Bacteroides are major members of the gut microbiota, and host-specific organisms within this genus have been used extensively to gain information on pollution sources. This study provides a broad view of the population structure of Bacteroides within sewage to contextualize the well-studied HF183 marker for a human-associated Bacteroides. The study also delineates host-specific sequence patterns across multiple hypervariable regions of the 16S rRNA gene to improve our ability to use sequence data to assess water quality. Here, we demonstrate that regions downstream of the HF183 marker are nonspecific but other potential human-associated markers are present. Furthermore, we show the most abundant Bacteroides in sewage is free living, rather than host associated, and specifically found in sewage. Quantitative PCR assays that target organisms specific to sewer pipes offer measures that are independent of the human microbiome for identifying sewage pollution in water.
KEYWORDS: Bacteroides, HF183, human fecal indicator, microbial source tracking, next-generation sequencing, population structure, qPCR, sewage
ABSTRACT
The identification of sewage contamination in water has primarily relied on the detection of human-associated Bacteroides using markers within the V2 region of the 16S rRNA gene. Despite the establishment of multiple assays that target the HF183 cluster (i.e., Bacteroides dorei) and other Bacteroides organisms (e.g., Bacteroides thetaiotaomicron), the potential for more human-associated markers in this genus has not been explored in depth. We examined the Bacteroides population structure in sewage and animal hosts across the V4V5 and V6 hypervariable regions. Using near-full-length cloned sequences, we identified the sequences in the V4V5 and V6 hypervariable regions that are linked to the HF183 marker in the V2 region and found these sequences were present in multiple animals. In addition, the V4V5 and V6 regions contained human fecal marker sequences for organisms that were independent of the HF183 cluster. The most abundant Bacteroides in untreated sewage was not human associated but pipe derived. Two TaqMan quantitative PCR (qPCR) assays targeting the V4V5 and V6 regions of this organism were developed. Validation studies using fecal samples from seven animal hosts (n = 76) and uncontaminated water samples (n = 30) demonstrated the high specificity of the assays for sewage. Freshwater Bacteroides were also identified in uncontaminated water samples, demonstrating that measures of total Bacteroides do not reflect fecal pollution. A comparison of two previously described human Bacteroides assays (HB and HF183/BacR287) in municipal wastewater influent and sewage-contaminated urban water samples revealed identical results, illustrating the assays target the same organism. The detection of sewage-derived Bacteroides provided an independent measure of sewage-impacted waters.
IMPORTANCE Bacteroides are major members of the gut microbiota, and host-specific organisms within this genus have been used extensively to gain information on pollution sources. This study provides a broad view of the population structure of Bacteroides within sewage to contextualize the well-studied HF183 marker for a human-associated Bacteroides. The study also delineates host-specific sequence patterns across multiple hypervariable regions of the 16S rRNA gene to improve our ability to use sequence data to assess water quality. Here, we demonstrate that regions downstream of the HF183 marker are nonspecific but other potential human-associated markers are present. Furthermore, we show the most abundant Bacteroides in sewage is free living, rather than host associated, and specifically found in sewage. Quantitative PCR assays that target organisms specific to sewer pipes offer measures that are independent of the human microbiome for identifying sewage pollution in water.
INTRODUCTION
Human fecal pollution in urban waters from untreated sewage contains pathogenic bacteria, virus, and protozoa that cause gastrointestinal diseases through the ingestion of polluted water (1, 2) or skin, eye, and respiratory infections through direct contact (1). Human sources of fecal pollution are considered a higher health risk than animal sources of fecal pollution (3, 4). The detection of traditional fecal indicator bacteria (FIB), such as fecal coliforms, Escherichia coli, and enterococci (5), does not distinguish human sources from animal sources of fecal pollution, because they commonly occur in all mammalian intestines (6). Many studies have demonstrated a lack of correlation between FIB levels and pathogen occurrence or adverse human health outcomes (7–10), because some sources of fecal pollution do not carry human pathogens.
Microbial source tracking (MST) methods, which rely on the quantification of levels of certain fecal microorganisms that are specific to a host (11), have been used for fecal source identification for a number of years (12). To date, the most characterized microorganisms used in MST belong to the genus Bacteroides within order Bacteroidales (see Table S1 in the supplemental material), one of the most predominant genera in the human gut. The best-studied human Bacteroides marker, the HF183 marker, is found in Bacteroides dorei and its closely related taxa (13) and is located in the V2 hypervariable region of the 16S rRNA gene. This marker was first reported by Bernhard and Field (14) as a PCR assay (i.e., HF183F/Bac708R) (Table S1) (14–16).
Because most human-associated Bacteroides markers have been developed using clone libraries that target Bacteroidales using the Bac708R primer (13), quantitative PCR (qPCR) assays have been limited to the V2-V4 hypervariable regions. Assays and their average specificities include HF183/SSHBac-R (91.1%) (17–21), HF183/BFDrev (76.8%) (22, 23), HB (90.9%) (24, 25), HF183/BacR287 (91.2%) (21, 23, 25), BacHum-UCD (77.9%) (15, 18, 20, 21, 26), BacH (92.6%) (15, 21, 27), HuBac (54.5%) (15, 16, 18, 28), Human-Bac1 (44.4%) (15, 29), and BacHuman (81.5%) (30). More detailed information on these assays is found in Table S1, and a primer map is shown in Fig. S1.
In addition to the assays that use the HF183 marker directly as a forward primer (i.e., HF183/SSHBac_R, HF183/BFDrev, HB, and HF183/BacR287), assays that use primers or probes that overlap the HF183 marker (Fig. S1), such as the BacHum-UCD and BacH assays, also reported low-level animal cross-reactivity, further demonstrating the human specificity of the HF183 marker. Human Bacteroides fecal marker PCR/qPCR assays have also been developed for the 16S rRNA gene and genomic sequences of Bacteroides thetaiotaomicron (Table S1) (22, 31, 32), another predominant species in human feces that typically shows up more often in human feces than in animal sources (31, 33). Overall, there is no one assay that is exclusively human specific, and animal source cross-reactions were reported for all the PCR/qPCR assays mentioned above, such as cat, dog, pig, chicken, turkey, cow, and deer (Table S1).
The goal of this study was to explore the potential of genus Bacteroides for MST, in addition to the widely applied HF183 marker, to expand methods for sewage detection and quantification. The characterization of the population structure using hypervariable regions beyond V2, and delineation of the linkage patterns among markers in different regions, may reveal additional host-preferred and host-specific Bacteroides organisms and help couple community sequencing data to marker assays. To explore the host specificity of this genus, we compared the population structure of Bacteroides in 27 sewage and 151 animal fecal samples using next-generation sequencing (NGS) across multiple variable regions. We also explored the human Bacteroides V2, V4V5, and V6 region sequence linkages and specificities by analyzing V2-V9 region sewage clone libraries. Several human-associated Bacteroides markers that were not related to the HF183 marker, including one from the V4V5 region and one from the V6 region, were identified from NGS data. We also identified a sewage-associated Bacteroides that appears to be specifically propagated in urban sewer systems and developed two TaqMan qPCR assays targeting the V4V5 region and the V6 region.
RESULTS
Bacteroides population structures in sewage, animal hosts, and freshwater samples.
We applied oligotyping to V6 region sequences of Bacteroides from 27 sewage influent samples and 151 animal fecal samples. In total, 1.48 × 107 Bacteroides reads (97.66% of total reads) were analyzed, including 1.96 × 106 reads from sewage samples and 1.29 × 107 reads from animal fecal samples. The oligotype (n = 1,730) distribution in each sample is shown in Fig. 1. Eighty-two oligotypes were exclusively found in sewage, and of these, 30 were among the 100 most abundant sewage oligotypes. The sewage oligotype patterns were consistent between U.S. and Spain sewage samples and were distinguishable from animal hosts (Fig. 1). The animal and sewage oligotype profiles were dissimilar in individual host groups (n = 8, adonis R2 = 0.419, P = 0.001) and in sewage compared to those of a pooled animal group (adonis R2 = 0.119, P = 0.001). A Bray-Curtis dissimilarity-based hierarchical cluster analysis of Bacteroides oligotypes demonstrated animal and sewage samples clustered by source, and sewage was the most distant sample group compared to all other animal groups (see Fig. S2). In addition, certain oligotypes were associated with specific hosts (Fig. 1 and S2). For example, 80 oligotypes were found only in cows, 11 were only in deer, 5 were only in dogs, and 4 were only in pigs, indicating that genus Bacteroides is a good target for animal fecal markers.
