Bacteriocins are regarded as potential alternatives for antibiotics in the absence of highly resistant bacteria. In particular, two-peptide (class IIb) bacteriocins exhibit the maximum activity through the synergy of two components, and their antimicrobial spectra are known to be relatively wide. However, there are few reports of synergistic activity of class IIb bacteriocins determined by isolation and purification of individual peptides. Our results clarified the interaction of each class IIb component peptide for GT and GS via the construction of homologous mutants, which were not dependent on the purification. These data may contribute to understanding the mechanisms of action by which class IIb bacteriocins exhibit wide antibacterial spectra.
KEYWORDS: Lactobacillus gasseri, antimicrobial peptide, bacteriocin, homologous expression, lactic acid bacteria, multiple transporter, synergistic effect
ABSTRACT
Lactobacillus gasseri LA327, isolated from the large intestine tissue in humans, is a bacteriocinogenic strain with two kinds of class IIb bacteriocin structural genes, i.e., those for gassericin T (GT) and acidocin LF221A (Acd LF221A). In this study, DNA sequencing of the genes for GT and Acd LF221A from L. gasseri LA327 revealed that the amino acid sequences for GT corresponded with those for GT genes, except for GatK (histidine kinase). However, Acd LF221A genes had analogues which differed in at least one amino acid residue, to encode a class IIb bacteriocin designated gassericin S (GS). The LA327 strain retained antimicrobial activity after the deletion of the GT structural genes (gatAX); however, both GS and GT activities were lost by deletion of the putative ABC transporter gene (gatT). This indicates that the LA327 strain produces GS and GT and that GS secretion is performed via GT genes with the inclusion of gatT. Homologous expression using deletion mutants of GS and GT, each containing a single peptide, elucidated that GS (GasAX) and GT (GatAX) showed synergistic activity as class IIb bacteriocins and that no synergistic activity was observed between GS and GT peptides. The molecular mass of GS was estimated to be theoretical ca. 5,400 Da by in situ activity assay after SDS-PAGE, clarifying that GS was actually expressed as an active class IIb bacteriocin. Furthermore, the stability of expressed GS to pH, heat, and protease was determined.
IMPORTANCE Bacteriocins are regarded as potential alternatives for antibiotics in the absence of highly resistant bacteria. In particular, two-peptide (class IIb) bacteriocins exhibit the maximum activity through the synergy of two components, and their antimicrobial spectra are known to be relatively wide. However, there are few reports of synergistic activity of class IIb bacteriocins determined by isolation and purification of individual peptides. Our results clarified the interaction of each class IIb component peptide for GT and GS via the construction of homologous mutants, which were not dependent on the purification. These data may contribute to understanding the mechanisms of action by which class IIb bacteriocins exhibit wide antibacterial spectra.
INTRODUCTION
Lactic acid bacteria (LAB) have a long history of consumption by humans and inhabit various ecosystems, including gastrointestinal (GI) tracts of humans and animals and other environments (for instance, vegetables, milks, and meats). In addition, lactobacilli are recognized as important members of the beneficial GI microbiota in humans and animals, along with bifidobacteria (1). In particular, Lactobacillus gasseri is one of the predominant species of lactobacilli in the human small intestine, and it has been isolated not only from the GI tract and feces but also from the oral and the vaginal cavities and mammary areola. It is considered a promising candidate for probiotics (2–5).
Many researchers have reported the beneficial effects of L. gasseri on hosts, including immunoregulation, the alleviation of allergic symptoms, prevention of bacterial and viral infections, the antitumor effect, and the inhibition of lipid absorption (6–11). Indeed, at least three yogurt brands and one supplement containing L. gasseri strains are commercially sold in Japan to provide the benefits mentioned above to consumers. It has been known that LAB may produce various antimicrobial agents, such as lactic acids, diacetyl, hydrogen peroxide, and bacteriocins, to survive in competitive microbial niches (12–14).
Bacteriocins are defined as ribosomally synthesized antimicrobial peptides and proteins from microorganisms (15). LAB bacteriocins especially are regarded as a potential alternative to antibiotics due to several advantageous properties, such as their potency (as determined in vitro and in vivo), variety of antimicrobial spectra (both narrow and broad), low toxicity, and ease of the bioengineering (16). Presently, bacteriocins from LAB are classified into three major classes: ribosomally produced and posttranslationally modified peptides (RiPPs) (less than 10 kDa, class I), unmodified bacteriocins less than 10 kDa (class II), and unmodified bacteriocins greater than 10 kDa (class III). Furthermore, class I is subclassified into six subclasses: class Ia (lanthipeptides), class Ib (head-to-tail cyclized peptides), class Ic (sactibiotics), class Id [linear azol(in)e-containing peptides], class Ie (glycocins), and class If (lasso peptides). Additionally, class II is subclassified into four subclasses: class IIa (pediocin-like bacteriocins), class IIb (two-peptide bacteriocins), class IIc (leaderless bacteriocins), and class IId (non-pediocin-like, single-peptide bacteriocins) (17).
Previously, we reported that gassericin A (GA), a head-to-tail cyclized bacteriocin (class IId) from L. gasseri LA39 (18), and gassericin T (GT), a putative two-peptide bacteriocin (class IIb), consisted of GatA and GatX from L. gasseri SBT2055 and LA158 (19, 20). In addition, Bogovic-Matijašić et al. (21) reported that acidocin LF221A (Acd LF221A) and acidocin LF221B (Acd LF221B) were produced by L. gasseri LF221; the latter was an analogue of GatX (the second peptide of GT) containing alanine at position 50 instead of glycine in GatX, and the former was potentially a novel bacteriocin derived from L. gasseri. Recently, we isolated L. gasseri LA327 harboring the structural genes for GT (gatAX) and Acd LF221A (acd 221αA), detected by PCR from human large intestine tissue. In addition to GT, Acd LF221A, and Acd LF221B, gassericin K7A and gassericin K7B from L. gasseri K7 and gassericin E from L. gasseri EV1461 have been reported as class IIb bacteriocins produced by L. gasseri (22–24). The individual peptides of Acd LF221A (Acd 221A), Acd LF221B (Acd 221B), gassericin K7A (GasK7A_β), gassericin K7B (GasK7B_β), and gassericin E (GaeA) have antibacterial activity, and no increase of the activity was recognized by combination assay with Acd 221A and Acd 221B. However, complete purification of two peptides of class IIb bacteriocins produced by L. gasseri has never been achieved, and the synergistic activity produced by coexistence of each peptide, which should be an essential character for class IIb bacteriocin, has still not been demonstrated.
In this study, we focused on identification and characterization of two class IIb bacteriocins detected from L. gasseri LA327 and demonstration of the two-component action through homologous expression of these bacteriocins using genetically constructed mutant strains.
RESULTS
DNA sequencing and genetic analysis.
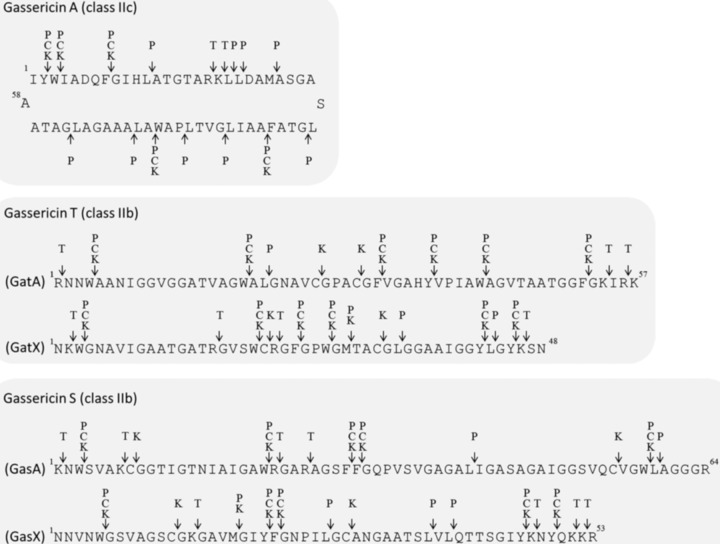
The nucleotide sequencing of the region surrounding gatAX and gasAX (like that for acidocin LF221A and gassericin K7A) on the chromosomal DNA of Lactobacillus gasseri LA327 highlighted 6,935-bp and 1,143-bp DNA sequences harboring nine and three open reading frames (ORFs) (DDBJ accession no. LC389592 and LC389591), respectively. Homology searching of the predicted amino acid sequences for peptides and proteins encoded in each ORF revealed that the nine products for the gat operon matched those of L. gasseri LA158 (GenBank accession no. AB710328), with the exception of one amino acid residue in GatK (the amino acid residue at position 168 is valine in LA327 instead of alanine in LA158 [data not shown]), and the three ORFs for the gas operon were not detected in LA158. Furthermore, the gas and gat operons were similar to those of acidocin LF221A (Acd LF221A) and gassericin K7A and to those of acidocin LF221B (Acd LF221B) and gassericin K7B, respectively (Tables 1 and 2). However, the sequences of two class IIb bacteriocins for the gas and gat operons in L. gasseri LA327 were slightly different (in the range of 0 to 2 amino acid residues) from those for Acd LF221A and Acd LF221B (GenBank accession no. AY295874.1 and AY297947.1) (Fig. 1A and B), so the new putative bacteriocin (derived from the gas operon) was designated as gassericin S (GasA and GasX).
