This study explored the role of CgHog1-mediated CgRds2 phosphorylation in response to osmotic stress in Candida glabrata. CgHog1 interacts with and phosphorylates CgRds2, a zinc cluster transcription factor, under osmotic stress. Phosphorylated CgRds2 plays an important role in increasing glycerophospholipid composition and membrane integrity, thereby enhancing cell growth and survival.
KEYWORDS: glycerophospholipid, membrane, mitogen-activated protein kinase, MAPK, phosphorylation, stress response, transcription factor
ABSTRACT
Under stress conditions, Hog1 is required for cell survival through transiently phosphorylating downstream targets and reprogramming gene expression. Here, we report that Candida glabrata Hog1 (CgHog1) interacts with and phosphorylates CgRds2, a zinc cluster transcription factor, in response to osmotic stress. Additionally, we found that deletion of CgRDS2 led to decreases in cell growth and cell survival by 23.4% and 39.6%, respectively, at 1.5 M NaCl, compared with levels of the wild-type strain. This is attributed to significant downregulation of the expression levels of glycerophospholipid metabolism genes. As a result, the content of total glycerophospholipid decreased by 30.3%. Membrane integrity also decreased 47.6% in the Cgrds2Δ strain at 1.5 M NaCl. In contrast, overexpression of CgRDS2 increased the cell growth and cell survival by 10.2% and 6.3%, respectively, owing to a significant increase in the total glycerophospholipid content and increased membrane integrity by 27.2% and 12.1%, respectively, at 1.5 M NaCl, compared with levels for the wild-type strain. However, a strain in which the CgRDS2 gene encodes the replacement of Ser64 and Thr97 residues with alanines (Cgrds22A), harboring a CgRds2 protein that was not phosphorylated by CgHog1, failed to promote glycerophospholipid metabolism and membrane integrity at 1.5 M NaCl. Thus, the above results demonstrate that CgHog1-mediated CgRds2 phosphorylation enhanced glycerophospholipid composition and membrane integrity to resist osmotic stress in C. glabrata.
IMPORTANCE This study explored the role of CgHog1-mediated CgRds2 phosphorylation in response to osmotic stress in Candida glabrata. CgHog1 interacts with and phosphorylates CgRds2, a zinc cluster transcription factor, under osmotic stress. Phosphorylated CgRds2 plays an important role in increasing glycerophospholipid composition and membrane integrity, thereby enhancing cell growth and survival.
INTRODUCTION
Owing to frequent changes in the natural environment, microorganisms are often subjected to various stresses, including nutrient limitation (1), osmotic stress (2), oxidative stress (3), and heat stress (4). To adapt to these environmental challenges, microorganisms have evolved specific mechanisms to survive and produce chemicals, such as those controlled by the TOR signal pathway (5, 6), the Snf1 pathway (7), and the high-osmolarity glycerol (HOG) pathway (8, 9). In particular, the HOG pathway plays an important role in resisting osmotic stress through transiently and acutely regulating the transcription process in cells (10).
As the central regulator of the HOG pathway, the protein kinase Hog1 mainly regulates three processes: (i) gene expression (11), (ii) cell cycle (12), and (iii) signal transduction (13). First, Hog1 regulates gene expression via two main mechanisms: the first involves direct interaction with gene promoters or RNA polymerase II, and the second involves indirect regulation through interaction with transcription factors. With regard to the first mechanism, previous studies have shown that recruitment of Hog1 to promoters is essential to coordinate chromatin remodeling and assembly of the transcriptional machinery under stress conditions (14). Moreover, Hog1 can physically interact with RNA polymerase II and cross-link to transcribed regions, allowing for efficient transcriptional elongation (15). With regard to the second mechanism, Hog1 has been shown to interact with and directly phosphorylate transcription factors, which alters their activity and enables them to control osmostress-responsive genes (14, 16). Second, Hog1 regulates the cell cycle in response to osmotic stress. In the G1/S transition, Hog1 controls Sic1 degradation and downregulates the expression of G1- and S-phase cyclins (Cln1, Cln2, and Clb5) (17). In the G2/M phase, Hog1 regulates Swe1 accumulation and consequently reduces Clb2-Cdc28 activity, thereby leading to G2-phase arrest (18). Third, Hog1 is involved in signal transduction. The core of this pathway comprises mitogen-activated protein kinase (MAPK) kinase (Ssk2, Ssk22, and Ste11), which phosphorylates the unique MAPK kinase Pbs2 that, in turn, phosphorylates and activates the MAPK Hog1 (19). This cascade of phosphorylation ensures signal fidelity in response to osmotic stress.
The cell membrane is an essential barrier that protects cells from environmental stress (20–22). Previous studies have indicated that some stress response pathways are involved in regulating membrane composition (23, 24). For example, the TOR signaling pathway operates through two protein complexes, TORC1 and TORC2, both of which are involved in regulating plasma membrane homeostasis (25). TORC1 is required for the expression of Pma1 protein and the full activity of the proton pump to resist pH stress (5). TORC2 phosphorylates and stimulates the downstream protein kinase Ypk1/Ypk2 to regulate membrane lipids and proteins (26, 27). In addition, the Snf1 pathway is involved in regulation of cell membrane biosynthesis, given that deletion of the subunits of the SNF1 complex resulted in hypersensitivity to cell membrane stress (28, 29). Moreover, the Snf1 pathway has been proposed to have an additional function of coordinating membrane transport proteins, which is an important strategy for achieving the fast rearrangement of the plasma membrane to resist stress, apart from coordinating transcriptional gene expression (30, 31). Although several studies have demonstrated a link between the HOG pathway and lipid metabolism, we here report a new mechanism by which Candida glabrata Hog1 (CgHog1), an ortholog of Hog1, phosphorylates a zinc cluster transcription factor, CgRds2, to alter the glycerophospholipid composition in response to osmotic stress. In Saccharomyces cerevisiae, the absence of Rds2 increased sensitivity to the antifungal drug ketoconazole and the cell wall-perturbing agent calcofluor white (32). Deletion of YPL133C, the open reading frame of RDS2, resulted in impaired growth on a nonfermentable carbon source through regulation of the expression levels of gluconeogenesis genes. (33). Although these studies showed the involvement of Rds2 in the stress response, the exact role and regulation mechanism of Rds2 have not been identified. Therefore, in this study, we also reported the important role of CgRds2 in resisting osmotic stress and revealed the underlying mechanism in Candida glabrata.
RESULTS
CgRds2 is essential for cell growth under osmotic stress.
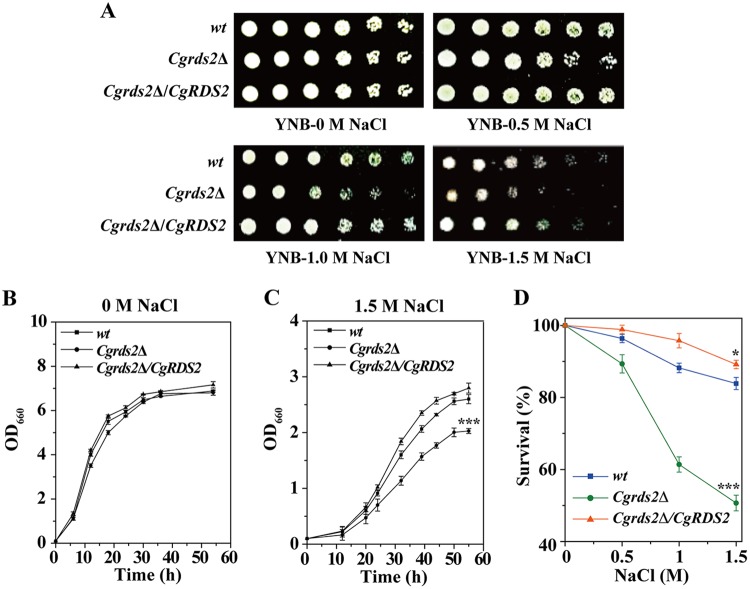
To identify novel genes required for cell growth under osmotic stress in C. glabrata, we performed genetic screening by deleting 15 genes that were reported to play potential roles in response to stress conditions (see Table S1 in the supplemental material). Then, 15 gene deletion mutants were spotted on yeast nitrogen base (YNB) medium with or without 1.5 M NaCl to observe their sensitivity to osmotic stress (Fig. S1). Deletion of CgRDS2 caused the most significant growth defect at 1.5 M NaCl, whereas overexpression of CgRDS2 enhanced growth compared to that of the wild-type strain (Fig. 1A). Similar results were obtained when the growth experiment was performed using a liquid medium. At 0 M NaCl, the final biomass of the Cgrds2Δ and Cgrds2Δ/CgRDS2 strains was similar to that of the wild-type strain (Fig. 1B), whereas at 1.5 M NaCl, the final biomass of the Cgrds2Δ strain was 23.4% lower than that of the wild-type strain but increased by 10.2% in the Cgrds2Δ/CgRDS2 strain (Fig. 1C). In addition, cell survival for all three strains was determined over broad concentration ranges of NaCl (Fig. 1D). At 1.5 M NaCl, 83.9% of the wild-type cells survived, while the Cgrds2Δ and Cgrds2Δ/CgRDS2 strains exhibited 50.7% and 89.2% cell survival, representing a 39.6% decrease and 6.3% increase compared to levels in wild-type cells, respectively (Fig. 1D). These results suggest that CgRds2 plays an important role in the growth of C. glabrata at 1.5 M NaCl.
FIG 1.
CgRds2 is essential for cell growth at 1.5 M NaCl. (A) Growth profiles of the wild-type (wt), Cgrds2Δ, and Cgrds2Δ/CgRDS2 strains grown on YNB medium at different concentrations of NaCl. (B and C) Growth curves of the wild-type (wt), Cgrds2Δ, and Cgrds2Δ/CgRDS2 strains at 0 M NaCl and 1.5 M NaCl. (D) Cell survival of all three strains at different concentrations of NaCl. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. *, P < 0.05; ***, P < 0.001.
CgHog1 interacts with CgRds2 under osmotic stress.
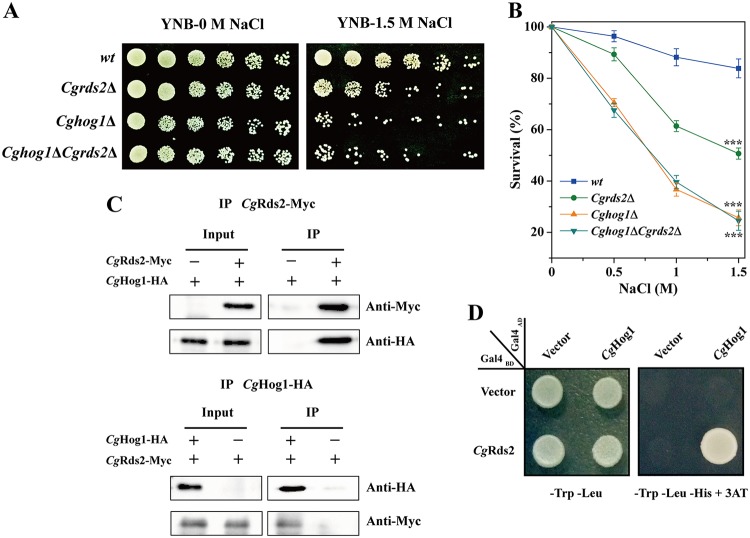
Hog1, a protein kinase of the HOG pathway, is known to be essential for cell survival by rapidly activating a number of downstream genes under osmotic stress (2, 14). Thus, we wondered whether there was a relationship between CgHog1 and CgRds2 at 1.5 M NaCl. To address this possibility, the genetic interaction was evaluated using a spot assay, revealing similar phenotypes among all strains at 0 M NaCl. In contrast, at 1.5 M NaCl, the Cghog1Δ strain was much more sensitive than the Cgrds2Δ strain, and the phenotype of the Cghog1Δ Cgrds2Δ double mutant was similar to that of the single mutant Cghog1Δ strain (Fig. 2A). Moreover, the Cghog1Δ, Cgrds2Δ, and Cghog1Δ Cgrds2Δ strains displayed 25.7%, 50.7%, and 24.5% cell survival at 1.5 M NaCl, representing 69.4%, 39.6%, and 70.8% reductions compared to the level of the wild-type strain (83.9%), respectively (Fig. 2B). These results suggest that the genes CgHOG1 and CgRDS2 are epistatic.
