We demonstrate here the genetic diversity of K. pneumoniae isolates from dairy cows and the mixed phylogenetic lineages between bovine and human isolates. The ferric uptake operon kfuABC genes were more prevalent in strains from clinical mastitis cows. Furthermore, we report the emergence of an IncN-type plasmid carrying the blaCTX-M-1 and mph(A) genes among dairy farms in the United States. Our study evaluated the genomic diversity of the bovine and human isolates, and the findings uncovered different profiles of virulence determinants among bovine and human K. pneumoniae isolates at the genome population level.
KEYWORDS: antimicrobial resistance, genomic epidemiology, Klebsiella pneumoniae, virulence
ABSTRACT
Klebsiella pneumoniae is a leading cause of severe infections in humans and dairy cows, and these infections are rapidly becoming untreatable due to the emergence of multidrug-resistant (MDR) strains. However, little is known about the relationship between bovine and human K. pneumoniae isolates at the genome population level. Here, we investigated the genomic structures, pangenomic profiles, virulence determinants, and resistomes of 308 K. pneumoniae isolates from humans and dairy cows, including 96 newly sequenced cow isolates. We identified 177 functional protein families that were significantly different across human and bovine isolates; genes expressing proteins related to metal ion (iron, zinc, and calcium) metabolism were significantly more prevalent among the bovine isolates. Siderophore systems were found to be prevalent in both the bovine and the human isolates. In addition, we found that the Klebsiella ferric uptake operon kfuABC was significantly more prevalent in clinical mastitis cases than in healthy cows. Furthermore, on two dairy farms, we identified a unique IncN-type plasmid, pC5, coharboring blaCTX-M-1 and mph(A) genes, which confer resistance to cephalosporins and macrolides, respectively. We provide here the complete annotated sequence of this plasmid.
IMPORTANCE We demonstrate here the genetic diversity of K. pneumoniae isolates from dairy cows and the mixed phylogenetic lineages between bovine and human isolates. The ferric uptake operon kfuABC genes were more prevalent in strains from clinical mastitis cows. Furthermore, we report the emergence of an IncN-type plasmid carrying the blaCTX-M-1 and mph(A) genes among dairy farms in the United States. Our study evaluated the genomic diversity of the bovine and human isolates, and the findings uncovered different profiles of virulence determinants among bovine and human K. pneumoniae isolates at the genome population level.
INTRODUCTION
Klebsiella pneumoniae is a Gram-negative, rod-shaped, encapsulated, facultative anaerobe that is commonly found in the mouth, skin, and intestines of humans and animals. As an opportunistic pathogen in humans, K. pneumoniae primarily attacks immunocompromised individuals and has become a leading cause of community-acquired and hospital-acquired infections in humans (1). K. pneumoniae is a clinically important species and causes serious nosocomial infections, such as septicemia, pneumonia, urinary tract infection, surgical site infection, and soft tissue infection (2). Nosocomial infections caused by K. pneumoniae are a leading cause of morbidity and mortality in immunocompromised people, with fatality rates being upwards of 50% despite the use of appropriate antibiotic therapy (3, 4).
In veterinary medicine, K. pneumoniae is of interest because it commonly causes clinical mastitis in dairy herds. Mastitis is a highly prevalent disease in dairy cows and creates an incontestable economic burden in the dairy industry worldwide, in addition to being a serious animal welfare issue because of the pain associated with the infection (5). The economic losses associated with mastitis are predominantly due to decreased milk production, discarded milk, treatment costs, and premature culling (6, 7). Coliform organisms are among the highest-incidence organisms on farms with otherwise excellent udder health management. The implementation of programs for mastitis control has reduced the prevalence of important contagious pathogens, and currently, approximately 40% of all clinical mastitis cases are associated with opportunistic Gram-negative bacteria, such as Klebsiella spp., Escherichia coli, Pseudomonas spp., and Pasteurella spp. (8, 9, 10). Among the major mastitis pathogens causing mastitis, coliform organisms such as Klebsiella spp. and E. coli cause the most severe clinical cases, with greater milk loss and an average cost of more than $221.03 per case (11). Studies have shown differences in the pathogenicity of Klebsiella spp. as a mastitis pathogen and other Gram-negative organisms (12, 13). Klebsiella spp. cause longer intramammary infections than E. coli (14), more severe clinical episodes than Serratia spp. and E. coli (15, 16), and a greater milk production loss and risk of culling than intramammary infections caused by E. coli (17). Therefore, due to the severity of bovine clinical mastitis, the low effectiveness of antibiotic treatments, and the lack of advancement in preventive measures against Klebsiella infection, K. pneumoniae is a particularly concerning pathogen.
Whole-genome analysis is the ideal approach to building a robust phylogeny in recently emerged pathogens to explore their diverse backgrounds through the identification of single nucleotide polymorphisms (SNPs) or other genetic variants. A recent pangenomic analysis of K. pneumoniae isolates from humans and bovines found that genes named regulators of mucoid phenotype A (rmpA) and A2 (rmpA2) as well as a cluster of siderophore genes (kfuABC and kvgAS) are associated with invasive infection in humans (18). However, the analysis of the genetic determinants of the bovine isolates was limited, and the relatedness of the human and bovine K. pneumoniae isolates was not explored. A more comprehensive study with a larger number of isolates from cows would allow for exploration of the relatedness of human isolates and bovine isolates.
Here, we assembled a collection of K. pneumoniae strains recovered from 143 mastitic cows with various degrees of disease severity and sequenced the genomes of 96 distinct isolates to compare their genomic structures, virulence factors, and antimicrobial resistance (AMR) genes with those of the K. pneumoniae isolates studied by Holt et al. (18). We investigated the pangenomic gene functions of 308 isolates in total and identified a group of virulence genes that had a different distribution between K. pneumoniae isolates collected from dairy cows and humans. Furthermore, we found a resistance plasmid not previously identified in the United States and clarified its genetic context according to the closed plasmid structure.
RESULTS
Diversity and AMR profile of K. pneumoniae on dairy farms.
