mavingui et al. 2012) could be mapped to the complete assembly (fig. 3A, supplementary table S1A, Supplementary Material online). Analysis of these alignments identified 70 SNPs and 4 small indels in the contigs of the published assembly with reference to the complete circular genome as reference (supplementary table S1B, Supplementary Material online). The average GC content of the genome is 34.4%, which is within the typical range for Wolbachia genomes (table 1). DNA methylation in bacteria often serves a critical part of the Restriction–Modification (RM) systems, where it prevents self-DNA digestion by restriction enzymes used by the bacteria to inactivate infecting phages (Murray et al. 2012). PacBio sequencing enables direct identification of the methylation status of bacteria (Murray et al. 2012). Because there are no previous reports on the methylation status of any Wolbachia, we considered it of interest to perform an analysis on wAlbB. Analysis with RS_Modification_and_Motif_Analysis.1 pipeline did not identify DNA modifications such as m4C or m6A suggesting that these modifications are absent from the wAlbB genome (supplementary fig. S2, Supplementary Material online).
Table 1.
Key Characteristics of All Complete Wolbachia Genomes
| Name | Host Organism | Supergroup | RefSeq Assembly Accession | Size (Mb) | GC% | Proteins | rRNA | tRNA | Other RNA | Total Genes | Pseudogenes (%) | Ankyrin Proteins | BUSCO Score |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| wAu | Drosophila simulans | A | GCF_000953315.1 | 1.26 | 35.2 | 1,099 | 3 | 34 | 4 | 1,265 | 125 (9.9) | 21 | 83.8 |
| wHa | D. simulans | A | GCF_000376605.1 | 1.29 | 35.3 | 1,126 | 3 | 35 | 4 | 1,263 | 95 (7.5) | 26 | 83.3 |
| wMel | D. melanogaster | A | GCF_000008025.1 | 1.27 | 35.2 | 1,100 | 3 | 34 | 4 | 1,270 | 129 (10.2) | 19 | 82.9 |
| wRi | D. simulans | A | GCF_000022285.1 | 1.45 | 35.2 | 1,254 | 3 | 34 | 4 | 1,403 | 108 (7.7) | 25 | 83.3 |
| wAlbB | Ae. albopictus (Aa23 cell line) | B | GCF_004171285.1 | 1.48 | 34.4 | 1,205 | 3 | 34 | 4 | 1,434 | 188 (13.1) | 34 | 81.9 |
| wNo | D. simulans | B | GCF_000376585.1 | 1.30 | 34.0 | 1,065 | 3 | 34 | 4 | 1,231 | 125 (10.2) | 41 | 82.4 |
| wPip | C. quinquefasciatus | B | GCF_000073005.1 | 1.48 | 34.2 | 1,257 | 3 | 34 | 4 | 1,402 | 104 (7.4) | 43 | 81.9 |
| wTpre | Trichogramma pretiosum | B | GCF_001439985.1 | 1.13 | 33.9 | 827 | 3 | 35 | 4 | 1,106 | 237 (21.4) | 10 | 82.4 |
| wOo | Onchocerca ochengi | C | GCF_000306885.1 | 0.96 | 32.1 | 651 | 3 | 34 | 4 | 759 | 67 (8.8) | 1 | 74.7 |
| wOv | Onchocerca volvulus | C | GCF_000530755.1 | 0.96 | 32.1 | 649 | 3 | 34 | 4 | 763 | 73 (9.6) | 1 | 75.6 |
| wBm | Brugia malayi | D | GCF_000008385.1 | 1.08 | 34.2 | 839 | 3 | 34 | 4 | 1,047 | 167 (16.0) | 12 | 80.6 |
| wFol | Folsomia candida | E | GCF_001931755.1 | 1.80 | 34.8 | 1,513 | 3 | 35 | 4 | 1,658 | 103 (6.2) | 83 | 81.0 |
| wCle | Cimex lectularius | F | GCF_000829315.1 | 1.25 | 36.3 | 981 | 3 | 34 | 4 | 1,246 | 224 (18.0) | 33 | 80.6 |
Note.—For a standardized analysis across all genomes, the number of ankyrin proteins in each genome was determined in this study, using an identical pipeline based on Pfam domain annotations. Similarly, the BUSCO scores (reported as % complete BUSCOs) for each of the genomes was also calculated using identical parameters (−lineage = proteobacteria_odb9, 221 BUSCO groups). The other genome characteristics were obtained from the RefSeq database using the accession numbers indicated.
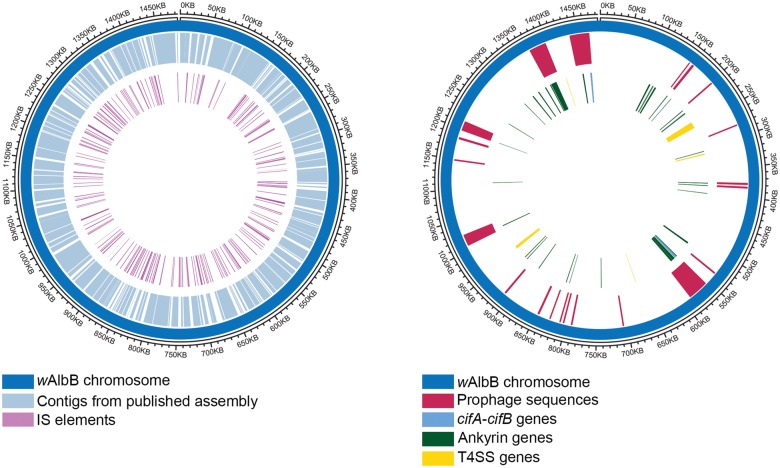
Fig. 3.
—Circos plot representation of features on the circular wAlbB genome. The wAlbB genome is represented as the outer blue circle, with the coordinates marked on the outermost circle. (A) The completed wAlbB genome contains all the contigs from the published draft genome (depicted in light blue on the 1st inner circle) revealing the gaps (white regions, 1st inner circle). The IS elements (purple, innermost circle) are distributed all over the genome, and tend to be located near the gaps, close to the termini of the contigs from the published draft. (B) Positions of prophage associated genes are indicated in the first inner circle. Prophage regions and WO-like islands are shown clockwise from the origin: wAlbB_WO-like_island _01, wAlbB_WO-like_island _02, wAlbB_ProphageRegion _01, wAlbB_WO-like_island _03, and wAlbB_WO-like_island _04. Locations of cifA-cifB gene pairs (light blue), and genes encoding ankyrin proteins (green) and T4SS components (yellow) are indicated in the second inner circle.