FIG 2.
CgHog1 interacts with CgRds2 at 1.5 M NaCl. (A) Growth profiles of the wild-type (wt), Cgrds2Δ, Cghog1Δ, and Cghog1Δ Cgrds2Δ strains grown on YNB medium with or without 1.5 M NaCl. (B) Cell survival of all four strains at different concentrations of NaCl. (C) Coimmunoprecipitation assay to detect the interaction between CgHog1 and CgRds2 in vivo. (D) Yeast two-hybrid assay to confirm the interaction between CgHog1 and CgRds2. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. ***, P < 0.001.
Furthermore, Hog1 makes contact with downstream proteins via complex formation and controls their expression (14, 34). Thus, the possibility that CgHog1 and CgRds2 physically interact was addressed by testing whether CgHog1 was immunoprecipitated with CgRds2 using cell extracts expressing Myc-tagged CgRds2 and hemagglutinin (HA)-tagged CgHog1 at 1.5 M NaCl. The results showed that CgHog1-HA did coprecipitate with CgRds2-Myc and vice versa (Fig. 2C). This interaction was also confirmed by a two-hybrid assay (Fig. 2D). These observations suggest that CgHog1 directly interacts with CgRds2 physically.
CgHog1 phosphorylates CgRds2 under osmotic stress.
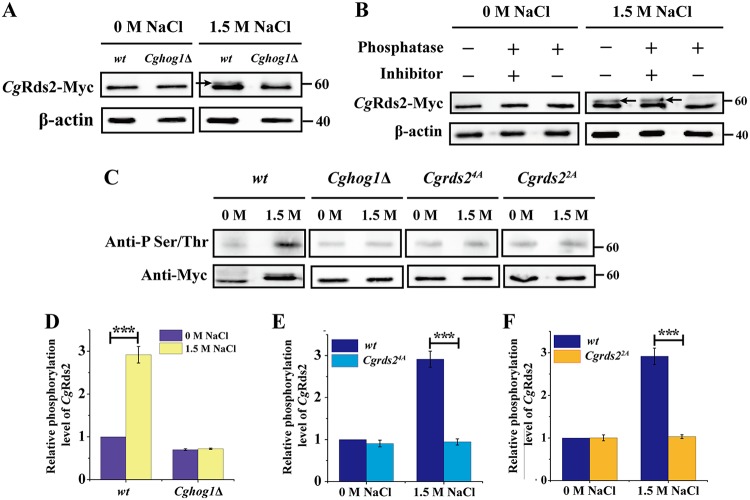
We next evaluated whether CgHog1 phosphorylated CgRds2 at 1.5 M NaCl. Cell extracts expressing Myc-tagged CgRds2 were prepared from the wild-type and Cghog1Δ strains and probed with a specific monoclonal antibody against the Myc epitope. As shown in Fig. 3A, CgRds2 was detected at the expected molecular mass (56 kDa) in the wild-type strain under both conditions, and there was a gel mobility shift of CgRds2 in the wild-type strain under 1.5 M NaCl. This shift of CgRds2 was further confirmed since it disappeared following treatment with alkaline phosphatase, and this effect was abolished with addition of the alkaline phosphatase inhibitor (Fig. 3B). These results suggest that the shift band of CgRds2 observed in the wild-type strain at 1.5 M NaCl was the result of phosphorylation. Furthermore, the shifted band was not observed in the Cghog1Δ strain (Fig. 3A), demonstrating that the phosphorylation of CgRds2 detected under 1.5 M NaCl was mostly dependent on CgHog1. To further verify these results, the phosphorylation level of CgRds2 by CgHog1 was detected by an anti-phosphoserine threonine antibody, which was used to detect the phosphorylated Ser-Pro/Thr-Pro (SP/TP) sites (Hog1 consensus phosphorylation sites), in the wild-type and Cghog1Δ strains. A weak phosphorylation band of CgRds2 was detected in both strains at 0 M NaCl, whereas at 1.5 M NaCl, the phosphorylation level of CgRds2 increased significantly (2.9-fold) in the wild-type strain but not in the Cghog1Δ strain (Fig. 3C and D). Together, these results demonstrate that CgHog1 phosphorylates CgRds2 at 1.5 M NaCl.
FIG 3.
CgHog1 phosphorylates CgRds2 at 1.5 M NaCl. (A) Immunoprecipitation of CgRds2-Myc was performed in the wild-type (wt) and Cghog1Δ strains at 0 M NaCl and 1.5 M NaCl, followed by Western blotting using anti-Myc antibody. The arrow indicates the phosphorylation band of CgRds2. (B) Extracts prepared from CgRds2-Myc-expressing wild-type cells, grown at 0 M NaCl and 1.5 M NaCl, were treated with alkaline phosphatase and phosphatase inhibitor as indicated. The arrows indicate the phosphorylation band of CgRds2. (C) Immunoprecipitation of phosphorylated CgRds2 was performed in the wild-type (wt), Cghog1Δ, Cgrds24A, and Cgrds22A strains at 0 M NaCl and 1.5 M NaCl, followed by Western blot analysis using anti-phosphoserine/threonine antibody. Quantification of relative phosphorylation levels of CgRds2 in the wild-type (wt), Cghog1Δ (D), Cgrds24A (E), and Cgrds22A (F) strains at 0 M NaCl and 1.5 M NaCl. β-Actin was used as a loading control. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. ***, P < 0.001.
To further identify the phosphorylation site(s) for CgHog1 in CgRds2, we investigated four sequences corresponding to the consensus phosphorylation sites for CgHog1 (Ser-Pro or Thr-Pro) (2, 17, 35) in the CgRds2 protein (Ser64, Thr97, Thr238, and Ser455) (Fig. S2). First, a mutant Cgrds24A strain was constructed, in which Cgrds2 encodes a mutated version of the CgRds2 protein in which all four putative Ser and Thr residues were replaced with Ala residues. The phosphorylation level of CgRds2 by CgHog1 in the Cgrds24A strain showed a significant decrease (69%) compared with that of the wild-type strain at 1.5 M NaCl although a slight decrease (9.3%) was also observed in the Cgrds24A strain compared with the level of the wild-type strain at 0 M NaCl (Fig. 3C and 3E). Next, to determine which site is critical for the phosphorylation of CgRds2 by CgHog1, four single-point mutants and all possible double-combination mutants were constructed. The results showed that single mutants of Ser64, Thr97, Thr238, and Ser455 did not alter the phosphorylation of CgRds2 by CgHog1 (data not shown). However, among the double-combination mutants, replacement of both Ser64 and Thr97 with alanines (here called Cgrds22A strain) mostly decreased (65%) the phosphorylation level of CgRds2 by CgHog1 compared with the level of the wild-type strain at 1.5 M NaCl, and there was no difference between the wild-type and Cgrds22A strains at 0 M NaCl (Fig. 3C and F). Given that the phosphorylation level of CgRds2 by CgHog1 in the Cgrds22A strain was similar to that of the Cgrds24A strain, we can conclude that the phosphorylation sites Ser64 and Thr97 are critical for the phosphorylation of CgRds2 by CgHog1 at 1.5 M NaCl.
Deletion of CgRDS2 decreased the expression level of glycerophospholipid genes under osmotic stress.
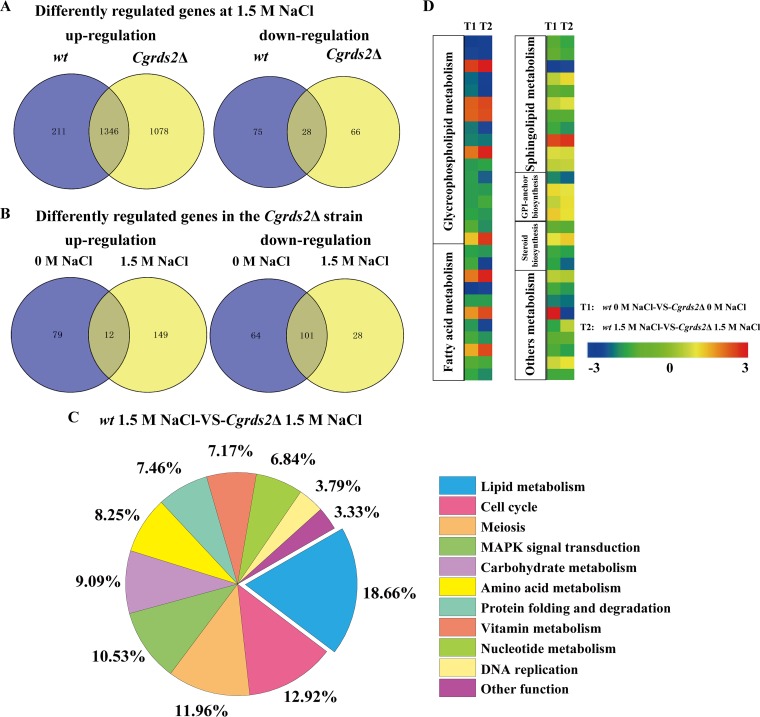
To elucidate the physiological role of CgRds2 in C. glabrata, transcriptome sequencing (RNA-seq) analysis was performed to compare gene expression levels in the wild-type and Cgrds2Δ strains. We first compared the gene expression levels of wild-type cells with or without 1.5 M NaCl treatment. Transcriptional profiling analysis revealed 1,660 genes whose expression was significantly modified (≥2.0-fold change; P ≤ 0.5): 1,557 were upregulated and 103 were downregulated. In the Cgrds2Δ strain, the expression levels of 2,518 genes displayed significant changes at 1.5 M NaCl compared with levels at 0 M NaCl. Of these, 2,424 were upregulated and 94 were downregulated. Specifically, a subset of 1,346 upregulated and 28 downregulated genes were common to both the wild-type and Cgrds2Δ strains (Fig. 4A). Gene Ontology (GO) analysis of commonly upregulated genes included processes such as oxidative phosphorylation (GO:0006119), vesicle-mediated transport (GO:0016192), protein folding (GO:0006457), and tRNA transcription (GO:0009304), whereas the commonly downregulated genes were enriched in the meiosis I (GO:0007127), ribosome biogenesis (GO:0042254), and arginine metabolic process (GO:0006525) (see Data Sets S1 and S2 in the supplemental material).
FIG 4.
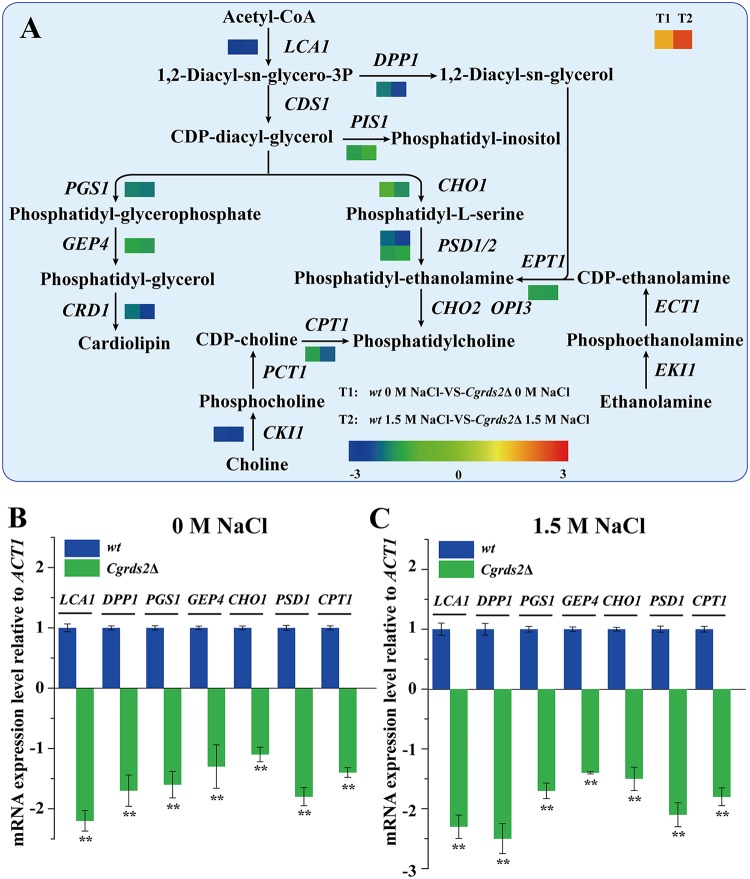
CgRds2 involves in regulating glycerophospholipid metabolism. (A) Venn diagram depicting the overlap of upregulated and downregulated genes between wild-type (wt) and Cgrds2Δ strains at 1.5 M NaCl compared with 0 M NaCl. (B) Venn diagram depicting the overlap of upregulated and downregulated genes between 0 M NaCl and 1.5 M NaCl in the Cgrds2Δ strain compared with the wild-type (wt) strain. (C) Statistical analysis of the metabolic pathways in which the differentially expressed genes were significantly enriched in the Cgrds2Δ strain compared with levels in the wild-type (wt) strain at 1.5 M NaCl. (D) Heat maps of differentially expressed genes involved in the lipid metabolism.
