This study highlights the abundance, distribution, and diversity of a poorly known microbial compartment in natural aquatic ecosystems, the Bdellovibrio and like organisms (BALOs). These obligate bacterial predators of other bacteria may have an important functional role. This study shows the relative quantitative importance of the three main families of this group, with the design of a new primer pair, and their diversity. While both the diversity and the abundances of these BALOs were globally low, it is noteworthy that the abundance of the Peredibacteraceae could reach important values.
KEYWORDS: Bdellovibrio and like organisms, abundance, diversity, lake, obligate predator
ABSTRACT
Microbes drive a variety of ecosystem processes and services, but many of them remain largely unexplored because of a lack of knowledge on both the diversity and functionality of some potentially crucial microbiological compartments. This is the case with and within the group of bacterial predators collectively known as Bdellovibrio and like organisms (BALOs). Here, we report the abundance, distribution, and diversity of three families of these obligate predatory Gram-negative bacteria in three perialpine lakes (Lakes Annecy, Bourget, and Geneva). The study was conducted at different depths (near-surface versus 45 or 50 m) from August 2015 to January 2016. Using PCR-denaturing gradient gel electrophoresis (PCR-DGGE) and cloning-sequencing approaches, we show that the diversity of BALOs is relatively low and very specific to freshwaters or even the lakes themselves. While the Peredibacteraceae family was represented mainly by a single species (Peredibacter starrii), it could represent up to 7% of the total bacterial cell abundances. Comparatively, the abundances of the two other families (Bdellovibrionaceae and Bacteriovoracaceae) were significantly lower. In addition, the distributions in the water column were very different between the three groups, suggesting various life strategies/niches, as follows: Peredibacteraceae dominated near the surface, while Bdellovibrionaceae and Bacteriovoracaceae were more abundant at greater depths. Statistical analyses revealed that BALOs seem mainly to be driven by depth and temperature. Finally, this original study was also the opportunity to design new quantitative PCR (qPCR) primers for Peredibacteraceae quantification.
IMPORTANCE This study highlights the abundance, distribution, and diversity of a poorly known microbial compartment in natural aquatic ecosystems, the Bdellovibrio and like organisms (BALOs). These obligate bacterial predators of other bacteria may have an important functional role. This study shows the relative quantitative importance of the three main families of this group, with the design of a new primer pair, and their diversity. While both the diversity and the abundances of these BALOs were globally low, it is noteworthy that the abundance of the Peredibacteraceae could reach important values.
INTRODUCTION
Over the last few years, studies on western European large and deep perialpine lakes have revealed that these ecosystems harbor a very diverse and dynamic auto- and heterotrophic prokaryotic community (1–7). These studies and others have also highlighted that both biotic and abiotic factors are likely to regulate these communities. Among these factors, inorganic nutrients, viruses, nanoflagellates, and other heterotrophic grazers (including ciliates and/or metazooplankton) have been identified as critical players in the dynamics of the abundance, community composition, or structure patterns (1, 5, 8–12). Clearly, viral lysis and nanoflagellate or ciliate grazing have been observed to be important biotic factors involved in bacterial mortality, affecting their abundance with a rate ranging from 10% to 60% of bacterial loss per day, but also in regulating their (community) structure and/or diversity (10, 13–15).
Other types of biotic interactions are known to exist but have been poorly investigated. They include the interactions between micro- and macroorganisms, the interactions between bacteria and other organisms, or the role of eukaryotic pathogens (e.g., fungi) which still remain scarce (12, 16, 17). Another type of biotic interaction that has been largely neglected in aquatic ecosystems is the bacterial predation by other bacteria. To the best of our knowledge, the diversity, abundance, dynamics, and functional role of these groups of predators (sometimes also referred to as parasitoids) have never been investigated in alpine lakes so far. Among these predatory bacteria that can belong to several phyla, a “group” is of particular interest toward which this study was directed, i.e., the Bdellovibrio and like organisms (BALOs).
BALOs are small bacteria (ranging in size from 0.2 to 0.5 µm to 0.5 to 2.5 µm [18]), very motile (moving at up to 160 µm/s [19]), and Gram negative. To ensure their survival, they hunt for other bacteria, typically Gram-negative cells, making them specific obligate predators. It is noteworthy, however, that recent studies revealed that BALOs can also prey on Gram-positive bacteria (20, 21) when they have enough time to adapt to such new types of prey. It seems that the adaptation time is related to the synthesis of necessary enzymes, which grant the predator the capacity to degrade the Gram-positive cell wall. Furthermore, BALOs are ubiquitous and widely distributed in different ecosystems, like salt waters, freshwater, sewage, soil, and sediments, and they have also been isolated from different animals, such as mammals, including in human guts and feces (22–25). So far, their abundance and taxonomic diversity across these various habitats have been unexplored or at least underestimated, largely because of the use of culturing approaches. The use of culture-independent methods, for instance, metagenomics, has indeed confirmed that the diversity of cultivated BALOs represents only a small fraction of their diversity (26).
Bdellovibrio and like organisms are a polyphyletic group and can be found within two different classes, the Alphaproteobacteria within the genus Micavibrio and the Oligoflexia (formerly classified in the Deltaproteobacteria) that includes five families, Bdellovibrionaceae, Peredibacteraceae, Bacteriovoracaceae, Pseudobacteriovoracaceae, and Halobacteriovoraceae (23, 27–29). The actual BALO classification is primarily based on the following four criteria: (i) the 16S rRNA gene sequence (30), (ii) the sequence of the gene encoding the β-subunit of bacterial RNA polymerase (rpoB) (31), (iii) the GC content (%), and (iv) the sodium chloride requirement for growth. Following this, species and/or strain types have been proposed to represent each family. For instance, Bdellovibrio bacteriovorus HD100 and Bdellovibrio exovorus JSS depict the Bdellovibrionaceae family. For the Peredibacteraceae, it is Peredibacter starrii A3.12. For the Bacteriovoracaceae, Bacteriovorax stolpii UKi2 is the type strain. Finally, Halobacteriovoraceae is represented by two type species exclusively found in salty ecosystems, Halobacteriovorax marinus SJ and Halobacteriovorax litoralis JS5 (27). Comparatively, the genus Micavibrio may only represent a minor group within BALOs and is most often represented by M. admirandus or M. aeruginosavorus, which are both epibiotic predators (23).
BALOs have been reported to play an essential role in bacterial ecology by shaping the bacterial community (32). The general assumption states that BALOs act most likely as an ecological balancer in their environment (20, 22). BALOs are organized in distinct populations under seasonal and spatial segregation; therefore, their actions may be continuously modified (33). The understanding of the ecology of this bacterial community remains largely unknown in many aquatic environments, especially in natural systems such as large and deep lakes, for which there are almost no data available. Interestingly, a few years ago, the work of Roux et al. (34) in Lake Bourget, followed by the study by Zhong et al. (35) for Lakes Annecy and Bourget (France), showed that there is a significant single-stranded DNA virus community in these lakes, the Microviridae, which are abundant and diverse. This community displays boom-bust dynamics, but the correlation between the abundance of these viruses and the abundance of total heterotrophic bacteria remained challenging to establish (35). However, some viruses within the Microviridae are known to infect BALOs, such as B. bacteriovorus (36). Thus, the presence of a relatively abundant and diverse community of single-stranded DNA (ssDNA) viruses in perialpine lakes could suggest that there is an abundant and diverse community of cellular hosts, including the BALOs. If so, these bacteria, by their potential trophic interactions with other populations of bacteria, could play a significant role in the functioning of the microbial compartment (37, 38). This is the reason why we decided to examine the existence, as determined by abundance, distribution, and diversity, of these bacterial predators in perialpine lakes.
Thus, the objective of this pioneering work dealing with freshwater BALOs was to reveal the existence of these bacteria in typical and representative perialpine lakes (Lakes Annecy, Bourget, and Geneva) and to address the following questions: (i) can BALOs be readily detected in these ecosystems? (ii) What are the structure and diversity of the BALOs? (iii) What is the quantitative importance of the leading groups among this community of predatory bacteria? (iv) What are the relationships between the population of the BALOs and heterotrophic bacteria? (v) What environmental factors appear to be important in the regulation of these interactions?
RESULTS
Primer selection.
Among the 12 primer sets tested by PCR or quantitative PCR (qPCR) and checked using cloning-sequencing, we chose one primer set for the phylogenetic analysis and another one for qPCR analysis for each BALO family (Table 1 and Table S1 in the supplemental material). All selected primers were highly specific since all sequences obtained were characterized by more than 96% identity with different cultured or uncultured bacteria of BALO families found in databases (not shown). Between the two primers that we designed to quantify the 16S rRNA gene sequence of the Peredibacteraceae family by qPCR, the Per699F (CTGCCTGGACGACTATTGAC) and Per974R (CGGGTTCGTAGGAGTTCAAG) primer pair was the best.
TABLE 1.