Annotation of the genome using the NCBI prokaryotic annotation pipeline (Angiuoli et al. 2008) identified 1,205 protein-coding genes, an increase of 250 genes over the published version (supplementary table S1C, Supplementary Material online). The genome also encodes 34 tRNA genes that include cognates for all amino acids, three rRNA (16S, 23S, and 5S), plus another three non-coding RNAs (6S RNA, RNAse P RNA, and a signal recognition particle sRNA small type), and one tmRNA gene. A total of 188 pseudogenes were identified resulting from one or more of the following causes: frameshift (n = 39), incomplete (n = 150), internal stop (n = 23), or multiple problems (n = 21). The percentage of pseudogenes is comparable across all the completed Wolbachia genomes from various supergroups (table 1).
The completeness of genome annotation was evaluated using the BUSCO pipeline (Simão et al. 2015), which measures the proportion of expected gene content from highly conserved, single-copy orthologs (BUSCO groups). The analysis was carried out against 221 BUSCO groups derived from 1,520 proteobacterial species. The 1,205 protein coding genes in the wAlbB genome contain 179 complete and single copy BUSCO groups, 2 complete and duplicated BUSCO groups, 6 fragmented BUSCOs, and 34 missing BUSCOs, resulting in a 81% BUSCO completeness score. For comparison, the BUSCO scores were also calculated for the other completed Wolbachia genomes (supplementary fig. S3, Supplementary Material online) and similar completeness scores were obtained for all genomes analyzed (table 1).
IS Elements Comprise 13% of the wAlbB Genome
ISs are one of the simplest transposable elements, usually encoding only a transposase flanked by short direct- or inverted-repeats, and can play a major role in genome evolution even in a short time scale (Siguier et al. 2015). IS elements have been classified into ∼20 families based on sequence similarities (Siguier et al. 2006). Wolbachia genomes often harbor numerous IS elements and their identification is essential for a comprehensive study of Wolbachia genome evolution (Cerveau et al. 2011). To annotate IS elements in the wAlbB genome, the ISsaga web service (Varani et al. 2011) was used to query the ISfinder database (Siguier et al. 2006) and 218 IS elements were found distributed throughout the genome (fig. 3A, supplementary table S2, Supplementary Material online), including 45 partial IS elements containing pseudogenized transposase (supplementary table S2, Supplementary Material online). The IS982 and IS481 family IS elements were the most abundant, with 96 and 75 copies, respectively. The median size for IS elements is 873 bp, and they range in length from 66 to 1,683 bp, adding up to total of 191,182 bp or about 13% of the entire wAlbB genome which is similar to that described for wRi (11%) (Cerveau et al. 2011). Mapping the contigs from the published draft genome (Mavingui et al. 2012) to the completed genome revealed that break points and/or gaps overlap with IS elements (fig. 3A). This indicates that the repetitive nature of IS elements hinder genome assembly using short-read data, and this problem can be overcome by using long-read technologies such as PacBio.
Movement of IS elements can cause insertions/deletions in a genome, sometimes leading to pseudogene formation. For example, the published contig NZ_CAGB01000139.1 (17,533 bp) was found to map to two regions of ∼540 and ∼17,000 bp on the complete genome, with a 1,203 bp insertion caused by an IS481 element, resulting in the formation of two pseudogenes (DEJ70_01295, DEJ70_01305) derived from the rsmD gene. PCR amplification and Sanger sequencing across this region confirmed this insertion and pseudogenization of the rsmD gene (supplementary fig. S4, Supplementary Material online).
Orthology Analysis and Identification of a Core Proteome Across Completed Wolbachia Genomes
Orthology relationships between the wAlbB proteins and the RefSeq protein sequences from all other complete Wolbachia genomes (table 1) were analyzed using the OrthoFinder program, which identifies families of homologous proteins and assigns them to orthogroups (Emms and Kelly 2015). A total of 1,604 orthogroups were derived from a combined set of 13,002 proteins (supplementary table S3, Supplementary Material online). Of these, 1,171 orthogroups (supplementary table S3A, Supplementary Material online) comprised of 12,569 proteins are shared by two or more genomes (“shared orthogroups”), while the remaining 433 orthogroups (supplementary table S3B, Supplementary Material online) are unique to each Wolbachia analyzed. For wAlbB, 1,192 of the 1,205 protein-coding genes (99%) were assigned to 918 shared orthogroups, while 13 wAlbB proteins could not be assigned to any such group (supplementary table S3, Supplementary Material online). Similarly, for all the other genomes analyzed, more than 93.5% of proteins could be assigned to a shared orthogroup, and the proportion of potentially genome-specific orthogroups was found to be low (<6.5%). The only outlier was wFol, with 14% of its 1,403 proteins found to be specific (supplementary table S3, Supplementary Material online).
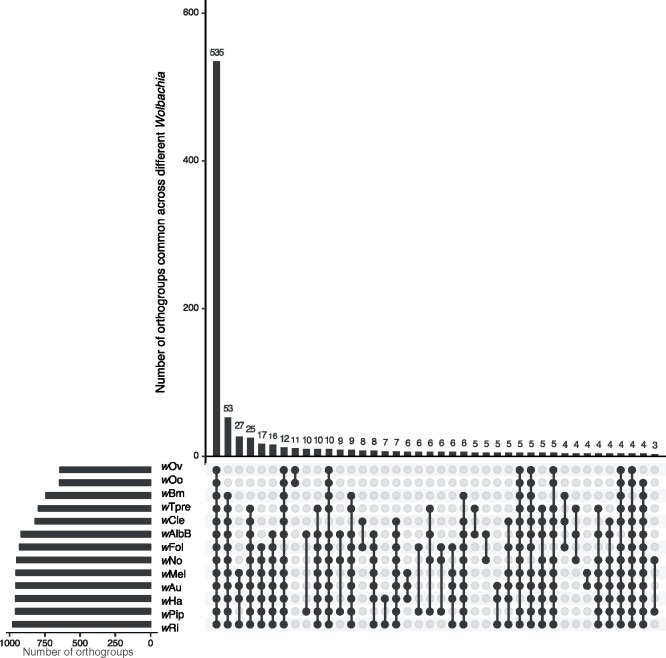
The core proteome, defined as the set of proteins present in all genomes analyzed, consists of 535 orthogroups (fig. 4), and 519 of these orthogroups contain single-copy 1:1 orthologs (supplementary table S3, Supplementary Material online). Outside the core proteome, the number of shared orthologous groups decreased substantially (fig. 4).
