Abstract
Riboswitches are RNA sensors that affect post-transcriptional processes through their ability to bind to small molecules. Thiamine pyrophosphate (TPP) riboswitch class is the most widespread riboswitch occurring in all three domains of life. Even though it controls different genes involved in the synthesis or transport of thiamine and its phosphorylated derivatives in bacteria, archaea, fungi, and plants, the TPP aptamer has a conserved structure. In this study, we aimed at understanding differences in the structural dynamics of TPP riboswitches from Escherichia coli and Arabidopsis thaliana, based on their crystallographic structures (TPPswec and TPPswat, respectively) and dynamics in aqueous solution, both in apo and holo states. A combination of Molecular Dynamics Simulations and Network Analysis empowered to find out slight differences in the dynamical behavior of TPP riboswitches, although relevant for their dynamics in bacteria and plants species. Our results suggest that distinct interactions in the microenvironment surrounding nucleotide U36 of TPPswec (and U35 in TPPswat) are related to different responses to TPP. The network analysis showed that minor structural differences in the aptamer enable enhanced intramolecular communication in the presence of TPP in TPPswec, but not in TPPswat. TPP riboswitches of plants present subtler and slower regulation mechanisms than bacteria do.
Introduction
Riboswitches are natural ribonucleic acid (RNA) sensors that affect post-transcriptional processes through their ability to bind to small molecules such as vitamins, amino acids and nucleotides1,2. Riboswitches are highly conserved and structured elements located in the untranslated regions (UTRs) or introns of pre-mRNAs and can be found in all the three domains of life3–5.
Riboswitches have been shown to modulate gene expression by influencing transcription, translation, alternative splicing and RNA stability6–8. Typically, genes regulated by a given riboswitch are involved in biosynthesis, catabolism, signaling or transport of a metabolite that binds to the riboswitch, creating feedback regulation mechanisms to control its adequate levels9. When the threshold of a metabolic pathway increases, binding of the metabolite to the Riboswitch leads to a negative feedback mechanism resulting in the suppression of the involved genes. Riboswitches display high specificity for the substrate, conferring efficiency to carry out their activity even in the presence of other similar metabolites10.
The structure of riboswitches consists of a two-domain set: the sensory and the regulatory domains. The sensory domain is the aptamer, whose sequence and structure are highly conserved. It acts as a receptor for particular metabolites, whereas the binding of small molecules are transduced to genetic regulatory signals by the expression platform localized adjacent to the aptamer10. This is achieved by a structural rearrangement, resulting in an immediate RNA conformational change that alters mRNA translation.
To date, the only riboswitch described in eukaryotes was the TPP riboswitch, since most of the studies have been carried out in prokaryotic organisms11. In bacteria, such as Escherichia coli, two functionally different TPP riboswitches were reported. The first one is located in the 5′UTR region of the thiM gene, where it controls gene expression at the translation level11,12, while the second guides both translation and transcription of 5′ UTR region of the thiC gene11.
TPP riboswitches have been found in 5′ UTRs regions of genes encoding thiamine biosynthetic enzymes in fungi13,14, and algae15, in which they promote alternatively spliced transcripts. Conversely, in all species of plants previously studied, the TPP riboswitch resides in the 3′ UTRs region of the thiC gene. This difference in mRNA localization suggests a unique mode of action for plant riboswitches8,16.
The TPP aptamer has sequence and structures highly conserved, independently if the gene is involved at different stages of the synthesis or transport processes of thiamine in bacteria, archaea, fungi, and plants. Rfam database17 has a total of 9180 TPP riboswitch sequences (Rfam accession RF00059) from distinct organisms, and the consensus secondary structure of these entries is highly conserved. Structurally, the TPP aptamer consists of five stems. The P1 stem is responsible for connecting the aptamer domain to the expression platform. Stems P2 and P3 are involved in binding of the TPP pyrimidine ring while stems P4 and P5 bind to the pyrophosphate group.
Most of the observed differences among species are found in the P3 stem region. For instance, bacteria and archaea commonly have a P3a stem18 not observed in eukaryotic riboswitches, whereas in eukaryotic organisms, the P3 stem is significantly variable in length, sequence, and base pairings19.