TABLE 1.
Deduced peptides and proteins encoded in gat operon of Lactobacillus gasseri LA327
| ORF | Protein or peptide |
|||||
|---|---|---|---|---|---|---|
| Length (amino acids) | Mol wt | pI | Function | Homology search resulta (identity [residual ratio, %]) | Localization (no. of TMS)b | |
| gatP | 50 | 5,852 | 9.83 | Inducer peptide precursor | Lactacin F inducer peptide precursor of L. gasseri K7 (50/50, 100) | Soluble (0) |
| gatK | 435 | 50,444 | 5.2 | Histidine kinase | Histidine kinase of L. gasseri K7 (430/435, 99) | Membrane (6) |
| gatR | 265 | 30,280 | 5.36 | Response regulator | Chemotaxis protein CheY of L. gasseri K7 (265/265, 100) | Soluble (0) |
| gatT | 719 | 81,054 | 7.71 | ABC transporter permease component | Peptide ABC transporter ATP-binding protein of L. gasseri K7 (718/719, 99) | Membrane (6) |
| gatC | 197 | 21,938 | 10.13 | Putative accessory protein | Putative gassericin K7 B accessory protein (196/197, 99) | Membrane (1) |
| gatZ | 33 | 3,924 | 7.57 | Putative uncharacterized protein | Unknown protein of L. gasseri LF221 (33/33, 100) | Soluble (0) |
| gatA | 75 | 7,480 | 10.02 | GatA precursor | Putative complement factor Acd 221b of L. gasseri LF221 (75/75, 100) | Membrane (1) |
| gatX | 65 | 6,673 | 10.11 | GatX precursor | Acidocin LF221B Acd 221B of L. gasseri LF221 (64/65, 98) | Soluble (0) |
| gatI | 112 | 13,343 | 9.81 | Immunity protein | Putative immunity protein Aci 221B of L. gasseri LF221 (112/112, 100) | Membrane (4) |
Homology searching was done with the BLAST program (http://blast.ddbj.nig.ac.jp/top-j.html).
Deduced using the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM/).
TABLE 2.
Deduced peptides and proteins encoded in the gas operon of Lactobacillus gasseri LA327
| ORF | Protein or peptide |
|||||
|---|---|---|---|---|---|---|
| Length (amino acids) | Mol wt | pI | Function | Homology search resulta (identity [residual ratio, %]) | Localization (no. of TMS)b | |
| gasA | 79 | 7,662 | 10.27 | GasA precursor | Gassericin K7 A complementary factor (78/79, 99) | Membrane (1)c |
| gasX | 69 | 7,287 | 10.00 | GasX precursor | Acidocin LF221A Acd 221A (67/69, 97) | Membrane (1)c |
| gasI | 90 | 10,789 | 7.42 | Putative immunity protein | Gassericin K7 A immunity protein (90/90,100) | Membrane (2) |
Homology searching was done with the BLAST program (http://blast.ddbj.nig.ac.jp/top-j.html).
Deduced using the TMHMM program (http://www.cbs.dtu.dk/services/TMHMM/).
With putative mature peptide.
FIG 1.
(A) Comparison of deduced amino acid sequences for gassericin S (GasA and GasX) and acidocin LF221A (Acd 221α and Acd 221A). Arrows, underlining, and bold indicate predicted cleavage sites, putative transmembrane domains, and amino acid residue differences between GS and Acd LF221A, respectively. (B) Comparison of deduced amino acid sequences for gassericin T (GatA and GatX) and acidocin LF221B (Acd 221β and Acd 221B). Arrows, underlining, and bold indicate predicted cleavage sites, putative transmembrane domains, and amino acid residue differences between GT and Acd LF221B, respectively.
Verification of the GS production mechanism.
GT activity (15,754 arbitrary units [AU]/ml) in the culture supernatant (cell-free supernatant [CFS]) of L. gasseri LA158 was completely lost by deletion of the GT structural genes (gatAX) from chromosomal DNA of LA158 (LA158 ΔgatAX), and the bacteriocin activity of L. gasseri LA327 was retained and increased (from 16 AU/ml to 64 AU/ml) after gatAX was eliminated (LA327 ΔgatAX). However, the bacteriocin activity in the CFS of LA327 ceased after elimination of the gene for the putative ABC transporter (gatT) from the chromosomal DNA of LA327 (LA327 ΔgatT), suggesting that gassericin S (GS) is a bacteriocin secreted through GatT.
Synergistic activity among the peptides of GS and GT.
Bacteriocin activities (62 AU/ml and 123 AU/ml) were detected in the CFSs of coproducer strains for GasAX [L. gasseri LA158 ΔgatAX(pGS-AX) and LA158 ΔgatAX(pGS-AXI), respectively], and no activities were obtained in the CFSs of single-producer strains for GasA and GasX [L. gasseri LA158 ΔgatAX(pGS-AXΔX), LA158 ΔgatAX(pGS-AXIΔX), LA158 ΔgatAX(pGS-AXΔA), and LA158 ΔgatAX(pGS-AXIΔA)], even when concentrated 40-fold (Table 3). On the other hand, a mixture of equal amounts of GasA (GasAI) and GasX (GasXI) showed bacteriocin activity (Table 3).
TABLE 3.
Antibacterial activities of Gas peptides in alone and combination
| CFSa | Bacteriocin activityb (AU/ml) |
|---|---|
| GasA | NDc |
| GasX | ND |
| GasAI | ND |
| GasXI | ND |
| GasAX | 62 |
| GasAXI | 123 |
| GasA + GasX (1:1) | 62 |
| GasA + GasXI (1:1) | 31 |
| GasAI + GasX (1:1) | 62 |
| GasAI + GasXI (1:1) | 31 |
GasA, cell-free supernatant (CFS) of L. gasseri LA158 ΔgatAX(pGS-AXΔX); GasX, CFS of L. gasseri LA158 ΔgatAX(pGS-AXΔA); GasAI, CFS of L. gasseri LA158 ΔgatAX(pGS-AXIΔX); GasXI, CFS of L. gasseri LA158 ΔgatAX(pGS-AXIΔA); GasAX, CFS of L. gasseri LA158 ΔgatAX(pGS-AX); GasAXI, CFS of L. gasseri LA158 ΔgatAX(pGS-AXI).
n > 3. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2).
ND, not detected. Approximately 40-fold-concentrated CFS was used.
Similarly, the assay for combination activity of component peptides of GT (GatA and GatX) in the CFSs of single-producer strains for GatA and GatX (L. gasseri LA158 ΔgatX and LA158 ΔgatA) clarified that GatA displayed activity (62 AU/ml) when alone, and remarkably high activity (1,969 AU/ml) was observed with a combination of GatA and GatX (Table 4). In the combination activity assay between each GS and GT component peptide, a remarkably high activity level was not observed (Table 5).
TABLE 4.
Combined activity of GatA and GatX
GatA, CFS of L. gasseri LA158 ΔgatX; GatX, CFS of L. gasseri LA158 ΔgatA.
n > 3. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2).
ND, not detected. Approximately 40-fold-concentrated CFS was used.
TABLE 5.
