ABSTRACT
Macrophages are the primary host target cells of Mycobacterium tuberculosis (M. tb). As a subunit of immunoregulatory cytokines IL-27 and IL-35, Epstein–Barr virus-induced gene 3 (EBI3) has typically been explored as the secreted form and assessed in terms of its effects triggered by extracellular EBI3. However, little is known about intracellular EBI3 function. In the current study, we report that EBI3 production by macrophages is elevated in TB patients. We further demonstrate that increased EBI3 accumulates in virulent M. tb-treated murine macrophages. Eukaryotic translation elongation factor 1-alpha 1 (eEF1A1) binds to intracellular EBI3 to reduce Lys48 (K48)-linked ubiquitination of EBI3, leading to EBI3 accumulation. Moreover, the intracellular EBI3 inhibits caspase-3-mediated apoptosis in M. tb-treated macrophages. Herein, we propose a novel mechanism for accumulating intracellular EBI3 and its regulation of macrophage apoptosis in response to virulent M. tb.
Keywords: M. tb, macrophage, EBI3, eEF1A1, ubiquitination, apoptosis
Accumulation of EBI3 is induced by eEF1A1-medicated reduction of K48-linked ubiquitination and inhibits apoptosis in virulent M. tb-treated murine macrophages.
INTRODUCTION
Mycobacterium tuberculosis (M. tb) remains a global health threat that causes tuberculosis (TB), the leading cause of death by infectious disease worldwide. According to the World Health Organization (WHO), one-third of the world's population is infected with M. tb, and approximately 0.895 million new TB cases occurred in China (World Health Organization 2017). Macrophages are the primary target for M. tb, and M. tb modulates the macrophage-mediated microbicidal and phagocytic activity to facilitate the M. tb survival in cells (Pieters 2008). Several studies have reported that M. tb induces abnormal accumulation of iron, lipids, lipoproteins and inflammatory mediators in macrophages, which are associated with disorders of cell metabolism and maintenance of M. tb (Basaraba et al. 2008; Palanisamy et al. 2012; Singh et al. 2012; Zheng et al. 2015).
Epstein–Barr virus-induced gene 3 (EBI3) was originally characterized as a gene induced in Epstein–Barr virus (EBV)-transformed B cells (Devergne et al. 1998), and EBI3 was later reported to be a member of the interleukin-12 (IL)-12 family, which was associated with IL-27p28 or IL-12p35 to build immunoregulatory cytokines IL-27 and IL-35, respectively (Bohme et al. 2016). EBI3 knockout disturbs the secretion of IL-27 by murine macrophages during parasite infection (Bohme et al. 2016; Quirino et al. 2016) and also inhibits IL-35 production by tumor-associated macrophages (Lee et al. 2018). Both IL-27- and IL-35-producing macrophages play immunomodulatory roles, such like suppressing T cell responses during Trypanosoma cruzi infection (Bohme et al. 2016) or facilitating metastatic colonization (Lee et al. 2018). EBI3 gene polymorphisms are associated with susceptibility to pulmonary TB. Furthermore, EBI3 knockout (EBI3−/−) mice are more resistant to M. tb infection than wild-type (WT) mice (Zheng et al. 2015). However, the mechanism for elevated EBI3 induced by M. tb remains unclear.
Eukaryotic translation elongation factor 1-alpha 1 (eEF1A1) is one isoform of the alpha subunit of the elongation factor-1 complex. The other isoform is eEF1A2. The eEF1A1 isoform is ubiquitously expressed, while eEF1A2 is detected in brain, heart and skeletal muscle (Blanch et al. 2013). eEF1A1 is an important, well-characterized eukaryotic protein, that couples the hydrolysis of GTP to GDP with the delivery of aminoacyl tRNAs to the ribosome during protein translation. Recent studies have also identified a crucial role for eEF1A1 in viral infections (Matsuda, Yoshinari and Dreher 2004; Davis et al. 2007; Chen et al. 2016), indicating that eEF1A1 has multiple functions in the immune response.
During virulent M. tb infection, the bacteria evolve strategies to evade host immune attack. It has been reported that virulent M. tb preferentially induces necrosis rather than apoptosis in macrophages, resulting in cell lysis and infection spread (Behar, Divangahi and Remold 2010). In the present study, we report a novel mechanism of M. tb-induced and eEF1A1-mediated accumulation of intracellular EBI3 in macrophages, which is involved in inhibiting M. tb-induced apoptosis.
MATERIALS AND METHODS
Patient samples
Peripheral blood samples from 14 patients with newly diagnosed active pulmonary TB and 14 healthy donors came from Wuhan Medical Treatment Center (Wuhan, China) and Zhongnan Hospital of Wuhan University (Wuhan, China). Blood samples were collected from active TB (ATB) patients before medical treatment. A diagnosis of active TB was based on positive cultures for M. tb. TB patients co-infected with HIV or other diseases were excluded from the study. This study was approved by the ethics committee of the Wuhan University School of Basic Medical Science and the Wuhan Medical Treatment Center, and all patients and healthy donors in the study provided informed consent before blood donation.
Bacteria, cells and animals
M. tb H37Rv (strain American Type Culture Collection (ATCC) 93 009) and Mycobacterium bovis BCG (ATCC strain 35 734) were purchased from the Beijing Biological Product Institute (Beijing, China) (Sun et al. 2016; Tang et al. 2017). The mycobacterial strain was grown in Middlebrook 7H9 broth (BD Biosciences, NJ, USA) supplemented with 10% oleic acid-albumin-dextrose-catalase (OADC, BD Biosciences, NJ, USA) and 0.05% Tween-80.
Murine macrophage cell line RAW 264.7 (GDC143) was obtained from China Center for Type Culture Collection (CCTCC, Wuhan, China). EBI3−/− mice (B6.129 × 1-Ebi3tm1Rsb/J; 0 08691; Jackson Laboratories) were a gift from Professor Zhinan Yin (Jinan University, Guangzhou, China). C57BL/6 mice were purchased from Wuhan Centers for Disease Prevention and Control. To harvest murine resident peritoneal macrophages, thioglycolate-elicited macrophages were prepared by injecting mice with 3.5 mL of 3% sterile thioglycolate media (BD Biosciences, NJ, USA) (Tang et al. 2017). After 5 days, peritoneal cells were harvested by lavage and cultured overnight. After removing non-adherent cells, adherent macrophages were collected and stained with anti-F4/80 antibody for purity analysis. F4/80+ macrophages were greater than 90% of total cells (data not shown).