Fig. 4.
—Orthology analysis of proteins from all complete Wolbachia genomes identifies core Wolbachia proteome. The set of all 13,002 proteins from wAlbB and 12 other completed Wolbachia genomes were grouped into 1,171 orthogroups using the OrthoFinder software. Of these, 535 orthogroups were present across all the genomes analyzed, representing the core Wolbachia proteome, represented by the first bar in the Upset plot. Other orthogroups showed various patterns of distribution. Filled dots (black) denote presence and empty dots (gray) indicate absence of orthogroups in each Wolbachia. Genomes of Wolbachia from Drosophila melanogaster (wMel), D. simulans (wRi, wHa, wNo, wAu), C. quinquefasciatus (wPip), Brugia malayi (wBm), Onchocerca ochengi (wOo), Onchocerca volvulus (wOv), Folsomia candida (wFol), Trichogramma pretiosum (wTpre), and Cimex lectularius (wCle) were included in the analysis. The largest 40 intersections/overlaps between orthogroups across different Wolbachia are displayed.
Pfam and eggNOG Annotations
Analysis of Pfam protein domains (Finn et al. 2016) encoded in the wAlbB genome identified 995 genes encoding proteins containing at least one Pfam domain, representing 83% of the total genes present (supplementary table S4, Supplementary Material online). By far the most abundant domains arise from mobile genetic elements. For example, the DDE Transposase domain (“DDE_Tnp_1_3,” Pfam accession PF13612) was the most abundant domain, present in 82 proteins, followed by the “Retroviral Integrase” domain (“rve,” Pfam accession PF00665) present in 67 proteins. We also found 48 proteins containing the reverse transcriptase domain (“RVT_1,” Pfam accession PF00078), and 53 proteins with the Group II intron reverse transcriptase domain (“GIIM,” Pfam accession PF08388). Further annotation of gene function based on orthology using the eggNOG software (Huerta-Cepas et al. 2017) could assign a putative function to 1,044 (87%) of the 1,205 wAlbB protein-coding genes, with transposase, integrase, and reverse transcriptase functions again being the most abundant classes (supplementary table S5, Supplementary Material online).
wAlbB Genome Contains Degenerated WO Prophage
Prophages play an important role in Wolbachia biology for example the prophage WO from the Wolbachia strain wMel contributes to the CI in its Drosophila host (Masui et al. 2000, 2001; Beckmann et al. 2017; LePage et al. 2017). Availability of a complete genome made the search for any potential prophages in wAlbB feasible. The PHASTER webserver (Arndt et al. 2016) identified two pseudogene fragments (DEJ70_03825 and DEJ70_03855) of a phage capsid protein, formed by insertion of four different IS-elements and one Group II intron reverse transcriptase/maturase into its open reading frame. In addition, in a different region, essential phage proteins such as phage late control D protein, tail protein X, tail tape measure protein, and a major tail tube protein (encoded by DEJ70_05835, DEJ70_05840, DEJ70_05850, and DEJ70_05865, respectively), pseudogenized versions of a tail sheath protein (loci DEJ70_05870 and DEJ70_05880, as well as some of the genes found in Eukaryotic Association Modules of WO phages (Bordenstein and Bordenstein 2016), were found. This region showed highest nucleotide similarity to the WOVitA1 prophage, possibly indicative of its origin, and was named wAlbB_prophageRegion_01 (fig. 3B, supplementary table S6A, Supplementary Material online). Further BLAST comparisons of genome and proteins from wAlbB with those of WOVitA1 identified wAlbB orthologs for 40 of the 63 WOVitA1 genes, and seven pseudogenes corresponding to five other WOVitA1 genes, while 18 WOVitA1 genes were found to be completely absent in the wAlbB genome (supplementary table S6B, Supplementary Material online). The prophage genes absent in wAlbB include the components essential for a phage particle assembly, such as tail subunit I, baseplate subunits J, W, and V, phage portal protein and phage minor capsid protein, suggesting an inactive prophage. BLAST-based comparisons to various WO phages (see Materials and Methods section) identified four WO-like islands (named wAlbB_WO_like_island_01 through wAlbB_WO_like_island_04, supplementary table S6A, Supplementary Material online), and additional 19 prophage-associated loci (13 CDS, 6 pseudogenes), distributed over the genome (fig. 3B). The nucleotide sequences of these scattered loci were found to be most similar to those from wPip (strain Pel) and wNo. None of these additional loci can compensate for functions missing in WOVitA1-derived prophage regions. Overall, wAlbB has 111 prophage-associated loci (83 protein coding CDS, 28 pseudogenes), with a combined size of 116 kb, comprising about 8% of the entire wAlbB genome. Together these observations suggest that the prophages in wAlbB have undergone degeneration and are not active.
CI Genes in wAlbB
The wAlbB Wolbachia is known to cause CI in its Ae. albopictus host (Dobson et al. 2001). Genetic studies of CI in Drosophila and Culex hosts have identified a pair of genes, cifA and cifB (Beckmann et al. 2017; LePage et al. 2017; Bonneau et al. 2018), which are sometimes located within the WO prophage regions (Lindsey et al. 2018). A phylogenetic analysis of cifA and cifB across all Wolbachia has found them to co-occur as a pair of neighboring genes, and grouped them into four monophyletic “Types” (LePage et al. 2017; Lindsey et al. 2018). A search for cifA and cifB homologs in wAlbB identified two sets of cifA and cifB gene-pairs. The first pair is composed of a Type IV cifA (DEJ70_02760) and a Type IV cifB (DEJ70_02755) and is located in wAlbB_WO_like_island_01, while the second pair is composed of a Type III cifA (DEJ70_07090) and a Type III cifB (DEJ70_07095), situated in wAlbB_WO_like_island_04.