Transcriptional profiling also revealed 91 and 165 genes that were upregulated and downregulated, respectively, in the Cgrds2Δ strain compared with levels in the wild-type strain at 0 M NaCl (Fig. 4B). The set of upregulated genes was enriched in processes such as unsaturated fatty acid biosynthetic process (GO:0006636), protein processing (GO:0016485), ribosome biogenesis (GO:0042254), and rRNA processing (GO:0006364), whereas the set of downregulated genes was enriched in meiotic cell cycle (GO:0051321), membrane protein proteolysis (GO:0033619), and pyrimidine nucleoside metabolic process (GO:0006213). Furthermore, at 1.5 M NaCl, there were 161 upregulated and 129 downregulated genes in the Cgrds2Δ strain compared with levels in the wild-type strain (Fig. 4B). GO analysis indicated that the set of upregulated genes was enriched in glycolysis (GO:0006096), ribosome biogenesis (GO:0042254), and rRNA transcription (GO:0009303), whereas the set of downregulated genes was enriched in cell cycle (GO:0007049), MAPK signal transduction (GO:0007165), lipid metabolic process (GO:0006629), DNA replication (GO:0006260), and amino acid metabolic processes (e.g., arginine [GO:0006525], methionine [GO:0006555], and ornithine [GO:0006591]) (Data Sets S3 and S4).
On the basis of these results, we concluded that lipid metabolism was the most notable differentially regulated pathway, accounting for 18.66% of all affected genes in the Cgrds2Δ strain compared with levels in the wild-type strain at 1.5 M NaCl (Fig. 4C and Data Set S5). Thus, we next focused on comparing the expression levels of genes related to lipid metabolism between the wild-type and Cgrds2Δ strains at 1.5 M NaCl. As shown in Fig. 4D, there was significant enrichment in genes involved in glycerophospholipid metabolism process (GO:0006650), fatty acid metabolic process (GO:0006631), and sphingolipid metabolic process (GO:0006665). Further analysis revealed that the most altered genes were enriched in glycerophospholipid metabolism (Fig. 5A; Table 1). The mRNA levels of genes involved in glycerophospholipid metabolism were verified by quantitative reverse transcription-PCR (qRT-PCR) analysis. At 0 M NaCl, the mRNA levels of lysophospholipid acyltransferase (LCA1), phosphatidate phosphatase (DPP1), CDP-diacylglycerol-phosphatidyltransferase (PGS1), phosphatidylglycerophosphatase (GEP4), CDP-diacylglycerol-serine O-phosphatidyltransferase (CHO1), phosphatidylserine decarboxylase (PSD1), and diacylglycerol cholinephosphotransferase (CPT1) were 2.1-, 1.7-, 1.6-, 1.3-, 1.0-, 1.8-, and 1.4-fold lower in the Cgrds2Δ strain than those in the wild-type strain, respectively (Fig. 5B), whereas at 1.5 M NaCl, the mRNA levels of these genes were 2.3-, 2.5-, 1.7-, 1.5-, 1.5-, 2.1-, and 1.8-fold lower in the Cgrds2Δ strain than those in the wild-type strain (Fig. 5C). These data indicated that deletion of CgRDS2 strongly downregulated the transcription of glycerophospholipid genes.
FIG 5.
CgRds2 involves in regulating glycerophospholipid metabolism. (A) Measured changes of the expression of genes involved in glycerophospholipid metabolism in the Cgrds2Δ strain compared with levels in the wild-type (wt) strain at 0 M NaCl (left-hand squares, represented by T1) and 1.5 M NaCl (right-hand squares, represented by T2). (B and C) qRT-PCR verified the mRNA expression levels of the glycerophospholipid genes, calculated relative to the ACT1 level, at 0 M NaCl and 1.5 M NaCl. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. **, P < 0.01.
TABLE 1.
Differentially expressed genes associated with glycerophospholipid metabolism
| Gene name | S. cerevisiae homolog | Gene function | Log2 fold change in expression (Cgrds2Δ strain vs wild type) with: |
|
|---|---|---|---|---|
| 0 M NaCl | 1.5 M NaCl | |||
| CAGL0L04642g | LCA1 | Lysophospholipid acyltransferase | −2.177 | −2.336 |
| CAGL0M04367g | CKI1 | Choline kinase | −2.018 | −2.108 |
| CAGL0D04972g | TAZ1 | Monolysocardiolipin acyltransferase | 1.939 | 2.282 |
| CAGL0J06226g | PSD1 | Phosphatidylserine decarboxylase | −1.766 | −2.094 |
| CAGL0I03784g | CRD1 | Cardiolipin synthase | −1.712 | −2.202 |
| CAGL0G08690g | GBG1 | 1-Acylglycerone-phosphate reductase | 1.67 | 1.839 |
| CAGL0J11748g | PLB2 | Lysophospholipase | 1.663 | 1.794 |
| CAGL0H01177g | DPP1 | Phosphatidate phosphatase | −1.662 | −2.469 |
| CAGL0B04741g | PGS1 | CDP-diacylglycerol–glycerol-3-phosphate 3-phosphatidyltransferase | −1.608 | −1.678 |
| CAGL0J07040g | GDE1 | Glycerophosphodiester phosphodiesterase | 1.526 | 2.13 |
| CAGL0I08745g | PSD2 | Phosphatidylserine decarboxylase | −1.411 | −1.308 |
| CAGL0K09570g | CPT1 | Diacylglycerol cholinephosphotransferase | −1.4 | −1.828 |
| CAGL0L13068g | EPT1 | Ethanolaminephosphotransferase | −1.394 | −1.421 |
| CAGL0G03157g | PIS1 | CDP-diacylglycerol–inositol 3-phosphatidyltransferase | −1.387 | −1.136 |
| CAGL0H04389g | GEP4 | Phosphatidylglycerophosphatase | −1.31 | −1.434 |
| CAGL0C03069g | CHO1 | CDP-diacylglycerol–serine O-phosphatidyltransferase | −1.026 | −1.519 |
| CAGL0H06699g | GUT2 | Glycerol-3-phosphate dehydrogenase | 1.022 | 1.963 |
Phosphorylation of CgRds2 by CgHog1 is required for regulating glycerophospholipid genes.
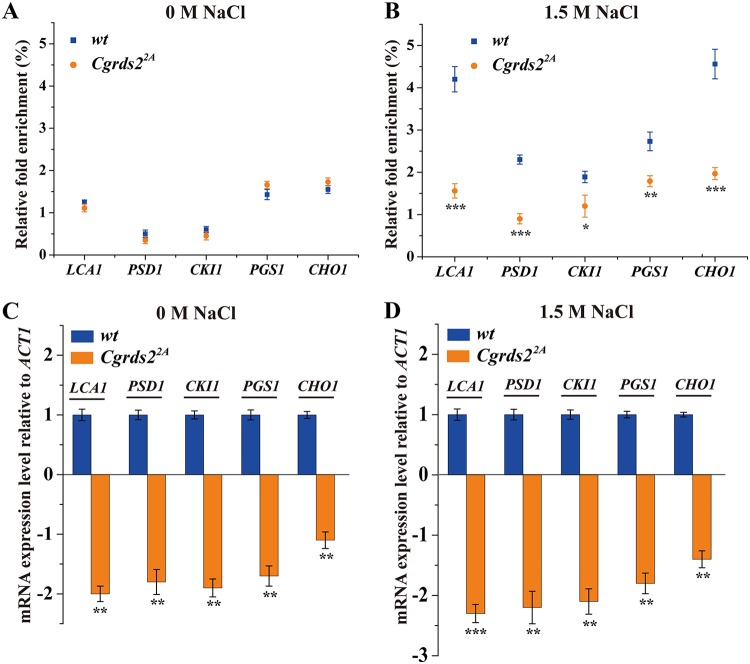
To determine whether the CgRds2 function is controlled by CgHog1, we assessed whether the phosphorylation of CgRds2 is required for its function in regulating genes involved in glycerophospholipid metabolism. First, a chromatin immunoprecipitation (ChIP) assay combined with qRT-PCR was performed to detect the binding of CgRds2 to glycerophospholipid genes in the wild-type and Cgrds22A strains, in which the phosphorylation state between CgRds2 and CgHog1 was abolished. The qRT-PCR analysis revealed that at 0 M NaCl, the binding levels of CgRds2 to the promoter regions of LCA1, PSD1, CKI1, PGS1, and CHO1 were not significantly different between the wild-type and Cgrds22A strains (Fig. 6A), whereas the binding levels were 62.0%, 60.9%, 36.5%, 34.4%, and 56.8% lower in the Cgrds22A strain than those of the wild-type strain at 1.5 M NaCl (Fig. 6B). These data demonstrate that the phosphorylation of CgRds2 by CgHog1 promotes the binding of CgRds2 to target promoters under osmotic stress. Subsequently, we investigated the transcription levels of LCA1, PSD1, CKI1, PGS1, and CHO1 in the wild-type and Cgrds22A strains, which were 2.0-, 1.8-, 1.9-, 1.7-, and 1.1-fold lower in the Cgrds22A strain than those of wild-type strain at 0 M NaCl (Fig. 6C) and were 2.3-, 2.2-, 2.1-, 1.8-, and 1.4-fold lower in the Cgrds22A strain than those of the wild-type at 1.5 M NaCl, respectively (Fig. 6D). These results suggest that phosphorylation of CgRds2 by CgHog1 is indeed important for enhancing the transcription levels of glycerophospholipid genes.
FIG 6.
Phosphorylation of CgRds2 by CgHog1 is required for regulating glycerophospholipid genes. (A and B) Association of CgRds2 with the core promoter of glycerophospholipid metabolism genes was determined by ChIP analysis combined with and qRT-PCR to measure occupancy in the wild-type (wt) and Cgrds22A strains at 0 M and 1.5 M NaCl. Relative fold enrichment was calculated by the formula provided in Materials and Methods. (C and D) Transcript levels of genes involved in glycerophospholipid metabolism were analyzed with RNA prepared from the wild-type (wt) and Cgrds22A strains at 0 M NaCl and 1.5 M NaCl. The data were normalized to the expression level of the ACT1 gene. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
CgRds2 regulates the glycerophospholipid composition in a CgHog1-mediated manner.