Construction of shRNA expression vectors and transfection
The pSilencer 1.0-U6 (Ambion Life Technologies, Carlsbad, CA, USA), which has a U6 promoter, was used as described in our previous work (Li et al. 2008). The 21-mer short hairpin RNAs (shRNAs) against Mus musculus eEF1A1 (GenBank accession no. NM_01 0106) mRNAs were designed. According to the target sequences, two pairs of oligonucleotides coding for each shRNA were designed. eEF1A1-Pair1: 5′-GGAGCTAA TTCTCGGGCTT CTTTCA-3′ (forward), 5′-AGCTTGAAAGAAGCC CGAGAATTAGCTCCGGCC-3′ (reverse); and eEF1A1-Pair2: 5′-AGCTTAGAAGCCCGAGAATTAGCTCCTTTTTT-3′ (forward), 5′-AATTAAA AAAGGAGCTAATT CTCGGGCTT CTA-3′ (reverse). Scramble (Scr)-shRNA-Pair1: 5′-TTCTCCG AACGTGTC ACGTTCA-3′ (forward), 5′-AGCTT GAACGTGA CACGTT CGGAG AAGGCC-3′ (reverse); Scr-shRNA-Pair2: 5′-AGCTTCGTGACA CGTTCGGAGAAT TTTT-3′ (forward), 5′-AATTAAAAATTCTCCGAACGTGTCACGA-3′ (reverse). Pairs of oligonucleotides were synthesized, annealed and inserted into the pSilencer vector. Recombinant vectors were transformed into E. coli DH5α. Each shRNA sequence contains a 9-bp loop sequence that separates the two complementary domains. Sequences for the complete shRNA insert templates are as follows: eEF1A1-shRNA 5′-GGAGCTAAT TCTCGGGCTT CTTTCAAGCTT AGAAGCC CGAGAATT AGCTCC TTTTTT-3′(sense); 5′-AGCTTGA AAGAAGCCC GAGAATTA GCTCCGGC CAATTAAAA AAGGAGC TAATTCT CGGGCT TCTA-3′(antisense). Scr-shRNA 5′-TTCTCCG AACGTGTCAC GTTCAAGCTT CGTGACACG TTCGGAGA ATTTTT-3′ (sense); 5′-CCGG AAGAGGCTT GCACAGT GCAAGTTCG AAGC ACTGTGCAAG CCTCTTAAAAA TTAA-3′ (antisense). RAW 264.7 cells were transfected with shRNA vectors by using jetPRIME reagent (Polypuls transfection, Strasbourg, France) according to the manufacturer's instructions. Briefly, 3 × 106 cells were transfected with 4 μg of plasmid DNA in 200 μL buffer containing 4μL jetPRIME reagent.
Enzyme-linked immunosorbent assay
Murine peritoneal macrophages (1 × 106/mL/well) in 1 mL of culture medium were treated with heat-inactivated M. tb H37Rv/BCG (iH37Rv/iBCG) for 2 h, 6 h, 18 h and 24 h (multiplicity of infection, MOI 1:10). EBI3 levels in culture supernatant were determined by the Mouse EBI3/IL-27B enzyme-linked immunosorbent assay (ELISA) Kit according the manufacturer's instructions (Shanghai Enzyme-linked Biotechnology Co., Ltd., Shanghai, China).
Flow cytometry
To detect EBI3 expression in human macrophages, blood from TB patients was directly treated with RBC lysis buffer (Beyotime, Shanghai, China) and then a single cell suspension was prepared in Cell Staining Buffer (Biolegend, CA, USA). Cells were incubated with human Fc Receptor Blocking Solution (Biolegend, CA, USA) composed of anti-human CD16, CD32 and CD64 antibodies for blocking FcRs. The cells were stained with FITC anti-CD14 antibody and then fixed in Fixation Buffer (Biolegend, CA, USA) in the dark for 20 min. After resuspending the fixed cells in Intracellular Staining Perm Wash Buffer (Biolegend, CA, USA), the cells were stained with PE anti-EBI3 antibody (Biolegend, CA, USA) for flow cytometry (FCM) analysis.
For apoptosis assessment, an Annexin V/Propidium Iodide (PI) assay was used to quantify cell death in vitro as described previously (Jongstra-Bilen et al. 2017; Fan et al. 2018). The murine peritoneal macrophages were stimulated with iH37Rv/iBCG (MOI 1:10) for 6 h, 18 h or 24 h. After washing with ice-cold phosphate-buffered saline (PBS), the cells can be gently scraped to dislodge by a cell scraper and stained for Annexin V and PI (Biolegend, CA, USA) antibodies. Stained cells were immediately analyzed by FCM.
To assess the extracellular EBI3 effects on iH37Rv-induced apoptosis in macrophages, the peritoneal EBI3−/- macrophages were stimulated with iH37Rv (MOI 1:10) in the presence of recombinant mouse EBI3 protein (50 pg/106 cells/ml, Abcam, Cambridge, UK) for 6 h, 18 h or 24 h. The cell apoptosis was determined by Annexin V/PI assay. All antibodies used in the FCM analysis were purchased from Biolegend (CA, USA).
Immunoblot and immunoprecipitation analysis
To detect EBI3 and eEF1A1 expression, cells were stimulated with iH37Rv and iBCG (MOI 1:10). In some experiments, cells were treated with proteasome inhibitor MG 132 (2 nM) for 7 h prior to iH37Rv stimulation. After the stimulation, cells were washed with ice-cold PBS and lysed in RIPA buffer (Beyotime, Shanghai, China). Supernatants were collected and subjected to SDS-PAGE. Proteins were then transferred to PVDF membranes, and the membranes were incubated with 2% BSA in TBST (Tris buffered saline with Tween 20), and then with anti-eEF1A1 (Abcam, Cambridge, UK), anti-EBI3 (Santa Cruz Biotech, CA, USA), anti-caspase-3 (Cell Signaling Technology, MA, USA), anti-caspase-8 (Cell Signaling Technology, MA, USA) and anti-caspase-9 (Cell Signaling Technology, MA, USA) antibodies. After addition of HRP-conjugated secondary antibody, chemiluminescent detection was performed using ECL Plus Western blotting reagents.
