Volatile sulfur compounds contribute to wine aromas that may be considered pleasant, such as “tropical,” “passionfruit,” and “guava,” as well as aromas that are considered undesirable, such as “rotten eggs,” “onions,” and “sewer.” During fermentation, wine yeasts release some of these compounds from odorless precursor molecules, a process that is most efficient when performed by yeasts that express active forms of the protein Irc7p. We show that most wine yeasts carry mutations that reduce activity of this protein, affecting the formation of volatile sulfur compounds that impart both pleasant and unpleasant aromas. The results provide winemakers with guidance on the choice of yeasts that can emphasize or deemphasize this particular contribution to wine quality.
KEYWORDS: aroma, lyase, sulfur, thiols, wine, yeasts
ABSTRACT
During alcoholic fermentation of grape sugars, wine yeasts produce a range of secondary metabolites that play an important role in the aroma profile of wines. In this study, we have explored the ability of a large number of wine yeast strains to modulate wine aroma composition, focusing on the release of the “fruity” thiols 3-mercaptohexan-1-ol (3-MH) and 4-mercapto-4-methylpentan-2-one (4-MMP) from their respective cysteinylated nonvolatile precursors. The role of the yeast gene IRC7 in thiol release has been well established, and it has been shown that a 38-bp deletion found in many wine strains cause them to express a truncated version of Irc7p that does not possess cysteine-S-conjugate β-lyase activity. In our data, we find that IRC7 allele length alone does not fully explain the capacity of a strain to release thiols. Screening of a large number of strains coupled with analysis of genomic sequence data allowed us to identify several previously undescribed single-nucleotide polymorphisms (SNPs) in IRC7 that, when coupled with allele length, more robustly explain the ability of a particular yeast strain to release thiols from their cysteinylated precursors. We also demonstrate that allelic variation of IRC7 not only affects the release of thiols but modulates the formation of negative volatile sulfur compounds from the amino acid cysteine. The results of this study provide winemakers with an improved understanding of the genetic determinants that affect wine aroma and flavor, which can be used to guide the choice of yeast strains that are fit for purpose.
IMPORTANCE Volatile sulfur compounds contribute to wine aromas that may be considered pleasant, such as “tropical,” “passionfruit,” and “guava,” as well as aromas that are considered undesirable, such as “rotten eggs,” “onions,” and “sewer.” During fermentation, wine yeasts release some of these compounds from odorless precursor molecules, a process that is most efficient when performed by yeasts that express active forms of the protein Irc7p. We show that most wine yeasts carry mutations that reduce activity of this protein, affecting the formation of volatile sulfur compounds that impart both pleasant and unpleasant aromas. The results provide winemakers with guidance on the choice of yeasts that can emphasize or deemphasize this particular contribution to wine quality.
INTRODUCTION
Volatile sulfur compounds (VSCs) are among the most impactful classes of aroma-active molecules in wine and can be considered a “double-edged sword” due to their potential to have positive and negative impacts on perceived wine quality (1). The production of negative VSCs, such as hydrogen sulfide (H2S), is a significant problem for the wine industry, since it imparts an undesirable “rotten egg”-like flavor, even at low concentrations. On the other hand, some sulfur-containing flavor compounds, such as the polyfunctional thiols 4-mercapto-4-methyl-pentan-2-one (4-MMP), 3-mercaptohexan-1-ol (3-MH), and 3-mercaptohexyl acetate (3-MHA), impart “passionfruit,” “grapefruit,” “guava,” and “box tree” aromas (2–4). These compounds have very low perception thresholds and are important contributors to the flavors of Sauvignon blanc and Chardonnay wines (5).
The formation and fate of VSCs in wine is mediated by several factors (6), although a prominent role is played by the Saccharomyces cerevisiae yeasts that are used to perform wine fermentation (1). The likelihood of a wine being negatively impacted by H2S, for example, can be greatly reduced through choice of yeast strain. Propensity to produce H2S has been linked to mutations in the sulfite reductase-encoding genes MET10 and MET5 (7, 8), and also to mutations in SKP2 and MET2 (9). Choice of yeast strain also modulates the concentration of 4-MMP, 3-MH, and 3-MHA in wine (10–13). Further opportunities to enhance polyfunctional thiol concentrations include sequential inoculation or cofermentation with non-Saccharomyces yeasts, such as Torulaspora delbrueckii and Pichia kluyveri (14–16), as well as the use of S. cerevisiae × Saccharomyces kudriavzevii hybrids (17).
It has been shown that 4-MMP and 3-MH exist in the grape berry as nonvolatile precursors, mainly as conjugates of glutathione (Glut-4-MMP and Glut-3-MH) and cysteine (Cys-4-MMP and Cys-3-MH) (18, 19). Dipeptide conjugates of 3-MH, S-3-cysteinyl glycinylhexan-1-ol (Cys-Gly-3-MH), and S-3-γ-glutamyl cysteinylhexan-1-ol (γ-Glu-Cys-3-MH) have also been identified (20–22). The acetate ester of 3-MH (3-MHA) is not formed from a nonvolatile precursor but results from acetylation of 3-MH by the yeast alcohol acetyltransferase ATF1 (23). Biotransformation of conjugated precursors into free thiols by yeast involves their uptake through amino acid and glutathione (GSH) transporters in the plasma membrane (21, 24, 25). Once inside the cytoplasm, Glut-3-MH is cleaved to Cys-Gly-3-MH in the yeast vacuole by action of the γ-glutamate transpeptidase ECM38, which might also catalyze the formation of Cys-3-MH from the precursor dipeptide γ-Glu-Cys-3-MH (21, 26). The carboxypeptidase responsible for the cleavage of the dipeptide Cys-Gly-3-MH into Cys-3-MH remains to be identified. On the other hand, the degradation pathway of Glut-4-MMP to yield 4-MMP remains uncharacterized, as the two main GSH degradation pathways appear to be ineffective in metabolizing Glut-4-MMP (21).
Once the cysteinylated precursors are formed through degradation of their glutathionylated or dipeptide precursors, or are assimilated from grape must, these are cleaved by yeast enzymes with carbon–sulfur β-lyase activity to release the free thiols. In S. cerevisiae, there are at least three genes, IRC7, STR3 and CYS3, encoding cysteine-S-conjugate β-lyase enzymes. Of these enzymes, Irc7p and Str3p have been directly shown to release polyfunctional thiols from cysteine conjugates (27, 28), while Cys3p and Str3p were recently shown to release 2-furfurylthiol from its cysteine conjugate (29). Interestingly, while Irc7p was found to be the key determinant of 4-MMP release from Cys-4-MMP and is a significant contributor to 3-MH release from Cys-3-MH (27), most strains of S. cerevisiae have a 38-bp deletion in the IRC7 coding sequence that renders the protein inactive (27, 30). It has been hypothesized that the presence or absence of this deletion might account for the strain variation observed in 4-MMP release, and a positive correlation between the expression level of the full-length and active version of IRC7 and 4-MMP production within industrial strains has been reported (31). Irc7p also shows cysteine desulfhydrase activity, thereby releasing ammonium and H2S (32), and this deletion has the potential to affect formation of negative VSCs during winemaking.
The biological role of IRC7 is not well established, as many functional studies have been conducted in strain backgrounds containing the inactive version of IRC7 (33–35). While IRC7 was originally suggested to be involved in DNA metabolism since the null mutant causes increased rates of recombination between homologous chromosomes (33), its role as cysteine desulfhydrase also points out to an involvement in the regulation of cysteine homeostasis (32).
In this study, across a large number of wine yeast strains, we show that while IRC7 allele length does modulate formation of 3-MH and 4-MMP from their cysteinylated precursors, this alone does not fully explain the variation observed. We identify several single-nucleotide polymorphisms (SNPs) in IRC7 that reduce activity of Irc7p against cysteine and cysteine conjugates, and, through allele replacement, demonstrate their impact upon the release of polyfunctional thiols and the formation of negative VSCs during wine fermentation.
RESULTS
It has been shown previously that a 38-bp deletion in IRC7 (between nucleotides 1023 and 1060) results in a truncated protein of 340 amino acids (short Irc7p or Irc7pS), that displays limited β-lyase activity compared with that of the 400-amino acid full-length and active protein (long Irc7p or Irc7pL) (27). We took advantage of the recently published whole-genome sequencing data from 212 strains of S. cerevisiae (36) to determine the presence of the short or long versions of IRC7 in a subset of 179 wine strains, comprising 94 commercial strains and 85 wine isolates from around the world. There was no difference between commercial strains and wine isolates with regard to the proportion that harbored copies of this short allele (Table 1). Thus, 56% of the sequenced strains were found to be homozygous for the short allele (IRC7S IRC7S), 27% were heterozygous (IRC7S IRC7L), and only 17% were homozygous for the long allele (IRC7L IRC7L).
TABLE 1.
Distribution of IRC7 genotypes across 179 Saccharomyces cerevisiae wine yeast strains
| Genotypea | Total no. of isolates (%) | No. of commercial strains (% [n = 94]) | No. of wine isolates (% [n = 85]) |
|---|---|---|---|
| IRC7S IRC7S | 100 (56) | 54 (57) | 46 (54) |
| IRC7S IRC7L | 48 (27) | 27 (29) | 21 (25) |
| IRC7L IRC7L | 31 (17) | 13 (14) | 18 (21) |
IRC7S, short allele of IRC7; IRC7L, long or full-length allele of IRC7.