We isolated 143 nonreplicate K. pneumoniae strains (see Fig. S1 and Data Set S1 in the supplemental material) from mastitic dairy cows from four farms located in upstate New York. Molecular typing of the isolates using pulsed-field gel electrophoresis (PFGE) identified 97 distinct PFGE groups, revealing a high genetic diversity within the isolate collection. Capsule locus typing of those 143 isolates showed that 127 (88.8%) were assigned to 44 known capsule loci (Fig. S1). The capsule loci KN3 (30/143, 21%), K13 (15/143, 11%), and KN1 (10/143, 7%) were the most prevalent in our sample set, accounting for approximately 39% of all infections. High genetic diversity was also reflected by the sequence types (STs) of those strains. Among the 96 newly sequenced strains, 46 possessed new alleles of the housekeeping genes used for multilocus sequence typing (MLST), and no predominant STs were found. These new alleles were submitted to the MLST database and were assigned 43 new STs.
Overall, the AMR profiles were diverse among these strains, and 40% (57/143) were resistant to one or more antimicrobial agents (Fig. S1; Data Set S1). The agent to which the highest prevalence of resistance was detected was streptomycin (29.4%, 42/143), followed by tetracycline (5.6%, 8/143) and gentamicin (4.2%, 6/143). Using the genomic data for the 96 newly sequenced isolates from mastitic cows, we detected 17 AMR genes with various prevalences (Fig. S2). The fosA (fosfomycin resistance), oqxAB (quinolones resistance), blaAmpH (β-lactamase), and blaSHV (β-lactamase) genes were common resistance genes found in our isolate collection. The resistance to streptomycin in our isolate collection was attributed to the strA and strB genes, located on an IncHI1B-type plasmid in 27 of the 96 strains. Analysis of the genetic environment revealed that the strA and strB genes were flanked by transposon Tn5393, the most common mobile element that mediates the transmission of strAB genes across species (19).
Identification of an IncN-type plasmid carrying blaCTX-M-1 and mph(A).
We found that four isolates were resistant to ceftiofur, with the MIC being 512 μg/ml. Among those, three strains that had genomic sequences were found to coharbor the blaCTX-M-1 (β-lactamase) and mph(A) (macrolide resistance) genes, which were located in the same contig in each strain. Comparison of the sequences by BLASTn analysis showed that all three contigs shared a high degree of similarity with the sequence of pL2-43 (GenBank accession no. KJ484641), an IncN-type plasmid found in an E. coli isolate recovered from a lamb in Switzerland in 2014 (20). Analysis of the plasmid sequences confirmed that they carried the same plasmid (named pC5) that coharbored the blaCTX-M-1 and mph(A) genes. The complete sequence of pC5 (GenBank accession no. MF953243) is 41,608 bp in size and carries 51 open reading frames (ORFs) (Fig. 1; Table S1). The blaCTX-M-1 gene is flanked by an ISEcp1 element in its upstream region, which is disrupted by an intact IS26 element (820 bp) in the opposite orientation. The sequences between the blaCTX-M-1 and mph(A) genes (1,572 bp) contain two ORFs: one ORF carries a truncated mrx gene (1,272 bp), and the other ORF is similar to ORF477 but is truncated by an inverted right repeat (IRR) of the ISEcp1 element (23 bp). The mph(A) gene was mediated by another IS26 element with the same orientation. We then compared the genetic background of pC5 with that of seven publicly available plasmid sequences with high similarity (coverage, >90%; identity, >99%), all of which were identified in E. coli strains in European countries (20, 21, 22, 23) (Table 1). They all maintained a typical IncN plasmid backbone scaffold, including a replicon and genes involved in plasmid stability, mutagenesis enhancement, antirestriction functions, and conjugative transfer (24). The difference among these plasmids was in the mosaic region that carried resistance genes and in the mobile genetic elements represented (Fig. 1). Plasmid pC5 shared an identical genetic neighborhood of the blaCTX-M-1 and mph(A) genes with pHHA45, pKC394, and pVI. However, in plasmid pL2-43, the reverse position of mph(A)-IS26 was the main structural difference from our plasmid.
FIG 1.
Plasmid structures of pC5 and reference plasmids. (A) Circular genetic map of plasmid pC5 (GenBank accession no. MF953243) and reference plasmids pL2-43 (GenBank accession no. KJ484641), pKC394 (GenBank accession no. HM138652), pH1038-142 (GenBank accession no. KJ484634), pVI (GenBank accession no. LT795508), pHHA45 (GenBank accession no. JX065630), pQNR2078 (GenBank accession no. HE613857), and pKC396 (GenBank accession no. HM138653). The map was drawn using the BLAST Ring Image Generator (BRIG) program (http://sourceforge.net/projects/brig/). (B) Schematic presentation of major structural features of pC5 in comparison with the reference plasmids pL2-43, pKC394, pH1038-142, pVI, pHHA45, pQNR2078, and pKC396. Areas shaded in gray indicate homologous regions of ≥99% nucleotide sequence identity in the plasmid scaffold regions. ORFs are portrayed by arrows to indicate the direction of transcription and colored on the basis of their predicted gene functions. Antimicrobial resistance genes are indicated by red boxes. Blue boxes denote mobile genetic elements. Green boxes indicate ORFs encoding hypothetical proteins. The figure is drawn to scale.
TABLE 1.
Summary of the features of the newly sequenced pC5 and seven related plasmids in the literature
| Isolate | Strain | Yr of isolation | Country | Origin | MLST | Plasmid | GenBank accession no. | Plasmid size (bp) | Inc type | Resistance gene(s) | Coveragea (%) | Identitya (%) | Source or reference |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C5 | K. pneumoniae | 2016 | USA | Cow | 2748 | pC5 | MF953243 | 41,608 | IncN | blaCTX-M-1, mph(A) | —b | — | This study |
| L-2 | E. coli | — | Switzerland | Lamb | 295 | pL2-43 | KJ484641 | 43,265 | IncN | blaCTX-M-1, mph(A) | 100 | 99 | 20 |
| 394 | E. coli | 2006 | Germany | Human | 131 | pKC394 | HM138652 | 53,207 | IncN | blaCTX-M-1, mph(A) | 99 | 99 | 21 |
| H-1038 | E. coli | 2010 | Switzerland | Human | New | pH1038-142 | KJ484634 | 142,875 | IncF-IncN | blaCTX-M-1, blaTEM-1, tetR-tet(A), strAB, catA1, sul3, aadA1, dfrA1 | 97 | 99 | 20 |
| KV7 | E. coli | — | UK | Pig | — | pVI | LT795508 | 39,744 | IncN | blaCTX-M-1, mph(A) | 93 | 99 | GenBankc |
| HHA45 | E. coli | 2006 | Denmark | Pig | — | pHHA45 | JX065630 | 39,510 | IncN | blaCTX-M-1, mph(A) | 93 | 99 | 22 |
| 2078 | E. coli | — | Germany | Horse | 86 | pQNR2078 | HE613857 | 42,379 | IncN | qnrB19 | 91 | 99 | 23 |
| 396 | E. coli | 2006 | Germany | Human | 131 | pKC396 | HM138653 | 44,216 | IncN | blaCTX-M-65 | 91 | 99 | 21 |
pC5 was the query plasmid used to investigate the sequence coverage and identity.
