Naturally produced and anthropogenically released DCM can reside in anoxic environments, yet little is known about the diversity of organisms, enzymes, and mechanisms involved in carbon-chlorine bond cleavage in the absence of oxygen. A proteogenomic approach identified two RDases and four corrinoid-dependent methyltransferases expressed by the DCM degrader “Candidatus Dichloromethanomonas elyunquensis” strain RM, suggesting that reductive dechlorination and methyl group transfer play roles in anaerobic DCM degradation. These findings suggest that the characterized DCM-degrading bacterium Dehalobacterium formicoaceticum and “Candidatus Dichloromethanomonas elyunquensis” strain RM utilize distinct strategies for carbon-chlorine bond cleavage, indicating that multiple pathways evolved for anaerobic DCM metabolism. The specific proteins (e.g., RDases and methyltransferases) identified in strain RM may have value as biomarkers for monitoring anaerobic DCM degradation in natural and contaminated environments.
KEYWORDS: anaerobic dichloromethane metabolism; “Candidatus Dichloromethanomonas elyunquensis,” genomics; methyltransferases; proteomics; reductive dehalogenases; Wood-Ljungdahl pathway
ABSTRACT
Dichloromethane (DCM) is susceptible to microbial degradation under anoxic conditions and is metabolized via the Wood-Ljungdahl pathway; however, mechanistic understanding of carbon-chlorine bond cleavage is lacking. The microbial consortium RM contains the DCM degrader “Candidatus Dichloromethanomonas elyunquensis” strain RM, which strictly requires DCM as a growth substrate. Proteomic workflows applied to DCM-grown consortium RM biomass revealed a total of 1,705 nonredundant proteins, 521 of which could be assigned to strain RM. In the presence of DCM, strain RM expressed a complete set of Wood-Ljungdahl pathway enzymes, as well as proteins implicated in chemotaxis, motility, sporulation, and vitamin/cofactor synthesis. Four corrinoid-dependent methyltransferases were among the most abundant proteins. Notably, two of three putative reductive dehalogenases (RDases) encoded within strain RM’s genome were also detected in high abundance. Expressed RDase 1 and RDase 2 shared 30% amino acid identity, and RDase 1 was most similar to an RDase of Dehalococcoides mccartyi strain WBC-2 (AOV99960, 52% amino acid identity), while RDase 2 was most similar to an RDase of Dehalobacter sp. strain UNSWDHB (EQB22800, 72% amino acid identity). Although the involvement of RDases in anaerobic DCM metabolism has yet to be experimentally verified, the proteome characterization results implicated the possible participation of one or more reductive dechlorination steps and methyl group transfer reactions, leading to a revised proposal for an anaerobic DCM degradation pathway.
IMPORTANCE Naturally produced and anthropogenically released DCM can reside in anoxic environments, yet little is known about the diversity of organisms, enzymes, and mechanisms involved in carbon-chlorine bond cleavage in the absence of oxygen. A proteogenomic approach identified two RDases and four corrinoid-dependent methyltransferases expressed by the DCM degrader “Candidatus Dichloromethanomonas elyunquensis” strain RM, suggesting that reductive dechlorination and methyl group transfer play roles in anaerobic DCM degradation. These findings suggest that the characterized DCM-degrading bacterium Dehalobacterium formicoaceticum and “Candidatus Dichloromethanomonas elyunquensis” strain RM utilize distinct strategies for carbon-chlorine bond cleavage, indicating that multiple pathways evolved for anaerobic DCM metabolism. The specific proteins (e.g., RDases and methyltransferases) identified in strain RM may have value as biomarkers for monitoring anaerobic DCM degradation in natural and contaminated environments.
INTRODUCTION
Dichloromethane (DCM) is an anthropogenic groundwater contaminant but is also produced naturally by macroalgae and as a product of bacterial chloroform organohalide respiration (1–3). DCM is toxic and contributes to stratospheric ozone depletion (4), and a better understanding of the environmental fate of DCM will aid in better management of risk to human health and the environment. Under oxic and nitrate-reducing conditions, methylotrophic bacteria featuring glutathione-dependent DCM-dehalogenases degrade DCM (5–7). DCM degradation has further been characterized under acetogenic (8, 9), methanogenic (10), and fermentative (3, 11, 12) conditions. Anaerobic DCM degradation associated with cell growth was demonstrated for the isolate Dehalobacterium formicoaceticum (11, 13), the recently described “Candidatus Dichloromethanomonas elyunquensis” strain RM (14), and Dehalobacter- and Dehalobacterium-related microorganisms in mixed cultures (3, 15). Efforts to isolate “Candidatus Dichloromethanomonas elyunquensis” strain RM have not been successful, presumably because strain RM relies on hydrogen-scavenging partner microorganisms (16).
Aspects of anaerobic DCM degradation have been resolved and acetate, formate/hydrogen, carbon dioxide, and inorganic chloride were observed as the major degradation products (3, 11–14). The DCM carbon constituted the methyl carbon of acetate, with the carboxyl group derived from CO2 (3, 13), consistent with the observation that Dehalobacterium formicoaceticum and “Candidatus Dichloromethanomonas elyunquensis” strain RM require CO2 during growth with DCM as the sole electron donor. The molar ratio of formate to acetate was 2:1 during DCM degradation by Dehalobacterium formicoaceticum (12, 13), and the activities of Wood-Ljungdahl pathway (WLP; also referred to as the reductive acetyl coenzyme A [acetyl-CoA] pathway) enzymes (carbon monoxide [CO] dehydrogenase, methylene-tetrahydrofolate [THF] dehydrogenase, methenyl-THF cyclohydrolase, formyl-THF synthase, formate dehydrogenase) were detected in cell extracts of Dehalobacterium formicoaceticum (11). Yet, the reactions leading to carbon-chlorine bond cleavage and chloride release remained elusive.
Previous biochemical studies on Dehalobacterium formicoaceticum revealed that the methylene group of DCM was transferred onto THF, which required substoichiometric amounts of ATP, but apparently did not require electrons for the removal of the two chlorine atoms (13). This enigmatic DCM dehalogenase activity in Dehalobacterium formicoaceticum has remained largely unexplored. The reaction was inhibited in a light-reversible fashion by propyl iodide, suggesting an involvement of a Co(I) corrinoid (13). The reducing equivalents generated by methylene-THF oxidation were likely used by methylene-THF reductase and CO dehydrogenase in the formation of acetate from methylene-THF and CO2. Metabolite analysis of Dehalobacterium formicoaceticum cultures grown with 13C-DCM revealed that the labeled carbon of DCM was incorporated into formate and the methyl group of acetate (13). Methylene-THF reductase activity was present, but it was suggested that the enzyme depends on an as-yet-unidentified endogenous electron donor (13). Therefore, in the postulated pathway of anaerobic DCM degradation in Dehalobacterium formicoaceticum, DCM is converted to methylene-THF, catalyzed by one or more yet-uncharacterized enzymes, and metabolized further by enzymes of the WLP.
The WLP was first described in homoacetogenic bacteria using hydrogen as a reductant for CO2 conversion to acetate (17). The WLP serves reductive or oxidative purposes in bacteria and archaea with diverse respiratory processes (18, 19), including methanogenesis (20), hydrogen generation (21), sulfate reduction (22, 23), and possibly anaerobic ammonium oxidation (24). The WLP was further discovered in organohalide-respiring bacteria such as Dehalobacter sp. strain UNSWDHB (25), and Dehalococcoides mccartyi strain 195 (26–29), strain VS (30), strain BAV1 (30), strain CBDB1 (31), and strain GT (32). A full set of WLP genes was detected on the genomes of the DCM degraders “Candidatus Dichloromethanomonas elyunquensis” strain RM (14, 33) and Dehalobacterium formicoaceticum sp. strain DMC (34), emphasizing that WLP enzymes play a role in anaerobic DCM metabolism.
To elucidate further details of the anaerobic DCM metabolic pathway, we applied proteogenomics to consortium RM harboring the DCM-degrading bacterium “Candidatus Dichloromethanomonas elyunquensis” strain RM, here referred to as strain RM. The metagenome of consortium RM, which included the draft genome of strain RM, served as a reference database to catalog protein expression by DCM-grown strain RM. Proteomic measurements revealed that strain RM expressed two putative RDases and several methyltransferases during growth in defined medium with DCM as the sole source of energy. The possible involvement of RDases in carbon-chlorine bond cleavage and methyltransferases in methyl group transfer reactions during anaerobic DCM degradation led to a refined model for anaerobic DCM degradation in strain RM.
RESULTS
Growth of consortium RM.
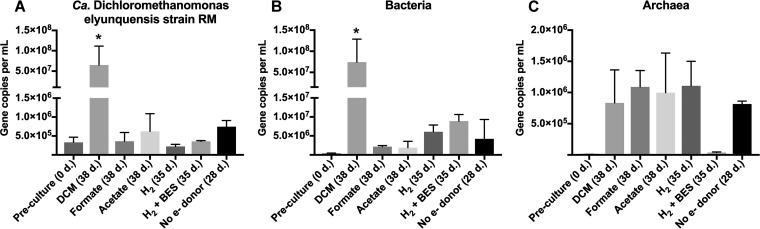
Quantitative real-time PCR (qPCR) enumeration revealed that strain RM 16S rRNA gene copy numbers increased significantly (P ≤ 0.05) from 3.3 × 105 (±1.4 × 105) (i.e., cells introduced with the inoculum) to 6.5 × 107 (±4.7 × 107) per ml in the presence of DCM over a 38-day incubation period (Fig. 1). Similar significant (P ≤ 0.05) increases in gene copy numbers were measured with a general bacterial 16S rRNA gene-targeted qPCR assay, indicating that strain RM was the predominant population in the consortium during the active phase of DCM degradation. In cultures without DCM but amended with acetate, formate, hydrogen, or hydrogen plus 2-bromoethanesulfonate (BES; an inhibitor of methanogenesis) and in cultures without electron donor addition, the 16S rRNA genes of strain RM did not increase (i.e., no growth occurred) but remained detectable over the up to 38-day incubation period (Fig. 1).
FIG 1.
