Abstract
Objective
This study aims to identify metabolites with altered levels of expression in patients with early and progressive stages of Alzheimer's disease (AD).
Methods
All participants of the study underwent genetic screening and were diagnosed using both neuropsychological assessment and amyloid imaging before metabolome analysis. According to these assessments, the patients were classified as normal (n = 15), with mild cognitive impairment (n = 10), and with AD (n = 15).
Results
Using a targeted metabolomic approach, we found that plasma levels of C3, C5, and C5‐DC acylcarnitines, arginine, phenylalanine, creatinine, symmetric dimethylarginine (SDMA) and phosphatidylcholine ae C38:2 were significantly altered in patients with early and progressive stages of AD. We created a predictive model based on the decision tree that included three main parameters: age, arginine and C5 plasma concentrations. The model distinguished AD patients from other participants with 60% sensitivity and 86.7% specificity. For healthy controls, the sensitivity was 85.7% and specificity was 61.5%. Multivariate ROC analysis to develop a decision tree showed that our model reached moderate diagnostic power in differentiating between older adults who are cognitively normal (AUC = 0.77) and those with AD (AUC = 0.72).
Interpretation
The plasma levels of arginine and valeryl carnitine, together with subject age, are promising as biomarkers for the diagnosis of AD in older adults.
Introduction
Alzheimer's disease (AD) is a growing public health problem worldwide. AD progresses slowly, taking decades before clinical symptoms manifest.1, 2, 3, 4, 5 The prodromal stage of AD provides a window of opportunity for treatment to delay the onset of the disease and slow the progress of neurodegeneration.6, 7 Thus, diagnostic methods for the disease during the prodromal stage are important. Several methods are based on the detection of amyloid‐β (Aβ) and neurofibrillary tangles,8 which begin to form decades before any symptoms of dementia appear.9 These first signs of AD have been detected using amyloid‐β positron‐emission tomography (PET) imaging. Amyloid imaging using radiotracers such as Pittsburgh compound B and 18F‐Florbetapir is a promising tool for diagnosing AD during the prodromal stage.3 The measurement of Aβ42 and tau protein levels in cerebrospinal fluid (CSF) provides another means of identifying AD at early stage.10, 11 However, these methods are invasive, time‐consuming, and expensive. Thus, the identification of blood‐based biomarkers capable of indicating early pathology preceding symptoms is crucial for AD.
Several molecules have been investigated recently as plasma markers of AD, including folate 12 and a variety of inflammation‐related proteins.13 The ratios of plasma amyloid‐β precursor protein (APP)669–711/amyloid‐β (Aβ)1–42 and Aβ 1–40/Aβ 1–42 are predictive of patient brain amyloid‐β status and correlate with the levels of Aβ1–42 in CSF.14 Several studies suggest that high blood cholesterol concentration is a risk factor for AD.15, 16 Studies of a blood‐based lipid biomarker panel show conflicting results, with a set of ten targeted phosphatidylcholine, lysophophatidylcholine, and acetylcarnitines showing 90% accuracy in predicting AD,17 while the same methods applied to a larger cohort failed to replicate these findings.18
The lack of reproducibility of metabolome study results is a major obstacle to obtaining reliable blood‐based biomarkers for AD. One contributor to such low reproducibility is the incorrect initial diagnosis of subjects.4 AD is typically diagnosed by three stages of progression: preclinical, characterized by brain pathology, including amyloid aggregation and neuronal changes but without significant clinical symptoms; mild cognitive impairment (MCI), marked by memory and cognitive problems; and Alzheimer's dementia, the final stage of the disease associated with memory loss and other cognitive problems.7, 19 However, the MCI diagnosis is only 50–70% accurate, even when assessed by an experienced specialist. The addition of amyloid imaging results to the clinical judgment improves the accuracy rate of diagnosis to 80% or higher.19, 20 Another diagnostic indicator of AD is the apolipoprotein E ε4 allele, which is the strongest risk factor for sporadic AD.21
Our study aims to identify potential diagnostic biomarkers of MCI and AD through the analysis of blood plasma metabolites of subjects carefully diagnosed using clinical judgment, amyloid imaging results, and apolipoprotein E status. After identifying several metabolites altered in MCI and AD patients, we develop a predictive model capable of distinguishing MCI and AD patients from normal subjects.
Materials and Methods
Diagnostic criteria and grouping
The cohort inclusion criteria are as follows: (1) a Hachinski Ischemic Score <4 and a Geriatric Depression Scale score < 6; (2) at least 6 grades of education; (3) age 55–90 years. All individuals were identified as normal controls (NC; n = 15), mild cognitive impairment (MCI; n = 10), or Alzheimer's disease (AD; n = 15) using clinical data, family information, and neuropsychological tests to ascertain meeting further inclusion criteria, as described below.