—, feature not identified.
GenBank direct submission.
Phylogeny of K. pneumoniae from cows and humans.
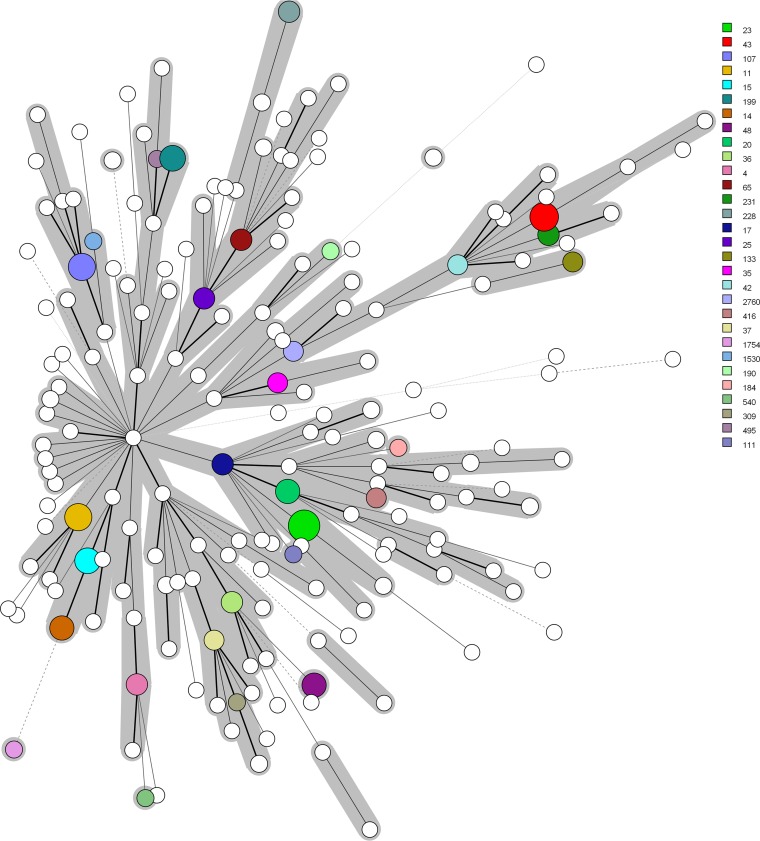
We compared 308 genomes from K. pneumoniae KpI isolates, including 123 isolates from dairy cows and 185 isolates from humans. We identified a total of 92,409 core genome SNPs among these 308 genomes and built a maximum likelihood (ML) phylogenetic tree based on these SNPs (Fig. 2). The results revealed a deep branching and scattered population structure that was broadly classified into 169 distinct phylogenetic lineages. The 123 bovine isolates had diverse population structures and did not form specific clusters. Furthermore, the population structure of the 123 bovine isolates was highly mixed with the 185 human isolates, and there were 27 lineages containing both human and bovine isolates. We assessed the clonality and clades of 294 isolates with assigned STs from the 308 strains as minimum-spanning trees (Fig. 3). Of these, we identified 190 distinct STs. The most prevalent STs were ST23 (n = 10 isolates), ST43 (n = 8), ST11 (n = 7), ST107 (n = 7), ST15 (n = 6), ST199 (n = 6), ST14 (n = 5), ST20 (n = 5), and ST48 (n = 5). Furthermore, ST34, ST35, ST37, ST43, ST65, ST107, ST133, ST290, ST294, ST309, and ST791 were found in both human and bovine isolates.
FIG 2.
Phylogeny of core genome SNPs in 308 genomes of K. pneumoniae KpI isolates from humans and dairy cows. The core genome SNPs were identified by mapping sequencing reads against a reference genome (K. pneumoniae strain NTUH-K2044, GenBank accession no. NC_012731.1). The RAxML program was used to calculate the phylogenetic tree to construct a maximum likelihood phylogeny.
FIG 3.
Minimum-spanning trees of 294 K. pneumoniae isolates from humans and dairy cows by MLST type. The 294 isolates that had assigned STs were derived from 308 K. pneumoniae isolates from humans and dairy cows. Each node within the tree represents a single ST. The size of the nodes is proportional to the number of K. pneumoniae isolates belonging to that ST. The length of the branch between each node is proportional to the number of distinct alleles of seven housekeeping genes that differ between the two linked nodes. The 30 STs with the highest numbers of isolates are labeled with different colors.
Pangenomic analysis identified distinct proteins between cow and human isolates.
We then investigated the functions of all genes across the 308 human and bovine isolates. In total, there were 1,705,306 open reading frames (ORFs) in the 308 genomes, with an average of 5,536 ORFs in each genome. All the ORFs could be classified into 68,420 clusters by sequence identity, which included 24,753 clusters harboring multiple sequences and 43,667 clusters with singleton genes. Function annotation against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database showed that 72% of the representative genes were assigned to 9,407 distinct KEGG hits with known functions among different functional categories (Fig. S3; Table S2). These functions included environmental information processing, genetic information processing, cellular processes, carbohydrate metabolism, amino acid metabolism, metabolism of cofactors and vitamins, energy metabolism, human disease-related functions, enzyme families, and nucleotide metabolism.