To search for potential proteins binding to intracellular EBI3, RAW 264.7 cells were stimulated with iH37Rv (MOI, 1:10) for 6 h. Cells were lysed, and the supernatant was incubated with anti-EBI3 antibody followed by incubation with protein A/G magnetic beads (MedChemExpress, NJ, USA). Precipitated samples were washed with PBS and then analyzed with SDS-PAGE. Protein bands were extracted from SDS gels and prepared for protein identification by liquid chromatography-mass spectrometry (LC-MS)/MS analysis (NanoLC Ultra 1D plus coupled with TripleTOF 5600 plus, AB Sciex, USA). To confirm binding of EBI3 to eEF1A1, precipitated samples were subjected to immunoprecipitation with anti-EBI3 antibody and then immunoblot analysis with anti-EBI3 and anti-eEF1A1 antibodies.
To assess the effects of eEF1A1 on K48-linked ubiquitination of EBI3, cells were transfected with eEF1A1 shRNA. After 24 h, the eEF1A1-silenced cells were stimulated with iH37Rv. Cells were lysed, and supernatants were incubated with anti-EBI3 antibody followed by incubation with protein A/G magnetic beads. Precipitated samples were subjected to immunoblot analysis with anti-EBI3 antibody and rabbit K48-linkage specific polyubiquitin antibody (Cell Signaling Technology, MA, USA).
Immunofluorescence analysis
Cells were stimulated with either iH37Rv or iBCG (MOI 1:10) for 6 h. Cells were fixed with ice-cold methanol for 10 min and then washed with PBS, followed by incubation with anti-eEF1A1, anti-EBI3 and anti-Rab4 (Proteintech Group, IL, USA) antibodies at 4˚C overnight. After washing, cells were incubated with Alexa Fluor 488-conjugated goat anti-rabbit IgG (Proteintech Group, IL, USA) and Alexa Fluor 594-conjugated goat anti-mouse IgG (Proteintech Group, IL, USA) for 1 h at room temperature, followed by staining with 4′,6-diamidino-2-phenylindole (DAPI) for 10 min. After washing, cells were observed by confocal microscopy.
Statistics analysis
Data were analyzed with SPSS software. Differences were considered to be statistically significant for P-values < 0.05. Experimental data were analyzed by ANOVA.
RESULTS
EBI3 production by macrophages is elevated in TB patients
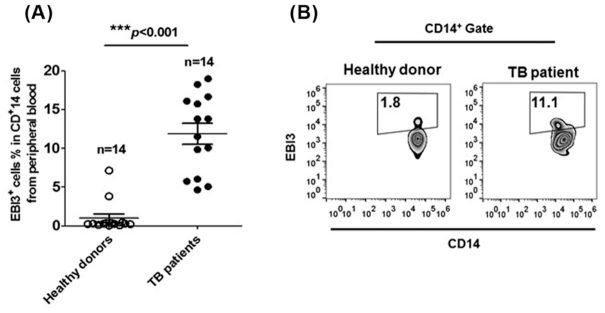
To assess the role of EBI3 during M. tb infection, we measured levels of EBI3 in human CD14+ macrophages from the peripheral blood of pulmonary TB patients. As shown in Fig. 1A and B, EBI3 levels in macrophages were significantly increased in TB patients than in healthy donors. These results suggest that EBI3 production by macrophages is upregulated during M. tb infection.
Figure 1.
EBI3 production by CD14+ macrophages is elevated in TB patients. The percentages of EBI3+ cells in CD14+ human monocytes from peripheral blood were determined by FCM. (A) Pooled data and (B) Representative dot plots. The data in (A) are shown as mean ± SD (n = 14).
EBI3 accumulation is increased in virulent M. tb-treated murine macrophages
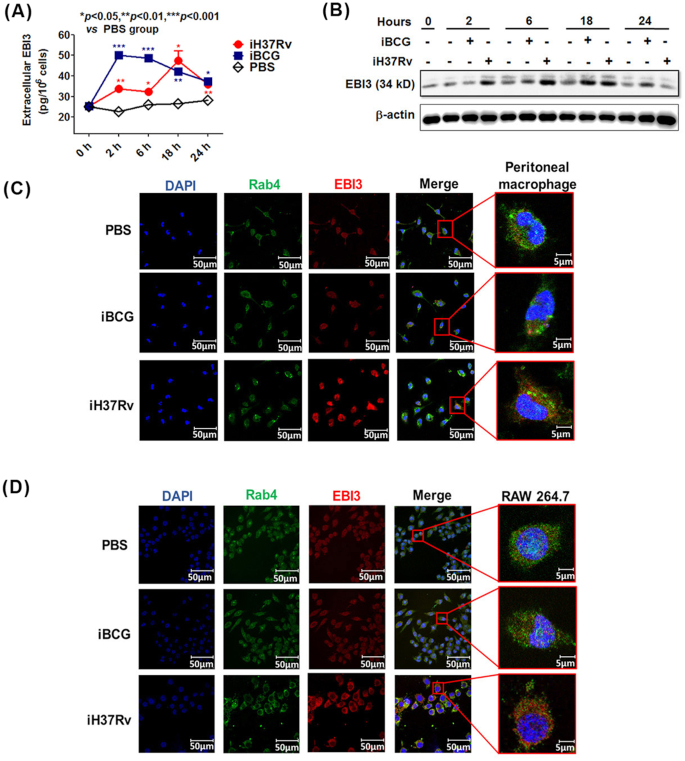
Next, we investigated the EBI3 production in murine peritoneal macrophages treated with M. tb. After stimulation with iH37Rv/iBCG, the cell culture supernatants were collected for measuring extracellular EBI3 levels. The cell pellets then were washed by PBS, and lysed to determine the intracellular EBI3. As shown in Fig. 2A, levels of extracellular EBI3 were increased after iH37Rv/iBCG treatment. The maximal EBI3 secretion (about 50 pg/106 cells/ml) by macrophages was observed after 2 h stimulation with iBCG but 18 h stimulation with iH37Rv, indicating that EBI3 secretion took longer to reach its peak level in the virulent strain iH37Rv group compared with the iBCG group.