FIG 1.
Oligotype patterns of the V6 region sequences of the Bacteroides 16S rRNA gene in sewage and seven animal hosts. Samples are grouped by host types. Colors correspond to different sequences in the profile, with the bar height representing the oligotype relative abundance.
We also examined freshwater samples using the Indicspecies package to identify potential freshwater Bacteroides sequences based on the relative abundances of unique V6 sequences in freshwater samples (n = 35) compared with those in sewage and animal samples (n = 178). Three unique Bacteroides V6 sequences were found only in freshwater samples (mean ± standard deviation [SD] relative abundance, 4.7% ± 9.3% of all Bacteroides), and 27 unique sequences were found in freshwater with comparatively low occurrence in sewage (relative abundance 37.4% ± 32.1% of all Bacteroides compared with 1.5% ± 0.74% in sewage) and with no occurrence in animal samples. BLAST results against the NCBI nucleotide database showed no identical matches between a human fecal source and the three freshwater-specific sequences; for the 27 “freshwater-preferred” sequences, only two were found to have identical matches with a human stool source, and another two were found to match with bioreactors using farm animal waste (e.g., cow and pig). This indicates that there is a potential for Bacteroides populations to occur in the freshwater environment in the absence of fecal contamination.
Identification of V4V5 and V6 regions downstream of the HF183 human Bacteroides marker.
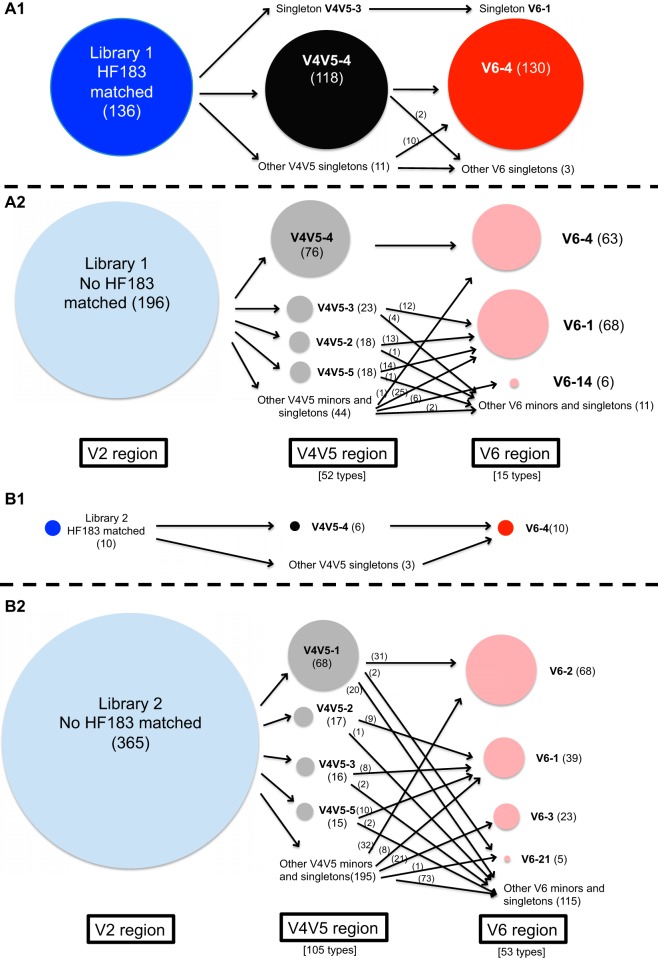
We utilized our sewage clone libraries to examine the specific marker sequences in the V4V5 and V6 regions of 16S rRNA gene that were downstream of the HF183 marker sequence. A total of 136 clones matching the HF183 marker (41% of sequences) were found in library 1, which targeted human Bacteroides by using the BacH_f primer to make the library. There was one primary V4V5 and one primary V6 sequence downstream of the HF183 organism (Fig. 2A1). Only 3% of sequences in library 2 (representing total Bacteroides from sewage) had the HF183 marker (Fig. 2B1), indicating that the HF183 marker cluster is a small fraction of Bacteroides in sewage. All HF183-positive clones in library 2 had the BacH_f primer site, supporting that library 1 was inclusive of HF183 cluster organisms.
FIG 2.
Associations of the V2, V4V5, and V6 regions of sewage Bacteroides clones. (A1) Associations of the three regions in HF183-matched clones in library 1. (A2) Associations in non-HF183 clones in library 1. (B1) Associations in HF183-matched clones in library 2. (B2) Associations in non-HF183-clones in library 2. The deep/light blue circles represent V2 region, black/gray circles represent V4V5 region, and red/pink circles represent V6 region. Circle sizes are directly proportional to the sequence read numbers (except the non-HF183-matched V2 region in library 2, which is smaller than the proportional area). The unique type numbers in V4V5 and V6 regions are annotated at the bottom of A2 and B2. Numbers within parentheses indicate clone numbers. Clones that have no NGS matches are not included.
We used the NGS data set of animal fecal samples to examine the host distribution of the primary sequences downstream of the HF183 marker (designated V4V5-4 and V6-4 according to their rank of abundance in sewage samples in corresponding NGS data sets) and found they occurred in multiple animals. The V4V5-4 sequence occurred in 40% of the samples, including cat, dog, cow, and deer, and the V6-4 sequence was found in 16.8% of the samples, including cat, dog, cow, pig, chicken, deer, raccoon, and rabbit, indicating the regions downstream of the HF183 marker are not specific to humans (see Fig. S3). We tested a subset of these samples for the HF183 marker by qPCR in cases where DNA material was available. Overall, 2 of 13 samples containing the V4V5-4 sequence were positive by the HF183/BacR287 assay. For available samples containing the V6-4 sequences, only one of three samples was positive by the HF183/BacR287 assay. These results support that the downstream region is not specific, as opposed to these animals carrying an HF183-positive organism.
Potential human and sewage markers in Bacteroides V4V5 and V6 regions that are not associated with the HF183 marker cluster.
We aimed to identify additional human- or sewage-associated Bacteroides markers in the V4V5 and V6 regions so that they could be used in PCR applications but, more importantly, could also be used as markers in sequencing data sets, since these regions are commonly sequenced. We applied the Indicspecies permutation test and identified markers from the V4V5 region and V6 region that were >90% sewage specific and sensitive for sewage. Within these, there were nearly 20-fold more V6 region markers than V4V5 markers that were 100% specific and sensitive to sewage (see Fig. S4). These results may be due to the higher variability in the V6 region, which provides more resolution and therefore more unique human- or sewage-associated sequences than the V4V5 region. Although the V4V5 region had fewer markers, there were two that had 100% specificity and sensitivity (V4V5-1 and V4V5-7), both of which did not appear in human gut microbiome data sets, suggesting they were resident organisms within the sewer pipes. There were seven markers identified in the V6 region, with only one of these associated with human feces. The most abundant sewer pipe-associated markers fell within a clade of Bacteroides graminisolvens (see Fig. S5). Human-associated and sewer pipe-associated markers and their specificities for the V4V5 region are listed in Table S2, and markers in the V6 region are found in Table S3.
Assay development and sensitivity for sewage detection.