Type IV Secretion System in wAlbB
Many symbiotic and pathogenic intracellular bacteria use a Type IV secretion system (T4SS) for successful infection, proliferation, and persistence within hosts. It is a diverse and versatile transporter system which spans the entire cell envelope functioning in conjugation, competence, and effector molecule (DNA and/or protein) translocation (Bhattacharya and Newton 2017; Grohmann et al. 2018). Genome analysis of wAlbB revealed the presence of a T4SS with 15 components organized in two operons and four individual genes (fig. 3B). Operon 1 contains virB8-1 (DEJ70_04590), virB9-1 (DEJ70_04585), virB10 (DEJ70_04580), virB11 (DEJ70_04575), and virD4 (DEJ70_04570). The vitamin B2 biosynthetic enzyme ribA encoded by DEJ70_04595, may be co-transcribed in this operon as observed in wBm (Li and Carlow 2012). Operon 2 contains virB3 (DEJ70_01260), virB4 (DEJ70_01265), virB6-1 (DEJ70_01270), virB6-2 (DEJ70_01275), virB6-3 (DEJ70_01280), and virB6-4 (DEJ70_01285). Three duplicated genes: virB4-2 (DEJ70_01565), virB8-2 (DEJ70_03190), and virB9-2 (DEJ70_06825) are found scattered elsewhere in the genome. Interestingly, virB2 (DEJ70_04445), which has been presumed absent from Wolbachia (Rancès et al. 2008), was also found in the wAlbB genome. Previous studies in other bacteria have shown that the T4SS is not constitutively expressed but tightly regulated by transcription factors (Félix et al. 2008), such as wBmxR1 and wBmxR2 in wBm (Li and Carlow 2012). In wAlbB one corresponding homolog, DEJ70_05760, with higher sequence similarity to wBmxR1 was found.
Analysis of Ankyrin Genes
Ankyrin repeat-containing (ANK) proteins are involved in protein–protein interactions and are rare in bacteria, but are found in Wolbachia, where they may be involved in host–Wolbachia interactions. The T4SS has been shown to be responsible for the secretion of the ankyrin repeat-containing protein AnkA in Anaplasma phagocytophilum (Lin et al. 2007), an intracellular bacterium closely related to Wolbachia. Based on genome-wide Pfam protein domain annotations (supplementary table S4, Supplementary Material online), 34 wAlbB proteins were found to contain at least one copy of an ankyrin repeat domain (table 1), with a total of 81 copies of various types of ankyrin domains (supplementary table S4, Supplementary Material online). The same analysis performed for all complete Wolbachia genomes revealed a similar number of ANK proteins across insect Wolbachia, and fewer ANKs in filarial Wolbachia (table 1).
KEGG Pathway Analysis Identifies Missing Genes in wAlbB
KAAS (Moriya et al. 2007) was used to obtain functional annotations of predicted protein sequences from the wAlbB genome. A total of 595 proteins were assigned a KO (KEGG Orthology) number. The KEGG pathway map and KO assignments for wAlbB were compared with those from the closely related Wolbachia wPip and wRi. Pairwise comparisons (wAlbB vs. wPip, wAlbB vs. wRi) of the KEGG annotated proteins revealed 5 proteins absent in wAlbB, namely DNA-3-methyladenine glycosylase (MPG, EC: 3.2.2.21), diacylglycerol kinase (DgkA, EC: 2.7.1.107), Cytochrome bd ubiquinol oxidase subunit I (CydA, EC: 1.10.3.14) and subunit II (CydB, EC: 1.10.3.14), and FtsI (EC: 3.4.6.4). The presence of these five proteins was determined in other Wolbachia genomes available in the KEGG database, namely wMel, wRi, wHa, wNo, wPip, wBm, wOo, and wCle (table 2). MPG was found in all other Wolbachia analyzed except in wAlbB, while the other four proteins were absent in wAlbB and in at least one additional species (table 2).
Table 2.
KAAS Server-Based KEGG Annotations Identifies Five Genes Missing in wAlbB
| wMel | wRi | wHa | wNo | wPip | wBm | wOo | wCle | |
|---|---|---|---|---|---|---|---|---|
| DgkA | WD_1163 | wRi_011390 | wHa_09720 | wNo_07140 | WP0909 | - | - | - |
| MPG | WD_1110 | wRi_012850 | wHa_09290 | wNo_05480 | WP0867 | wBm0254 | wOo_04680 | wCLE_011920 |
| CydA | WD_0740 | wRi_007360 | wHa_06280 | - | - | - | - | - |
| CydB | WD_0741 | wRi_007350 | wHa_06290 | - | - | - | - | - |
| FtsI | WD_1273 | wRi_012430 | wHa_10600 | - | - | - | - | wCLE_010110 |
Note.—Missing genes, denoted by a hyphen (-), were identified by comparing wAlbB KEGG annotations to wPip and wRi annotations at the KAAS server. Corresponding orthologs were then identified in other Wolbachia annotations available on the KAAS server.
Discussion
We have assembled the complete genome of wAlbB from Aa23 cells. The key factors facilitating this were the relatively pure high molecular weight DNA extracted from host cell-free Wolbachia, and long read PacBio sequencing at high coverage.
The long reads enabled sequencing over complex repeat regions which have been difficult to resolve with short read sequencing. High depth of coverage (∼500×) enabled multiple rounds of polishing of the assembly to remove any SNPs or indel errors that are occasionally observed in technologies such as PacBio or Nanopore (Watson and Warr 2019). Absence of errors was also confirmed using Illumina data (∼600× coverage) generated from the same DNA sample.
Wolbachia genomes range in size from ∼0.9 to 1.8 Mb. Currently, 42 genomes have been deposited in the GenBank database, however, only 12 genomes show complete status. The complete circular wAlbB chromosome is 1,484,007 bp in size making it one of the largest sequenced Wolbachia genomes to date. The complete assembly is 321 kb larger than the draft sequence (Mavingui et al. 2012) and contains all the contigs from the published genome. Many of the gaps in the published draft were observed to be flanked by IS elements, suggesting that the repetitive nature of IS elements hinders assembly. In addition, some contigs from the published draft mapped to multiple locations in the finished genome, indicating that they originate from repeated regions in the genome. Such repeated regions can be difficult to assemble using only short reads from Illumina or 454 platforms, but can be successfully assembled using PacBio long reads.
The genome of wAlbB contains 218 IS elements, belonging to 10 families, including one new family, scattered throughout the genome. The genomes of arthropod infecting Wolbachia have a large number of repetitive and mobile elements, particularly IS elements (Cerveau et al. 2011). A total of 11 distinct IS families was reported across wBm, wPel, wRi, and wMel genomes (Cerveau et al. 2011). The distribution and copy number of IS elements from different families varies between genomes (Cerveau et al. 2011). The wAlbB genome lacks members from IS6 and IS200/605 families, while IS982 (n = 96) and IS481 (n = 75) are present in higher abundance in comparison to other Wolbachia genomes (Cerveau et al. 2011). IS elements can cause disruption of genes, giving rise to pseudogenes. In wAlbB, many such examples were observed, for example pseudogenization of rsmD and dgkA genes.
