Salt marshes represent highly dynamic ecosystems where the atmosphere, continents, and the ocean interact. The bacterial distribution in this ecosystem is of great ecological concern, as it provides essential functions acting on ecosystem services. However, ecological processes mediating bacterial assembly are poorly understood for salt marshes, especially the ones located in estuaries. In this study, the distribution and assembly of bacterial communities in an estuarine marsh located in south Hangzhou Bay were investigated. The results revealed an intricate interplay between stochastic and deterministic processes mediating the assembly of bacterial communities in the studied gradient system. Collectively, our findings illustrate the main drivers of community assembly, taking into consideration changes in sediment abiotic variables and potential biotic interactions. Thus, we offer new insights into estuarine bacterial communities and illustrate the interplay of ecological processes shaping the assembly of bacterial communities in estuarine marsh ecosystems.
KEYWORDS: assembly mechanism, bacterial community, cooccurrence network, estuarine marsh
ABSTRACT
Bacterial communities play essential roles in estuarine marsh ecosystems, but the interplay of ecological processes underlying their community assembly is poorly understood. Here, we studied the sediment bacterial communities along a linear gradient extending from the water-land junction toward a high marsh, using 16S rRNA gene amplicon sequencing. Bacterial community compositions differed significantly between sediment transects. Physicochemical properties, particularly sediment nutrient levels (i.e., total nitrogen [TN] and available phosphorus [AP]), as well as sediment physical structure and pH (P < 0.05), were strongly associated with the overall community variations. In addition, the topological properties of bacterial cooccurrence networks varied with distance to the water-land junction. Both node- and network-level topological features revealed that the bacterial network of sediments farthest from the junction was less intense in complexity and interactions than other sediments. Phylogenetic null modeling analysis showed a progressive transition from stochastic to deterministic community assembly for the water-land junction sites toward the emerging terrestrial system. Taken together, data from this study provide a detailed outline of the distribution pattern of the sediment bacterial community across an estuarine marsh and inform the mechanisms and processes mediating bacterial community assembly in marsh soils.
IMPORTANCE Salt marshes represent highly dynamic ecosystems where the atmosphere, continents, and the ocean interact. The bacterial distribution in this ecosystem is of great ecological concern, as it provides essential functions acting on ecosystem services. However, ecological processes mediating bacterial assembly are poorly understood for salt marshes, especially the ones located in estuaries. In this study, the distribution and assembly of bacterial communities in an estuarine marsh located in south Hangzhou Bay were investigated. The results revealed an intricate interplay between stochastic and deterministic processes mediating the assembly of bacterial communities in the studied gradient system. Collectively, our findings illustrate the main drivers of community assembly, taking into consideration changes in sediment abiotic variables and potential biotic interactions. Thus, we offer new insights into estuarine bacterial communities and illustrate the interplay of ecological processes shaping the assembly of bacterial communities in estuarine marsh ecosystems.
INTRODUCTION
Salt marshes are highly productive ecosystems located at the intersection between land and open salt water or brackish water, which are responsible for several ecosystem services, such as water filtration, nutrient cycling, and carbon sequestration (1–3). Sediment bacterial communities act as primary regulators of biogeochemical processes and nutrient cycling at initial marsh development (4, 5), playing critical roles in marsh ecosystem function. Consequently, knowledge of sediment bacterial communities can improve our understanding of the ecology of salt marsh ecosystems, apart from providing baseline information on how to effectively manage and conserve the declining marsh biodiversity.
Salt marshes are landform ecosystems located at the open coast of continents. Of particular importance, estuarine marshes are those in which the land connects with the sea and freshwater meets salt water. These ecosystems are different from constantly salt open-coast marshes. Until recently, several studies have been focusing on the sediment bacterial distribution in open-coast marshes (6–8). For instance, Dini-Andreote et al. (8) revealed the bacterial community succession in an open-coast marsh chronosequence. However, information regarding estuarine marshes is still limited. In addition, microbe-microbe interactions specifically have received more and more attention until recently (9). Many studies have used cooccurrence network analysis as a tool to decipher the potential intraspecies or interspecies interactions in complex microbial assemblages, such as in the open ocean (10), and in lake (11) and soil (12) systems. Some central or keystone species could be identified within an ecological network, playing an irreplaceable role in maintaining the structure and function of the whole microbial community. These findings revealed that microbial communities generally have nonrandom cooccurrence patterns and modular structures, which strongly implies the essential role of biotic interactions in governing community assembly (13).
A surge in the literature has been providing increasing evidence that both deterministic and stochastic processes govern the assembly of bacterial communities (14–16). Deterministic processes include ecological selection influencing bacterial fitness and thereby determining community composition and structure (15). Various deterministic factors, such as environmental conditions (5, 8), carbon and nutrient availability (17), and plant diversity (6), have been shown to be important in governing bacterial community structure. Stochastic processes include probabilistic dispersal and random changes in species relative abundances (ecological drift) that are not the consequence of environmentally determined fitness (18, 19). The relative importance of each of these processes depends largely on the local environment, species traits, and spatial scale investigated (15), which has seldom been evaluated in estuarine marshes.
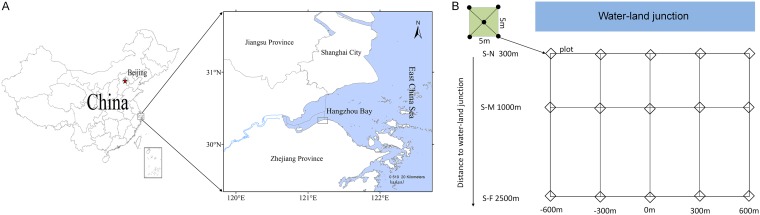
The estuarine marsh located in south Hangzhou Bay (Fig. 1A), a typical subtropical estuary in China, has been formed by the accumulation of sediments carried by the Yangtze and Qiantang Rivers and is extending at a velocity of 292 m/year (20). Here, we sampled this estuarine marsh along the distance from the water-land junction to explore the bacterial distribution pattern in surface sediments using 16S rRNA gene amplicon sequencing. In detail, the objectives of this study were to (i) assess the bacterial community distributions across the estuarine marsh, (ii) investigate the relationship between physicochemical factors and bacterial distributions, (iii) identify potential biotic interactions in the system by using cooccurrence network analysis, and (iv) disentangle the relative influences of community assembly processes underlying the bacterial communities in the estuarine marsh.
FIG 1.
Maps showing the sampling stations. (A) Sampling sites in the estuarine marsh in south Hangzhou Bay. (B) Surface sediment samples (S-N, S-M, and S-F) were collected along transects based on distance to the water-land junction (about 300 m, 1,000 m, and 2,500 m, respectively) (see Table 1 for a detailed description of the sampling sites).
RESULTS
Sediment physicochemical properties and bacterial community diversity.
The sampled gradient encompasses a wide-range variation of sediment physicochemical properties (P < 0.05 by analysis of variance [ANOVA]) (Table 1). Significant increases (P < 0.05) in nutrients (organic carbon [OC], ammonium, available phosphorus [AP], and total nitrogen [TN]) and silt and clay contents were detected along the distance to the water-land junction (Table 1). Sediment pH rose significantly (P < 0.05) from pH 6.6 (S-N, located about 300 m away from the water-land junction) to pH 8.0 (S-F, located about 2,500 m away from the water-land junction) as the distance to the water-land junction increased, while the electrical conductivity (EC) value decreased.