We then calculated the distribution of these KEGG assignments. There were 1,800 and 2,392 functional units found in all bovine isolates and human isolates, respectively. Among them, 2,684 functional units were found at a prevalence of >95% in both bovine and human isolates. Nonetheless, there were 177 functional units with significant differences between the human and bovine isolates (P < 0.0001, χ2 test) (Fig. S4; Table S3). Proteins related to metal ion (iron, zinc, and calcium) metabolism were significantly more prevalent in the bovine isolates; examples included the Fe3+ dicitrate transport protein FecA (81.3% versus 43.2%), the zinc protease PqqL (65.9% versus 7%), and the calcium permeable stress-gated cation channel protein TMEM63 (60.2% versus 44.3%). However, the reverse situation was true for proteins related to heavy metal ion-related transport; for example, 29.7% of the human isolates but only 2.4% of the bovine isolates carried the mercuric ion transport protein MerT.
Extensive diversity of virulence genes among bovine and human isolates.
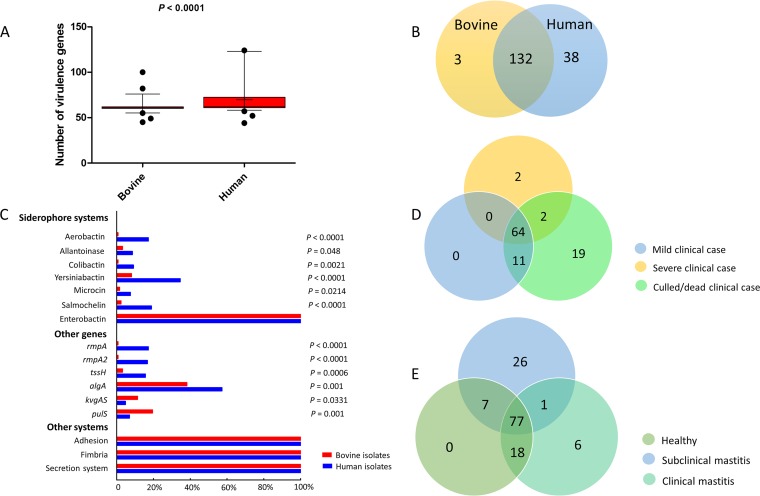
In total, we detected 173 virulence genes in the 308 isolates (Table S4). These included 135 and 170 virulence genes found in the bovine and human isolates, respectively, of which 132 genes were common to both types of isolates (Fig. 4). The enterobactin loci, adhesion-related gene clusters, secretion system-related gene clusters, and fimbria gene clusters were found in all isolates. Of note, the average number of virulence genes per isolate was significantly higher for the human isolates than for the bovine isolates (70 versus 62; P < 0.0001, Kruskal-Wallis test) (Fig. 4). The virulence genes associated with colibactin, aerobactin, allantoinase, yersiniabactin, microcin, salmochelin, and the rmpA and rmpA2 genes were rare (0.5% to 5%) among the bovine isolates, whereas they were found at increased frequency in the human isolates (9% to 35%) (Fig. 4). Furthermore, the combination of colibactin, aerobactin, allantoinase, yersiniabactin, microcin, salmochelin, and the rmpA and rmpA2 genes was frequent in K. pneumoniae isolates from humans. Among all 123 bovine isolates, we also investigated the distribution of virulence genes in healthy cows and mastitic cows. All the virulence genes found in the healthy group were present in either the subclinical mastitis or clinical mastitis cases. However, 26 virulence genes were exclusive to the subclinical mastitis cases and 6 were exclusive to the clinical mastitis cases (Fig. 4). Notably, we found that the Klebsiella ferric uptake operon kfuABC was significantly more prevalent (P < 0.05, χ2 test) in clinical mastitis cases than in subclinical mastitis cases and healthy cows (39%, 20%, and 11%, respectively; P = 0.0336, χ2 test).
FIG 4.
Virulence genes in 308 K. pneumoniae KpI isolates. (A) Number of virulence genes per isolate. Box plots indicate the average number of virulence genes in 123 bovine strains and 185 human strains. The means are shown as horizontal bars, with values of 62 and 70 for the bovine and human isolates, respectively. The P value was calculated using the Kruskal-Wallis test. (B) Distribution of virulence genes in 123 bovine isolates and 185 human isolates. Each number indicates the number of different virulence genes found within each group. (C) Frequency of gene clusters among 123 bovine isolates and 185 human isolates. The P values were calculated using the chi-square test. (D) Distribution of virulence genes in newly sequenced clinical K. pneumoniae KpI isolates. Each number indicates the number of different virulence genes found within each group. (E) Distribution of virulence genes in 123 bovine isolates. Each number indicates the number of different virulence genes found within each group.
DISCUSSION
The emergence and transmission of multidrug-resistant and virulent K. pneumoniae strains can cause severe, untreatable infections in humans and animals. However, due to insufficient data at the population level, the diversity of AMR genes of K. pneumoniae in cows is still unknown. In this study, we identified an IncN-type plasmid, pC5, coharboring the extended-spectrum β-lactamase (ESBL) gene blaCTX-M-1 and the macrolide phosphotransferase gene mph(A) in four distinct isolates on two farms. Ceftiofur is one of the most commonly used antibiotics in both adult and young dairy cattle in the United States, whereas macrolides are mostly used in young dairy cattle (<20 months of age) for the treatment of respiratory disease (https://www.aphis.usda.gov/animal_health/nahms/dairy/downloads/dairy07/Dairy07_is_AntibioticUse.pdf). The use of ceftiofur and/or macrolides may promote the dissemination of pC5-like plasmids. Seven plasmids that had been previously identified in E. coli either in humans or in animals in European countries shared a high similarity with pC5. In the United States, CTX-M-like-producing Enterobacteriaceae strains were first reported in 2003 (25). CTX-M-15 is the most common CTX-M enzyme found in Enterobacteriaceae in humans in the United States (26), whereas CTX-M-1 is rare. A previous study found 1 CTX-M-1-producing isolate among 208 ESBL-producing K. pneumoniae isolates from humans and 1 CTX-M-1-producing isolate among 163 ESBL-producing E. coli isolates from humans (27). The wide geographical and source distribution of pC5-like plasmids in European countries indicated that such IncN plasmids are responsible for the dissemination of blaCTX-M-1 and other resistance genes. Our study reports the emergence of this plasmid in the United States.