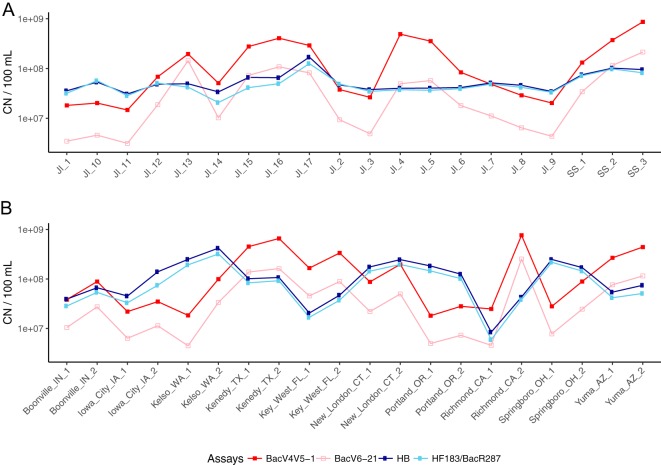
TaqMan qPCR assays were developed to target the most abundant Bacteroides in sewage, which were designated the BacV4V5-1 assay and BacV6-21 assay (Table 1). We tested 40 U.S. sewage influent samples, including 20 from Milwaukee and 20 from ten other U.S. cities, using the two assays and compared the results with those from the HB and HF183/BacR287 assays (Fig. 3). All four assays showed 100% sensitivity in sewage samples. In Milwaukee sewage samples, the BacV6-21 assay showed approximately the same magnitude of copy numbers (CN) (4.9 × 107 ± 5.8 × 107 CN/100 ml [mean ± SD]) as the HB assay (5.8 × 107 ± 3.3 × 107 CN/100 ml) and the HF183/BacR287 assay (5.1 × 107 ± 2.6 × 107 CN/100 ml), but was found to have greater fluctuation. The BacV4V5-1 marker was approximately 4-fold higher than the V6-21 marker, with CN equal to 1.9 × 108 ± 2.2 × 108 per 100 ml. In other U.S. cities, similar sewage sensitivities were detected for BacV4V5-1 and BacV6-21 assays, with the BacV6-21 assay showing 5.5 × 107 ± 6.67 × 107 CN/100 ml and the BacV4V5-1 assay showing 1.9 × 108 ± 2.3 × 108 CN/100 ml. The V4V5-1 marker mirrored the V6-21 marker fluctuation, suggesting they target the same organism (Pearson’s r = 0.931, P < 2.2e−16). Likewise, the HB assay and the HF183/BacR287 assay were tightly coupled (Pearson’s r = 0.990, P < 2.2e−16). The V4V5-1 and V6-21 markers were not correlated to either the HB or the HF183/BacR287 marker, with the Pearson’s r values ranging from −0.083 to −0.061.
TABLE 1.
BacV4V5-1 and BacV6-21 marker assays
| Assay marker | Primer or probe name and sequence (5′→3′) |
||
|---|---|---|---|
| Forward primer | Probe | Reverse primer | |
| V4V5-1 | Bac573f, AAGGGAGCGTAGGTTGACATA | Bac599p, FAM-CAGCTGTGAAAGTTTACGGCTC-NFQ-MGB | Bac673r, CGCCCACCTCTTGTACACT |
| V6-21 | Bac989f, GCTTGAATTGCAGAGGAATA | Bac1010p, FAM-AGTTGAAAGATTATGGCCGCA-NFQ-MGB | Bac1162r, GCAGTCTCACTAGAGTCCTCAG |
FIG 3.
Comparison of the BacV4V5-1, BacV6-21, HB, and HF183/BacR287 assay copy numbers (CN) in sewage samples. (A) Results from four assays showing CN in 20 sewage samples from Jones Island (JI) and South Shore (SS) wastewater treatment plants, Milwaukee, WI. (B) Results from four assays showing CN in 20 sewage samples from ten other U.S. cities, each tested at two different time points.
The four Bacteroides assays were also tested using freshwater samples that had no known evidence of human fecal pollution (n = 30). The HB, HF183/BacR287, and BacV6-21 assays all showed negative results. The BacV4V5-1 assay, however, showed low CN in two lake/harbor samples (200 ± 6 CN/100 ml [mean ± SD]) and six beach samples (180 ± 111 CN/100 ml). All qPCR results for the four assays are shown in Data Set S1.
Assay validation in animal fecal samples.
We validated the newly designed assays using samples from a total of 76 animals in the formats of individual and pooled samples, including from cat, dog, pig, cow, deer, gull, and chicken (Table 2). The BacV4V5-1 and BacV6-1 assays showed higher specificity than the two human-associated assays. The BacV4V5-1 assay gave a very low signal (5.2 CN/ng DNA) in one pig sample (pig pool 3) at 1 ng · μl−1, and was negative at 0.1 ng · μl−1 and 0.01 ng · μl−1 DNA template levels. The BacV6-21 assay was negative for all animals at all three dilutions of DNA template. In contrast, the HB and HF183/BacR287 assays showed sporadic cross-reactivity with animals. The HB assay cross-reacted with one dog pool sample, whereas the HF183/BacR287 assay was negative for this sample (25). Results for all three dilutions of DNA are detailed in Data Set S1.
TABLE 2.
Animal validation results of the Bacteroides assays
| Animal | Total no. (no. of pools containing two samples) | Results for: |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| BacV4V5-1 |
BacV6-21 |
HBa
|
HF183/BacR287a
|
||||||
| No. positive | Average CN per ng DNA/average CN per g feces | No. positive | Average CN per ng DNA/average CN per g feces | No. positive | Average CN per ng DNA/average CN per g feces | No. positive | Average CN per ng DNA/average CN per g feces | ||
| Cat | 13 (1) | 0 | 0 | 0 | 0 | 1 | 8/115,000 | 1 | 5/77,000 |
| Cow | 10 (2) | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Deer | 11 (1) | 0 | 0 | 0 | 0 | 3b ,c | 406/404,000 | 3b ,c | 364/362,000 |
| Dog | 13 (2) | 0 | 0 | 0 | 0 | 2b | 375/3,330,000 | 0 | 0 |
| Pig | 22 (2) | 2b ,c | 5/84,700 | 0 | 0 | 0 | 0 | 0 | 0 |
| Chicken | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Gull | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Partial results for the HB and HF183/BacR287 assay validation were generated in a previous study (25).
A pool was positive in each of these animal groups and was counted as two animals being positive.
The positive pooled samples were also run in format of individuals at 1 ng · μl−1 DNA material level (see Data Set S1 in the supplemental material for details).
Sensitivity of BacV4V5-1, BacV6-21, HB, and HF183/BacR287 assays for environmental water samples.
We tested the BacV4V5-1, BacV6-21, HB, and HF183/BacR287 assays using 20 sewage-contaminated local river water samples and 13 known agricultural contaminated local river water samples. Overall, the four assays were significantly correlated for these environmental water samples (Table 3). The BacV4V5-1 and BacV6-21 assay CN results showed very similar fluctuation patterns and were more highly correlated with each other than with the HB or HF183/BacR287 assay (Table 3). The BacV4V5-1 marker CN was 4.0- ± 1.4-fold (mean ± SD) higher than the BacV6-21 marker CN, which corresponded to levels in the U.S. sewage samples that were tested. The HB and HF183/BacR287 assays showed identical CN fluctuation patterns in sewage-contaminated river samples (see Fig. S6A) and in agricultural contaminated samples containing low levels of sewage (Fig. S6B) and were highly correlated with each other, indicating the equivalency of these two HF183 marker-based assays. In addition, all four assays clearly distinguished human contamination from that of ruminants in agricultural contaminated river samples, with none of the results from the assays proportionally increasing with increasing ruminant contamination (Fig. S6). Detailed CN data are shown in Data Set S1.
TABLE 3.
Pearson’s correlation of the four Bacteroides assays in 20 sewage-contaminated and 13 agricultural contaminated water samples
| Assay | Pearson’s r (P value) for: |
|||
|---|---|---|---|---|
| BacV4V5-1 | BacV6-21 | HB | HF183/BacR287 | |
| BacV4V5-1 | 1.000 | |||
| BacV6-21 | 0.995 (<2.2 × 10−16) | 1.000 | ||
| HB | 0.842 (7.9 × 10−10) | 0.817 (6.7 × 10−9) | 1.000 | |
| HF183/BacR287 | 0.824 (3.8 × 10−9) | 0.792 (3.9 × 10−8) | 0.995 (<2.2 × 10−16) | 1.000 |
DISCUSSION
Potential of genus Bacteroides to contain host and sewage markers.
The identification of human fecal pollution provides evidence to assess public health risks caused by waterborne diseases. The fecal anaerobic microorganism Bacteroides has been utilized as a target for human fecal source detection since the early 2000s, when the specificity of the HF183 cluster was identified (12). Our study further explores the host specificity patterns of this genus among 27 sewage and 151 animal fecal samples across seven hosts by using deep-sequencing data. We demonstrated the host-specific nature of Bacteroides populations, consistent with previous studies using the V4V5 and V6 variable regions (34) and the V2 region (18, 28). The oligotyping results from previous studies and this study suggest that the V6 region from genus Bacteroides could be used as markers for certain animals, such as cows and deer, since specific patterns were evident within these hosts. For example, dairy cow and beef cattle had dissimilar Bacteroides oligotypes, which may be caused by dietary differences (35, 36) (Fig. 1; see also Fig. S2 in the supplemental material). With the high variability among cattle, the development of more restrictive host animal fecal markers could be useful; for example, specific markers targeting dairy cows or cattle on forage diets common to certain regions may be more feasible than employing a “universal” cattle marker.