Combined activity of GS and GT components
| CFSa | Theoretical activity (AU/ml) | Actual activityb (AU/ml) |
|---|---|---|
| GasA + GatA | (NDc + 62)/2 | 31 |
| GasA + GatX | (ND + ND)/2 | ND |
| GasX + GatA | (ND + 62)/2 | 31 |
| GasX + GatX | (ND + ND)/2 | ND |
| GasA + GasX + GatA + GatX | (123 + 1,969)/2 | 985 |
GasA, CFS of L. gasseri LA158 ΔgatAX(pGS-AXIΔX); GasX, CFS of L. gasseri LA158 ΔgatAX(pGS-AXΔA); GatA, CFS of L. gasseri LA158 ΔgatX; GatX, CFS of L. gasseri LA158 ΔgatA. CFSs were used in equal amounts.
n > 3. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2).
ND, not detected.
Synergistic activities of GS (GasA plus GasX) and GT (GatA plus GatX) were detected with isobolograms (Fig. 2 and 3), while no synergistic activity was obtained for the combination of GS (GasA plus GasX) plus GT (GatA plus GatX) (Fig. 4).
FIG 2.
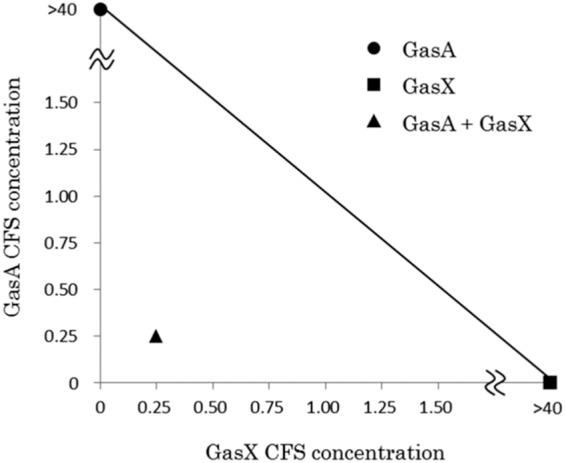
Isobologram assay of GS (GasA plus GasX). CFS, cell-free supernatant. Concentrations: 1, undiluted CFS; <1, diluted CFS; >1, concentrated CFS. All data for bacteriocin activities were measured with at least triplicates.
FIG 3.
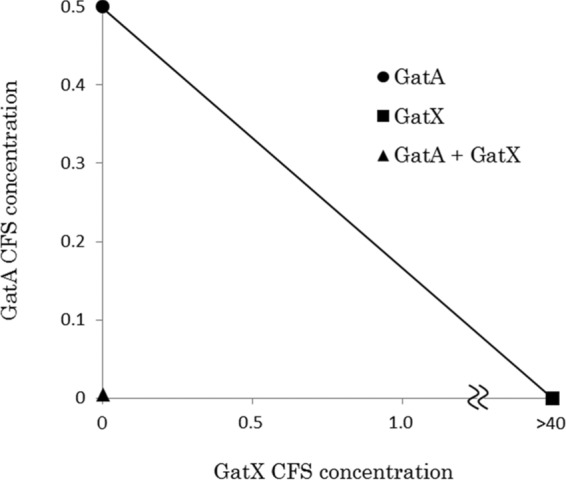
Isobologram assay of GT (GatA plus GatX). CFS, cell-free supernatant. Concentrations: 1, undiluted CFS; <1, diluted CFS; >1, concentrated CFS. All data for bacteriocin activities were measured with at least triplicates.
FIG 4.
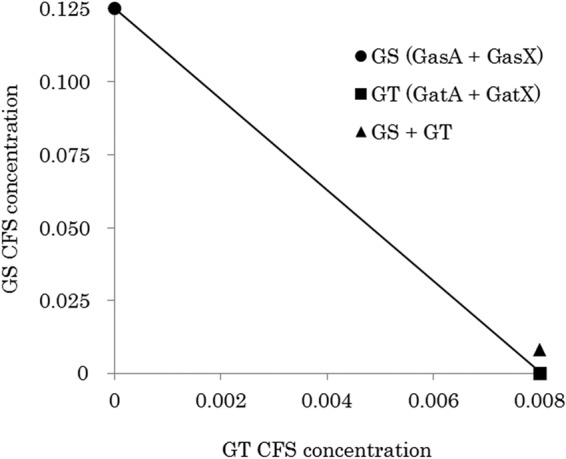
Isobologram assay of GS and GT. CFS, cell-free supernatant. Concentrations: 1, undiluted CFS; <1, diluted CFS; >1, concentrated CFS. All data for bacteriocin activities were measured with at least triplicates.
In situ activity assay for GS.
A clear zone derived from GS appeared in the in situ activity assay after SDS-PAGE, and the molecular mass of GS was found to be approximately 5,400 Da by using a standard curve constructed with the electrophoresis distance of each molecular marker (Fig. 5).
FIG 5.
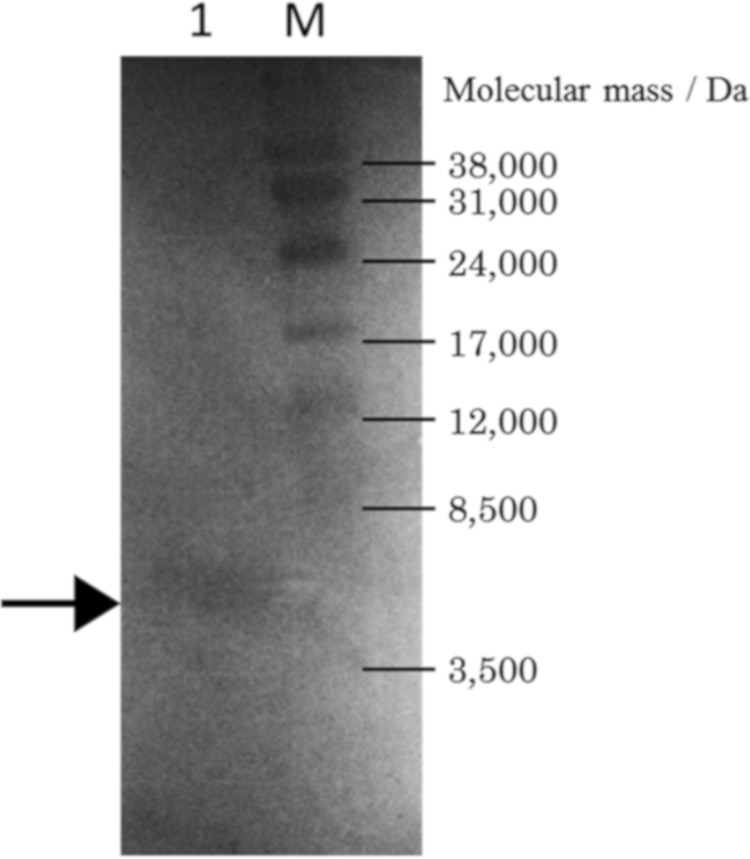
In situ activity assay of semipurified gassericin S. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2). Lane M, molecular weight markers; lane1, semipurified gassericin S. The arrow indicates the clear zone of gassericin S.
GS stability against pH, heat, and proteolysis.
The stability of GS under various pH, thermal, and proteolytic conditions was investigated. The bacteriocin activity of GS in the CFS was stable in a wide pH range (pH 2, 4, 7, and 10) (data not shown), and 25% of GS activity was retained under the most severe heating condition (121°C, 15 min) used in the tests (Table 6). The GS activity decreased to 25% under incubation at 37°C but was maintained at 4°C (Table 6). Through examination of the proteolysis resistance of GA, GT, and GS, it was determined that there were no bacteriocins showing tolerance against proteinase K. GS completely lost activity upon treatment with all proteases used in this study, and GT had a slight tolerance only for pepsin. On the other hand, the bacteriocin activity of GA, a circular bactericidal peptide, was detected after treatment with all proteases used, with the exception of proteinase K (Table 7).
TABLE 6.
Heat stability of gassericin Sa
| Condition | Bacteriocin activity,b AU/ml (residual rate, %) |
|---|---|
| Untreated | 123 |
| 121°C, 15 min | 31 (25) |
| 95°C, 5 min | 62 (50) |
| 70°C, 1 h | 62 (50) |
| 37°C, 1 mo | 31 (25) |
| 4°C, 1 mo | 123 (100) |
Determined with the CFS of L. gasseri LA158 ΔgatAX(pGS-AXI).
n > 3. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2).