The Mini‐Mental State Exam (MMSE) is a widely used test for the elderly with aging, MCI and AD in clinical practice; it includes tests of orientation, attention, memory or recall, registration, calculation, language and ability to follow simple commands. WMS is used to assess memory deficits, particularly in differentiating between MCI and normal aging.22, 23, 24 The ADAS‐cog was used as a diagnostic tool to further evaluate mild and moderate AD which was not performed on subjects in NC and MCI groups.25
The NC subjects were recruited from a pool of patient spouses, hospital volunteers, and individuals from the surrounding community. The NC group inclusion criteria are as follows: no significant impairment in cognitive function or daily living activities; a MMSE score of 24–30; a clinical dementia rating (CDR) of 0; a delayed recall of story A in the Logical Memory (LM) subtest of the Chinese version of the Wechsler Memory Scale Logical Memory III (WMS‐III) ≥9 for those with education ≥16 years and ≥5 for those with education 6–15 years; negative for the Apo Ɛ4 allele.
The MCI group inclusion criteria are as follows: MMSE score of 24–30; nondemented; CDR 0.5, with a mandatory requirement of the memory box score ≥ 0.5; delayed recall of story A from the LM subtest of the Chinese version WMS‐III ≤8 for those with education ≥16 years and ≤4 with education 6–15 years; carry at least one copy of the Apo Ɛ4 allele.
The AD inclusion criteria are as follows: meet the criteria of the Diagnostic and Statistical Manual of Mental Disorders, 4th edition and National Institute of Neurological and Communicative Disorders and Stroke/Alzheimer's Disease and Related Disorders Association (NINCDS‐ADRDA); carry at least one copy of the Apo Ɛ4 allele. Disease severity was graded according to the Clinical Dementia Rating (1, mild; 2, moderate) and MMSE to determine cognitive function. The protocol was approved by the institutional review board of Chang Gung Memorial Hospital (103‐3230B, 103‐6317C and 104‐1812C). The details of each evaluation are further described in the Data S1.
Genetic analysis of ApoE allele
Genomic DNA was extracted from EDTA blood samples and used for genotyping. The genetic polymorphism of the candidate genes was determined using polymerase chain reaction and verified by restriction fragment length polymorphism analysis.
Analysis of metabolites
The AbsoluteIDQ180 kit (Biocrates Life Science, Innsbruck, Austria) was used to determine metabolite concentrations in the serum of all subjects. The kit is based on a combination of LC‐MS/MS assay and direct flow injection assay, identifying and quantifying 185 endogenous metabolites in five different compound classes: 40 acylcarnitines, 21 amino acids, 19 biogenic amines, 1 sugar, 15 sphingomyelins, and 90 glycerophospholipids. The assay was performed using a Waters Acquity UPLC System with Xevo TQ‐S Mass Spectrometry (Department of Laboratory medicine at Chang Gung Memorial Hospital) according to the manufacturer's instruction. Briefly, the serum samples were thawed, vortexed and centrifuged at 13,000g. A total of 10 μL of the sample supernatant was loaded on a filter paper and dried under nitrogen flow. An amount of 20 μL of 5% phenyl‐isothiocyanate was added for derivatization. After 20‐min incubation, the filter spots were dried under nitrogen flow for 45 min. The metabolites were then extracted by the addition of 300 μL of methanol containing 5 mmol/L ammonium acetate and collected into 96‐well plates. The extracts were suitable for mass spectrometry analysis. The data were analyzed by principal components to find variables that correlated across the samples. Data were analyzed using a web‐based server MetaboAnalyst (www.metaboanalyst.ca) for metabolomic data analysis.26, 27
Statistical analysis
Continuous variables were presented as the mean ± standard deviation, and the differences between groups were analyzed using the Kruskal–Wallis test. Dunn's post hoc analysis was performed to determine differences between the three CDR groups (0, 0.5, or ≥1) for multiple testing. Statistically significant markers were identified by comparing the plasma concentrations between CDR groups. The subjects were randomly divided into the training group and validation group at a ratio of 1:1. Decision tree analysis was performed using the C5.0 algorithm of R program, a nonparametric technique. After selecting the candidate predictors in the training group, the prediction accuracy was evaluated in the validation group. Sensitivity and specificity were calculated between the NC, MCI, and AD groups. Multivariate receiver operating characteristic (ROC) curve analysis with leave‐one‐out‐cross validation was used to evaluate the diagnostic power of the selected markers at baseline and decision tree. All statistical tests were two‐sided (P < 0.05). Statistical analyses were performed using SAS 9.4 (Windows NT version, SAS Institute, Inc., Cary, NC, USA).
