Abstract
In this study, we found mcr-1.1 and mcr-1.5 genes carried by IncI2 plasmids in a subset of Escherichia coli isolates recovered from commercial broiler farms in Argentina. The comparative analysis of the sequences of these plasmids with those described in human clinical isolates suggests that this replicon-type is one of the main mcr-disseminator sources in Argentina.
Keywords: mcr-1 colistin resistance, IncI2 plasmids, Escherichia coli, poultry, animals
Introduction
Colistin is a last-resort antimicrobial against multidrug-resistant Gram-negative pathogens. A public health concern about colistin resistance has been risen due to a plasmid-mediated mechanism called mcr, described in enterobacteria of clinical and food-animal origin in several countries (Poirel et al., 2017). Fourteen allelic variants of mcr-1 have been reported lately, designated mcr-1.1 to mcr-1.14 (Partridge et al., 2018). mcr-1 genes were found in plasmids belonging to different incompatibility groups (IncI2, IncHI2, IncP, IncX4, IncFI, and IncFIB) (Poirel et al., 2017), which mediate their horizontal transfer to different bacterial species.
The global distribution of mcr genes in Escherichia coli emphasizes the importance of understanding the mechanisms involved in their spread. Rational use of colistin is urgently required to prevent the rapid dissemination of mcr to other bacteria and in different niches, including human hospitals and foodborne settings.
In a previous study, we have characterized 149 mcr-1-positive E. coli isolates recovered from 129 commercial broiler healthy chicken (aged 4–6 weeks) from farms located in several provinces of Argentina (Dominguez et al., 2017). A subset of 10 E. coli from that previous study was included in the present work. We describe a comparative analysis of the sequences of their mcr-harboring plasmid with those described in human clinical isolates from Argentina.
Materials and Methods
Ten mcr-1-positive E. coli isolates were included in this study from healthy chickens recovered from commercial farms located at Entre Rios and Buenos Aires provinces (Table 1). Susceptibility profiles were determined by the agar dilution method with the exception of colistin tested by broth microdilution method. The results were interpreted according to the Clinical and Laboratory Standards Institute guidelines (CLSI, 2017); colistin and tigecycline were interpreted by the 2018 European Committee on Antimicrobial Susceptibility Testing guidelines (http://www.eucast.org). mcr-1, ESBL, pAmpC, and PMQR-coding-genes were screened by PCR (Anchordoqui et al., 2015; Liu et al., 2016; Albornoz et al., 2017). The genetic relatedness of E. coli isolates was studied by PFGE of XbaI-digested genomic fragments. Isolates were also genotyped by multilocus sequence typing (MLST). The allelic numbers and STs were assigned online using http://mlst.warwick.ac.uk/mlst/dbs/Ecoli. Plasmid profile of the isolates was analyzed by S1-PFGE. Sodium azide-resistant E. coli J53 was used as a recipient strain in conjugation experiments to study the transferability of the resistance genes. Plasmids were extracted from mcr-transconjugants strains using the Qiagen Large-Construct kit (Qiagen) and sequenced using Illumina's MiSeq system. The obtained reads were assembled using CLC Genomics Workbench software (CLCbio, Qiagen), annotated using RAST server (http://rast.nmpdr.org/rast.cgi) and the sequences (gaps were not filled) compared in a pairwise fashion using BRIG (Alikhan et al., 2011). The contigs were also analyzed by ResFinder, PlasmidFinder, and VirulenceFinder tools available from the Center for Genomic Epidemiology website (https://cge.cbs.dtu.dk/services).
Table 1.
Antimicrobial susceptibility profiles, sequence types (ST) and resistance determinant of E. coli isolates.