16S rRNA gene copies of “Candidatus Dichloromethanomonas elyunquensis” strain RM (A), bacteria (B), and archaea (C) per ml of culture suspension sampled after 28 to 38 days (d.) of incubation, as determined by qPCR. The preculture (0 d.), used as inoculum for all culture conditions, was grown with DCM. Values represent averages ± the standard deviations of results obtained from three replicate incubations. Significant differences (P ≤ 0.05) of 16S rRNA gene copies among different culture conditions are indicated with an asterisk (*).
Methanogenic archaea dominated in cultures amended with formate or hydrogen, starting with 5.5 × 103 ± 3.6 × 103 16S rRNA gene copies per ml and yielding 1.1 × 106 ± 2.6 × 105 (formate) and 1.1 × 106 ± 3.9 × 105 (hydrogen) gene copies per ml, respectively (Fig. 1). The 16S rRNA gene copies detected for methanogenic archaea were substantially lower (3.7 × 104 ± 1.1 × 104) in hydrogen/BES-amended cultures.
Protein expression patterns.
DCM is the only known growth substrate for strain RM. Cultures that received acetate, formate, hydrogen/BES, or no electron donor addition served as controls to identify proteins expressed in response to DCM metabolism by strain RM. Using the metagenome of consortium RM (see Data Set S1 in the supplemental material) as the reference database, a total of 2,048 different proteins were detected under the different incubation conditions (Data Set S2). In DCM-amended cultures, about one-third of the proteins were assigned to the draft genome of strain RM, while about two-thirds of proteins were assigned to the metagenome of consortium RM (excluding the draft genome of strain RM) and corresponded to other community members (Data Set S2). In control incubations without DCM, no more than 13% of proteins could be assigned to strain RM (Data Set S2), which can be attributed to strain RM cells transferred with the inoculum. qPCR demonstrated that no growth of strain RM occurred without DCM. Proteins identified for other community members were associated with methanogenesis, acetogenesis, chemotaxis, vitamin/cofactor synthesis, motility, and sporulation (Data Set S2). During anaerobic growth with DCM, strain RM expressed proteins involved in chemotaxis, vitamin/cofactor synthesis, motility, sporulation and the WLP (Data Set S2). Interestingly, among the most abundant proteins were two putative reductive dehalogenases (RDases) and four corrinoid-dependent methyltransferases (Data Set S2).
RDases.
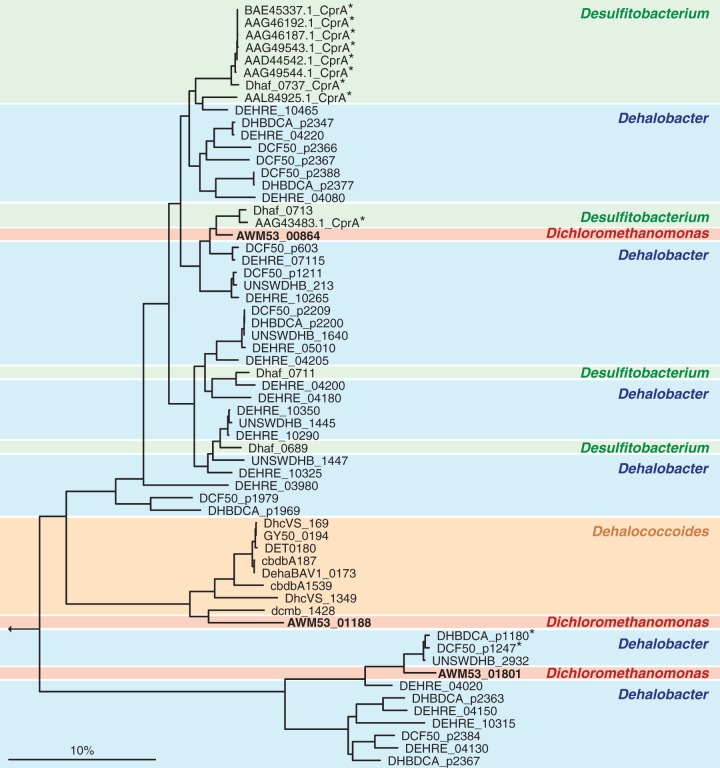
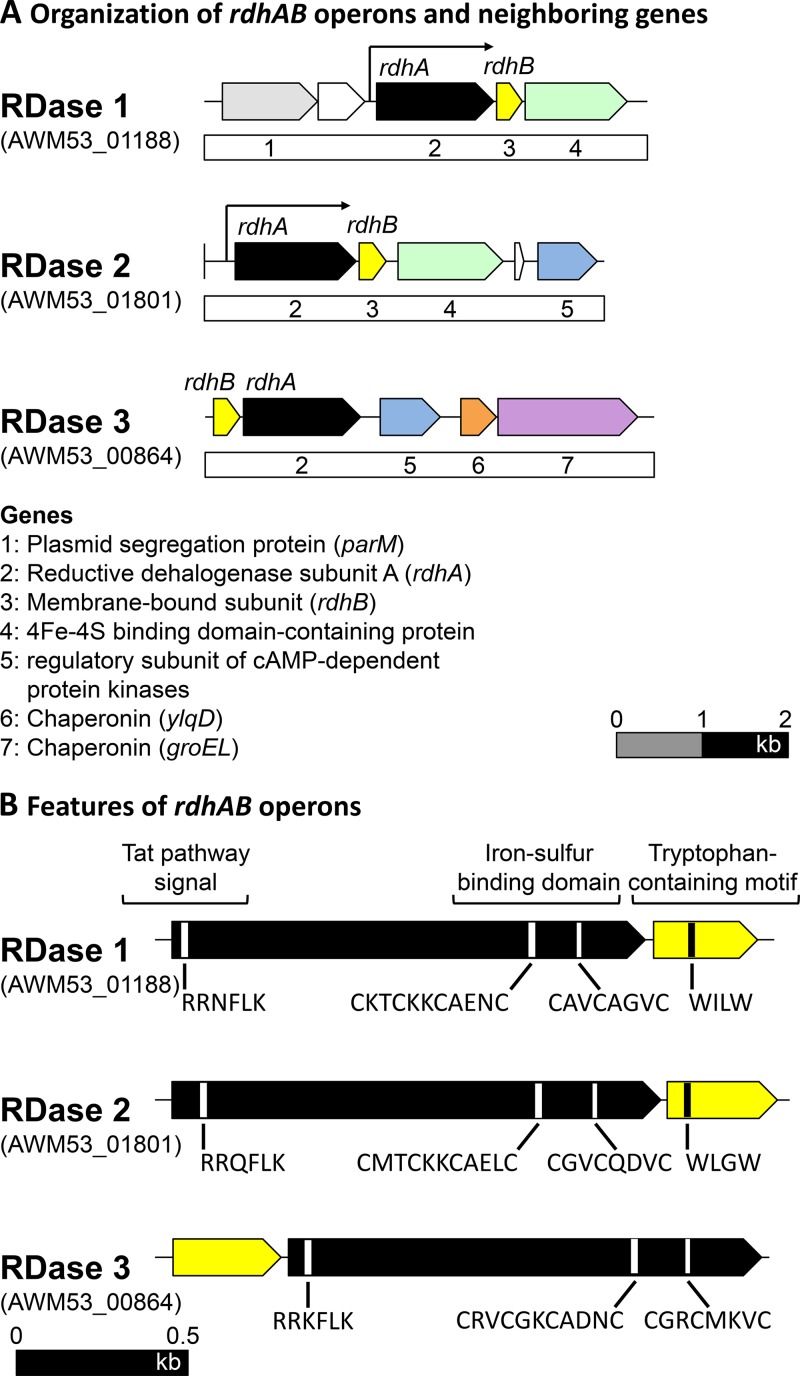
Three RDase genes with a G+C content of 47.0 to 47.4% were identified on the draft genome of strain RM (14). The three RDases, here referred to as RDase 1 (AWM53_01188), RDase 2 (AWM53_01801), and RDase 3 (AWM53_00864), only shared 29 to 36% amino acid identity among each other (Fig. 2). RDase 1 was most similar to an RDase of Dehalococcoides mccartyi strain WBC-2 (AOV99960, 52% amino acid identity), whereas RDase 2 was most similar to an RDase of Dehalobacter sp. strain UNSWDHB (EQB22800, 72% amino acid identity), and RDase 3 was most similar to an RDase of Dehalobacter sp. strain FTH1 (WP_026156428, 72% amino acid identity; Table 1). Functional prediction of these three genes was further supported by assignment of each gene to pfam13489 and TIGR02486 RDase ortholog families (Table S1). The gene neighborhoods encoding RDase 1 and RDase 2 revealed RDase-characteristic features, including a B gene encoding a putative membrane anchor located 19 and 16 bp upstream, respectively, of the RDase A gene encoding the catalytic subunit. Also detected were genes encoding typical RDase accessory proteins such as chaperons and regulatory proteins (Fig. 3A). The RDase A genes encoding RDase 1 and RDase 2 both contained the characteristic sequences of a twin-arginine translocation (Tat) signal near the N terminus and two iron-sulfur cluster-binding motifs closer toward the C terminus (Fig. 3B), which are all characteristic RDase features. The RDase 3-encoding region also comprised RDase A and RDase B genes but the RDase B gene was detected 14 bp downstream of the RDase A gene (Fig. 3). The RDase A gene of RDase 3 also encoded a predicted Tat motif and two iron-sulfur cluster-binding regions (Fig. 3B). All three RDase B membrane anchor proteins featured three predicted transmembrane helices. The analysis further revealed one predicted N-terminal transmembrane helix in RDase 1 and RDase 2 (consistent with an N-terminal Tat signal peptide), while no clear transmembrane helix was predicted in RDase 3. The signal peptide analysis program SignalP (35) detected a canonical Tat signal cleavage site in RDase 1, which is apparently lacking in RDase 2, leaving open the possibility that RDase 2 remains membrane-anchored even without the involvement of an RDase B protein (36).
FIG 2.
Phylogenetic relationships of putative RDases of “Candidatus Dichloromethanomonas elyunquensis” strain RM with RDases of organisms capable of organohalide respiration. RDases from Desulfitobacterium spp. (green), Dehalobacter spp. (blue), Dehalococcoides mccartyi (orange), and “Candidatus Dichloromethanomonas elyunquensis” strain RM (red) are color-coded. Biochemically characterized RDases are marked with an asterisk (*). The bar represents 10% estimated sequence divergence.