Figure 2.
Increased EBI3 accumulates in virulent M. tb-treated murine macrophages. (A) and (B) Murine peritoneal macrophages were stimulated with iH37Rv or iBCG for 2 h, 6 h, 18 h and 24 h. (A) Extracellular EBI3 level in cell culture supernatant was determined by ELISA. The data are shown as mean ± SD (n = 3). (B) Intracellular EBI3 was determined by immunoblot analysis. The iH37Rv/iBCG-treated cell pellets were washed with PBS, and employed to determine the intracellular EBI3. (C) Peritoneal macrophages and (D) RAW 264.7 cells were stimulated with iH37Rv or iBCG for 6 h. Intracellular EBI3 levels were determined by confocal microscopy.
Levels of intracellular EBI3 in macrophages were determined by immunoblot analysis. As shown in Fig. 2B, after iH37Rv treatment, levels of intracellular EBI3 were rapidly elevated after 2 h stimulation and reached their maximum level after 6–18 h stimulation. Compared with the iH37Rv group, an increase in intracellular EBI3 was delayed in iBCG-treated macrophages (Fig. 2B). Levels of intracellular EBI3 were slightly increased after 6 h stimulation and achieved their peak level after 18 h stimulation in the iBCG group (Fig. 2B). After 24 h stimulation, intracellular EBI3 was greatly reduced in both iH37Rv and iBCG groups. These results imply that intracellular EBI3 in macrophages accumulates after iH37Rv stimulation. Our results are consistent with the previous study, in which elevated mRNA and protein levels of EBI3 were observed in macrophages subjected to M. tb H37Rv treatment compared with iBCG treatment (Zheng et al. 2015).
To further investigate whether iH37Rv induces accumulation of EBI3 in macrophages, intracellular EBI3 was visualized by confocal analysis (Fig. 2C and D). We used the endosome marker Rab4 to determine the location of EBI3 in M. tb-treated macrophages (Fig. 2C and D). As expected, markedly increased intracellular EBI3 was found in both peritoneal macrophages and the macrophage cell line RAW 264.7 after iH37Rv treatment. Intracellular EBI3 was located within endosomes as well as outside endosomes in the iH37Rv group (Fig. 2C and D). These results confirm that EBI3 expression is upregulated and accumulates in macrophages after iH37Rv treatment.
Accumulation of intracellular EBI3 inhibits apoptosis in iH37Rv-treated macrophages
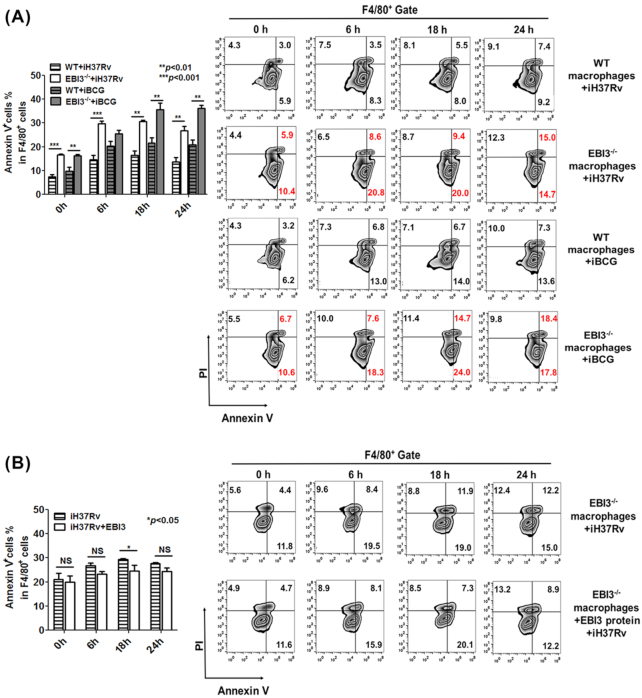
To assess the effects of intracellular EBI3 accumulation on iH37Rv-treated macrophages, we used murine EBI3−/− cells. EBI3−/− peritoneal macrophages were treated with iH37Rv, and apoptosis was assessed. As shown in Fig. 3A, both PI+AnnexinV+ and PI−AnnexinV+ cells were increased in EBI3−/− macrophages upon stimulation with iH37Rv, indicating that EBI3 is involved in hindering apoptosis in iH37Rv-treated macrophages. Consistent with other reports (Riendeau and Kornfeld 2003; Guo et al. 2012), our results showed that iBCG more strongly induced apoptosis in WT macrophages than iH37Rv did (Fig. 3A). However, the higher level of apoptosis in iBCG-treated EBI3−/− macrophages was also observed compared with iBCG-treated WT cells (Fig. 3A). These results suggest that EBI3 inhibits apoptosis induced by both virulent and avirulent mycobacteria in macrophages.
Figure 3.
Accumulated intracellular EBI3 inhibits apoptosis in iH37Rv-treated macrophages.(A) Apoptosis was increased in iH37Rv/iBCG-treated EBI3−/− macrophages. WT and EBI3−/− peritoneal macrophages were treated with iH37Rv/iBCG (MOI 1:10) for 6 h, 18 h and 24 h. Apoptotic cells were analyzed by FCM. Left panel, pool data. The data are shown as mean ± SD (n = 3). Right panel, representative dot plots. (B) Extracellular EBI3 had a limited effect on apoptosis in iH37Rv-treated EBI3−/− macrophages. The recombinant mouse EBI3 protein (50 pg/106 cells/ml) was added to the EBI3−/− macrophages, and the cells were stimulated with iH37Rv (MOI 1:10). Apoptosis in the EBI3−/− macrophages was determined by FCM.