Results
Demographic and metabolome characteristics
The study cohort included 40 participants: 15 normal controls, 10 subjects in the MCI group, and 15 in the AD group. The PET scan was abnormal (>1) for the MCI and AD subjects and normal (=1) for normal controls. The mean age of the AD group was significantly higher than that of the NC group (Table S1). Analysis of the dataset of 180 metabolites revealed significant differences in the distribution of C3, C5, C5‐DC/C6‐OH, arginine, phenylalanine, creatinine, SDMA, and PC ae C38:2 between the groups (Table S1). Significant differences were observed in the distribution of C3 and C5 between the MCI and AD groups; arginine, phenylalanine, and SDMA between the MCI and NC groups; and creatinine distribution between all groups.
Validation of the selected characteristics
The age and significantly different metabolome characteristics were further analyzed using multivariate ROC (Fig. 1). The results show good discrimination power for these characteristics between groups (AUC = 0.59–0.76). We also conducted enforced ROC analysis with two additional biomarker ratios (Cit/Arg and Kyn/Trp) and found that the addition of these extra variables decreased the discrimination power (Fig. 1).
Figure 1.
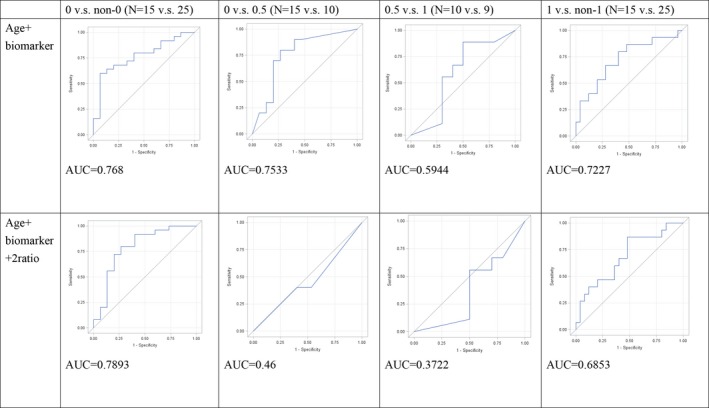
Results of multivariate ROC analysis. ROC analysis plots for pairwise comparison based on CDR values using age and significant metabolite levels. Results for the combination of age and individual metabolites are shown in the top row. The bottom row contains plots obtained after adding Cit/Arg and Kyn/Trp ratio biomarkers. The curves show the best delimitation of AUC for each combination. Designation 0 versus non‐0 refers to CDR and indicates the power of that particular marker combination to discriminate normal controls from other participants. Similarly, 0 versus 0.5 refers to discrimination of normal controls from MCI patients (0.5 vs. 1, discrimination between MCI and AD; 1 vs. non‐1, discrimination between AD and non‐AD participants).
Creating a decision tree and verification of prediction accuracy
We used decision tree analysis to find the best candidate predictors of AD and MCI. A 1:1 random assignment was performed before creating the decision tree. The distribution of the selected characteristics shows that the random assignment is similar between the training and validation groups (Table S2). The only exception is C3 concentration, which was excluded from further analysis.
Application of the C5.0 decision tree algorithm yielded good discrimination of Alzheimer disease in the training group (accuracy, 85%). We found that arginine concentration, age, and C5 concentration were the best discriminative variables for Alzheimer's disease classification (Fig. 2).
Figure 2.
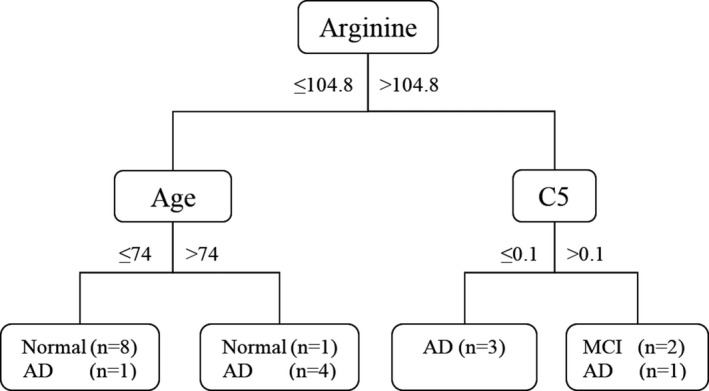
Decision tree model generated in the training group. C5.0 classification tree for the best candidate predictors of AD and MCI. Boxes at the end of each classification path contain the number of participants satisfying the selected conditions in each of the diagnostic group. The decision values are given as years of age and μmol/L for arginine and C5 blood plasma concentrations. MCI, mild cognitive impairment; AD, Alzheimer's disease
The root node was arginine concentration. Participants with arginine concentration under 104.8 μmol/L were divided into two groups according to age. The group under the age of 74 mainly consisted of normal controls, while the group over 74 years mainly consisted of AD patients. The second node for the branch of arginine over 104.8 μmol/L was C5 concentration. All participants with C5 concentration under 0.1 μmol/L were AD patients and those with C5 over 0.1 μmol/L were either MCI or AD.