Existing and selected primers used in this studya
| Primer set name | Primer name | Direction | Sequence (5′–3′) | Amplicon size (bp) | Type of analysis | Targeted group | Reference or source |
|---|---|---|---|---|---|---|---|
| Couple Bde1 | BbeF216 | Forward | TTTCGCTCTAAGATGAGTCCGCGT | 492 | Bdellovibrionaceae | Van Essche (73) | |
| BbeR707 | Reverse | TTCGCCTCCGGTATTCCTGTTGAT | |||||
| Couple Bde2 | Bde347F | Forward | GGAGGCAGCAGTAGGGAATA | 203 | qPCR | Bdellovibrionaceae | |
| Bde549R | Reverse | GCTAGGATCCCTCGTCTTACC | |||||
| Couple Bde3 | Bde529F | Forward | GGTAAGACGAGGGATCCT | 479 | PCR-DGGE & cloning-sequencing | Bdellovibrionaceae | Davidov et al. (26) |
| Bde1007R | Reverse | TCTTCCAGTACATGTCAAG | |||||
| Couple Bx4 | BxF519 | Forward | CAGCAGCCGCGGTAATAC | 159 | qPCR | Bacteriovoracaceae | Zheng et al. (74) |
| BxR677 | Reverse | CGGATTTTACCCCTACATGC | |||||
| Couple Bx5 | Bx1442R | Reverse | GCCACGGCTTCAGGTAAG | 767 | Bacteriovoracaceae | Davidov et al. (26) | |
| Bx676F | Forward | ATTTCGCATGTAGGGGTA | |||||
| Couple Per6 | Per676F | Forward | ATTTCACGTGTAGGGGTA | 768 | PCR-DGGE & cloning-sequencing | Peredibacteraceae | |
| Per1443R | Reverse | AGTCACGTCTTAAAATGAAA | |||||
| Couple Bct8 | 63F | Forward | CAGGCCTAACACATGCAAGTC | 1,316 | Nested PCR | Bacteria | Marchesi et al. (75) |
| 1378R | Reverse | CGGTGTGTACAAGGCCCGGGAACG | Bacteria | Heuer et al. (76) | |||
| Couple Bct7 | 519F | Forward | CAGCMGCCGCGGTAATWC | 389 | qPCR | Bacteria | Kandel et al. (33) |
| 907R | Reverse | CCGTCAATTCMTTTRAGTTT | Bacteria | ||||
| Couple Bde9 | Bd824F | Forward | ACTTGTTGTTGGAGGTAT | 399 | Bdellovibrionaceae | Pasternak et al. (77) | |
| Bd1222R | Reverse | TTGTAGCACGTGTGTAG | |||||
| Couple Bx10 | Bx341F | Forward | CTACGGGAGGCAGCAG | 332 | PCR-DGGE & cloning-sequencing | Bacteriovoracaceae | |
| Bx672R | Reverse | TACCCCTACATGCGAAATTCC | |||||
| Couple Per11 | Per699F | Forward | CTGCCTGGACGACTATTGAC | 276 | qPCR | Peredibacteraceae | This study |
| Per974R | Reverse | CGGGTTCGTAGGAGTTCAAG | |||||
| Couple Per12 | Per521F | Forward | GAAACTGCGTCTGAAACTGC | 234 | Peredibacteraceae | ||
| Per754R | Reverse | GCGTCACTGAAGGAGTCAAT |
Bold type corresponds to the primers used in this study.
Abundances and distribution of the BALOs.
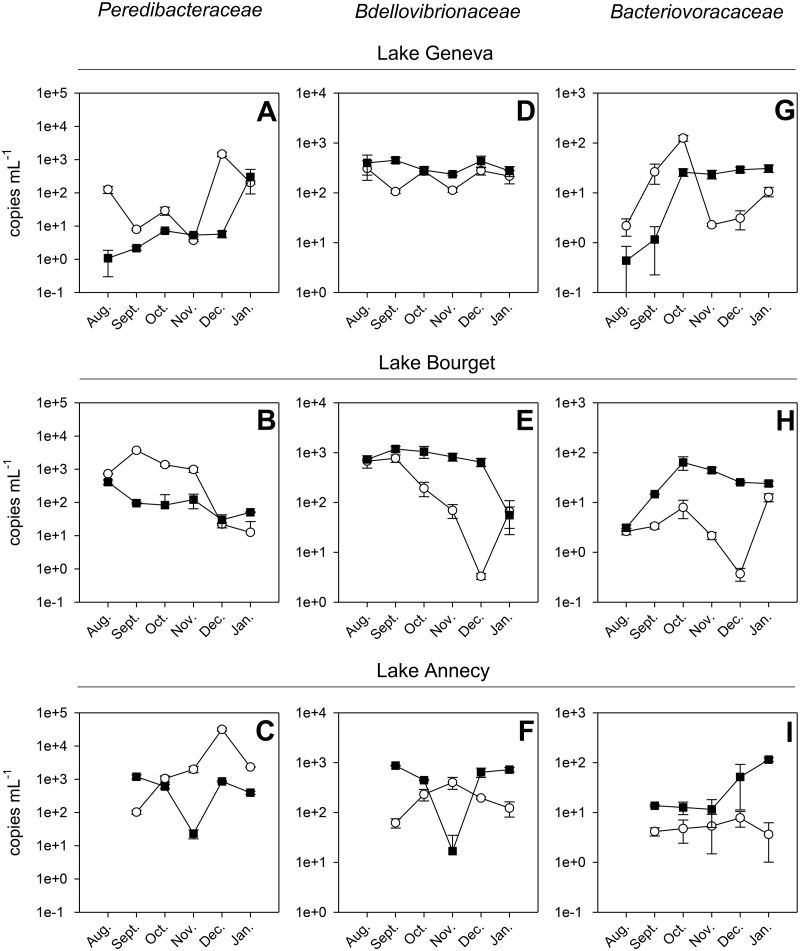
In the analysis of the absolute abundances of the different BALO families (in copies per milliliter, measured by qPCR) disregarding the month and the depth, the most represented family of BALOs in the three lakes was the Peredibacteraceae, with an abundance reaching up to 1.62 × 105 gene copies per ml. In contrast, Bdellovibrionaceae and Bacteriovoracaceae were on average 10,000 times lower in abundance than was the Peredibacteraceae, with maximum concentrations reaching 4 and 1.25 × 101 copies per ml, respectively. Compared to total bacteria also quantified using qPCR or flow cytometry (FCM), the Peredibacteraceae represented up to 7.12% of the total bacteria, while Bacteriovoracaceae and Bdellovibrionaceae accounted for less than 0.05% of the bacterial community. The highest concentrations were always recorded in the free-living bacterial fraction. No evident seasonal variations were recorded here. When discriminating the three families at the two distinct depths, i.e., the surface (2 m, 2.5 m, or 3 m, depending on the lake) versus deeper waters (45 m or 50 m, depending on the lake), (i) Peredibacteraceae were the most abundant (with 100 to 10,000 times more copies per ml than Bdellovibrionaceae and Bacteriovoracaceae), (ii) Peredibacteraceae were generally more abundant at the surface compared than in deeper waters. On the other hand, we observed an opposite trend for Bdellovibrionaceae and Bacteriovoracaceae, in particular for Lake Bourget (Fig. 1).
FIG 1.
Dynamics of abundances for the different BALOs, Peredibacteraceae (A to C), Bdellovibrionaceae (D to F), and Bacteriovoracaceae (G to I), obtained at two contrasting depths in the three lakes. Each sample was analyzed in duplicate. Open circles correspond to surface water (2 m for Lake Bourget, 2.5 m for Lake Geneva, and 3 m for Lake Annecy), whereas filled squares correspond to deep water (45 m for Lake Annecy and 50 m for Lakes Bourget and Geneva).
Relationships between BALOs, total bacteria, and other environmental data.
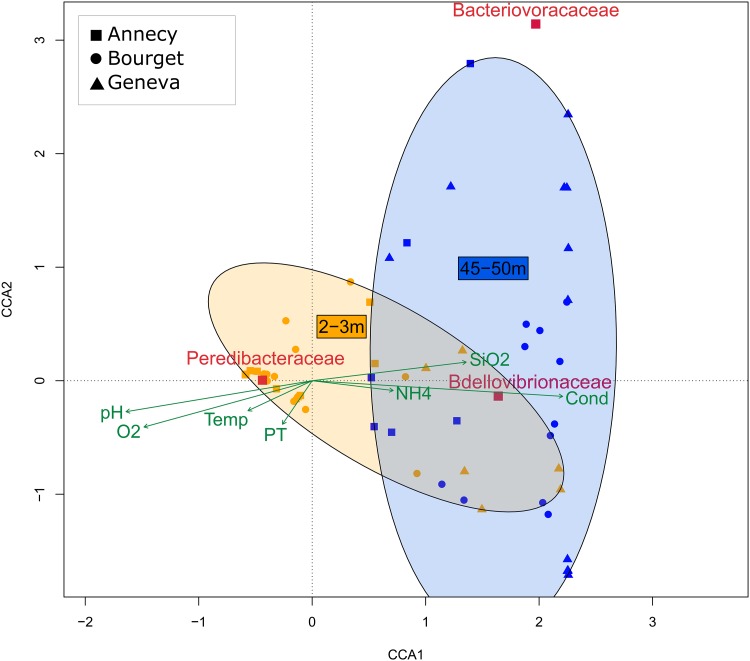
Using the relative abundances of BALO families and environmental data obtained from the in situ surveys of perialpine lakes (e.g., see http://www6.inra.fr/soere-ola/), a canonical correspondence analysis (CCA) was conducted to assess the relationships between BALOs and their biotic and abiotic environments (Fig. 2). The first two axes of the CCA (CCA1 and CCA2) explained 53.1% of the total variance. Bdellovibrionaceae displayed clear links with conductivity (P < 0.05) and ammonium concentration (P < 0.05), whereas Peredibacteraceae displayed clear links with pH (P < 0.05), dissolved oxygen (P < 0.05), and temperature (P < 0.05). Compared to the two other families, no significant relationships were found for the Bacteriovoracaceae with any of the environmental factors tested here. Moreover, the two distinct water layers, i.e., the surface (<3 m, depending on the lake) versus deeper waters (>45 m, depending on the lake), could be separated (Fig. 2). The analysis suggested that Bacteriovoracaceae are more abundant in deep waters (P < 0.05) and driven by ecological factors specific to this part of the water column. In contrast, Peredibacteraceae were more abundant in near-surface waters (P < 0.05) and driven by environmental factors more specific to this layer (such as dissolved O2, chlorophyll a, and higher temperatures). As for Bdellovibrionaceae, the repartition seemed to be less specific to the surface than the deeper waters (P > 0.05).