Moreover, as shown in Fig. 3B, apoptosis in the iH37Rv-treated EBI3−/− macrophages was slightly inhibited by extracellular EBI3 at the concentration of 50 pg/106 cells/ml (the maximal EBI3 secretion measured above). These results indicate that extracellular EBI3 has fairly limited effects on apoptosis, and accumulation of intracellular EBI3 inhibits apoptosis in iH37Rv-treated macrophages.
eEF1A1 binds to intracellular EBI3 in iH37Rv-treated macrophages
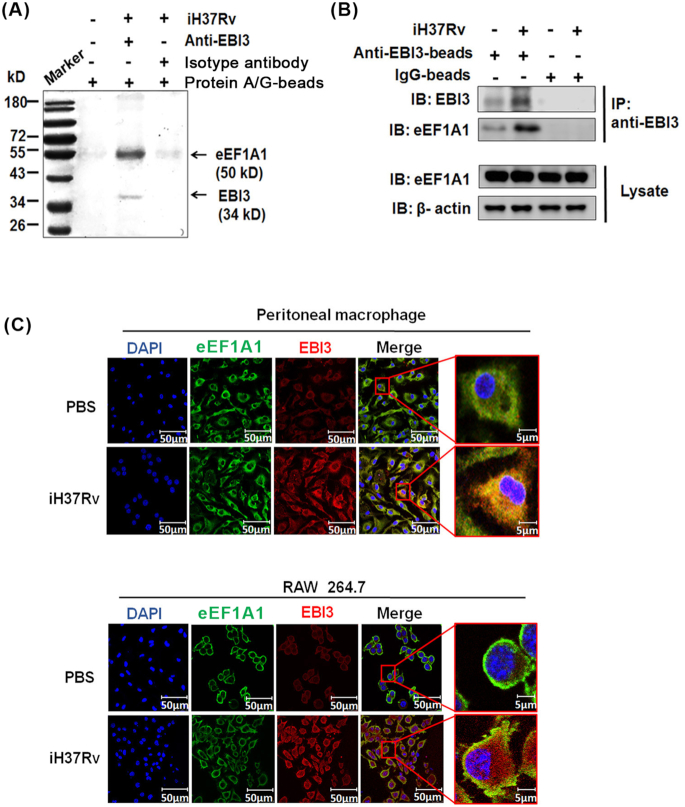
To identify potential proteins binding to intracellular EBI3, we performed immunoprecipitation with an anti-EBI3 antibody, followed by LC-MS/MS analysis. RAW 264.7 cells were stimulated with iH37Rv, and the cell lysate was incubated with anti-EBI3 antibody and protein A/G-beads. Results showed a 40–55 kD protein pulled-down by anti-EBI3 antibody in higher abundance in the iH37Rv group compared with other groups (Fig. 4A and Table 1). The pulled-down protein was then identified as eEF1A1 (Fig. 4A and B; Table 1). As shown in Fig. 4B, the expression level of eEF1A1 did not change after iH37Rv treatment (lower panel), but the binding of eEF1A1 to EBI3 was increased in the iH37Rv-treated cells (upper panel).
Figure 4.
eEF1A1 binds to EBI3 in iH37Rv-treated macrophages. (A) eEF1A1 was identified as EBI3 binding protein. RAW 264.7 cells were stimulated with iH37Rv for 6 h, and the cell lysate was incubated with anti-EBI3 antibody and protein A/G magnetic beads. Precipitated samples were analyzed by SDS-PAGE. The specific band was identified by LC-MS/MS analysis. eEF1A1 peptides were confirmed by peptide mass fingerprinting (PMF). (B) The binding between eEF1A1 and EBI3 was determined by pull-down and immunoblot analysis. Cell lysates were incubated with anti-EBI3 antibody and protein A/G magnetic beads. Precipitated samples were analyzed by anti-EBI3 and anti-eEF1A1 antibodies. (C)Peritoneal macrophages and RAW 264.7 cells were stimulated with iH37Rv for 6 h. Intracellular EBI3 and eEF1A1 were determined by confocal microscopy.
Table 1.
eEF1A1 was confirmed by LC-MS/MS.
| Protein | NCBI accession number | Molecular weight (kD) | Position | Identified peptides | Confidence |
|---|---|---|---|---|---|
| Elongation factor 1-alpha 1 | AAH04005.1 | 50 | 21–30 | STTTGHLIYK | 99.00% |
| 38–44 | TIEKFEK | 99.00% | |||
| 155–165 | MDSTEPPYSQK | 99.00% | |||
| 173–180 | EVSTYIKK | 99.00% | |||
| 256–266 | IGGIGTVPVGR | 99.00% | |||
| 431–439 | QTVAVGVIK | 99.00% |
Binding between EBI3 and eEF1A1 was further confirmed by immunofluorescence analysis. As shown in Fig. 4C, the expression of EBI3 was increased after iH37Rv treatment, while the eEF1A1 expression in iH37Rv-treated macrophages did not change. Importantly, colocalization of EBI3 and eEF1A1 was markedly increased in iH37Rv-treated peritoneal macrophages and RAW 264.7 cells compared with PBS control groups (Fig. 4C). These results suggest that total intracellular eEF1A1 may not change after iH37Rv treatment, but the eEF1A1 binding to EBI3 is increased.
eEF1A1 mediates EBI3 accumulation in iH37Rv-treated macrophages via reduction of Lys48 (K48)-linked ubiquitination of EBI3
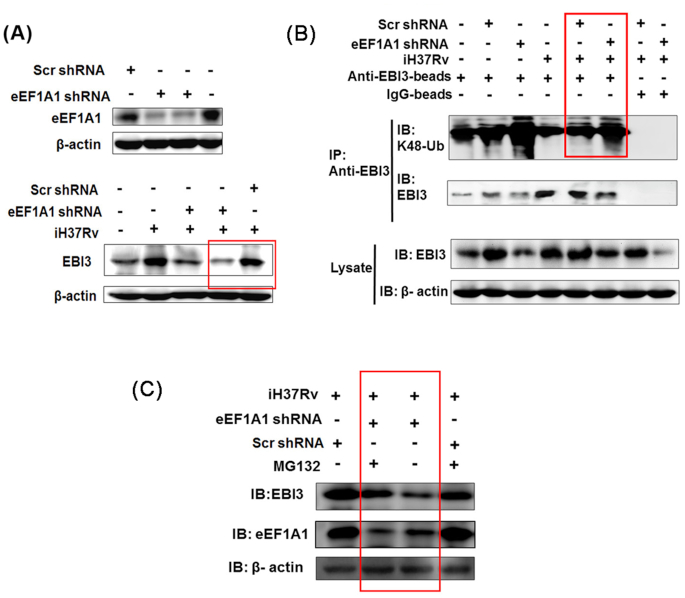
Next, we tested whether eEF1A1 was involved in intracellular accumulation of EBI3. When eEF1A1 expression in RAW 264.7 cells was silenced by shRNA (Fig. 5A, upper panel), intracellular EBI3 level was greatly reduced in the iH37Rv treatment group compared with scr shRNA + iH37Rv group (Fig. 5A, lower panel). These results indicate that eEF1A1 is involved in the intracellular accumulation of EBI3 in iH37Rv-treated macrophages.