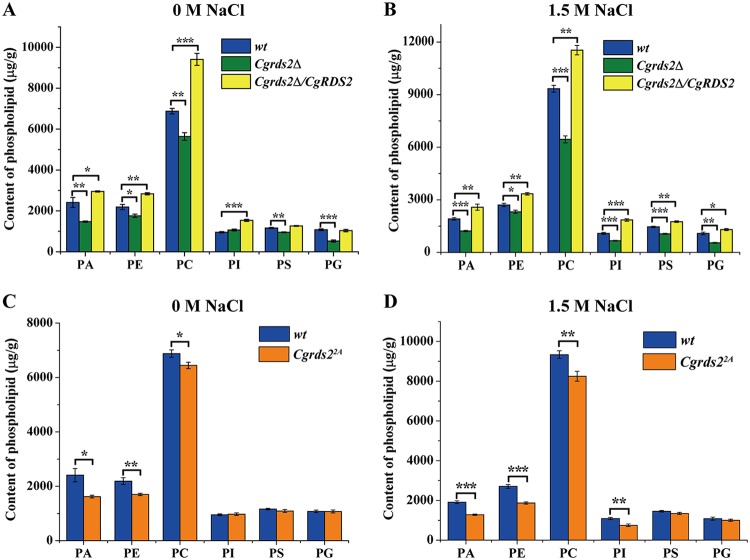
On the basis of the above results, we investigated the glycerophospholipid composition in the wild-type, Cgrds2Δ, and Cgrds2Δ/CgRDS2 strains, using mass spectrometry of methanol-chloroform-extracted cells. At 0 M NaCl, the contents of phosphatidic acid (PA), phosphatidylethanolamine (PE), phosphocholine (PC), phosphatidylserine (PS), and phosphatidylglycerol (PG) decreased by 38.9%, 19.6%, 18.0%, 17.9%, and 51.2% in the Cgrds2Δ strain compared with those of the wild-type strain, respectively, while the level of phosphatidylinositol (PI) was unchanged. In the Cgrds2Δ/CgRDS2 strain, the PA, PE, PC, and PI contents increased by 22.4%, 29.6%, 36.8%, and 60.8%, respectively, whereas the PS and PG contents were unchanged (Fig. 7A; Table S2). At 1.5 M NaCl, the PA, PE, PC, PI, PS, and PG contents in the Cgrds2Δ strain decreased by 36.1%, 14.4%, 31.0%, 38.9%, 27.2% and, 49.2%, respectively. In the Cgrds2Δ/CgRDS2 strain, the contents of PA, PE, PC, PI, PS, and PG increased by 34.9%, 23.5%, 23.6%, 70.4%, 20.6%, and 20.4% (Fig. 7B; Table S2). Taken together, these data indicate that CgRds2 is a crucial regulator of the glycerophospholipid composition.
FIG 7.
CgRds2 regulates glycerophospholipid composition in a CgHog1-mediated manner. (A and B) Glycerophospholipid composition changes in the wild-type (wt), Cgrds2Δ, and Cgrds2Δ/CgRDS2 strains at 0 M NaCl and 1.5 M NaCl. (C and D) Glycerophospholipid composition changes in the wild-type (wt) and Cgrds22A strains at 0 M NaCl and 1.5 M NaCl. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
Furthermore, to investigate whether CgHog1-mediated phosphorylation is important for CgRds2 in regulating glycerophospholipid profiling, we also measured the glycerophospholipid composition levels in the Cgrds22A strain. At 0 M NaCl, PA, PE, and PC contents were 32.7%, 22.3%, and 10.3% lower than those in the wild-type strain, respectively, and the levels of PI, PS, and PG were unchanged (Fig. 7C; Table S2). At 1.5 M NaCl, the contents of PA, PE, PC, and PI were 32.9%, 30.9%, 11.6%, and 31.3% lower than those in the wild-type strain, while the PS and PG contents remained unchanged (Fig. 7D; Table S2). These results revealed that the role of CgRds2 in regulating glycerophospholipid composition may be partly controlled by CgHog1-mediated phosphorylation.
CgRds2 alters membrane integrity in a CgHog1-mediated manner.
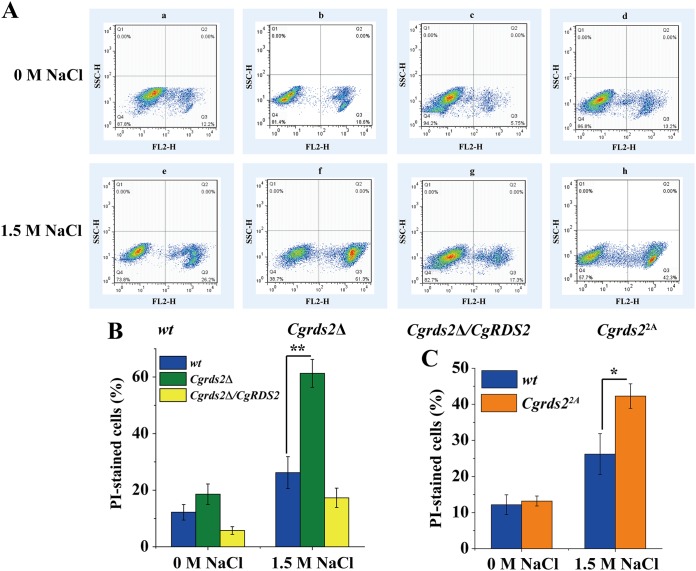
To investigate the role of CgRds2 in membrane integrity, cells of the wild-type, Cgrds2Δ, and Cgrds2Δ/CgRDS2 strains were treated to 1.5 M NaCl for 4 h and then subjected to propidium iodide uptake analysis. There was no significant difference in the percentages of propidium iodide-stained cells among the three strains at 0 M NaCl, whereas at 1.5 M NaCl, the proportion of stained cells increased significantly, by 1.3-fold, among Cgrds2Δ cells but decreased by 33.9% among Cgrds2Δ/CgRDS2 cells compared with levels of the wild-type strain (Fig. 8A and B). These results suggest that CgRds2 contributes to increasing membrane integrity under osmotic stress. Furthermore, to explore whether CgHog1-mediated phosphorylation is essential for CgRds2 function in regulating membrane integrity, the same experiments were performed in the Cgrds22A strain. At 0 M NaCl, the percentage of propidium iodide-stained cells in the Cgrds22A strain was similar to that of the wild-type strain, whereas the proportion of stained cells in the Cgrds22A strain was 61.2% higher than that of the wild-type under 1.5 M NaCl (Fig. 8A and C). These results suggest that CgHog1-mediated phosphorylation partially contributed to the effect of CgRds2 on increasing membrane integrity under osmotic stress.
FIG 8.
CgRds2 alters membrane integrity in a CgHog1-mediated manner. (A) Flow cytometry analysis of membrane integrity in the wild-type (wt), Cgrds2Δ, Cgrds2Δ CgRDS2, and Cgrds22A strains at 0 M NaCl and 1.5 M NaCl. Q3 region, the percentage of PI-stained cells, suggesting that cell membrane was destroyed by osmotic stress; Q4 region, the percentage of PI-unstained cells, suggesting that cell membrane was intact in osmotic stress. (B and C) Quantification of membrane integrity in the wild-type (wt), Cgrds2Δ, Cgrds2Δ/CgRDS2, and Cgrds22A strains at 0 M NaCl and 1.5 M NaCl. All data are presented as mean values of three independent experiments. Error bars indicate the standard deviations. *, P < 0.05; **, P < 0.01.
To observe the damage of cell membrane visually, transmission electron microscopy (TEM) analysis was performed in all strains (wild-type, Cgrds2Δ, Cgrds2Δ/CgRDS2, and Cgrds22A strains) with or without 1.5 M NaCl. We observed 60 cells of each strain at random and found that at 0 M NaCl, cells of all strains exhibited a regular cell structure with undamaged cell membrane (Fig. S3a to d). However, when treated with 1.5 M NaCl for 4 h, 33.3% of the Cgrds2Δ cells exhibited damaged cell membranes characterized by rough surface, disappearance, and separation from the cell wall (Fig. S3h to j), compared to those of the wild-type strain (Fig. S3e to g), whereas there was no change in the Cgrds2Δ/CgRDS2 strain (Fig. S3k to m). Meanwhile, 21.6% of the Cgrds22A cells also showed similar phenotypes to those of the Cgrds2Δ strain (Fig. S3n to p).
DISCUSSION
In this study, we demonstrated that under 1.5 M NaCl, CgHog1 interacts with and phosphorylates CgRds2 in C. glabrata. Subsequently, the phosphorylated CgRds2 induces the expression of genes involved in glycerophospholipid metabolism to consequently increase the content of glycerophospholipid composition. Additionally, membrane integrity was also increased. Thus, CgHog1-mediated CgRds2 phosphorylation alters glycerophospholipid composition and membrane integrity to resist osmotic stress in C. glabrata.
Hog1 is essential for cell survival through rapidly phosphorylating downstream targets under osmotic stress. In Saccharomyces cerevisiae, many transcription factors have been reported to be directly phosphorylated by Hog1, such as Sko1 (36), Hot1 (37), Smp1 (34), and Rtg1/Rtg3 (38). Recent studies have also identified additional proteins phosphorylated by Hog1 to coordinate the metabolic pathway under stress conditions. For example, yeast Cip1 was shown to be phosphorylated by Hog1 to inhibit Cdk1-G1 cyclins, resulting in cell cycle delay (2). The Hog1-mediated phosphorylation of Mrc1 protein was found to protect genomic integrity through preventing conflicts between DNA replication and transcription (35). In mammals, p38, the homolog of Hog1, has highly conserved functions in response to stress through phosphorylating target proteins involved in the cell cycle (39), transcription process (40), mitochondrion metabolism (41), and signal transduction (42). Through this study, we provide further evidence that in C. glabrata, CgHog1 interacts with and phosphorylates CgRds2, a zinc cluster transcription factor, to respond to osmotic stress. This may be achieved by two mechanisms. First, Hog1 phosphorylates target transcription factors to promote their binding to downstream genes (34). Second, Hog1 phosphorylates and activates target transcription factors, and these active transcription factors recruit Hog1 to the downstream genes (37). Our results indicate that CgHog1 phosphorylates CgRds2 to resist stress through the first mechanism, given that CgHog1-mediated phosphorylation was essential for CgRds2 binding to downstream genes.
Transcriptome analysis showed that deletion of CgRDS2 significantly changes the global transcription level of genes at 1.5 M NaCl. These genes are involved in multiple pathways, three of which deserve special attention. First, CgRds2 induced glycerophospholipid gene expression and increased the content of glycerophospholipid composition under treatment of 1.5 M NaCl. A previous genome-wide location analysis of S. cerevisiae demonstrated that Rds2 could bind to several genes related to fatty acid metabolism (43). However, to our knowledge, no report has identified that C. glabrata Rds2 is involved in regulating lipid metabolism; thus, the present study provides new insight into the physiological function of CgRds2. Glycerophospholipid is the main lipid of the membrane bilayer and plays an important role in response to environmental stress (44, 45). For example, cells lacking PLB1, which is critical for the remodeling of membrane phospholipids, showed sensitivity to osmotic stress (46). Moreover, overexpression of OLE1, encoding delta-9 desaturase to generate unsaturated fatty acids in Saccharomyces cerevisiae, activated the MAPK HOG pathway, thereby enhancing the tolerance to osmotic stress (8). Second, CgRds2 was found to regulate genes involved in carbohydrate metabolism. Several previous studies demonstrated that cells lacking RDS2 had impaired growth on a nonfermentable carbon source (ethanol or glycerol) through downregulating the expression levels of gluconeogenesis genes (PCK1, FBP1, PFK27, VID24, and GID8) (33). Soontorngun et al. showed that Snf1-mediated Rds2 phosphorylation was required for the binding of Rds2 to the FBP1 promoter after a shift from glucose to nonfermentable sources in S. cerevisiae (33). Consistently, we found that CgHog1-mediated phosphorylation was important for the CgRds2 function of regulating glycerophospholipid genes. Thus, there may be two sets of phosphorylation sites in the CgRds2 protein: some targeted by CgHog1 that lead to activation of lipid metabolism to resist osmotic stress and some targeted by CgSnf1 that alter carbohydrate metabolism in response to a glucose starvation condition. It will be worth examining the phosphorylation sites in the CgRds2 protein that are specific for CgSnf1 andidentifying the association between CgHog1 and CgSnf1 pathways in regulating the expression of CgRds2 under stress conditions. Third, deletion of CgRDS2 was found to repress genes linked to the typical osmotic stress response at 1.5 M NaCl (see Table S3 in the supplemental material). Many previous studies indicated that S. cerevisiae cells respond to osmotic stress through three main pathways: glycerol metabolism, trehalose metabolism, and ion transport (47). In our study, certain genes involved in these pathways which were partly controlled by the transcription factor CgRds2 were also changed under osmotic stress in C. glabrata. For example, the expression levels of genes encoding Na+-exporting P-type ATPase (ENA1), glycerol-3-phosphate dehydrogenase (GPD1), and alpha, alpha-trehalose-phosphate synthase (TPS1) were downregulated by 2.11-, 2.54-, and 2.27-fold, respectively, in the Cgrds2Δ strain compared to levels of the wild-type strain at 1.5 M NaCl. However, there was no significant change in the expression of FPS1, encoding aquaglyceroporins, which plays a critical role in mediating the export of accumulated glycerol in yeast. This is consistent with the fact that Fps1 closed upon hyperosmotic shock in the hog1Δ strain, indicating the existence of another mechanism to regulate the Fps1-mediated glycerol efflux (48).