Prophages represent another class of highly mobile elements that can have a significant impact on Wolbachia biology (Gavotte et al. 2006; Bordenstein and Bordenstein 2016; Beckmann et al. 2017; LePage et al. 2017). In a previous study of wild-caught, Ae. albopictus mosquitoes carrying either only wAlbA or both wAlbA and wAlbB, phage particles could be detected and quantified using qPCR (Chauvatcharin et al. 2006). In the current wAlbB genome, although 40 of the 63 genes from the WOVitA1 prophage (Bordenstein and Bordenstein 2016) could be found, many genes encoding essential components for a phage particle assembly were absent, while a few others were pseudogenized due to insertion of IS elements. In addition, the prophage-related genes were found to be scattered over the wAlbB genome, unlike in an intact prophage. Therefore, the ancestral temperate prophage has undergone degradation in the wAlbB genome, a phenomenon also observed in other Wolbachia such as wRec from Drosophila recens (Metcalf et al. 2014). It is therefore possible that the phage particles previously reported in Ae. albopictus mosquitoes (Chauvatcharin et al. 2006) were derived only from wAlbA, or the wAlbB phage was degraded during its in vitro culture in Aa23 cells. Interestingly, orthologs of the two WO prophage proteins CifA and CifB involved in CI (Beckmann et al. 2017; LePage et al. 2017; Bonneau et al. 2018) are encoded in wAlbB by two pairs of genes, and are situated in WO-like islands, consistent with observations in other Wolbachia prophage systems (Bordenstein and Bordenstein 2016; LePage et al. 2017; Lindsey et al. 2018). These gene-pairs might be remnants from an earlier integrated prophage which has since undergone degradation. Similarly, in another Wolbachia, wRec, approximately 75% of its prophage gene content has been lost (Metcalf et al. 2014), but still retains an intact cifA, cifB gene pair (Lindsey et al. 2018).
Orthology analysis of all annotated proteins from wAlbB and 12 other complete Wolbachia genomes identified the core Wolbachia proteome comprising 535 orthogroups. The majority of these (n = 519) contain single-copy, 1:1 orthologs which are ideally suited for phylogenomic analysis. Further analysis of this core proteome could shed light on the unique intracellular lifestyle of Wolbachia and provide insight into essential Wolbachia genes that may be targeted in an anti-symbiotic approach to new drug discovery in filarial parasites. Conversely, studies on orthogroups unique to a particular genome (e.g. 13 proteins present only in wAlbB) may lead to the discovery of proteins which are involved in the adaptation of Wolbachia to a particular host/cell niche.
The T4SS of wAlbB is encoded by two operons and a few genes scattered throughout the genome. Their organization and sequence are conserved across various Wolbachia, likely due to their important roles in its biology, such as secretion of effectors that influence host:bacteria interactions. Several candidate effectors of the T4SS in wMel were identified recently (Rice et al. 2017), including one which interacts with the host cytoskeleton (Sheehan et al. 2016). Ankyrins are established T4SS effectors of intracellular bacteria. The genome of wAlbB encodes 34 ANK proteins. In the closely related Anaplasma and Ehrlichia, an ANK protein has been shown to regulate transcription, suppress host innate immunity, inhibit host cell apoptosis, and reduce reactive oxygen species (Rikihisa and Lin 2010; Liu et al. 2012). However, most ANK genes contain many copies of short open reading frames of unknown function (Wu et al. 2004). The T4SS could also be involved in lateral gene transfer events between Wolbachia and its host (Dunning Hotopp et al. 2007). For example, VirB6, an essential trans-membrane channel component of the T4SS in many bacteria, has been shown to direct DNA export through the T4SS in Agrobacterium tumefaciens (Jakubowski et al. 2004). Interestingly, all Wolbachia genomes, including wAlbB, encode 4 VirB6 paralogs.
Comparing the KEGG pathway maps and KO assignments in wAlbB with those in closely related wPip and wRi identified five proteins that were absent in wAlbB, namely DgkA, MPG, CydA, CydB, and FtsI. In the wAlbB genome, dgkA is pseudogenized due to insertion of an IS982 family transposase, while the gene is intact in wMel, wRi, wHa, wNo, and wPip, but is absent in wBm, wOo, and wCle. DgkA phosphorylates diacylglycerol to generate phosphatidic acid in glycerolipid and glycerophospholipid metabolism pathways and plays an important role in microbial stress responses (Yamashita et al. 1993). MPG is involved in the DNA base excision repair pathway by recognizing a variety of base lesions, mainly caused by alkylating agents, resulting in release of the damaged base in free form from alkylated DNA and initiation of repair (Costa de Oliveira et al. 1987). MPG is present in all the Wolbachia examined here, except wAlbB. The genes cydA and cydB are present in wMel, wRi, wHa, and wNo, yet absent in wBm, wOo, wCle, and wAlbB. The proteins CydA and CydB are members of a family of integral membrane proteins involved in catalyzing terminal electron transfer in eubacterial and archaeal respiration. Their high oxygen affinity enables them to scavenge and reduce oxygen to water, preventing damage to oxygen-sensitive enzymes, and permitting growth in microaerobic and anaerobic environments and survival under a number of stress conditions (Borisov et al. 2011). FtsI is a class B3 penicillin-binding protein (PBP B3) that functions as a transpeptidase in peptidoglycan metabolism, essential for bacterial cell wall synthesis and cell division (Cayô et al. 2011). Although the wAlbB genome does not have ftsI, it does encode a class PBP B2 transpeptidase, MrdA, while other Wolbachia, wMel, wRi, wHa, and wCle, have both FtsI and MrdA, indicating potential redundancy.
The availability of a complete circular genome from wAlbB will provide further insight into phylogenetic relationships between the different Wolbachia supergroups, and enable further biochemical, molecular, and genetic analyses on wAlbB and related Wolbachia. The annotation and analysis of mobile elements highlight their considerable effect on genome evolution and gene content in intracellular symbionts, suggesting that such elements could be repurposed as tools for genetic manipulation of Wolbachia. The genome also provides an important baseline for further studies of Wolbachia interactions with its host that may advance practical applications such as the use of Wolbachia for pest and disease control.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
The authors are grateful to Dr Sarah Bordenstein for her contribution to the analyses of WO prophage and helpful comments on the manuscript. We also thank Dr Mylene Weill and the following for helpful discussions and comments on the manuscript: Brian Anton, Rich Roberts, Peter Weigele, Rick Morgan, Tom Evans, Bill Jack, Jeremy Foster, Barton Slatko, Emilie Lefoulon, Youseuf Suliman, and Catherine Poole; and the DNA sequencing core at New England Biolabs for Illumina sequencing. The authors are also grateful for the continued encouragement from Don Comb. This work was funded by New England Biolabs.