Figure 5.
eEF1A1 reduces K48-linked EBI3 ubiquitination in iH37Rv-treated macrophages. (A) EBI3 expression was reduced in eEF1A1-silenced macrophages upon iH37Rv stimulation. RAW 264.7 cells were transfected with eEF1A1 shRNA. After 24 h of transfection, cells were stimulated with iH37Rv for 6 h. Upper panel, after 24 h of transfection, eEF1A1 expression was determined by immunoblot analysis. Lower panel, after 6 h of iH37Rv stimulation, EBI3 expression was determined in eEF1A1-slienced cells. (B) eEF1A1 inhibited K48-linked ubiquitination of EBI3 in iH37Rv-treated cells. RAW 264.7 cells were transfected with eEF1A1 shRNA and stimulated with iH37Rv. Supernatant from the cell lysate was incubated with anti-EBI3 antibody followed by incubation with protein A/G magnetic beads. The precipitated samples were subjected to immunoblot analysis with anti-EBI3 antibody and K48-linkage specific polyubiquitin antibody. (C) eEF1A1-silienced RAW 264.7 cells were treated with proteasome inhibitor MG132 (2 nM) prior to iH37Rv treatment. The EBI3 production were measured by immunoblot analysis with anti-EBI3 antibody.
It has been reported that eEF1A1 participates in stabilization of intracellular viral proteins via reduction of the K48-linked protein ubiquitination during the viral infection (Chen et al. 2016). K48-linked ubiquitination is associated with proteasomal degradation (Mallette and Richard 2012). We then determined the K48-linked ubiquitination of EBI3 in eEF1A1-silenced RAW 264.7 cells upon iH37Rv stimulation. As shown in Fig. 5B, K48-linked ubiquitination of EBI3 was markedly reduced after stimulation with iH37Rv (1st lane vs. 4th lane). After eEF1A1 was silenced, K48 ubiquitination of EBI3 was greatly increased in iH37Rv-treated RAW 264.7 cells (Fig. 5B, 6th lane vs. 5th lane). When the cells were treated with proteasome inhibitor MG132, the EBI3 production were obviously recovered in the eEF1A1-silenced macrophages upon iH37Rv stimulation (Fig. 5C). These results demonstrate that reduction of K48 ubiquitination of EBI3 is mediated by eEF1A1 in iH37Rv-treated cells, which leads to EBI3 accumulation.
Intracellular EBI3 inhibits caspase-3 activation in iH37Rv-treated macrophages
Because elevated EBI3 inhibits apoptosis induced by virulent M. tb, we explored the potential mechanism for regulating apoptosis via intracellular EBI3. As shown in Fig. 6, activation of caspase-8, caspase-9 and caspase-3 was determined in iH37Rv-treated macrophages. When apoptosis is triggered, inactive monomeric procaspases require dimerization, and often cleavage, for activation (McIlwain, Berger and Mak 2013). As shown in Fig. 6, cleaved caspase-8 and caspase-3 increased in EBI3−/− macrophages after 6–24 h of iH37Rv treatment compared with the levels in the WT macrophage group. Cleaved caspase-9 also slightly increased in EBI3−/− macrophages after 6 h and 18 h of iH37Rv treatment. These results suggest that EBI3 is involved in the inhibition of caspase-3 activation in iH37Rv-treated macrophages.
Figure 6.
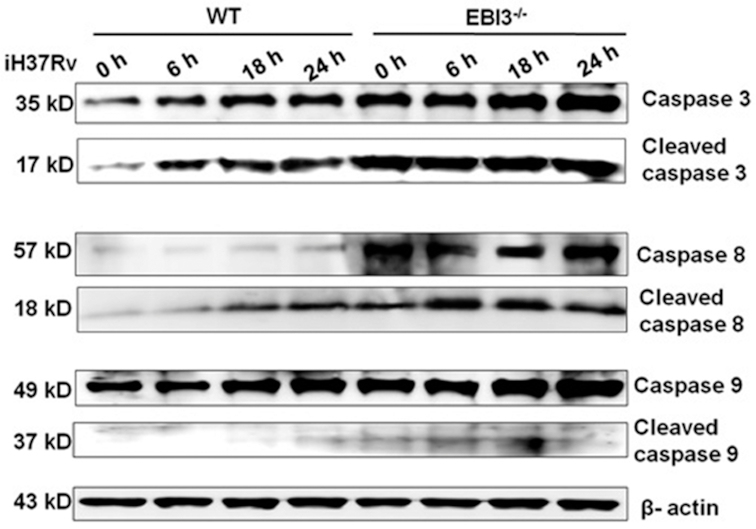
Intracellular EBI3 inhibits caspase-3 activation in iH37Rv-treated macrophages. Activation levels of caspase-3, caspase-8 and caspase-9 were increased in iH37Rv-treated EBI3−/− macrophages. WT and EBI3−/− macrophages were treated with iH37Rv for 6 h, 18 h and 24 h. Activation of caspase-3, caspase-8 and caspase-9 was determined by immunoblot analysis.
DISCUSSION
EBI3 has immunomodulatory effects in models of autoimmune and infectious diseases. Using EBI3−/− mice, several studies have reported that EBI3 regulates NK cell response and is involved in the establishment of mouse cytomegalovirus (MCMV) latency (Jensen et al. 2017), suppresses T helper type 1, type 17 and type 2 immune responses against Trypanosoma cruzi parasitemia (Bohme et al. 2016), and is associated with M. tb loads in mouse lungs (Zheng et al. 2015). As a subunit of IL-27 and IL-35, EBI3 has typically been explored as the secreted form and assessed in terms of its effects triggered by extracellular EBI3 binding to its receptors (IL-12Rβ1, IL-12Rβ2, IL-23R, gp130 and WSX-1) (Vignali and Kuchroo 2012). In the present study, we report accumulation of intracellular EBI3 in M. tb-treated murine macrophages, which contributes to the inhibition of virulent M. tb-induced apoptosis in macrophages and might, thus, facilitate the mycobacterial infection.