Multiple markers within the V4V5 and V6 regions were identified as being specific to sewage. Many of these did not match human microbiome organisms, but appeared to originate within the sewer pipes, and included the most abundant Bacteroides in sewage. Bacteroides in mammalian guts is responsible for the breakdown of complex polysaccharides (37–40). In addition, studies have been focused on free-living Bacteroides species, which also have the ability to degrade complex organic matter, such as polysaccharides (41–43). The sewer pipe-derived Bacteroides organism represented by the V4V5-1 and V6-2 (and V6-21) markers closely matched B. graminisolvens based on near-full-length clone sequences (Fig. S5). This organism was isolated from a methanogenic reactor at a cattle farm, where it was implicated in breakdown of hemicellulose (41), and has been detected in a microbial fuel cell reactor, where it performed similar functions (i.e., degrading carbohydrates) (44). Just as Bacteroides has coevolved and been selected for in the human gut (45), it appears that Bacteroides organisms in the urban sewer infrastructure may have been selected for or evolved in sewer pipes as a result of the nutrition available to them from human waste, where they provide further breakdown of material not completely utilized in the gut. Most notable is the ubiquitous occurrence of identical V4V5 and V6 marker sequences in all of the cities studied. It is unknown if these organisms were originally deposited as minor members of the human gut microbiome or if they arose from an environmental source. Given the short transit time in some of the systems studied (i.e., 6 to 24 h) (46), coupled with the high abundance patterns in relation to what is found in human fecal material, the sewer pipe-derived Bacteroides organisms appear to be residents in the system.
We designed assays targeting the most abundant Bacteroides represented by the V4V5-1 marker. The most common marker for this organism in the V6 region was V6-2 (Fig. 2B). However, this sequence was also found in animals. Therefore, we targeted a smaller subpopulation for qPCR assays, represented by the V6-21 marker for the V6 region assay. The sewer pipe-associated marker assays strengthen the capability to detect human sources of pollution, because they are sewer derived and have essentially no cross-reactivity with animal sources, unlike gut-derived organisms, for which the distinguishing members of the community are more often host-preferred organisms versus those that are strictly host specific (6). Furthermore, sewer pipe-associated markers may not be subjected to differences in the human microbiome in different regions, as is observed with some of the human-derived markers (47, 48). Further testing of urban sewer systems worldwide is needed to determine the applicability of the markers in areas where the HF183 marker is low or absent. Importantly, since this organism appears to be free living rather than host associated, further validation studies of uncontaminated water are needed to determine if this organism is exclusively found in sewer systems and similar environments (manure detention ponds, anaerobic digesters, etc.).
In addition, we demonstrated the presence of Bacteroides in a freshwater environment, which differed from Bacteroides in sewage and in the seven animal fecal sources. Bacteroides in freshwater has previously been reported on Cladophora mats (49, 50), which is consistent with the organism’s ability to breakdown complex polysaccharides. In general, there was a single dominant sequence type in an apparently uncontaminated sample, and different samples had different sequence types (Data Set S2, Tab 2), indicating the freshwater Bacteroides population may be very diverse and specific to the location. Freshwater Bacteroides may be detected when using universal Bacteroides marker assays (18), causing false-positive results for fecal pollution detection. In addition, high levels of these organisms may interfere with Bacteroides assays that employ closely related primer sequences.
The HF183 assays and the sewer-associated Bacteroides assays target two independent Bacteroides organisms but are overall correlated.
The high correlation between the two HF183-based assays (Table 3) indicated that they amplify the same Bacteroides organism. In our animal validation results, the HF183/BacR287 assay showed better specificity (93.2%) than the HB assay (90.5%) because of cross-reactivity with certain dog samples in the latter assay. Overall, these two assays showed nearly identical sensitivity patterns among sewage and sewage- and agriculture-contaminated environmental water samples, demonstrating they are interchangeable for the purpose of human fecal source detection. However, their application needs to be considered cautiously when employing the HB marker if dog waste is suspected, and specific testing using a canine marker or verification using a second human marker should be considered.
The BacV4V5-1 and BacV6-21 markers (targeting a sewer pipe-derived Bacteroides) had consistent ratios in sewage and sewage-contaminated environmental water samples (i.e., the CN was approximately 4.0- ± 1.0-fold higher in the BacV4V5-1 assay than in the BacV6-21 assay) and were highly correlated in environmental waters. The linkage of the V4V5-1 and V6-21 markers in clone libraries (Fig. 2B2) verified that these two assays target the same Bacteroides organism. In water samples where sewage was present, all four assays were correlated, demonstrating that they all detect sewage similarly.
Next-generation sequencing revealed potential cross-reactions of human fecal marker with animals.
Having access to a large V4V5 NGS data set allowed us to examine other established human-associated Bacteroides assays targeting the V4 region. We compared the HumanBac-1 (29) and HuBac (28) assays, which both target the V4 region of the Bacteroides 16S rRNA gene, with our V4V5 NGS data set; exact primer and probe matches in both assays were found in cat, dog, pig, cow, deer, and rabbit, suggesting true animal cross-reactions occur, which explains the comparatively low human specificity of these assays (Table S1).
Cross-reaction of human fecal markers with animals can be influenced by multiple complex factors, such as similarities in gut microbial communities as a result of dietary factors (25, 35, 51) and possible animal ingestion of human waste (52). We have previously noted that employing markers from two different bacterial groups, such as Bacteroides and Lachnospiraceae, can increase confidence in results where cross-reactivity is suspected (25). For a well-designed fecal marker qPCR assay (e.g., one optimized for avoiding dimers, hairpin structures, annealing temperature, etc.), NGS could not only be used to verify the assay’s host specificity but also identify closely related sequences that might interfere. Deep sequencing has also been proven to be valuable for the identification of host-associated markers on the scale of the whole microbial community without the effort of constructing a sequence clone library. However, linking different regions to the same organism is difficult without continuous more-extended sequence data, since some variable regions appear to be less discriminatory and found in multiple host types. Therefore, sequencing databases for common regions of 16S rRNA genes in sewage and animal fecal samples from a wide geographical range (e.g., across the United States) with key host information (e.g., animal diet and cohabitation) could help to verify the applicability of markers and interpret site-specific data. Data like these could be shared between research laboratories and would be extremely useful in assay validation in silico, therefore providing substantial evidence of specificity and sensitivity (25).
Combining NGS and qPCR for water quality assessments.
qPCR is indispensable for rapidly quantifying sources of fecal pollution such as human or cattle waste. However, most contamination scenarios are complex, especially in urban environments where there may be sewage contamination mixed with nonpoint sources from storm water that add a significant fecal indicator bacterium burden (24). In addition, there are known sensitivity and specificity issues with individual fecal bacterium markers; the microbiomes of many animals have not been characterized, and so cross-reactivity has not been completely characterized (53). NGS data create a high-resolution inventory of organisms present, and with falling sequencing costs, NGS may be feasible to employ in the future for directly characterizing fecal pollution sources. Computational or machine learning approaches such as Source Tracker (53) or random forest (54) can use sequence abundance patterns of the whole community, or of taxonomic groups, to identify pollution signals within a water sample. These methods rely on signatures of sequences that include their relative abundance patterns within the community, and sequences shared between sources generally do not also share overall relative abundance patterns within the signature (54). Furthermore, fecal bacterium sequences within these data sets that do not match a characterized source could be used to indicate extraneous sources that may be contributing fecal indicator bacteria but are not considered a significant human health risk (i.e., bird or pet waste and urban wildlife). Anchoring the relative abundance derived from sequencing with qPCR for host-associated markers will enable quantification. As the complexity of fecal pollution signals is unraveled, combining NGS with qPCR methods for source tracking may become common metrics in the future for assessing microbial water quality.
MATERIALS AND METHODS
Sample collection and DNA extraction.
Influent sewage samples used for qPCR in this study were from Jones Island (JI) and South Shore (SS) wastewater treatment plants (WWTPs) in Milwaukee, WI (n = 20), along with samples from ten other U.S. cities representing geographical regions of the United States that were sampled in two different seasons over a year (n = 20) (55). Sewage-contaminated river water samples (n = 20) were collected during a 2016 Milwaukee combined sewer overflow (CSO) event. Agricultural contaminated water samples were collected from the Milwaukee River (n = 13) after rains in the spring and early summer in 2014 and 2015; these samples also had evidence of sewage contamination but at 3 to 4 orders of magnitude lower than ruminant contamination as determined using a ruminant marker (56). Freshwater samples that had no evidence of human fecal contamination, i.e., had zero or extremely low colony counts of FIB and were negative in HB and human Lachnospiraceae qPCR assays (25), were collected from Lake Michigan (n = 20) and Milwaukee area beaches (n = 10).