Despite the fact that both prokaryotic and eukaryotic TPP aptamers are structurally similar and play roles in the regulation of genes involved in thiamine biosynthesis, smaller structural differences may affect the way TPP binds and influences riboswitches in species of bacteria and plants. To analyze these differences, we performed a computational study using molecular dynamics simulations and correlation network analysis to compare the TPP riboswitch dynamical behavior in two of theses organisms. Our results suggest that distinct interactions in the microenvironment surrounding nucleotide U36 of TPPswec (and U35 in TPPswat) are related to different responses to TPP. We also showed that minor structural differences in TPP riboswitch made the regulatory mechanism of A. thaliana subtler and slower than E. coli.
Results
RNA content analysis
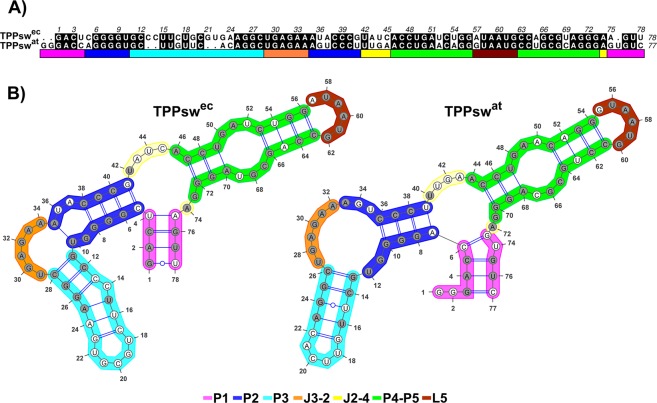
Nucleotide sequence alignment between of Escherichia coli (TPPswec) and Arabidopsis thaliana (TPPswat) revealed they have 68% of identity (Fig. 1A), along with highly conserved secondary structures (Fig. 1B). According to the SimTree server20, the secondary structure analysis indicated a normalized score of 0.68 in a scale from 0 to 1, where 1 indicates a perfect match and 0 no match at all. Both sequence and secondary structure share similar conservation.
Figure 1.
Sequence alignment and secondary structures of TPPswec and TPPswat. (A) Sequence alignment between TPPswec and TPPswat. Black filled positions of the alignment represent conserved residues. (B) Secondary structure of TPPswec and TPPswat. Conserved nucleotides were colored in grey. Stems, loops, and junctions were identified according to the figure legend caption.
A detailed inspection of the secondary structure content showed that the aptamers domains display identical J3-2 junctions and an equal number of pairings in the P1, P4, and P5 helices. On the other hand, the P3 helix was the least conserved substructure, presenting four nucleotides and an additional base pair in TPPswec. It is noteworthy that although the number of nucleotides in the P1 helix of TPPswec is smaller than in TPPswat, the number of base pairing remains the same, being four for each case. Differences can also be found in the J2-4 junction and P2 helix, in which an additional pairing in TPPswec is formed. Altogether, the 2D riboswitch structures are highly conserved between species, although minor differences are observed mainly concerning the content of base pairings, being TPPswec two pairings longer than TPPswat.