FIG 2.
Canonical correspondence analysis (CCA) showing the distribution of relative abundances of each group of BALOs (Bacteriovoracaceae, Bdellovibrionaceae, and Peredibacteraceae) quantified by qPCR, according to ecological variables measured during samplings. Temp, temperature; PT, total phosphorus; NH4, ammonium; SiO2, silicate; Cond, conductivity; O2, dissolved oxygen.
Genetic structure.
The DGGE analysis revealed only a limited number of bands no matter the BALO family considered, and no seasonal patterns were recorded. Only 1 to 5 major bands could be detected, suggesting a low genotypic diversity. A maximum of three bands was detected for Bacteriovoracaceae. One major band with three minor (regarding intensity) bands were observed for Bdellovibrionaceae. Two bands were generally observed for Peredibacteraceae (see Fig. S1 in the supplemental material).
Diversity.
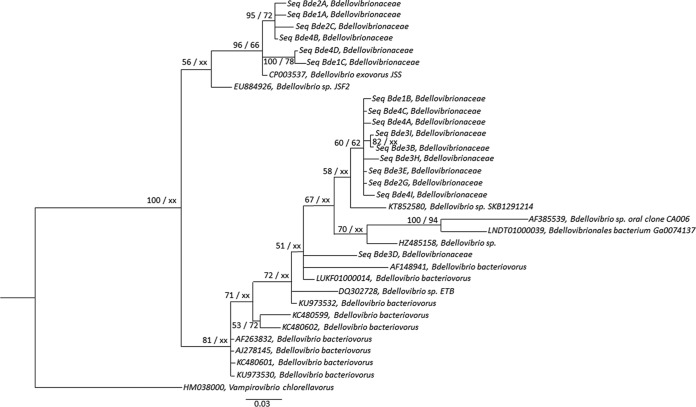
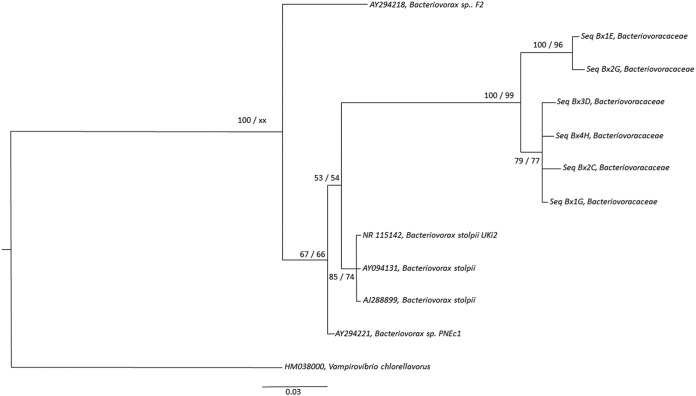
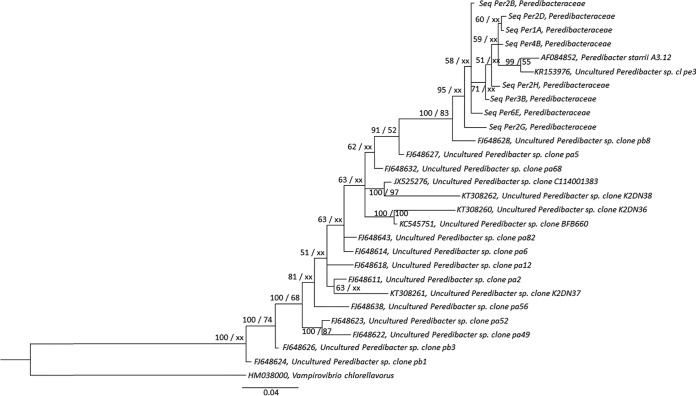
A cloning-sequencing approach was chosen as a first attempt to assess the genetic diversity of the BALOs. Phylogenetic trees were constructed from 30 sequences based on the 16S rRNA gene of each BALO family arising from all the studied lakes. After cleaning, conducting a BLAST search, chimera checking, clustering, and aligning, we obtained 16 centroid sequences for the Bdellovibrionaceae family, 6 centroid sequences for the Bacteriovoracaceae, and 8 centroid sequences for the Peredibacteraceae. Our results clearly show (in agreement with the DGGE results) that the diversity of each BALO family was relatively low. The phylogenetic tree of the Bdellovibrionaceae (Fig. 3) reveals the presence of two distinct clusters. One is related to Bdellovibrio exovorus JSS (6 centroid sequences), and the other corresponds to Bdellovibrio bacteriovorus and its substrains (10 centroid sequences). For the Bacteriovoracaceae also (Fig. 4), two clusters emerged. Our sequences are related to the species Bacteriovorax stolpii, but they may constitute two other species. The tree suggests that our sequences fall into two distinct strains, with two centroid sequences forming one species and four centroid sequences forming the other. For the Peredibacteraceae sequences (Fig. 5), a single cluster emerged. All our sequences looked to be closely related to Peredibacter starrii. These trees suggest that perialpine lakes hold the usual BALO members found in other ecosystems with maybe some new species when considering the genus Bacteriovorax.
FIG 3.
Phylogenetic analysis of 16 centroid sequences of Bdellovibrionaceae from Lakes Annecy, Bourget, and Geneva based on 16S rRNA gene Sanger sequencing obtained after curation and clustering, along with 16 other sequences retrieved from Arb-SILVA (42), including two type species, Bdellovibrio bacteriovorus and Bdellovibrio exovorus. All sequences were aligned using MUSCLE (66) via MEGA6 (60). The alignment was trimmed at both ends to eliminate gaps and then curated with Gblocks (68), resulting in 241 positions from 245 positions. The best-fit model of nucleotide substitution was selected using jModelTest-2.1.1 (69) through an Akaike model selection strategy, resulting in a TIM1+I+G model. Phylogenetic tree was constructed by the maximum likelihood method using PhyML-3.1 (71), and Bayesian inference (GTR+I+G) was conducted using MrBayes 3.2.6 (72) with 5 million generations and a burn-in value of 25%. Posterior probability (PP) values followed by bootstrap values are added to the left of a node when possible (PP/BS). Bootstraps below 50 were deleted. Accession numbers are listed to the left of some organism names. Vampirovibrio chlorellavorus was used as an outgroup to root the Bdellovibrionaceae tree.
FIG 4.
Phylogenetic tree of 16S rRNA Bacteriovoracaceae lineage. The tree is based on maximum likelihood analysis using PhyML-3.1 (71) and Bayesian inference (GTR+G) analysis using MrBayes 3.2.6 (72) with 2 million generations and a burn-in value of 25%. The tree encompasses 6 centroid sequences of Bacteriovoracaceae from Lakes Annecy, Bourget, and Geneva based on Sanger sequencing obtained after curation and clustering, along with 5 other sequences retrieved from Arb-SILVA (42), including one type species, Bacteriovorax stolpii. All sequences were aligned using MUSCLE (66) via MEGA6 (60). The alignment was trimmed at both ends to eliminate gaps and then curated with Gblocks (68), resulting in 262 positions from 292 positions. The best-fit model of nucleotide substitution was selected using jModelTest-2.1.1 (69) through an Akaike model selection strategy, resulting in a GTR+G model. Posterior probability (PP) values followed by bootstrap values are added to the left of a node when possible (PP/BS). Bootstraps below 50 were deleted. Accession numbers are listed to the left of some organism names. Vampirovibrio chlorellavorus was used as an outgroup.
FIG 5.
Bayesian tree generated from 16S rRNA gene data set of Lakes Annecy, Bourget, and Geneva with 8 curated and clustered Peredibacteraceae centroid sequences originating from Sanger sequencing, along with 19 other sequences retrieved from the NCBI (GenBank) (67), including one type species, Peredibacter starrii. These sequences were aligned using MUSCLE (66) via the MEGA6 software (60), and the alignment was curated using Gblocks (68), resulting in 544 positions from 570 positions. A Bayesian tree was built using MrBayes 3.2.6 (72) with 5 million generations and a burn in value of 25% using a GTR+I+G model. As for the ML tree, PhyML-3.1 (71) was used based on a jModelTest-2.1.1 (69) best substitution model, i.e., TrN+I+G. Posterior probability (PP) values followed by bootstrap values are added to the left of a node when possible (PP/BS). Bootstraps below 50 were deleted. Accession numbers are listed to the left of some organism names. Vampirovibrio chlorellavorus was used as an outgroup to root the Peredibacteraceae tree.