| Isolates | Provinces* | MIC (μg/ml)** | MLST (ST)*** | mcr-1 allele | ESBL/pAmpC genes | PMQR genes | Plasmids ID | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| COL | AMS | CAZ | CTX | FEP | ATM | NAL | CIP | AMK | GEN | TGC | FOS | MIN | |||||||
| M22607 | BA | 16 (R) | 16 (I) | 0.5 (S) | 4 (R) | 2 (S) | 2 (S) | ≥64 (R) | 32 (R) | 2 (S) | 0.5 (S) | 0.12 (S) | ≤0.25 (S) | 2 (S) | 617 | mcr-1.5 | CTX-M-14 | None | pGN2424 |
| M22608 | BA | 16 (R) | 1 (S) | 0.12 (S) | 0.03 (S) | 0.03 (S) | ≤0.06 (S) | 16 (S) | 0.25 (S) | 2 (S) | 0.5 (S) | 0.12 (S) | ≤0.25 (S) | 4 (S) | 1,141 | mcr-1.1 | None | qnrB | pGN2415 |
| M22609 | BA | 8 (R) | 64 (R) | 4 (S) | 64 (R) | 8 (I) | 32 (R) | ≥64 (R) | 64 (R) | 1 (S) | 0.25 (S) | 0.25 (S) | 64 (S) | 4 (S) | 410 | mcr-1.1 | CTX-M-2 | None | pGN2416 |
| M22610 | BA | 16 (R) | 64 (R) | 4 (S) | 64 (R) | 8 (I) | 32 (R) | ≥64 (R) | 16 (R) | 2 (S) | 0.5 (S) | 0.25 (S) | ≤0.25 (S) | 32 (R) | 155 | mcr-1.5 | CTX-M-2 | None | pGN2417 |
| M22611 | BA | 16 (R) | 16 (I) | 16 (R) | 8 (R) | 0.12 (S) | 8 (I) | 2 (S) | 0.008 (S) | 2 (S) | 0.5 (S) | 0.12 (S) | ≤0.25 (S) | 4 (S) | 1,286 | mcr-1.5 | CMY-2 | None | pGN2418 |
| M22612 | BA | 16 (R) | 64 (R) | 4 (S) | 64 (R) | 8 (I) | 32 (R) | 16 (S) | 0.5 (S) | 2 (S) | 4 (S) | 0.12 (S) | ≤0.25 (S) | 0.5 (S) | 1,011 | mcr-1.5 | CTX-M-2 | qnrB | pGN2419 |
| M22613 | ER | 8 (R) | 2 (S) | 0.12 (S) | 0.03 (S) | 0.03 (S) | ≤0.06 (S) | 16 (S) | 0.5 (S) | 2 (S) | 0.5 (S) | 0.12 (S) | 0.5 (S) | 0.25 (S) | 10 | mcr-1.5 | None | qnrB | pGN2420 |
| M22614 | ER | 16 (R) | 4 (S) | 0.25 (S) | 0.12 (S) | 0.06 (S) | 0.12 (S) | ≥64 (R) | 0.5 (S) | 2 (S) | 0.5 (S) | 0.25 (S) | 0.5 (S) | 1 (S) | 155 | mcr-1.5 | None | None | pGN2421 |
| M22615 | ER | 32 (R) | 2 (S) | 0.12 (S) | 0.03 (S) | 0.03 (S) | ≤0.06 (S) | 16 (S) | 0.25 (S) | 4 (S) | 1 (S) | 0.12 (S) | 0.5 (S) | 4 (S) | 1,408 | mcr-1.5 | None | qnrB | pGN2422 |
| M22616 | ER | 8 (R) | 16 (I) | 1 (S) | 32 (R) | 4 (I) | 8 (I) | ≥64 (R) | 0.5 (S) | 2 (S) | 16 (R) | 0.12 (S) | 0.5 (S) | 0.5 (S) | Unknown ST | mcr-1.5 | CTX-M-14 | None | pGN2423 |
BA, Buenos Aires; ER, Entre Ríos.
COL, colistin; AMS, ampicillin-sulbactam; CAZ, ceftazidime; CTX, cefotaxime; FEP, cefepime; ATM, aztreonam; NAL, nalidixic acid; CIP, ciprofloxacin; AMK, amikacin; GEN, gentamicin; TGC, tigecycline; FOS, fosfomycin; MIN, minocycline. R, resistant; I, intermediate, S, susceptible by the agar dilution method with the exception of colistin tested by broth microdilution method.
MLST, Multilocus Sequence Typing.
Results and Discussion
All E. coli isolates were positive for mcr-1 and exhibited resistance to colistin. Some isolates also exhibited a multidrug-resistant (MDR) phenotype including resistance to expanded-spectrum cephalosporins, quinolones, gentamicin, and minocycline, but all of them were susceptible to amikacin, tigecycline, and fosfomycin. mcr-1-positive isolates were determined to carry ESBL (5/10, blaCTX−M−2 or blaCTX−M−14), pAmpC (1/10, blaCMY−2), and PMQR (4/10, qnrB) genes (Table 1).