Our study sequenced 96 strains which belonged to different PFGE groups, and 46 of these were new STs. This indicates that there is no dominant strain responsible for infections with K. pneumoniae in cows. We built a representative pool of genomes from bovine and human strains comprising more than 300 genomes. Both the human and bovine isolates shared a large number of functional modules to form a core genome that may contribute to the maintenance of bacterial life. However, environmental changes and selection pressures have led to differences in some specific functions among the human and bovine isolates. For example, we found that proteins involved in metal ion (iron, zinc, and calcium) metabolism were significantly more prevalent in bovine isolates. Iron, such as Fe3+, is sequestered by transferrin and lactoferrin (28). The concentration of iron in vivo is critical for the control of cellular metabolism in bacteria, and insufficient iron abolishes bacterial growth (29). To grow successfully in host tissues, bacteria must be able to obtain metal ions from host transport proteins.
The pathogenicity of clinical K. pneumoniae strains is caused by various virulence determinants. We found that human isolates carried more virulence genes than bovine isolates. Virulence genes related to siderophore biosynthesis, fimbria proteins, adhesion, and secretion systems were found to be prevalent in both bovine and human isolates. The most common siderophore in cows is enterobactin. However, some siderophore systems (e.g., aerobactin, colibactin, allantoinase, yersiniabactin, microcin, and salmochelin systems) were significantly more prevalent in human isolates. Furthermore, previous studies showed that the ferric uptake operon kfuABC genes were significantly associated with tissue invasion in humans (18, 30). Our study confirmed that kfuABC is also associated with clinical mastitis cases in dairy cows. The secreted siderophores could grab iron from transferrin, lactoferrin, or ferritin and support the growth and infection of K. pneumoniae in hosts (31). Our results provide information related to the genomic structure, the virulence profile, and the resistome of K. pneumoniae isolates from cows and humans.
MATERIALS AND METHODS
Sampling design and bacterial strains.
This work was conducted between January and August 2017 at four commercial dairy farms, which were eligible for inclusion in the study if they met the following criteria: they had a herd size of >500 lactating cows, they participated in monthly Dairy Herd Improvement Association (DHIA) testing, they had an accurate recording of clinical mastitis cases, and they were willing to participate. All the farms were located in Cayuga County of New York State, and the farms were located inside a 15-mi-diameter circular region. Each dairy farm produced its own supply of forages, corn, grass, and alfalfa silage and purchased concentrated feed (corn, soybean meal, etc.) from a local vendor. Additionally, veterinary care was provided by a different veterinary clinic for each one of the four farms. Ceftiofur is routinely used on all four dairy farms. A known presence of Klebsiella clinical mastitis was also required to ensure that an appropriate number of Klebsiella isolates could be collected from each farm. Medians for this sample of herds were 1,057 lactating cows (range, 524 to 1,466 lactating cows), 12,150 kg of milk/cow per year (range, 11,773 to 13,091 kg of milk/cow per year), and a bulk tank somatic cell count of 140,500 cells/ml (range, 100,000 to 267,000 cells/ml).
Clinical scores (mild, severe, and culled/dead) for mastitis were assessed after diagnosis according to the severity scoring system previously described (32). Milk samples were collected from one affected quarter of mastitis cases aseptically on the farm and cultured using an on-farm culture system (AccuMast; FERA Animal Health, Ithaca, NY) in the diagnostic laboratory of the farm. Plates with Klebsiella-positive colonies were sent to the laboratory for further analysis.
Antimicrobial susceptibility testing.
All Klebsiella isolates were examined for susceptibility to antimicrobial agents using the Bauer-Kirby disk diffusion method following the Clinical and Laboratory Standards Institute (CLSI) recommendations (33). A double-disk synergy test for screening ESBL-producing isolates was carried out as described previously (34). An additional determination of the ceftiofur MIC was carried out for those ESBL-producing isolates. E. coli ATCC 25922 was used as the control strain.
PFGE.
Pulsed-field gel electrophoresis (PFGE) was conducted in accordance with the Pulse Net protocol (www.cdc.gov/pulsenet) provided by the Centers for Disease Control and Prevention (CDC). The PFGE patterns were analyzed and clustered using BioNumerics (version 7.6) software (Applied Maths, Kortrijk, Belgium). Different PFGE clusters were represented in alphabetical order. Every band difference within a PFGE cluster resulted in adding a numerical order to the pulsed-field cluster.
Capsule locus typing.
Isolation of genomic DNA from every strain was performed using a PowerSoil DNA isolation kit (MO BIO Laboratories, Inc, Carlsbad, CA) according to the manufacturer’s instructions. A PCR targeting the capsule polysaccharide synthesis (cps) region in K. pneumoniae was used to identify the capsular types in this study (35). For the strains that had whole-genome sequencing data, we also used the Kaptive tool to identify the whole-capsule synthesis locus (K-locus), based on assembly scaffolds (36).
Whole-genome sequencing.
In total, 96 K. pneumoniae isolates with distinguishable PFGE profiles were subjected to whole-genome sequencing. DNA libraries were sequenced on a MiSeq 2000 platform (Illumina, Inc., San Diego, CA). The quality of the original reads was evaluated using the FASTQC tool. The FASTX-trimmer and Trimmomatic tools were used to do the trimming of sequencing reads (37). The sequencing reads were assembled into contigs using the SPAdes (version 3.10) algorithm (38), and plasmid sequences were assembled using the plasmidSPAdes algorithm (39). Moreover, the genomes of 185 K. pneumoniae KpI isolates from humans hospitalized with septicemia, pneumonia, and wound infection from several countries and 49 K. pneumoniae KpI isolates from bovines were accessed from a previous study (18). The bovine isolates from the study of Holt et al. (18) included 20 isolates from cows with clinical mastitis, 10 isolates from cows with subclinical mastitis, and 19 fecal or rumen isolates.
Phylogenetic analysis.