TABLE 7.
Proteolysis sensitivity of bacteriocins produced by Lactobacillus gasseri
| Protease | Residual activity of bacteriocin, AU/ml (rate, %)a |
||
|---|---|---|---|
| GA | GT | GS | |
| None | 985 | 31,508 | 246 |
| Pepsin | 492 (50) | 492 (1.6) | NDb |
| Trypsin | 62 (6.3) | ND | ND |
| Alpha-chymotrypsin | 246 (25) | ND | ND |
| Proteinase K | ND | ND | ND |
n > 3. Indicator, L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2). GA, gassericin A (CFS of L. gasseri LA39); GT, gassericin T (CFS of L. gasseri LA158); GS, gassericin S (CFS of L. gasseri LA327).
ND, not detected.
DISCUSSION
In this study, we determined the complete sequence of a gassericin S (GS) gene cluster constituted by three ORFs containing the structural genes (gasAX) and the predicted immunity gene (gasI). Although the genes encoding bacteriocin precursors, immunity peptides/proteins (frequently found near the structural gene [25]), and transporter proteins are required for bacteriocin synthesis (26), no genes related to regulation and transport of GS were recognized in the surrounding areas of the gas operon. Deletion of the gassericin T (GT) transporter gene (gatT) clarified that GatT works as a multiple transporter for GT and GS, like EnkT which is responsible for the secretion of three enterocins from Enterococcus faecium NKR-5-3 (27). The complete isolation and purification of class IIb bacteriocins from Lactobacillus gasseri were not achieved, and synergy between the component peptides has never been reported. In this study, we constructed recombinant strains expressing a single peptide of GS and GT and demonstrated the existence of synergistic activities for each gassericin (Fig. 2 and 3), indicating that GS and GT are class IIb bacteriocins.
Majhenič et al. (28) reported that acidocins LF221A and LF221B, which have two- and one-amino-acid variants of the second peptides (GasX and GatX) for GS and GT, respectively, inhibited the growth of Lactobacillus sakei NCDO2714 when isolated. Although difference in the indicator strains used may be related to the different results, they are more likely due to the variation of the amino acid residues in acidocin LF221A and LF221B. Influences of single-amino-acid variation between the second peptides of enterocin C and enterocin 1017 on their antimicrobial spectra have been clarified (29). However, inhibition against Lactobacillus delbrueckii subsp. bulgaricus LMG 6901T (JCM 1002T) of the second peptide for gassericin K7B, 100% homologous to the GT second peptide, was reported (23), indicating that the structure of the gassericin K7B second peptide (active) may be different from that of the GT second peptide (inactive) in this study, owing to the test solutions and/or expression hosts used, in spite of the same indicator strain and amino acid sequences being used. Component peptides of the class IIb bacteriocin carnobacteriocin XY showed a high α-helix content in trifluoroethanol solution, while neither peptide showed remarkable secondary structure in aqueous conditions (30). Furthermore, our previous attempts to isolate and purify individual GT peptides from the CFS of L. gasseri LA158 failed because of polymerization of the peptides in 60% 2-propanol at the final step for high-pressure liquid chromatography (HPLC) purification (20). Therefore, the organic solvents used for bacteriocin extraction may be involved in the finding that individual activity of a second peptide of each class IIb bacteriocin from L. gasseri has been reported. In contrast, the activity exhibited by only a first peptide of gassericin E (one-amino-acid variants of the GT first peptide, W22L) (24) supports that the GT first peptide has antibacterial activity alone. Moreover, in this study, a putative immunity protein, GasI, promoted the production of GS, while production of individual peptides for GS was not affected by gasI expression (Table 3). Ra et al. (31) reported that deletion of nisI, encoding the immunity protein of nisin A, led to a decrease in nisin production. These results indicate that the resistance of GasI may be unnecessary for production of each inactive single peptide for GS, while GasI supports active GS production.
It is not rare that multiple bacteriocins are produced by a single strain, but many questions about the advantages of this still remain. In this study, synergistic activity of a cross-combination of GT and GS was not shown (Fig. 4). Similarly, no synergistic activity was observed in a cross-combination of plantaricin EF and plantaricin JK (32), but high activity was obtained in the case of the hemilateral peptide of lactococcin G and the complementary peptide of enterocin 1017 and lactococcin Q (33–35). It seems that presentation of cross-combination activity is dependent on amino acid sequence similarity, as demonstrated by the low similarity of GS and GT (ca. 26% as calculated by BLAST [https://blast.ncbi.nlm.nih.gov/Blast.cgi]) and the high similarity of lactococcin G and enterocin 1017 or lactococcin Q (ca. 88% and 57%, respectively).
After tests at various pHs and temperatures, the bacteriocin activity of GS produced by L. gasseri LA158 ΔgatAX(pGS-AXI) remained, even when the CFS was exposed to pH 2 to 10 (data not shown) and heated at 121°C for 15 min (Table 6). However, the activity decreased remarkably with longer incubation at 37°C, which differed from holding at 4°C (Table 6). In our previous study, the bacteriocin activity of GT produced by LA158 similarly decreased during incubation at 37°C, but the rate of decay of the activity was weakened after heat treatment of the CFS containing GT at 99°C for 10 min (data not shown), suggesting that the degradation of GT and GS activity may be caused by an extracellular protease(s) derived from LA158, used as the GS expression host.
In proteolysis sensitivity testing, the activities of GT and GS were greatly reduced and then completely disappeared with all proteases used in the test, and GA had a high protease tolerance, as expected from reports that peptides and proteins having cyclic structures are generally stable when exposed to heat, denaturant, and protease treatments (36). These characteristics would be available for application at various stages through the combination of GA and GT/GS. For instance, we reported that GA and GT prevented the growth of Staphylococcus aureus isolated from mastitis milk and breast organs (37, 38). In particular, GA is expected to sustain bacteriocin activity, due to its high stability, not only as a food preservative but also as a mastitis therapeutic agent, and GT and GS may be used as safe food preservatives that will be degraded in the human intestine after intake because of their high sensitivity to proteinases. Although most cleavage sites of pepsin (FYWML) overlap with those of alpha-chymotrypsin (FYW) and proteinase K (FYWMC), the effects of each protease treatment on the activities of GA, GT, and GS were different (Table 7; Fig. 6), indicating that the amounts of protease units and tertiary structures of gassericins, rather than cleavage sites, may be important for maintaining bacteriocin activities. The elucidation of the antibacterial spectra, mode of action, and immunity system for GS is still in progress.
FIG 6.
Predicted cleavage sites for each protease on gassericins A, T, and S. Arrows indicate the predicted cleavage sites for each protease (P, pepsin; C, alpha- chymotrypsin; K, proteinase K; T, trypsin).
MATERIALS AND METHODS
Bacterial strains, plasmids, and media.
The strains and plasmids used in this study are summarized in Table 8. The gassericin T (GT) producer Lactobacillus gasseri LA158 was isolated from the feces of a six-month-old female human infant. The gassericin S (GS) and GT producer L. gasseri LA327 was isolated from the large intestine of an adult human. L. gasseri strains and the indicator strain, Lactobacillus delbrueckii subsp. bulgaricus JCM 1002T(pSYE2), were cultivated in MRS broth (Becton Dickson, MD, USA) and cultured at 37°C. The intermediate-expression host strains Lactococcus lactis subsp. cremoris MG1363 and Lactococcus lactis subsp. lactis IL-1401 were cultivated in M17 medium (Oxoid, Hants, UK) supplemented with 0.5% glucose (GM17) and cultured at 30°C. For recombinant strains harboring plasmid vectors, erythromycin (Em) was used as a screening agent in GM17 broth and MRS broth at a final concentration of 25 µg ml−1. All bacteria were stored at −80°C in their respective media with 30% (wt/vol) glycerol and cultivated with 10% inoculation more than twice before use.