TABLE 1.
Enzymes expressed by “Candidatus Dichloromethanomonas elyunquensis” strain RM during growth with DCMa
| Locus tag (AWM53) | Gene | BLAST results |
Predicted localizationb | Expressed | ||
|---|---|---|---|---|---|---|
| Enzyme | Taxon | Identity (%) | ||||
| 01188 | rdhA | Reductive dehalogenase | Dehalococcoides mccartyi WBC-2 | 52 | Periplasm | Yes |
| 01801 | rdhA | Reductive dehalogenase | Dehalobacter sp. UNSWDHB | 72 | Membrane | Yes |
| Dehalobacter sp. DCA | 71 | |||||
| 00864 | rdhA | Reductive dehalogenase | Dehalobacter sp. FTH1 | 72 | Cytoplasm | ND |
| 01380 | mtrH | Tetrahydromethanopterin S-methyltransferase subunit H | Dehalobacterium formicoaceticum | 91 | Cytoplasm | Yes |
| 01379 | mtaA | Methylcobamide:CoM methyltransferase MtaA | Dehalobacterium formicoaceticum | 94 | Cytoplasm | Yes |
| 02085 | mtsA | Methylated-thiol coenzyme M methyltransferase | Dehalobacterium formicoaceticum | 92 | Cytoplasm | Yes |
| 02086 | mtbC | Corrinoid methyltransferase | Dehalobacterium formicoaceticum | 86 | Cytoplasm | Yes |
| 01363 | fhs | Formyl-THF ligase | Syntrophobotulus glycolicus | 85 | Membrane | Yes |
| 00605 | fchA | Methenyl-THF cyclohydrolase | Desulfosporosinus youngiae | 69 | Membrane | Yes |
| 00606 | folD | Bifunctional: 5,10-methylene-THF dehydrogenase, 5,10-methylene-THF cyclohydrolase | Desulfosporosinus acidiphilus | 72 | Cytoplasm | Yes |
| 00219 | metF | Methylene-THF reductase [NAD(P)H] | Dehalobacter sp. FTH1 | 66 | Cytoplasm | Yes |
| 00225 | acsE | Carbon monoxide dehydrogenase | Dehalobacter sp. FTH1 | 93 | Cytoplasm | Yes |
| 00220 | cooS | Carbon monoxide dehydrogenase catalytic subunit | Dehalobacter restrictus DSM 9455 | 80 | Membrane | Yes |
| 00226 | acsB | Acetyl-CoA decarbonylase/synthase | Dehalobacter sp. FTH1 | 85 | Membrane | Yes |
The most closely related proteins and predicted localization (i.e., periplasm, membrane, and cytoplasm) of strain RM enzymes are also shown. ND, not detected.
Based on PSORT and SignalP 4.1.
FIG 3.
Organization of RDase operons, with a putative catalytic subunit of the RDase (rdhA) and a putative membrane anchor of the RDase (rdhB) (A) and features of rdhAB operons (B) of “Candidatus Dichloromethanomonas elyunquensis” strain RM. (A) Protein-coding genes (colored) and hypothetical genes (white) are illustrated. The RDases expressed when grown with DCM are indicated with arrows. The scale bar represents 2 kb. (B) The RDase subunit A genes encode twin-arginine translocation (Tat) pathway signals and iron-sulfur binding domains. The RDase subunit B genes show a characteristic tryptophan-containing motif. The scale bar represents 0.5 kb.
The proteomic measurements detected RDase 1 and RDase 2 in biomass grown with DCM, while RDase 3 peptides were never observed (Data Set S2). RDase 1 was among the most abundant proteins (normalized spectral counts [nSpc]: 611; Data Set S2) in cultures grown with DCM. In cultures without DCM and amended with acetate, formate, or hydrogen and in cultures without any electron donor amendment, RDase 1 occurred in lower abundance (nSpc: 19-266; Data Set S2). The detection of strain RM proteins in the absence of DCM is due to strain RM cells introduced with the inoculum, which was grown with DCM. RDase 2 was also among the most abundant proteins (nSpc: 470; Data Set S2) in cultures grown with DCM. RDase 2 was absent or detected in low abundance in cultures without DCM (i.e., cultures that received acetate, formate, hydrogen, or no electron donor amendment; Data Set S2).
Methyltransferases.
In the draft genome of strain RM, 96 genes were predicted to encode methyltransferases (Data Set S3). Of these, 14 were expressed and four were among the most abundant proteins in DCM-grown cultures (nSpc: 994-3401; Data Set S2). The four most abundant methyltransferases of strain RM shared high identity with proteins from Dehalobacterium formicoaceticum (86 to 94% amino acid identity; Table 1), Dehalobacter sp. strain UNSWDHB, Dehalobacter sp. strain CF, and Dehalobacter sp. strain DCA (86 to 93% amino acid identity; Table S2). Of note, other proteins in the nonredundant NCBI database showed substantially lower identities, not exceeding 45%, and were associated with the candidate divisions MSBL1 and BRC1, as well as a Planctomycetes sp. and Thermacetogenium phaeum DSM 12270 (Table S2). While these proteins were consistently placed in methyltransferase ortholog families (Table S1), the lack in similar and characterized proteins prohibits specific functional assignments. Taken together, these findings suggest that the expressed methyltransferases are unique to certain members of the Peptococcaceae implicated in chlorinated compound metabolism. High expression of the two RDases and the four methyltransferases was corroborated in replicate experiments with DCM-grown biomass from subsequent transfer cultures (data not shown), suggesting their functional involvement in anaerobic DCM metabolism by strain RM.
Wood-Ljungdahl pathway-associated enzymes.
In addition to the two RDases and the methyltransferases, strain RM expressed enzymes of the WLP when grown with DCM (Data Set S2 and Table S1). The proteomic analysis revealed a complete set of WLP enzymes comprising formate dehydrogenase (K00123/K05299), formyl-THF synthetase (COG2759), methenyl-THF cyclohydrolase (COG3404), 5,10-methylene-THF dehydrogenase (K01491), 5,10-methylene-THF reductase (K00297), 5-methyltetrahydrofolate corrinoid/iron sulfur protein methyltransferase (K15023), CO dehydrogenase (K00198), and acetyl-CoA synthase (K14138) (Data Set S2 and Table S1).
The WLP proteins were most similar to enzymes of Dehalobacter sp. strain FTH1, Dehalobacter restrictus DSM 9455, Desulfitobacterium metallireducens, Syntrophobotulus glycolicus, Desulfosporosinus youngiae, Desulfosporosinus acidiphilus, and Carboxydothermus islandicus, with amino acid sequence identities ranging from 55 to 93% (Table 1). The enzyme 5,10-methylene-THF reductase, catalyzing the conversion of 5,10-methylene-THF to 5-methyl-THF, had the highest amino acid sequence identity of 55% to a 5,10-methylene-THF reductase of Carboxydothermus islandicus.
Proteins associated with energy metabolism, biosynthesis, or responses to environmental cues.
The genome of strain RM harbors 21 hydrogenase genes (14). Proteomic analysis detected peptides of two subunits (HndA and HndC) of a NADP+-reducing hydrogenase (AWM53_00304, AWM53_00305, AWM53_00306; Data Set S2) to be highly abundant in cultures grown with DCM, suggesting that these hydrogenases might be involved in hydrogen formation during growth on DCM.
In the presence of DCM, strain RM also expressed proteins implicated in electron transfer reactions. Specifically, an electron transfer flavoprotein-ubiquinone oxidoreductase (AWM53_00047), the electron transport complex protein RnfC (AWM53_00217) and the putative electron transport protein YccM (AWM53_01799) were detected in DCM-grown RM cultures, suggesting the potential for respiratory energy conservation in addition to substrate-level phosphorylation via the WLP. The genome of strain RM further revealed an almost complete biosynthesis pathway for the electron carrier menaquinone (i.e., 8 of 9 genes detected; Data Set S3). Peptides consistent with expression of two menaquinone biosynthesis genes were detected in the proteome (AWM53_00989 and AWM53_00991; Data Set S2). Taken together, these findings suggest that chemiosmotic energy conservation might occur in strain RM. In support of this assertion, proteomics detected a putative potassium-stimulated, pyrophosphate-energized sodium pump (AWM53_02050; Data Set S2) and also identified F0F1 ATP synthase subunits, including AtpA (AWM53_01065; Data Set S2), AtpC (AWM53_01062), AtpD (AWM53_01063), and AtpG (AWM53_01064).
Also expressed were proteins associated with chemotaxis and motility (i.e., chemotaxis proteins CheA, CheW, and PomA [AWM53_00569, AWM53_00534, AWM53_00568, and AWM53_00582; Data Set S2], the flagellar hook protein FlgE [AWM53_00538], the flagellar motor switch protein FliN [AWM53_00556], and a two-component regulator propeller [AWM53_01511]), suggesting that the DCM degrader is motile and can respond to environmental cues (Data Set S2).
DISCUSSION
The proteomic approach applied in this study revealed enzymes likely involved in carbon-chlorine bond cleavage and DCM metabolism. These data sets provide a basis for further targeted experimental efforts aimed at elucidating specific functional roles for these proteins. The proteomic approach provided relative protein abundance information across various samples, thus revealing a protein expression pattern of strain RM during growth with DCM. The expression of methyltransferases, WLP enzymes, and two RDases provided information for proposing possible DCM degradation pathways in “Candidatus Dichloromethanomonas elyunquensis” strain RM. Strain RM, the DCM degrader in the microbial consortium RM, requires DCM as a growth substrate (14, 16). The consumption of DCM by strain RM yielded 2.6 × 107 ± 0.5 × 107 cells per µmol Cl– released, with a doubling time of around 4.6 days for strain RM (14). Like typical obligate organohalide-respiring bacteria, its growth is slow and has a low yield, e.g., the doubling times and growth yields of the Dehalococcoides mccartyi strains ranged from 0.8 to 3 days and 6.3 × 107 to 3.1 × 108 cells per µmol Cl– released (37), respectively, when grown on chlorinated solvents. Although strain RM has a complete WLP, growth with formate or hydrogen as electron donor and CO2 as electron acceptor was not observed, presumably due to the lack of hydrogen-oxidizing hydrogenases or energy conservation modules required for growth via reductive acetogenesis. The detection of strain RM proteins in incubations without DCM is explained by cellular proteins, introduced with the inoculum, which had been grown with DCM.