After selecting the candidate predictors in the training group, we applied the predictive model to the validation group to build a confusion matrix (Table 1). The sensitivity, specificity, and accuracy of the model were calculated (Table 2). We found that the accuracy was 60%. The group prediction of the model showed the best result for the NC group, with 85.7% of the group classified correctly. In the AD group, 60% of the patients were classified correctly, with the remaining 40% in the NC group. The poorest result was observed for the MCI group, as only 37.5% of patients were classified correctly; the remaining 62.5% were distributed nearly evenly between the NC and AD groups.
Table 1.
Confusion matrix for validation group
| Predict | Golden standard | Total | ||
|---|---|---|---|---|
| Normal | MCI | AD | ||
| Normal | 6 (30%) | 3 (15%) | 2 (10%) | 11 |
| MCI | 1 (5%) | 3 (15%) | 0 (0%) | 4 |
| AD | 0 (0%) | 2 (10%) | 3 (15%) | 5 |
| Total | 7 | 8 | 5 | 20 |
Golden standard refers to validation group. Columns represent actual normal, MCI, and AD participants. Rows represent classification based on the decision tree model. Accuracy is 60%. MCI, mild cognitive impairment; AD, Alzheimer's disease.
Table 2.
Prediction of metabolome classification for validation group
| Normal versus non‐normal | MCI versus non‐MCI | AD versus non‐AD | |
|---|---|---|---|
| Sensitivity | 85.7% | 37.5% | 60% |
| Specificity | 61.5% | 91.7% | 86.7% |
The values are calculated using the confusion matrix obtained with the decision tree model. MCI, mild cognitive impairment; AD, Alzheimer's disease.
To further evaluate the model, we performed multivariate ROC analysis for the decision tree (Fig. 3). We found that the model was best suited for discriminating AD patients (AUC = 0.75). Comparison of the latter ROC results with those obtained for the collective biomarkers showed that using the selected markers (arginine, age, and C5) for creating the decision tree increased the ability of the model to discriminate AD patients from other subjects (compare Fig. 3 to Fig. 1). Overall, the results of ROC analysis suggest that the decision tree developed, while having only moderate diagnostic power, has potential with further investigations to differentiate cognitively normal older people from those with AD.
Figure 3.
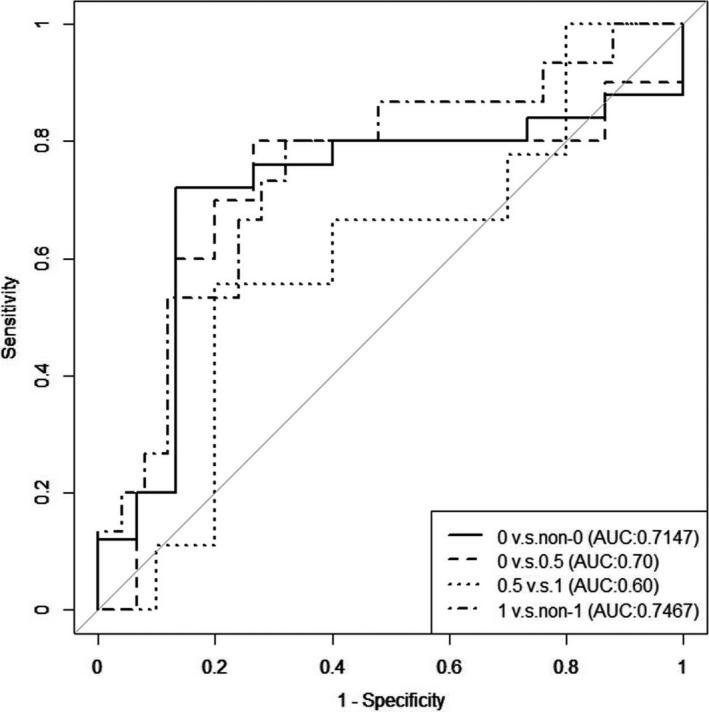
Results of multivariate ROC analysis for decision tree construction. The figure shows plots of ROC analysis for pairwise comparison of the groups based on CDR value using the combination of three markers: age, arginine concentration, and C5 concentration. The curves show the best delimitation of AUC for each comparison. The compared pairs are the same as those described in Figure 1.