The core genome SNPs were identified by mapping sequence reads against a reference genome (K. pneumoniae strain NTUH-K2044, GenBank accession no. NC_012731.1) using the Burrows-Wheeler aligner (40). The plasmid sequences in K. pneumoniae strain NTUH-K2044 were excluded. The Genome Analysis Toolkit (GATK) was used for base quality score recalibration, indel realignment, and duplicate removal (41) and also for SNP and indel discovery, along with genotyping, using standard hard filtering parameters or variant quality score recalibration according to GATK best practices recommendations (42, 43). The alleles at all filtered SNP sites were concatenated to form a multiple-sequence alignment of all 308 K. pneumoniae KpI genomes. To construct a maximum likelihood phylogeny of the K. pneumoniae isolates, the RAxML program was used to calculate the phylogenetic tree from the combined alignment file with 1,000 bootstraps (44). The lineages of the phylogenetic tree were defined using the BAPS (version 6.0) program (45). We used Interactive Tree of Life (iTOL) (46) and FigTree (http://tree.bio.ed.ac.uk/software/figtree/) software to visualize and edit the phylogenetic tree.
Genome annotation and taxonomy.
For each de novo assembled genome, coding sequences were predicted using the Prodigal (version 2.6) program (47) and annotated using the rapid prokaryotic genome annotation tool Prokka (48). We concatenated all ORFs in all genomes into a single file. We then clustered all the ORFs into clusters that represented a group of highly similar proteins by using the UCLUST algorithm in the USEARCH program with a 90% identity cutoff (49). We extracted the seed sequence for every cluster group in order to do functional annotation, using the DIAMOND program (50), against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. The descriptive statistics of functional genes in bovine and humans were calculated in JMP Pro (version 12) software (SAS Institute Inc., Cary, NC) using the chi-square test.
Identification of virulence and AMR genes.
We detected known virulence genes and sequence conservation using a direct read mapping approach with the SRST2 program with a coverage cutoff of 90% (51). The reference sequence of virulence genes and alleles was retrieved from the K. pneumoniae BIGSdb database (https://bigsdb.pasteur.fr/), the virulence factor database (VFDB) (52), and the PathogenFinder database (53). Taxonomic investigation of virulence among genomic sequences was performed with the MG-RAST (version 4.0.2) metagenomics analysis server (http://metagenomics.anl.gov) by uploading the raw reads of sequencing to the server. The associations of virulence genes with clinical cases were calculated in SAS (version 9.3) software (SAS Institute Inc., Cary, NC).
The AMR genes were identified by use of the SRST2 program (51) according to the ResFinder database (54). The MLST of newly sequenced K. pneumoniae isolates was identified using the MLST (version 1.8) program (55). The new alleles were submitted to the MLST database (https://bigsdb.pasteur.fr/klebsiella/klebsiella.html) to assign new STs. Plasmid replicon types were detected using the PlasmidFinder (version 1.3) program (56).
Accession number(s).
The genome assemblies of newly sequenced strains and the plasmid sequence of pC5 (GenBank accession no. MF953243) have been deposited in the NCBI database under BioProject no. PRJNA414542 and PRJNA400493, respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by Agriculture and Food Research Initiative competitive grant no. 2013-67015-21233 from the USDA National Institute of Food and Agriculture.
We thank the team of curators at the Institut Pasteur MLST system (Paris, France) for importing novel alleles, profiles, and/or isolates at https://bigsdb.pasteur.fr/, Svetlana Ferreira Lima for help in whole-genome sequencing, Helen Korzec and Martin Zinicola for collecting the samples, Erika Korzune Ganda for help in data analysis, Qi Sun and Minghui Wang for assistance with genomic analysis, Julie Siler for help in antimicrobial susceptibility testing, Martin Wiedmann for providing the Salmonella sp. strain H9812 used in PFGE typing, Eva Heinz for help in capsule locus identification, and Daryl Henderson for help in preparing the manuscript.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/AEM.02654-18.
REFERENCES
- 1.Magill SS, Edwards JR, Bamberg W, Beldavs ZG, Dumyati G, Kainer MA, Lynfield R, Maloney M, McAllister-Hollod L, Nadle J, Ray SM, Thompson DL, Wilson LE, Fridkin SK, Emerging Infections Program Healthcare-Associated Infections and Antimicrobial Use Prevalence Survey Team. 2014. Multistate point-prevalence survey of health care-associated infections. N Engl J Med 370:1198–1208. doi: 10.1056/NEJMoa1306801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee W-H, Choi H-I, Hong S-W, Kim K-S, Gho YS, Jeon SG. 2015. Vaccination with Klebsiella pneumoniae-derived extracellular vesicles protects against bacteria-induced lethality via both humoral and cellular immunity. Exp Mol Med 47:e183. doi: 10.1038/emm.2015.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Borer A, Saidel-Odes L, Riesenberg K, Eskira S, Peled N, Nativ R, Schlaeffer F, Sherf M. 2009. Attributable mortality rate for carbapenem-resistant Klebsiella pneumoniae bacteremia. Infect Control Hosp Epidemiol 30:972–976. doi: 10.1086/605922. [DOI] [PubMed] [Google Scholar]
- 4.Ipekci T, Seyman D, Berk H, Celik O. 2014. Clinical and bacteriological efficacy of amikacin in the treatment of lower urinary tract infection caused by extended-spectrum beta-lactamase-producing Escherichia coli or Klebsiella pneumoniae. J Infect Chemother 20:762–767. doi: 10.1016/j.jiac.2014.08.007. [DOI] [PubMed] [Google Scholar]
- 5.Medrano-Galarza C, Gibbons J, Wagner S, de Passillé AM, Rushen J. 2012. Behavioral changes in dairy cows with mastitis. J Dairy Sci 95:6994–7002. doi: 10.3168/jds.2011-5247. [DOI] [PubMed] [Google Scholar]