TABLE 8.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Descriptiona | Reference(s) or source |
|---|---|---|
| Strains | ||
| Lactobacillus gasseri | ||
| LA39(JCM 11657) | Producer of gassericin A | Laboratory collection |
| LA158(JCM 11046) | Producer of gassericin T, plasmid free | Laboratory collection |
| LA327 | Producer of gassericin T and gassericin S, plasmid free | Laboratory collection |
| LA158 ΔgatAX | Host for recombinant plasmids, plasmid free, derivative of Lactobacillus gasseri LA158 lacking the gatAX operon | This study |
| LA158 ΔgatX | GatA producer, plasmid free, derivative of Lactobacillus gasseri LA158 lacking the gatX operon | This study |
| LA158 ΔgatA | GatX producer, Lactobacillus gasseri LA158 lacking the gatA operon | This study |
| LA327 ΔgatAX | Producer of gassericin S, plasmid free, derivative of Lactobacillus gasseri LA327 lacking the gatAX operon | This study |
| LA327 ΔgatT | Plasmid free, derivative of Lactobacillus gasseri LA327 lacking the gatT operon | This study |
| Lactobacillus delbrueckii subsp. bulgaricus JCM1002T |
Indicator for bacteriocin activity assay | JCMb |
| Lactococcus lactis subsp. cremoris MG1363 |
Intermediate host for recombinant plasmids, plasmid free, derivative of Lactococcus lactis subsp. cremoris NCDO 712 | 48 |
| Lactococcus lactis subsp. lactis IL1403 |
Intermediate host for recombinant plasmids, plasmid free | 49 |
| Plasmids | ||
| pSYE2 | Emr; pSY1 derivative with erythromycin resistance gene (emrA) of pAMβ1 from Enterococcus faecalis | 50 |
| pIL253-P32 | Emr; pIL253 derivative with P32 promotor | 51 |
| pTERM09 | Emr; pSY1 derivative with ts mutation in the repA gene | 40, 43 |
| pGS-AX | Emr; pIL253-P32 derivative carrying gasAX | This study |
| pGS-AXI | Emr; pIL253-P32 derivative carrying gasAXI | This study |
| pGS-AXΔA | Emr; pGS-AX derivative with gasA eliminated | This study |
| pGS-AXΔX | Emr; pGS-AX derivative with gasX eliminated | This study |
| pGS-AXIΔA | Emr; pGS-AXI derivative with gasA eliminated | This study |
| pGS-AXIΔX | Emr; pGS-AXI derivative with gasX eliminated | This study |
Emr, erythromycin resistance.
JCM, Japan Collection of Microorganisms, Tsukuba, Japan.
DNA sequencing and genetic analysis.
The nucleotide sequences surrounding two bacteriocin structural genes (gatAX and gasAX) presumed to encode GT and Acd LF221A were determined with primer walking, using the chromosomal DNA of L. gasseri LA327 prepared by the method of Luchansky et al. (39) as the template. The DNA sequencing was performed using the dideoxy chain termination method with a Prism 3100 Genetic Analyzer (Applied Biosystems, CA, USA) and a BigDye Terminator v1.1 cycle sequencing kit (Applied Biosystems) according to the manufacturer’s protocols.
Open reading frames (ORFs), promoters, and terminators on the determined DNA sequence were deduced using the GENETYX-MAC software (Genetyx Corporation, Tokyo, Japan) and an online genetic analysis site, SoftBerry (http://www.softberry.com/). Predicted amino acid sequences of peptides and proteins encoded by the DNA sequences of the ORFs were subjected to homology searching using the BLAST program in the DDBJ database (http://blast.ddbj.nig.ac.jp/top-j.html). The cellular localization and transmembrane segments (TMS) of each peptide and protein were deduced using the online program TMHMM (http://www.cbs.dtu.dk/services/TMHMM/).
Electrotransformation of bacteria.
Electrotransformation of L. gasseri strains, L. lactis subsp. lactis IL-1403, and L. lactis subsp. cremoris MG1363 was performed as described by Ito et al. (40) and Holo and Nes (41). Transformants were selected by using 25 μg ml−1 Em.
Deletion of the GT structural genes (gatAX) and ABC-type transporter gene (gatT) from L. gasseri.
The primers used in this study are summarized in Table 9. The cloning vector, pTERM13 (40), is derived from pSY1 (GenBank accession no. E05087), which carries a replication protein gene (repA) identical to that of pWVO1 (42). The temperature-sensitive (ts) mutation known for pWVO1 repA (43) was transplanted to repA of pTERM13 to obtain a novel ts vector, pTERM09. In order to remove gatAX and gatT from the chromosomes of L. gasseri LA158 and LA327, a double-crossover (DCO) substitution was used. The 5′-flanking 763-bp and the 3′-flanking 770-bp sequences of gatAX, or the 5′-flanking 792-bp and the 3′-flanking 791-bp sequences of gatT, were amplified and joined by using the splice-overlap extension PCR method with the primers ΔgatAX pr1, ΔgatAX pr2, ΔgatAX pr3, and ΔgatAX pr4, as well as ΔgatT pr1, ΔgatT pr2, ΔgatT pr3, and ΔgatT pr4. Similarly, gatA and gatX were removed from chromosome of L. gasseri LA158. After the 5′-flanking 763-bp, and the 3′-flanking 767-bp sequences of gatA, or the 5′-flanking 770-bp and the 3′-flanking 770-bp sequences of gatX, were amplified with primers ΔgatA pr1, ΔgatA pr2, ΔgatA pr3, and ΔgatA pr4, as well as ΔgatX pr1, ΔgatX pr2, ΔgatX pr3, and ΔgatX pr4, they were joined using the aforementioned method. The PCR was done using Phusion DNA polymerase (New England Biolabs, MA, USA). The joined fragment was cloned into the unique SmaI site of pTERM09 to construct pLGΔgatAX and pLGΔgatT. These recombinants were electrotransformed to L. gasseri LA158 and/or LA327, and transformants were selected at the permissive temperature of 32°C on MRS agar plates containing Em. Integration of the recombinants into the chromosomal gat locus of L. gasseri LA158 and/or LA327 was done by cultivating the transformant at 39°C, and DCO resolution of pLGΔgatAX and pLGΔgatT from the chromosome was induced by cultivating the integrant at 32°C in MRS medium without Em. Colony-direct PCR with ΔgatAX pr1 and ΔgatAX pr4 or ΔgatT pr1 and ΔgatT pr4 was used to screen gatAX- and gatT-deletant clones, from which 1.5-kb and 1.6-kb fragments, respectively, were amplified. Each relevant fragment was sequenced to confirm the correctness of the deletion. The gatAX and gatT deletants thus obtained were designated LA158 ΔgatAX, LA327 ΔgatAX, and LA327 ΔgatT.
TABLE 9.
Primers used in this study
| Primer | Sequence (5′to 3′) | Purpose |
|---|---|---|
| ΔgatA pr1 | AGTACAGTCTCTTCGGTTGG | Removing gatA from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatA pr2 | AGCCATATTAATTCCAATAAAGACCTCCTA | Removing gatA from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatA pr3 | TAGGAGGTCTTTATTGGAATTAATATGGCT | Removing gatA from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatA pr4 | CTTAATTTCTGAGTTTTTCC | Removing gatA from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatX pr1 | AAACTTTTCTGTACCTACAG | Removing gatX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatX pr2 | AAATCTCTATATAAAATTAATTCCCTACTT | Removing gatX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatX pr3 | AAGTAGGGAATTAATTTTATATAGAGATTT | Removing gatX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatX pr4 | CATATTTCTGAGGTGATACA | Removing gatX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatAX pr2 | AAATCTCTATATAAAAATAAAGACCTCCTA | Removing gatAX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatAX pr3 | TAGGAGGTCTTTATTTTTATATAGAGATTT | Removing gatAX from the chromosomes of Lactobacillus gasseri LA158 and LA327 |
| ΔgatT pr1 | AAAAAGCAAGATCCAAATGCACA | Removing gatT from the chromosomes of Lactobacillus gasseri LA327 |
| ΔgatT pr2 | CTTCACTAAGTCATAATATTGAATACTATT | Removing gatT from the chromosomes of Lactobacillus gasseri LA327 |
| ΔgatT pr3 | AATAGTATTCAATATTATGACTTAGTGAAG | Removing gatT from the chromosomes of Lactobacillus gasseri LA327 |
| ΔgatT pr4 | AGTTTCAGGCAATTTAAATCC | Removing gatT from the chromosomes of Lactobacillus gasseri LA327 |
| pIL F4 | CGGTTACTTTGGATTTTTGTGAG | Confirming PCR |
| pIL R4 | TGCACTGATTGGTGTATCATTTC | Confirming PCR |
| gasA-Sal Fw | ACGCGTCGACCTAAATTAGTCACTTTTCCTCTTAAG | Amplifying gasAX and gasAXI located in the chromosome of Lactobacillus gasseri LA327 |
| gasX-Xba Rv | GCTCTAGACTATCCATATTCCCGTCATATAC | Amplifying gasAX located in the chromosome of Lactobacillus gasseri LA327 |
| gasI-Xba Rv | GCTCTAGAGATTATTACCAAATTGAACCTAAGAAC | Amplifying gasAXI located in the chromosome of Lactobacillus gasseri LA327 |
| pGSΔA Fw | GAGGTGGAAGATAATGATCG | Removing gasA from pGS-AX and pGS-AXI |
| pGSΔA Rv | CTTAAGAGGAAAAGTGACTAATTTAG | Removing gasA from pGS-AX and pGS-AXI |
| pGSΔX Fw | TTGGTTCTACAAACTACTAGTGG | Removing gasX from pGS-AX and pGS-AXI |
| pGSΔX RV | TTTCGATCATTATCTTCCACCTC | Removing gasX from pGS-AX and pGS-AXI |
Construction of GS-producing strains for homologous expression.