Discussion
To search for potential AD biomarkers, we used a targeted metabolomic approach to assay dozens of blood plasma metabolites simultaneously. We observed that acylcarnitines, amino acids, biogenic amines, and phosphatidylcholine PC ae C38:2 differ between normal controls and MCI and AD patients. Post hoc pairwise comparison showed that C5 and C3 acylcarnitine concentrations are significantly lower in AD compared to MCI, while arginine, phenylalanine, creatinine, and SDMA concentrations are elevated in MCI compared to normal controls. Our predictive model based on age and plasma concentrations of arginine and C5 distinguished AD patients from other participants with 60% sensitivity and 86.7% specificity. For healthy controls, we observed a sensitivity of 85.7% and specificity of 61.5%. Our results suggest that plasma levels of arginine and valeryl carnitine, along with subject age, can be used with acceptable diagnostic power to differentiate between cognitively normal and AD adults.
The age distribution differed significantly between the groups is not surprising, as nearly every study reports an association between increasing patient age and AD progression.28 However, age is not often used in combination with metabolic parameters for creating predictive models. In our study, combining subject age with significant metabolome characteristics (C3, C5, C5‐DC, arginine, phenylalanine, creatinine, SDMA and PC ae C38:2) in multivariate ROC analysis provided discriminated normal controls (AUC = 0.77) and AD (AUC = 0.72) with good accuracy but with poor accuracy for MCI (AUC = 0.59). Creating a decision tree with three significant factors‐ age, C5 and arginine concentrations slightly increased the predictive power for AD.
In previous studies, plasma and serum levels of a large number of metabolites were evaluated for MCI and AD patients and for healthy older adults who converted to MCI/AD.29, 30, 31 The elevated levels of arginine in MCI and AD subjects observed in our study are consistent with previous reports of high levels of l‐arginine in stable MCI subjects and in those who converted to AD. These patients demonstrated significant alterations in l‐arginine and polyamine metabolism.32 These metabolic pathways are closely linked, as polyamine synthesis starts with the conversion of arginine to ornithine by arginase.33 Several groups have found altered levels of arginase expression in AD brains, which, along with reduced ornithine decarboxylase and polyamine levels, suggest a link between arginine metabolism and AD.34, 35 The activity and protein expression of nitric oxide synthase and arginase, along with tissue concentrations of L‐arginine and its downstream metabolites, are shown to be altered with AD.36 In CVN‐AD mice, which produce low levels of immune‐mediated nitric oxide (comparable to that of humans), treatment with the arginase and ornithine decarboxylase inhibitor difluoromethylornithine (DFMO) reduced their AD‐like symptoms.37 Likewise, another research group showed that people with superior memory, regarded as the opposite of AD patients, had lower levels of L‐arginine.30 Together, these findings suggest that altered arginine metabolism plays an essential role in AD pathogenesis.
NO initiates brain lesion development during AD pathogenesis.38 The production of NO is reflected by an increase in the citrulline/arginine ratio, which increases during the development of clinical dementia. Neopterin concentrations were observed to correlate with the Cit/Arg ratio only among demented subjects.39 Thus, we included the Cit/Arg ratio in our analysis. While the previous findings indicate that the Cit/Arg ratio should be elevated in patients, we observed no difference in this ratio between groups in our study.
We report here that the serum concentrations of two acylcarnitines (C3 and C5) were reduced in AD patients and elevated in MCI patients. Previous studies have reported low levels of some acylcarnitines in both AD and MCI patients.30, 40 These results may differ because of the different methodological approaches and analytical tools used in the metabolomic analyses. Preanalysis sample preparation and storage can also contribute to result variations, as some metabolites may be unstable after one or two freeze‐thaw cycles.41 Another possible explanation is differences in the cohort size and in demographic and clinical characteristics of the subjects between these studies.