Three groups of membrane lipids play an important role in influencing the physical properties of the membrane: (i) phospholipids, (ii) sphingolipids, and (iii) sterols, (49). First, phospholipids constitute the bulk of the membrane lipids. Both the head group and acyl chain could regulate the membrane properties (50, 51). PE has a conical head group, which could increase the negative membrane curvature (52). PC always contains an unsaturated acyl chain, which contributes to the increase of membrane fluidity (53). Despite their low abundance, PS and PI play important roles in balancing the membrane surface charge (54, 55). In this study, we demonstrated a role of CgRds2 in regulating glycerophospholipid composition and membrane integrity at 1.5 M NaCl in C. glabrata, further suggesting a correlation between phospholipid and physical properties of the membrane. In addition, we found that CgHog1-mediated phosphorylation was important for the CgRds2 function in regulating glycerophospholipid metabolism. This is consistent with the idea that some conserved protein kinases were involved in regulating membrane lipid organization. For example, the TORC2 protein kinase complex was shown to combine with Ypk1/Ypk2 kinases to regulate plasma membrane sphingolipid levels (56). Second, sphingolipid is a cylindrical phospholipid, containing a serine backbone combined with saturated or trans-unsaturated acyl chains, which increases the membrane density (57). A previous study also showed a correlation between sphingolipid content and membrane integrity (58). Third, sterols, abundant apolar membrane lipids, can increase membrane thickness and impermeability by reducing the flexibility of the surrounding acyl chains (59). In our previous study, we also showed that increasing the ergosterol content by overexpression of CgMED15 may enhance membrane fluidity, thereby promoting the resistance to low pH (60). Importantly, to manipulate the ratio of these membrane lipids, compensatory mechanisms exist in yeast. For example, deficiency of inositol, a major regulator of phospholipid biosynthesis in yeast, decreased the PI levels, which led to changes in the content of other membrane lipids derived from PI, including sphingolipids and glycosylphosphatidylinositol (GPI) anchors, thereby activating the HOG pathway (61).
C. glabrata is a well-established microorganism that is widely used for the industrial production of organic acids (62–64). However, accumulation of organic acids causes acidification of the fermentation broth, which ultimately inhibits cell growth and decreases metabolite production (65, 66). To maintain a suitable pH of the medium, some alkaline reagents must be added, which leads to osmotic stress (67). In this study, we screened the novel genes required for cell growth under osmotic stress in C. glabrata and revealed the important roles of transcription factor CgRds2 in resisting osmotic stress: (i) CgRds2 is a new target of CgHog1, (ii) CgRds2 is a critical regulator of glycerophospholipid metabolism, and (iii) CgHog1 phosphorylates CgRds2 to resist osmotic stress by regulating the glycerophospholipid composition and membrane integrity. Therefore, this study may provide a potential strategy for the resistance of osmotic stress in C. glabrata during organic acids fermentation.
MATERIALS AND METHODS
Strains, media, and culture conditions.
The yeast strains and plasmids used in this study are listed in Table 2. Escherichia coli JM109 was used for cloning and plasmid propagation. All yeast strains used in this study were derived from a Candida glabrata CgHTUΔ (his3Δ trp1Δ ura3Δ) strain. The deletion strains were constructed by homologous recombination of a CgHIS3 or CgTRP1 marker in the CgRDS2 or CgHOG1 locus. The marker genes were amplified from the C. glabrata strain ATCC 2001 genome and fused between the upstream and downstream regions of the CgRDS2 or CgHOG1 gene open reading frame by fusion PCR. PCR products were transformed in the Candida glabrata strain CgHTUΔ as described previously (60), and the deletion strains were confirmed by genomic PCR and DNA sequencing. Overexpression strains were constructed using the pY26 plasmid carrying the target genes. The plasmids were then transformed into the corresponding deletion mutants and confirmed by colony PCR.
TABLE 2.
Strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| C. glabrata strains | ||
| ATCC 2001 | 60 | |
| HTUΔ | his3Δ trp1Δ ura3Δ | 60 |
| Cgrds2Δ strain | his3Δ trp1Δ ura3Δ CgRDS2::CgHIS3 | This study |
| Cgrds2Δ/CgRDS2 strain | his3Δ trp1Δ ura3Δ rds2Δ CgRDS2::CgHIS3 pY26-PTEF/CgRDS2 | This study |
| Cghog1Δ strain | his3Δ trp1Δ ura3Δ CgHOG1::CgTRP1 | This study |
| Cghog1Δ Cgrds2Δ strain | his3Δ trp1Δ ura3Δ CgHOG1::CgTRP1 CgRDS2::CgHIS3 | This study |
| Cgrds2Δ/CgRds2-Myc strain | his3Δ trp1Δ ura3Δ CgRDS2::CgHIS3 pY26-PTEF/CgRds2-Myc | This study |
| Cgrds24A strain | his3Δ trp1Δ ura3Δ CgRDS2::CgRDS2-S64A T97A T238A S455A | This study |
| Cgrds22A strain | his3Δ trp1Δ ura3Δ CgRDS2::CgRDS2-S64A T97A | This study |
| Cgrds2Δ/CgRds22A-Myc strain | his3Δ trp1Δ ura3Δ CgRDS2::CgHIS3 pY26-PTEF/CgRDS2-S64A T97A-Myc | This study |
| Cghog1Δ Cgrds2Δ/CgRds2-Myc/CgHog1-HA strain | his3Δ trp1Δ ura3Δ CgHOG1::CgTRP1 CgRDS2::CgHIS3 pY26-PTEF/CgRds2-Myc pY26-PGPD/CgHog1-HA | This study |
| S. cerevisiae AH109 | trp1Δ leu2Δ ura3Δ his3Δ gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-LacZ MEL1 | Clontech |
| Plasmids | ||
| pY26 | 2 μm, Amp, URA3, PGPD, PTEF | Turbo |
| pGBKT7 | Kan, TRP1, GAL4 DNA-binding domain fusion | Clontech |
| pGADT7 | Amp, LEU2, GAL4 DNA-binding domain fusion | Clontech |
Site-directed mutagenesis was performed by PCR using mutagenic primers listed in Table 3 and plasmid pY26-CgRDS2 as the template. The DpnI-digested PCR product of 3 μl was used to transform 80 μl of Escherichia coli JM109 chemically competent cells, and colonies after transformation were incubated for DNA sequencing until all the designed mutants were obtained. Subsequently the plasmid of each mutant was extracted and named pY26-CgRDS24A or pY26-CgRDS22A. The CgRDS24A or CgRDS22A gene was amplified from the plasmid pY26-CgRDS24A or pY26-CgRDS22A, respectively, and integrated into the CgHTUΔ genome using homologous recombination as mentioned above.
TABLE 3.
Primers used in this study
| Primer function and name | Sequence (5′–3′) |
|---|---|
| Deletion | |
| L-CgRDS2-F | ATTCGAAGGCCCACTGTA |
| L-CgRDS2-R | ACCCTCTTAACAAACGCCATGTCAAAAATATGATGCTGTGCTTAG |
| CgHIS3(CgRDS2)-F | CACAGCATCATATTTTTGACATGGCGTTTGTTAAGAGGGT |
| CgHIS3(CgRDS2)-R | ACTTGTCTATGCATATGTGTCTATGCTAGGACACCCTTAGT |
| R-CgRDS2-F | CTAAGGGTGTCCTAGCATAGACACATATGCATAGACAAGTTATATACA |
| R-CgRDS2-R | CCACTATTAGTGGCCCTAAATAAGT |
| L-CgHOG1-F | CAGAGGTCTTCATTCTGATCACTCTC |
| L-CgHOG1-R | AGTAACGAATCAAATGACATTATTTCTTTCTGGTGTTCCTATTTAATTAA |
| CgTRP1(CgHOG1)-F | CACCAGAAAGAAATAATGTCATTTGATTCGTTACTCGACA |
| CgTRP1(CgHOG1)-R | AATTAAATTTTATATGTAAGTCATTGTTTCTTTGCATTTTGTACAT |
| R-CgHOG1-F | GCAAAGAAACAATGACTTACATATAAAATTTAATTATTCGTCTTCCT |
| R-CgHOG1-R | TGCATTAATGAGTTGCAATTATTACTAA |
| Overexpression | |
| CgRDS2-F | AAGGAAAAAAGCGGCCGCATGGAAGAACCAGCAGC |
| CgRDS2-R | GGAAGATCTTTAGTTGGAATGATCTCTTGTAGGA |
| CgRDS22A-F | AAGGAAAAAAGCGGCCGCATGGAAGAACCAGCAGC |
| CgRDS22A-R | GGAAGATCTTTAGTTGGAATGATCTCTTGTAGGA |
| CgRDS22A-Myc-F | AAGGAAAAAAGCGGCCGCATGGAAGAACCAGCAGC |
| CgRDS22A-Myc-R | GGAAGATCTTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCGTTGGAATGATCTCTTGTAG |
| Point mutation | |
| CgRds2/S64A-F | AATAGTATGGCGCCAATAGG |
| CgRds2/S64A-R | CCTATTGGCGCCATACTATT |
| CgRds2/T97A-F | GGTACAGAAGCACCAATATC |
| CgRds2/T97A-R | GATATTGGTGCTTCTGTACC |
| CgRds2/T238A-F | TATGACGATGCACCCACATC |
| CgRds2/T238A-R | GATGTGGGTGCATCGTCATA |
| CgRds2/S455A-F | ATCCCACTAGCTCCTACAAG |
| CgRds2/S455A-R | CTTGTAGGAGCTAGTGGGAT |
| Yeast two-hybrid | |
| BD-CgRDS2-F | GGGCCTCGAGCCCGGGTCGACATGGAAGAACCAGCAGCCAA |
| BD-CgRDS2-R | CTATAGGGCTCTAGAGTCGACTTAGTTGGAATGATCTCTTGTAGGAGA |
| AD-CgHOG1-F | AAAGAGATCGAATTAGGATCCATGGCTACTAATGAAGAGTTCATAAGAAC |
| AD-CgHOG1-R | TTCTCTGCTAGCAGAGGATCCTTATTGTTGGAATTCATTTGCAAAA |
| Coimmunoprecipitation | |
| pY26/PTEF-CgRDS2-F | AAGGAAAAAAGCGGCCGCATGGAAGAACCAGCAGC |
| pY26/PTEF-CgRDS2-R | GGAAGATCTTTACAGATCCTCTTCAGAGATGAGTTTCTGCTCGTTGGAATGATCTCTTGTAG |
| pY26/PGPD-CgHOG1-F | CAGGAATTCGATATCAAGCTTATGGCTACTAATGAAGAGTTCATAAGAAC |
| pY26/PGPD-CgHOG1-R | GTCGACGGTATCGATAAGCTTTTAAGCGTAGTCTGGGACGTCGTATGGGTATTGTTGGAATTCATTTGCAA |
| RT-PCR and ChIP | |
| CgLCA1-F | AGACCTGGCTATTATATGA |
| CgLCA1-R | CCGAATGACAACTTGATA |
| CgDPP1-F | GAGTTAATGATGTGATGTTATTC |
| CgDPP1-R | TCAGTCTTCCAATCCAAT |
| CgPGS1-F | CAAGTATAGCACCGAATC |
| CgPGS1-R | CAGTAGTTATCTTCTTCTTCA |
| CgGEP4-F | AGAGACGAGATTATTGAAT |
| CgGEP4-R | CTTACTGAAGGCACTATT |
| CgCHO1-F | GATTGTGAGTTGCCTGAG |
| CgCHO1-R | ACCTAAGAATATGAAGAAGTGT |
| CgPSD1-F | AAGATGAGGAGGCAACTG |
| CgPSD1-R | TTAGGTGAGACTTCGGTTAG |
| CgCPT1-F | CACAATAAGAAGAATGACA |
| CgCPT1-R | GTTAGATGACCAACAATC |
| CgCKI1-F | GAGCAGGAATCTACTAAGG |
| CgCKI1-R | TGTATCTATGTCATCTACCAAT |
E. coli JM109 cells were grown in LB medium (2% tryptone, 2% NaCl, 1% yeast extract) and incubated at 37°C with shaking at 200 rpm. Ampicillin (100 mg/liter) was added for the selection of cells carrying the relevant plasmid. Unless otherwise stated, yeast strains were grown in yeast nitrogen base (YNB) (0.67% yeast nitrogen base, 2% glucose) medium supplemented with essential nutrients and incubated at 30°C with shaking at 200 rpm.