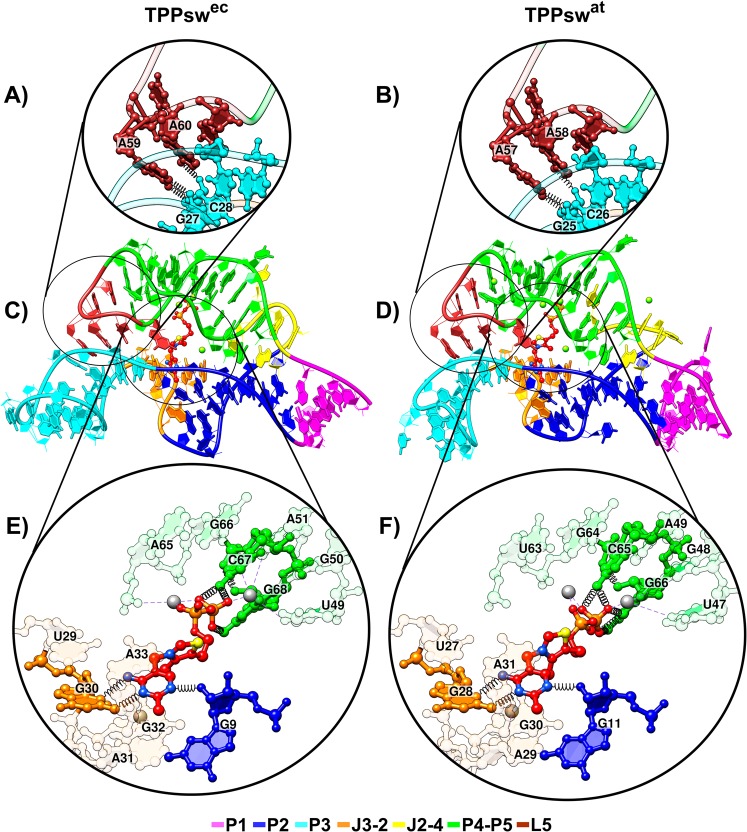
Both prokaryotic and eukaryotic TPP aptamers share a notable common 3D structure and organization. The superposition between TPPswec and TPPswat crystal structures resulted in heavy atoms root-mean-square deviation (RMSD) of approximately 0.63 Å (Fig. 2C,D). The insertion and relative position of TPP is also similar presenting a 0.69 Å RMSD. The aptamer consists of a switch helix (P1) and two sensor arms (P2/P3 and P4/P5), forming a long-range tertiary rearrangement capable of stabilizing the interaction between the L5 loop and the P3 stem (Fig. 2A,B). In both crystal structures, the P2/P3 arm helps accommodate the TPP pyrimidine ring while residues from the P4/P5 arm interact with the TPP pyrophosphate group (Fig. 2E,F).
Figure 2.
Tertiary structures of TPPswec and TPPswat. Cartoon and stick representation of P3-L5 substructures (A,B), whole aptamer (C,D), and TPP binding site (E,F). Stems, loops, and junctions are figure caption. Black springs and gray dashed lines indicate hydrogen bonds formation and interactions with the Mg+2 ion respectively (A,B,E,F). (A,B) Nucleotides A59(57) and A60(58) from loop L5 connect with residues G27(25) and C28(26) from the P3 minor groove. (E,F) The aminopyrimidine ring of TPP formed hydrogen bonds with G30(28) and the 2-OH′ of G9(11). Direct contacts to nonbridging oxygens of β-phosphate of TPP were also established via N4 of C67(65) and N1 of G68(66). All other pyrophosphate-RNA contacts were mediated through two Mg2+ ions (colored in grey).
Global and local stability of the aptamer domain
The structural stability of riboswitches in aqueous solution, in both apo and holo configurations, was evaluated by comparing the average RMSD (Table 1 and Supplementary Fig. S1) and root-mean-square fluctuation (RMSF) (Fig. 3) values calculated over the trajectory production stage, taking as references the structures obtained after equilibration.
Table 1.
Root mean square deviations (Å) of free and bound states of TPPswec and TPPswat as a whole and for substructures.
| Free (Å) | Bound (Å) | |||
|---|---|---|---|---|
| TPPswec | TPPswat | TPPswec | TPPswat | |
| Whole | 2.83 (0.45) | 3.18 (0.34) | 2.62 (0.41) | 2.73 (0.37) |
| P1 | 5.91 (1.56) | 4.97 (0.67) | 6.44 (1.71) | 5.22 (0.71) |
| P2 | 1.70 (0.33) | 2.37 (0.40) | 2.81 (0.80) | 2.06 (0.27) |
| P3-L3 | 2.99 (0.66) | 3.16 (0.48) | 2.88 (0.58) | 2.87 (0.50) |
| P4-P5 | 1.70 (0.25) | 2.03 (0.17) | 1.65 (0.27) | 1.76 (0.35) |
| J3-2 | 1.37 (0.29) | 1.78 (0.26) | 1.85 (0.42) | 1.94 (0.43) |
| J2-4 | 3.42 (0.38) | 2.96 (0.40) | 3.74 (0.26) | 1.63 (0.61) |
| L5 | 1.43 (0.29) | 1.41 (0.31) | 1.53 (0.41) | 1.52 (0.48) |
| TPP | — | — | 1.38 (0.30) | 1.41 (0.36) |
The number in parenthesis indicates the RMSD standard deviation. “—” no RMSD values.
Figure 3.