Low levels of several acylcarnitines are reported in AD patients.30 Acylcarnitines are important for the β‐oxidation of fatty acids in mitochondria, suggesting that their deficiency may alter energy metabolism in the brain.42 Consistent with this hypothesis is the observation that AD patients have significantly lower than normal carnitine shuttle activity.43 Disruption of the carnitine shuttle may be one cause of mitochondrial dysfunctions associated with neurodegenerative diseases including AD.44 In addition to fatty acid metabolism, acylcarnitines are involved in other brain processes that may play a role in AD pathology, including acetylcholine and phospholipid synthesis, gene expression modulation, cholinergic neurotransmission, elimination of oxidative products, and protection from excitotoxicity.42, 45 Acylcarnitines are reported to exert neuroprotective effects and show promising results as potential therapeutic agents.42, 46, 47
We should note that numerous acylcarnitines are commonly detected in metabolomic assays; thus, the acylcarnitines reported as present in low levels in other studies actually may be different types of acylcarnitine. Surprisingly, we observed no significantly low plasma level for any acylcarnitine in MCI patients. In addition to methodological differences and heterogeneity of the samples, these discrepant results may be explained by the complexity of AD, with several subtypes characterized by different mechanisms and pathology progression.48, 49 However, inconsistency of the results can also be explained by the limitations of our study. First, the sample size, especially the number of MCI patients, was small and should be increased in the future studies. Second, the effects habits such as smoking or alcohol consumption before and during the study period should be evaluated. Third, we focused only on normal controls and stable MCI and AD patients; longitudinal follow‐up study would allow for the identification of metabolites that changed in concentration during the conversion from normal to MCI and from MCI to AD. The present work is a pilot study and should be built upon taking all these limitations into account.
In conclusion, we want to emphasize the importance of accurate diagnosis of subjects before using a metabolomic approach. Although we have not identified biomarkers specific for MCI, our results suggest that plasma levels of arginine and valeryl carnitine along with the subjects’ age can be used for creating a predictive model with moderate accuracy for differentiating cognitively normal older adults and those with AD.
Conflict of Interest
None.
Supporting information
Data S1: The details and related reference of clinical evaluation, neuropsychological assessment and [18F] AV‐45 PET imaging are descripted in supplemental materials.
Table S1. Cohort demographics and metabolome characteristics
Table S2. Characteristic of training and validation groups.
Acknowledgments
This study was sponsored by a grant (CMRPG3F0511) from Chang Gung Memorial Hospital. The authors appreciate the Imaging Core Laboratory, Institute for Radiological Research, Chang Gung University/Chang Gung Memorial Hospital, Linkou for the imaging analysis. The metabolomics analysis using LC‐MS and/or NMR spectroscopy was carried out at the Metabolomics Core Laboratory, Healthy Aging Research Center, Chang Gung University and Clinical Metabolomics Core Laboratory, Chang Gung Memorial Hospital.
Funding Information
This study was sponsored by a grant (CMRPG3F0511) from Chang Gung Memorial Hospital.
Funding Statement
This work was funded by Chang Gung Memorial Hospital grant CMRPG3F0511.
References
- 1. Fargo K. Alzheimer's association report: 2014 Alzheimers disease facts and figures. Alzheimer's Dement 2014;10:e47–e92. [DOI] [PubMed] [Google Scholar]
- 2. Brookmeyer R, Johnson E, Ziegler‐Graham K, Arrighi HM. Forecasting the global burden of Alzheimer's disease. Alzheimer's Dement 2007;3:186–191. [DOI] [PubMed] [Google Scholar]
- 3. Dubois B, Feldman HH, Jacova C, et al. Revising the definition of Alzheimer's disease: a new lexicon. Lancet Neurol 2010;9:1118–1127. [DOI] [PubMed] [Google Scholar]
- 4. O'Bryant SE, Mielke MM, Rissman RA, et al. Blood‐based biomarkers in Alzheimer disease: current state of the science and a novel collaborative paradigm for advancing from discovery to clinic. Alzheimer's Dement 2017;13:45–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Crous‐Bou M, Minguillón C, Gramunt N, Molinuevo JL. Alzheimer's disease prevention: from risk factors to early intervention. Alzheimers Res Ther 2017;9:71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sevigny J, Chiao P, Bussière T, et al. The antibody aducanumab reduces Aβ plaques in Alzheimer's disease. Nature 2016;537:50–56. [DOI] [PubMed] [Google Scholar]