- 6.Bar D, Tauer LW, Bennett G, Gonzalez RN, Hertl JA, Schukken YH, Schulte HF, Welcome FL, Grohn YT. 2008. The cost of generic clinical mastitis in dairy cows as estimated by using dynamic programming. J Dairy Sci 91:2205–2214. doi: 10.3168/jds.2007-0573. [DOI] [PubMed] [Google Scholar]
- 7.Hertl JA, Grohn YT, Leach JD, Bar D, Bennett GJ, Gonzalez RN, Rauch BJ, Welcome FL, Tauer LW, Schukken YH. 2010. Effects of clinical mastitis caused by gram-positive and gram-negative bacteria and other organisms on the probability of conception in New York State Holstein dairy cows. J Dairy Sci 93:1551–1560. doi: 10.3168/jds.2009-2599. [DOI] [PubMed] [Google Scholar]
- 8.Botrel MA, Haenni M, Morignat E, Sulpice P, Madec JY, Calavas D. 2010. Distribution and antimicrobial resistance of clinical and subclinical mastitis pathogens in dairy cows in Rhone-Alpes, France. Foodborne Pathog Dis 7:479–487. doi: 10.1089/fpd.2009.0425. [DOI] [PubMed] [Google Scholar]
- 9.Erskine RJ, Eberhart RJ, Hutchinson LJ, Spencer SB, Campbell MA. 1988. Incidence and types of clinical mastitis in dairy herds with high and low somatic cell counts. J Am Vet Med Assoc 192:761–765. [PubMed] [Google Scholar]
- 10.Olde Riekerink RG, Barkema HW, Kelton DF, Scholl DT. 2008. Incidence rate of clinical mastitis on Canadian dairy farms. J Dairy Sci 91:1366–1377. doi: 10.3168/jds.2007-0757. [DOI] [PubMed] [Google Scholar]
- 11.Cha E, Bar D, Hertl JA, Tauer LW, Bennett G, Gonzalez RN, Schukken YH, Welcome FL, Grohn YT. 2011. The cost and management of different types of clinical mastitis in dairy cows estimated by dynamic programming. J Dairy Sci 94:4476–4487. doi: 10.3168/jds.2010-4123. [DOI] [PubMed] [Google Scholar]
- 12.Bannerman DD. 2009. Pathogen-dependent induction of cytokines and other soluble inflammatory mediators during intramammary infection of dairy cows. J Anim Sci 87:10–25. doi: 10.2527/jas.2008-1187. [DOI] [PubMed] [Google Scholar]
- 13.Rajala-Schultz PJ, Gröhn YT. 2001. Comparison of economically optimized culling recommendations and actual culling decisions of Finnish Ayrshire cows. Prev Vet Med 49:29–39. [DOI] [PubMed] [Google Scholar]
- 14.Todhunter DA, Smith KL, Hogan JS, Schoenberger PS. 1991. Gram-negative bacterial infections of the mammary gland in cows. Am J Vet Res 52:184–188. [PubMed] [Google Scholar]
- 15.Erskine RJ, Bartlett PC, VanLente JL, Phipps CR. 2002. Efficacy of systemic ceftiofur as a therapy for severe clinical mastitis in dairy cattle. J Dairy Sci 85:2571–2575. doi: 10.3168/jds.S0022-0302(02)74340-3. [DOI] [PubMed] [Google Scholar]
- 16.Roberson JR, Warnick LD, Moore G. 2004. Mild to moderate clinical mastitis: efficacy of intramammary amoxicillin, frequent milk-out, a combined intramammary amoxicillin, and frequent milk-out treatment versus no treatment. J Dairy Sci 87:583–592. doi: 10.3168/jds.S0022-0302(04)73200-2. [DOI] [PubMed] [Google Scholar]
- 17.Grohn YT, Gonzalez RN, Wilson D, Hertl JA, Bennett G, Schulte H, Schukken YH. 2005. Effect of pathogen-specific clinical mastitis on herd life in two New York State dairy herds. Prev Vet Med 71:105–125. doi: 10.1016/j.prevetmed.2005.06.002. [DOI] [PubMed] [Google Scholar]
- 18.Holt KE, Wertheim H, Zadoks RN, Baker S, Whitehouse CA, Dance D, Jenney A, Connor TR, Hsu LY, Severin J, Brisse S, Cao H, Wilksch J, Gorrie C, Schultz MB, Edwards DJ, Nguyen KV, Nguyen TV, Dao TT, Mensink M, Minh VL, Nhu NT, Schultsz C, Kuntaman K, Newton PN, Moore CE, Strugnell RA, Thomson NR. 2015. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A 112:E3574. doi: 10.1073/pnas.1501049112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sundin GW, Bender CL. 1996. Dissemination of the strA-strB streptomycin-resistance genes among commensal and pathogenic bacteria from humans, animals, and plants. Mol Ecol 5:133–143. [DOI] [PubMed] [Google Scholar]
- 20.Wang J, Stephan R, Power K, Yan Q, Hachler H, Fanning S. 2014. Nucleotide sequences of 16 transmissible plasmids identified in nine multidrug-resistant Escherichia coli isolates expressing an ESBL phenotype isolated from food-producing animals and healthy humans. J Antimicrob Chemother 69:2658–2668. doi: 10.1093/jac/dku206. [DOI] [PubMed] [Google Scholar]
- 21.Cullik A, Pfeifer Y, Prager R, von Baum H, Witte W. 2010. A novel IS26 structure surrounds blaCTX-M genes in different plasmids from German clinical Escherichia coli isolates. J Med Microbiol 59:580–587. doi: 10.1099/jmm.0.016188-0. [DOI] [PubMed] [Google Scholar]
- 22.Dolejska M, Villa L, Hasman H, Hansen L, Carattoli A. 2013. Characterization of IncN plasmids carrying blaCTX-M-1 and qnr genes in Escherichia coli and Salmonella from animals, the environment and humans. J Antimicrob Chemother 68:333–339. doi: 10.1093/jac/dks387. [DOI] [PubMed] [Google Scholar]
- 23.Schink AK, Kadlec K, Schwarz S. 2012. Detection of qnr genes among Escherichia coli isolates of animal origin and complete sequence of the conjugative qnrB19-carrying plasmid pQNR2078. J Antimicrob Chemother 67:1099–1102. doi: 10.1093/jac/dks024. [DOI] [PubMed] [Google Scholar]
- 24.Paterson ES, More MI, Pillay G, Cellini C, Woodgate R, Walker GC, Iyer VN, Winans SC. 1999. Genetic analysis of the mobilization and leading regions of the IncN plasmids pKM101 and pCU1. J Bacteriol 181:2572–2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moland ES, Black JA, Hossain A, Hanson ND, Thomson KS, Pottumarthy S. 2003. Discovery of CTX-M-like extended-spectrum beta-lactamases in Escherichia coli isolates from five U.S. states. Antimicrob Agents Chemother 47:2382–2383. doi: 10.1128/AAC.47.7.2382-2383.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lewis JS II, Herrera M, Wickes B, Patterson JE, Jorgensen JH. 2007. First report of the emergence of CTX-M-type extended-spectrum beta-lactamases (ESBLs) as the predominant ESBL isolated in a U.S. health care system. Antimicrob Agents Chemother 51:4015–4021. doi: 10.1128/AAC.00576-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G, Huang T, Surendraiah PK, Wang K, Komal R, Zhuge J, Chern CR, Kryszuk AA, King C, Wormser GP. 2013. CTX-M beta-lactamase-producing Klebsiella pneumoniae in suburban New York City, New York, USA. Emerg Infect Dis 19:1803–1810. doi: 10.3201/eid1911.121470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wandersman C, Delepelaire P. 2004. Bacterial iron sources: from siderophores to hemophores. Annu Rev Microbiol 58:611–647. doi: 10.1146/annurev.micro.58.030603.123811. [DOI] [PubMed] [Google Scholar]