A total number of 76 animal fecal samples, including from 22 pigs, 13 dogs, 12 cats, 11 deer, 10 cows, 4 gulls, and 4 chickens, were collected for qPCR assay validation. Among these animal fecal samples, 46 were extracted in a previous study (25) but rediluted for qPCR experiments in this study. Fecal sample processing and storage were as described previously (25).
All sewage, animal fecal, and environmental water sample details, including their associated studies and qPCR results, are listed in Data Set S1 in the supplemental material.
NGS data used for oligotyping, clone comparisons, and marker identification.
To examine the overall population structure of Bacteroides populations, sequence data generated from two previous studies (25, 34) of the V6 region 16S rRNA gene from 27 sewage samples and 151 animal fecal samples, including hosts of cat, dog, pig, cow, deer, raccoon, and chicken, were analyzed using oligotyping (57). All raw sequences were trimmed using cutadapt software (58) and assembled using PEAR (59) software. Sequences were then classified using GAST (60) with a comparison to SILVA reference database version 132 to parse out Bacteroides sequences. Oligotyping was run using parameters -s (the minimum number of samples with an oligotype present) equal to 9 (5% of total sample), -M (the minimum substantive abundance) equal to 85, and -c (number of base locations) equal to 33. The output of the oligotype count matrix was plotted using ggplot2 package (61) in R (version 3.5.1) (62). The statistical analysis of sewage and animal oligotypes was performed with the adonis function in the vegan package (63) in R.
For clone comparisons and marker identification (described below), V4V5 and V6 sequence data sets from previous studies (25, 34, 55) were obtained from the Visualization and Analysis of Microbial Population Structures platform (VAMPS; https://vamps2.mbl.edu) (64) with reference to SILVA database version 119. A taxbyseq file, which described whole community unique sequences, taxonomy, and abundance in each sample, was used. The total number of sequences for each sample was normalized to the median total sequence count for all samples (V4V5 region data set, 89,341; V6 region data set, 741,189). Singletons were removed to form the whole community NGS data sets. The genus Bacteroides data were then extracted. The samples, their usage in this study, the associated studies, and SRA study accession numbers are listed in Data Set S2, Tab 1.
Sewage clone libraries.
Two sewage clone libraries were generated using four sewage influent samples collected from different U.S. cities (Milwaukee, Palo Alto, Laramie, and Key West) in August 2012 (55). The first clone library (library 1) was constructed using a human Bacteroides group forward primer (BacH_f) (27) and a universal 16S rRNA gene reverse primer (1492R); the BacH_f primer was chosen to form human Bacteroides amplicons that were long enough to cover the V2 region. The second clone library (library 2) was generated using the universal 8F primer and a new reverse primer, designated 1030R (5′-CCACCTTCCTCACATCTTACGA-3′), which was designed to broadly target Bacteroides. The Probe Match function in the Ribosomal Database Project (RDP) (65) demonstrated that the 1030R primer matched 34,100 of 35,602 Bacteroides sequences. The PCR products were cloned into the pCR2.1 vector using the TOPO TA Cloning kit (Invitrogen, Carlsbad, CA), and plasmids were extracted as previously detailed (46). Sanger sequencing was performed with M13F, 331F, and M13R primers using the ABI BigDye Terminator kit (Applied Biosystems, Foster City, CA) on an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA) (46). In all, 332 sequences were generated in library 1 and 375 sequences were generated in library 2. Nearly 64.6% of the Bacteroides sequence types in the V4V5 data set and 35.5% in the V6 NGS data set were represented in library 2.
Linkage of the HF183 marker representing the V2 region with the V4V5 region and the V6 region of Bacteroides.
The sewage clone libraries were compared with the HF183 marker sequence to identify clones containing this marker, and then the corresponding unique V4V5 and V6 sequence types in the clones were identified in the NGS data sets; both comparisons were performed using BLAST+ (66). The V4V5 and V6 sequence types for each HF183 clone were compiled in Excel using the VLOOKUP function.
Freshwater Bacteroides population identification.
We used freshwater samples with no or low levels of fecal pollution (n = 35) that were previously sequenced for the V6 region to identify environmental Bacteroides (Data Set S2, Tab 1). These were compared to the sewage and animal fecal samples used in oligotyping. The “uncontaminated” samples were collected under baseflow conditions (i.e., no rain in the previous 48 h) from Lake Michigan nearshore and offshore surface water (n = 6) as grab samples (67) and from the Milwaukee, Kinnickinnic, and Menomonee Rivers (n = 29) using automated Teledyne ISCO 3700 full-size portable sequential samplers (56). To identify freshwater-preferred Bacteroides sequences, the R package Indicspecies (68) was applied with a setting of 999 permutations. These V6 region Bacteroides sequences are listed in Data Set S2, Tab 2.
Bacteroides marker identification.
We used sewage, sewage-contaminated water, and animal fecal samples previously sequenced for the V4V5 region (25, 55) and V6 region (25, 34) for marker identification (Data Set S2, Tab 1). For the V6 region, 22,006 unique Bacteroides sequences were present from 40 sewage and sewage-contaminated water samples and 156 animal fecal samples; for the V4V5 region, 22,104 unique Bacteroides sequences were present from 195 sewage and 60 animal samples (see Data Set S2, Tab 3 and Tab 4 for the 100 most abundant Bacteroides V4V5 and V6 region sequences in sewage). These unique Bacteroides sequences were named according to their abundance ranks in sewage samples in the data set, e.g., V4V5-1 and V6-1 are the most abundant V4V5 and V6 unique Bacteroides sequences in sewage samples, respectively.
To identify sewage-associated Bacteroides markers in the V4V5 and V6 regions, we used a subset of samples from the NGS data sets where both V4V5 and V6 regions were sequenced, which included 16 sewage and 51 animal fecal samples. These data were analyzed using R package Indicspecies (68), and the numbers of indicators for each region that were >90% host specific and sensitive were compared. To identify candidate sewage-associated Bacteroides markers, candidates were chosen by the criteria that they were >90% sensitive and 100% specific from Indicspecies results. To identify the probable source of these marker candidates (i.e., whether they are human derived or likely residents of the sewer pipes), we compared these marker candidate sequences with the National Center for Biotechnology Information (NCBI) nucleotide database and published V3V5 (69–71), V4V6 (72), V6 (73), and V6V8 (74) region human stool sequences using BLAST+. The most abundant Bacteroides in sewage was specific to sewage but did not appear to be of fecal origin. This organism was chosen for qPCR assay development, with the corresponding markers identified as V4V5-1 (V4V5 region) and V6-21 (V6 region). Candidate markers and their specificity, the probable source, and the sequence are shown in Tables S2 and S3.
Phylogenetic placement of sewer pipe-associated markers.
Near-full-length Bacteroides clones containing the matched V4V5 and/or the V6 markers identified by Indicspecies were used to construct a maximum likelihood tree in MEGA7 (75), based on Kimura 2 parameters (76) with gamma distribution and invariant sites (K2 + G + I) with bootstrapping for 1,000 replications.
Design of sewage-specific Bacteroides 16S rRNA gene fecal marker assays.
Primers and probes were designed based on 16S rRNA gene sequence alignment using animal fecal and sewage samples and visualized in MegAlign Pro program in DNASTAR software (version Lasergene 12). The marker sequences, a B. dorei 16S rRNA gene reference sequence (GenBank accession number AB242142) and sewage clone library sequences containing the V4V5-1 and V6-21 marker sequences, were included in the alignment. In addition, published near-full-length animal fecal Bacteroides clone sequences were included in the alignment of sequences from pigs (77), dogs (78), cows (36), chickens (79, 80), and mice (81) to discriminate from possible animal sources in the assay design. Primers and probes were named according to their base pair locations when aligned to an E. coli reference sequence (GenBank accession number J01859), with a comparison of universal 16S rRNA gene primers. Details are shown in Table 1. The amplicons of the two assays and their reference clone GenBank accession numbers are listed in Table S4.
qPCR experiments.