All isolates exhibited different PFGE profiles, indicating that they were genetically unrelated (Supplementary Figure 1). Supporting the PFGE results, most of the isolates had different STs. Only two of them, recovered in both provinces (Entre Rios and Buenos Aires), were ST155. Four isolates belonged to clonal complex 10 (CC10): one ST10, two single locus variants (ST1141 and ST1286), and one double locus variants (ST617) of ST10 (Table 1). E. coli CC10 isolates are globally recovered from food-producing animals and human samples, however, it is particularly frequent in livestock animals as susceptible or multidrug-resistant isolates (ESBL and/or pAmpC producers) (El Garch et al., 2017). One isolate was ST410, this ST was previously found in a mcr-1-positive clinical E. coli isolate in Argentina (Tijet et al., 2017), which was defined as a hyperepidemic clone and the possible founder of the disseminated CC23 (Turrientes et al., 2014).
A diverse plasmid content was found in the isolates by S1-PFGE. All isolates harbored a ca. 61-kb plasmid, present also in all the mcr-1-transconjugant strains conferring them only resistance to colistin (Supplementary Figure 2).
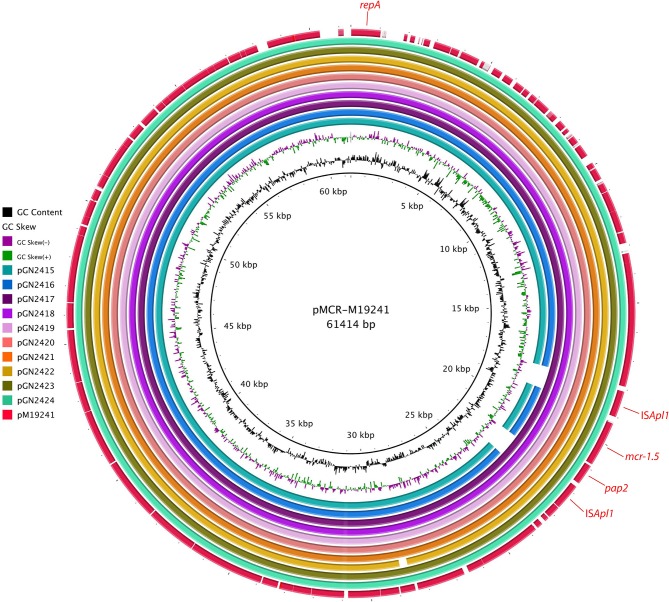
Assembling of short reads yields between 6 and 8 contigs from 8 isolates, while 2 isolates rendered 4 contigs. In all cases the calculated total length, ca. 61 kb, was in agreement with the plasmid sizes estimated by S1-PFGE. Eight of the ten plasmids analyzed had the same variant described as mcr-1.5, found in clinical isolates of Argentina (Tijet et al., 2017), and the remaining two contained the mcr-1.1 variant. Blast-based query revealed that all plasmids belonged to the IncI2 incompatibility group and none of them carried additional resistance or virulence genes. A comparison of the plasmids with pMCR-M19241 (obtained from human clinical isolate, GenBank KY471311) (Tijet et al., 2017) shows that eight of them (pGN2417 to pGN2424) contained two copies of the insertion sequence ISApI1 flanking the mcr-1.5/pap2 fragment, which might facilitate the transfer of mcr-1 between DNA molecules. ISApI1 was not present in pGN2415 and pGN2416 (Figure 1). Plasmids had a typical backbone responsible for its replication, maintenance, and transfer (Sun et al., 2016). Main differences observed between the plasmids were mainly due to reorganization of the pilV shufflon (data not shown).
Figure 1.
BRIG alignment of mcr-1-positive plasmids recovered from E. coli isolates. The location of mcr-1.5 and the repA plasmid replication gene are indicated. The comparisons are made relative to plasmid pMCR-M19241 (GenBank KY471311), recovered from a clinical E. coli isolate from Argentina.
mcr-1-encoding IncI2 plasmids have previously been reported in studies from Asia (mainly in China and Japan), Europe and the U.S (Ohsaki et al., 2017). Moreover, different plasmid types harboring mcr-1 have been reported in South-America, in isolates recovered from food animals, clinical samples and environmental reservoirs (Delgado-Blas et al., 2016; Fernandes et al., 2017; Monte et al., 2017; Rossi et al., 2017; Saavedra et al., 2017). Our results, together with previous studies (Tijet et al., 2017), suggest that in Argentina the spread of mcr-1 in E. coli isolates from animals and humans could be mainly mediated by IncI2-type plasmids.