- 29.Huang SH, Wang CK, Peng HL, Wu CC, Chen YT, Hong YM, Lin CT. 2012. Role of the small RNA RyhB in the Fur regulon in mediating the capsular polysaccharide biosynthesis and iron acquisition systems in Klebsiella pneumoniae. BMC Microbiol 12:148. doi: 10.1186/1471-2180-12-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ma LC, Fang CT, Lee CZ, Shun CT, Wang JT. 2005. Genomic heterogeneity in Klebsiella pneumoniae strains is associated with primary pyogenic liver abscess and metastatic infection. J Infect Dis 192:117–128. doi: 10.1086/430619. [DOI] [PubMed] [Google Scholar]
- 31.Skaar EP. 2010. The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog 6:e1000949. doi: 10.1371/journal.ppat.1000949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wenz JR, Garry FB, Barrington GM. 2006. Comparison of disease severity scoring systems for dairy cattle with acute coliform mastitis. J Am Vet Med Assoc 229:259–262. doi: 10.2460/javma.229.2.259. [DOI] [PubMed] [Google Scholar]
- 33.Clinical and Laboratory Standards Institute. 2015. Performance standards for antimicrobial susceptibility testing; 25th informational supplement. M100-S25 Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 34.Luzzaro F, Mantengoli E, Perilli M, Lombardi G, Orlandi V, Orsatti A, Amicosante G, Rossolini GM, Toniolo A. 2001. Dynamics of a nosocomial outbreak of multidrug-resistant Pseudomonas aeruginosa producing the PER-1 extended-spectrum beta-lactamase. J Clin Microbiol 39:1865–1870. doi: 10.1128/JCM.39.5.1865-1870.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pan YJ, Lin TL, Chen YH, Hsu CR, Hsieh PF, Wu MC, Wang JT. 2013. Capsular types of Klebsiella pneumoniae revisited by wzc sequencing. PLoS One 8:e80670. doi: 10.1371/journal.pone.0080670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wyres KL, Wick RR, Gorrie C, Jenney A, Follador R, Thomson NR, Holt KE. 2016. Identification of Klebsiella capsule synthesis loci from whole genome data. Microb Genom 2:e000102. doi: 10.1099/mgen.0.000102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Lesin VM, Nikolenko SI, Pham S, Prjibelski AD, Pyshkin AV, Sirotkin AV, Vyahhi N, Tesler G, Alekseyev MA, Pevzner PA. 2012. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Antipov D, Hartwick N, Shen M, Raiko M, Lapidus A, Pevzner PA. 2016. plasmidSPAdes: assembling plasmids from whole genome sequencing data. Bioinformatics 32:3380–3387. doi: 10.1093/bioinformatics/btw493. [DOI] [PubMed] [Google Scholar]
- 40.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.DePristo MA, Banks E, Poplin R, Garimella KV, Maguire JR, Hartl C, Philippakis AA, del Angel G, Rivas MA, Hanna M, McKenna A, Fennell TJ, Kernytsky AM, Sivachenko AY, Cibulskis K, Gabriel SB, Altshuler D, Daly MJ. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet 43:491–498. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Van der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy-Moonshine A, Jordan T, Shakir K, Roazen D, Thibault J, Banks E, Garimella KV, Altshuler D, Gabriel S, DePristo MA. 2013. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr Protoc Bioinformatics 43:11.10.1–11.10.33. doi: 10.1002/0471250953.bi1110s43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Stamatakis A. 2006. RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]
- 45.Cheng L, Connor TR, Siren J, Aanensen DM, Corander J. 2013. Hierarchical and spatially explicit clustering of DNA sequences with BAPS software. Mol Biol Evol 30:1224–1228. doi: 10.1093/molbev/mst028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Letunic I, Bork P. 2016. Interactive Tree of Life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hyatt D, Chen GL, Locascio PF, Land ML, Larimer FW, Hauser LJ. 2010. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 11:119. doi: 10.1186/1471-2105-11-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seemann T. 2014. Prokka: rapid prokaryotic genome annotation. Bioinformatics 30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 49.Edgar RC. 2010. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. doi: 10.1093/bioinformatics/btq461. [DOI] [PubMed] [Google Scholar]
- 50.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. doi: 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 51.Inouye M, Dashnow H, Raven LA, Schultz MB, Pope BJ, Tomita T, Zobel J, Holt KE. 2014. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med 6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen L, Zheng D, Liu B, Yang J, Jin Q. 2016. VFDB 2016: hierarchical and refined dataset for big data analysis—10 years on. Nucleic Acids Res 44:D694–D697. doi: 10.1093/nar/gkv1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cosentino S, Voldby Larsen M, Moller Aarestrup F, Lund O. 2013. PathogenFinder—distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zankari E, Hasman H, Cosentino S, Vestergaard M, Rasmussen S, Lund O, Aarestrup FM, Larsen MV. 2012. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother 67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Larsen MV, Cosentino S, Rasmussen S, Friis C, Hasman H, Marvig RL, Jelsbak L, Sicheritz-Ponten T, Ussery DW, Aarestrup FM, Lund O. 2012. Multilocus sequence typing of total-genome-sequenced bacteria. J Clin Microbiol 50:1355–1361. doi: 10.1128/JCM.06094-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Carattoli A, Zankari E, Garcia-Fernandez A, Larsen MV, Lund O, Villa L, Aarestrup FM, Hasman H. 2014. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother 58:3895–3903. doi: 10.1128/AAC.02412-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.