Expression and involvement of RDases.
Characteristic features of RDases involved in organohalide respiration include two iron-sulfur cluster-binding regions, a Tat motif that directs periplasmic localization, and an associated B protein with predicted membrane anchoring function (38). Analysis of the strain RM genome revealed that the three RDases comprise these characteristic features, suggesting that these RDases might be involved in organohalide respiration. Understanding of the structure-function relationships of RDases is in its infancy, and the prediction of substrate range based on RDase sequence similarity is ambiguous (39, 40). RDases that only share a maximum of 29.8% pairwise identity (range, 20.9 to 29.8%) can dechlorinate the same substrate, i.e., tetrachloroethene (41). On the other hand, even minor sequence variations can result in distinct RDase substrate specificity (39, 42), thus making substrate range prediction based on sequence similarity a daunting task.
Peptides of RDase 1 and RDase 2, but never of RDase 3, were detected, suggesting that strain RM can regulate RDase gene expression (43–45). The possibility that strain RM controls RDase gene expression is supported by the observation that all three RDase gene clusters of strain RM comprise accessory genes with putative regulatory function. Of course, it is possible that regulation does not occur and that the RDases are constitutively expressed; only detailed biochemical studies can confirm the specific roles of RDase 1 and RDase 2 in anaerobic DCM metabolism. The finding that RDase 3 was not detected in DCM-grown cultures (although nondetection of a protein in proteomic measurements does not prove its absence) may suggest that strain RM metabolizes additional chlorinated compounds.
Assuming an initial reductive dechlorination step, the first DCM metabolite would be chloromethane (CM); however, CM production was never observed in consortium RM (12, 14, 16). While it is possible that CM is rapidly consumed or remains enzyme-bound, thus evading detection, such scenarios are not supported by experimental data because strain RM did not grow with CM (16). Thus, reductive dechlorination of DCM to CM is unlikely, and DCM does not appear to be the substrate for the expressed RDases. An intriguing possibility is the formation of chlorinated WLP intermediates (i.e., chlorinated THF derivatives [Fig. S1]). The formation of a bond between DCM and THF would yield chlorinated WLP intermediates, which could be the substrates for the expressed RDases. To date, no evidence exists for the proposed modification of the WLP pathway, although the formation of chlorinated-THF intermediates appears feasible. Attempts to identify chlorinated-THF intermediates in DCM-grown biomass using liquid chromatography-mass spectrometry (LC-MS) approaches to detect folate intermediates were not successful (46). THF intermediates are labile and likely present in low concentrations, which makes their detection very challenging or impossible. The methodology employed in a previous study (46) was most successful when cell densities were high and compounds that blocked folate metabolism were used to increase the concentration of the analytes. This technique has recently been supplanted by an approach that uses derivatization agents to trap unstable methylene-THF intermediates as stable methyl-THF intermediates (47). As the derivatization step may lead to dechlorination, this method was not used in the present study. Even though chlorinated THF intermediates could not be detected, their formation cannot be excluded at this time due to a lack of sufficiently sensitive detection methods.
Potential involvement of methyltransferases in chlorinated compound metabolism.
Of the 96 methyltransferase genes annotated on the strain RM genome, 4 were highly expressed during growth with DCM. Highly similar methyltransferase genes were found on the genomes of Dehalobacterium formicoaceticum, Dehalobacter sp. strain UNSWDHB, Dehalobacter sp. strain CF, and Dehalobacter sp. strain DCA. Dehalobacterium formicoaceticum degrades DCM, but strain CF and strain DCA do not, suggesting that these methyltransferases have functions not directly involving DCM. A proteomic study of Dehalobacter sp. strain UNSWDHB grown with CF did not report methyltransferase expression (25), and proteomic studies with other members of the Peptococcaceae are lacking. Detailed biochemical experiments with cell extracts or purified proteins will be needed to clarify the role of methyltransferases and methyl-group carriers such as THF.
Possible pathways for anaerobic DCM degradation.
RDase 1 and RDase 2 expressed by strain RM might be involved in anaerobic DCM degradation by performing reductive dechlorination of yet-unknown chlorinated DCM metabolites of the WLP. In such a scenario, membrane-anchored RDases should face the cytoplasm as WLP intermediates occur in the cytoplasm rather than the periplasmic space. Although the presence of Tat signals in the expressed RDases makes cytoplasmic facing unlikely, Tat signals are known to sometimes direct alternate cellular localization, especially in cases where the signal peptide is not cleaved off (48, 49). Accordingly, while RDase 2 has a canonical Tat signal, it lacks a clear cleavage site, making its ultimate cellular location unpredictable. The proteomic analysis revealed the expression of ABC transporters and, thus active transport of DCM metabolites between cytoplasm and periplasm might be possible. In organohalide-respiring bacteria, RDases act as terminal reductases transferring electrons from an electron donor (e.g., hydrogen) to the chlorinated electron acceptor. The situation in strain RM is different because DCM is the electron donor (i.e., DCM is the only energy source), and DCM metabolism does not generate an obvious reductive dechlorination end product (i.e., CM and methane). If the RDases are involved in DCM degradation, they may be only responsible for cleaving DCM C-Cl bonds (50) without a direct link to energy conservation.
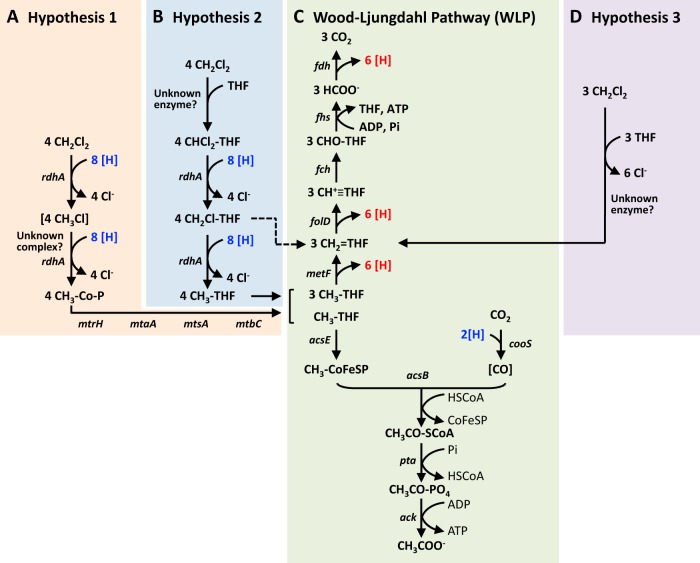
Based on the proteogenomic findings and prior work with Dehalobacterium formicoaceticum, hypothetical pathways for anaerobic DCM degradation in strain RM emerge (Fig. 4A and B, hypotheses 1 and 2). One possible scenario (Fig. 4A, hypothesis 1) is the initial reductive dechlorination of DCM catalyzed by one of the expressed RDases. Since CM was never detected and consortium RM did not grow with CM, CM might remain bound to an enzyme complex comprising the RDase and a methyltransferase (Fig. 4A). This RDase-methyltransferase complex might have specificity for DCM and may not initiate methyl group transfer directly from CM, thus explaining why CM cannot be metabolized by strain RM. In a subsequent enzymatic step, after removal of the second chlorine substituent, the methyl group of CM would be transferred to a corrinoid protein by another methyltransferase to yield a methylated corrinoid protein. Subsequent reactions would involve methyl group transfer from the methylated corrinoid protein to THF, yielding CH3-THF (Fig. 4A), which enters the WLP (Fig. 4C).
FIG 4.
Proposed DCM degradation pathway in “Candidatus Dichloromethanomonas elyunquensis” and in Dehalobacterium formicoaceticum. Two hypothetical schemes (A = hypothesis 1, B = hypothesis 2) for reactions initiating DCM degradation are proposed for strain RM and one hypothetical scheme (D = hypothesis 3) is proposed for Dehalobacterium formicoaceticum (13). Each hypothetical scheme connects with the WLP (C). According to hypothesis 3 (D), 3 mol of DCM is transformed to CH2=THF and subsequently disproportionated, with two-thirds being oxidized to formate and one third being reduced to acetate (13). The dashed arrow indicates reactions that remain to be elucidated. Brackets indicate enzyme-bound intermediates. Abbreviations: [H], electrons/reducing equivalents; Cl−, chloride ions; CH2Cl2, dichloromethane; CH3Cl, chloromethane; CH3-Co-P, methylated corrinoid protein; THF, tetrahydrofolate; CO2, carbon dioxide; HCOO−, formate; CHO-THF, formyl-tetrahydrofolate; CH+≡THF, methenyl-tetrahydrofolate; CH2=THF, methylene-tetrahydrofolate; CH3-THF, methyl-tetrahydrofolate; CH3-CoFeSP, methylated corrinoid/iron-sulfur protein; CH3CO-SCoA, acetyl-CoA; CO, carbon monoxide; CH3CO-PO4, acetyl phosphate; CH3COO−, acetate; rdhA, reductive dehalogenase; mtrH, mtaA, mtsA, and mtbC, corrinoid-dependent methyltransferases; fdh, formate dehydrogenase; fhs, formate-THF ligase; fch, methenyl-THF cyclohydrolase; folD, 5,10-methylene-THF dehydrogenase; metF, 5,10-methylene-THF reductase; acsE, methyltransferase; cooS, CO dehydrogenase; acsB, acetyl-CoA synthase; pta, phosphate acetyltransferase; ack, acetate kinase.
Another possible DCM degradation pathway (Fig. 4B, hypothesis 2) may involve a THF-DCM intermediate. This compound, CHCl2-THF, would then undergo reductive dechlorination to a THF-CM intermediate (i.e., CH2Cl-THF) (Fig. 4B). Such a postulated THF-CM intermediate could be the substrate for the second RDase (Fig. 4B) to directly generate CH3-THF that is further metabolized via the WLP (Fig. 4C). However, spontaneous dechlorination of CH2Cl-THF leading to the formation of 5,10-methylene-THF (CH2=THF) is also a possibility (see dotted arrow between Fig. 4B and C), although this would not account for the expression of two distinct RDases. In contrast to hypothesis 1, this pathway would not rely on methyltransferase reactions (Fig. 4A), which is inconsistent with the high expression of four methyltransferases.