Plasmids.
The CgRDS2 gene was amplified from C. glabrata genomic DNA using the primers listed in Table 3. The CgRDS2 gene was cloned into the vector pY26 using the NotI and BglII restriction sites, and the construct was named pY26-CgRDS2. Clones were confirmed by DNA sequencing.
The CgHOG1-HA and CgRDS2-Myc constructs were amplified from C. glabrata genomic DNA using the primers listed in Table 3. CgRDS2-Myc was cloned into the vector pY26 using the NotI and BglII restriction sites, and the construct was named pY26-CgRDS2-Myc. CgHOG1-HA was cloned into the plasmid pY26-CgRDS2-Myc using the HindIII restriction site and a ClonExpress II One Step Cloning kit (C112-01; Vazyme), and the construct was named pY26-CgRDS2-Myc-CgHOG1-HA. Similarly, clones were confirmed by DNA sequencing.
The CgHOG1 and CgRDS2 genes were amplified from C. glabrata genomic DNA using the primers listed in Table 3. The CgRDS2 gene was cloned into the vector pGBKT7 using the SalI restriction site and a ClonExpress II One Step Cloning kit (C112-01; Vazyme), and the construct was named pGBKT7-CgRDS2. The CgHOG1 gene was cloned into the vector pGADT7 using the BamHI restriction site and a ClonExpress II One Step Cloning kit (C112-01; Vazyme), and the construct was named pGADT7-CgHOG1.
The CgRDS22A-Myc construct was amplified from the plasmid pY26-CgRDS22A using the primers listed in Table 3. CgRDS22A-Myc was cloned into the vector pY26 using the NotI and BglII restriction sites, and the construct was named pY26- CgRDS22A-Myc. Clones were confirmed by DNA sequencing.
Spot assay.
Yeast cells were cultivated in logarithmic phase and diluted to an absorbance at 660 nm (A660) of 1.0 in phosphate-buffered saline (PBS). Aliquots (4 μl) of 10-fold serial dilutions were spotted onto YNB agar plates with or without the indicated concentration of NaCl. Growth was assessed after incubation for 2 to 4 days at 30°C.
Growth and survival analysis.
To analyze the growth of C. glabrata strains at 1.5 M NaCl, logarithmic-phase cells were diluted into fresh YNB medium at 0 M or 1.5 M NaCl at an initial A660 of 0.1. Cultures were taken at regular time intervals, and the A660 values were recorded. The A660 was calibrated against the dry weight of cells (DCW) on the basis of a standard curve where an A660 of 1 is equal to a DCW of 0.23 g/liter (60). To analyze cell survival, yeast cells were cultivated in logarithmic phase and then treated with various concentrations of NaCl for 1 h at 30°C with shaking at 200 rpm. Cells were then centrifuged and washed with sterile water three times. After dilution, cells were plated on YNB medium plates with the same number from each concentration of NaCl and incubated at 30°C for 2 to 4 days. Then the surviving colonies were counted. Data are presented as a percentage relative to that of untreated cells of the corresponding strain.
Coimmunoprecipitation.
Cells expressing CgRds2-Myc and CgHog1-HA were grown to logarithmic phase in YNB medium. Cells were then treated with 1.5 M NaCl for 30 min and harvested at 4°C. Pellets were resuspended in lysis buffer (45 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 2 mM dithiothreitol [DTT]) with protease and phosphatase inhibitors, followed by the addition of glass beads and sonication at 4°C. Protein extracts were clarified by centrifugation at 6,000 × g for 10 min at 4°C. Proteins were incubated with primary antibody (see below) at 4°C overnight and then incubated with protein A agarose beads (Sangon Biotech) at 4°C for 6 to 8 h. Beads were washed six times with lysis buffer and one time with PBS and then boiled in SDS loading buffer for 10 min. The binding proteins were resolved by 10% SDS-PAGE and detected by Western blotting.
Two-hybrid analysis.
Two-hybrid analysis was carried out by using pGADT7 (Gal4AD) as the activation domain (AD) plasmid and pGBKT7 (Gal4BD) as the DNA-binding domain (BD) plasmid. The plasmid pGADT7-CgHOG1 was cotransformed with pGBKT7-CgRDS2 using the AH109 reporter strain. Positive clones were selected and further tested as follows. The transformed yeast strains were grown until mid-log phase in YNB medium, diluted on synthetic dextrose (SD)-Leu-Trp plates and SD-Leu-Trp-His selective plates with the histidine biosynthesis inhibitor 1,2,4-aminotrizole (3-AT), and incubated for 2 to 4 days at 30°C.
Alkaline phosphatase treatment.
Samples used for the alkaline phosphatase treatment were processed as described for Western blotting, except that after the lysis buffer washes, PBS buffer was added to the immunoprecipitated proteins bound to the beads. Samples were treated with alkaline phosphatase (D7027; Beyotime) in the presence or absence of alkaline phosphatase inhibitor (P1081; Beyotime) and incubated for 1 h at 30°C with occasional shaking. The untreated samples were used as controls. The immunoprecipitated proteins were then washed twice with the wash buffer without protease inhibitors and released from the beads by boiling them in SDS loading buffer.
In vivo phosphorylation assay.
The CgRds2 and Cgrds22A genes were tagged with Myc. Cells were grown to logarithmic phase in YNB medium and then left untreated or treated with 1.5 M NaCl for 30 min, followed by harvesting at 4°C. Pellets were resuspended in lysis buffer (45 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 10% glycerol, 1% Triton X-100, 2 mM DTT) supplemented with protease and phosphatase inhibitors (P1060; Beyotime). Glass beads were added, and cells were lysed by sonication at 4°C. Protein extracts were clarified by centrifugation at 6,000 × g for 10 min at 4°C. Proteins were incubated overnight with an anti-Myc antibody (AM926, 1:1,000; Beyotime) and then incubated with protein A agarose beads (Sangon Biotech) at 4°C overnight. Beads were washed six times in lysis buffer and one time in PBS, boiled in SDS loading buffer for 10 min, and analyzed by 10% SDS-PAGE. The binding proteins were detected by Western blotting with an anti-phosphoserine threonine antibody (612548, 1:1,000; BD Biosciences)).
Western blotting.
Whole proteins were extracted and resolved by SDS-PAGE and transferred to a polyvinylidene difluoride (PVDF) membrane. The membrane was blocked in 5% nonfat milk at room temperature for 2 h, followed by incubation with primary antibody at 4°C overnight. Primary antibodies were used to detect HA (AH158, 1:1,000; Beyotime), Myc (AM926, 1:1,000; Beyotime), β-actin (AF0003, 1:1,000; Beyotime), and phosphorylated SP/TP sites (612548, 1:1,000; BD Biosciences). After being washed with Tris-buffered saline plus Tween 20 (TBST), the membrane was incubated with secondary antibody goat anti-mouse IgG conjugated with horseradish peroxidase (HRP) (A0216, 1:1,000; Beyotime) at room temperature for 2 h. Signals were developed using Clarity Western ECL Substrate (Bio-Rad) and visualized using a ChemiDoc XRS+ imaging system (Bio-Rad). The image was quantified by ImageJ software.
Transcriptome sequencing analysis.
C. glabrata cells were cultured to logarithmic phase and then reinoculated into fresh YNB medium at an initial A660 of 0.1. After incubation for 6 h, cells were harvested and released to YNB medium with or without 1.5 M NaCl for 30 min. Then, cells were recollected and washed twice with phosphate-buffered saline by resuspension and centrifugation at 3,500 × g for 10 min at 4°C. Total RNA was isolated by using a MiniBEST universal RNA extraction kit (TaKaRa Bio, Shiga, Japan). The concentration and quality of total RNA were determined by microspectrophotometry using an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Frozen samples were sent to the Majorbio Institute for global gene analysis.
qRT-PCR analysis.
C. glabrata cells were cultured as described for transcriptome sequencing analysis. Total RNA was extracted using a MiniBEST universal RNA extraction kit, and 1 μg was used to synthesize cDNA with a PrimeScript II 1st-strand cDNA synthesis kit (TaKaRa Bio). The cDNA mixture was diluted to approximately 100 ng/μl and used for quantitative real-time PCR (qRT-PCR) with TB Green Premix Ex Taq (TaKaRa Bio) on an iQ5 continuous fluorescence detector system (Bio-Rad, Hercules, CA). Data were normalized to values of the actin gene. The primers used for qRT-PCR are given in Table 3.
ChIP assay.
Cells were grown to logarithmic phase and then left untreated or treated with 1.5 M NaCl for 30 min. Cells were cross-linked with 1% formaldehyde for 20 min at room temperature. Glycine was added to a final concentration of 330 mM, and the incubation continued for 15 min. Cells were collected, washed four times with cold Tris-buffered saline (TBS) (20 mM Tris-HCl [pH 7.5], 150 mM NaCl), and kept at −20°C for further processing. Cell pellets were resuspended in 0.3 ml of cold lysis buffer (50 mM HEPES-KOH [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% sodium deoxycholate, 0.1% SDS, 1 mM phenylmethylsulfonyl fluoride [PMSF]) supplemented with 1% Triton X-100 and lysed by a bead beater. Then the cross-linked chromatin was sonicated to yield an average DNA fragment size of 350 bp (range, 100 to 850 bp). Finally, the sample was clarified by centrifugation at 12,000 × g for 5 min at 4°C. An aliquot of chromatin solution was used for immunoprecipitation (IP), input (IN), and control immunoprecipitation (CIP), and the remaining samples were stored at −20°C. The IP and CIP samples were incubated with anti-Myc monoclonal antibody and anti-IgG monoclonal antibody, respectively, which precoupled to magnetic beads (9006; Cell Signaling Technology). After 2 h at 4°C on a rotator, beads were washed twice with lysis buffer, twice with lysis buffer plus 500 mM NaCl, twice with washing buffer (10 mM Tris-HCl [pH 8.0], 0.25 M LiCl, 1 mM EDTA, 0.5% N-P40, 0.5% sodium deoxycholate), and once with TE buffer (10 mM Tris-HCl [pH 8.0], 1 mM EDTA). Chromatin was eluted, and cross-linking was reversed by incubation at 65°C overnight. After extraction with phenol-chloroform-isoamyl alcohol (25:24:1, vol/vol/vol), DNA was ethanol precipitated for 4 h at −20°C and resuspended in TE buffer. Quantitative real-time PCR was used to analyze DNA with TB Green Premix Ex Taq (TaKaRa Bio) on an iQ5 continuous fluorescence detector system (Bio-Rad, Hercules, CA). Relative fold enrichment was calculated by the following formula: fold change = (IP intensity − CIP intensity)/IN intensity. Primers used in this study are listed in the Table 3.
Glycerophospholipid extraction and measurement.
Logarithmic-phase C. glabrata cells were harvested and released into fresh YNB medium with or without 1.5 M NaCl for 4 h. Cells were harvested, washed twice with PBS, and freeze-dried. Fifty milligrams of dried cells was resuspended in methanol-chloroform-distilled water solution (1:2:1, vol/vol/vol). The sample extraction was carried out as described previously (63). The extracted glycerophospholipids were dried under a nitrogen stream and dissolved in methanol-isopropanol (1:1, vol/vol).
Mass spectrometry analysis of glycerophospholipid.