DISCUSSION
The present study aimed at investigating the existence of some predatory bacteria referred to as BALOs in three large and deep French and western European alpine lakes. To our knowledge, the existence and quantitative importance of BALOs in such lake ecosystems have not been studied previously, and so different methods were tested and optimized to assess their diversity and abundance. First of all, we designed a new specific qPCR primer set to target the Peredibacteraceae, since no primer existed yet for this recently discovered freshwater BALO family. Then, we quantified and assessed the diversity of three representative groups of BALOs, the Peredibacteraceae, the Bdellovibrionaceae, and the Bacteriovoracaceae, and compared these results to others obtained from a variety of environments. Even if our study revealed unambiguously the presence of BALOs in large perialpine lakes, we are well aware of the limits associated with some of the methodologies chosen and used here (i.e., the DGGE and the cloning-sequencing approaches), as well as the number of analyzed samples (i.e., only a few depths and a few months). In addition, the primers we used were not degenerate. Therefore, our approach may be too stringent to recover a higher BALOs diversity. In fine, this study serves as a pioneering analysis revealing a part of the diversity, distribution, and dynamics of the BALO bacterial community in some freshwater ecosystems.
Probable role and diversity of BALOs in perialpine lakes.
BALOs were found in each lake and at each depth investigated, whatever the period of the year sampled. While this study did not assess the role of BALOs in the microbial loop and lake functioning, it is already known from other studies that these bacteria are likely to be important bioagents of mortality (38). It is noteworthy, however, that very few studies have focused on the role and effect of such predatory bacteria on the bacterial community of natural or man-made environments, and the understanding of bacterial mortality has been mainly and mostly based on the study of viruses and protists so far (33). Unlike viruses and protists, BALO predation is not dependent on the physiology or size of the prey (19). Additionally, BALOs are ubiquitous in nature (38). Thus, predation by BALOs adds a new dimension to the recycling of organic matter through the microbial loop. Both viruses and BALOs recycle nutrients via the microbial loop; however, the recycling mechanisms are different. Viral lysis results in the release of the entire intracellular contents of the prey into the environment, while BALOs consume most of the prey content, hence releasing few nutrients in the environment. Having said that, BALOs yield a higher energetic value since they are filled with nutrients; therefore, when other organisms graze on them, the nutrient uptake efficiency is higher (38). Regarding their diversity, BALOs form highly heterogeneous groups with a large phylogenetic diversity (26). We managed to detect in perialpine lakes the usual BALOs already found in the current bibliography, although we used a fingerprinting approach for which many biases are associated. We are aware indeed that DGGE bands only reflect the microorganism populations found at relatively high concentrations. Additionally, bands can comigrate in the DGGE gel; thereby, the numbers of bands can be over- or underestimated (11). Definitely, a high-throughput sequencing approach will reveal better the hidden diversity of these BALOs. Therefore, highly specific primers for each BALO family should be designed with a fair amount of degeneracy in order to limit nontarget region binding but at the same time maximize taxon detection (39). In our study, we used nondegenerate primers for the PCR-DGGE and a cloning-sequencing approach; therefore, we might have missed some BALOs. Another very important point that we should emphasize about is the taxonomy assignment of BALOs present in 16S rRNA gene databases. Since the early 2000, BALOs taxonomy has changed. Baer et al. (40) reclassified Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax. Then, Davidov and Jurkevitch (30) reclassified Bacteriovorax starrii as Peredibacter starrii, hence creating a new family, the Peredibacteraceae. At the same time, Baer et al. (41) proposed to reclassify saltwater Bdellovibrio spp. as Bacteriovorax marinus and Bacteriovorax litoralis. At last, Koval et al. (27) redirected saltwater BALOs into a new genus, Halobacteriovax, creating a new family, the Halobacteriovoraceae. As a result, these adjustments have caused a few confusions in 16S rRNA gene databases. Typically, when working with Arb-SILVA SSUParc release number 132 (42), we encountered Peredibacter and Halobacteriovax spp. grouped in the Bacteriovoracaceae. Furthermore, some sequences were assigned to Bdellovibrio. At the beginning of the discovery of BALOs, any found sequence was cataloged under the Bdellovibrionaceae family. Lately, some efforts were made to assign correctly these sequences, but there is much work to be done. Today again, one cannot determine whether some sequences belong to Bdellovibrio, Bacteriovorax, Peredibacter, or Halobacteriovax.
Peredibacteraceae are the most abundant BALO family in perialpine lakes.
The Bdellovibrionaceae displayed little diversity in perialpine lakes. This result is in agreement with the study by Li and Williams (43) who also reported that the population structure of the Bdellovibrionaceae differed from one lake to another. The Bacteriovoracaceae seem to be more diverse in salt water than in freshwater (30). While the Peredibacteraceae may not be a much-diversified family according to our study, these bacteria were found in higher concentrations than both the Bdellovibrionaceae and Bacteriovoracaceae. Indeed, the Peredibacteraceae isolated from freshwater and soil and described by Pineiro et al. (31) constituted the most abundant family for all the conditions studied (within the three lakes, depths, different fractions, and different sampling periods). This result suggests that the Peredibacteraceae are well adapted to perialpine lake ecosystems, either by being a generalist or a versatile hunter regarding the heterotrophic bacteria present or by preying on bigger preys, thus growing faster and making more descendants. The number of preys present in the environment and the differential use of these preys (33) affect the abundance of one population to another. Environmental factors, such as temperature and salinity, can also affect the distribution and abundance of BALO families, and here, the Peredibacteraceae were more correlated to temperature than were the Bdellovibrionaceae or the Bacteriovoracaceae. In addition, it is known that the presence of a variety of predators, such as protists (i.e., the nanoflagellates or the ciliates), metazooplankton, and bacteriophages can affect the survival and growth of bacteria within the ecosystem. In fact, these microorganisms can play a significant role in controlling bacterial populations (21).
Low abundance of BALOs may not be indicative of a weak functional role.
The study of the abundances of the Bdellovibrionaceae and the Bacteriovoracaceae in aquaculture systems reported concentrations between 103 and 106 cells per ml (33). These results combined with our findings suggest that these two families might have a low impact on the community of heterotrophic bacteria in perialpine lakes. However, recent studies have also shown that a low abundance of BALOs is not necessarily evidence of a lower functional impact on prey dynamics (38, 44, 45). Hence, despite the very low abundances of the Bdellovibrionaceae and the Bacteriovoracaceae we found, their functional role may not be negligible. Moreover, the number of native BALOs in the environment is reported to be low (19). In fact, it has been suggested that BALOs rarely dominate continuously from a numerical point of view but form reasonably abundant populations that fluctuate over time (33). For example, the formation of a bacterial hot spot may alter the structure and abundance of BALOs in an ecosystem at any time. This led Williams et al. (38) to hypothesize about the “seed bank” theory. The theory implies that when some conditions are met, BALOs could switch from a state of inactive and sparse to a state where they are highly active and abundant to the point of becoming dominant for a limited period of time. The results from our previous study about ssDNA viruses and their boom and bust dynamics reinforce this idea (35). Relatively closed ecosystems, such as ponds, are usually rich in organic matter, resulting in high concentrations of heterotrophic bacteria that can favor the growth of BALO populations. For example, the number of heterotrophic bacteria in shrimp ponds is 10 to 100 times greater than that in natural coastal waters. BALOs react to high prey biomass densities, thus increasing their abundance (46), and can become invasive since they have very high adaptability to different environments (47).
BALOs and environmental factors.