Conclusion
These findings point toward effective dissemination of the mcr-1 gene in Argentina by efficient horizontal transfer almost exclusively by IncI2 type plasmids (Tijet et al., 2017). Our recent study suggested that a group of mcr-1-positive plasmids with same backbones are present in poultry farm E. coli isolates as well as human clinical E. coli isolates. Here, we report comparative genomics of over 10 representative mcr-1-bearing plasmids. These findings expand the scenery of mcr-1-harboring plasmids in Argentina.
Author Contributions
JD, DF, SG, AC, MF-M, and RM participated in the design of the study. JD, DF, and NT performed the experiments. JD, DF, SG, AC, and RM analyzed the data. JD and MF-M collected E. coli strains. JD, DF, NT, SG, AC, MF-M, and RM wrote the paper. All authors contributed to the critical revision of the manuscript and have seen and approved the final draft. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
Funding. This work was supported by a grant from the Instituto Nacional de Tecnología Agropecuaria (INTA, PNSA PE-1115056) and Fondo para la Investigación Científica y Tecnológica (FONCYT), PICT-1366-2010, and PICT-3154-2016. This work was also supported by Public Health Ontario.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fcimb.2019.00041/full#supplementary-material
The genetic relatedness of mcr-1-positive E. coli isolates. PFGE: Gel order; Line 2, M22607; 3, M22608; 4, M22609; 5, M22610; 6, M22611; 7, M22612; 8, M22613; 9, M22614; 10, M22615; 11, M22616; Lanes 1 and 12, S. Branderup.
Plasmid profile. S1-PFGE: Gel order; Line 2, M22608; 3, M22608-TC; 4, M22609; 5, M22609-TC; 6, M22610; 7, M22610-TC; 8, M22611; 9, M22611-TC; 10, M22612; 11, M22612-TC; 12, M22613; 13, M22613-TC; 14, M22615; 15, M22615-TC; 16, M22616; 17, M22616-TC; 19, M22607; 20, M22607-TC; 21, M22614; 22, M22624-TC; Lanes 1, 18, and 23, S. Branderup. TC: transconjugants obtained using E. coli J53 AZR as the recipient strain. Red arrows highlight plasmids containing the mcr-1 gene.
References
- Albornoz E., Lucero C., Romero G., Quiroga M. P., Rapoport M., Guerriero, et al. (2017). Prevalence of plasmid-mediated quinolone resistance genes in clinical enterobacteria from Argentina. Microb. Drug Resist. 23, 177–187. 10.1089/mdr.2016.0033 [DOI] [PubMed] [Google Scholar]
- Alikhan N. F., Petty N. K., Ben Zakour N. L., Beatson S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anchordoqui M. S., De Belder D., Lucero C., Rapoport M., Faccone D., Rodriguez, et al. (2015). In vivo horizontal dissemination of the blaKPC−2 gene carried on diverse genetic platforms among clinical isolates of Enterobacteriaceae. J. Glob. Antimicrob. Resist. 3, 210–213. 10.1016/j.jgar.2015.05.001 [DOI] [PubMed] [Google Scholar]
- CLSI (2017). Performance standards for antimicrobial susceptibility testing, 27th Edn, in CLSI Supplement M100 (Wayne, PA: Clinical an Laboratory Standards Institute; ). [Google Scholar]
- Delgado-Blas J. F., Ovejero C. M., Abadia-Patiño L., Gonzalez-Zorn B. (2016). Coexistence of mcr-1 and blaNDM−1 in Escherichia coli from Venezuela. Antimicrob. Agents Chemother. 60, 6356–6358. 10.1128/AAC.01319-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominguez J. E., Figueroa Espinosa R. A., Redondo L. M., Cejas D., Gutkind G. O., Chacana, et al. (2017). Plasmid-mediated colistin resistance in Escherichia coli recovered from healthy poultry. Rev. Argent. Microbiol. 49, 297–298. 10.1016/j.ram.2017.02.001 [DOI] [PubMed] [Google Scholar]
- El Garch F., Sauget M., Hocquet D., Lechaudee D., Woehrle F., Bertrand X. (2017). mcr-1 is borne by highly diverse Escherichia coli isolates since 2004 in food-producing animals in Europe. Clin. Microbiol. Infect. 23, 51.e1–e4. 10.1016/j.cmi.2016.08.033 [DOI] [PubMed] [Google Scholar]