Data deposition: This project has been deposited at the NCBI GenBank under the BioProject accession PRJNA454708, the PacBio raw data under the accessions SRR7784284, SRR7784285, SRR7784286, SRR7784287, and the Illumina raw data under the accession SRR7623731. The assembled genome and annotations have been deposited at the NCBI GenBank database under the accession CP031221.
Literature Cited
- Aliota MT, Peinado SA, Velez ID, Osorio JE.. 2016. The wMel strain of Wolbachia reduces transmission of Zika virus by Aedes aegypti. Sci Rep. 6:28792.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aliota MT, Walker EC.. 2016. The wMel strain of Wolbachia reduces transmission of chikungunya virus in Aedes aegypti. PLoS Negl Trop Dis. 10(4):e0004677.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrews ES, Crain PR, Fu Y, Howe DK, Dobson SL.. 2012. Reactive oxygen species production and Brugia pahangi survivorship in Aedes polynesiensis with artificial Wolbachia infection types. PLoS Pathog. 8(12):e1003075.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angiuoli SV, et al. 2008. Toward an online repository of Standard Operating Procedures (SOPs) for (meta)genomic annotation. OMICS J Integr Biol. 12(2):137–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arndt D, et al. 2016. PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44(W1):W16–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldo L, et al. 2006. Multilocus sequence typing system for the endosymbiont Wolbachia pipientis. Appl Environ Microbiol. 72(11):7098–7110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bazzocchi C, et al. 2008. Combined ivermectin and doxycycline treatment has microfilaricidal and adulticidal activity against Dirofilaria immitis in experimentally infected dogs. Int J Parasitol. 38(12):1401–1410. [DOI] [PubMed] [Google Scholar]
- Beckmann JF, Ronau JA, Hochstrasser M.. 2017. A Wolbachia deubiquitylating enzyme induces cytoplasmic incompatibility. Nat Microbiol. 2:17007.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharya T, Newton ILG.. 2017. Mi Casa es Su Casa: how an intracellular symbiont manipulates host biology. Environ Microbiol. [Epub ahead of print] doi: 10.1111/1462-2920.13964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonneau M, et al. 2018. Culex pipiens crossing type diversity is governed by an amplified and polymorphic operon of Wolbachia. Nat Commun. 9(1):319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bordenstein SR, Bordenstein SR.. 2016. Eukaryotic association module in phage WO genomes from Wolbachia. Nat Commun. 7:13155.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borisov VB, Gennis RB, Hemp J, Verkhovsky MI.. 2011. The cytochrome bd respiratory oxygen reductases. Biochim Biophys Acta 1807(11):1398–1413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchfink B, Xie C, Huson DH.. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59.. [DOI] [PubMed] [Google Scholar]
- Carrington LB, et al. 2018. Field- and clinically derived estimates of Wolbachia-mediated blocking of dengue virus transmission potential in Aedes aegypti mosquitoes. Proc Natl Acad Sci. 115(2):361–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cayô R, et al. 2011. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother. 55(12):5907–5913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerveau N, Leclercq S, Leroy E, Bouchon D, Cordaux R.. 2011. Short- and long-term evolutionary dynamics of bacterial insertion sequences: insights from Wolbachia endosymbionts. Genome Biol Evol. 3:1175–1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chauvatcharin N, Ahantarig A, Baimai V, Kittayapong P.. 2006. Bacteriophage WO‐B and Wolbachia in natural mosquito hosts: infection incidence, transmission mode and relative density. Mol Ecol. 15(9):2451–2461. [DOI] [PubMed] [Google Scholar]
- Chin C-S, et al. 2013. Nonhybrid, finished microbial genome assemblies from long-read SMRT sequencing data. Nat Methods 10(6):563. [DOI] [PubMed] [Google Scholar]
- Chouin-Carneiro T, et al. 2016. Differential susceptibilities of Aedes aegypti and Aedes albopictus from the Americas to Zika virus. PLoS Negl Trop Dis. 10(3):e0004543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conway JR, Lex A, Gehlenborg N, Hancock J.. 2017. UpSetR: an R package for the visualization of intersecting sets and their properties. Bioinformatics 33(18):2938–2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Bouchon D, Grève P.. 2011. The impact of endosymbionts on the evolution of host sex-determination mechanisms. Trends Genet. 27(8):332–341. [DOI] [PubMed] [Google Scholar]
- Costa de Oliveira R, Laval J, Boiteux S.. 1987. Induction of SOS and adaptive responses by alkylating agents in Escherichia coli mutants deficient in 3-methyladenine-DNA glycosylase activities. Mutat Res Repair Rep. 183:11–20. [DOI] [PubMed] [Google Scholar]
- Dobson SL, Marsland EJ, Rattanadechakul W.. 2001. Wolbachia-induced cytoplasmic incompatibility in single- and superinfected Aedes albopictus (Diptera: Culicidae). J Med Entomol. 38(3):382–387. [DOI] [PubMed] [Google Scholar]
- Dobson SL, Marsland EJ, Veneti Z, Bourtzis K, O'Neill SL.. 2002. Characterization of Wolbachia host cell range via the in vitro establishment of infections. Appl Environ Microbiol. 68(2):656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodson BL, et al. 2014. Wolbachia enhances West Nile virus (WNV) infection in the mosquito Culex tarsalis. PLoS Negl Trop Dis. 8(7):e2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eid J, et al. 2008. Real-time DNA sequencing from single polymerase molecules. Science. 323:133–138. [DOI] [PubMed] [Google Scholar]
- Emms DM, Kelly S.. 2015. OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol. 16:157.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Félix C, et al. 2008. Characterization and transcriptional analysis of two gene clusters for Type IV secretion machinery in Wolbachia of Armadillidium vulgare. Res Microbiol. 159(6):481–485. [DOI] [PubMed] [Google Scholar]