TABLE 1.
Physicochemical properties of the sediment transectsa
| Transectb | Mean pH ± SD | Mean ammonium concn (mg kg−1) ± SD | Mean AP concn (mg kg−1) ± SD | Mean AK concn (mg kg−1) ± SD | Mean TN content (%) ± SD | Mean OC content (g kg−1) ± SD | Mean Fed content (g kg−1) ± SD | Mean EC (mS/cm) ± SD | Mean clay content (%) ± SD | Mean silt content (%) ± SD | Mean sand content (%) ± SD |
|---|---|---|---|---|---|---|---|---|---|---|---|
| S-N | 6.6 ± 0.2 C | 68.5 ± 8.8 C | 21.1 ± 8.3 C | 336.8 ± 49.0 B | 0.2 ± 0.0 C | 3.2 ± 0.4 C | 9.5 ± 0.8 C | 1.2 ± 0.2 B | 2.2 ± 0.5 C | 12.8 ± 3.3 C | 85.1 ± 3.8 A |
| S-M | 7.7 ± 0.1 B | 101.4 ± 7.1 B | 63.5 ± 4.8 B | 281.1 ± 46.2 A | 0.4 ± 0.04 B | 5.5 ± 0.5 B | 16.9 ± 0.9 B | 1.6 ± 0.2 A | 8.4 ± 0.2 B | 57.0 ± 2.1 B | 34.7 ± 2.2 B |
| S-F | 8.0 ± 0.1 A | 134.2 ± 9.8 A | 89.4 ± 14.2 A | 141.2 ± 40.2 A | 0.6 ± 0.1 A | 6.8 ± 0.6 A | 18.2 ± 0.7 A | 0.4 ± 0.1 C | 8.9 ± 0.2 A | 60.0 ± 0.7 A | 31.1 ± 0.5 C |
Values are means ± standard deviations (n = 5). Different letters within the same column denote significant differences (P < 0.05) between the sediments. AP, available phosphorus; AK, available potassium; TN, total nitrogen; OC, organic carbon; Fed, free Fe2O3; EC, electrical conductivity.
S-N, sampling transect located about 300 m away from the water-land junction, without vegetation; S-M, sampling transect located about 1,000 m away from the water-land junction, mainly covered with Scirpus mariqueter; S-F, sampling transect located about 2,500 m away from the water-land junction, mainly covered with Phragmites australis.
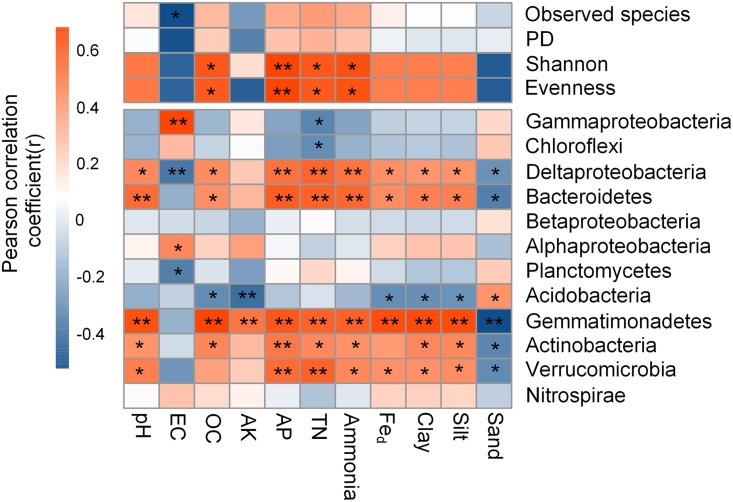
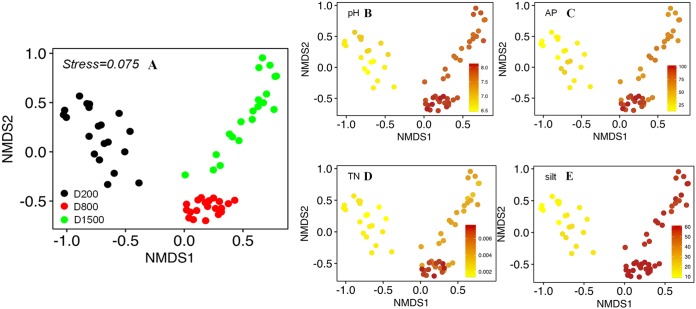
The α-diversity indexes, including observed species, evenness, Shannon’s index, and phylogenetic diversity (PD), were highest in the S-F sediments (see Fig. S1 in the supplemental material). Generally, evenness and Shannon’s index progressively increased with distance to the water-land junction, whereas the PD index and observed species first decreased and then increased. The α-diversity indexes (Shannon’s index and evenness) showed significant positive correlations with nutrient levels (OC, ammonium, AP, and TN) (Fig. 2). Nonmetric multidimensional scaling (NMDS) analysis revealed that bacterial communities were significantly different (P values listed in Table S2) according to the sampling transect (Fig. 3A).
FIG 2.
Correlation between the α-diversity indexes, dominant phyla/classes, and physicochemical properties. Correlation coefficients ranged from negative to positive and are indicated by color intensity, changing from dark blue to red, respectively, as illustrated in the key. **, P < 0.01; *, P < 0.05. For abbreviations, see Table 1.
FIG 3.
(A) Nonmetric multidimensional scaling (NMDS) plots were used to visualize the differences in bacterial community compositions based on the Bray-Curtis distance among the sediment transects. (B to E) NMDS plots displaying four environmental variables, pH (B), AP (C), TN (D), and silt content (E), that mainly explain variations in bacterial community composition. The parameter values are color-coded in the corresponding heat map legends.
Distribution of taxa and phylotypes.
Significant differences in bacterial community composition at the phylum level (class for Proteobacteria) were detected among sediment transects (Table S1 and Fig. S2). The relative abundances of Chloroflexi, Bacteroidetes, Acidobacteria, and Gemmatimonadetes significantly increased with distance to the water-land junction (P < 0.05). Gammaproteobacteria and Planctomycetes had hump-shaped curves, peaking in the S-M sediments at 18.49% ± 1.19% and 7.43% ± 1.59%, respectively. Conversely, the relative abundance of Betaproteobacteria first decreased and then increased along the distance to the water-land junction (Table S1 and Fig. S2).
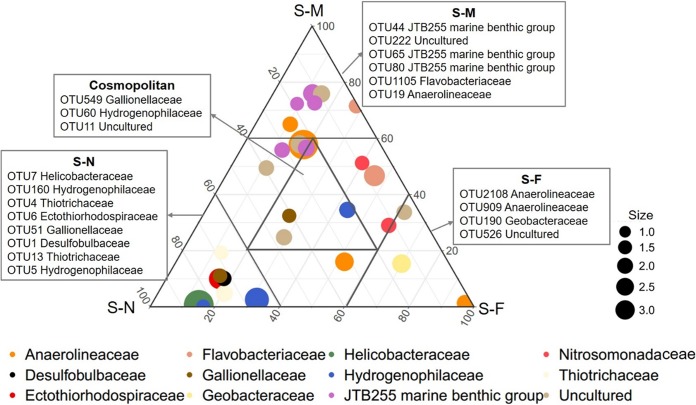
At a finer taxonomic resolution, we selected the top 30 most abundant operational taxonomic units (OTUs) in the sediments to reflect the consistent distributional patterns of dominant taxa across the marsh (Fig. 4). In the S-N sediments, the OTUs mainly affiliated with Thiotrichaceae, Hydrogenophilaceae, Helicobacteraceae, Ectothiorhodospiraceae, and Desulfobulbaceae were more abundant, whereas OTUs affiliated with the JTB255 marine benthic group (JTB255-MBG) were found to be relatively abundant in the S-M (located about 1,000 m away from the water-land junction) sediments. The OTUs affiliated with Anaerolineaceae, Nitrosomonadaceae, and Geobacteraceae were more abundant in the S-F sediments. OTU549, OTU60, and OTU11 were found to be cosmopolitan OTUs across the three sediment transects (Fig. 4). Spearman’s correlation analysis showed that the most abundant OTUs were extensively correlated with physicochemical properties (Fig. S3).