Molecular cloning was performed using standard methods (44). Recombinant plasmids of gasAX and gasAXI (pGS-AX and pGS-AXI, respectively) were constructed in order to obtain the GS producers. After gasAX and gasAXI were amplified by PCR using corresponding primers (gasA-Sal Fw and gasX-Xba Rv for gasAX and gasA-Sal Fw and gasI-Xba Rv for gasAXI), PCR was performed using chromosomal DNA from L. gasseri LA327 as the template DNA, TaKaRa Ex Taq polymerase (EC 2.7.7.7; TaKaRa Bio Inc., Shiga, Japan), and a T100 thermal cycler (Bio-Rad Laboratories, Watford, UK). Each PCR product then was ligated with expression vector pIL253-P32 using T4 DNA ligase (EC 6.5.1.1; TaKaRa Bio Inc.) via double digestion using SalI and XbaI (EC 3.1.21.4; TaKaRa Bio Inc.). Furthermore, to construct the GasA and GasX producers, recombinant plasmids of gasA, gasX, gasAI, and gasXI (pGS-A, pGS-X, pGS-AI, and pGS-XI, respectively) were constructed by deletion of the gasA or gasX region from pGS-AX and pGS-AXI. Following this, inverse PCR using outward-facing primers for deletion of each region (primer pair pGSΔA Fw and pGSΔA Rv for deletion of gasA and primer pair pGSΔX Fw and pGSΔX Rv for deletion of gasX) was performed. Inverse PCR products were phosphorylated using T4 polynucleotide kinase (EC 2.7.1.78; TaKaRa Bio Inc.) and continuously self-ligated using T4 DNA ligase (TaKaRa Bio Inc.). Each recombinant plasmid was transformed into an intermediate strain, Lactococcus lactis subsp. cremoris MG1363, and then subsequently into the final expression host strain, L. gasseri LA158 ΔgatAX.
Bacteriocin activity assay by the agar well diffusion method.
The CFS of each L. gasseri strain was prepared by centrifugation (8,000 × g, 10 min, 4°C) of the MRS cultures and filtrated through a 0.45-µm membrane filter (Advantec, Tokyo, Japan). Bacteriocin activity was assayed using the agar well diffusion method (45). Samples (CFSs) were twofold serially diluted using 0.85% phosphate-buffered saline (PBS). MRS agar plates (90 mm in diameter and 4 mm thick, 15 ml) containing 1.5% (wt/vol) agar (Oxoid) were overlaid with a soft agar lawn (10 ml 0.75% agar, 10 µg ml−1 Em) and inoculated with a one-tenth-diluted overnight culture (250 µl) of the indicator strain L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2). Wells (6 mm in diameter) were cut off from the plates, and then 65 µl of the twofold serially diluted (1:1 to 1:1,023) samples was added to each well. The plates were incubated for 18 h at 37°C. The unit of bacteriocin activity (arbitrary unit [AU]) was defined as the reciprocal of the highest dilution inhibiting the growth of the indicator strain. The assay for each sample was performed at least in triplicate.
Bacteriocin activity assay of Gas and Gat producers and verification of synergistic activities of GS and/or GT.
Bacteriocin activities of Gas producers were assayed by the agar well diffusion method as described above. After the bacteriocin activities in the CFSs of GasAX and GasAXI producers were tested, those of GasA, GasX, GasAI, and GasXI producers were examined alone and in combination (GasA or -AI and GasX or -XI, 1:1), and then the activities of Gat producers (producing GatA and/or GatX) were similarly assayed. Furthermore, the CFSs of GatA, GatX, GasA, and GasX producers were subjected to the assay in combination (six patterns of two peptides and the combination of four peptides).
In order to verify the synergistic activities of GS (GasA plus GasX), GT (GatA plus GatX), and GS (GasA plus GasX) plus GT (GatA plus GatX), isobologram assays were performed with the minimum concentration of the CFS of each bacteriocin peptide, which shows the inhibitory zone on the agar well diffusion assay, and the concentration of each undiluted (original) CFS (65 μl) was defined as 1.00.
Preparation of crude GS and in situ activity assay.
The CFS of the GS producer L. gasseri LA158 ΔgatAX(pGS-AXI), was concentrated approximately 40 times by ultrafiltration (Amicon Ultra-100K centrifugal filter units; Merck Millipore, Tullagreen, Ireland), and then the concentrate (crude GS) was subjected to SDS-PAGE following Laemmli’s method (46) with a 4.5% spacer gel and 20% separating gel. The Amersham ECL Rainbow low-range marker (GE Healthcare, Tokyo, Japan), with marker range of 3,500 to 40,000 Da, was used as a molecular marker. For detection of bacteriocin activity on the gel, an in situ activity assay was performed as described by Daba et al. (47). Briefly, the half-cut gel was put in a petri dish after immobilization with 20% isopropanol–10% acetate and was washed with Milli-Q water; MRS agar containing the indicator strain was then stratified and incubated at 37°C for 24 h. The bacteriocin activity of GS was observed as a clear zone, and the molecular mass was estimated by electrophoretic mobility.
Assay of pH stability of GS.
The pHs of the CFSs from the GS producer L. gasseri LA158 ΔgatAX (pGS-AXI) and the non-bacteriocin producer (control) L. gasseri LA158 ΔgatAX were adjusted to 2, 4, 7, and 10 with 1 N HCl and 1 N NaOH at room temperature. Following this, the bacteriocin activities of these CFSs were assayed by the agar well diffusion method, using L. delbrueckii subsp. bulgaricus JCM 1002T(pSYE2) as the indicator.
Assay of heat stability of GS.
The CFS of the GS producer L. gasseri LA158 ΔgatAX(pGS-AXI) was dispensed at 100 µl in a 0.2-ml PCR tube (Bio-Rad) and autoclaved (121°C for 15 min) or heated (95°C for 5 min and 70°C for 1 h). For confirmation of the preservation stability, CFS containing GS was incubated at 37°C and 4°C for 1 month and then utilized in the bacteriocin activity assay.
Assay of proteolytic sensitivity of GS.
Protease solutions (0.2%, wt/vol) were prepared by dissolving pepsin (EC 3.4.23.1) (Wako Pure Chemical Industries, Osaka, Japan) in 0.2 M HCl-KCl buffer (pH 2) and dissolving trypsin (4,200 USP units/mg; EC 3.4.21.4) (Wako), alpha-chymotrypsin (40 to 50 units/mg; EC 3.4.21.1) (MP Biomedicals, Illkirch, France), and proteinase K (590 units/ml; EC 3.4.21.64) (Wako) in 0.2 M sodium phosphate buffer (pH 8). The CFSs of the GA producer (L. gasseri LA39), the GT producer (L. gasseri LA158), and the GS producer [L. gasseri LA158 ΔgatAX(pGS-AXI)] were mixed with each protease solution in an equal amount and were incubated at 37°C for 5 h in a water bath. These treated CFSs were utilized in the bacteriocin activity assay.