- Fenn K, et al. 2006. Phylogenetic relationships of the Wolbachia of nematodes and arthropods. PLoS Pathog. 2(10):e94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenollar F, Maurin M, Raoult D.. 2003. Wolbachia pipientis growth kinetics and susceptibilities to 13 antibiotics determined by immunofluorescence staining and real-time PCR. Antimicrob Agents Chemother. 47(5):1665–1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenollar F, Scola BL, et al. 2003. Culture and phenotypic characterization of a Wolbachia pipientis isolate. J Clin Microbiol. 41(12):5434–5441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44(D1):D279–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flusberg BA, et al. 2010. Direct detection of DNA methylation during single-molecule, real-time sequencing. Nat Methods 7(6):461–465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao F, Luo H, Zhang C-T.. 2012. DoriC 5.0: an updated database of oriC regions in both bacterial and archaeal genomes. Nucleic Acids Res. 41(D1):D90–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavotte L, et al. 2006. A survey of the bacteriophage WO in the endosymbiotic bacteria Wolbachia. Mol Biol Evol. 24(2):427–435. [DOI] [PubMed] [Google Scholar]
- Gerenday A, Fallon AM.. 1996. Cell cycle parameters in Aedes albopictus mosquito cells. In Vitro Cell Dev Biol Anim. 32(5):307–312. [DOI] [PubMed] [Google Scholar]
- Grard G, et al. 2014. Zika virus in Gabon (Central Africa)—2007: a new threat from Aedes albopictus? PLoS Negl Trop Dis. 8(2):e2681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gratz NG. 2004. Critical review of the vector status of Aedes albopictus. Med Vet Entomol. 18(3):215–227. [DOI] [PubMed] [Google Scholar]
- Grohmann E, Christie PJ, Waksman G, Backert S.. 2018. Type IV secretion in Gram-negative and Gram-positive bacteria. Mol Microbiol. 107(4):455–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Gu L, Eils R, Schlesner M, Brors B.. 2014. circlize Implements and enhances circular visualization in R. Bioinform Oxf Engl. 30:2811–2812. [DOI] [PubMed] [Google Scholar]
- Hertig M. 1936. The Rickettsia, Wolbachia pipientis (gen. et sp.n.) and associated inclusions of the mosquito, Culex pipiens. Parasitology 28(04):453–486. [Google Scholar]
- Hilgenboecker K, Hammerstein P, Schlattmann P, Telschow A, Werren JH.. 2008. How many species are infected with Wolbachia? A statistical analysis of current data. FEMS Microbiol Lett. 281(2):215–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotopp JCD, et al. 2007. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science 317(5845):1753–1756. [DOI] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2017. Fast genome-wide functional annotation through orthology assignment by eggNOG-Mapper. Mol Biol Evol. 34(8):2115–2122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta-Cepas J, et al. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 44(D1):D286–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Koga R, Xue P, Fukatsu T, Rasgon JL.. 2011. Wolbachia infections are virulent and inhibit the human malaria parasite Plasmodium falciparum in Anopheles gambiae. PLoS Pathog. 7(5):e1002043.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes GL, Vega-Rodriguez J, Xue P, Rasgon JL.. 2012. Wolbachia strain wAlbB enhances infection by the rodent malaria parasite Plasmodium berghei in Anopheles gambiae mosquitoes. Appl Environ Microbiol. 78(5):1491–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioannidis P, et al. 2007. New criteria for selecting the origin of DNA replication in Wolbachia and closely related bacteria. BMC Genomics 8(1):182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski SJ, Krishnamoorthy V, Cascales E, Christie PJ.. 2004. Agrobacterium tumefaciens VirB6 domains direct the ordered export of a DNA substrate through a Type IV secretion system. J Mol Biol. 341(4):961–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston KL, et al. 2014. Repurposing of approved drugs from the human pharmacopoeia to target Wolbachia endosymbionts of onchocerciasis and lymphatic filariasis. Int J Parasitol Drugs Drug Resist. 4(3):278–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, et al. 2010. Wolbachia stimulates immune gene expression and inhibits Plasmodium development in Anopheles gambiae. PLoS Pathog. 6(10):e1001143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambris Z, Cook PE, Phuc HK, Sinkins SP.. 2009. Immune activation by life-shortening Wolbachia and reduced filarial competence in mosquitoes. Science 326(5949):134–136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kittayapong P, Baisley KJ, Baimai V, O’Neill SL.. 2000. Distribution and diversity of Wolbachia infections in Southeast Asian mosquitoes (Diptera: Culicidae). J Med Entomol. 37(3):340–345. [DOI] [PubMed] [Google Scholar]
- Kotsakiozi P, et al. 2017. Population genomics of the Asian tiger mosquito, Aedes albopictus: insights into the recent worldwide invasion. Ecol Evol. 7(23):10143–10157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krzywinski M, et al. 2009. Circos: an information aesthetic for comparative genomics. Genome Res. 19(9):1639–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langworthy NG, et al. 2000. Macrofilaricidal activity of tetracycline against the filarial nematode Onchocerca ochengi: elimination of Wolbachia precedes worm death and suggests a dependent relationship. Proc R Soc Lond B Biol Sci. 267(1448):1063–1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefoulon E, et al. 2016. Breakdown of coevolution between symbiotic bacteria Wolbachia and their filarial hosts. PeerJ 4:e1840.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LePage DP, et al. 2017. Prophage WO genes recapitulate and enhance Wolbachia-induced cytoplasmic incompatibility. Nature 543(7644):243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lex A, Gehlenborg N, Strobelt H, Vuillemot R, Pfister H.. 2014. UpSet: visualization of intersecting sets. IEEE Trans Vis Comput Graph. 20(12):1983–1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. 2018. Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R.. 2009. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 25(14):1754–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Carlow CKS.. 2012. Characterization of transcription factors that regulate the Type IV secretion system and riboflavin biosynthesis in Wolbachia of Brugia malayi. PLoS One 7(12):e51597.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M, Dulk‐Ras AD, Hooykaas PJJ, Rikihisa Y.. 2007. Anaplasma phagocytophilum AnkA secreted by Type IV secretion system is tyrosine phosphorylated by Abl-1 to facilitate infection. Cell Microbiol. 9(11):2644–2657. [DOI] [PubMed] [Google Scholar]
- Lindsey ARI, et al. 2018. Evolutionary genetics of cytoplasmic incompatibility genes cifA and cifB in prophage WO of Wolbachia. Genome Biol Evol. 10(2):434–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Bao W, Lin M, Niu H, Rikihisa Y.. 2012. Ehrlichia Type IV secretion effector ECH0825 is translocated to mitochondria and curbs ROS and apoptosis by upregulating host MnSOD. Cell Microbiol. 14(7):1037–1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo N, et al. 2007. Taxonomic status of the intracellular bacterium Wolbachia pipientis. Int J Syst Evol Microbiol. 57(3):654–657. [DOI] [PubMed] [Google Scholar]