Alternatively, only a single reductive dechlorination step occurs and RDase 1 and RDase 2 may have overlapping substrate ranges (i.e., are functionally equivalent), or one of the RDases is not functional. Studies with the organohalide-respiring bacterium Dehalococcoides mccartyi demonstrated that several RDases were expressed in the presence of a single chlorinated electron acceptor (51), yet only a single RDase was involved in electron acceptor reduction. The second chlorine atom might be removed by spontaneous cleavage of Cl-C bonds which might occur with CH2Cl-THF. Central to all of the proposed hypothetical pathways in strain RM is that the Cl compound is funneled into the WLP. Consistent with this observation is that the proteomic analysis detected a full set of WLP-associated enzymes (Fig. 4C).
Pathway diversity of anaerobic DCM degradation.
In addition to strain RM, another DCM degrader, Dehalobacterium formicoaceticum expressed WLP-associated enzymes involved in anaerobic DCM degradation (11). In contrast to strain RM, genome analysis of Dehalobacterium formicoaceticum did not reveal RDases (34), suggesting mechanistically distinct DCM degradation pathways in these two organisms. In support of this hypothesis, a recent isotope fractionation study revealed distinctly different dual carbon-chlorine isotope correlations associated with DCM degradation by strain RM versus Dehalobacterium formicoaceticum (52). Interestingly, the measured carbon kinetic isotope effect (AKIEC) for DCM degradation by strain RM falls in the range indicative of reductive dechlorination, while the AKIEC for DCM degradation by Dehalobacterium formicoaceticum falls in the range of an SN2-type nucleophilic substitution reaction (52). Based on physiological experiments and enzyme activities detected in cell extracts, a hypothetical DCM degradation pathway in Dehalobacterium formicoaceticum was proposed (11, 13). The postulated scenario for Dehalobacterium formicoaceticum (hypothesis 3; Fig. 4D) (11, 13) involves a substitutive dehalogenation reaction catalyzed by an unknown enzyme, leading to the formation of CH2=THF, which is funneled into the WLP (Fig. 4C).
Expanding pathway diversity might be crucial for microorganisms occupying diverse ecological niches that compete for the same substrate(s). Horizontal gene transfer appears important for DCM degradation in oxic, DCM-contaminated environments (53, 54). This process might also be a driving force for the acquisition of RDase genes in strain RM, since the G+C content of the RDases of strain RM (47.0 to 47.4%) are all above the average G+C content of the genome (43.5%) (33). The acquisition of RDase genes might bestow ecological advantages on the host, such as the utilization of alternative carbon and energy sources, the consumption of substrates at low concentrations, or metabolic activity under different redox conditions.
Implications.
Microbial DCM degradation in anoxic environments contributes to DCM removal, is of environmental relevance for destroying an ozone-depleting substance (4), and has implications for bioremediation of sites impacted with polychlorinated methanes. The specific genes/proteins identified in this proteogenomic study of “Candidatus Dichloromethanomonas elyunquensis” strain RM (e.g., putative RDases and methyltransferases) may serve as biomarkers for monitoring anaerobic DCM degradation in various environmental systems, both in pristine and in DCM-contaminated habitats. Furthermore, the proposed hypothetical DCM pathways (Fig. 4) based on detailed proteogenomic analyses of strain RM presented in this study will facilitate future research endeavors to biochemically resolve the anaerobic DCM degradation pathways.
MATERIALS AND METHODS
Chemicals.
All chemicals were obtained from Sigma Chemical Co. (St. Louis, MO), unless specified otherwise. High performance liquid chromatography (HPLC)-grade water and other solvents were obtained from Burdick & Jackson (Muskegon, MI), 99% formic acid was purchased from EM Science (Darmstadt, Germany), and sequencing-grade trypsin was acquired from Promega (Madison, WI).
Cultivation.
A DCM-degrading consortium was obtained from pristine freshwater sediment collected from the Rio Mameyes in Luquillo, Puerto Rico (latitude 18°21′43.9″, longitude −65°46′8.4″), in October 2009 (12). The culture was maintained under static conditions at 20°C in 160-ml glass serum bottles containing 100 ml of defined, anoxic, 30 mM bicarbonate and 10 mM HEPES-buffered (pH 7.2) mineral salt medium reduced with 0.2 mM Na2S and l-cysteine (55) amended with vitamins (56). The vessel headspace consisted of a N2/CO2 (80/20, vol/vol) atmosphere. DCM was added using a 10-µl Hamilton microliter syringe (Reno, NV) to reach initial aqueous phase concentrations of 0.6 to 1.2 mM. Following DCM consumption, cultures received up to three additional DCM feedings. During the active DCM consumption phase, the DCM degrader strain RM dominated the consortium (>87% of the community based on qPCR data, i.e., related to bacterial and methanogenic gene copy numbers).
For proteomic and qPCR analysis, parallel biological replicate cultures were amended with DCM, acetate, formate, hydrogen, or hydrogen plus BES, respectively. Vessels without substrate addition served as negative controls. Initial substrate concentrations were 0.6 to 1.2 mM DCM, 2 mM formate, 2 mM acetate, 4.1 mM (nominal concentration) hydrogen, or 4.1 mM hydrogen plus 1 mM BES. Cultures amended with DCM received one additional feeding, whereas all other cultures (except the no-substrate controls) received two additional feedings. For DNA and protein analysis, all cultures were sacrificed after 28 to 39 days of incubation when the amended substrates (e.g., hydrogen, formate, and DCM) had been consumed.
Analytical methods.
Concentrations of DCM and the formation of potential transformation products (i.e., CM and methane) were monitored by manual headspace injections (0.1 ml) into an Agilent 7890 gas chromatograph (GC; Santa Clara, CA) equipped with a DB-624 column (60 m length, 0.32 mm i.d., 1.8 µm film thickness) and a flame ionization detector (FID). The GC inlet was maintained at 200°C, the oven temperature was kept at 60°C for 2 min followed by an increase to 200°C at a rate of 25°C min−1, and the FID detector was operated at 280°C as described previously (57). Hydrogen was measured with a Peak Performer 1 reducing compound photometer (Peak Laboratories, Mountain View, CA). Formate and acetate were quantified with an Agilent 1200 series HPLC system as described previously (16).
DNA extraction.
Biomass was collected from a 100- to 160-ml culture suspension using Sterivex cartridges (Millipore, Billerica, MA). The membrane filters were removed from the Sterivex cartridges and cut into strips with a sterile razor, and the pieces were placed in a 2-ml plastic tube. A total volume of 1.8 ml of DNA extraction buffer (0.1 M Tris-HCl [pH 8], 0.1 M Na-EDTA [pH 8], 0.1 M NaH2PO4 [pH 8], 1.5 M NaCl, 5% cetyltrimethylammonium bromide [CTAB]) was added to each tube. Samples were vortex mixed for 2 min at the maximum speed, frozen at –80°C and thawed at 65°C three times, and then incubated on a shaker table at 50 rpm for 30 min at 37°C. Sodium dodecyl sulfate (SDS; 20%, wt/vol; 60 μl), 20 μl of proteinase K (10 mg/ml), and 200 μl of lysozyme (50 mg/ml) were added to each sample. During a 2-h incubation period in a water bath at 65°C, the tubes were gently inverted every 15 min. The tubes were centrifuged at room temperature (6,000 × g; 5 min), and the supernatants from individual tubes were collected in sterile 15-ml plastic tubes. The membrane filters were extracted two more times (first extraction: 1 ml of DNA extraction buffer with 75 μl of SDS and 20 μl of proteinase K; second extraction: 0.37 ml of DNA extraction buffer with 75 μl of SDS and 10 μl of proteinase K). The samples were incubated for 10 min in a water bath at 65°C and centrifuged at room temperature (6,000 × g; 5 min). The respective supernatants were pooled before an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added to each 15-ml plastic tube, and the tubes were gently mixed and centrifuged at room temperature (3,000 × g; 10 min). The aqueous (top) layer from each tube was withdrawn and equally divided into three 2-ml plastic tubes. To each tube, a 0.6 volume of isopropanol was added, followed by gentle mixing. Precipitation of the DNA proceeded overnight at room temperature. The precipitated DNA was collected by centrifugation at room temperature (13,000 × g; 45 min), the isopropanol supernatant was removed and replaced with 1 ml of 70% ice-cold ethanol. After a final centrifugation step (13,000 × g; 30 min), the ethanol was removed, and the DNA was dried for 30 min inside a laminar flow hood. The DNA pellets in each tube were suspended in 20 μl of TE buffer (10 mM Tris-HCl, 1 mM Na-EDTA [pH 8]), yielding 60 μl of DNA solution per sample. The DNA was quantified using the Qubit dsDNA BR Assay (Life Technologies) according to the manufacturer’s manual and stored at –80°C until analysis.
Quantitative real-time PCR and statistics.
16S rRNA gene-targeted qPCR assays using primers and linear hybridization (TaqMan) probes enumerated the gene abundances of total bacteria, methanogenic archaea, and strain RM (Table 2). A single 16S rRNA gene was found on the draft genome of strain RM, suggesting gene copy numbers directly convert to cell numbers. Calibration curves (the concentrations ranged from 4.67 × 108 to 46.7 copies) were obtained by using serial 10-fold dilutions of plasmid DNA carrying a cloned 16S rRNA gene. qPCR was conducted using an ABI 7500 Fast Real Time PCR System equipped with 7500 software v2.0.3 (Applied Biosystems, Carlsbad, CA) as described previously (58). Briefly, every 20 μl-reaction contained 10 μl of 2× TaqMan Universal PCR Master Mix (Applied Biosystems), 2 μl of DNA template, and forward and reverse primers and probe at final concentrations of 300 nM each. Reactions were initially held for 2 min at 50°C and 10 min at 95°C, following 40 cycles of denaturation at 95°C for 15 sec and annealing and extension at 60°C for 1 min.
TABLE 2.