Our CCA revealed some significant relationships between the BALOs and some environmental factors likely to be important to better understand the ecology of the predators. On one hand, we found that the Peredibacteraceae displayed clear links with pH, dissolved oxygen, and temperature and distributed more preferentially in near-surface waters, where waters are warmer and richer in phytoplankton biomass and bacterial prey (Fig. S1). According to Davidov and Jurkevitch (30), the optimal temperature range of Peredibacter starrii is 20 to 30°C. In the summer, the abundance of the Peredibacteraceae was clearly higher than in the winter or autumn. On another hand, the Bdellovibrionaceae seemed to be more sensitive to conductivity and ammonium concentrations. At last, no significant relationships were found for the Bacteriovoracaceae with any of the environmental factors tested in our study, while this group was also found to be more abundant in deep water. Unlike Peredibacter, Bacteriovorax stolpii can handle a wider range of temperatures. The optimal temperature range for growth of this species is 15 to 35°C (40). In general, environmental factors are undeniably a driving force in bacterial structure, specifically salinity and temperature (48). However, in the literature, only two factors, i.e., temperature and salinity, have been shown to induce a shift in BALO structure (33), while other factors seemed to play a minor role in BALO structure. For instance, Chen et al. (49) observed that growth and predation activity of estuarine BALOs were reduced when the temperature dropped below 10°C. In parallel, the same trend occurred when salinity reached more than 30 ppt. Excluding temperature and salinity, BALOs may not be directly correlated to conductivity, pH, oxygen, ammonium, chlorophyll, or other measurements but might be dependent on the presence of prey bacteria. This is what Chauhan et al. (50) found with clear positive correlations between BALOs and allochthonous prey bacterial abundances. Van Essche et al. (25) suggested that BALOs can prey in microaerophilic or anaerobic habitats. They identified a cytochrome oxidase complex (Cyt bb3) in Bdellovibrio bacteriovorus strain HD100 that eases microaerophilic respiration. Additionally, Sockett and Lambert (51) indicated that Bdellovibrio spp. can utilize other substrates than oxygen, such as nitrite or nitric oxide, for respiration. Burnham et al. (52) reported that Bdellovibrio bacteriovorus 15143 lyses extracellularly the blue-green alga Phormidium luridum. The secreted enzymes from the predator inhibited 75% of the algal photosynthesis. Therefore, when chlorophyll a measurements are low in the environment, one can expect that among other reasons, BALO enzymes are here and there. To conclude, since the existence and the abundance of prey are more likely to impact BALOs than are any other parameters, the next very important step will be to study which heterotrophic bacteria in perialpine lakes are associated with the main BALO species.
Provenance and form of BALOs.
While studying an estuarine ecosystem, Williams (53) observed that BALOs might be of allochthonous origin. BALOs would largely derive from river runoff and a wastewater treatment plant likely to constitute hot spots of concentration. In addition, it was reported that BALOs prefer benthic habitats over the pelagic zone, typically sediments or biofilms formed on small rocks near shore. These past observations could explain the relatively low abundance of BALOs in perialpine lakes sampled in open water, far from the main tributaries and the littoral zone. It is noteworthy, however, that if and when certain conditions are met, such as temperature warming and heterotrophic bacterial blossoming, the abundance of BALOs will most likely start to increase.
We also want to remind the reader that we used two types of filters to obtain information on both attached (2-µm pore size) and free-living (0.2-µm pore size) cells. The attached form implies BALOs undergoing periplasmic (bdelloplast) or epibiotic cycles but also BALOs physically attached to any type of particle. We hypothesized that 2-µm-pore-size filters would result in more BALOs than the <2-µm fraction, because a bdelloplast can contain at least three progenies, and epibiotic BALOs divide into two cells. Nevertheless, the 0.2-µm-pore-size filter yielded the highest concentrations of BALOs, suggesting at first glance that the free-living form is dominant. However, we prefer to point out that this result is to be taken with caution since we cannot eliminate the possibility that the filtration step as well as the extraction protocol using the GenElute kit may not have mechanically separated aggregates and then lysed the bdelloplasts, leading to false conclusions.
Conclusions and perspectives.
Our results have revealed the presence of bacterial predators belonging to the three main families of BALOs in perialpine lakes, with, at least for the Peredibacteraceae, concentrations reaching relatively high values. These results lead to the conclusion that these bacteria are likely to play a significant role in the functioning of these ecosystems. However, their role remains to be determined. A first perspective of this work is to investigate the interactions between prey and predator, for instance, throughout the use of approaches such as next-generation sequencing (NGS) to better capture their diversity and build interaction networks. We assume that using NGS approaches such as 16S rRNA gene metabarcoding combined with high-throughput sequencing will cover more in-depth the diversity of BALOs and considerably improve our knowledge regarding these communities. One needs to keep in mind that such methods can also fail to detect taxa at low densities (54). A second perspective is to investigate the action of different environmental factors on prey-predator relationships. To reach this goal, experimental approaches should be carried out with isolates from different strains of BALO families from perialpine lakes with a spectrum of prey bacteria cocultured in microcosms. Experiments in micro- or mesocosms could be proposed under different conditions, following the experimental approach proposed by Williams et al. (38), who used qPCR and SIP after the addition of radiolabeled prey under different conditions. The catalog and analysis of the diversity of the various BALOs in -cosms would allow a direct correlation of the different environmental factors characteristic of the lacustrine environment (such as prey quantity, prey diversity, types of nutrients, etc.) with the distribution of the various BALOs. The study of the abundance, structure, and diversity of BALOs within other matrices, such as biofilms and sediments within the perialpine lake environment, constitutes another exciting issue, as does the analysis of the possible functional importance of the last group of BALOs not studied in this work, e.g., Micavibrio.
MATERIALS AND METHODS
Study sites and sampling strategy.
Sampling was conducted at the reference station of the three largest natural deep lakes in France and western Europe, i.e., Lakes Annecy, Bourget, and Geneva. Different trophic statuses characterize these ecosystems: mesotrophic for Lake Geneva, oligomesotrophic for Lake Bourget, and oligotrophic for Lake Annecy (55, 56). The samples were taken at different depths, characteristics of the epi- or the metahypolimnion, i.e., 2 or 2.5 m versus 50 m for Lakes Bourget and Geneva, and 3 m versus 45 m for Lake Annecy. These samples were taken on average once per month for each lake (except for Lake Annecy) between August 2015 and January 2016. For each depth and sampling site, 1 liter of water was filtered successively through two types of 47-mm-diameter polycarbonate filters, a 2-µm-pore-size (to obtain the bacterial community attached to particles, epibiotic BALOs attached to prey, and periplasmic BALOs within prey), and 0.2-µm-pore-size (to retrieve only the so-called free-living bacteria, typically the BALO free attack phase). Filters were frozen and kept at −20°C. Both physical and chemical descriptors, as well as total bacterial counts, were obtained as reported in previous studies (4, 7, 56, 57). Physical descriptors, nutrients, chlorophyll a, and other environmental factors, including total bacterial abundance using flow cytometry, were obtained as previously described (2, 4, 9–11).
DNA extraction and PCR primers.
DNA extraction was conducted from filters using the GenElute bacterial genomic DNA kit. Different cultures of BALOs, referred to as HD100 and 109J for Bdellovibrio bacteriovorus and A3.12 for Peredibacter starrii (provided by E. Jurkevitch), were used as positive controls for PCR assays (see below) and were centrifuged (10 min, 4°C, 13,000 × g) in order to collect the pellet and extract the DNA. DNA concentrations were quantified and quality controlled using a NanoDrop 1000 spectrophotometer and Qubit 3.0 fluorometer (Thermo Fisher Scientific) with three replicates for each sample.
Different primers for either PCR or qPCR were selected for their specificity for the 16S rRNA gene of the three BALOs families, i.e., the Bdellovibrionaceae, Bacteriovoracaceae, and Peredibacteraceae. A total of 12 primers (Table 1) were tested using different PCR and qPCR protocols (Table S2). As no qPCR primers for quantifying Peredibacteraceae were available in any previous studies, we designed and tested new primers using the NCBI/Primer-BLAST online tool (58), the FastPCR software (59), and the MEGA 6 software (60). One hundred twenty existing sequences of 16 rRNA genes of Peredibacteraceae were aligned using ClustalW within MEGA6. A consensus sequence was obtained and used to find specific primers with NCBI/Primer-BLAST, with a high stringency (i.e., primer with at least 3 total mismatches to unintended targets, including at least 2 mismatches within the last 6 bp at the 3′ end; targets with 7 or more mismatches to the primer were ignored; and the target had a maximum size of 350 bp). Each primer pair designed was then verified by qPCR amplification and cloning-sequencing.
qPCR reactions were performed using the QuantiTect SYBR green PCR kit and with the Rotor-Gene Q thermocycler. Standard curves were established in triplicate using serial dilutions of Escherichia coli plasmids containing 16S rRNA gene sequences of each of the three families. Linear standard curves were obtained within the range of 101 to 106 plasmid copies per reaction. The efficacy was 0.99 with an R2 value of 0.998 and a slope value of −3.32. The specificity of reactions was confirmed by both melting-curve analyses and agarose gel electrophoresis to identify unspecific PCR products. The plasmid copy numbers were calculated using the following formula (61): copy number = (DNA amount [ng] × 6.022 × 1023)/(length [bp] × 109 × 650).
FCM.
To obtain total bacterial counts, without PCR bias, we used a FACSCalibur flow cytometer, as previously described (2, 4) (see the supplemental material). Note also that we compared these abundances with qPCR data obtained using the universal primer set for total bacterial counts and obtained a fairly good relationship (r = 0.654, not shown).
DGGE.
The BALO community was analyzed by denaturing gradient gel electrophoresis (DGGE) following the manufacturer’s protocol instruction manual (DGGE-2001; C.B.S.-Scientific Company, Inc.). One-millimeter-thick polyacrylamide gel (6% [wt/vol] acrylamide in 1× TAE buffer [40 mM Tris, 20 mM sodium acetate, 1 mM EDTA] [pH adjusted to 7.4]) was prepared with a linear formamide/urea gradient ranging from 40% to 55% after several tests to find the best gradient. It was overlaid with a nondenaturing stacking gel. Each well was loaded with 15 ng PCR product and 5 μl loading buffer. Electrophoresis was conducted for 16 h at 120 V and 60°C. Subsequently, the gels were stained in darkness for 40 min in 1× TAE buffer with 2× SYBR gold solution, as specified by the manufacturer. The DGGE profiles were only analyzed visually because of the low number of bands obtained and the lack of apparent important diversity (Fig. S1). Each band was then cut under UV light with the Gel Doc XR+ system (Bio-Rad) and conserved in 30 µl of TAE buffer at −20°C. DNA extraction from the DGGE band was performed by incubating tubes 20 min at −80°C and 20 min at −4°C, and then by centrifugation (10 min, 13,000 rpm, 4°C). The supernatant was conserved at 4°C for PCR.