To assess variation among different strains in their ability to release the polyfunctional thiols 3-MH and 4-MMP, a series of small-scale fermentations were carried out in synthetic grape juice supplemented with grape-like concentrations of the cysteine precursors of 3-MH and 4-MMP (SGJ-thiol). The release of these polyfunctional thiols was analyzed in a subset of 80 strains, representative of each of the three IRC7 genotypes observed in the larger population of sequenced strains (Table S1). Results showed a 20-fold difference across the yeasts studied in their ability to release 3-MH (from 131 to 2,508 ng·liter−1), and an even greater difference, 30-fold, in the case of 4-MMP (from 171 to 5,298 ng·liter−1). The release of both polyfunctional thiols was highly correlated (R2 = 0.78, P < 0.001). Overall there was no difference between commercial strains and wine isolates in their ability to release either thiol from their cysteinylated precursors (Table 2). However, for the subset of 39 commercial strains, those recommended by yeast manufacturers for white winemaking released significantly more 3-MH and 4-MMP than did strains recommended either for red winemaking or for both red and white winemaking (Table 2). When the isolates were grouped according to genome sequencing-derived clades described by Borneman et al. (36), we observed that strains from the three major wine yeast clades released significantly less 4-MMP than did wine isolates from outside these clades (designated “Other”; Table S2).
TABLE 2.
Release of 3-MH and 4-MMP by 80 wine yeast strains in SGJ-thiol medium according to their origin
| Origin or recommended usage | No. of strains | 3-MH (ng·liter−1)b | 4-MMP (ng·liter−1)b |
|---|---|---|---|
| Origin | |||
| Wine isolates | 41 | 727 (181–2,508) | 902 (150–5,298) |
| Commercial | 39 | 696 (131–2,481) | 844 (170–4,690) |
| Recommended usagea | |||
| White | 14 | 1,016 (412–2,481) A | 1749 (170–4,690) A |
| Red | 13 | 590 (131–839) B | 618 (201–1,904) B |
| White/red | 12 | 547 (170–1,290) B | 512 (189–3,579) B |
Based upon commercial yeast manufacturers’ published materials.
Results are expressed as the mean for each of the groups, and the range of production within each group is shown in parentheses. Means with the same letter are not significantly different from each other (Tukey’s HSD test, P < 0.05).
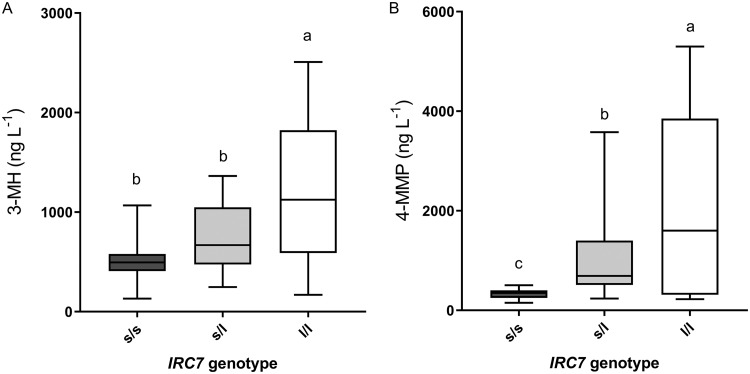
When thiol release data were grouped according to IRC7 genotype, strains that were homozygous for the IRC7L allele produced, on average, 6 and 2.5 times more 4-MMP and 3-MH, respectively, than did strains homozygous for the IRC7S allele (Fig. 1A and B). Within each genotype, particularly in the IRC7L IRC7L and IRC7S IRC7L groups, there was substantial variation in the release of these compounds between individual strains, as recently described (30). Of note, 5 of the 6 strains belonging to the Vin7 clade are homozygous for the IRC7L allele yet produced levels of 4-MMP equivalent to those of the IRC7S IRC7S group (Table S2). Our results suggest that a substantial portion of variation between yeasts for capacity to release thiols is not explained solely by IRC7 allele length.
FIG 1.
Release of 3-MH (A) and 4-MMP (B) by 80 wine strains in SGJ-thiol medium, grouped according to their IRC7 genotype. s/s refers to the homozygous short allele (IRC7S IRC7S) and l/l to the homozygous long or full-length allele (IRC7L IRC7L), while s/l refers to heterozygous strains (IRC7S IRC7L). Means with the same letter are not significantly different from each other (Tukey’s honestly significant difference [HSD] test, P < 0.05). Totals of 43, 20, and 17 strains from the s/s, s/l, and l/l IRC7 genotypes, respectively, were included.
Identification of IRC7 sequence variants that affect polyfunctional thiol release.
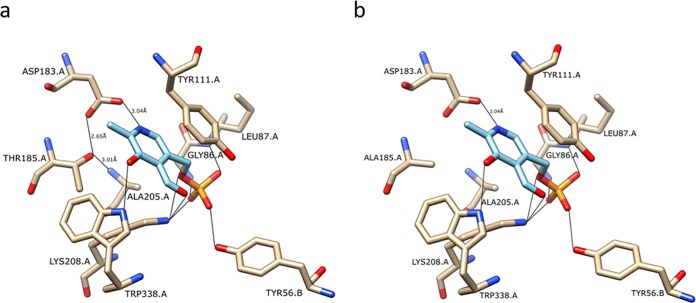
Upon closer examination of the IRC7 open reading frame (ORF) across the 179 sequenced strains (36), we identified a series of single-nucleotide polymorphisms (SNPs). The most common of these SNPs leads to substitution of threonine in position 185 to alanine, which is linked with the long allele of IRC7 (IRC7L T185A). We found that 80% of the strains in the IRC7S IRC7L and IRC7L IRC7L groups exhibited this allele (data not shown). Other mutations were found in a smaller proportion of strains, and, in most cases, these were linked with the IRC7L T185A allele (Table S1). A structural model of S. cerevisiae Irc7pL was constructed, and all observed mutated amino acids were mapped against the model to facilitate prediction of impact on protein function. Only the T185A mutation was proximal to the active site of Irc7pL (Fig. 2). The presence of this SNP appeared to have a deleterious effect on the release of both thiols, particularly that of 4-MMP (Table 3). Strains in the IRC7L IRC7L group that were homozygous for T185A produced 80% less 4-MMP (715 ng·liter−1) than did strains without this mutation (4,618 ng·liter−1).
FIG 2.
Model of the Irc7pL active site in the presence of PLP cofactor (light blue). The model shows amino acid residues from two subunits (A and B) of Irc7pL that form the catalytic dimer. In the model on the left (a), the hydroxyl group of Thr185 forms a hydrogen bond with the carboxylate group of Asp183, fixing it for optimal interaction with the nitrogen of the pyridine ring of PLP. In the model on the right (b), mutation on the Thr185 to Ala results in the loss of the hydrogen bond between this amino acid and Asp183.
TABLE 3.
Release of 3-MH and 4-MMP by 80 wine yeast strains in SGJ-thiol medium according to their IRC7 genotype
| Genotypea | Amino acid in position 185b | No. of strains | 3-MH (ng·liter−1)c | 4-MMP (ng·liter−1)c |
|---|---|---|---|---|
| IRC7S IRC7S | T185 | 43 | 495 ± 169 C | 324 ± 87 C |
| IRC7S IRC7L | 20 | 755 ± 330 | 1,107 ± 973 | |
| T185 | 5 | 1,074 ± 229 B | 2,505 ± 960 B | |
| T185/A185 | 14 | 635 ± 297 C | 666 ± 295 C | |
| A185 | 1 | 840 BC | 275 C | |
| IRC7L IRC7L | 17 | 1,236 ± 764 | 2,027 ± 1,925 | |
| T185 | 5 | 2,158 ± 388 A | 4,618 ± 930 A | |
| T185/A185 | 2 | 1,326 ± 448 B | 2,109 ± 719 B | |
| A185 | 10 | 757 ± 468 BC | 715 ± 708 C |
IRC7S indicates the short allele of IRC7, while IRC7L indicates the long or full-length allele of IRC7.
In the IRC7S IRC7L group, the presence of an alanine in position 185 (A185) is associated with IRC7L.
Means with the same letter are not significantly different from each other (Tukey’s HSD test, P < 0.05).
Functional in vitro characterization of variant IRC7L alleles.