- Fernandes M. R., Sellera F. P., Esposito F., Sabino C. P., Cerdeira L., Lincopan N. (2017). Colistin-resistant mcr-1-positive Escherichia coli on public beaches, an infectious threat emerging in recreational waters. Antimicrob. Agents Chemother. 61:e00234–17. 10.1128/AAC.00234-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y. Y., Wang Y., Walsh T. R., Yi L. X., Zhang R., Spencer J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. 10.1016/S1473-3099(15)00424-7 [DOI] [PubMed] [Google Scholar]
- Monte D. F., Mem A., Fernandes M. R., Cerdeira L., Esposito F., Galvão J. A., et al. (2017). Chicken meat as a reservoir of colistin-resistant Escherichia coli strains carrying mcr-1 genes in South America. Antimicrob. Agents Chemother. 61:e02718–16. 10.1128/AAC.02718-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohsaki Y., Hayashi W., Saito S., Osaka S., Taniguchi Y., Koide S., et al. (2017). First detection of an Escherichia coli strain harboring the mcr-1 gene in retail domestic chicken meat in Japan. Jpn. J. Infect. Dis. 70, 590–592. 10.7883/yoken.JJID.2016.572 [DOI] [PubMed] [Google Scholar]
- Partridge S. R., Di Pilato V., Doi Y., Feldgarden M., Haft D. H., Klimke, et al. (2018). Proposal for assignment of allele numbers for mobile colistin resistance (mcr) genes. J. Antimicrob. Chemother. 73, 2625–2630. 10.1093/jac/dky262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poirel L., Jayol A., Nordmann P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. 10.1128/CMR.00064-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi F., Girardello R., Morais C., Cury A. P., Farage Martins L., Da Silva A. M., et al. (2017). Plasmid-mediated in carbapenem-susceptible Escherichia coli ST156 causing a blood infection: an unnoticeable spread of colistin resistance in Brazil? Clinics 72, 642–644. 10.6061/clinics/2017(10)09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saavedra S. Y., Diaz L., Wiesner M., Correa A., Arévalo S. A., Reyes J., et al. (2017). Genomic and molecular characterization of clinical isolates of Enterobacteriaceae harboring mcr-1 in Colombia, 2002 to 2016. Antimicrob. Agents Chemother. 61:e00841–17. 10.1128/AAC.00841-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J., Li X. P., Yang R. S., Fang L. X., Huo W., Li S. M., et al. (2016). Complete nucleotide sequence of an IncI2 plasmid coharboring blaCTX-M-55 and mcr-1. Antimicrob. Agents Chemother. 60, 5014–5017. 10.1128/AAC.00774-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tijet N., Faccone D., Rapoport M., Seah C., Pasterán F., Ceriana P., et al. (2017). Molecular characteristics of mcr-1-carrying plasmids and new mcr-1 variant recovered from polyclonal clinical Escherichia coli from Argentina and Canada. PLoS ONE 12:e0180347. 10.1371/journal.pone.0180347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turrientes M. C., González-Alba J. M., Del Campo R., Baquero M. R., Cantón R., Baquero F., et al. (2014). Recombination blurs phylogenetic groups routine assignment in Escherichia coli: setting the record straight. PLoS ONE 9:e105395. 10.1371/journal.pone.0105395 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The genetic relatedness of mcr-1-positive E. coli isolates. PFGE: Gel order; Line 2, M22607; 3, M22608; 4, M22609; 5, M22610; 6, M22611; 7, M22612; 8, M22613; 9, M22614; 10, M22615; 11, M22616; Lanes 1 and 12, S. Branderup.
Plasmid profile. S1-PFGE: Gel order; Line 2, M22608; 3, M22608-TC; 4, M22609; 5, M22609-TC; 6, M22610; 7, M22610-TC; 8, M22611; 9, M22611-TC; 10, M22612; 11, M22612-TC; 12, M22613; 13, M22613-TC; 14, M22615; 15, M22615-TC; 16, M22616; 17, M22616-TC; 19, M22607; 20, M22607-TC; 21, M22614; 22, M22624-TC; Lanes 1, 18, and 23, S. Branderup. TC: transconjugants obtained using E. coli J53 AZR as the recipient strain. Red arrows highlight plasmids containing the mcr-1 gene.