DNA purification, cloning and sequencing.
The DNA of each DGGE band was eluted from the gel slice, after its excision, by adding 100 µl sterile 1× TAE buffer and heating at 95°C for 15 min. Three microliters of eluted DNA served as the template in a 22-µl PCR mixture using the corresponding primer set. The PCRs were performed under the same conditions as in the first PCR stage described above. The amplicons were first verified by electrophoresis in a 1.5% agarose gel, purified using the illustra GFX PCR DNA and Gel Band purification kit (GE Healthcare), and finally cloned into pCR4-TOPO vectors using the TOPO TA Cloning kit (Invitrogen). Randomly selected clones were sent to GATC Biotech (Germany) for sequencing.
Sequence processing, alignment, and phylogenetic analysis.
Sequenced DNA from Sanger sequencing required different steps in order to be cleaned. The same workflow was applied to the sequences of each BALO family. First, sequences shorter than 100 bp were discarded. Second, the remnant of E. coli vector at the 5′ and 3′ ends was detected and removed using NCBI BLASTn (62). Then, actual BALO sequences were trimmed at the 3′ end to remove the poor-quality bases. Next, sequences that matched other species than BALOs or unknown bacteria, i.e., “uncultured bacterium,” were also discarded. Afterward, sequences were dereplicated using the OBITOOLS command “OBIUNIQ” (63), checked for chimera sequences using “VSEARCH uchime_denovo” (64), and clustered at a 97% identity threshold using the command “CLUSTER FAST” of Usearch (65). Next, using the MEGA6 software (58), centroid sequences were aligned using MUSCLE (66) and trimmed to equal length. Poorly aligned sequences were discarded. Later, for each family, a reference database was constructed using Arb-SILVA (42), and when not enough sequences were found, the NCBI nucleotide database (67) was used to complete the database. Each sequence of the database was subjected to a BLAST search and confirmed to belong to the chosen BALO family. Sequences from the reference database and cleaned Sanger sequences were then aligned using MUSCLE and trimmed equally. The alignment was curated using Gblocks (68). The best substitution model was selected using jModelTest-2.1.10 (69) with the Akaike information criterion (AIC) (70). Next, maximum likelihood phylogeny was constructed using PhyML-3.1 (71) with 100 bootstrap replicates, and Bayesian phylogeny inference was made with MrBayes 3.2.6 (72). For each family, the same outgroup species was used, i.e., Vampirovibrio chlorellavorus (GenBank accession no. HM038000.1), based on Kandel et al. (33).
Data analysis of abundance in relation to environmental data.
A canonical correspondence analysis (CCA) was performed, taking into account only the most significant and nonredundant ecological variables to highlight the relationships between the relative abundances of the three families of BALOs (obtained by qPCR) with environmental factors. The CCA was tested using the vegan package in R with the following ecological descriptors: temperature, total phosphorus, orthophosphates, nitrates, ammonium, silicon dioxide, dissolved oxygen, chlorophyll a, pH, and conductivity, and only pH, dissolved oxygen, temperature, total phosphorus, conductivity, and ammonium were conserved after forward selection of the variables. A statistical test of the relationship between the abundance of each family with environmental factors was performed with the permutational multivariate analysis of variance (PERMANOVA) test from vegan.
Data availability.
All sequences are available in the GenBank database with the following accession numbers: MH537943, MH537944, MH537945, MH537946, MH537947, MH537948, MH537949, MH537950, MH537951, MH537952, MH537953, MH537954, MH537955, MH537956, MH537957, MH537958, MH537959, MH537960, MH537961, MH537962, MH537963, MH537964, MH537965, MH537966, MH537967, MH537968, MH537969, and MH537970 (https://www.ncbi.nlm.nih.gov/nuccore/?term=MH537943:MH537970[accn]).
Supplementary Material
ACKNOWLEDGMENTS
We thank Edouard Jurkevitch, who kindly provided us with the different BALO strains. We also thank Cécile Chardon for her help with molecular biology. Yves Desdevises is greatly acknowledged for his numerous pieces of advice and critical reading of several versions of the manuscript. The English of the manuscript was corrected by the Oxford proofreading company.
We declare no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02494-18.
REFERENCES
- 1.Comte J, Jacquet S, Viboud S, Fontvieille D, Millery A, Paolini G, Domaizon I. 2006. Microbial community structure and dynamics in the largest natural French lake (Lake Bourget). Microb Ecol 52:72–89. doi: 10.1007/s00248-004-0230-4. [DOI] [PubMed] [Google Scholar]
- 2.Personnic S, Domaizon I, Dorigo U, Berdjeb L, Jacquet S. 2009. Seasonal and spatial variability of virio-, bacterio-, and picophytoplanktonic abundances in three peri-alpine lakes. Hydrobiologia 627:99–116. doi: 10.1007/s10750-009-9718-8. [DOI] [Google Scholar]
- 3.Debroas D, Humbert JF, Enault F, Bronner G, Faubladier M, Cornillot E. 2009. Metagenomic approach studying the taxonomic and functional diversity of the bacterial community in a mesotrophic lake (Lac du Bourget - France). Environ Microbiol 11:2412–2424. doi: 10.1111/j.1462-2920.2009.01969.x. [DOI] [PubMed] [Google Scholar]
- 4.Berdjeb L, Ghiglione JF, Jacquet S. 2011. Bottom-up versus top-down control of hypo-and epilimnion free-living bacterial community structures in two neighboring freshwater lakes. Appl Environ Microbiol 77:3591–3599. doi: 10.1128/AEM.02739-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berdjeb L, Pollet T, Chardon C, Jacquet S. 2013. Spatio-temporal changes in the structure of archaeal communities in two deep freshwater lakes. FEMS Microbiol Ecol 86:215–230. doi: 10.1111/1574-6941.12154. [DOI] [PubMed] [Google Scholar]
- 6.Domaizon I, Savichtcheva O, Debroas D, Arnaud F, Villar C, Pignol C, Alric B, Perga ME. 2013. DNA from lake sediments reveals the long-term dynamics and diversity of Synechococcus assemblages. Biogeosciences 10:2515–2564. doi: 10.5194/bgd-10-2515-2013. [DOI] [Google Scholar]
- 7.Parvathi A, Zhong X, Ram ASP, Jacquet S. 2014. Dynamics of auto- and heterotrophic picoplankton and associated viruses in Lake Geneva. Hydrol Earth Syst Sci 18:1073–1087. doi: 10.5194/hess-18-1073-2014. [DOI] [Google Scholar]
- 8.Domaizon I, Viboud S, Fontvieille D. 2003. Taxon-specific and seasonal variations in flagellates grazing on heterotrophic bacteria in the oligotrophic Lake Annecy–importance of mixotrophy. FEMS Microbiol Ecol 46:317–329. doi: 10.1016/S0168-6496(03)00248-4. [DOI] [PubMed] [Google Scholar]
- 9.Personnic S, Domaizon I, Sime-Ngando T, Jacquet S. 2009. Seasonal variations of microbial abundances and virus- versus flagellate-induced mortality of picoplankton in three peri-alpine lakes. J Plankton Res 31:1161–1177. doi: 10.1093/plankt/fbp057. [DOI] [Google Scholar]
- 10.Thomas R, Berdjeb L, Sime-Ngando T, Jacquet S. 2011. Viral abundance, production, decay rates and life strategies (lysogeny versus lysis) in Lake Bourget (France). Environ Microbiol 13:616–630. doi: 10.1111/j.1462-2920.2010.02364.x. [DOI] [PubMed] [Google Scholar]
- 11.Berdjeb L, Pollet T, Domaizon I, Jacquet S. 2011. Effect of grazers and viruses on bacterial community structure and production in two contrasting trophic lakes. BMC Microbiol 11:88–106. doi: 10.1186/1471-2180-11-88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perga ME, Domaizon I, Guillard J, Hamelet V, Anneville O. 2013. Are cyanobacterial blooms trophic dead ends? Oecologia 172:551–562. doi: 10.1007/s00442-012-2519-1. [DOI] [PubMed] [Google Scholar]
- 13.Jacquet S, Domaizon I, Personnic S, Pradeep Ram AS, Hedal M, Duhamel S, Sime-Ngando T. 2005. Estimates of protozoan- and viral-mediated mortality of bacterioplankton in Lake Bourget (France). Freshwater Biol 50:627–645. doi: 10.1111/j.1365-2427.2005.01349.x. [DOI] [Google Scholar]
- 14.Sime-Ngando T, Colombet J, Personnic S, Domaizon I, Dorigo U, Perney P, Hustache JC, Viollier E, Jacquet S. 2008. Short-term variations in abundances and potential activities of viruses, bacteria and nanoprotists in Lake Bourget. Ecol Res 23:851–861. doi: 10.1007/s11284-007-0448-y. [DOI] [Google Scholar]