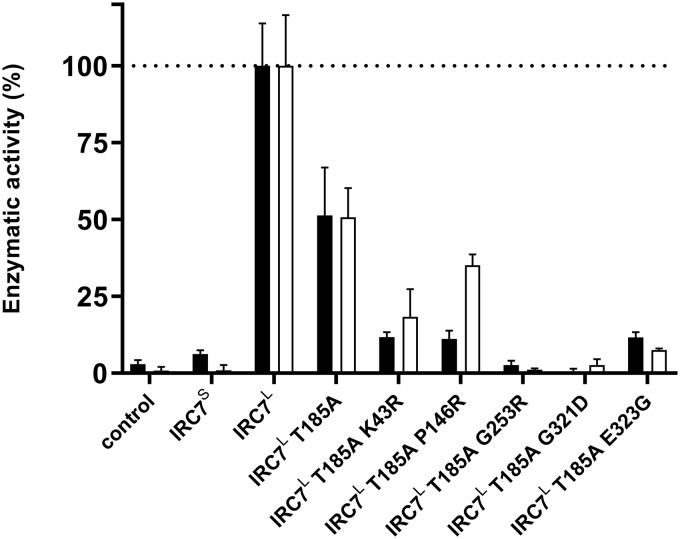
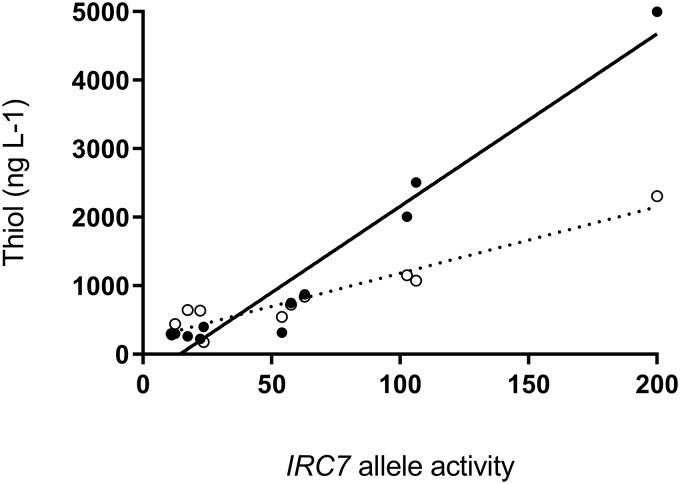
While none of the additional observed SNPs induced changes to protein sequences near functional sites, they may explain why some of the strains in the IRC7L IRC7L group produced levels of both thiols similar to IRC7S IRC7S strains. To investigate whether this was the case, each of the most common SNPs was introduced individually in a multicopy plasmid containing IRC7L (pHVX2-IRC7L), and these were transformed into BY4742 ΔIRC7. Then, activity of protein extracts with the physiological substrate cysteine and with Cys-4-MMP was assayed (Fig. 3). Activity against Cys-3-MH was too small to be quantified by enzymatic methods. For both assayed substrates, the T185A mutation (pHVX2-IRC7L T185A) resulted in a 50% decrease in the activity of the protein. The presence of additional mutations in the IRC7L T185A construct resulted in further decreases in activity. In particular, the G253R and G321D mutations completely abolished the activity of Irc7p, while some residual activity could still be observed for the K43R, P146R, and E323G mutations. Enzymatic activity of the constructed IRC7 alleles against Cys-4-MMP (Fig. 4) was highly correlated with end-of-ferment thiol concentrations for the corresponding diploid wine strains that exhibited the same alleles (R2 = 0.95, P < 0.0001).
FIG 3.
Enzymatic activity toward Cys-4-MMP (black) and cysteine (white) in protein extracts from BY4742 ΔIRC7 yeast cells overexpressing the multicopy plasmid pHVX2 with different alleles of IRC7. Each result is expressed as the mean of three independent replicates. For both substrates, activity was expressed as a percentage relative to cells harboring the pHVX2-IRC7L construct (100%) overexpressing the long or full-length version of IRC7. Enzymatic activity for this allele was found to be 214 ± 36 and 89.6 ± 12.4 nmol·min−1 µg·prot−1 for cysteine and Cys-4-MMP respectively.
FIG 4.
Correlation between the activity of combinations of different alleles of IRC7 and the release of 4-MMP (filled circles) and 3-MH (empty circles) in SGJ-thiol medium by diploid strains exhibiting these alleles. Activity was evaluated in protein extracts in triplicate from cells overexpressing different alleles of IRC7 using Cys-4-MMP as the substrate. The IRC7L allele was given an arbitrary value of 100, and diploid strains with the IRC7L IRC7L genotype were given a value of 200. The correlation coefficient (R2) for 4-MMP and 3-MH was found to be 0.95 and 0.90, respectively.
Impact of variant IRC7L alleles on VSC and thiol formation during wine fermentation.
To confirm the role of IRC7 variant alleles in modulating formation of VSCs during fermentation, each was integrated into the haploid wine strain AWRI1631 at the IRC7 chromosomal location, under the control of the native promoter. Replacing the native IRC7S allele of AWRI1631 with IRC7L resulted in a dramatic increase in the levels of 4-MMP (22-fold) after fermentation of SGJ-thiol, while more modest increases were observed for both 3-MH and 3-MHA (3- and 6-fold, respectively) (Table 4). The conversion yields for the AWRI1631 IRC7L strain were found to be 12.1% and 0.78%, for 4-MMP and 3-MH, respectively. The restoration of the fully active Irc7pL also caused an increase in the levels of H2S released during fermentation. In this background, the T185A mutation significantly decreased the amounts of 4-MMP and 3-MH (62% and 42% less) released relative to those released by the AWRI1631 IRC7L strain. In combination with T185A, the G253R mutation had the greatest impact upon formation of polyfunctional thiols, which resulted in an IRC7L allele that functioned equivalently to the IRC7S allele.
TABLE 4.
Effect of chromosomal integration of different IRC7 alleles in the haploid strain AWRI1631 in sulfur compound formation during fermentation of 90 ml of SGJ-thiol medium
| AWRI1631 construct | Sulfur compounda
|
|||
|---|---|---|---|---|
| H2S tube (µg) | 4-MMP (ng·liter−1) | 3-MH (ng·liter−1) | 3-MHA (ng·liter−1) | |
| No yeast | ND | 107 | ND | ND |
| IRC7S | 122 BC | 662 G | 324 F | 9.6 E |
| IRC7L | 178 A | 14,672 A | 961 A | 61.4 A |
| IRC7L T185A | 135 B | 5,469 B | 564 C | 28.2 C |
| IRC7L T185A K43R | 121 BC | 4,549 D | 563 C | 25.1 CD |
| IRC7L T185A P146R | 121 BC | 3,559 E | 531 CD | 20.6 DE |
| IRC7L T185A G253R | 94 C | 652 G | 367 EF | 7.6 F |
| IRC7L T185A G321D | 116 BC | 2,303 F | 453 DE | 16.5 E |
| IRC7L T185A E323G | 123 BC | 4,849 C | 740 B | 40.8 B |
Each results is expressed as the mean of three independent replicates; standard deviations were typically 10% and never exceeded 20%. The conversion yields of the AWRI1631 IRC7L strain for Cys-3-MH and Cys-4-MMP were 0.78% and 12.1%, respectively. H2S released during fermentation (H2S tube) was measured with silver nitrate selective gas detector tubes. Means with the same letter are not significantly different from each other (Tukey’s HSD test, P < 0.05). ND, not detected.
Since we observed a significant increase in the levels of H2S released throughout fermentation by restoration of Irc7p activity, we investigated the effect of the three main alleles of IRC7 on the formation of other VSCs. We used increasing concentrations of cysteine to enhance the possible impact of Irc7p on the formation of sulfur compounds, since it has a relatively high Km for cysteine (3.8 mM, 460 mg·liter−1) (32). We observed a general increase in the formation of the different VSCs analyzed with increasing concentrations of cysteine, particularly for H2S released during fermentation (H2S tube) and ethyl thioacetate (EtSAc) (Table 5). The magnitude of these changes was also dependent upon the allele of IRC7 tested. Thus, the presence of the IRC7L allele was responsible for significant increases in the formation of the different sulfur compounds, while fermentation with the IRC7L T185A allele displayed an intermediate phenotype between the short and full-length versions of IRC7. Differences between the different alleles were more noticeable for H2S tube, H2S, and EtSAc for the whole range of concentration of cysteine, and in the case of ethanethiol (EtSH), methyl thioacetate (MeSAc), and carbon disulfide (CS2), only for the highest concentration of cysteine. On the other hand, the formation of methanethiol (MeSH) was less responsive to the levels of cysteine or the allele of IRC7 used.
TABLE 5.