Accession number(s).
The 6,935-bp and 1,143-bp DNA sequences containing nine genes (gat operon) and three genes (gas operon), respectively, were deposited in the DDBJ database under accession numbers LC389592 and LC389591, respectively.
ACKNOWLEDGMENTS
We thank Yoshiyuki Ito for advice on experimental design and recombinant construction.
This research received no specific grant from any funding agency in the public, commercial, or not-for-profit sector.
REFERENCES
- 1.Zoetendal EG, von Wright A, Vilpponen-Salmela T, Ben-Amor K, Akkermans AD, de Vos WM. 2002. Mucosa-associated bacteria in the human gastrointestinal tract are uniformly distributed along the colon and differ from the community recovered from feces. Appl Environ Microbiol 68:3401–3407. doi: 10.1128/AEM.68.7.3401-3407.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reuter G. 2001. The Lactobacillus and Bifidobacterium microflora of the human intestine: composition and succession. Curr Issues Intest Microbiol 2:43–53. [PubMed] [Google Scholar]
- 3.Dall Bello F, Hertel C. 2006. Oral cavity as natural reservoir for intestinal lactobacilli. Syst Appl Microbiol 29:69–76. doi: 10.1016/j.syapm.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 4.Marin ML, Arroyo R, Jimenez E, Gomez A, Fernandez L, Rodriguez JM. 2009. Cold storage of human milk: effect on its bacterial composition. J Pediatr Gastroenterol Nutr 49:343–348. doi: 10.1097/MPG.0b013e31818cf53d. [DOI] [PubMed] [Google Scholar]
- 5.Matsumiya Y, Kato N, Watanabe K, Kato H. 2002. Molecular epidemiological study of vertical transmission of vaginal Lactobacillus species from mothers to newborn infants in Japanese, by arbitrarily primed polymerase chain reaction. J Infect Chemother 8:43–49. doi: 10.1007/s101560200005. [DOI] [PubMed] [Google Scholar]
- 6.Miyazaki K, Kwase M, Kubota A, Yoda K, Harata G, Hosoda M, He F. 2015. Heat-killed Lactobacillus gasseri can enhance immunity in the elderly in a double-blind, placebo-controlled clinical study. Benef Microbes 6:441–449. doi: 10.3920/BM2014.0108. [DOI] [PubMed] [Google Scholar]
- 7.Ghadimi D, Folster-Holst R, de Vrese M, Winkler P, Heller KJ, Schrezenmeir J. 2008. Effect of probiotic bacteria and their genomic DNA on TH1/TH2-cytokine production by peripheral blood mononuclear cells (PBMCs) of healthy and allergic subjects. Immunobiology 213:677–692. doi: 10.1016/j.imbio.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 8.Nishiyama K, Seto Y, Yoshioka K, Kakuda T, Takai S, Yamamoto Y, Mukai T. 2014. Lactobacillus gasseri SBT2055 reduces infection by and colonization of Campylobacter jejuni. PLoS One 9:e108827. doi: 10.1371/journal.pone.0108827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kassaa IA, Hober D, Hamze M, Caloone D, Dewilde A, Chihib NE, Drider D. 2015. Vaginal Lactobacillus gasseri CMUL57 can inhibit herpes simplex type 2 but not coxsackievirus B4E2. Arch Microbiol 197:657–664. doi: 10.1007/s00203-015-1101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Motevaseli E, Shirzad M, Akrami SM, Mousavi AS, Mirsalehian A, Modarressi MH. 2013. Normal and tumour cervical cells respond differently to vaginal lactobacilli, independent of pH and lactate. J Med Microbiol 62:1065–1072. doi: 10.1099/jmm.0.057521-0. [DOI] [PubMed] [Google Scholar]
- 11.Ogawa A, Kobayashi T, Sakai F, Kadooka Y, Kawasaki Y. 2015. Lactobacillus gasseri SBT2055 suppresses fatty acid release through enlargement of fat emulsion size in vitro and promotes fecal fat excretion in healthy Japanese subjects. Lipids Health Dis 14:20–29. doi: 10.1186/s12944-015-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Atassi F, Servin AL. 2010. Individual and co-operative roles of lactic acid and hydrogen peroxide in the killing activity of enteric strain Lactobacillus johnsonii NCC933 and vaginal strain Lactobacillus gasseri KS120.1 against enteric, uropathogenic and vaginosis-associated pathogens. FEMS Microbiol Lett 304:29–38. doi: 10.1111/j.1574-6968.2009.01887.x. [DOI] [PubMed] [Google Scholar]
- 13.Kommineni S, Bretl DJ, Lam V, Chakraborty R, Hayward M, Simpson P, Cao Y, Bousounis P, Kristich CJ, Salzman NH. 2015. Bacteriocin production augments niche competition by enterococci in the mammalian gastrointestinal tract. Nature 526:719–722. doi: 10.1038/nature15524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mishra C, Lambert J. 1996. Production of anti-microbial substances by probiotics. Asia Pac J Clin Nutr 5:20–24. [PubMed] [Google Scholar]
- 15.Klaenhammer TR. 1993. Genetics of bacteriocins produced by lactic acid bacteria. FEMS Microbiol Rev 12:39–85. doi: 10.1111/j.1574-6976.1993.tb00012.x. [DOI] [PubMed] [Google Scholar]
- 16.Cotter PD, Ross RP, Hill C. 2013. Bacteriocins—a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105. doi: 10.1038/nrmicro2937. [DOI] [PubMed] [Google Scholar]
- 17.Alvarez-Sieiro P, Montalbán-López M, Mu D, Kuipers OP. 2016. Bacteriocins of lactic acid bacteria: extending the family. Appl Microbiol Biotechnol 100:2939–2951. doi: 10.1007/s00253-016-7343-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kawai Y, Saito T, Kitazawa H, Itoh T. 1998. Gassericin A; an uncommon cyclic bacteriocin produced by Lactobacillus gasseri LA39 linked at N- and C-terminal ends. Biosci Biotechnol Biochem 62:2438–2440. doi: 10.1271/bbb.62.2438. [DOI] [PubMed] [Google Scholar]
- 19.Kawai Y, Saitoh B, Takahashi O, Kitazawa H, Saito T, Nakajima H, Itoh T. 2000. Primary amino acid and DNA sequences of gassericin T, a lactacin F-family bacteriocin produced by Lactobacillus gasseri SBT2055. Biosci Biotechnol Biochem 64:2201–2208. doi: 10.1271/bbb.64.2201. [DOI] [PubMed] [Google Scholar]
- 20.Yasuta N, Arakawa K, Kawai Y, Chujo T, Nakamura K, Suzuki H, Ito Y, Nishimura J, Makino Y, Shigenobu S, Saito T. 2014. Genetic and biochemical evidence for gassericin T production from Lactobacillus gasseri LA158. Milk Sci 63:9–17. doi: 10.11465/milk.63.9. [DOI] [Google Scholar]
- 21.Bogovic-Matijašić BB, Rogelj I, Nes IF, Holo H. 1998. Isolation and characterization of two bacteriocins of Lactobacillus acidophilus LF221. Appl Microbiol Biotechnol 49:606–612. doi: 10.1007/s002530051221. [DOI] [PubMed] [Google Scholar]
- 22.Zorič-Peternel M, Čanžek-Majhenič A, Holo H, Nes IF, Salehian Z, Berlec A, Rogelj I. 2010. Wide-inhibitory spectra bacteriocins produced by Lactobacillus gasseri K7. Probiotics Antimicrob Proteins 2:233–240. doi: 10.1007/s12602-010-9044-5. [DOI] [PubMed] [Google Scholar]
- 23.Mavrič A, Tompa G, Trmčić A, Rogelj I, Bogovic-Matijašić B. 2014. Bacteriocins of Lactobacillus gasseri K7—monitoring of gassericin K7 A and B genes’ expression and isolation of an active component. Process Biochem 49:1251–1259. doi: 10.1016/j.procbio.2014.04.022. [DOI] [Google Scholar]