FIG 4.
Ternary plot showing the distribution of the 30 most abundant OTUs in the sediment transects. The symbol size indicates the relative abundance of each OTU. The ranking in each box indicates the relative abundance of the OTU (shown at the family level) from high to low. Each dot is colored at the family level. See Table 1 for transect notation.
Physicochemical properties affecting the bacterial community.
Physicochemical properties showed significant correlations with bacterial β-diversity (based on the Bray-Curtis distance at the OTU level; R2 = 0.87 and P < 0.01 [Adonis]). Mantel tests identified TN, AP, silt content, and pH as four dominant factors that correlated with bacterial community structure (Table S3), which is clearly shown by the NMDS visualization (Fig. 3B to E). A correlation analysis confirmed a significant relationship between bacterial community composition at the phylum level and the above-mentioned factors (P < 0.05) (Fig. 2). The distance-based multivariate linear model (DistLM) further demonstrated that TN, AP, silt content, and pH had high rankings for contribution to community variation (Table 2). Sequential tests showed that the combination of the above-described significant parameters explained 85.47% of the variation (Table 2). Other factors might also affect the bacterial composition but not in a consistent and predictable manner.
TABLE 2.
Distance-based multivariate linear model of bacterial community variability against the physicochemical properties of the sediment samples after 999 permutationsa
| Variable | Pseudo-F | P | % variation explained | Cumulative % variation explained |
|---|---|---|---|---|
| Marginal tests | ||||
| TN | 6.018 | 0.001 | 31.64 | |
| AP | 5.999 | 0.001 | 31.58 | |
| Silt | 5.458 | 0.002 | 29.57 | |
| Sand | 5.420 | 0.002 | 29.42 | |
| Clay | 5.139 | 0.005 | 28.33 | |
| pH | 5.107 | 0.004 | 28.20 | |
| Fed | 4.813 | 0.005 | 27.02 | |
| Ammonia | 4.468 | 0.005 | 25.58 | |
| OC | 3.792 | 0.010 | 22.58 | |
| EC | 3.746 | 0.007 | 22.37 | |
| AK | 2.774 | 0.017 | 17.58 | |
| Sequential tests | ||||
| TN | 6.018 | 0.001 | 31.64 | 31.64 |
| EC | 6.671 | 0.001 | 24.42 | 56.07 |
| Silt | 2.022 | 0.012 | 6.82 | 62.89 |
| OC | 1.170 | 0.275 | 3.89 | 66.78 |
| pH | 1.167 | 0.266 | 3.81 | 70.59 |
| Fed | 1.055 | 0.441 | 3.43 | 74.02 |
| AK | 0.999 | 0.495 | 3.25 | 77.26 |
| Clay | 0.941 | 0.515 | 3.08 | 80.34 |
| Sand | 1.046 | 0.421 | 3.40 | 83.74 |
| AP | 0.191 | 0.952 | 0.91 | 84.65 |
| Ammonia | 0.140 | 0.937 | 0.82 | 85.47 |
For marginal tests, each variable was analyzed individually (ignoring other variables). For sequential tests (forward selection of variables), the percentage of variation explained by each variable added to the model was conditional on the variables already in the model. Data in boldface type are significantly correlated (P < 0.05). The abbreviations of physicochemical properties are the same as those in Table 1.
Phylotype cooccurrence within transects across the marsh.
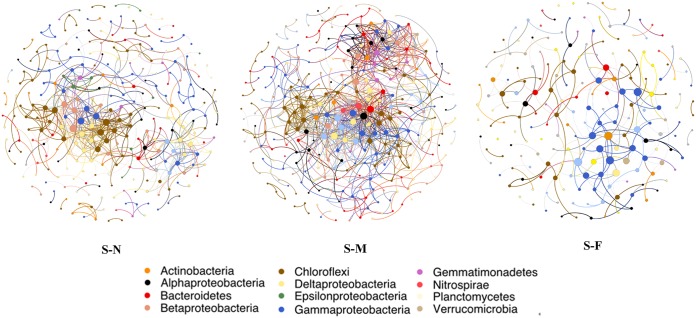
A cooccurrence network for each transect was individually generated to explore bacterial interactions within the bacterial community (Table 3 and Fig. 5). The network roughly followed a scale-free degree distribution (Fig. S4), implicating a scale-free network structure and a nonrandom cooccurrence pattern. The highest correlation values were observed in the bacterial communities of the S-M sediments, whereas the lowest value was observed in the S-F sediments (Table 3). Patterns in topological features, such as the clustering coefficient, density, and average degree, were similar to the patterns observed for the edge numbers (Table 3). In contrast, the average path length of the network was lowest in the S-M sediments, whereas modularity was highest in the S-F sediments. Each network had several main subnetworks (modules containing more than 20 nodes), while other subnetworks had one or a few elements (Fig. S5). The most densely connected node in each main module was defined as a “hub.” Four nodes belonging to Rhodobacterales, subgroup 23, unclassified Deltaproteobacteria, and Anaerolineales from the S-N network were categorized as module hubs. Four nodes allocated into module hubs in the S-M network belonged to unclassified Nitrospinae, unclassified Betaproteobacteria, unclassified Gammaproteobacteria, and Xanthomonadales, while Nitrosomonadales were the only hub in the S-F network.
TABLE 3.
Topological properties of the networks within each sediment transecta
| Network metric | Value for sediment transect |
||
|---|---|---|---|
| S-N | S-M | S-F | |
| Node | 282 | 306 | 198 |
| Edge | 560 | 1,135 | 208 |
| Modularity (no. of modules) | 0.993 (65) | 1.134 (34) | 1.751 (75) |
| Density | 0.014 | 0.024 | 0.011 |
| Avg degree | 3.972 | 7.418 | 2.101 |
| Clustering coefficient | 0.375 | 0.394 | 0.257 |
| Avg path length | 5.207 | 4.333 | 5.235 |
See Table 1 for transect notation.
FIG 5.
Network analyses showing the cooccurrence pattern for each sediment transect. Each dot represents a bacterial phylotype (an OTU clustered at 97%). The size of each node is proportional to the number of connections (that is, degree). Each node is labeled at the phylum level, except for the phylum Proteobacteria, which is shown at the class level. The edges show the correlations between two nodes. A connection represents a strong (r > 0.8) and significant (P < 0.01) correlation. See Table 1 for transect notation.
Ecological processes governing the assembly of bacterial communities.