The qPCR reaction conditions, volumes, and methods for establishing the standard curve and testing inhibitions were followed as described by Templar et al. (24). Each run included a sewage-positive control and a no DNA control. The annealing temperatures were optimized by running a gradient qPCR for 1:100 (vol/vol) diluted sewage DNA (n = 4) from 59°C to 64°C. The same sewage samples were then tested in different dilution ratios at different annealing temperatures to make sure no amplification efficiency was lost (25). The amplification program included one cycle at 50°C for 2 min, followed by one cycle at 95°C for 10 min, and then 40 cycles of 95°C for 15 s followed by 1 min at 64°C for the BacV4V5-1 assay and 60°C for the BacV6-21 assay.
For assay validation to test for cross-reactivity, cat, dog, pig, cow, and deer fecal samples were tested in the formats of individual samples (i.e., from a single animal) and pooled samples (i.e., from two single animals of the same type). Pooled samples were retested individually unless there was insufficient material. Gull and chicken fecal samples were tested only as individuals. Each animal fecal sample was tested at DNA template concentrations of 1 ng · μl−1, 0.1 ng · μl−1, and 0.01 ng · μl−1, and the animal qPCR results were converted to the units of copy number (CN) per 1 ng of input DNA, CN per 0.1 ng of input DNA, and CN per 0.01 ng of input DNA. For sewage samples, DNA templates were diluted 1:100 (vol/vol). For environment water samples, DNA templates were tested without dilution. All the sewage and environmental water results were expressed as CN/100 ml filtrated sample. Amplification after cycle 35 was considered negative for all samples. A subset of 40 samples, including sewage, animal feces, and environmental water samples, were tested for inhibition using salmon sperm DNA as the internal control as previously described (24). No inhibition was observed in these samples. Statistical analysis of qPCR assay correlations was performed using cor and cor.test functions in R. The qPCR assay slopes, y intercepts, r2, and efficiency values are shown in Table S5.
Accession number(s).
The partial 16S rRNA gene sequences of the sewage clone libraries were deposited in the NCBI GenBank database. The library 1 sequences were deposited under accession numbers MH515295 to MH515584 and MH515940 to MH515981, and library 2 sequences were deposited under accession numbers MH515585 to MH515939 and MH515982 to MH516001. All V4V5 region NGS sequences of sewage and animal samples were from BioProjects PRJNA261344 (55) and PRJNA433408 (25). The V6 region NGS sequences of sewage and animal were from NCBI Sequence Read Archive (SRA) SRP041262 (34) and BioProject PRJNA433407 (25); V6 region NGS sequences of baseflow lake samples were from SRA SRP056973 (67), and the baseflow river sample sequences were deposited in NCBI SRA SRP168560.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hillary Morrison at the Bay Paul Center, Marine Biological Laboratory (MBL), University of Chicago, and the Great Lakes Genomics Center (GLGC), University of Wisconsin–Milwaukee, for offering expertise in NGS and Sanger sequencing, respectively. We thank Adélaïde Roguet for help with raw sequence processing and Melinda J. Bootsma for qPCR assistance in this study.
Funding for this study was provided by National Institutes of Health (NIH), grant number R01 AI091829.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02696-18.
REFERENCES
- 1.Griffin DW, Donaldson KA, Paul JH, Rose JB. 2003. Pathogenic human viruses in coastal waters. Clin Microbiol Rev 16:129–143. doi: 10.1128/CMR.16.1.129-143.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Donovan E, Unice K, Roberts JD, Harris M, Finley B. 2008. Risk of gastrointestinal disease associated with exposure to pathogens in the water of the Lower Passaic River. Appl Environ Microbiol 74:994–1003. doi: 10.1128/AEM.00601-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sinton LW, Finlay RK, Hannah DJ. 1998. Distinguishing human from animal faecal contamination in water: a review. N Z J Mar Freshwater Res 32:323–348. doi: 10.1080/00288330.1998.9516828. [DOI] [Google Scholar]
- 4.Soller JA, Schoen ME, Bartrand T, Ravenscroft JE, Ashbolt NJ. 2010. Estimated human health risks from exposure to recreational waters impacted by human and non-human sources of faecal contamination. Water Res 44:4674–4691. doi: 10.1016/j.watres.2010.06.049. [DOI] [PubMed] [Google Scholar]
- 5.EPA. 2012. Recreational water quality criteria. U.S. Environmental Protection Agency, Washington, DC. [Google Scholar]
- 6.McLellan SL, Eren AM. 2014. Discovering new indicators of fecal pollution. Trends Microbiol 22:697–706. doi: 10.1016/j.tim.2014.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemarchand K, Lebaron P. 2003. Occurrence of Salmonella spp. and Cryptosporidium spp. in a French coastal watershed: relationship with fecal indicators. FEMS Microbiol Lett 218:203–209. doi: 10.1016/S0378-1097(02)01135-7. [DOI] [PubMed] [Google Scholar]
- 8.Harwood VJ, Levine AD, Scott TM, Chivukula V, Lukasik J, Farrah SR, Rose JB. 2005. Validity of the indicator organism paradigm for pathogen reduction in reclaimed water and public health protection. Appl Environ Microbiol 71:3163–3170. doi: 10.1128/AEM.71.6.3163-3170.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Colford JM, Wade TJ, Schiff KC, Wright CC, Griffith JF, Sandhu SK, Burns S, Sobsey M, Lovelace G, Weisberg SB. 2007. Water quality indicators and the risk of illness at beaches with nonpoint sources of fecal contamination. Epidemiology 18:27–35. doi: 10.1097/01.ede.0000249425.32990.b9. [DOI] [PubMed] [Google Scholar]
- 10.Ercumen A, Pickering AJ, Kwong LH, Arnold B, Parvez SM, Alam M, Sen D, Islam S, Kullmann C, Chase C, Ahmed R, Unicomb L, Luby S, Colford JM Jr. 2017. Animal feces contribute to domestic fecal contamination: evidence from E. coli measured in water, hands, food, flies and soil in Bangladesh. Environ Sci Technol 51:8725–8734. doi: 10.1021/acs.est.7b01710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harwood VJ, Staley C, Badgley BD, Borges K, Korajkic A. 2014. Microbial source tracking markers for detection of fecal contamination in environmental waters: relationships between pathogens and human health outcomes. FEMS Microbiol Rev 38:1–40. doi: 10.1111/1574-6976.12031. [DOI] [PubMed] [Google Scholar]
- 12.Bernhard AE, Field KG. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl Environ Microbiol 66:1587–1594. doi: 10.1128/AEM.66.4.1587-1594.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahmed W, Hughes B, Harwood VJ. 2016. Current status of marker genes of Bacteroides and related taxa for identifying sewage pollution in environmental waters. Water 8:231. doi: 10.3390/w8060231. [DOI] [Google Scholar]
- 14.Bernhard AE, Field KG. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl Environ Microbiol 66:4571–4574. doi: 10.1128/AEM.66.10.4571-4574.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ahmed W, Goonetilleke A, Powell D, Gardner T. 2009. Evaluation of multiple sewage-associated Bacteroides PCR markers for sewage pollution tracking. Water Res 43:4872–4877. doi: 10.1016/j.watres.2009.08.042. [DOI] [PubMed] [Google Scholar]
- 16.Shanks OC, White K, Kelty CA, Sivaganesan M, Blannon J, Meckes M, Varma M, Haugland RA. 2010. Performance of PCR-based assays targeting Bacteroidales genetic markers of human fecal pollution in sewage and fecal samples. Environ Sci Technol 44:6281–6288. doi: 10.1021/es100311n. [DOI] [PubMed] [Google Scholar]
- 17.Seurinck S, Defoirdt T, Verstraete W, Siciliano SD. 2005. Detection and quantification of the human-specific HF183 Bacteroides 16S rRNA genetic marker with real-time PCR for assessment of human faecal pollution in freshwater. Environ Microbiol 7:249–259. doi: 10.1111/j.1462-2920.2004.00702.x. [DOI] [PubMed] [Google Scholar]
- 18.Kildare BJ, Leutenegger CM, McSwain BS, Bambic DG, Rajal VB, Wuertz S. 2007. 16S rRNA-based assays for quantitative detection of universal, human-, cow-, and dog-specific fecal Bacteroidales: a Bayesian approach. Water Res 41:3701–3715. doi: 10.1016/j.watres.2007.06.037. [DOI] [PubMed] [Google Scholar]