Primers and TaqMan probes used to quantify 16S rRNA genes
| Primer/probe | Sequence (5′–3′) | Target(s) | Reference(s) or source |
|---|---|---|---|
| Set 1 | |||
| Bac1055YF | ATGGYTGTCGTCAGCT | Bacteria | 58, 78 |
| Bac1392R | ACGGGCGGTGTGTAC | Bacteria | 79 |
| Bac1115-probe | FAM-CAACGAGCGCAACCC-MGB | Bacteria | 79, 80 |
| Set 2 | |||
| DhbRM114F | CCGCAACGAGCGCAAC | Strain RM | This study |
| DhbRM1168R | TTGTCACCGGCAGTCTATCCA | Strain RM | This study |
| Dhc-H-PR2-probe | FAM-CAGTTACCAGCACGTAAA-MGB | Strain RM | This study |
| Set 3 | |||
| Mtgen835F | GGGRAGTACGKYCGCAAG | Methanogens | This study |
| Mtgen918R | GAVTCCAATTRARCCGCA | Methanogens | This study |
| Mtgen831-probe | FAM-CCAATTCCTTTAAGTTTCA-MGB | Methanogens | This study |
Significant differences (P ≤ 0.05) of 16S rRNA gene copies among culture setups were determined by analysis of variance (ANOVA) tests with multiple-comparison Bonferroni post hoc tests using GraphPad Prism (version 7.0d; GraphPad Software, La Jolla, CA).
Sample preparation for proteomics.
Biomass from 100-ml culture suspensions was collected on 0.22-μm membranes using Sterivex cartridges and immediately stored at –20°C. Total cellular proteins were extracted from cells collected on membrane filters using a detergent-based protein extraction protocol (59). Briefly, the filters were cut into small pieces, suspended in SDS lysis buffer (59), and processed as described previously (60). The supernatant was separated from the filter pieces, chilled 100% trichloroacetic acid (TCA) was added to a final concentration of 25% (vol/vol), and the samples were kept at –20°C overnight. Samples were centrifuged at 21,000 × g for 20 min, and the protein pellets were processed as described previously (60) and solubilized in 6 M guanidine buffer (6 M guanidine; 10 mM dithiothreitol in Tris-CaCl2 buffer (10 mM Tris; pH 7.8) with a 3-h incubation at 60°C (60). From each sample, 25-µl aliquots were retained for protein estimation. The remainder of the protein sample was digested with sequencing-grade trypsin (Promega, Madison, WI), the resulting peptides were desalted, and the solvent was exchanged as described previously (61). The amount of protein extracted from each sample was calculated using an RC/DC protein estimation kit (Bio-Rad Laboratories, Hercules, CA) according to the manufacturer’s instructions. Bovine serum albumin (supplied with the RC/DC kit) was used as a standard.
NanoLC-MS/MS analysis.
Technical (DCM) or biological (acetate, formate, hydrogen with or without BES, control) duplicates were analyzed by Nano-2D-LC-MS/MS. Peptides were loaded onto an in-house-prepared biphasic resin-packed column (SCX [Luna; Phenomenex, Torrance, CA]) and a C18 column (Aqua; Phenomenex, Torrance, CA) according to published procedures (61, 62) and then subjected to an offline wash for 15 min (63). The sample column was aligned with an in-house C18 packed nanospray tip (New Objective, Woburn, MA) connected to a Proxeon (Odense, Denmark) nanospray source as described previously (63). Peptides were eluted after chromatographic separation and ionized by nano-electrospray ionization (nano-ESI) for subsequent measurements via 24-h Multi-Dimensional Protein Identification Technology (MuDPIT) approach (61–63). Measurements were carried out using an LTQ-Orbitrap-Elite mass spectrometer (Thermo Fisher Scientific, Germany) linked to the Ultimate 3000 HPLC system (Dionex) and operated in data-dependent mode using Thermo Xcalibur software v2.1.0. Each full scan (1 microscan) was followed by fragmentation via collision-activated dissociation using 35% collision energy of the 20 most abundant parent ions (1 microscan) with a mass exclusion width of 0.2 m/z and a dynamic exclusion duration of 60 s.
Proteomic data analysis and annotation.
For protein identifications, the raw spectra were searched against a customized database with the Myrimatch v2.1 algorithm (64) using described parameters (65) with minor modifications. Common contaminant peptide sequences from trypsin and keratin were concatenated to the database. Reverse database sequences were included as decoy sequences to calculate false discovery rates (<0.1%). The database was constructed using the metagenome of the microbial consortium RM comprising the DCM degrader strain RM (GenBank accession no. SRP065755) (33). Metagenomic binning enabled the assembly of a draft genome of strain RM (33). This Whole Genome Shotgun project has been deposited at DDBJ/ENA/GenBank under the accession number LNDB00000000. The version described here is version LNDB01000000. Gene sequences were converted into amino acid sequences. The final database contained information about all protein-coding genes from the microbial consortium RM, including those of strain RM (Data Set S3). Using this database, protein abundance data were analyzed for the microbial consortium RM (all proteins) or selectively examined for proteins specific to strain RM. Static cysteine and dynamic oxidation modifications were not considered, and identification of at least two peptides per protein sequence (one unique and one nonunique) was a prerequisite for protein identifications. Spectral counts of identified peptides were normalized as described previously (66) to obtain the normalized spectral abundance factor, also referred to as normalized spectral counts (nSpc). The average nSpc from all runs of the same sample were used for downstream data analysis.
Sequence-based analyses.
Relationships and identities of RDases, methyltransferases and WLP-associated enzymes were examined using amino acid sequences and the Basic Local Alignment Search Tool (BLAST) (67). Functional predictions of genes of interest were further explored by annotation with Pfam (68), TIGRFAMs (69), KEGG (70), and COG (71), performed using Integrated Microbial Genomes (IMG) (72) and the eggNOG annotation servers (73). The RDase phylogenetic tree was calculated using amino acid sequences that were aligned in Geneious (74) with the ClustalW (75) plugin tool using a gap opening of 10 (range, 0 to 100) and a gap extension of 0.1 (range, 0 to 100). The RDase tree was calculated by maximum-likelihood analysis (PhyML) using the ARB software (76). Amino acid identities of the RDases of strain RM were calculated using the genome blast function implemented in the IMG database and comparative analysis system (72). Transmembrane helices of RDases were determined using the IMG server. Protein localization predictions were performed using PSORT Prediction (77). The presence of N-terminal signal peptides was investigated using SignalP 4.1 (35).
Supplementary Material
ACKNOWLEDGMENTS
This study was supported by The Chemours Company and in part by the Strategic Environmental Research and Development Program (project ER-2312). S.K. is supported by an Emmy-Noether fellowship (grant 326028733) from the German Research Foundation (Deutsche Forschungsgemeinschaft).
We thank Jun Yan for providing the RDase amino acid sequence database and helpful comments on the manuscript.
This manuscript has been authored by UT-Battelle, LLC, under contract DE-AC05-00OR22725 with the U.S. Department of Energy (DOE). DOE will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
The authors declare no competing interest or conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02768-18.
REFERENCES
- 1.Grostern A, Duhamel M, Dworatzek S, Edwards EA. 2010. Chloroform respiration to dichloromethane by a Dehalobacter population. Environ Microbiol 12:1053–1060. doi: 10.1111/j.1462-2920.2009.02150.x. [DOI] [PubMed] [Google Scholar]
- 2.Gribble GW. 2010. Naturally occurring organohalogen compounds: a comprehensive update. Springer Verlag, Vienna, Austria. doi: 10.1002/chin.201022254. [DOI] [Google Scholar]
- 3.Lee M, Low A, Zemb O, Koenig J, Michaelsen A, Manefield M. 2012. Complete chloroform dechlorination by organochlorine respiration and fermentation. Environ Microbiol 14:883–894. doi: 10.1111/j.1462-2920.2011.02656.x. [DOI] [PubMed] [Google Scholar]
- 4.Hossaini R, Chipperfield MP, Montzka SA, Leeson AA, Dhomse SS, Pyle JA. 2017. The increasing threat to stratospheric ozone from dichloromethane. Nat Commun 8:15962. doi: 10.1038/ncomms15962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nikolausz M, Kappelmeyer U, Nijenhuis I, Ziller K, Kästner M. 2005. Molecular characterization of dichloromethane-degrading Hyphomicrobium strains using 16S rDNA and DCM dehalogenase gene sequences. Syst Appl Microbiol 28:582–587. doi: 10.1016/j.syapm.2005.03.011. [DOI] [PubMed] [Google Scholar]
- 6.Braus-Stromeyer SA, Hermann R, Cook AM, Leisinger T. 1993. Dichloromethane as the sole carbon source for an acetogenic mixed culture and isolation of a fermentative dichloromethane-degrading bacterium. Appl Environ Microbiol 59:3790–3797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leisinger T, Braus-Stromeyer SA. 1995. Bacterial growth with chlorinated methanes. Environ Health Perspect 103:33–36. doi: 10.2307/3432475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egli C, Tschan T, Scholtz R, Cook AM, Leisinger T. 1988. Transformation of tetrachloromethane to dichloromethane and carbon dioxide by Acetobacterium woodii. Appl Environ Microbiol 54:2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mägli A, Rainey FA, Leisinger T. 1995. Acetogenesis from dichloromethane by a two-component mixed culture comprising a novel bacterium. Appl Environ Microbiol 61:2943–2949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Freedman DL, Gossett JM. 1991. Biodegradation of dichloromethane and its utilization as a growth substrate under methanogenic conditions. Appl Environ Microbiol 57:2847–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mägli A, Wendt M, Leisinger T. 1996. Isolation and characterization of Dehalobacterium formicoaceticum gen. nov. sp. nov., a strictly anaerobic bacterium utilizing dichloromethane as source of carbon and energy. Arch Microbiol 166:101–108. doi: 10.1007/s002030050362. [DOI] [Google Scholar]