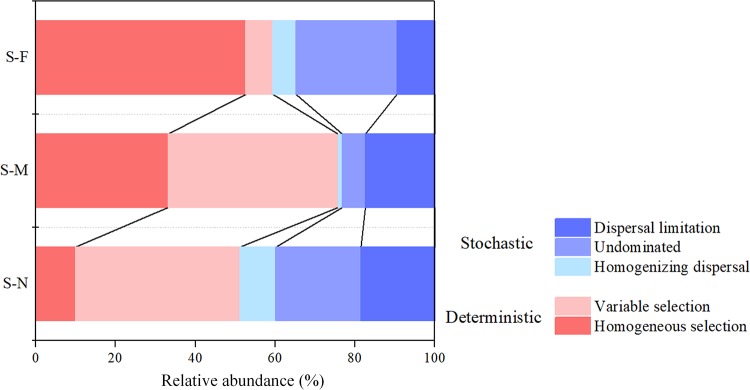
The beta nearest-taxon index (βNTI) was used to provide deep insight into the potential roles of deterministic and stochastic processes in governing bacterial community assembly. The result showed a distance-dependent variation in the relative influence of deterministic and stochastic processes: the assembly of bacterial communities shifted from stochasticity in the S-N sediments (|βNTI| < 2 [i.e., −2 < βNTI < 2]) to strong variable selection in the S-M sediments (βNTI > 2) and then to strong homogeneous selection in the S-F sediments (βNTI < −2). We further quantified the relative contributions of major ecological processes that structure the bacterial community in the sediments. Generally, homogeneous selection was obviously increased for structuring the bacterial community assembly along the distance to the water-land junction and was highest in the S-F sediments (52.6%) (Fig. 6). In contrast, the importance of dispersal limitation and variable selection processes exhibited an opposite pattern: they were lowest in the S-F sediments (9.5% and 6.8%, respectively) (Fig. 6).
FIG 6.
Summary of ecological processes that govern the bacterial community assembly of each sediment transect. Percentages are given for the relative contribution of each process to the community assembly, as indicated by different colors. See Table 1 for transect notation.
DISCUSSION
Sediment bacterial diversity varies with distance to the water-land junction.
Based on the space-for-time assumption, we would expect that diversity would typically increase from early to late stages of succession due to an increase in potential niches, resource diversity and availability, and habitat heterogeneity. In this study, the bacterial α-diversity values (evenness and Shannon index) were significantly lower in the S-N sediments, which were bare (see Fig. S1 in the supplemental material), while they were significantly correlated with OC, ammonia, AP, and TN (Fig. 2). It may have been the case that plant richness was a key factor. Higher plant richness is normally connected with higher plant productivity because of niche complementarity and more resource supply, resulting in increases in the nutrient pools that are available to bacteria, which favor higher bacterial diversity (21). The specific rhizosphere bacteria and symbiotic bacteria that benefited from denser vegetation litter and/or root exudates could also boost bacterial diversity (21). In addition, the bacterial diversities of S-M and S-F sediments differed from each other (Fig. 3; Fig. S1). Scirpus mariqueter and Phragmites australis were functionally different, with different growth habits and resource use strategies, which led to differences in the sediment environments and bacterial communities.
Dominant bacterial assemblages are altered with distinct habitats.
The bacterial communities in this region were mainly dominated by Gammaproteobacteria, Deltaproteobacteria, and Chloroflexi (Fig. S2), which were also dominant in the sediment of Pearl Estuary (22). Additionally, distinct bacterial community compositions were observed among sediment transects (Fig. 4). Thiotrichaceae (OTU13), Hydrogenophilaceae (OTU160 and OTU5), Ectothiorhodospiraceae (OTU6), and Desulfobulbaceae (OTU1), which have received much attention due to their wide distribution and critical role in sulfur cycles, were more abundant in the S-N sediments (Fig. 4). Sulfur-oxidizing bacteria (SOB) and sulfate-reducing bacteria (SRB) were considered to be coabundant in coastal sediments (22). Moreover, many SRB were observed to couple with each other; they may collaborate to perform the same process (22, 23). The JTB255 marine benthic group (JTB255-MBG), which represents one of the core groups of the global sediment microbiome (24), was more abundant in the S-M sediments (Fig. 4). So far, the exact environmental function of the JTB255-MBG is unknown. A recent study indicated that the JTB255-MBG might play a significant role in dark carbon fixation in coastal surface sediments (25). According to Fig. S3, we suspect that its abundance is related to the sediment EC value. The OTUs (OTU9, OTU23, OTU5, and OTU190) affiliated with Anaerolineaceae were more abundant in the S-F sediments, which is consistent with previous results showing that soil in the Phragmites australis zone harbored more Anaerolineaceae (26). Anaerolineaceae, as the representative group of the Chloroflexi, could degrade carbohydrates and other cellular materials (e.g., amino acids) (27). The plant litter of Phragmites australis contained higher contents of cellulose and lignin, which may account for the relatively high relative abundance of Anaerolineaceae. They would potentially weaken mineralization and accelerate humification, resulting in higher contents of soil organic carbon, which would stimulate plant growth in turn (28). In addition, The OTU affiliated with Geobacteraceae was also more abundant in the S-F sediments (Fig. 4). Geobacteraceae have been reported to be involved in Fe(III) reduction in sedimentary environments (29, 30). The presence of Phragmites australis could change the redox characteristics of the sediments and contribute to the processes of iron oxidation and reduction (31, 32).
Local physicochemical properties shape the bacterial distribution.
Soils/sediments are considered the most heterogeneous in terrestrial ecosystems, due to variable labile organic matter inputs by plants within and across ecosystems. It has been suggested that most bacteria are restricted to specific niches by different environmental forces (33). Previous studies have shown that the bacterial community structure in estuarine marshes is correlated with salinity, nutrients, pH, and texture, etc. (5, 8). Our results showed that soil properties shifted in response to the plant species (Table 1), and sediment TN and AP concentrations were the main factors that were significantly correlated with the soil bacterial community structure (Table 2 and Fig. 2 and 3; Table S3). Nutrient levels could affect bacterial growth and limit bacterial diversity in the sediments (5, 8, 34), and nitrogen and phosphate are critical for the primary productivity and structure of dominant bacterial taxa in the marine ecosystem (35), which have a large impact on bacterial community compositions in sediments and soils (17, 34). An in-depth study will improve the understanding of their relative influences on the bacterial community. Numerous studies have highlighted texture and pH as important factors controlling soil and sediment bacterial communities across different ecosystems and types (8, 12). In this study, sedimentary silt content and pH also apparently affected the bacterial community (Table 2 and Fig. 2 and 3; Table S3). Sediment pH may act as an integrating variable that provides an integrated index of sediment conditions. Moreover, pH and texture could directly impose a physiological constraint on sediment/soil bacteria, altering competitive outcomes or reducing the net growth of individual taxa that are unable to survive (36). In total, these measured factors were shown to be important in shaping bacterial communities. However, this study was unable to explain all the variations. Further research is needed to evaluate the effects of other factors that were not considered in this study, such as various contaminants.
Sediment bacterial cooccurrence network varies with distance to the water-land junction.
The network in this study showed similar cooccurrence patterns with microorganisms and macroorganisms previously reported, suggesting that nonrandom community assembly may be a general characteristic across all life domains (37). It further confirmed that many bacterial taxa exhibit predictable biogeographical patterns, which is similar to our results (Fig. 3 and 4). Regarding overall network topology, the results showed that the bacterial community network for the S-M sediment is more connected than those of the other sediments (Table 3 and Fig. 5), which indicated that it was a more efficient system and was faster at transmitting information, energy, and resources (38). In contrast, the S-F network was relatively simple and had the highest modularity (Table 3 and Fig. 5). This implied that the S-F bacterial community is probably the most dynamic community with the largest number of taxa with no common cooccurrence across sites (39, 40). Furthermore, the lack of complex network systems in the S-F sediment might be due to bacteria being in dormant or inactive states (41), which would imply that there had not been a reduction in bacterial diversity (42, 43). Indeed, although the S-F sediment had the lowest network complexity among the sediments, it harbored the highest diversity level, which was statistically comparable to those of the other sediments (Fig. S1). However, this needs to be tested in future studies.