Analysis of glycerophospholipid mixtures was carried out utilizing ultrahigh-performance liquid chromatography tandem mass spectrometry (UPLC-MS; Waters, USA) and a CORTECS UPLC hydrophilic interaction liquid chromatography (HILIC) column (2.1 by 150 mm; inner diameter, 1.6-μm) with gradient elution at 45°C and a rate of infusion of 0.3 ml·min−1. The mobile-phase gradient was formed by buffer A (acetonitrile) and buffer B (11 mM ammonium acetate). The A/B ratios were 95:5, 95:5, 70:30, 60:40, and 95:5 at run times of 0, 2, 15, 17, and 17.10 min, respectively. The capillary voltage was set at +3.5 kV or −3.5 kV for the positive or negative mode, respectively. Data analysis was based on the following commercial standards at a concentration of 1 mg·ml−1: 16:0 PA (830855; Avanti Polar Lipids), 16:0 PC (850355; Avanti Polar Lipids), 16:0 PS (840037; Avanti Polar Lipids), 16:0 PG (840455; Avanti Polar Lipids), 16:0 PE (850705; Avanti Polar Lipids), and 16:0 PI (850141; Avanti Polar Lipids). The mass amounts of glycerophospholipid were calculated by the following equation: content of glycerophospholipid = a1c0v/a0m, where a1 is the peak area of the samples, a0 is the peak area of the standard, c0 is the concentration of the standard, v is the total volume of the sample, and m is the mass of freeze-dried cells.
Cell membrane integrity analysis.
Logarithmic-phase C. glabrata cells were harvested and released into fresh YNB medium with or without 1.5 M NaCl for 4 h. Samples were centrifuged, washed twice with PBS, and diluted to an A660 of 0.5. Diluted sample (500 μl) was incubated with 5 μl of propidium iodide (Sangon Bio, Shanghai City, China) for 5 min at room temperature in the dark and then harvested, washed twice with PBS, and resuspended in the same volume of PBS. The cell number and fluorescence intensity (excitation, 536 nm; emission, 617 nm) of cell suspensions were measured by flow cytometry analysis using a FACSCalibur apparatus (BD Biosciences, Shanghai City, China). More than 20,000 events were analyzed for each sample and at a rate of 600 to 1,000 events/s. The data were acquired and analyzed using CellQuest software.
TEM analysis.
Logarithmic-phase C. glabrata cells were harvested and released into fresh YNB medium with or without 1.5 M NaCl for 4 h. Samples were centrifuged and washed twice with PBS buffer (pH 7.4). The cells were resuspended in 5% (vol/vol) glutaraldehyde in 0.1 M PBS and prefixed overnight at 4°C. The cell pellets were washed three times with 0.1 M PBS and then postfixed with 1% osmic acid in 0.1 M PBS. After 2 h of incubation at room temperature, the fixed samples were washed six times with 0.1 M PBS, dehydrated with ethyl alcohol series, and embedded in epoxy resin. Ultrathin sections were stained with uranyl acetate and lead citrate and examined using a transmission electron microscope (TEM) (H-7650; Hitachi, Japan).
Statistical analysis.
Experimental data are shown as the means ± standard errors of the means (SEM). All quantitative data were analyzed using Student's t test or one-way analysis of variance (ANOVA). Each experiment was repeated at least three times.
Accession number(s).
The RNA-seq raw reads were submitted to NCBI under BioProject number PRJNA488115, and the Sequence Read Archive (SRA) entries are SRX4774478, SRX4774479, SRX4774480, SRX4774481.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Karl Kuchler from the Medical University Vienna for the generous gift of the C. glabrata ATCC 2001 and ATCC 55 strains.
This work was supported by the National Natural Science Foundation of China (grants 21676118 and 21706095), the Jiangsu Province 333 High-level Talents Cultivating Project (BRA2016365), and the national first-class discipline program of Light Industry Technology and Engineering (LITE2018-08).
C.W., X.C., and L.L. designed the research; C.W., G.Z., and R.Y. performed the research; C.W. and X.C. analyzed data; and C.W. and L.L. wrote the paper.
We declare that we have no competing financial interests.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02822-18.
REFERENCES
- 1.Duc C, Pradal M, Sanchez I, Noble J, Tesniere C, Blondin B. 2017. A set of nutrient limitations trigger yeast cell death in a nitrogen-dependent manner during wine alcoholic fermentation. PLoS One 12:e0184838. doi: 10.1371/journal.pone.0184838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang YL, Tseng SF, Huang YC, Shen ZJ, Hsu PH, Hsieh MH, Yang CW, Tognetti S, Canal B, Subirana L, Wang CW, Chen HT, Lin CY, Posas F, Teng SC. 2017. Yeast Cip1 is activated by environmental stress to inhibit Cdk1-G1 cyclins via Mcm1 and Msn2/4. Nat Commun 8:56. doi: 10.1038/s41467-017-00080-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berterame NM, Martani F, Porro D, Branduardi P. 2018. Copper homeostasis as a target to improve Saccharomyces cerevisiae tolerance to oxidative stress. Metab Eng 46:43–50. doi: 10.1016/j.ymben.2018.02.010. [DOI] [PubMed] [Google Scholar]
- 4.Baldi S, Bolognesi A, Meinema AC, Barral Y. 2017. Heat stress promotes longevity in budding yeast by relaxing the confinement of age-promoting factors in the mother cell. Elife 6:e28329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mahmoud S, Planes MD, Cabedo M, Trujillo C, Rienzo A, Caballero-Molada M, Sharma SC, Montesinos C, Mulet JM, Serrano R. 2017. TOR complex 1 regulates the yeast plasma membrane proton pump and pH and potassium homeostasis. FEBS Lett 591:1993–2002. doi: 10.1002/1873-3468.12673. [DOI] [PubMed] [Google Scholar]
- 6.Blenis J. 2017. TOR, the gateway to cellular metabolism, cell growth, and disease. Cell 171:10–13. doi: 10.1016/j.cell.2017.08.019. [DOI] [PubMed] [Google Scholar]
- 7.Li LT, Kaplan J, Ward DM. 2017. The glucose sensor Snf1 and the transcription factors Msn2 and Msn4 regulate transcription of the vacuolar iron importer gene CCC1 and iron resistance in yeast. J Biol Chem 292:15577–15586. doi: 10.1074/jbc.M117.802504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nasution O, Lee YM, Kim EJ, Lee YJ, Kim WK, Choi WJ. 2017. Overexpression of OLE1 enhances stress tolerance and constitutively activates the MAPK HOG pathway in Saccharomyces cerevisiae. Biotechnol Bioeng 114:620–631. doi: 10.1002/bit.26093. [DOI] [PubMed] [Google Scholar]
- 9.Ji H, Zhuge B, Zong H, Lu X, Fang H, Zhuge J. 2016. Role of CgHOG1 in stress responses and glycerol overproduction of Candida glycerinogenes. Curr Microbiol 73:827–833. doi: 10.1007/s00284-016-1132-7. [DOI] [PubMed] [Google Scholar]
- 10.Silva A, Cavero S, Begley V, Solé C, Böttcher R, Chávez S, Posas F, de Nadal E. 2017. Regulation of transcription elongation in response to osmostress. PLoS Genet 13:e1007090. doi: 10.1371/journal.pgen.1007090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chang N, Yao S, Chen D, Zhang L, Huang J, Zhang L. 2018. The Hog1 positive regulated YCT1 gene expression under cadmium tolerance of budding yeast. FEMS Microbiol Lett 365:fny170. doi: 10.1093/femsle/fny170. [DOI] [PubMed] [Google Scholar]
- 12.Correia I, Alonso-Monge R, Pla J. 2017. The Hog1 MAP Kinase promotes the recovery from cell cycle arrest induced by hydrogen peroxide in Candida albicans. Front Microbiol 7:2133. doi: 10.3389/fmicb.2016.02133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Carapia-Minero N, Castelan-Vega JA, Perez NO, Rodriguez-Tovar AV. 2018. The phosphorelay signal transduction system in Candida glabrata: an in silico analysis. J Mol Model 24:13. doi: 10.1007/s00894-017-3545-z. [DOI] [PubMed] [Google Scholar]
- 14.Sole C, Nadal-Ribelles M, Kraft C, Peter M, Posas F, de Nadal E. 2011. Control of Ubp3 ubiquitin protease activity by the Hog1 SAPK modulates transcription upon osmostress. EMBO J 30:3274–3284. doi: 10.1038/emboj.2011.227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Proft M, Mas G, de Nadal E, Vendrell A, Noriega N, Struhl K, Posas F. 2006. The stress-activated Hog1 kinase is a selective transcriptional elongation factor for genes responding to osmotic stress. Mol Cell 23:241–250. doi: 10.1016/j.molcel.2006.05.031. [DOI] [PubMed] [Google Scholar]
- 16.Martins TS, Pereira C, Canadell D, Vilaca R, Teixeira V, Moradas-Ferreira P, de Nadal E, Posas F, Costa V. 2018. The Hog1p kinase regulates Aft1p transcription factor to control iron accumulation. Biochim Biophys Acta Mol Cell Biol Lipids 1863:61–70. doi: 10.1016/j.bbalip.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Gonzalez-Novo A, Jimenez J, Clotet J, Nadal-Ribelles M, Cavero S, de Nadal E, Posas F. 2015. Hog1 targets Whi5 and Msa1 transcription factors to downregulate cyclin expression upon stress. Mol Cell Biol 35:1606–1618. doi: 10.1128/MCB.01279-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clotet J, Escote X, Adrover MA, Yaakov G, Gari E, Aldea M, de Nadal E, Posas F. 2006. Phosphorylation of Hsl1 by Hog1 leads to a G2 arrest essential for cell survival at high osmolarity. EMBO J 25:2338–2346. doi: 10.1038/sj.emboj.7601095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vazquez-Ibarra A, Subirana L, Ongay-Larios L, Kawasaki L, Rojas-Ortega E, Rodriguez-Gonzalez M, de Nadal E, Posas F, Coria R. 2018. Activation of the Hog1 MAPK by the Ssk2/Ssk22 MAP3Ks, in the absence of the osmosensors, is not sufficient to trigger osmostress adaptation in Saccharomyces cerevisiae. FEBS J 285:1079–1096. doi: 10.1111/febs.14385. [DOI] [PubMed] [Google Scholar]
- 20.Dong SJ, Yi CF, Li H. 2015. Changes of Saccharomyces cerevisiae cell membrane components and promotion to ethanol tolerance during the bioethanol fermentation. Int J Biochem Cell Biol 69:196–203. doi: 10.1016/j.biocel.2015.10.025. [DOI] [PubMed] [Google Scholar]
- 21.Besada-Lombana PB, Fernandez-Moya R, Fenster J, Da Silva NA. 2017. Engineering Saccharomyces cerevisiae fatty acid composition for increased tolerance to octanoic acid. Biotechnol Bioeng 114:1531–1538. doi: 10.1002/bit.26288. [DOI] [PubMed] [Google Scholar]
- 22.Renne MF, de Kroon A. 2018. The role of phospholipid molecular species in determining the physical properties of yeast membranes. FEBS Lett 592:1330–1345. doi: 10.1002/1873-3468.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knupp J, Martinez-Montanes F, Van Den Bergh F, Cottier S, Schneiter R, Beard D, Chang A. 2017. Sphingolipid accumulation causes mitochondrial dysregulation and cell death. Cell Death Differ 24:2044–2053. doi: 10.1038/cdd.2017.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khandelwal NK, Chauhan N, Sarkar P, Esquivel BD, Coccetti P, Singh A, Coste AT, Gupta M, Sanglard D, White TC, Chauvel M, d'Enfert C, Chattopadhyay A, Gaur NA, Mondal AK, Prasad R. 2018. Azole resistance in a Candida albicans mutant lacking the ABC transporter CDR6/ROA1 depends on TOR signaling. J Biol Chem 293:412–432. doi: 10.1074/jbc.M117.807032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hatakeyama R, Kono K, Yoshida S. 2017. Ypk1 and Ypk2 kinases maintain Rho1 at the plasma membrane by flippase-dependent lipid remodeling after membrane stresses. J Cell Sci 130:1169–1178. doi: 10.1242/jcs.198382. [DOI] [PubMed] [Google Scholar]