- 12.Justicia-Leon SD, Ritalahti KM, Mack EE, Löffler FE. 2012. Dichloromethane fermentation by a Dehalobacter sp. in an enrichment culture derived from pristine river sediment. Appl Environ Microbiol 78:1288–1291. doi: 10.1128/AEM.07325-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mägli A, Messmer M, Leisinger T. 1998. Metabolism of dichloromethane by the strict anaerobe Dehalobacterium formicoaceticum. Appl Environ Microbiol 64:646–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kleindienst S, Higgins SA, Tsementzi D, Chen G, Konstantinidis KT, Mack EE, Löffler FE. 2017. “Candidatus Dichloromethanomonas elyunquensis” gen. nov., sp. nov., a dichloromethane-degrading anaerobe of the Peptococcaceae family. Syst Appl Microbiol 40:150–159. doi: 10.1016/j.syapm.2016.12.001. [DOI] [PubMed] [Google Scholar]
- 15.Trueba-Santiso A, Parladé E, Rosell M, Lliros M, Mortan SH, Martínez-Alonso M, Gaju N, Martín-González L, Vicent T, Marco-Urrea E. 2017. Molecular and carbon isotopic characterization of an anaerobic stable enrichment culture containing Dehalobacterium sp. during dichloromethane fermentation. Sci Total Environ 581-582:640–648. doi: 10.1016/j.scitotenv.2016.12.174. [DOI] [PubMed] [Google Scholar]
- 16.Chen G, Kleindienst S, Griffiths DR, Mack EE, Seger ES, Löffler FE. 2017. Mutualistic interaction between dichloromethane- and chloromethane-degrading bacteria in an anaerobic mixed culture. Environ Microbiol 19:4784–4796. doi: 10.1111/1462-2920.13945. [DOI] [PubMed] [Google Scholar]
- 17.Ljungdahl LG. 1986. The autotrophic pathway of acetate synthesis in acetogenic bacteria. Annu Rev Microbiol 40:415–450. doi: 10.1146/annurev.mi.40.100186.002215. [DOI] [PubMed] [Google Scholar]
- 18.Ragsdale SW, Pierce E. 2008. Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim Biophys Acta 1784:1873–1898. doi: 10.1016/j.bbapap.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fuchs G. 1994. Variations of the acetyl-CoA pathway in diversely related microorganisms that are not acetogens, p 507–520. In Drake H. (ed), Acetogenesis. Springer, New York, NY. doi: 10.1007/978-1-4615-1777-1_19. [DOI] [Google Scholar]
- 20.Ladapo J, Whitman WB. 1990. Method for isolation of auxotrophs in the methanogenic archaebacteria: role of the acetyl-CoA pathway of autotrophic CO2 fixation in Methanococcus maripaludis. Proc Natl Acad Sci U S A 87:5598–5602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Svetlitchnyi V, Dobbek H, Meyer-Klaucke W, Meins T, Thiele B, Römer P, Huber R, Meyer O. 2004. A functional Ni-Ni-[4Fe-4S] cluster in the monomeric acetyl-CoA synthase from Carboxydothermus hydrogenoformans. Proc Natl Acad Sci U S A 101:446–451. doi: 10.1073/pnas.0304262101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spormann A, Thauer R. 1988. Anaerobic acetate oxidation to CO2 by Desulfotomaculum acetoxidans. Arch Microbiol 150:374–380. doi: 10.1007/BF00408310. [DOI] [Google Scholar]
- 23.Schauder R, Preuss A, Jetten M, Fuchs G. 1988. Oxidative and reductive acetyl CoA/carbon monoxide dehydrogenase pathway in Desulfobacterium autotrophicum. Arch Microbiol 151:84–89. doi: 10.1007/BF00444674. [DOI] [Google Scholar]
- 24.Schouten S, Strous M, Kuypers MMM, Rijpstra WIC, Baas M, Schubert CJ, Jetten MSM, Sinninghe DJS. 2004. Stable carbon isotopic fractionations associated with inorganic carbon fixation by anaerobic ammonium-oxidizing bacteria. Appl Environ Microbiol 70:3785–3788. doi: 10.1128/AEM.70.6.3785-3788.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jugder BE, Ertan H, Wong YK, Braidy N, Manefield M, Marquis CP, Lee M. 2016. Genomic, transcriptomic and proteomic analyses of Dehalobacter UNSWDHB in response to chloroform. Environ Microbiol Rep doi: 10.1111/1758-2229.12444. [DOI] [PubMed] [Google Scholar]
- 26.Seshadri R, Adrian L, Fouts DE, Eisen JA, Phillippy AM, Methe BA, Ward NL, Nelson WC, Deboy RT, Khouri HM, Kolonay JF, Dodson RJ, Daugherty SC, Brinkac LM, Sullivan SA, Madupu R, Nelson KE, Kang KH, Impraim M, Tran K, Robinson JM, Forberger HA, Fraser CM, Zinder SH, Heidelberg JF. 2005. Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes. Science 307:105–108. doi: 10.1126/science.1102226. [DOI] [PubMed] [Google Scholar]
- 27.Johnson DR, Brodie EL, Hubbard AE, Andersen GL, Zinder SH, Alvarez-Cohen L. 2008. Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase. Appl Environ Microbiol 74:2864–2872. doi: 10.1128/AEM.02208-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Men Y, Feil H, Verberkmoes NC, Shah MB, Johnson DR, Lee PK, West KA, Zinder SH, Andersen GL, Alvarez-Cohen L. 2012. Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses. ISME J 6:410–421. doi: 10.1038/ismej.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnson DR, Nemir A, Andersen GL, Zinder SH, Alvarez-Cohen L. 2009. Transcriptomic microarray analysis of corrinoid responsive genes in Dehalococcoides ethenogenes strain 195. FEMS Microbiol Lett 294:198–206. doi: 10.1111/j.1574-6968.2009.01569.x. [DOI] [PubMed] [Google Scholar]
- 30.McMurdie PJ, Behrens SF, Müller JA, Goke J, Ritalahti KM, Wagner R, Goltsman E, Lapidus A, Holmes S, Löffler FE, Spormann AM. 2009. Localized plasticity in the streamlined genomes of vinyl chloride respiring Dehalococcoides. PLoS Genet 5:e1000714. doi: 10.1371/journal.pgen.1000714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kube M, Beck A, Zinder SH, Kuhl H, Reinhardt R, Adrian L. 2005. Genome sequence of the chlorinated compound-respiring bacterium Dehalococcoides species strain CBDB1. Nat Biotechnol 23:1269–1273. doi: 10.1038/nbt1131. [DOI] [PubMed] [Google Scholar]
- 32.Zhuang W-Q, Yi S, Bill M, Brisson VL, Feng X, Men Y, Conrad ME, Tang YJ, Alvarez-Cohen L. 2014. Incomplete Wood–Ljungdahl pathway facilitates one-carbon metabolism in organohalide-respiring Dehalococcoides mccartyi. Proc Natl Acad Sci U S A 111:6419–6424. doi: 10.1073/pnas.1321542111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kleindienst S, Higgins SA, Tsementzi D, Konstantinidis KT, Mack EE, Löffler FE. 2016. Draft genome of a strictly anaerobic dichloromethane-degrading bacterium. Genome Announc 4:e00037–e00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen G, Murdoch RW, Mack EE, Seger ES, Löffler FE. 2017. Complete genome sequence of Dehalobacterium formicoaceticum strain DMC, a strictly anaerobic dichloromethane-degrading bacterium. Genome Announc 5:e00897–e00817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Petersen TN, Brunak S, von Heijne G, Nielsen H. 2011. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 8:785. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 36.Ferrandez Y, Condemine G. 2008. Novel mechanism of outer membrane targeting of proteins in Gram-negative bacteria. Mol Microbiol 69:1349–1357. doi: 10.1111/j.1365-2958.2008.06366.x. [DOI] [PubMed] [Google Scholar]
- 37.Löffler FE, Yan J, Ritalahti KM, Adrian L, Edwards EA, Konstantinidis KT, Müller JA, Fullerton H, Zinder SH, Spormann AM. 2013. Dehalococcoides mccartyi gen. nov., sp. nov., obligately organohalide-respiring anaerobic bacteria relevant to halogen cycling and bioremediation, belong to a novel bacterial class, Dehalococcoidia classis nov., order Dehalococcoidales ord. nov. and family Dehalococcoidaceae fam. nov., within the phylum Chloroflexi. Int J Syst Evol Microbiol 63:625–635. doi: 10.1099/ijs.0.034926-0. [DOI] [PubMed] [Google Scholar]
- 38.Dobbek H, Leys D. 2016. Insights into reductive dehalogenase function obtained from crystal structures, p 485–495. In Adrian L, Löffler FE (ed), Organohalide-respiring bacteria. Springer, Berlin, Germany. doi: 10.1007/978-3-662-49875-0_20. [DOI] [Google Scholar]
- 39.Hug LA. 2016. Diversity, evolution, and environmental distribution of reductive dehalogenase genes, p 377–393. In Adrian L, Löffler FE (ed), Organohalide-respiring bacteria. Springer, Berlin, Germany. doi: 10.1007/978-3-662-49875-0_16. [DOI] [Google Scholar]
- 40.Buttet GF, Holliger C, Maillard J. 2013. Functional genotyping of Sulfurospirillum spp. in mixed cultures allowed the identification of a new tetrachloroethene reductive dehalogenase. Appl Environ Microbiol 79:6941–6947. doi: 10.1128/AEM.02312-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hug LA, Maphosa F, Leys D, Löffler FE, Smidt H, Edwards EA, Adrian L. 2013. Overview of organohalide respiration and introduction of a simple classification system for reductive dehalogenases. Philos Trans R Soc B 368:20120322. doi: 10.1098/rstb.2012.0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhao S, Rogers MJ, He J. 2017. Microbial reductive dehalogenation of trihalomethanes by a Dehalobacter-containing coculture. Appl Microbiol Biotechnol 101:5481–5492. doi: 10.1007/s00253-017-8236-2. [DOI] [PubMed] [Google Scholar]
- 43.Maillard J, Genevaux P, Holliger C. 2011. Redundancy and specificity of multiple trigger factor chaperones in Desulfitobacteria. Microbiology 157:2410–2421. doi: 10.1099/mic.0.050880-0. [DOI] [PubMed] [Google Scholar]