Effect of chromosomal integration of three IRC7 alleles in the haploid strain AWRI1631 in VSC formation during fermentation of 90 ml of a synthetic grape juice (SGJ) supplemented with different concentrations of cysteine
| Medium | AWRI1631 construct | VSC (µg·liter−1)a
|
H2S tube (µg)b | |||||
|---|---|---|---|---|---|---|---|---|
| H2S | MeSH | EtSH | CS2c | MeSAc | EtSAc | |||
| SGJ-Cys 17 mg·liter−1 | IRC7S | 2.6 ± 0.3 B | 4.6 ± 0.5 A | <LOD | <LOD | 12.3 ± 0.7 A | 1.6 ± 0.1 B | 118 ± 8 B |
| IRC7L T185A | 2.1 ± 0.4 B | 4.0 ± 0.5 A | <LOD | <LOD | 16.8 ± 2.6 A | 2.3 ± 0.6 AB | 132 ± 9 B | |
| IRC7L | 4.0 ± 1.0 A | 4.7 ± 0.6 A | <LOD | <LOD | 16.9 ± 1.7 A | 2.6 ± 0.3 A | 175 ± 28 A | |
| SGJ-Cys 50 mg·liter−1 | IRC7S | 3.7 ± 0.6 B | 4.7 ± 0.7 A | <LOD | <LOD | 17.2 ± 3.6 A | 3.7 ± 1.1 B | 140 ± 11 C |
| IRC7L T185A | 7.6 ± 2.6 AB | 4.1 ± 0.4 A | <LOD | <LOD | 20.1 ± 1.2 A | 6.8 ± 1.8 AB | 228 ± 19 B | |
| IRC7L | 7.5 ± 1.7 A | 4.3 ± 0.8 A | <LOD | <LOD | 21.0 ± 2.8 A | 9.4 ± 2.6 A | 326 ± 58 A | |
| SGJ-Cys 200 mg·liter−1 | IRC7S | 11.6 ± 1.2 C | 4.5 ± 0.4 B | 0.5 ± 0.1 C | <LOD | 14.7 ± 1.5 B | 7.3 ± 0.7 C | 280 ± 22 C |
| IRC7L T185A | 20.9 ± 3.3 B | 5.5 ± 1.0 AB | 1.9 ± 0.3 B | <LOD | 23.2 ± 1.2 B | 24.8 ± 2.7 B | 630 ± 90 B | |
| IRC7L | 51.8 ± 3.0 A | 8.6 ± 2.3 A | 8.4 ± 2.9 A | 1.2 ± 0.3 | 45.5 ± 8.3 A | 61.8 ± 13.5 A | 1,315 ± 221 A | |
Results are expressed as the mean of three independent replicates. Means with the same letter are not significantly different from each other (Tukey’s HSD test, P < 0.05).
H2S released during fermentation (H2S tube) was measured with silver nitrate selective gas detector tubes.
LOD, limit of detection.
DISCUSSION
Previous research identified Irc7p as being primarily responsible for release of the polyfunctional thiol 4-MMP from its cysteinylated precursor during fermentation (27), and subsequent work linked variation between strains in this capacity to the presence or absence of the full-length version of this protein (26, 30, 31). Overexpression of IRC7 was also recently implicated in production of H2S in a solid medium containing cysteine (32). In this study, we have expanded upon this knowledge by providing evidence that many wine strains harbor a variety of mutations in IRC7 that can have a profound effect on the activity of the encoded protein. This not only alters the capacity of these strains to release polyfunctional thiols from their cysteinylated-conjugated precursors but also their ability to produce other VSCs during wine fermentation.
The most common IRC7 SNP observed across the 179 wine yeasts studied was the substitution of a threonine in position 185 with alanine, which was present in around 80% of the strains with at least one copy of IRC7L. Thr185 is a highly conserved amino acid in pyridoxal-5′-phosphate (PLP)-dependent carbon-sulfur lyases, not only in fungi but also in prokaryotes and mammals (37). The hydroxyl group of Thr185 forms a strong hydrogen bond with the carboxylate group of Asp183, thereby tethering Asp183 in an optimal position to interact with the pyridine nitrogen of the PLP cofactor (37). As a result of the mutation to Ala, the hydrogen bond is then lost. This mutation results in a 50% loss of enzymatic activity, which corresponds to a similar decrease in the formation of the positive and negative VSCs during fermentation of a synthetic grape juice. In our model of the catalytic Irc7pL dimer, the other common and deleterious mutations linked to the IRC7L T185A allele (K43R, P146R, G253R, G321D, and E323G), were not located near the active site of the protein. Irc7pL forms a homotetramer (32), as do other enzymes of the family of cysteine/methionine metabolism PLP-dependent enzymes (37, 38). Gly253 is a highly conserved amino acid in this family, and in the sequence is located next to an Arg252, which is involved in tetramer contact (37). Therefore, the mutation of the small and neutral Gly253 to Arg might create a disruption in tetramer formation due to the nearby presence of two positively charged amino acids.
It is striking that, in addition to the previously described prevalence of the inactive IRC7S allele among wine strains, a significant percentage of those with one or two copies of the IRC7L allele harbor other inactivating mutations. This indicates that S. cerevisiae has a strong selective pressure toward the inactivation of Irc7p, at least among wine strains, whereas other members of the Saccharomyces sensu stricto group have the full-length version of the gene (32). IRC7 shows evidence of being horizontally transferred into an ancestral yeast from an unknown bacterium related to Yersinia pestis (39) prior to the whole-genome duplication event. Only a few other yeast species from the Zygosaccharomyces, Lachancea, and Torulaspora genera harbor IRC7 orthologs. This suggests that among descendants of the ancestral yeast that acquired IRC7, there appear to be many lineages where this gene has been lost altogether. Overall, it seems reasonable to speculate that Irc7p carbon-sulfur β-lyase activity confers benefit to some species/strains, but not others.
It is important to note that most functional studies have been conducted by deletion of IRC7S in different S. cerevisiae laboratory strain backgrounds (33–35). Even though we, and others, have shown that the Irc7pS has no activity toward substrates such as cysteine or Cys-4-MMP, the truncated protein and/or the genomic locus itself may be functionally important in yeast. Indeed, deletion of the short IRC7 allele in the diploid laboratory strain W303 increased the levels of mitotic recombination (33). Reduced risk of deleterious recombination events may explain both the common occurrence of inactivating truncations/mutations among wine strains and the absence of naturally occurring structural variants, based upon the assumption that Irc7pL activity has some fitness cost under environmental conditions these yeasts encounter.
What might these conditions be? IRC7 is under nitrogen catabolite repression (NCR) control, and increased transcription of IRC7 is observed in poor nitrogen sources, such as methionine or proline (40). For this reason, complementation of grape must with an optimal nitrogen source, such as ammonium, decreases 3-MH production (25). Yeasts existing in environments lacking preferred nitrogen sources would benefit from cysteine desulfhydrase activity of Irc7pL as a means to recover ammonium, so this seems an unlikely driver of the observed allelic variation in S. cerevisiae wine yeasts.
IRC7 is not only regulated by NCR but also by the copper-sensing transcription factor Mac1p, which activates the expression of various genes, including IRC7, under copper-deficient conditions. Conversely, IRC7 expression is inhibited under high copper conditions (41). The physiological explanation for IRC7 being regulated by copper concentrations has not been elucidated but might be related to its putative biological role in cysteine homeostasis (32). In response to high concentrations of copper, transcription of the cysteine-rich metallothionein CUP1 is heavily induced (41). Cup1p acts to sequester excess copper, which is toxic (42). Under these conditions, it might be necessary to repress the cysteine desulfhydrase activity of Irc7p to ensure that sufficient intracellular cysteine is available for synthesis of Cup1p in high quantities. One of the well-described domestication signatures of wine yeasts is tolerance to copper (43), which is a component of common vineyard sprays and is used during winemaking to sequester negative VSCs (1). It is possible that through long-term exposure to copper, wine strains of S. cerevisiae have accumulated IRC7 mutations as a means to ensure cysteine availability in order to respond to acute copper exposure.
Regardless of the evolutionary drivers for Irc7p inactivation, allelic variation among S. cerevisiae strains at the IRC7 locus has profound impact upon winemaking outcomes. In our experimental fermentation conditions, in which we spiked a synthetic grape juice with grape-like concentrations of both cysteinylated precursors of 4-MMP and 3-MH, the IRC7 genotype seemed to explain much of the variation observed between wine strains for the release of these fruity thiols. Both our allele replacement experiments in the haploid AWRI1631 background and our survey of 80 wine yeast strains found that Irc7pL was responsible for the release of at least 95% of 4-MMP from the Cys-4-MMP precursor during fermentation, confirming previous results (27). In the case of 3-MH, our results show that Irc7pL is responsible for a significant part of the 3-MH released from Cys-3-MH, ranging from 66% in the haploid background to approximately 75% in the screening of strains. When combined with enzymatic assays, our results agree with previous studies (27, 28) that Irc7pL is more active with Cys-4-MMP than with Cys-3-MH. All this evidence points toward other yeast β-lyases playing a small part in the release of 3-MH from its cysteinylated precursor. In this sense, Str3p has been shown to catalyze the release of 3-MH from Cys-3-MH (28).
The potential for contribution of cysteine as a precursor to VSCs in wine is generally disregarded, as other more abundant sources of sulfur exist. Nevertheless, we observed differences in H2S and EtSAc concentrations according to IRC7 allele during fermentation of model grape juice with cysteine at levels observed in natural musts (44). These observations were consistent with those of Kinzurik et al. (45), who demonstrated the role of H2S made by yeast during fermentation in the formation of EtSH and EtSAc. Differences in VSC concentration according to the IRC7 allele were exacerbated when the concentration of cysteine was increased to 50 and 200 mg liter−1, which led to a general increase of most of the VSCs analyzed.
The cysteine-containing tripeptide GSH is naturally present in grape juice in concentrations ranging from 1 up to 102 mg liter−1 (46), and the International Organization of Vine and Wine (OIV) has recently allowed the addition of GSH to must or wine (20 mg liter−1). In this context, research has shown that supplementation of must with GSH enhances the production of H2S and other negative VSCs, which increases the intensity of sulfidic off flavors in wine (47, 48). Another source of GSH is nutrients that are commonly added to active dry yeast rehydration medium to improve yeast fermentation performance, which might act as a source for H2S production during fermentation (49). The mechanisms of H2S release due to GSH addition are not yet understood, but it has been suggested that this might occur via degradation of GSH to the individual amino acids and by subsequent degradation of cysteine to H2S by cysteine desulfhydrase activity (49, 50). With this in mind, the relative impact of the wine yeast IRC7 genotype in modulating VSC formation might be most important when the concentrations of cysteine and/or GSH are elevated, in combination with low-nitrogen or low-phenolic grape musts (47, 48).