In our study, iBCG-induced higher level of extracellular EBI3 than iH37Rv-induced at 2 h and 6 h (Fig. 2A), whereas iBCG-induced lower level of intracellular EBI3 than iH37Rv-induced at 2 h and 6 h (Fig. 2B). The dynamic changes of EBI3 secretion induced by iH37Rv and iBCG were different. Some reports also showed that the cytokine production (like IL-1β and IL-12) by dendritic cells and macrophages after virulent M. tb treatment might be different compared with BCG treatment (Dwivedi et al. 2012; Mendoza-Coronel and Castanon-Arreola 2016). The differences in the cytokine production probably were due to differences in bacterial genes and proteins between the virulent M. tb and BCG (Brosch et al. 2001; Gunawardena et al. 2013).
Our findings are consistent with a previous study by Zheng et al (Zheng et al. 2015). They reported that the human EBI3 gene rs4740 polymorphism was associated with susceptibility to pulmonary TB (Zheng et al. 2015). More importantly, we report for the first time a mechanism for increased EBI3 accumulation within the macrophages upon M. tb stimulation (Fig. 2). We demonstrate that increased EBI3 accumulation in iH37Rv-treated macrophages is due to eEF1A1-mediated reduction of K48-linked EBI3 ubiquitination (Figs. 4 and 5). However, there are no reports about EBI3 deficiency effects on macrophages' function like phagocytosis or clearing M. tb. How M. tb manipulates the macrophage function via EBI3 in host need to be clarified in further study.
eEF1A1, a well-characterized eukaryotic protein, has multiple roles in protein synthesis. In addition to its function of delivering aminoacyl tRNAs to the ribosome, eEF1A1 exhibits chaperone-like activity (Caldas, El Yaagoubi and Richarme 1998; Abbas, Kumar and Herbein 2015). eEF1A1 has also been shown to protect non-structural (NS) proteins NS3 and NS5 of hepatitis C virus from being degraded by the ubiquitin-proteasome system (Lee et al. 2018). Herein, we demonstrate that M. tb H37Rv promotes the eEF1A1-EBI3 interaction resulting in EBI3 accumulation in macrophages (Fig. 5). Moreover, the eEF1A1 binding to EBI3 results in reduction of K48 ubiquitination of EBI3, which leads to less apoptosis in the macrophages upon iH37Rv stimulation (Figs. 3–5).
Virulent M. tb favors necrosis over apoptosis in infected macrophages for evading host immunity. Several reports indicate that both live Mycobacterium and heat killed Mycobacterium induce macrophages apoptosis (Lee et al. 2006; Subramaniam et al. 2016). However, live Mycobacterium residing in an intracellular compartment is also associated with apoptosis (Lee et al. 2006). In our current study, we demonstrate that increased accumulation of intracellular EBI3 hinders iH37Rv-induced apoptosis via inhibiting the caspase-3 activation (Fig. 3 and Fig. 6), partially revealing the intracellular EBI3 effects on apoptosis in macrophages during M. tb infection. The live virulent M. tb need to be used to explore the mechanism of EBI3-mediated inhibition of apoptosis in future. Collectively, in the present study, we show that eEF1A1 participates in the reduction of K48-linked ubiquitination of EBI3 upon stimulation with virulent M. tb. Furthermore, accumulation of intracellular EBI3 hinders apoptosis in macrophages.
ACKNOWLEDGEMENTS
We thank the participants in this study.
AUTHOR CONTRIBUTIONS: XL. Z. and Q. P designed the research and supervised the experiments; JH. D. and HY. C. performed the experiments; C. H. and JM. Y. helped with the experiments; Z.Y. gave suggestions for the research; XL. Z. and Q. P drafted manuscript and wrote the manuscript.
FUNDING
This work was supported by grants from the National Key R&D Program of China (2018YFA0507603 to XL. Z.), National Natural Science Foundation of China (81471910 and 31770145 to Q. P.; 31221061, 31370197 and 21572173 to XL. Z.), National Science and Technology Major Project (2017ZX10201301-006 to XL. Z.), National Outstanding Youth Foundation of China (81025008 to XL. Z.), the Major Projects of Technological Innovation of Hubei Province (2016ACA150 to XL. Z.), Natural Science Foundation Project of Hunan Province (2016CFA062 to XL. Z.), the Outstanding Youth Foundation of Hubei Province (2018CFA037 to Q. P.) and the Wuhan Youth Science and Technology Chenguang Plan (2016070204010127 to Q. P.).