- 15.Meunier A, Jacquet S. 2015. Do phages impact microbial dynamics, prokaryotic community structure and nutrient dynamics in Lake Bourget? Biol Open 4:1528–1537. doi: 10.1242/bio.013003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Humbert JF, Dorigo U, Cecchi P, Le Berre B, Debroas D, Bouvy M. 2009. Comparison of the structure and composition of bacterial communities from temperate and tropical freshwater ecosystems. Environ Microbiol 11:2339–2350. doi: 10.1111/j.1462-2920.2009.01960.x. [DOI] [PubMed] [Google Scholar]
- 17.Mangot JF, Debroas D, Domaizon I. 2011. Perkinsozoa, a well-known marine protozoan flagellate parasite group, newly identified in lacustrine systems: a review. Hydrobiologia 659:37–48. doi: 10.1007/s10750-010-0268-x. [DOI] [Google Scholar]
- 18.Rendulic S. 2004. A predator unmasked: life cycle of Bdellovibrio bacteriovorus from a genomic perspective. Science 303:689–692. doi: 10.1126/science.1093027. [DOI] [PubMed] [Google Scholar]
- 19.Chauhan A, Cherrier J, Williams HN. 2009. Impact of sideways and bottom-up control factors on bacterial community succession over a tidal cycle. Proc Natl Acad Sci U S A 106:4301–4306. doi: 10.1073/pnas.0809671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iebba V, Totino V, Santangelo F, Gagliardi A, Ciotoli L, Virga A, Ambrosi C, Pompili M, De Biase RV, Selan L, Artini M, Pantanella F, Mura F, Passariello C, Nicoletti M, Nencioni L, Trancassini M, Quattrucci S, Schippa S. 2014. Bdellovibrio bacteriovorus directly attacks Pseudomonas aeruginosa and Staphylococcus aureus cystic fibrosis isolates. Front Microbiol 5:280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pantanella F, Iebba V, Mura F, Dini L, Totino V, Neroni B, Bonfiglio G, Trancassini M, Passariello C, Schippa S. 2018. Behaviour of Bdellovibrio bacteriovorus in the presence of Gram-positive Staphylococcus aureus. New Microbiol 41:145–152. [PubMed] [Google Scholar]
- 22.Oyedara OO, De Luna-Santillana E. d J, Olguin-Rodriguez O, Guo X, Mendoza-Villa MA, Menchaca-Arredondo JL, Elufisan TO, Garza-Hernandez JA, Garcia Leon I, Rodriguez-Perez MA. 2016. Isolation of Bdellovibrio sp. from soil samples in Mexico and their potential applications in control of pathogens. Microbiologyopen 5:992–1002. doi: 10.1002/mbo3.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rotem O, Pasternak Z, Shimoni E, Belausov E, Porat Z, Pietrokovski S, Jurkevitch E. 2015. Cell-cycle progress in obligate predatory bacteria is dependent upon sequential sensing of prey recognition and prey quality cues. Proc Natl Acad Sci U S A 112:E6028–E6037. doi: 10.1073/pnas.1515749112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schwudke D, Strauch E, Krueger M, Appel B. 2001. Taxonomic studies of predatory bdellovibrios based on 16S rRNA analysis, ribotyping and the hit locus and characterization of isolates from the gut of animals. Syst Appl Microbiol 24:385–394. doi: 10.1078/0723-2020-00042. [DOI] [PubMed] [Google Scholar]
- 25.Van Essche M, Quirynen M, Sliepen I, Loozen G, Boon N, Van Eldere J, Teughels W. 2011. Killing of anaerobic pathogens by predatory bacteria. Mol Oral Microbiol 26:52–61. doi: 10.1111/j.2041-1014.2010.00595.x. [DOI] [PubMed] [Google Scholar]
- 26.Davidov Y, Friedjung A, Jurkevitch E. 2006. Structure analysis of a soil community of predatory bacteria using culture-dependent and culture-independent methods reveals a hitherto undetected diversity of Bdellovibrio-and-like organisms. Environ Microbiol 8:1667–1673. doi: 10.1111/j.1462-2920.2006.01052.x. [DOI] [PubMed] [Google Scholar]
- 27.Koval SF, Williams HN, Stine OC. 2015. Reclassification of bacteriovorax marinus as Halobacteriovorax marinus gen. nov., comb. nov. and bacteriovorax litoralis as Halobacteriovorax litoralis comb. nov.; description of Halobacteriovoraceae fam. nov. in the class Deltaproteobacteria. Int J Syst Evol Microbiol 65:593–597. doi: 10.1099/ijs.0.070201-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hahn MW, Schmidt J, Koll U, Rohde M, Verbarg S, Pitt A, Nakai R, Naganuma T, Lang E. 2017. Silvanigrella aquatica gen. nov., sp. nov., isolated from a freshwater lake located in the Black Forest, Germany, description of Silvanigrellaceae fam. nov., Silvanigrellales ord. nov., reclassification of the order Bdellovibrionales in the class Oligoflexia, reclassification of the families Bacteriovoracaceae and Halobacteriovoraceae in the new order Bacteriovoracales ord. nov., and reclassification of the family Pseudobacteriovoracaceae in the order Oligoflexiales. Int J Syst Evol Microbiol 67:2555–2568. doi: 10.1099/ijsem.0.001965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCauley EP, Haltli B, Kerr RG. 2015. Description of Pseudobacteriovorax antillogorgiicola gen. nov., sp. nov., a bacterium isolated from the gorgonian octocoral Antillogorgia elisabethae, belonging to the family Pseudobacteriovoracaceae fam. nov., within the order Bdellovibrionales. Int J Syst Evol Microbiol 65:522–530. doi: 10.1099/ijs.0.066266-0. [DOI] [PubMed] [Google Scholar]
- 30.Davidov Y, Jurkevitch E. 2004. Diversity and evolution of Bdellovibrio-and-like organisms (BALOs), reclassification of Bacteriovorax starrii as Peredibacter starrii gen. nov., comb. nov., and description of the Bacteriovorax-Peredibacter clade as Bacteriovoracaceae fam. nov. Int J Syst Evol Microbiol 54:1439–1452. doi: 10.1099/ijs.0.02978-0. [DOI] [PubMed] [Google Scholar]
- 31.Pineiro SA, Sahaniuk GE, Romberg E, Williams HN. 2004. Predation pattern and phylogenetic analysis of Bdellovibrionaceae from the Great Salt Lake, Utah. Curr Microbiol 48:113–117. doi: 10.1007/s00284-003-4136-z. [DOI] [PubMed] [Google Scholar]
- 32.Shemesh Y, Jurkevitch E. 2004. Plastic phenotypic resistance to predation by Bdellovibrio and like organisms in bacterial prey. Environ Microbiol 6:12–18. [DOI] [PubMed] [Google Scholar]
- 33.Kandel PP, Pasternak Z, van Rijn J, Nahum O, Jurkevitch E. 2014. Abundance, diversity and seasonal dynamics of predatory bacteria in aquaculture zero discharge systems. FEMS Microbiol Ecol 89:149–161. doi: 10.1111/1574-6941.12342. [DOI] [PubMed] [Google Scholar]
- 34.Roux S, Enault F, Robin A, Ravet V, Personnic S, Theil S, Colombet J, Sime-Ngando T, Debroas D. 2012. Assessing the diversity and specificity of two freshwater viral communities through metagenomics. PLoS One 7:e33641. doi: 10.1371/journal.pone.0033641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhong X, Guidoni B, Jacas L, Jacquet S. 2015. Structure and diversity of ssDNA Microviridae viruses in two peri-alpine lakes (Annecy and Bourget, France). Res Microbiol 166:644–654. doi: 10.1016/j.resmic.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 36.Brentlinger KL, Hafenstein S, Novak CR, Fane BA, Borgon R, McKenna R, Agbandje-McKenna M. 2002. Microviridae, a family divided: isolation, characterization, and genome sequence of φMH2K, a bacteriophage of the obligate intracellular parasitic bacterium Bdellovibrio bacteriovorus. J Bacteriol 184:1089–1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pérez J, Moraleda-Muñoz A, Marcos-Torres FJ, Muñoz-Dorado J. 2016. Bacterial predation: 75 years and counting! Environ Microbiol 18:766–779. doi: 10.1111/1462-2920.13171. [DOI] [PubMed] [Google Scholar]
- 38.Williams HN, Lymperopoulou DS, Athar R, Chauhan A, Dickerson TL, Chen H, Laws E, Berhane T-K, Flowers AR, Bradley N, Young S, Blackwood D, Murray J, Mustapha O, Blackwell C, Tung Y, Noble RT. 2016. Halobacteriovorax, an underestimated predator on bacteria: potential impact relative to viruses on bacterial mortality. ISME J 10:491–499. doi: 10.1038/ismej.2015.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Elbrecht V, Hebert PDN, Steinke D. 2018. Slippage of degenerate primers can cause variation in amplicon length. Sci Rep 8:10999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Baer ML, Ravel J, Chun J, Hill RT, Williams HN. 2000. A proposal for the reclassification of Bdellovibrio stolpii and Bdellovibrio starrii into a new genus, Bacteriovorax gen. nov. as Bacteriovorax stolpii comb. nov. and Bacteriovorax starrii comb. nov., respectively. Int J Syst Evol Microbiol 50:219–224. doi: 10.1099/00207713-50-1-219. [DOI] [PubMed] [Google Scholar]
- 41.Baer ML, Ravel J, Piñeiro SA, Guether-Borg D, Williams HN. 2004. Reclassification of salt-water Bdellovibrio sp. as Bacteriovorax marinus sp. nov. and Bacteriovorax litoralis sp. nov. Int J Syst Evol Microbiol 54:1011–1016. doi: 10.1099/ijs.0.02458-0. [DOI] [PubMed] [Google Scholar]