Different wine yeast strains produce divergent profiles of aroma compounds, and the choice of a yeast strain can have a significant impact on the final flavor and aroma of wine. In this study, we have comprehensively demonstrated the vast range of thiol-releasing capacities across a large number of wine yeasts. Furthermore, we have identified new molecular markers that can be used to predict this capacity. We also provide further evidence of the intrinsic link between polyfunctional thiol release and the potential for production of negative VSCs from organic sulfur precursors. This knowledge will guide strain development and choice of yeast to balance these flavor impacts.
MATERIALS AND METHODS
Chemicals.
Analytical reagents were purchased from Sigma-Aldrich (Castle Hill, NSW, Australia) unless otherwise indicated. 4-Mercapto-4-methylpentan-2-one (4-MMP) and S-4-(4-methylpentan-2-one)-l-cysteine (Cys-4-MMP) were synthesized as described previously (10), and [2H10]4-MMP and 3-mercaptohexan-1-ol (3-MH), 3-mercaptohexyl acetate (3-MHA), and [2H5]-3-MHA were also synthesized as described previously (51). The synthesis of [2H10]3-MH was carried out according to a previously described method (52). S-3-(Hexan-1-ol)-l-cysteine (Cys-3-MH), as a mixture of diastereomers, was prepared as described previously (53).
Microorganisms and culture conditions.
All yeast strains were obtained from The Australian Wine Research Institute (AWRI) culture collection. Yeast cultures were maintained on solid yeast extract-peptone-dextrose (YPD) agar plates (2% glucose, 2% peptone, 1% yeast extract, and 2% agar).
DNA manipulation, genome analysis and construction of plasmids.
Standard procedures for the isolation and manipulation of DNA were used (54). Primers used in this study as summarized in Table 6. Genomic DNA from yeast strains was isolated using the Gentra Puregene Yeast/Bact. kit B (Qiagen, Australia).
TABLE 6.
Primers used in this study
| Name | Commentsb | Sequence (5′–3′)a |
|---|---|---|
| TC1 | Cloning IRC7S or IRC7L using EcoRI (underlined), forward | GACTGAATTCATGATTGATCGTACCGAGTTATCG |
| TC2 | Cloning IRC7S using XhoI (underlined), reverse | TAGCCTCGAGCTAGCCACCCCATGAAAATCCCA |
| TC3 | Cloning IRC7L using XhoI (underlined), reverse | GACTCTCGAGTCAGATTTGTAAAGGATTCAAAG |
| TC4 | Sequencing primer (pHVX2 plasmid), forward | TCAAACAGAATTGTCCGAATCG |
| TC5 | Sequencing primer (pHVX2 plasmid), reverse | CCGAACATAGAAATATCGAATGG |
| TC6 | Site-directed mutagenesis (IRC7L K43R), forward | GGGGTCAACCATCATTCTTAgAAAACTTAGTGATTTAGAAC |
| TC7 | Site-directed mutagenesis (IRC7L M106I), forward | GCTGGTGATCATATCTTGATtACTGATAGTGTCTACGTGC |
| TC8 | Site-directed mutagenesis (IRC7L P146R), forward | GATATAGAAAAACTAGTTAAGCgAAATACAACCGTCATTTTCCTC |
| TC9 | Site-directed mutagenesis (IRC7L G253R), forward | GTCAATTAGCATTGCGAaGAATGCGTACATTGCAC |
| TC10 | Site-directed mutagenesis (IRC7L W274R), forward | CCTGGATTTGGCTGCTcGGCTCGGAAATCGAGAT |
| TC11 | Site-directed mutagenesis (IRC7L G321D), forward | GAGGAAAATGACGGTTGTATTTcGCTTAACTAGTTTTTCTATATC |
| TC12 | Site-directed mutagenesis (IRC7L E323G), forward | CACAAGAGCTGGTCTGGgGAAAATGGTAGAAGGGA |
| TC13 | Deletion of IRC7 using CORE cassette, forward | ATGATTGATCGTACCGAGTTATCGAAGTTTGGTATTACTACGCAACTGTCgagcccccgatttagagctt |
| TC14 | Deletion of IRC7 using CORE cassette, reverse | TCAGATTTGTAAAGGATTCAAAGAAATTTCTTCTGCGAGACGTTCAAAGCCctcgaggtcgacggtatcgat |
| TC15 | Confirmation deletion IRC7 (5′-UTR IRC7), forward | ATTGTCGCCTCTTGTTTTGAG |
| TC16 | Confirmation deletion IRC7 (5′), reverse | AAGACTCTCCTCCGTGCGTC |
| TC17 | Confirmation deletion IRC7 (3′-UTR IRC7), reverse | GGAGTGAGATCACATGTCATATTG |
| TC18 | Confirmation deletion IRC7 (3′), forward | TGAGCATGCCCTGCCCCTA |
For primers TC6 through TC12, the reverse primers used for site-directed mutagenesis are the reverse complement of the corresponding forward primers; the nucleotide to be changed is indicated in lowercase in the sequence. For primers TC13 through TC14, the sequence annealing the CORE cassette of the pCORE5 plasmid is indicated in lowercase.
UTR, untranscribed region.
Detection of IRC7 short and long alleles, as well as of single nucleotide polymorphisms (SNPs) in its open reading frame (ORF), was performed by visualizing read alignments in a subset of 179 sequenced wine yeast strains (BioProject accession number PRJNA303109), data aligned to the S288c strain genome, and using the Integrated Genomics Viewer (IGV) (63). The presence of the long or short allele of IRC7 was also confirmed by PCR as described previously (27).
The IRC7 ORF was amplified by PCR using Phusion High-Fidelity DNA polymerase (New England Biolabs), with genomic DNA from the commercial wine strains AWRI838 and AWRI1688 and laboratory strain BY4742 (MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0) as the templates. Primers TC1 and TC2 were used to amplify IRC7 short version (IRC7S) from BY4742 (1,023 bp), while primers TC1 and TC3 were used to amplify IRC7 long or full-length version (IRC7L) from both wine strains (1,203 bp); the restriction sites EcoRI (5′) and XhoI (3′) were introduced in the PCR. The fragments were purified and digested, and then cloned into the EcoRI and XhoI sites of the S. cerevisiae expression plasmid pHVX2 (55) to yield the following constructs: pHVX2-IRC7S, pHVX2-IRC7L, and pHVX2-IRC7L T185A (Table 7).
TABLE 7.
Strains and plasmids generated in this study
| IRC7 allele (donor strain) | Mutation(s) | Strainb | Plasmidc |
|---|---|---|---|
| IRC7S (BY4742) | 1023_1060dela | AWRI1631 IRC7S | pHVX2-IRC7S |
| IRC7L (AWRI838) | AWRI1631 IRC7L | pHVX2-IRC7L | |
| IRC7L T185A (AWRI1688) | 553A→G | AWRI1631 IRC7L T185A | pHVX2-IRC7L T185A |
| IRC7L T185A K43R (AWRI1493) | 128A→G, 553A→G | AWRI1631 IRC7L T185A K43R | pHVX2-IRC7L T185A K43R |
| IRC7L T185A P146R (AWRI722) | 437C→G, 553A→G | AWRI1631 IRC7L T185A P146R | pHVX2-IRC7L T185A P146R |
| IRC7L T185A G253R (AWRI1899) | 553A→G, 757G→A | AWRI1631 IRC7L T185A G253R | pHVX2-IRC7L T185A G253R |
| IRC7L T185A G321D (AWRI723) | 553A→G, 962G→A | AWRI1631 IRC7L T185A G321D | pHVX2-IRC7L T185A G321D |
| IRC7L T185A E323G (AWRI935) | 553A→G, 968A→G | AWRI1631 IRC7L T185A E323G | pHVX2-IRC7L T185A E323G |
The deletion of 38 bp in the IRC7S allele results in a protein of 340 amino acids (Gly340*), truncated compared to the full-length allele (IRC7L), which encodes a protein of 400 amino acids.
IRC7 alleles from different donor strains were integrated at the IRC7 chromosomal location of the haploid wine strain AWRI1631.
The multicopy episomal plasmid pHVX2 (55) was used to overexpress IRC7 alleles from different donor strains, under the control of the constitutive PGK1 promoter.
In order to introduce some of the most common SNPs found in IRC7 alleles from different wine yeast strains, the QuikChange site-directed mutagenesis kit (Agilent, Mulgrave, Australia) was used with the pHVX2-IRC7L T185A plasmid as the template (Table 7). For each of the IRC7 SNPs, both mutagenesis primers were designed so they contained the desired mutation and were annealed to the same sequence on opposite strands of the plasmid (primers TC6 to TC12). The appropriate substitutions, as well as the absence of unwanted mutations, were confirmed by Sanger sequencing using primers TC4 and TC5.