The most important module nodes or hubs in a given network may represent the most influential members in the bacterial community, which might be essential for community stability (43). The OTU affiliated with Rhodobacterales, which belongs to fundamentally aquatic bacteria that frequently thrive in marine environments (44), was the most important taxon in the S-N network. Rhodobacterales are deeply involved in sulfur and carbon biogeochemical cycling and symbiosis with aquatic micro- and macroorganisms (44). Their critical role in the S-N network may be related to the frequent exchange of substances between seawater and the S-N sediments. In addition, the keystone species of the S-M and S-F networks (Nitrospinae and Nitrosomonadales) are reported to be involved in nitrogen metabolism (45, 46), which indicated the pivotal role of estuarine marshes in nitrogen cycling, which is similar to coastal marshes. But their specific function in maintaining community structure and nitrogen cycling needs further study.
Homogeneous selection increases along the distance to the water-land junction.
Generally, the deterministic process was pivotal across the three sediment transects (Fig. 6). This observation was also supported by the nonrandom assembly patterns in the network, indicating the deterministic processes, including competitive interactions, nonoverlapping niches, or historical effects, shaping community composition (37). However, a greater influence of stochasticity was observed in the S-N sediments (Fig. 6), which are identified as being newly formed by sand accumulation and sedimentation according to the space-for-time strategy (47). This implied that stochastic processes played a critical role in microbial primary succession of diverse systems (14–16). At the initial sites, being an intersect between the land and sea, dispersal may likely contribute to the increase of the stochastic community assembly (Fig. 6), providing more opportunities for successful immigration. The most influential members in the S-N network shown by network analysis were fundamentally aquatic, which confirmed our speculation. Thus, random dispersal through both aerial and seawater vectors may be an important ecological factor affecting the estuarine sediments closer to the sea. However, the greater influence of stochasticity in the S-N sediments is contrary to reports that stochastic processes are less important in low-resource environments (47). This difference possibly resulted from different environmental gradients and geographic scales among the studied areas. The increased resource supply can increase stochasticity under physicochemical conditions that do not impose strong selection (48). However, the bacterial distribution was obviously shaped by physicochemical factors in this study (Table 3 and Fig. 2 and 3; Table S3). Thus, the increased resource supply did not increase stochasticity here. On the contrary, in terrestrial (more-mature) sites, the soil status might reduce variation, leading to a more deterministic assembly. For instance, homogeneous selection was associated with 33.2% and 52.6% of the variation in community assembly in the S-M and S-F sediments, respectively (Fig. 6). This may partly be attributed to the root exudates and plant residue. The S-M and S-F sediments are plant covered, while S-N sediments are nearly bare, with little vegetation. The plants may select bacterial communities and functions that are beneficial to them by the rhizosphere effect, which follows niche-based theory (49). Correspondingly, the influence of variable selection, which is also referred to as heterogeneous selection, showed an opposite pattern and was lowest in the S-F sediments (Fig. 6). Based on the existing results, we further hypothesize that the deterministic/stochastic continuum during primary succession will influence how community assembly processes operate during secondary succession (i.e., following disturbance). However, because our approaches typically focus on the sequencing of pools of 16S rRNA genes, it is much more difficult to link traits and taxonomy than it is for macroorganisms. Methodological developments that allow for the characterization of microbial community structure at finer spatial scales are steps in the right direction to overcome these challenges (50).
In conclusion, significant differences in bacterial diversity and composition were observed between sediment transects in an estuarine marsh. These were mainly correlated with changes in physicochemical properties (sediment nutrients, texture, and pH). Additionally, the bacterial interactions varied with the sediment transects, while they were less complex in sediments farthest from the junction. This study also provides novel insights about the relative roles of deterministic and stochastic processes in driving estuarine sediment bacterial communities. Although both ecological processes played important roles in the assembly of bacterial communities, homogeneous selection increased apparently with distance to the water-land junction. However, the only spring samples in this study cannot reflect the temporally dynamic pattern of sediment bacterial communities across space. Time series sampling on at least a seasonal scale should be conducted to confirm the observed pattern in future works. Moreover, in-depth investigations on sediment bacterial communities on a larger scale and with greater depth are expected to further expand our understanding of bacterial community assembly in estuarine marsh ecosystems.
MATERIALS AND METHODS
Study site and sampling.
Surface sediment samples were collected on 27 March 2017 from the estuarine marsh in south Hangzhou Bay (30°21′56″N, 121°12′39″E) (Fig. 1A). The water in the Hangzhou Bay estuary is characterized by a mixture of seawater and freshwater (20). The forelands of the marsh are largely unvegetated at close locations from the water-land junction (up to about 500 m). In this area, three transects were established in parallel (i.e., S-N, S-M, and S-F) to the forefront of the marsh. These sites were located ca. 300, 1,000, and 2,500 m from the water-land junction, respectively (Fig. 1B). The three transects had distinct vegetation types, i.e., bare sites, sites dominated by Scirpus mariqueter, and sites dominated by Phragmites australis, respectively. Each sampling site within a transect was separated from each other by ca. 300 m. Five surface sediments (5 to 20 cm) were collected along each transect (Fig. 1B). Soils collected from the five individual sediment cores at 5-m intervals were mixed to represent one composite sediment sample. A total of 15 composite sediment samples were collected and taken to the laboratory (<24 h) in coolers filled with ice. Each sample was handpicked to remove plant debris, stones, and earthworms. Samples were stored at 4°C for physicochemical analyses and at −80°C for DNA isolation.
Physicochemical analyses of sediment samples.
We determined soil pH, electrical conductivity (EC), organic carbon (OC) content, available phosphorus (AP), physical texture (sand, silt, and clay contents), and available potassium (AK). These analyses were carried out in collaboration with the Agricultural Chemistry Committee of China (51). Total OC (TOC) in the water extracts was measured using a Multi N/C TOC analyzer (Analytic Jena AG, Jena, Germany). The free sesquioxides were extracted with an NaHCO3 solution (52). Sediment samples were shaken for 1 h in 1 M KCl (5 g soil in a 50-ml KCl solution) and then filtered to analyze the ammonia concentration with a continuous-flow analyzer system (Seal Analytical GmbH, Norderstedt, Germany). All measured physicochemical properties were obtained from three independent replicates per sediment sample.
DNA extraction and sequencing.