- Masui S, Kamoda S, Sasaki T, Ishikawa H.. 2000. Distribution and evolution of bacteriophage WO in Wolbachia, the endosymbiont causing sexual alterations in arthropods. J Mol Evol. 51(5):491–497. [DOI] [PubMed] [Google Scholar]
- Masui S, et al. 2001. Bacteriophage WO and virus-like particles in Wolbachia, an endosymbiont of arthropods. Biochem Biophys Res Commun. 283(5):1099–1104. [DOI] [PubMed] [Google Scholar]
- Mavingui P, et al. 2012. Whole-genome sequence of Wolbachia strain wAlbB, an endosymbiont of tiger mosquito vector Aedes albopictus. J Bacteriol. 194(7):1840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metcalf JA, Jo M, Bordenstein SR, Jaenike J, Bordenstein SR.. 2014. Recent genome reduction of Wolbachia in Drosophila recens targets phage WO and narrows candidates for reproductive parasitism. PeerJ 2:e529.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M.. 2007. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35(Web Server):W182–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousson L, et al. 2012. The native Wolbachia symbionts limit transmission of dengue virus in Aedes albopictus. PLoS Negl Trop Dis. 6(12):e1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray IA, et al. 2012. The methylomes of six bacteria. Nucleic Acids Res. 40(22):11450–11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill SL, et al. 1997. In vitro cultivation of Wolbachia pipientis in an Aedes albopictus cell line. Insect Mol Biol. 6:33–39. [DOI] [PubMed] [Google Scholar]
- Pan X, et al. 2018. The bacterium Wolbachia exploits host innate immunity to establish a symbiotic relationship with the dengue vector mosquito Aedes aegypti. ISME J. 12(1):277–288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM.. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26(6):841–842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rancès E, Voronin D, Tran-Van V, Mavingui P.. 2008. Genetic and functional characterization of the Type IV secretion system in Wolbachia. J Bacteriol. 190:5020–5030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raquin V, et al. 2015. Native Wolbachia from Aedes albopictus blocks chikungunya virus infection In Cellulo. PLoS One 10(4):e0125066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasgon JL, Ren X, Petridis M.. 2006. Can Anopheles gambiae be infected with Wolbachia pipientis? Insights from an in vitro system. Appl Environ Microbiol. 72(12):7718–7722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice DW, Sheehan KB, Newton ILG.. 2017. Large-scale identification of Wolbachia pipientis effectors. Genome Biol Evol. 9(7):1925–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rikihisa Y, Lin M.. 2010. Anaplasma phagocytophilum and Ehrlichia chaffeensis Type IV secretion and Ank proteins. Curr Opin Microbiol. 13(1):59–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saucereau Y, et al. 2017. Comprehensive proteome profiling in Aedes albopictus to decipher Wolbachia-arbovirus interference phenomenon. BMC Genomics 18(1):635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan KB, Martin M, Lesser CF, Isberg RR, Newton ILG.. 2016. Identification and characterization of a candidate Wolbachia pipientis Type IV effector that interacts with the actin cytoskeleton. mBio 7:e00622–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siguier P, Gourbeyre E, Varani A, Ton-Hoang B, Chandler M.. 2015. Everyman’s guide to bacterial insertion sequences. Microbiol Spectrum 3(2):MDNA3-0030-2014. doi:10.1128/microbiolspec.MDNA3-0030-2014 [DOI] [PubMed]
- Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M.. 2006. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34(90001):D32–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM.. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31(19):3210–3212. [DOI] [PubMed] [Google Scholar]
- Sinkins SP. 2004. Wolbachia and cytoplasmic incompatibility in mosquitoes. Insect Biochem Mol Biol. 34(7):723–729. [DOI] [PubMed] [Google Scholar]
- Sinkins SP, Braig HR, Oneill SL.. 1995. Wolbachia pipientis: bacterial density and unidirectional cytoplasmic incompatibility between infected populations of Aedes albopictus. Exp Parasitol. 81(3):284–291. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Bandi C, Hoerauf A.. 2005. Wolbachia bacterial endosymbionts of filarial nematodes Advances in parasitology. Vol. 60 p. 245–284. [DOI] [PubMed] [Google Scholar]
- Varani AM, Siguier P, Gourbeyre E, Charneau V, Chandler M.. 2011. ISsaga is an ensemble of web-based methods for high throughput identification and semi-automatic annotation of insertion sequences in prokaryotic genomes. Genome Biol. 12(3):R30.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voronin D, Cook DAN, Steven A, Taylor MJ.. 2012. Autophagy regulates Wolbachia populations across diverse symbiotic associations. Proc Natl Acad Sci. 109(25):E1638–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker BJ, et al. 2014. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9(11):e112963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker T, et al. 2011. The wMel Wolbachia strain blocks dengue and invades caged Aedes aegypti populations. Nature 476(7361):450–453. [DOI] [PubMed] [Google Scholar]
- Watson M, Warr A.. 2019. Errors in long-read assemblies can critically affect protein prediction. Nat. Biotechnol. 37:124. [DOI] [PubMed] [Google Scholar]
- Werren JH, Baldo L, Clark ME.. 2008. Wolbachia: master manipulators of invertebrate biology. Nat Rev Microbiol. 6(10):741–751. [DOI] [PubMed] [Google Scholar]
- Wu M, et al. 2004. Phylogenomics of the reproductive parasite Wolbachia pipientis wMel: a streamlined genome overrun by mobile genetic elements. PLoS Biol. 2(3):e69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xi Z, Khoo CCH, Dobson SL.. 2005. Wolbachia establishment and invasion in an Aedes aegypti laboratory population. Science 310(5746):326–328. [DOI] [PubMed] [Google Scholar]
- Yamashita Y, Takehara T, Kuramitsu HK.. 1993. Molecular characterization of a Streptococcus mutans mutant altered in environmental stress responses. J Bacteriol. 175(19):6220–6228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zouache K, et al. 2009. Persistent Wolbachia and cultivable bacteria infection in the reproductive and somatic tissues of the mosquito vector Aedes albopictus. PLoS One 4(7):e6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.