Chromosomal integration of IRC7 alleles in AWRI1631.
Different IRC7 alleles were inserted into the haploid wine yeast strain AWRI1631 via homologous recombination (54) to replace the corresponding IRC7 wild-type locus (IRC7S) so that no extraneous DNA was left behind (Table 7). First, the CORE-ΔIRC7 cassette (3 kb) was amplified from the pCORE5 plasmid (56) by PCR using Phusion DNA polymerase and primers TC13 and TC14. These primers were designed so they have at their 3′ end 20 bp complementary to flanking sequences of the CORE cassette and, at their 5′ end, 50 bp complementary to either the start (TC13) or the end (TC14) of the IRC7 coding regions. The amplified CORE-ΔIRC7 cassette was then purified and transformed into AWRI1631, and positive transformants were selected in solid medium containing 100 mg·liter−1 of nourseothricin (clonNAT; Werner BioAgents, Germany). Confirmation of the replacement of the AWRI1631 IRC7 wild-type locus by the CORE-ΔIRC7 cassette was confirmed by PCR using primers TC15 and TC16 (5′ integration), and TC17 and TC18 (3′ integration), yielding the AWRI1631-ΔIRC7-CORE strain.
Then, a fragment of 1.9 kb, encompassing the IRC7 open reading frame plus a fragment of 280 bp before the start codon and one of 300 bp after the stop codon was amplified from genomic DNA of several strains of interest (Table 7) by PCR using Phusion DNA polymerase and primers TC15 and TC17. Positive transformants were selected in galactose plates, and integration of the each of the IRC7 alleles was confirmed by PCR and Sanger sequencing.
Laboratory-scale fermentation in a synthetic grape juice.
Laboratory-scale fermentations were performed in triplicate in a synthetic grape juice (SGJ) medium (21) with the following variations: sugar content was 100 g·liter−1 glucose and 100 g·liter−1 fructose, and 250 mg·liter−1 yeast-assimilable nitrogen was added. The concentrations of the sulfur-containing amino acids methionine and cysteine were set to 6 and 17 mg·liter−1, respectively. Depending on the experiment, SGJ was supplemented with additional amounts of cysteine, so the total concentration of cysteine was 50 or 200 mg·liter−1; or with 100 µg liter−1 of the cysteinylated precursors Cys-4-MMP and Cys-3-MH (SGJ-thiol). SGJ was filtered through 0.22 μm Stericup filters (Millipore). Yeast starter cultures were prepared by growing cells aerobically in YPD medium for 24 h to the stationary phase at 28°C. Then 1 × 106 cells ml−1 were inoculated into 50% diluted SGJ medium and grown for another 24 h at 22°C. The acclimatized cells were inoculated into 90 ml of SGJ at a density of 1 × 106 cells ml−1. Fermentations were conducted at 22° in 100-ml Schott bottles fitted with stir bars and stirred at 200 rpm using a magnetic stirrer. The lids of the Schott bottles were fitted with H2S detector tubes (Air-Met Scientific, South Australia) to measure the release of H2S during fermentation (H2S tube). Fermentation progress was followed by CO2 weight loss, measured every 24 h. After fermentation, the ferments were cold settled at 4°C for 5 days and then sampled for VSC and nonvolatile compound analysis.
Assay of carbon–sulfur β-lyase activity.
Strains were grown overnight in YPD medium at 28°C, and a volume of 8 ml of the culture was centrifuged for 2 min at 4,000 × g. Cells were washed twice with water, and the pellet was resuspended in 400 µl of cold buffer containing 100 mM HEPES (pH 7.5), 20 µM pyridoxal-5′-phosphate (PLP), 200 µM EDTA, 10% glycerol, and the protease inhibitors leupeptin (2 µg ml−1) and PMSF (1 mM). Glass beads were added to the suspension, and cells were repeatedly vortexed for 30 s, then allowed to rest for another 30 s, for a total of 10 min at 4°C. The suspension was centrifuged for 30 min at 16,000 × g at 4°C, and the supernatant was used for enzyme assays. Protein concentrations were measured using the Bio-Rad protein assay (catalog number 5000006), with bovine serum albumin as a standard.
Reactions were carried out at 30°C in a total volume of 200 µl in 96-well microplates (UV-Star UV-transparent microplates; Greiner Bio-One) containing 100 mM HEPES buffer (pH 7.5), 20 µM PLP, 25 µM EDTA, and 2 mM concentration of the substrates l-cysteine or Cys-4-MMP. The reaction was started by adding 10 µg of the protein extracts. The release of pyruvate as a result of β-lyase activity (57) was analyzed via linked enzymatic conversion to lactate by l-lactic dehydrogenase enzyme (5 U µl−1). Concomitant conversion of NADH (400 µM) to NAD+ by this enzyme was monitored by measurement of absorbance at 340 nm (molar absorptivity = 6,220 liter mol−1) every 5 min for 60 min.
Analysis of principal nonvolatile compounds.
The concentrations of sugars, ethanol, glycerol, and organic acids were measured by high-performance liquid chromatography using a Bio-Rad HPX-87H column (7).
Analysis of the thiols 3-MH, 3-MHA, and 4-MMP.
Analysis of 3-MH and 4-MMP occurred once the ferments achieved dryness and were clarified by centrifugation (2,500 × g for 5 min) to remove yeast cells. An aliquot (20 ml) of the clarified ferment samples was derivatized with 4,4′-dithiodipyridine, followed by solid-phase extraction (SPE) and analysis by high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) as described previously (58). Extracts were stored at −20°C until analysis.
Analysis of volatile sulfur compounds.
The volatile sulfur compounds (VSCs) H2S, methanethiol (MeSH), dimethyl sulfide, diethyl sulfide, dimethyl disulfide, diethyl disulfide, ethanethiol (EtSH), carbon disulfide (CS2), methyl thioacetate (MeSAc), ethyl thioacetate (EtSAc), were determined using an Agilent 355 sulfur chemiluminescence detector coupled to an Agilent 6890A gas chromatograph (Forest Hill, VIC, Australia). The analysis was carried out as described previously (59). H2S released during fermentation was detected in the headspace using silver nitrate selective gas detector tubes (Komyo, Kitagawa, Japan) (49).
Statistical analysis.
Minitab 17 statistical software (Minitab Inc., PA) was used for statistical analysis. Data were analyzed by one-way analysis of variance (ANOVA) using Tukey’s honestly significant difference (HSD) test (P < 0.05), and P values were determined by a two-tailed Student's t test.
Irc7p protein modeling.
Irc7p7 was modelled using the structures for Escherichia coli cystathionine β-lyase (60) (PDB accession number 1CL2) and Trichomonas vaginalis methionine γ-lyase (PDB accession number 1E5F) as comparative references. The A and B chains of 1CL2 were superposed to 1E5F. The sequences for Irc7p, 1CL2, and 1E5F were aligned with Clustal X (http://www.clustal.org/clustal2). Ten models were generated with UCSF Modeller v9.15 (61) and evaluated with PROCHECK (62). UCSF Chimera v1.10.2 (http://www.cgl.ucsf.edu/chimera) was used for visualizing models and modeling mutations.
Data availability.
The best model of Irc7p7 was uploaded to the Protein Model DataBase under accession number PM0081794.
Supplementary Material
ACKNOWLEDGMENTS
The Australian Wine Research Institute (AWRI), a member of the Wine Innovation Cluster in Adelaide, is supported by Australia’s grape growers and winemakers through their investment body, Wine Australia, with matching funds from the Australian Government.
We thank Simon Schmidt for his valuable help reviewing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02684-18.