- 19.Ahmed W, Yusuf R, Hasan I, Goonetilleke A, Gardner T. 2010. Quantitative PCR assay of sewage-associated Bacteroides markers to assess sewage pollution in an urban lake in Dhaka, Bangladesh. Can J Microbiol 56:838–845. doi: 10.1139/W10-070. [DOI] [PubMed] [Google Scholar]
- 20.Van De Werfhorst LC, Sercu B, Holden PA. 2011. Comparison of the host specificities of two Bacteroidales quantitative PCR assays used for tracking human fecal contamination. Appl Environ Microbiol 77:6258–6260. doi: 10.1128/AEM.00239-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nshimyimana JP, Cruz MC, Thompson RJ, Wuertz S. 2017. Bacteroidales markers for microbial source tracking in Southeast Asia. Water Res 118:239–248. doi: 10.1016/j.watres.2017.04.027. [DOI] [PubMed] [Google Scholar]
- 22.Haugland RA, Varma M, Sivaganesan M, Kelty C, Peed L, Shanks OC. 2010. Evaluation of genetic markers from the 16S rRNA gene V2 region for use in quantitative detection of selected Bacteroidales species and human fecal waste by qPCR. Syst Appl Microbiol 33:348–357. doi: 10.1016/j.syapm.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 23.Green HC, Haugland RA, Varma M, Millen HT, Borchardt MA, Field KG, Walters WA, Knight R, Sivaganesan M, Kelty CA, Shanks OC. 2014. Improved HF183 quantitative real-time PCR assay for characterization of human fecal pollution in ambient surface water samples. Appl Environ Microbiol 80:3086–3094. doi: 10.1128/AEM.04137-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Templar HA, Dila DK, Bootsma MJ, Corsi SR, McLellan SL. 2016. Quantification of human-associated fecal indicators reveal sewage from urban watersheds as a source of pollution to Lake Michigan. Water Res 100:556–567. doi: 10.1016/j.watres.2016.05.056. [DOI] [PubMed] [Google Scholar]
- 25.Feng S, Bootsma M, McLellan SL. 2018. Human-associated Lachnospiraceae genetic markers improve detection of fecal pollution sources in urban waters. Appl Environ Microbiol 84:e00309-18. doi: 10.1128/AEM.00309-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silkie SS, Nelson KL. 2009. Concentrations of host-specific and generic fecal markers measured by quantitative PCR in raw sewage and fresh animal feces. Water Res 43:4860–4871. doi: 10.1016/j.watres.2009.08.017. [DOI] [PubMed] [Google Scholar]
- 27.Reischer GH, Kasper DC, Steinborn R, Farnleitner AH, Mach RL. 2007. A quantitative real-time PCR assay for the highly sensitive and specific detection of human faecal influence in spring water from a large alpine catchment area. Lett Appl Microbiol 44:351–356. doi: 10.1111/j.1472-765X.2006.02094.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Layton A, McKay L, Williams D, Garrett V, Gentry R, Sayler G. 2006. Development of Bacteroides 16S rRNA gene TaqMan-based real-time PCR assays for estimation of total, human, and bovine fecal pollution in water. Appl Environ Microbiol 72:4214–4224. doi: 10.1128/AEM.01036-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Okabe S, Okayama N, Savichtcheva O, Ito T. 2007. Quantification of host-specific Bacteroides-Prevotella 16S rRNA genetic markers for assessment of fecal pollution in freshwater. Appl Microbiol Biotechnol 74:890–901. doi: 10.1007/s00253-006-0714-x. [DOI] [PubMed] [Google Scholar]
- 30.Lee DY, Weir SC, Lee H, Trevors JT. 2010. Quantitative identification of fecal water pollution sources by TaqMan real-time PCR assays using Bacteroidales 16S rRNA genetic markers. Appl Microbiol Biotechnol 88:1373–1383. doi: 10.1007/s00253-010-2880-0. [DOI] [PubMed] [Google Scholar]
- 31.Carson CA, Christiansen JM, Benson VW, Baffaut C, Jerri V, Broz RR, Kurtz WB, Rogers WM, Fales WH, Yampara-Iquise H, Davis JV. 2005. Specificity of a Bacteroides thetaiotaomicron marker for human feces. Appl Environ Microbiol 71:4945–4949. doi: 10.1128/AEM.71.8.4945-4949.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yampara-Iquise H, Zheng G, Jones JE, Carson CA. 2008. Use of a Bacteroides thetaiotaomicron-specific α-1-6, mannanase quantitative PCR to detect human faecal pollution in water. J Appl Microbiol 105:1686–1693. doi: 10.1111/j.1365-2672.2008.03895.x. [DOI] [PubMed] [Google Scholar]
- 33.Kreader CA. 1995. Design and evaluation of Bacteroides DNA probes for the specific detection of human fecal pollution. Appl Environ Microbiol 61:1171–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fisher JC, Murat Eren A, Green HC, Shanks OC, Morrison HG, Vineis JH, Sogin ML, McLellan SL. 2015. Comparison of sewage and animal fecal microbiomes by using oligotyping reveals potential human fecal indicators in multiple taxonomic groups. Appl Environ Microbiol 81:7023–7033. doi: 10.1128/AEM.01524-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shanks OC, Kelty CA, Archibeque S, Jenkins M, Newton RJ, McLellan SL, Huse SM, Sogin ML. 2011. Community structures of fecal bacteria in cattle from different animal feeding operations. Appl Environ Microbiol 77:2992–3001. doi: 10.1128/AEM.02988-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Durso LM, Harhay GP, Smith TPL, Bono JL, DeSantis TZ, Harhay DM, Andersen GL, Keen JE, Laegreid WW, Clawson ML. 2010. Animal-to-animal variation in fecal microbial diversity among beef cattle. Appl Environ Microbiol 76:4858–4862. doi: 10.1128/AEM.00207-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hespell RB, Whitehead TR. 1990. Physiology and genetics of xylan degradation by gastrointestinal tract bacteria. J Dairy Sci 73:3013–3022. doi: 10.3168/jds.S0022-0302(90)78988-6. [DOI] [PubMed] [Google Scholar]
- 38.Xu J, Bjursell MK, Himrod J, Deng S, Carmichael LK, Chiang HC, Hooper LV, Gordon JI. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074–2076. doi: 10.1126/science.1080029. [DOI] [PubMed] [Google Scholar]
- 39.Flint HJ, Scott KP, Duncan SH, Louis P, Forano E. 2012. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 3:289–306. doi: 10.4161/gmic.19897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wexler AG, Goodman AL. 2017. An insider’s perspective: Bacteroides as a window into the microbiome. Nat Microbiol 2:17026. doi: 10.1038/nmicrobiol.2017.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nishiyama T, Ueki A, Kaku N, Watanabe K, Ueki K. 2009. Bacteroides graminisolvens sp. nov., a xylanolytic anaerobe isolated from a methanogenic reactor treating cattle waste. Int J Syst Evol Microbiol 59:1901–1907. doi: 10.1099/ijs.0.008268-0. [DOI] [PubMed] [Google Scholar]
- 42.Hatamoto M, Kaneshige M, Nakamura A, Yamaguchi T. 2014. Bacteroides luti sp. nov., an anaerobic, cellulolytic and xylanolytic bacterium isolated from methanogenic sludge. Int J Syst Evol Microbiol 64:1770–1774. doi: 10.1099/ijs.0.056630-0. [DOI] [PubMed] [Google Scholar]
- 43.Ismaeil M, Yoshida N, Katayama A. 2018. Bacteroides sedimenti sp. nov., isolated from a chloroethenes-dechlorinating consortium enriched from river sediment. J Microbiol 56:619–627. doi: 10.1007/s12275-018-8187-z. [DOI] [PubMed] [Google Scholar]
- 44.Kim JR, Beecroft NJ, Varcoe JR, Dinsdale RM, Guwy AJ, Slade RCT, Thumser A, Avignone-Rossa C, Premier GC. 2011. Spatiotemporal development of the bacterial community in a tubular longitudinal microbial fuel cell. Appl Microbiol Biotechnol 90:1179–1191. doi: 10.1007/s00253-011-3181-y. [DOI] [PubMed] [Google Scholar]