Conflicts of interest. None declared
REFERENCE
- Abbas W, Kumar A, Herbein G. The eEF1A proteins: at the crossroads of oncogenesis, apoptosis, and viral infections. Front Oncol. 2015;5:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basaraba RJ, Bielefeldt-Ohmann H, Eschelbach EK et al.. Increased expression of host iron-binding proteins precedes iron accumulation and calcification of primary lung lesions in experimental tuberculosis in the guinea pig. Tuberculosis (Edinb). 2008;88:69–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behar SM, Divangahi M, Remold HG. Evasion of innate immunity by Mycobacterium tuberculosis: is death an exit strategy? Nat Rev Microbiol. 2010;8:668–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanch A, Robinson F, Watson IR et al.. Eukaryotic translation elongation factor 1-alpha 1 inhibits p53 and p73 dependent apoptosis and chemotherapy sensitivity. PLoS One. 2013;8:e66436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bohme J, Rossnagel C, Jacobs T et al.. Epstein-Barr virus-induced gene 3 suppresses T helper type 1, type 17 and type 2 immune responses after Trypanosoma cruzi infection and inhibits parasite replication by interfering with alternative macrophage activation. Immunology. 2016;147: 338–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brosch R, Pym AS, Gordon SV et al.. The evolution of mycobacterial pathogenicity: clues from comparative genomics. Trends Microbiol. 2001;9:452–8. [DOI] [PubMed] [Google Scholar]
- Caldas TD, El Yaagoubi A, Richarme G. Chaperone properties of bacterial elongation factor EF-Tu. J Biol Chem. 1998;273:11478–82. [DOI] [PubMed] [Google Scholar]
- Chen Z, Ye J, Ashraf U et al.. MicroRNA-33a-5p modulates Japanese encephalitis virus replication by targeting eukaryotic translation elongation factor 1A1. J Virol. 2016;90:3722–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis WG, Blackwell JL, Shi PY et al.. Interaction between the cellular protein eEF1A and the 3'-terminal stem-loop of West Nile virus genomic RNA facilitates viral minus-strand RNA synthesis. J Virol. 2007;81:10172–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devergne O, Cahir McFarland ED, Mosialos G et al.. Role of the TRAF binding site and NF-kappaB activation in Epstein-Barr virus latent membrane protein 1-induced cell gene expression. J Virol. 1998;72:7900–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwivedi VP, Bhattacharya D, Chatterjee S et al.. Mycobacterium tuberculosis directs T helper 2 cell differentiation by inducing interleukin-1beta production in dendritic cells. J Biol Chem. 2012;287:33656–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan L, Wu X, Jin C et al.. MptpB promotes Mycobacteria survival by inhibiting the expression of inflammatory mediators and cell apoptosis in macrophages. Front Cell Infect Microbiol. 2018;8:171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardena HP, Feltcher ME, Wrobel JA et al.. Comparison of the membrane proteome of virulent Mycobacterium tuberculosis and the attenuated Mycobacterium bovis BCG vaccine strain by label-free quantitative proteomics. J Proteome Res. 2013;12:5463–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo S, Xue R, Li Y et al.. The CFP10/ESAT6 complex of Mycobacterium tuberculosis may function as a regulator of macrophage cell death at different stages of tuberculosis infection. Med Hypotheses. 2012;78:389–92. [DOI] [PubMed] [Google Scholar]
- Jensen H, Chen SY, Folkersen L et al.. EBI3 regulates the NK cell response to mouse cytomegalovirus infection. Proc Natl Acad Sci USA. 2017;114:1625–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jongstra-Bilen J, Zhang CX, Wisnicki T et al.. Oxidized low-density lipoprotein loading of macrophages downregulates TLR-induced proinflammatory responses in a gene-specific and temporal manner through transcriptional control. J Immunol. 2017;99:2149–57. [DOI] [PubMed] [Google Scholar]
- Lee CC, Lin JC, Hwang WL et al.. Macrophage-secreted interleukin-35 regulates cancer cell plasticity to facilitate metastatic colonization. Nat Commun. 2018;9:3763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J, Remold HG, Ieong MH et al.. Macrophage apoptosis in response to high intracellular burden of Mycobacterium tuberculosis is mediated by a novel caspase-independent pathway. J Immunol. 2006;176:4267–74. [DOI] [PubMed] [Google Scholar]
- Li D, Li Y, Wu X et al.. Knockdown of Mgat5 inhibits breast cancer cell growth with activation of CD4+ T cells and macrophages. J Immunol. 2008;180:3158–65. [DOI] [PubMed] [Google Scholar]
- Mallette FA, Richard S. K48-linked ubiquitination and protein degradation regulate 53BP1 recruitment at DNA damage sites. Cell Res. 2012;22:1221–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda D, Yoshinari S, Dreher TW. eEF1A binding to aminoacylated viral RNA represses minus strand synthesis by TYMV RNA-dependent RNA polymerase. Virology. 2004;321: 47–56. [DOI] [PubMed] [Google Scholar]
- McIlwain DR, Berger T, Mak TW. Caspase functions in cell death and disease. Cold Spring Harb Perspect Biol. 2013;5:a008656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Coronel E, Castanon-Arreola M. Comparative evaluation of in vitro human macrophage models for mycobacterial infection study. Pathog Dis. 2016;74:ftw052. [DOI] [PubMed] [Google Scholar]
- Palanisamy GS, Kirk NM, Ackart DF et al.. Uptake and accumulation of oxidized low-density lipoprotein during mycobacterium tuberculosis infection in guinea pigs. PLoS One. 2012;7:e34148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pieters J. Mycobacterium tuberculosis and the macrophage: maintaining a balance. Cell Host Microbe. 2008;3:399–7. [DOI] [PubMed] [Google Scholar]
- Quirino GFS, Nascimento MSL, Davoli-Ferreira M et al.. Interleukin-27 (IL-27) mediates susceptibility to visceral leishmaniasis by suppressing the IL-17-neutrophil response. Infect Immun. 2016;84:2289–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riendeau CJ, Kornfeld H. THP-1 cell apoptosis in response to mycobacterial infection. Infect Immun. 2003;71:254–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Jamwal S, Jain R et al.. Mycobacterium tuberculosis-driven targeted recalibration of macrophage lipid homeostasis promotes the foamy phenotype. Cell Host Microbe. 2012;12: 669–81. [DOI] [PubMed] [Google Scholar]
- Subramaniam M, In LL, Kumar A et al.. Cytotoxic and apoptotic effects of heat killed Mycobacterium indicus pranii(MIP) on various human cancer cell lines. Sci Rep. 2016;6:19833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X, Pan Q, Yuan C et al.. A single ssDNA aptamer binding to mannose-capped lipoarabinomannan of Bacillus Calmette-Guerin enhances immunoprotective effect against tuberculosis. J Am Chem Soc. 2016;138:11680–9. [DOI] [PubMed] [Google Scholar]
- Tang XL, Yuan CH, Ding Q et al.. Selection and identification of specific glycoproteins and glycan biomarkers of macrophages involved in Mycobacterium tuberculosis infection. Tuberculosis (Edinb). 2017;104:95–106. [DOI] [PubMed] [Google Scholar]
- Vignali DA, Kuchroo VK. IL-12 family cytokines: immunological playmakers. Nat Immunol. 2012;13:722–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization. Global Tuberculosis Report 2017. http://www.who.int/tb/publications/global_report/en/(2017, date last accessed). [Google Scholar]
- Zheng R, Liu H, Song P et al.. Epstein-Barr virus-induced gene 3 (EBI3) polymorphisms and expression are associated with susceptibility to pulmonary tuberculosis. Tuberculosis (Edinb). 2015;95:497–504. [DOI] [PubMed] [Google Scholar]