A total of 60 sediment samples (3 sampling transects × 5 sampling sites × 4 replicates) were subjected to total DNA isolation using a MoBio PowerSoil DNA isolation kit (MoBio Laboratories, Carlsbad, CA, USA), using 0.5 g of initial sediment, according to the manufacturer’s instructions. The DNA purity and concentration were analyzed using a NanoDrop spectrophotometer (NanoDrop Technologies Inc., Wilmington, DE, USA). The primer set 515F (5′-GTGYCAGCMGCCGCGGTAA-3′) and 806R (5′-GGACTACNVGGGTWTCTAAT-3′) was used to amplify the V4 region of the bacterial 16S rRNA gene (53), using specific barcodes per sample. An aliquot of 10 ng of purified DNA from each sample was amplified in triplicate in a 20-μl PCR mixture under the following thermocycling conditions: 94°C for 3 min and 30 cycles of 95°C for 30 s, 50°C for 30 s, and 72°C for 45 s, with a final extension step of 72°C for 10 min. PCR products of each individual sample were pooled and purified using a PCR fragment purification kit (TaKaRa Biotech, Tokyo, Japan). Equimolar amounts of the PCR products were pooled and subjected to sequencing. The sequencing libraries were constructed using the TruSeq DNA sample prep kit (Illumina, San Diego, CA, USA), according to the manufacturer’s instructions. Sequencing was performed in an Illumina MiSeq instrument, using a 2 × 300-bp read configuration.
Sequence processing.
Raw sequencing data were analyzed using QIIME v.1.8 (54) and USEARCH v.6.1 (55). Sequences were demultiplexed and filtered using the following parameters: a quality score of >25, no ambiguous base calls, and a sequence length of >200 bp (56). The bacterial sequences were clustered into operational taxonomic units (OTUs) at 97% sequence similarity using the UCLUST method (57). Sequences were taxonomically assigned against the SILVA_128 database (https://www.arb-silva.de/documentation/release-128/), and a phylogenic tree was generated from the filtered alignment using FastTree (58). Sequences identified as archaea, chloroplasts, and mitochondria were removed from the analysis. Singletons were also removed from the OTU table prior to diversity analysis. After quality trimming, a total of 3,120,126 high-quality reads remained in the data set, which yielded a total of 50,487 OTUs. To correct for variable sampling efforts, the data were randomly rarefied at the same sequence depth (at 16,600 sequences per sample). Both alpha and beta diversity analyses were conducted using QIIME. We calculated the number of observed species, Shannon’s index, evenness, and phylogenetic diversity per sample and used the Bray-Curtis distance for β-diversity.
Quantifying community assembly processes.
To evaluate the relative influences of ecological processes governing the sediment bacterial community assembly, we calculated the weighted beta nearest-taxon index (βNTI), using the Picante package (59). βNTI values higher than 2 or lower than −2 indicate the influence of the deterministic process structuring the community, whereas βNTI values between −2 and 2 indicate the influence of stochastic processes (see reference 60 for details). In addition, the βNTI in combination with a modified Raup-Crick index (RCbray) was further used to quantify the relative influence of major ecological processes governing the sediment bacterial communities (60). βNTI values higher than 2 or lower than −2 indicate the influence of variable selection and homogeneous selection, respectively (14). If the |βNTI| is <2 but with an RCbray value lower than −0.95 or higher than +0.95, the community assembly is governed by homogenizing dispersal or dispersal limitation, respectively. In addition, a |βNTI| of <2 and an |RCbray| of <0.95 suggest that the community assembly is not dominated by any single process, as described above (61).
Cooccurrence network analysis.
To improve statistical confidence, we selected only those OTUs that were detected in more than 80% of the samples for network analysis. Subsequently, all possible pairwise Spearman’s rank correlations (r) between those OTUs were calculated (9). Only statistically significant (r > 0.8; P < 0.01) correlations were incorporated into network analyses. Network visualization, module detection, and topology resulting from network analysis were performed using the Gephi interactive platform (WebAtlas, Paris, France) (62). Modularity values of >0.4 indicate that the partition produced by the modularity algorithm can be used to detect distinct communities within the network. It shows that there are nodes in the network that are more densely connected than the rest of the network; in addition, it indicates that the module density is noticeably higher than the overall average (63).
Statistical analysis.
One-way analysis of variance (ANOVA) was used to test for significant differences in physicochemical properties, α-diversity, and bacterial composition across the sampling transects using SPSS statistical software (SPSS, Chicago, IL, USA). Permutational multivariate analysis of variance with the Adonis function was performed based on Bray-Curtis dissimilarity in order to evaluate the differences in community compositions between sediment transects.
A map containing a description of the sampling sites was constructed using ARCGIS 10.1. A nonparametric multidimensional scaling (NMDS) analysis was performed to illustrate the similarity between different sediment bacterial groups based on the Bray-Curtis distances. The color-mapped NMDS plots visualized the relationship between each major driving physicochemical factor and the sediment bacterial community. Pearson correlation was employed to explore the relationship between α-diversity, composition, and sediment physicochemical properties. Mantel tests demonstrated the correlations for environmental variability and the variation in bacterial β-diversity (Bray-Curtis distances) between sediment transects. Distance-based multivariate linear modeling, which uses the Bray-Curtis distances between samples, was performed by using the DISTLM_forward3 program (64). The results were used to investigate the correlation between bacterial β-diversity and environmental variations. “Marginal tests” assessed the contribution of each variable taken alone, and “sequential tests” with forward selection evaluated the cumulative contribution of the variables to community variation. All statistical analyses were performed with R statistical software (http://www.r-project.org) unless otherwise indicated.
Accession number(s).
Raw sequences were submitted to the NCBI Sequence Read Archive with the accession number SRP156339.
Supplementary Material
ACKNOWLEDGMENTS
The work was supported by the National Natural Science Foundation of China (41601517), the Natural Science Foundation of Ningbo University (XYL17023), and the K. C. Wong Magna Fund of Ningbo University.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02602-18.