- 24.Maldonado-Barragán A, Caballero-Guerrero B, Martín V, Ruiz-Barba JL, Rodríguez JM. 2016. Purification and genetic characterization of gassericin E, a novel co-culture inducible bacteriocin from Lactobacillus gasseri EV1461 isolated from the vagina of a healthy woman. BMC Microbiol 16:37. doi: 10.1186/s12866-016-0663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nes IF, Diep DB, Havarstein LS, Brurberg MB, Eijsink V, Holo H. 1996. Biosynthesis of bacteriocins in lactic acid bacteria. Antonie Van Leeuwenhoek 70:113–128. [DOI] [PubMed] [Google Scholar]
- 26.Nissen-Meyer J, Rogne P, Oppegård C, Haugen HS, Kristiansen PE. 2009. Structure-function relationships of the non-lanthionine-containing peptide (class II) bacteriocins produced by gram-positive bacteria. Curr Pharm Biotechnol 10:19–37. doi: 10.2174/138920109787048661. [DOI] [PubMed] [Google Scholar]
- 27.Ishibashi N, Himeno K, Masuda Y, Perez RH, Iwatani S, Zendo T, Wilaipun P, Leelawatcharamas V, Nakayama J, Sonomoto K. 2014. Gene cluster responsible for secretion of and immunity to multiple bacteriocins, the NKR-5-3 enterocins. Appl Environ Microbiol 80:6647–6655. doi: 10.1128/AEM.02312-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Majhenič AC, Venema K, Allison GE, Bogovic-Matijašić BB, Rogelj I, Klaenhammer TR. 2004. DNA analysis of the genes encoding acidocin LF221 A and acidocin LF221 B, two bacteriocins produced by Lactobacillus gasseri LF221. Appl Microbiol Biotechnol 63:705–714. doi: 10.1007/s00253-003-1424-2. [DOI] [PubMed] [Google Scholar]
- 29.Maldonado-Barragán A, Caballero-Guerrero B, Jiménez E, Jiménez-Díaz R, Ruiz-Barba JL, Rodríguez JM. 2009. Enterocin C, a class IIb bacteriocin produced by E. faecalis C901, a strain isolated from human colostrum. Int J Food Microbiol 133:105–112. doi: 10.1016/j.ijfoodmicro.2009.05.008. [DOI] [PubMed] [Google Scholar]
- 30.Acedo JZ, Towle KM, Lohans CT, Miskolzie M, McKay RT, Doerksen TA, Vederas JC, Martin-Visscher LA. 2017. Identification and three-dimensional structure of carnobacteriocin XY, a class IIb bacteriocin produced by Carnobacteria. FEBS Lett 591:1349–1359. doi: 10.1002/1873-3468.12648. [DOI] [PubMed] [Google Scholar]
- 31.Ra R, Beerthuyzen MM, de Vos WM, Saris PEJ, Kuipers OP. 1999. Effects of gene disruptions in the nisin gene cluster of Lactococcus lactis on nisin production and producer immunity. Microbiology 145:1227–1233. doi: 10.1099/13500872-145-5-1227. [DOI] [PubMed] [Google Scholar]
- 32.Anderssen EL, Diep DB, Nes IF, Eijsink VGH, Nissen-Meyer J. 1998. Antagonistic activity of Lactobacillus plantarum C11: two new two-peptide bacteriocins, plantaricin EF and JK, and the induction factor, plantaricin A. Appl Environ Microbiol 64:2269–2272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Oppegård C, Fimland G, Thorbæk L, Nissen-Meyer J. 2007. Analysis of the two-peptide bacteriocins lactococcin G and enterocin 1071 by site-directed mutagenesis. Appl Environ Microbiol 73:2931–2938. doi: 10.1128/AEM.02718-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Oppegård C, Rogne P, Emanuelsen L, Kristiansen PE, Fimland G, Nissen-Meyer J. 2007. The two-peptide class II bacteriocins: structure, production, and mode of action. J Mol Microbiol Biotechnol 13:210–219. doi: 10.1159/000104750. [DOI] [PubMed] [Google Scholar]
- 35.Zendo T, Koga S, Shigeri Y, Nakayama J, Sonomoto K. 2006. Lactococcin Q, a novel two-peptide bacteriocin produced by Lactococcus lactis QU 4. Appl Environ Microbiol 72:3383–3389. doi: 10.1128/AEM.72.5.3383-3389.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Craik DJ, Daly NL, Saska I, Trabi M, Rosengren KJ. 2003. Structures of naturally occurring circular proteins from bacteria. J Bacteriol 185:4011–4021. doi: 10.1128/JB.185.14.4011-4021.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Itoh T, Fujimoto Y, Kawai Y, Toba T, Saito T. 1995. Inhibition of food-borne pathogenic bacteria by bacteriocins from Lactobacillus gasseri. Lett Appl Microbiol 21:137–141. doi: 10.1111/j.1472-765X.1995.tb01025.x. [DOI] [PubMed] [Google Scholar]
- 38.Kawai Y, Saito T, Uemura J, Itoh T. 1997. Rapid detection method for bacteriocin and distribution of bacteriocin-producing strains in Lactobacillus acidophilus group lactic acid bacteria isolated from human feces. Biosci Biotechnol Biochem 61:179–182. doi: 10.1271/bbb.61.179. [DOI] [PubMed] [Google Scholar]
- 39.Luchansky JB, Tennant MC, Klaenhammer TR. 1991. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J Dairy Sci 74:3293–3302. doi: 10.3168/jds.S0022-0302(91)78515-9. [DOI] [PubMed] [Google Scholar]
- 40.Ito Y, Kawai Y, Honme Y, Arakawa K, Sasaki T, Saito T. 2009. Conjugative plasmid from Lactobacillus gasseri LA39 that carries genes for production of and immunity to the circular bacteriocin gassericin A. Appl Environ Microbiol 75:6340–6351. doi: 10.1128/AEM.00195-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Holo H, Nes IF. 1989. High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Leenhouts KJ, Tolner B, Bron S, Kok J, Venema G, Seegers JF. 1991. Nucleotide sequence and characterization of the broad-host-range lactococcal plasmid pWVO1. Plasmid 26:55–66. doi: 10.1016/0147-619X(91)90036-V. [DOI] [PubMed] [Google Scholar]
- 43.Biswas I, Gruss A, Ehrlich SD, Maguin E. 1993. High-efficiency gene inactivation and replacement system for gram-positive bacteria. J Bacteriol 175:3628–3635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sambrook JF, Russell DW (ed). 2001. Molecular cloning: a laboratory manual, 3rd ed Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [Google Scholar]
- 45.Toba T, Yoshioka E, Itoh T. 1991. Lacticin, a bacteriocin produced by Lactobacillus delbrueckii subsp. lactis. Lett Appl Microbiol 12:43–45. doi: 10.1111/j.1472-765X.1991.tb00499.x. [DOI] [Google Scholar]
- 46.Leammli UK. 1970. Cleavage of structural proteins during the assembly of the head of bacteriopharge T4. Nature 227:680–685. [DOI] [PubMed] [Google Scholar]
- 47.Daba H, Pandian S, Gosselin JF, Simard RE, Huang J, Lacroix C. 1991. Detection and activity of a bacteriocin produced by Leuconostoc mesenteroides. Appl Environ Microbiol 57:3450–3455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gasson JM. 1983. Plasmid complements of Streptococcus lactis NCDO 712 and other lactic streptococci after protoplast-induced curing. J Bacteriol 154:1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chopin A, Chopin MC, Moillo-Batt A, Langella P. 1984. Two plasmid-determined restriction and modification systems in Streptococcus lactis. Plasmid 11:260–263. doi: 10.1016/0147-619X(84)90033-7. [DOI] [PubMed] [Google Scholar]
- 50.Satoh E, Ito Y, Sasaki Y, Sasaki T. 1997. Application of the extracellular α-amylase gene from Streptococcus bovis 148 to construction of a secretion vector for yogurt starter strains. Appl Environ Microbiol 63:4593–4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kemperman R, Jonker M, Nauta A, Kuipers OP, Kok J. 2003. Functional analysis of the gene cluster involved in production of the bacteriocin circularin A by Clostridium beijerinckii ATCC 25752. Appl Environ Microbiol 69:5839–5848. doi: 10.1128/AEM.69.10.5839-5848.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]