- 44.Mac Nelly A, Kai M, Svatoš A, Diekert G, Schubert T. 2014. Functional heterologous production of reductive dehalogenases from Desulfitobacterium hafniense strains. Appl Environ Microbiol 80:4313–4322. doi: 10.1128/AEM.00881-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morita Y, Futagami T, Goto M, Furukawa K. 2009. Functional characterization of the trigger factor protein PceT of tetrachloroethene-dechlorinating Desulfitobacterium hafniense Y51. Appl Microbiol Biotechnol 83:775–781. doi: 10.1007/s00253-009-1958-z. [DOI] [PubMed] [Google Scholar]
- 46.Lu W, Kwon YK, Rabinowitz JD. 2007. Isotope ratio-based profiling of microbial folates. J Am Soc Mass Spectrom 18:898–909. doi: 10.1016/j.jasms.2007.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen L, Ducker GS, Lu W, Teng X, Rabinowitz JD. 2017. An LC-MS chemical derivatization method for the measurement of five different one-carbon states of cellular tetrahydrofolate. Anal Bioanal Chem 409:5955–5964. doi: 10.1007/s00216-017-0514-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hatzixanthis K, Palmer T, Sargent F. 2003. A subset of bacterial inner membrane proteins integrated by the twin-arginine translocase. Mol Microbiol 49:1377–1390. [DOI] [PubMed] [Google Scholar]
- 49.Goosens VJ, Otto A, Glasner C, Monteferrante CC, van der Ploeg R, Hecker M, Becher D, van Dijl JM. 2013. Novel twin-arginine translocation pathway-dependent phenotypes of Bacillus subtilis unveiled by quantitative proteomics. J Proteome Res 12:796–807. doi: 10.1021/pr300866f. [DOI] [PubMed] [Google Scholar]
- 50.Payne KA, Quezada CP, Fisher K, Dunstan MS, Collins FA, Sjuts H, Levy C, Hay S, Rigby SE, Leys D. 2015. Reductive dehalogenase structure suggests a mechanism for B12-dependent dehalogenation. Nature 517:513–516. doi: 10.1038/nature13901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Waller AS, Krajmalnik-Brown R, Löffler FE, Edwards EA. 2005. Multiple reductive-dehalogenase-homologous genes are simultaneously transcribed during dechlorination by Dehalococcoides-containing cultures. Appl Environ Microbiol 71:8257–8264. doi: 10.1128/AEM.71.12.8257-8264.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen G, Shouakar-Stash O, Phillips E, Justicia-Leon SD, Gilevska T, Sherwood Lollar B, Mack EE, Seger ES, Löffler FE. 2018. Dual carbon-chlorine isotope analysis indicates distinct anaerobic dichloromethane degradation pathways in two members of Peptococcaceae. Environ Sci Technol 52:8607–8616. doi: 10.1021/acs.est.8b01583. [DOI] [PubMed] [Google Scholar]
- 53.Vuilleumier S, Chistoserdova L, Lee M-C, Bringel F, Lajus A, Zhou Y, Gourion B, Barbe V, Chang J, Cruveiller S, Dossat C, Gillett W, Gruffaz C, Haugen E, Hourcade E, Levy R, Mangenot S, Muller E, Nadalig T, Pagni M, Penny C, Peyraud R, Robinson DG, Roche D, Rouy Z, Saenampechek C, Salvignol G, Vallenet D, Wu Z, Marx CJ, Vorholt JA, Olson MV, Kaul R, Weissenbach J, Médigue C, Lidstrom ME. 2009. Methylobacterium genome sequences: a reference blueprint to investigate microbial metabolism of C1 compounds from natural and industrial sources. PLoS ONE 4:e5584. doi: 10.1371/journal.pone.0005584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller EE, Bringel F, Vuilleumier S. 2011. Dichloromethane-degrading bacteria in the genomic age. Res Microbiol 162:869–876. doi: 10.1016/j.resmic.2011.01.008. [DOI] [PubMed] [Google Scholar]
- 55.Löffler FE, Sanford RA, Tiedje JM. 1996. Initial characterization of a reductive dehalogenase from Desulfitobacterium chlororespirans Co23. Appl Environ Microbiol 62:3809–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wolin EA, Wolin MJ, Wolfe RS. 1963. Formation of methane by bacterial extracts. J Biol Chem 238:2882–2886. [PubMed] [Google Scholar]
- 57.Amos BK, Christ JA, Abriola LM, Pennell KD, Löffler FE. 2007. Experimental evaluation and mathematical modeling of microbially enhanced tetrachloroethene (PCE) dissolution. Environ Sci Technol 41:963–970. [DOI] [PubMed] [Google Scholar]
- 58.Ritalahti KM, Amos BK, Sung Y, Wu Q, Koenigsberg SS, Löffler FE. 2006. Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microbiol 72:2765–2774. doi: 10.1128/AEM.72.4.2765-2774.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chourey K, Jansson J, VerBerkmoes N, Shah M, Chavarria KL, Tom LM, Brodie EL, Hettich RL. 2010. Direct cellular lysis/protein extraction protocol for soil metaproteomics. J Proteome Res 9:6615–6622. doi: 10.1021/pr100787q. [DOI] [PubMed] [Google Scholar]
- 60.Chourey K, Nissen S, Vishnivetskaya T, Shah M, Pfiffner S, Hettich RL, Löffler FE. 2013. Environmental proteomics reveals early microbial community responses to biostimulation at a uranium- and nitrate-contaminated site. Proteomics 13:2921–2930. [DOI] [PubMed] [Google Scholar]
- 61.Thompson MR, VerBerkmoes NC, Chourey K, Shah M, Thompson DK, Hettich RL. 2007. Dosage-dependent proteome response of Shewanella oneidensis MR-1 to acute chromate challenge. J Proteome Res 6:1745–1757. doi: 10.1021/pr060502x. [DOI] [PubMed] [Google Scholar]
- 62.Brown SD, Thompson MR, Verberkmoes NC, Chourey K, Shah M, Zhou J, Hettich RL, Thompson DK. 2006. Molecular dynamics of the Shewanella oneidensis response to chromate stress. Mol Cell Proteomics 5:1054–1071. [DOI] [PubMed] [Google Scholar]
- 63.Sharma R, Dill BD, Chourey K, Shah M, VerBerkmoes NC, Hettich RL. 2012. Coupling a detergent lysis/cleanup methodology with intact protein fractionation for enhanced proteome characterization. J Proteome Res 11:6008–6018. doi: 10.1021/pr300709k. [DOI] [PubMed] [Google Scholar]
- 64.Tabb DL, Fernando CG, Chambers MC. 2007. MyriMatch: highly accurate tandem mass spectral peptide identification by multivariate hypergeometric analysis. J Proteome Res 6:654–661. doi: 10.1021/pr0604054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Xiong W, Giannone RJ, Morowitz MJ, Banfield JF, Hettich RL. 2014. Development of an enhanced metaproteomic approach for deepening the microbiome characterization of the human infant gut. J Proteome Res 14:133–141. doi: 10.1021/pr500936p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paoletti AC, Parmely TJ, Tomomori-Sato C, Sato S, Zhu D, Conaway RC, Conaway JW, Florens L, Washburn MP. 2006. Quantitative proteomic analysis of distinct mammalian Mediator complexes using normalized spectral abundance factors. Proc Natl Acad Sci U S A 103:18928–18933. doi: 10.1073/pnas.0606379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. 1990. Basic local alignment search tool. J Mol Biol 215:403–410. [DOI] [PubMed] [Google Scholar]
- 68.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, Potter SC, Punta M, Qureshi M, Sangrador-Vegas A, Salazar GA, Tate J, Bateman A. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44:D279–D285. doi: 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Haft DH, Loftus BJ, Richardson DL, Yang F, Eisen JA, Paulsen Ian T, White O. 2001. TIGRFAMs: a protein family resource for the functional identification of proteins. Nucleic Acids Res 29:41–43. doi: 10.1093/nar/29.1.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. 2016. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res 44:D457–D462. doi: 10.1093/nar/gkv1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Tatusov RL, Galperin MY, Natale DA, Koonin EV. 2000. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Markowitz VM, Chen IMA, Palaniappan K, Chu K, Szeto E, Grechkin Y, Ratner A, Jacob B, Huang J, Williams P, Huntemann M, Anderson I, Mavromatis K, Ivanova NN, Kyrpides NC. 2012. IMG: the integrated microbial genomes database and comparative analysis system. Nucleic Acids Res 40:D115–D122. doi: 10.1093/nar/gkr1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Huerta-Cepas J, Szklarczyk D, Forslund K, Cook H, Heller D, Walter MC, Rattei T, Mende DR, Sunagawa S, Kuhn M, Jensen LJ, von Mering C, Bork P. 2016. eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res 44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Drummond AJ, Ashton B, Cheung M, Heled J, Kearse M, Moir R, Stones-Havas S, Thierer T, Wilson A. 2009. Geneious v4.7, http://www.geneious.com/.
- 75.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22:4673–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ludwig W, Strunk O, Westram R, Richter L, Meier H, Yadhukumar, Buchner A, Lai T, Steppi S, Jobb G, Förster W, Brettske I, Gerber S, Ginhart AW, Gross O, Grumann S, Hermann S, Jost R, König A, Liss T, Lüssmann R, May M, Nonhoff B, Reichel B, Strehlow R, Stamatakis A, Stuckmann N, Vilbig A, Lenke M, Ludwig T, Bode A, Schleifer K-H. 2004. ARB: a software environment for sequence data. Nucleic Acids Res 32:1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Nakai K, Horton P. 1999. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem Sci 24:34–36. [DOI] [PubMed] [Google Scholar]
- 78.Ferris MJ, Muyzer G, Ward DM. 1996. Denaturing gradient gel electrophoresis profiles of 16S rRNA-defined populations inhabiting a hot spring microbial mat community. Appl Environ Microbiol 62:340–346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Lane DJ. 1991. 16S/23S rRNA sequencing, p 115–148. In Stackebrandt E, Goodfellow M (ed), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Chichester, United Kingdom. [Google Scholar]
- 80.Harms G, Layton AC, Dionisi HM, Gregory IR, Garrett VM, Hawkins SA, Robinson KG, Sayler GS. 2002. Real-time PCR quantification of nitrifying bacteria in a municipal wastewater treatment plant. Environ Sci Technol 37:343–351. doi: 10.1021/es0257164. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.