REFERENCES
- 1.Duarte CM, Middelburg JJ, Caraco N. 2005. Major role of marine vegetation on the oceanic carbon cycle. Biogeosciences 2:1–8. doi: 10.5194/bg-2-1-2005. [DOI] [Google Scholar]
- 2.Bowen JL, Morrison HG, Hobbie JE, Sogin ML. 2012. Salt marsh sediment diversity: a test of the variability of the rare biosphere among environmental replicates. ISME J 6:2014–2023. doi: 10.1038/ismej.2012.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deegan LA, Johnson DS, Warren RS, Peterson BJ, Fleeger JW, Fagherazzi S, Wollheim WM. 2012. Coastal eutrophication as a driver of salt marsh loss. Nature 490:388–392. doi: 10.1038/nature11533. [DOI] [PubMed] [Google Scholar]
- 4.Madsen EL. 2011. Microorganisms and their roles in fundamental biogeochemical cycles. Curr Opin Biotechnol 22:456–464. doi: 10.1016/j.copbio.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 5.Liu J, Chen X, Shu HY, Lin XR, Zhou QX, Bramryd T, Shu WS, Huang LN. 2018. Microbial community structure and function in sediments from e-waste contaminated rivers at Guiyu area of China. Environ Pollut 235:171–179. doi: 10.1016/j.envpol.2017.12.008. [DOI] [PubMed] [Google Scholar]
- 6.Blum LK, Roberts MS, Garland JL, Mills AL. 2004. Distribution of microbial communities associated with the dominant high marsh plants and sediments of the United States East Coast. Microb Ecol 48:375–388. doi: 10.1007/s00248-003-1051-6. [DOI] [PubMed] [Google Scholar]
- 7.Crain CM, Albertson LK, Bertness MD. 2008. Secondary succession dynamics in estuarine marshes across landscape-scale salinity gradients. Ecology 89:2889–2899. [DOI] [PubMed] [Google Scholar]
- 8.Dini-Andreote F, de Cassia Pereira e Silva M, Triado-Margarit X, Casamayor EO, van Elsas JD, Salles JF. 2014. Dynamics of bacterial community succession in a salt marsh chronosequence: evidences for temporal niche partitioning. ISME J 8:1989–2001. doi: 10.1038/ismej.2014.54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Faust K, Raes J. 2012. Microbial interactions: from networks to models. Nat Rev Microbiol 10:538–550. doi: 10.1038/nrmicro2832. [DOI] [PubMed] [Google Scholar]
- 10.Mo Y, Zhang W, Yang J, Lin Y, Yu Z, Lin S. 2018. Biogeographic patterns of abundant and rare bacterioplankton in three subtropical bays resulting from selective and neutral processes. ISME J 12:2198–2210. doi: 10.1038/s41396-018-0153-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jones AC, Hambright KD, Caron DA. 2018. Ecological patterns among bacteria and microbial eukaryotes derived from network analyses in a low-salinity lake. Microb Ecol 75:917–929. doi: 10.1007/s00248-017-1087-7. [DOI] [PubMed] [Google Scholar]
- 12.Ma B, Wang HZ, Dsouza M, Lou J, He Y, Dai ZM, Brookes PC, Xu JM, Gilbert JA. 2016. Geographic patterns of co-occurrence network topological features for soil microbiota at continental scale in eastern China. ISME J 10:1891–1901. doi: 10.1038/ismej.2015.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hu A, Ju F, Hou L, Li J, Yang X, Wang H, Mulla SI, Sun Q, Bürgmann H, Yu CP. 2017. Strong impact of anthropogenic contamination on the co-occurrence patterns of a riverine microbial community. Environ Microbiol 19:4993–5009. doi: 10.1111/1462-2920.13942. [DOI] [PubMed] [Google Scholar]
- 14.Dini-Andreote F, Stegen JC, van Elsas JD, Salles JF. 2015. Disentangling mechanisms that mediate the balance between stochastic and deterministic processes in microbial succession. Proc Natl Acad Sci U S A 112:E1326–E1332. doi: 10.1073/pnas.1414261112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhou JZ, Ning DL. 2017. Stochastic community assembly: does it matter in microbial ecology? Microbiol Mol Biol Rev 81:e00002-17. doi: 10.1128/MMBR.00002-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shi Y, Li Y, Xiang X, Sun R, Yang T, He D, Zhang K, Ni Y, Zhu YG, Adams JM, Chu H. 2018. Spatial scale affects the relative role of stochasticity versus determinism in soil bacterial communities in wheat fields across the North China Plain. Microbiome 6:27. doi: 10.1186/s40168-018-0409-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.LeBrun ES, King RS, Back JA, Kang S. 2018. Microbial community structure and function decoupling across a phosphorus gradient in streams. Microb Ecol 75:64–73. doi: 10.1007/s00248-017-1039-2. [DOI] [PubMed] [Google Scholar]
- 18.Hubbell SP. 2001. The unified neutral theory of biodiversity and biogeography, vol 32 Princeton University Press, Princeton, NJ. [Google Scholar]
- 19.Chase JM, Myers JA. 2011. Disentangling the importance of ecological niches from stochastic processes across scales. Philos Trans R Soc Lond B Biol Sci 366:2351–2363. doi: 10.1098/rstb.2011.0063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao X, Wu M, Gu B, Chen Y, Liang X. 2013. Nutrient retention in plant biomass and sediments from the salt marsh in Hangzhou Bay estuary, China. Environ Sci Pollut Res Int 20:6382–6391. doi: 10.1007/s11356-013-1698-6. [DOI] [PubMed] [Google Scholar]
- 21.Ren BH, Hu YM, Chen BD, Zhang Y, Thiele J, Shi RJ, Liu M, Bu RC. 2018. Soil pH and plant diversity shape soil bacterial community structure in the active layer across the latitudinal gradients in continuous permafrost region of northeastern China. Sci Rep 8:5619. doi: 10.1038/s41598-018-24040-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu J, Yang H, Zhao M, Zhang XH. 2014. Spatial distribution patterns of benthic microbial communities along the Pearl Estuary, China. Syst Appl Microbiol 37:578–589. doi: 10.1016/j.syapm.2014.10.005. [DOI] [PubMed] [Google Scholar]
- 23.Bowman JP, McCuaig RD. 2003. Biodiversity, community structural shifts, and biogeography of prokaryotes within Antarctic continental shelf sediment. Appl Environ Microbiol 69:2463–2483. doi: 10.1128/AEM.69.5.2463-2483.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mußmann M, Pjevac P, Krüger K, Dyksma S. 2017. Genomic repertoire of the Woeseiaceae/JTB255, cosmopolitan and abundant core members of microbial communities in marine sediments. ISME J 11:1276–1281. doi: 10.1038/ismej.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dyksma S, Bischof K, Fuchs BM, Hoffmann K, Meier D, Meyerdierks A, Pjevac P, Probandt D, Richter M, Stepanauskas R, Mußmann M. 2016. Ubiquitous Gammaproteobacteria dominate dark carbon fixation in coastal sediments. ISME J 10:1939–1953. doi: 10.1038/ismej.2015.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Holt JG, Kreig NR, Sneath PHA, Stanley JT, Williams ST (ed). 1994. Bergey’s manual of determinative bacteriology, 9th ed Lippincott Williams & Wilkins, Baltimore, MD. [Google Scholar]
- 27.Liang B, Wang LY, Zhou Z, Mbadinga SM, Zhou L, Liu JF, Yang SZ, Gu JD, Mu BZ. 2016. High frequency of Thermodesulfovibrio spp. and Anaerolineaceae in association with Methanoculleus spp. in a long-term incubation of n-alkanes-degrading methanogenic enrichment culture. Front Microbiol 7:1431. doi: 10.3389/fmicb.2016.01431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan JF, Wang L, Hu Y, Tsang YF, Zhang YN, Wu JH, Fu XH, Sun Y. 2018. Plant litter composition selects different soil microbial structures and in turn drives different litter decomposition pattern and soil carbon sequestration capability. Geoderma 319:194–203. doi: 10.1016/j.geoderma.2018.01.009. [DOI] [Google Scholar]
- 29.Rooney-Varga JN, Anderson RT, Fraga JL, Ringelberg D, Lovley DR. 1999. Microbial communities associated with anaerobic benzene degradation in a petroleum-contaminated aquifer. Appl Environ Microbiol 65:3056–3063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snoeyenbos-West OL, Nevin KP, Anderson RT, Lovley DR. 2000. Enrichment of Geobacter species in response to stimulation of Fe(III) reduction in sandy aquifer sediments. Microb Ecol 39:153–167. [DOI] [PubMed] [Google Scholar]