- 45.Ley RE, Lozupone CA, Hamady M, Knight R, Gordon JI. 2008. Worlds within worlds: evolution of the vertebrate gut microbiota. Nat Rev Microbiol 6:776–788. doi: 10.1038/nrmicro1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vandewalle JL, Goetz GW, Huse SM, Morrison HG, Sogin ML, Hoffmann RG, Yan K, McLellan SL. 2012. Acinetobacter, Aeromonas and Trichococcus populations dominate the microbial community within urban sewer infrastructure. Environ Microbiol 14:2358–2552. doi: 10.1111/j.1462-2920.2012.02757.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Koskey AM, Fisher JC, Eren AM, Ponce-Terashima R, Reis MG, Blanton RE, McLellan SL. 2014. Blautia and Prevotella sequences distinguish human and animal fecal pollution in Brazil surface waters. Environ Microbiol Rep 6:696–704. doi: 10.1111/1758-2229.12189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mayer RE, Reischer GH, Ixenmaier SK, Derx J, Blaschke AP, Ebdon JE, Linke R, Egle L, Ahmed W, Blanch AR, Byamukama D, Savill M, Mushi D, Cristóbal HA, Edge TA, Schade MA, Aslan A, Brooks YM, Sommer R, Masago Y, Sato MI, Taylor HD, Rose JB, Wuertz S, Shanks OC, Piringer H, Mach RL, Savio D, Zessner M, Farnleitner AH. 2018. Global distribution of human-associated fecal genetic markers in reference samples from six continents. Environ Sci Technol 52:5076–5084. doi: 10.1021/acs.est.7b04438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olapade OA, Depas MM, Jensen ET, McLellan SL. 2006. Microbial communities and fecal indicator bacteria associated with Cladophora mats on beach sites along Lake Michigan shores. Appl Environ Microbiol 72:1932–1938. doi: 10.1128/AEM.72.3.1932-1938.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whitman RL, Byappanahalli MN, Spoljaric AM, Przybyla-Kelly K, Shively DA, Nevers MB. 2014. Evidence for free-living Bacteroides in Cladophora along the shores of the Great Lakes. Aquat Microb Ecol 72:117–126. doi: 10.3354/ame01688. [DOI] [Google Scholar]
- 51.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. 2005. Diversity of the human intestinal microbial flora. Science 308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Alm EW, Daniels-Witt QR, Learman DR, Ryu H, Jordan DW, Gehring TM, Santo Domingo J. 2018. Potential for gulls to transport bacteria from human waste sites to beaches. Sci Total Environ 615:123–130. doi: 10.1016/j.scitotenv.2017.09.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Brown CM, Staley C, Wang P, Dalzell B, Chun CL, Sadowsky MJ. 2017. A high-throughput DNA-sequencing approach for determining sources of fecal bacteria in a Lake Superior estuary. Environ Sci Technol 51:8263–8271. doi: 10.1021/acs.est.7b01353. [DOI] [PubMed] [Google Scholar]
- 54.Roguet A, Eren AM, Newton RJ, McLellan SL. 2018. Fecal source identification using random forest. Microbiome 6:185. doi: 10.1186/s40168-018-0568-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Newton RJ, McLellan SL, Dila DK, Vineis JH, Morrison HG, Murat Eren A, Sogin ML. 2015. Sewage reflects the microbiomes of human populations. mBio 6:e02574-14. doi: 10.1128/mBio.02574-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olds HT, Corsi SR, Dila DK, Halmo KM, Bootsma MJ, McLellan SL. 2018. High levels of sewage contamination released from urban areas after storm events: a quantitative survey with sewage specific bacterial indicators. PLoS Med 15:e1002614. doi: 10.1371/journal.pmed.1002614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Eren AM, Maignien L, Sul WJ, Murphy LG, Grim SL, Morrison HG, Sogin ML. 2013. Oligotyping: differentiating between closely related microbial taxa using 16S rRNA gene data. Methods Ecol Evol 4:1111–1119. doi: 10.1111/2041-210X.12114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Martin M. 2011. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17:10–12. doi: 10.14806/ej.17.1.200. [DOI] [Google Scholar]
- 59.Zhang J, Kobert K, Flouri T, Stamatakis A. 2014. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30:614–620. doi: 10.1093/bioinformatics/btt593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. 2008. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genet 4:e1000255. doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wickham H. 2016. ggplot2: elegant graphics for data analysis. Springer-Verlag, New York, NY. [Google Scholar]
- 62.R Core Team. 2018. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 63.Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, Mcglinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Henry M, Stevens H, Szoecs E, Wagner H, Oksanen MJ. 2018. vegan: community ecology package. R package version 2.4-5. R Foundation for Statistical Computing, Vienna, Austria: https://CRAN.R-project.org/package=vegan. [Google Scholar]
- 64.Huse SM, Mark Welch DB, Voorhis A, Shipunova A, Morrison HG, Eren AM, Sogin ML. 2014. VAMPS: a website for visualization and analysis of microbial population structures. BMC Bioinformatics 15:41. doi: 10.1186/1471-2105-15-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Cole JR, Wang Q, Fish JA, Chai B, McGarrell DM, Sun Y, Brown CT, Porras-Alfaro A, Kuske CR, Tiedje JM. 2014. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res 42:D633–D642. doi: 10.1093/nar/gkt1244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. 2009. BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Fisher JC, Newton RJ, Dila DK, McLellan SL. 2015. Urban microbial ecology of a freshwater estuary of Lake Michigan. Elementa (Wash D C) 3:000064. doi: 10.12952/journal.elementa.000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.De Cáceres M, Legendre P. 2009. Associations between species and groups of sites: indices and statistical inference. Ecology 90:3566–3574. [DOI] [PubMed] [Google Scholar]
- 69.Labus JS, Hollister EB, Jacobs J, Kirbach K, Oezguen N, Gupta A, Acosta J, Luna RA, Aagaard K, Versalovic J, Savidge T, Hsiao E, Tillisch K, Mayer EA. 2017. Differences in gut microbial composition correlate with regional brain volumes in irritable bowel syndrome. Microbiome 5:49. doi: 10.1186/s40168-017-0260-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Strati F, Cavalieri D, Albanese D, De Felice C, Donati C, Hayek J, Jousson O, Leoncini S, Renzi D, Calabrò A, De Filippo C. 2017. New evidences on the altered gut microbiota in autism spectrum disorders. Microbiome 5:24. doi: 10.1186/s40168-017-0242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zwittink RD, Renes IB, van Lingen RA, van Zoeren-Grobben D, Konstanti P, Norbruis OF, Martin R, Groot Jebbink LJM, Knol J, Belzer C. 2018. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur J Clin Microbiol Infect Dis 37:475–483. doi: 10.1007/s10096-018-3193-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Madan JC, Koestler DC, Stanton BA, Davidson L, Moulton LA, Housman ML, Moore JH, Guill MF, Morrison HG, Sogin ML, Hampton TH, Karagas MR, Palumbo PE, Foster JA, Hibberd PL, O’Toole GA. 2012. Serial analysis of the gut and respiratory microbiome in cystic fibrosis in infancy: interaction between intestinal and respiratory tracts and impact of nutritional exposures. mBio 3:e00251-12. doi: 10.1128/mBio.00251-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP, Egholm M, Henrissat B, Heath AC, Knight R, Gordon JI. 2009. A core gut microbiome in obese and lean twins. Nature 457:480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Smith-Brown P, Morrison M, Krause L, Davies PSW. 2016. Dairy and plant based food intakes are associated with altered faecal microbiota in 2 to 3 year old Australian children. Sci Rep 6:32385. doi: 10.1038/srep32385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kimura M. 1980. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120. [DOI] [PubMed] [Google Scholar]
- 77.Leser TD, Amenuvor JZ, Jensen TK, Lindecrona RH, Boye M, Møller K. 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl Environ Microbiol 68:673–690. doi: 10.1128/AEM.68.2.673-690.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Suchodolski JS, Camacho J, Steiner JM. 2008. Analysis of bacterial diversity in the canine duodenum, jejunum, ileum, and colon by comparative 16S rRNA gene analysis. FEMS Microbiol Ecol 66:567–578. doi: 10.1111/j.1574-6941.2008.00521.x. [DOI] [PubMed] [Google Scholar]
- 79.Zhu XY, Zhong T, Pandya Y, Joerger RD. 2002. 16S rRNA-based analysis of microbiota from the cecum of broiler chickens. Appl Environ Microbiol 68:124–137. doi: 10.1128/AEM.68.1.124-137.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bjerrum L, Engberg RM, Leser TD, Jensen BB, Finster K, Pedersen K. 2006. Microbial community composition of the ileum and cecum of broiler chickens as revealed by molecular and culture-based techniques. Poult Sci 85:1151–1164. doi: 10.1093/ps/85.7.1151. [DOI] [PubMed] [Google Scholar]
- 81.Nozu R, Ueno M, Hayashimoto N. 2016. Composition of fecal microbiota of laboratory mice derived from Japanese commercial breeders using 16S rRNA gene clone libraries. J Vet Med Sci 78:1045–1050. doi: 10.1292/jvms.15-0454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.