REFERENCES
- 1.Swiegers JH, Pretorius IS. 2007. Modulation of volatile sulfur compounds by wine yeast. Appl Microbiol Biotechnol 74:954–960. doi: 10.1007/s00253-006-0828-1. [DOI] [PubMed] [Google Scholar]
- 2.Darriet P, Tominaga T, Lavigne V, Boidron J-N, Dubourdieu D. 1995. Identification of a powerful aromatic component of Vitis vinifera L. var. sauvignon wines: 4-mercapto-4-methylpentan-2-one. Flavour Fragr J 10:385–392. doi: 10.1002/ffj.2730100610. [DOI] [Google Scholar]
- 3.Tominaga T, Furrer A, Henry R, Dubourdieu D. 1998. Identification of new volatile thiols in the aroma of Vitis vinifera L. var. Sauvignon blanc wines. Flavour Fragr J 13:159–162. doi:. [DOI] [Google Scholar]
- 4.Tominaga T, Darriet P, Dubourdieu D. 1996. Identification de l’acétate de 3-mercaptohexanol, composé à forte odeur de buis, intervenant dans l’arôme des vins de Sauvignon. Vitis 35:207–210. [Google Scholar]
- 5.Capone DL, Barker A, Williamson PO, Francis IL. 2018. The role of potent thiols in Chardonnay wine aroma. Aust J Grape Wine Res 24:38–50. doi: 10.1111/ajgw.12294. [DOI] [Google Scholar]
- 6.Smith ME, Bekker MZ, Smith PA, Wilkes EN. 2015. Sources of volatile sulfur compounds in wine. Aust J Grape Wine Res 21:705–712. doi: 10.1111/ajgw.12193. [DOI] [Google Scholar]
- 7.Cordente AG, Heinrich A, Pretorius IS, Swiegers JH. 2009. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res 9:446–459. doi: 10.1111/j.1567-1364.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- 8.Linderholm A, Dietzel K, Hirst M, Bisson LF. 2010. Identification of MET10-932 and characterization as an allele reducing hydrogen sulfide formation in wine strains of Saccharomyces cerevisiae. Appl Environ Microbiol 76:7699–7707. doi: 10.1128/AEM.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang C, Roncoroni M, Gardner RC. 2014. MET2 affects production of hydrogen sulfide during wine fermentation. Appl Microbiol Biotechnol 98:7125–7135. doi: 10.1007/s00253-014-5789-1. [DOI] [PubMed] [Google Scholar]
- 10.Howell KS, Swiegers JH, Elsey GM, Siebert TE, Bartowsky EJ, Fleet GH, Pretorius IS, de Barros Lopes MA. 2004. Variation in 4-mercapto-4-methyl-pentan-2-one release by Saccharomyces cerevisiae commercial wine strains. FEMS Microbiol Lett 240:125–129. doi: 10.1016/j.femsle.2004.09.022. [DOI] [PubMed] [Google Scholar]
- 11.Swiegers JH, Kievit RL, Siebert T, Lattey KA, Bramley BR, Francis IL, King ES, Pretorius IS. 2009. The influence of yeast on the aroma of Sauvignon blanc wine. Food Microbiol 26:204–211. doi: 10.1016/j.fm.2008.08.004. [DOI] [PubMed] [Google Scholar]
- 12.Dubourdieu D, Tominaga T, Masneuf I, Peyrot Des Gachons C, Murat ML. 2006. The role of yeasts in grape flavor development during fermentation: the example of Sauvignon blanc. Am J Enol Vitic 57:81. [Google Scholar]
- 13.King ES, Swiegers JH, Travis B, Francis IL, Bastian SE, Pretorius IS. 2008. Coinoculated fermentations using Saccharomyces yeasts affect the volatile composition and sensory properties of Vitis vinifera L. cv. Sauvignon blanc wines. J Agric Food Chem 56:10829–10837. doi: 10.1021/jf801695h. [DOI] [PubMed] [Google Scholar]
- 14.Belda I, Ruiz J, Beisert B, Navascues E, Marquina D, Calderon F, Rauhut D, Benito S, Santos A. 2017. Influence of Torulaspora delbrueckii in varietal thiol (3-SH and 4-MSP) release in wine sequential fermentations. Int J Food Microbiol 257:183–191. doi: 10.1016/j.ijfoodmicro.2017.06.028. [DOI] [PubMed] [Google Scholar]
- 15.Anfang N, Brajkovich M, Goddard MR. 2009. Co-fermentation with Pichia kluyveri increases varietal thiol concentrations in Sauvignon blanc. Aust J Grape Wine Res 15:1–8. doi: 10.1111/j.1755-0238.2008.00031.x. [DOI] [Google Scholar]
- 16.Renault P, Coulon J, Moine V, Thibon C, Bely M. 2016. Enhanced 3-sulfanylhexan-1-ol production in sequential mixed fermentation with Torulaspora delbrueckii/Saccharomyces cerevisiae reveals a situation of synergistic interaction between two industrial strains. Front Microbiol 7:293. doi: 10.3389/fmicb.2016.00293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Deroite A, Legras JL, Rigou P, Ortiz-Julien A, Dequin S. 2018. Lipids modulate acetic acid and thiol final concentrations in wine during fermentation by Saccharomyces cerevisiae × Saccharomyces kudriavzevii hybrids. AMB Express 8:130. doi: 10.1186/s13568-018-0657-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Capone DL, Sefton MA, Hayasaka Y, Jeffery DW. 2010. Analysis of precursors to wine odorant 3-mercaptohexan-1-ol using HPLC-MS/MS: resolution and quantitation of diastereomers of 3-S-cysteinylhexan-1-ol and 3-S-glutathionylhexan-1-ol. J Agric Food Chem 58:1390–1395. doi: 10.1021/jf903720w. [DOI] [PubMed] [Google Scholar]
- 19.Fedrizzi B, Pardon KH, Sefton MA, Elsey GM, Jeffery DW. 2009. First identification of 4-S-glutathionyl-4-methylpentan-2-one, a potential precursor of 4-mercapto-4-methylpentan-2-one, in Sauvignon blanc juice. J Agric Food Chem 57:991–995. doi: 10.1021/jf802799w. [DOI] [PubMed] [Google Scholar]
- 20.Capone DL, Pardon KH, Cordente AG, Jeffery DW. 2011. Identification and quantitation of 3-S-cysteinylglycinehexan-1-ol (Cysgly-3-MH) in Sauvignon blanc grape juice by HPLC-MS/MS. J Agric Food Chem 59:11204–11210. doi: 10.1021/jf202543z. [DOI] [PubMed] [Google Scholar]
- 21.Cordente AG, Capone DL, Curtin CD. 2015. Unravelling glutathione conjugate catabolism in Saccharomyces cerevisiae: the role of glutathione/dipeptide transporters and vacuolar function in the release of volatile sulfur compounds 3-mercaptohexan-1-ol and 4-mercapto-4-methylpentan-2-one. Appl Microbiol Biotechnol 99:9709–9722. doi: 10.1007/s00253-015-6833-5. [DOI] [PubMed] [Google Scholar]
- 22.Bonnaffoux H, Roland A, Remond E, Delpech S, Schneider R, Cavelier F. 2017. First identification and quantification of S-3-(hexan-1-ol)-gamma-glutamyl-cysteine in grape must as a potential thiol precursor, using UPLC-MS/MS analysis and stable isotope dilution assay. Food Chem 237:877–886. doi: 10.1016/j.foodchem.2017.05.116. [DOI] [PubMed] [Google Scholar]
- 23.Swiegers JH, Willmott R, Hill-Ling A, Capone DL, Pardon KH, Elsey GM, Howell KS, de Barros Lopes MA, Sefton MA, Lilly M, Pretorius IS. 2006. Modulation of volatile thiol and ester aromas by modified wine yeast, p 113–116. In Bredie WLP, Petersen MA (ed), Developments in food science, vol 43 Elsevier, Amsterdam, the Netherlands. [Google Scholar]
- 24.Subileau M, Schneider R, Salmon JM, Degryse E. 2008. New insights on 3-mercaptohexanol (3MH) biogenesis in Sauvignon Blanc wines: Cys-3MH and (E)-hexen-2-al are not the major precursors. J Agric Food Chem 56:9230–9235. doi: 10.1021/jf801626f. [DOI] [PubMed] [Google Scholar]
- 25.Subileau M, Schneider R, Salmon JM, Degryse E. 2008. Nitrogen catabolite repression modulates the production of aromatic thiols characteristic of Sauvignon blanc at the level of precursor transport. FEMS Yeast Res 8:771–780. doi: 10.1111/j.1567-1364.2008.00400.x. [DOI] [PubMed] [Google Scholar]
- 26.Santiago M, Gardner RC. 2015. Yeast genes required for conversion of grape precursors to varietal thiols in wine. FEMS Yeast Res 15:fov034. doi: 10.1093/femsyr/fov034. [DOI] [PubMed] [Google Scholar]
- 27.Roncoroni M, Santiago M, Hooks DO, Moroney S, Harsch MJ, Lee SA, Richards KD, Nicolau L, Gardner RC. 2011. The yeast IRC7 gene encodes a beta-lyase responsible for production of the varietal thiol 4-mercapto-4-methylpentan-2-one in wine. Food Microbiol 28:926–935. doi: 10.1016/j.fm.2011.01.002. [DOI] [PubMed] [Google Scholar]
- 28.Holt S, Cordente AG, Williams SJ, Capone DL, Jitjaroen W, Menz IR, Curtin C, Anderson PA. 2011. Engineering Saccharomyces cerevisiae to release 3-mercaptohexan-1-ol during fermentation through overexpression of an S. cerevisiae gene, STR3, for improvement of wine aroma. Appl Environ Microbiol 77:3626–3632. doi: 10.1128/AEM.03009-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zha M, Sun B, Yin S, Mehmood A, Cheng L, Wang C. 2018. Generation of 2-furfurylthiol by carbon-sulfur lyase from the Baijiu yeast Saccharomyces cerevisiae G20. J Agric Food Chem 66:2114–2120. doi: 10.1021/acs.jafc.7b06125. [DOI] [PubMed] [Google Scholar]
- 30.Belda I, Ruiz J, Navascues E, Marquina D, Santos A. 2016. Improvement of aromatic thiol release through the selection of yeasts with increased beta-lyase activity. Int J Food Microbiol 225:1–8. doi: 10.1016/j.ijfoodmicro.2016.03.001. [DOI] [PubMed] [Google Scholar]