- 7. Sperling RA, Aisen PS, Beckett LA, et al. Toward defining the preclinical stages of Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement 2011;7:280–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Caselli RJ, Beach TG, Knopman DS, Graff‐Radford NR. Alzheimer disease: scientific breakthroughs and translational challenges. Mayo Clin Proc 2017;92:978–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Perrin RJ, Fagan AM, Holtzman DM. Multimodal techniques for diagnosis and prognosis of Alzheimer's disease. Nature 2009;461:916–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Liu Z, Chen HH, Li TL, et al. A cross‐sectional study on cerebrospinal fluid biomarker levels in cognitively normal elderly subjects with or without a family history of Alzheimer's disease. CNS Neurosci Ther 2013;19:38–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kuo H‐C, Yen H‐C, Huang C‐C, et al. Cerebrospinal fluid biomarkers for neuropsychological symptoms in early stage of late‐onset Alzheimer's disease. Int J Neurosci 2015;125:747–754. [DOI] [PubMed] [Google Scholar]
- 12. Ikonomovic MD, Klunk WE, Abrahamson EE, et al. Post‐mortem correlates of in vivo PiB‐PET amyloid imaging in a typical case of Alzheimer's disease. Brain 2008;131:1630–1645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Zhou WW, Lu S, Su YJ, et al. Decreasing oxidative stress and neuroinflammation with a multifunctional peptide rescues memory deficits in mice with Alzheimer disease. Free Radic Biol Med 2014;74:50–63. [DOI] [PubMed] [Google Scholar]
- 14. Nakamura A, Kaneko N, Villemagne VL, et al. High performance plasma amyloid‐β biomarkers for Alzheimer's disease. Nature 2018;554:249–254. [DOI] [PubMed] [Google Scholar]
- 15. Yadav RS, Tiwari NK. Lipid integration in neurodegeneration: an overview of Alzheimer's Disease. Mol Neurobiol 2014;50:168–176. [DOI] [PubMed] [Google Scholar]
- 16. Morris JK, Honea RA, Vidoni ED, et al. Is Alzheimer's disease a systemic disease?. Biochim Biophys. Acta ‐ Mol Basis Dis 2014;1842:1340–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Mapstone M, Cheema AK, Fiandaca MS, et al. Plasma phospholipids identify antecedent memory impairment in older adults. Nat Med 2014;20:415–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Casanova R, Varma S, Simpson B, et al. Blood metabolite markers of preclinical Alzheimer's disease in two longitudinally followed cohorts of older individuals. Alzheimer's Dement 2016;12:815–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. McKhann GM, Knopman DS, Chertkow H, et al. The diagnosis of dementia due to Alzheimer's disease: recommendations from the National Institute on Aging‐Alzheimer's Association workgroups on diagnostic guidelines for Alzheimer's disease. Alzheimer's Dement 2011;7:263–269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dubois B, Feldman HH, Jacova C, et al. Research criteria for the diagnosis of Alzheimer's disease: revising the NINCDS‐ADRDA criteria. Lancet Neurol 2007;6:734–746. [DOI] [PubMed] [Google Scholar]
- 21. Martínez‐Morillo E, Hansson O, Atagi Y, et al. Total apolipoprotein E levels and specific isoform composition in cerebrospinal fluid and plasma from Alzheimer's disease patients and controls. Acta Neuropathol 2014;127:633–643. [DOI] [PubMed] [Google Scholar]
- 22. Qui S, Chang GH, Panagia M, et al. Fusion of deep learning models of MRI scans, mini–mental state examination, and logical memory test enhances diagnosis of mild cognitive impairment. Alzheimer's & Dementia 2018;10:737–749. doi.org/10.1016/j.dadm.2018.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wind AW, Schellevis FG, Van Staveren G. Limitations of the mini‐mental state examination in diagnosing dementia in general practice. Int J Geriatr Psychiatry 1997;12:101–108. [DOI] [PubMed] [Google Scholar]
- 24. Edmonds EC, Delano‐Wood L, Clark LR, et al. Susceptibility of the conventional criteria for mild cognitive impairment to false‐positive diagnostic errors. Alzheimer's & Dementia 2015;11:415–424. 10.1016/j.jalz.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Podhorna J, Krahnke T, Shear M, Harrison J. Alzheimer's disease assessment scale‐cognitive subscale variants in mild cognitive impairment and mild Alzheimer's disease: change over time and the effect of enrichment strategies. Alzheimer's Research & Therapy 2016;8:8 10.1186/s13195-016-0170-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xia J, Mandal R, Sinelnikov IV, et al. MetaboAnalyst 2.0‐a comprehensive server for metabolomic data analysis. Nucleic Acids Res 2012;40:127–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xia J, Wishart DS. Web‐based inference of biological patterns, functions and pathways from metabolomic data using metaboanalyst. Nat Protoc 2011;6:743–760. [DOI] [PubMed] [Google Scholar]