- 31.Han XF, Zhang WG, Chen MR, Yu LZ. 2003. Influence of plant on geochemical cycling of iron and the magnetic properties of intertidal sediments in the Yangtze Estuary. Acta Sedimentol Sin 3:495–499. (In Chinese.) [Google Scholar]
- 32.Liu JY, Zhang WG, Dong Y, Ge Y, Bai XX, Nguyen TTH, Chen MR. 2016. Magnetic properties of iron plaque on roots of Phragmites australis in tidal flat of the Yangtze Estuary. Acta Sci Circumstantiae 36:3354–3362. (In Chinese.) [Google Scholar]
- 33.Lozupone CA, Knight R. 2007. Global patterns in bacterial diversity. Proc Natl Acad Sci U S A 104:11436–11440. doi: 10.1073/pnas.0611525104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Guo XP, Lu DP, Niu ZS, Feng JN, Chen YR, Tou FY, Liu M, Yang Y. 2018. Bacterial community structure in response to environmental impacts in the intertidal sediments along the Yangtze Estuary, China. Mar Pollut Bull 126:141–149. doi: 10.1016/j.marpolbul.2017.11.003. [DOI] [PubMed] [Google Scholar]
- 35.Gilbert JA, Field D, Swift P, Newbold L, Oliver A, Smyth T, Somerfield PJ, Huse S, Joint I. 2009. The seasonal structure of microbial communities in the western English Channel. Environ Microbiol 11:3132–3139. doi: 10.1111/j.1462-2920.2009.02017.x. [DOI] [PubMed] [Google Scholar]
- 36.Lauber CL, Strickland MS, Bradford MA, Fierer N. 2008. The influence of soil properties on the structure of bacterial and fungal communities across land-use types. Soil Biol Biochem 40:2407–2415. doi: 10.1016/j.soilbio.2008.05.021. [DOI] [Google Scholar]
- 37.Horner-Devine MC, Silver JM, Leibold MA, Bohannan BJM, Colwell RK, Fuhrman JA, Green JL, Kuske CR, Martiny JBH, Muyzer G, Ovreås L, Reysenbach A-L, Smith VH. 2007. A comparison of taxon co-occurrence patterns for macro- and microorganisms. Ecology 88:1345–1353. [DOI] [PubMed] [Google Scholar]
- 38.Deng Y, Jiang YH, Yang Y, He Z, Luo F, Zhou JZ. 2012. Molecular ecological network analyses. BMC Bioinformatics 13:113. doi: 10.1186/1471-2105-13-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Fan KK, Cardona C, Li YT, Shi Y, Xiang XJ, Shen CC, Wang HF, Gilbert JA, Chu HY. 2017. Rhizosphere-associated bacterial network structure and spatial distribution differ significantly from bulk soil in wheat crop fields. Soil Biol Biochem 113:275–284. doi: 10.1016/j.soilbio.2017.06.020. [DOI] [Google Scholar]
- 40.Layeghifard M, Hwang DM, Guttman DS. 2017. Disentangling interactions in the microbiome: a network perspective. Trends Microbiol 25:217–228. doi: 10.1016/j.tim.2016.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fierer N, Lennon JT. 2011. The generation and maintenance of diversity in microbial communities. Am J Bot 98:439–448. doi: 10.3732/ajb.1000498. [DOI] [PubMed] [Google Scholar]
- 42.Jones SE, Lennon JT. 2010. Dormancy contributes to the maintenance of microbial diversity. Proc Natl Acad Sci U S A 107:5881–5886. doi: 10.1073/pnas.0912765107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kearns PJ, Angell JH, Howard EM, Deegan LA, Stanley RH, Bowen JL. 2016. Nutrient enrichment induces dormancy and decreases diversity of active bacteria in salt marsh sediments. Nat Commun 7:12881. doi: 10.1038/ncomms12881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pujalte MJ, Lucena T, Ruvira MA, Arahai DR, Macián MC. 2014. The family Rhodobacteraceae, p 439–512. In Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (ed), The prokaryotes: alphaproteobacteria and betaproteobacteria, 4th ed Springer-Verlag, Berlin, Germany. [Google Scholar]
- 45.Feng GF, Sun W, Zhang FL, Karthik L, Li ZY. 2016. Inhabitancy of active Nitrosopumilus-like ammonia-oxidizing archaea and Nitrospira nitrite-oxidizing bacteria in the sponge Theonella swinhoei. Sci Rep 6:24966. doi: 10.1038/srep24966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vila-Costa M, Bartrons M, Catalan J, Casamayor EO. 2014. Nitrogen-cycling genes in epilithic biofilms of oligotrophic high-altitude lakes (central Pyrenees, Spain). Microb Ecol 68:60–69. doi: 10.1007/s00248-014-0417-2. [DOI] [PubMed] [Google Scholar]
- 47.Yan W, Ma H, Shi G, Li Y, Sun B, Xiao X, Zhang Y. 2017. Independent shifts of abundant and rare bacterial populations across east Antarctica glacial foreland. Front Microbiol 8:1534. doi: 10.3389/fmicb.2017.01534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chase JM. 2010. Stochastic community assembly causes higher biodiversity in more productive environments. Science 328:1388–1391. doi: 10.1126/science.1187820. [DOI] [PubMed] [Google Scholar]
- 49.Mendes LW, Kuramae EE, Navarrete AA, van Veen JA, Tsai SM. 2014. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J 8:1577–1587. doi: 10.1038/ismej.2014.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nemergut DR, Schmidt SK, Fukami T, O’Neill SP, Bilinski TM, Stanish LF, Knelman JE, Darcy JL, Lynch RC, Wickey P, Ferrenberg S. 2013. Patterns and processes of microbial community assembly. Microbiol Mol Biol Rev 77:342–356. doi: 10.1128/MMBR.00051-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Agricultural Chemistry Committee of China. 1983. Conventional methods of soil and agricultural chemistry analysis. Science Press, Beijing, China: (In Chinese.) [Google Scholar]
- 52.Wang H, Ibekwe AM, Ma J, Wu L, Lou J, Wu Z, Liu R, Xu J, Yates SR. 2014. A glimpse of Escherichia coli O157:H7 survival in soils from eastern China. Sci Total Environ 476–477:49–56. doi: 10.1016/j.scitotenv.2014.01.004. [DOI] [PubMed] [Google Scholar]
- 53.Parada AE, Needham DM, Fuhrman JA. 2016. Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ Microbiol 18:1403–1414. doi: 10.1111/1462-2920.13023. [DOI] [PubMed] [Google Scholar]
- 54.Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R. 2011. UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27:2194–2200. doi: 10.1093/bioinformatics/btr381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG. 2013. Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10:57–59. doi: 10.1038/nmeth.2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 58.Price MN, Dehal PS, Arkin AP. 2009. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 26:1641–1650. doi: 10.1093/molbev/msp077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Stegen JC, Lin X, Konopka AE, Fredrickson JK. 2012. Stochastic and deterministic assembly processes in subsurface microbial communities. ISME J 6:1653–1664. doi: 10.1038/ismej.2012.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stegen JC, Lin X, Fredrickson JK, Chen X, Kennedy DW, Murray CJ, Rockhold ML, Konopka A. 2013. Quantifying community assembly processes and identifying features that impose them. ISME J 7:2069–2079. doi: 10.1038/ismej.2013.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Stegen JC, Lin X, Fredrickson JK, Konopka AE. 2015. Estimating and mapping ecological processes influencing microbial community assembly. Front Microbiol 6:370. doi: 10.3389/fmicb.2015.00370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Bastian M, Heymann S, Jacomy M. 2009. Gephi: an open source software for exploring and manipulating networks, p 361–362. In Proceedings of the 3rd International AAAI Conference on Weblogs and Social Media. Association for the Advancement of Artificial Intelligence, Menlo Park, CA. [Google Scholar]
- 63.Newman ME. 2006. Modularity and community structure in networks. Proc Natl Acad Sci U S A 103:8577–8582. doi: 10.1073/pnas.0601602103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Anderson MJ. 2013. DISTLM forward: a FORTRAN computer program to calculate a distance-based multivariate analysis for a linear model using forward selection. Department of Statistics, University of Auckland, Auckland, New Zealand. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.