- 31.Dufour M, Zimmer A, Thibon C, Marullo P. 2013. Enhancement of volatile thiol release of Saccharomyces cerevisiae strains using molecular breeding. Appl Microbiol Biotechnol 97:5893–5905. doi: 10.1007/s00253-013-4739-7. [DOI] [PubMed] [Google Scholar]
- 32.Santiago M, Gardner RC. 2015. The IRC7 gene encodes cysteine desulphydrase activity and confers on yeast the ability to grow on cysteine as a nitrogen source. Yeast 32:519–532. doi: 10.1002/yea.3076. [DOI] [PubMed] [Google Scholar]
- 33.Alvaro D, Lisby M, Rothstein R. 2007. Genome-wide analysis of Rad52 foci reveals diverse mechanisms impacting recombination. PLoS Genet 3:e228. doi: 10.1371/journal.pgen.0030228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasution O, Lee J, Srinivasa K, Choi IG, Lee YM, Kim E, Choi W, Kim W. 2015. Loss of Dfg5 glycosylphosphatidylinositol-anchored membrane protein confers enhanced heat tolerance in Saccharomyces cerevisiae. Environ Microbiol 17:2721–2734. doi: 10.1111/1462-2920.12649. [DOI] [PubMed] [Google Scholar]
- 35.Harsch MJ, Gardner RC. 2013. Yeast genes involved in sulfur and nitrogen metabolism affect the production of volatile thiols from Sauvignon Blanc musts. Appl Microbiol Biotechnol 97:223–235. doi: 10.1007/s00253-012-4198-6. [DOI] [PubMed] [Google Scholar]
- 36.Borneman AR, Forgan AH, Kolouchova R, Fraser JA, Schmidt SA. 2016. Whole genome comparison reveals high levels of inbreeding and strain redundancy across the spectrum of commercial wine strains of Saccharomyces cerevisiae. G3 (Bethesda) 6:957–971. doi: 10.1534/g3.115.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clausen T, Huber R, Laber B, Pohlenz HD, Messerschmidt A. 1996. Crystal structure of the pyridoxal-5′-phosphate dependent cystathionine beta-lyase from Escherichia coli at 1.83 Å. J Mol Biol 262:202–224. doi: 10.1006/jmbi.1996.0508. [DOI] [PubMed] [Google Scholar]
- 38.Messerschmidt A, Worbs M, Steegborn C, Wahl MC, Huber R, Laber B, Clausen T. 2003. Determinants of enzymatic specificity in the Cys-Met-metabolism PLP-dependent enzymes family: crystal structure of cystathionine gamma-lyase from yeast and intrafamiliar structure comparison. Biol Chem 384:373–386. doi: 10.1515/BC.2003.043. [DOI] [PubMed] [Google Scholar]
- 39.Hall C, Brachat S, Dietrich FS. 2005. Contribution of horizontal gene transfer to the evolution of Saccharomyces cerevisiae. Eukaryot Cell 4:1102–1115. doi: 10.1128/EC.4.6.1102-1115.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Scherens B, Feller A, Vierendeels F, Messenguy F, Dubois E. 2006. Identification of direct and indirect targets of the Gln3 and Gat1 activators by transcriptional profiling in response to nitrogen availability in the short and long term. FEMS Yeast Res 6:777–791. doi: 10.1111/j.1567-1364.2006.00060.x. [DOI] [PubMed] [Google Scholar]
- 41.Gross C, Kelleher M, Iyer VR, Brown PO, Winge DR. 2000. Identification of the copper regulon in Saccharomyces cerevisiae by DNA microarrays. J Biol Chem 275:32310–32316. doi: 10.1074/jbc.M005946200. [DOI] [PubMed] [Google Scholar]
- 42.Jensen LT, Howard WR, Strain JJ, Winge DR, Culotta VC. 1996. Enhanced effectiveness of copper ion buffering by CUP1 metallothionein compared with CRS5 metallothionein in Saccharomyces cerevisiae. J Biol Chem 271:18514–18519. [DOI] [PubMed] [Google Scholar]
- 43.Marsit S, Dequin S. 2015. Diversity and adaptive evolution of Saccharomyces wine yeast: a review. FEMS Yeast Res 15:fov067. doi: 10.1093/femsyr/fov067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bell SJ, Henschke PA. 2005. Implications of nitrogen nutrition for grapes, fermentation and wine. Aust J Grape Wine Res 11:242–295. doi: 10.1111/j.1755-0238.2005.tb00028.x. [DOI] [Google Scholar]
- 45.Kinzurik MI, Herbst-Johnstone M, Gardner RC, Fedrizzi B. 2016. Hydrogen sulfide production during yeast fermentation causes the accumulation of ethanethiol, S-ethyl thioacetate and diethyl disulfide. Food Chem 209:341–347. doi: 10.1016/j.foodchem.2016.04.094. [DOI] [PubMed] [Google Scholar]
- 46.Cheynier V, Souquet JM, Moutounet M. 1989. Glutathione content and glutathione to hydroxycinnamic acid ratio in Vitis vinifera grapes and musts. Am J Enology Vitic 40:320. [Google Scholar]
- 47.Wegmann-Herr P, Ullrich S, Schmarr H-G, Durner D. 2016. Use of glutathione during white wine production–impact on S-off-flavors and sensory production. BIO Web Conf 7:02031. doi: 10.1051/bioconf/20160702031. [DOI] [Google Scholar]
- 48.Rauhut D. 2008. Usage and formation of sulphur compounds, p 181–209. In König H, Unden G, Fröhlich J (ed), Biology of microorganisms on grapes, in must and in wine. Springer, Mainz, Germany. [Google Scholar]
- 49.Winter G, Henschke PA, Higgins VJ, Ugliano M, Curtin CD. 2011. Effects of rehydration nutrients on H2S metabolism and formation of volatile sulfur compounds by the wine yeast VL3. AMB Express 1:36. doi: 10.1186/2191-0855-1-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Winter G, Curtin C. 2012. In situ high throughput method for H2S detection during micro-scale wine fermentation. J Microbiol Methods 91:165–170. doi: 10.1016/j.mimet.2012.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Kotseridis Y, Ray JL, Augier C, Baumes R. 2000. Quantitative determination of sulfur containing wine odorants at sub-ppb levels. 1. Synthesis of the deuterated analogues. J Agric Food Chem 48:5819–5823. [DOI] [PubMed] [Google Scholar]
- 52.Pardon KH, Graney SD, Capone DL, Swiegers JH, Sefton MA, Elsey GM. 2008. Synthesis of the individual diastereomers of the cysteine conjugate of 3-mercaptohexanol (3-MH). J Agric Food Chem 56:3758–3763. doi: 10.1021/jf8000444. [DOI] [PubMed] [Google Scholar]
- 53.Wakabayashi H, Wakabayashi M, Eisenreich W, Engel KH. 2004. Stereochemical course of the generation of 3-mercaptohexanal and 3-mercaptohexanol by beta-lyase-catalyzed cleavage of cysteine conjugates. J Agric Food Chem 52:110–116. doi: 10.1021/jf0305478. [DOI] [PubMed] [Google Scholar]
- 54.Ausubel F, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K. 1994. Current protocols in molecular biology. John Wiley & Sons, New York, NY. [Google Scholar]
- 55.Volschenk H, Viljoen M, Grobler J, Petzold B, Bauer F, Subden RE, Young RA, Lonvaud A, Denayrolles M, van Vuuren HJ. 1997. Engineering pathways for malate degradation in Saccharomyces cerevisiae. Nat Biotechnol 15:253–257. doi: 10.1038/nbt0397-253. [DOI] [PubMed] [Google Scholar]
- 56.Kutyna DR, Cordente AG, Varela C. 2014. Genetic engineering of industrial Saccharomyces cerevisiae strains using a selection/counter-selection approach. Methods Mol Biol 1152:157–168. doi: 10.1007/978-1-4939-0563-8_9. [DOI] [PubMed] [Google Scholar]
- 57.Swiegers JH, Capone DL, Pardon KH, Elsey GM, Sefton MA, Francis IL, Pretorius IS. 2007. Engineering volatile thiol release in Saccharomyces cerevisiae for improved wine aroma. Yeast 24:561–574. doi: 10.1002/yea.1493. [DOI] [PubMed] [Google Scholar]
- 58.Capone DL, Ristic R, Pardon KH, Jeffery DW. 2015. Simple quantitative determination of potent thiols at ultratrace levels in wine by derivatization and high-performance liquid chromatography-tandem mass spectrometry (HPLC-MS/MS) analysis. Anal Chem 87:1226–1231. doi: 10.1021/ac503883s. [DOI] [PubMed] [Google Scholar]
- 59.Siebert TE, Solomon MR, Pollnitz AP, Jeffery DW. 2010. Selective determination of volatile sulfur compounds in wine by gas chromatography with sulfur chemiluminescence detection. J Agric Food Chem 58:9454–9462. doi: 10.1021/jf102008r. [DOI] [PubMed] [Google Scholar]
- 60.Clausen T, Huber R, Messerschmidt A, Pohlenz HD, Laber B. 1997. Slow-binding inhibition of Escherichia coli cystathionine beta-lyase by l-aminoethoxyvinylglycine: a kinetic and X-ray study. Biochemistry 36:12633–12643. doi: 10.1021/bi970630m. [DOI] [PubMed] [Google Scholar]
- 61.Webb B, Sali A. 2016. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinformatics 54:5.6.1–5.6.37. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 1993. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 63.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinformatics 14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The best model of Irc7p7 was uploaded to the Protein Model DataBase under accession number PM0081794.