- 26.Bourgoint C, Rispal D, Berti M, Filipuzzi I, Helliwell SB, Prouteau M, Loewith R. 2018. Target of rapamycin complex 2-dependent phosphorylation of the coat protein Pan1 by Akl1 controls endocytosis dynamics in Saccharomyces cerevisiae. J Biol Chem 293:12043–12053. doi: 10.1074/jbc.RA117.001615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leskoske KL, Roelants FM, Marshall MNM, Hill JM, Thorner J. 2017. The stress-sensing TORC2 complex activates yeast AGC-family protein kinase Ypk1 at multiple novel sites. Genetics 207:179–195. doi: 10.1534/genetics.117.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rippert D, Backhaus K, Rodicio R, Heinisch JJ. 2017. Cell wall synthesis and central carbohydrate metabolism are interconnected by the SNF1/Mig1 pathway in Kluyveromyces lactis. Eur J Cell Biol 96:70–81. doi: 10.1016/j.ejcb.2016.12.004. [DOI] [PubMed] [Google Scholar]
- 29.Ramírez-Zavala B, Mottola A, Haubenreißer J, Schneider S, Allert S, Brunke S, Ohlsen K, Hube B, Morschhäuser J. 2017. The Snf1-activating kinase Sak1 is a key regulator of metabolic adaptation and in vivo fitness of Candida albicans. Mol Microbiol 104:989–1007. doi: 10.1111/mmi.13674. [DOI] [PubMed] [Google Scholar]
- 30.Llopis-Torregrosa V, Ferri-Blazquez A, Adam-Artigues A, Deffontaines E, van Heusden GPH, Yenush L. 2016. Regulation of the yeast Hxt6 hexose transporter by the Rod1 alpha-arrestin, the Snf1 protein kinase, and the Bmh2 14-3-3 protein. J Biol Chem 291:14973–14985. doi: 10.1074/jbc.M116.733923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fujita S, Sato D, Kasai H, Ohashi M, Tsukue S, Takekoshi Y, Gomi K, Shintani T. 2018. The C-terminal region of the yeast monocarboxylate transporter Jen1 acts as a glucose signal-responding degron recognized by the α-arrestin Rod1. J Biol Chem 293:10926–10936. doi: 10.1074/jbc.RA117.001062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moreno I, Tutrone N, Sentandreu R, Valentin E. 2008. Saccharomyces cerevisiae Rds2 transcription factor involvement in cell wall composition and architecture. Int Microbiol 11:57–63. [PubMed] [Google Scholar]
- 33.Soontorngun N, Larochelle M, Drouin S, Robert F, Turcotte B. 2007. Regulation of gluconeogenesis in Saccharomyces cerevisiae is mediated by activator and repressor functions of Rds2. Mol Cell Biol 27:7895–7905. doi: 10.1128/MCB.01055-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.de Nadal E, Casadome L, Posas F. 2003. Targeting the MEF2-like transcription factor Smp1 by the stress-activated Hog1 mitogen-activated protein kinase. Mol Cell Biol 23:229–237. doi: 10.1128/MCB.23.1.229-237.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duch A, Felipe-Abrio I, Barroso S, Yaakov G, Garcia-Rubio M, Aguilera A, de Nadal E, Posas F. 2013. Coordinated control of replication and transcription by a SAPK protects genomic integrity. Nature 493:116–119. doi: 10.1038/nature11675. [DOI] [PubMed] [Google Scholar]
- 36.Proft M, Pascual-Ahuir A, de Nadal E, Ariño J, Serrano R, Posas F. 2001. Regulation of the Sko1 transcriptional repressor by the Hog1 MAP kinase in response to osmotic stress. EMBO J 20:1123–1133. doi: 10.1093/emboj/20.5.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bai C, Tesker M, Engelberg D. 2015. The yeast Hot1 transcription factor is critical for activating a single target gene, STL1. Mol Biol Cell 26:2357–2374. doi: 10.1091/mbc.E14-12-1626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ruiz-Roig C, Noriega N, Duch A, Posas F, de Nadal E. 2012. The Hog1 SAPK controls the Rtg1/Rtg3 transcriptional complex activity by multiple regulatory mechanisms. Mol Biol Cell 23:4286–4296. doi: 10.1091/mbc.E12-04-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Joaquin M, Gubern A, Gonzalez-Nunez D, Josue Ruiz E, Ferreiro I, de Nadal E, Nebreda AR, Posas F. 2012. The p57 CDKi integrates stress signals into cell-cycle progression to promote cell survival upon stress. EMBO J 31:2952–2964. doi: 10.1038/emboj.2012.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gao YQ, Gao Y, Hu L, Liang Y, Li L, Xiao JH. 2018. p38 regulates mRNA stability of cytokines stimulated by Zymosan A. Int J Clin Exp Med 11:1105–1114. [Google Scholar]
- 41.Debattisti V, Gerencser AA, Saotome M, Das S, Hajnoczky G. 2017. ROS Control mitochondrial motility through p38 and the motor adaptor Miro/Trak. Cell Rep 21:1667–1680. doi: 10.1016/j.celrep.2017.10.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Miura H, Kondo Y, Matsuda M, Aoki K. 2018. Cell-to-cell heterogeneity in p38-mediated cross-inhibition of JNK causes stochastic cell death. Cell Rep 24:2658–2668. doi: 10.1016/j.celrep.2018.08.020. [DOI] [PubMed] [Google Scholar]
- 43.Soontorngun N, Baramee S, Tangsombatvichit C, Thepnok P, Cheevadhanarak S, Robert F, Turcotte B. 2012. Genome-wide location analysis reveals an important overlap between the targets of the yeast transcriptional regulators Rds2 and Adr1. Biochem Biophys Res Commun 423:632–637. doi: 10.1016/j.bbrc.2012.05.151. [DOI] [PubMed] [Google Scholar]
- 44.Ye C, Sutter BM, Wang Y, Kuang Z, Tu BP. 2017. A metabolic function for phospholipid and histone methylation. Mol Cell 66:180–193. doi: 10.1016/j.molcel.2017.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tan Z, Khakbaz P, Chen Y, Lombardo J, Yoon JM, Shanks JV, Klauda JB, Jarboe LR. 2017. Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables. Metab Eng 44:1–12. doi: 10.1016/j.ymben.2017.08.006. [DOI] [PubMed] [Google Scholar]
- 46.Barman A, Gohain D, Bora U, Tamuli R. 2018. Phospholipases play multiple cellular roles including growth, stress tolerance, sexual development, and virulence in fungi. Microbiol Res 209:55–69. doi: 10.1016/j.micres.2017.12.012. [DOI] [PubMed] [Google Scholar]
- 47.Tekarslan-Sahin SH, Alkim C, Sezgin T. 2018. Physiological and transcriptomic analysis of a salt-resistant Saccharomyces cerevisiae mutant obtained by evolutionary engineering. Bosnian J Basic Med 18:55–65. doi: 10.17305/bjbms.2017.2250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Muir A, Roelants FM, Timmons G, Leskoske KL, Thorner J. 2015. Down-regulation of TORC2-Ypk1 signaling promotes MAPK-independent survival under hyperosmotic stress. Elife 4:e09336. doi: 10.7554/eLife.09336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Holthuis JCM, Menon AK. 2014. Lipid landscapes and pipelines in membrane homeostasis. Nature 510:48–57. doi: 10.1038/nature13474. [DOI] [PubMed] [Google Scholar]
- 50.Mioka T, Fujimura-Kamada K, Mizugaki N, Kishimoto T, Sano T, Nunome H, Williams DE, Andersen RJ, Tanaka K. 2018. Phospholipid flippases and Sfk1p, a novel regulator of phospholipid asymmetry, contribute to low permeability of the plasma membrane. Mol Biol Cell 29:1203–1218. doi: 10.1091/mbc.E17-04-0217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Velly H, Bouix M, Passot S, Penicaud C, Beinsteiner H, Ghorbal S, Lieben P, Fonseca F. 2015. Cyclopropanation of unsaturated fatty acids and membrane rigidification improve the freeze-drying resistance of Lactococcus lactis subsp lactis TOMSC161. Appl Microbiol Biotechnol 99:907–918. doi: 10.1007/s00253-014-6152-2. [DOI] [PubMed] [Google Scholar]
- 52.Putta P, Rankenberg J, Korver RA, van Wijk R, Munnik T, Testerink C, Kooijman EE. 2016. Phosphatidic acid binding proteins display differential binding as a function of membrane curvature stress and chemical properties. Biochim Biophys Acta 1858:2709–2716. doi: 10.1016/j.bbamem.2016.07.014. [DOI] [PubMed] [Google Scholar]
- 53.Morisada S, Ono Y, Kodaira T, Kishino H, Ninomiya R, Mori N, Watanabe H, Ohta A, Horiuchi H, Fukuda R. 2018. The membrane-bound O-acyltransferase Ale1 transfers an acyl moiety to newly synthesized 2-alkyl-sn-glycero-3-phosphocholine in yeast. FEBS Lett 592:1829–1836. doi: 10.1002/1873-3468.13103. [DOI] [PubMed] [Google Scholar]
- 54.Hirama T, Lu SM, Kay JG, Maekawa M, Kozlov MM, Grinstein S, Fairn GD. 2017. Membrane curvature induced by proximity of anionic phospholipids can initiate endocytosis. Nat Commun 8:1393. doi: 10.1038/s41467-017-01554-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Graber ZT, Shi Z, Baumgart T. 2017. Cations induce shape remodeling of negatively charged phospholipid membranes. Phys Chem Phys 19:15285–15295. doi: 10.1039/C7CP00718C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guerreiro JF, Muir A, Ramachandran S, Thorner J, Sa-Correia I. 2016. Sphingolipid biosynthesis upregulation by TOR complex 2-Ypk1 signaling during yeast adaptive response to acetic acid stress. Biochem J 473:4311–4325. doi: 10.1042/BCJ20160565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slotte JP. 2013. Biological functions of sphingomyelins. Prog Lipid Res 52:424–437. doi: 10.1016/j.plipres.2013.05.001. [DOI] [PubMed] [Google Scholar]
- 58.Bojsen R, Torbensen R, Larsen CE, Folkesson A, Regenberg B. 2013. The synthetic amphipathic peptidomimetic LTX109 is a potent fungicide that disturbs plasma membrane integrity in a sphingolipid dependent manner. PLoS One 8:e69483. doi: 10.1371/journal.pone.0069483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Parsons JB, Rock C. 2013. Bacterial lipids: metabolism and membrane homeostasis. Prog Lipid Res 52:249–276. doi: 10.1016/j.plipres.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Qi Y, Liu H, Yu J, Chen X, Liu L. 2017. Med15B regulates acid stress response and tolerance in Candida glabrata by altering membrane lipid composition. Appl Environ Microbiol 83:e01128-17. doi: 10.1128/AEM.01128-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jesch SA, Gaspar ML, Stefan CJ, Aregullin MA, Henry SA. 2010. Interruption of inositol sphingolipid synthesis triggers Stt4p-dependent protein kinase C signaling. J Biol Chem 285:41947–41960. doi: 10.1074/jbc.M110.188607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Chen X, Dong X, Wang Y, Zhao Z, Liu L. 2015. Mitochondrial engineering of the TCA cycle for fumarate production. Metab Eng 31:62–73. doi: 10.1016/j.ymben.2015.02.002. [DOI] [PubMed] [Google Scholar]
- 63.Chen X, Xu G, Xu N, Zou W, Zhu P, Liu L, Chen J. 2013. Metabolic engineering of Torulopsis glabrata for malate production. Metab Eng 19:10–16. doi: 10.1016/j.ymben.2013.05.002. [DOI] [PubMed] [Google Scholar]
- 64.Chen X, Wu J, Song W, Zhang L, Wang H, Liu L. 2015. Fumaric acid production by Torulopsis glabrata: engineering the urea cycle and the purine nucleotide cycle. Biotechnol Bioeng 112:156–167. doi: 10.1002/bit.25334. [DOI] [PubMed] [Google Scholar]
- 65.Yan D, Lin X, Qi Y, Liu H, Chen X, Liu L, Chen J. 2016. Crz1p regulates pH homeostasis in Candida glabrata by altering membrane lipid composition. Appl Environ Microbiol 82:6920–6929. doi: 10.1128/AEM.02186-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wu J, Chen X, Cai L, Tang L, Liu L. 2015. Transcription factors Asg1p and Hal9p regulate pH homeostasis in Candida glabrata. Front Microbiol 6:843. doi: 10.3389/fmicb.2015.00843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yanez R, Marques S, Girio FM, Roseiro JC. 2008. The effect of acid stress on lactate production and growth kinetics in Lactobacillus rhamnosus cultures. Process Biochem 43:356–361. doi: 10.1016/j.procbio.2007.12.014. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.