- 28. Craig LA, Hong NS, McDonald RJ. Revisiting the cholinergic hypothesis in the development of Alzheimer's disease. Neurosci Biobehav Rev 2011;35:1397–1409. [DOI] [PubMed] [Google Scholar]
- 29. Goedert M, Spillantini MG. REVIEWS a century of Alzheimer’ s disease. Science (80‐.) 2006;314:777–781. [DOI] [PubMed] [Google Scholar]
- 30. Mapstone M, Lin F, Nalls MA, et al. What success can teach us about failure: the plasma metabolome of older adults with superior memory and lessons for Alzheimer's disease. Neurobiol Aging 2017;51:148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. González‐Domínguez R, García A, García‐Barrera T, et al. Metabolomic profiling of serum in the progression of Alzheimer's disease by capillary electrophoresis‐mass spectrometry. Electrophoresis 2014;35:3321–3330. [DOI] [PubMed] [Google Scholar]
- 32. Graham SF, Chevallier OP, Elliott CT, et al. Untargeted metabolomic analysis of human plasma indicates differentially affected polyamine and L‐Arginine metabolism in mild cognitive impairment subjects converting to alzheimer's disease. PLoS ONE 2015;10:1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Minois N, Carmona‐Gutierrez D, Madeo F. Polyamines in aging and disease. Aging (Albany. NY) 2011;3:716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Colton CA, Mott RT, Sharpe H, et al. Expression profiles for macrophage alternative activation genes in AD and in mouse models of AD. J Neuroinflammation 2006;3:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hansmannel F, Sillaire A, Kamboh MI, et al. NIH Public Access. 2011;21:1013–1021. [Google Scholar]
- 36. Liu P, Fleete MS, Jing Y, et al. Altered arginine metabolism in Alzheimer's disease brains. Neurobiol Aging 2014. Sep;35:1992–2003. [DOI] [PubMed] [Google Scholar]
- 37. Kan MJ, Lee JE, Wilson JG, et al. Arginine deprivation and immune suppression in a mouse model of Alzheimer's disease. J Neurosci 2015;35:5969–5982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Aliev G, Palacios HH, Lipsitt AE, et al. Nitric oxide as an initiator of brain lesions during the development of Alzheimer disease. Neurotox Res 2009;16:293–305. [DOI] [PubMed] [Google Scholar]
- 39. Coppus AM, Fekkes D, Verhoeven WM, et al. Plasma levels of nitric oxide related amino acids in demented subjects with down syndrome are related to neopterin concentrations. Amino Acids 2010;38:923–928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ciavardelli D, Piras F, Consalvo A, et al. Medium‐chain plasma acylcarnitines, ketone levels, cognition, and gray matter volumes in healthy elderly, mildly cognitively impaired, or Alzheimer's disease subjects. Neurobiol Aging 2016;43:1–12. [DOI] [PubMed] [Google Scholar]
- 41. Breier M, Wahl S, Prehn C, et al. Targeted metabolomics identifies reliable and stable metabolites in human serum and plasma samples. PLoS ONE 2014;9:e89728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Jones LL, McDonald DA, Borum PR. Acylcarnitines: role in brain. Prog Lipid Res 2010;49:61–75. [DOI] [PubMed] [Google Scholar]
- 43. Stempler S, Yizhak K, Ruppin E. Integrating transcriptomics with metabolic modeling predicts biomarkers and drug targets for Alzheimer's disease. PLoS ONE 2014;9:e105383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hussain Bhat A, Dar KB, Anees S, et al. Oxidative stress, mitochondrial dysfunction and neurodegenerative diseases; a mechanistic insight. Biomed Pharmacother 2015;74:101–110. [DOI] [PubMed] [Google Scholar]
- 45. Onofrj M, Ciccocioppo F, Varanese S, et al. Acetyl‐L‐carnitine: from a biological curiosity to a drug for the peripheral nervous system and beyond. Expert Rev Neurother 2013;13:925–936. [DOI] [PubMed] [Google Scholar]
- 46. Pettegrew JW, Klunk WE, Panchalingam K, et al. Clinical and neurochemical effects of acetyl‐L‐carnitine in Alzheimer's disease. Neurobiol Aging 1995;16:1–4. [DOI] [PubMed] [Google Scholar]
- 47. Zanelli SA, Solenski NJ, Rosenthal RE, Fiskum G. Mechanisms of ischemic neuroprotection by acetyl‐L‐carnitine. Ann N Y Acad Sci 2005;1053:153–161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Jahn H. Memory loss in alzheimer's disease. Dialogues Clin Neurosci. 2013;15:445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Montgomery SA, Thal LJ, Amrein R. Meta‐analysis of double blind randomized controlled clinical trials of acetyl‐L‐carnitine versus placebo in the treatment of mild cognitive impairment and mild Alzheimer's disease. Int Clin Psychopharmacol 2003;18:61–71. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data S1: The details and related reference of clinical evaluation, neuropsychological assessment and [18F] AV‐45 PET imaging are descripted in supplemental materials.
Table S1. Cohort demographics and metabolome characteristics
Table S2. Characteristic of training and validation groups.