- 42.Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Glo FO, Yarza P, Peplies J, Glockner FO. 2013. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41:D590–D596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li N, Williams HN. 2015. 454 pyrosequencing reveals diversity of Bdellovibrio and like organisms in fresh and salt water. Antonie Van Leeuwenhoek 107:305–311. doi: 10.1007/s10482-014-0327-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Richards GP, Fay JP, Dickens KA, Parent MA, Soroka DS, Boyd EF. 2012. Predatory bacteria as natural modulators of Vibrio parahaemolyticus and Vibrio vulnificus in seawater and oysters. Appl Environ Microbiol 78:7455–7466. doi: 10.1128/AEM.01594-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Welsh RM, Zaneveld JR, Rosales SM, Payet JP, Burkepile DE, Thurber RV. 2016. Bacterial predation in a marine host-associated microbiome. ISME J 10:1540–1544. doi: 10.1038/ismej.2015.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen CQ, Lai XT, Xue M, Huang YL, Li HX, Zhou SN. 2009. Molecular typing and identification of Bdellovibrio-and-like organisms isolated from seawater shrimp ponds and adjacent coastal waters. J Appl Microbiol 106:1154–1162. doi: 10.1111/j.1365-2672.2008.04081.x. [DOI] [PubMed] [Google Scholar]
- 47.Yu R, Zhang S, Chen Z, Li C. 2017. Isolation and application of predatory Bdellovibrio-and-like organisms for municipal waste sludge biolysis and dewaterability enhancement. Front Environ Sci Eng 11:10. [Google Scholar]
- 48.Aguirre M, Abad D, Albaina A, Cralle L, Goñi-Urriza MS, Estonba A, Zarraonaindia I. 2017. Unraveling the environmental and anthropogenic drivers of bacterial community changes in the Estuary of Bilbao and its tributaries. PLoS One 12:e0178755. doi: 10.1371/journal.pone.0178755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen H, Laws EA, Martin JL, Berhane T-K, Gulig PA, Williams HN. 2018. Relative contributions of Halobacteriovorax and bacteriophage to bacterial cell death under various environmental conditions. mBio 9:e01202-18. doi: 10.1128/mBio.01202-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chauhan A, Fortenberry GZ, Lewis DE, Williams HN. 2009. Increased diversity of predacious Bdellovibrio-like organisms (BLOs) as a function of eutrophication in Kumaon Lakes of India. Curr Microbiol 59:1–8. doi: 10.1007/s00284-009-9385-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sockett RE, Lambert C. 2004. Bdellovibrio as therapeutic agents: a predatory renaissance? Nat Rev Microbiol 2:669–675. doi: 10.1038/nrmicro959. [DOI] [PubMed] [Google Scholar]
- 52.Burnham JC, Stetak T, Locher G. 1976. Extracellular lysis of the bluegreen alga Phormidium luridum by Bdellovibrio bacteriovorus. J Phycol 12:306–313. doi: 10.1111/j.1529-8817.1976.tb02849.x. [DOI] [Google Scholar]
- 53.Williams H. 1988. A study of the occurrence and distribution of bdellovibrios in estuarine sediment over an annual cycle. Microb Ecol 15:9–20. doi: 10.1007/BF02012949. [DOI] [PubMed] [Google Scholar]
- 54.de Dios Caballero J, Vida R, Cobo M, Maiz L, Suarez L, Galeano J, Baquero F, Canton R, Delcampo R. 2017. Individual patterns of complexity in cystic fibrosis lung microbiota including predator bacteria, over a 1-year period. mBio 8:e00959-17. doi: 10.1128/mBio.00959-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacquet S, Anneville O, Domaizon I. 2012. Evolution de paramètres clés indicateurs de la qualité des eaux et du fonctionnement écologique des grands lacs péri-alpins (Léman, Annecy, Bourget): Etude comparative de trajectoires de restauration post-eutrophisation. Arch Des Sci 65:191–208. [Google Scholar]
- 56.Jacquet S, Domaizon I, Anneville O. 2014. The need for ecological monitoring of freshwaters in a changing world: a case study of Lakes Annecy, Bourget, and Geneva. Environ Monit Assess 186:3455–3476. doi: 10.1007/s10661-014-3630-z. [DOI] [PubMed] [Google Scholar]
- 57.Zhong X, Berdjeb L, Jacquet S. 2013. Temporal dynamics and structure of picocyanobacteria and cyanomyoviruses in two large and deep peri-alpine lakes. FEMS Microbiol Ecol 86:312–326. doi: 10.1111/1574-6941.12166. [DOI] [PubMed] [Google Scholar]
- 58.Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S, Madden TL. 2012. Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kalendar R, Khassenov B, Ramanculov E, Ivanov KI. 2017. FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319. [DOI] [PubMed] [Google Scholar]
- 60.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. 2013. MEGA6: Molecular Evolutionary Genetics Analysis version 6. 0. Mol Biol Evol 30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Whelan JA, Russell NB, Whelan MA. 2003. A method for the absolute quantification of cDNA using real-time PCR. J Immunol Methods 278:261–269. [DOI] [PubMed] [Google Scholar]
- 62.Altschul SF, Gish W, Pennsylvania T, Park U. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 63.Boyer F, Mercier C, Bonin A, Le Bras Y, Taberlet P, Coissac E. 2016. OBITOOLS: a unix-inspired software package for DNA metabarcoding. Mol Ecol Resour 16:176–182. doi: 10.1111/1755-0998.12428. [DOI] [PubMed] [Google Scholar]
- 64.Rognes T, Flouri T, Nichols B, Quince C, Mahé F. 2016. VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 66.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Benson DA, Cavanaugh M, Clark K, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. 2013. GenBank. Nucleic Acids Res 41:36–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Castresana J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552. doi: 10.1093/oxfordjournals.molbev.a026334. [DOI] [PubMed] [Google Scholar]
- 69.Darriba D, Taboada GL, Doallo R, Posada D. 2012. jModelTest 2: more models, new heuristics and parallel computing CircadiOmics: integrating circadian genomics, transcriptomics, proteomics. Nat Methods 9:772. doi: 10.1038/nmeth.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akaike H. 1973. Information theory and an extension of the maximum likelihood principle, p 267–281. In Petrov BN, Csaki F (ed), Proceedings of the 2nd International Symposium on Information Theory. Akademiai Kiado, Budapest, Hungary. [Google Scholar]
- 71.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 72.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3. 2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Van Essche M, Sliepen I, Loozen G, Van Eldere J, Quirynen M, Davidov Y, Jurkevitch E, Boon N, Teughels W. 2009. Development and performance of a quantitative PCR for the enumeration of Bdellovibrionaceae. Environ Microbiol Rep 1:228–233. doi: 10.1111/j.1758-2229.2009.00034.x. [DOI] [PubMed] [Google Scholar]
- 74.Zheng G, Wang C, Williams HN, Pineiro SA. 2008. Development and evaluation of a quantitative real-time PCR assay for the detection of saltwater Bacteriovorax. Environ Microbiol 10:2515–2526. doi: 10.1111/j.1462-2920.2008.01676.x. [DOI] [PubMed] [Google Scholar]
- 75.Marchesi JR, Sato T, Weightman AJ, Martin TA, Fry JC, Hiom SJ, Wade WG. 1998. Design and evaluation of useful bacterium-specific PCR primers that amplify genes coding for bacterial 16S rRNA. Appl Environ Microbiol 64:795–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Heuer H, Krsek M, Baker P, Smalla K, Wellington EM. 1997. Analysis of Actinomycete communities by specific amplification of genes encoding 16S rRNA and gel-electrophoretic separation in denaturing gradients. Appl Environ Microbiol 63:3233–3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pasternak Z, Pietrokovski S, Rotem O, Gophna U, Lurie-Weinberger MN, Jurkevitch E. 2013. By their genes ye shall know them: genomic signatures of predatory bacteria. ISME J 7:756–769. doi: 10.1038/ismej.2012.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequences are available in the GenBank database with the following accession numbers: MH537943, MH537944, MH537945, MH537946, MH537947, MH537948, MH537949, MH537950, MH537951, MH537952, MH537953, MH537954, MH537955, MH537956, MH537957, MH537958, MH537959, MH537960, MH537961, MH537962, MH537963, MH537964, MH537965, MH537966, MH537967, MH537968, MH537969, and MH537970 (https://www.ncbi.nlm.nih.gov/nuccore/?term=MH537943:MH537970[accn]).