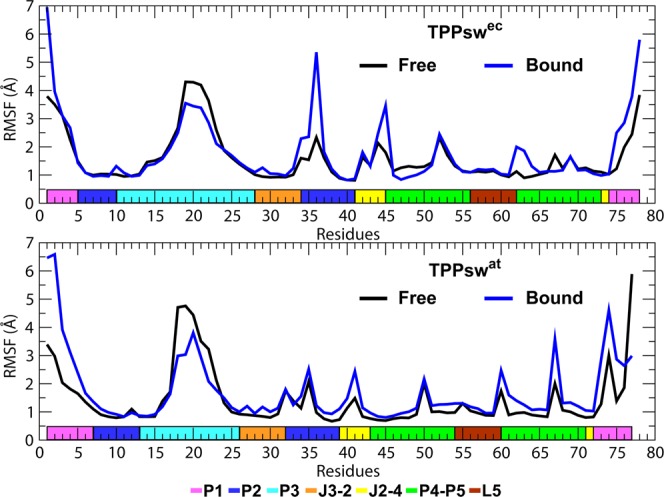
RMSF for the TPPswec and TPPswat heavy atoms in free and bound forms. The aptamer substructures are depicted at the lower margin of the plots and colored according to the figure caption.
During Molecular Dynamics (MD) simulations, both apo and holo forms presented deviations around 3 Å. No noticeable differences have been observed between the RMSD values obtained for both TPPswec and TPPswat upon TPP complexation. Indeed, both average RMSD values per region and the overall pattern of fluctuations were similar for all systems. These values are consistent with RMSD values seen in other studies reporting simulations of small RNA aptamers21–24 which ranged between 3–4 Å approximately.
The inspection at some particular regions of the RNA aptamer evidenced slight differences in their dynamical behavior. For example, the P1 region presented higher RMSD values than that of all other substructures in both species. Also, this region was more flexible in the holo forms. It is worth mentioning that the nucleotides composing the P1 helix are located in the terminal regions of the aptamer, showing broader movements during MD simulations. Unlike the P1 segment, the P3 helix displayed higher RMSD values in the apo forms. In addition, the comparison of the fluctuations between apo and holo riboswitches evidenced that TPP binding resulted in the stabilization of the P3 helix but contributed at the same time to the disruption of the P1 helix, according to the RMSF analysis (Fig. 3). Interestingly, the nucleotide U36, located in the P2 helix of TPPswec, displayed more significant fluctuations in the holo form than in the apo form (holo: 5.4 Å; apo: 2.3 Å) (Fig. 3). No significant changes were observed in the P2 substructure in apo and holo form of TPPswat.
Monitoring P3-L5 interaction
Despite being remotely located from the TPP binding site, the interaction between P3-L5 substructures is essential for metabolite binding25,26. The conformation of the P3-L5 region is kept in formation via two non-Watson-Crick (WC) base pairs. Nucleotides A59(57) and A60(58) from loop L5 connect with residues G27(25) and C28(26) from the P3 minor groove (Fig. 2A,B)27. Numbers in parenthesis indicate the nucleotide position in the TPPswat systems.
By monitoring the formation of non-WC hydrogen bonds between P3 and L5 we observed that, notably, the G27(25)·A59(57) interaction in apo TPPswec was the less stable with 68% occupancy during the simulation (Table 2). Upon TPP binding, the occupancy increased to 74% while no corresponding trend was observed upon comparison of the apo and holo TPPswat systems (presenting ~81% occupancy in both cases). Also, the C28(26)·A60(58) was more stable and presented occupancies higher than 84% in all systems. In addition, the base pair G27·A59 of TPPswec appears to be influenced by the presence of the ligand, which contributed to its stabilization.
Table 2.
Occupancy of hydrogen bonds between non-Watson-Crick pairs involved in the P3-L5 interaction of free and bound states of TPPswec and TPPswat.
| Nucleotides | frames (%) | ||||
|---|---|---|---|---|---|
| Free | Bound | ||||
| P3 | L5 | TPPswec | TPPswat | TPPswec | TPPswat |
| G27(25) | A59(57) | 68 | 82 | 74 | 81 |
| C28(26) | A60(58) | 93 | 89 | 89 | 84 |
TPP-RNA interaction
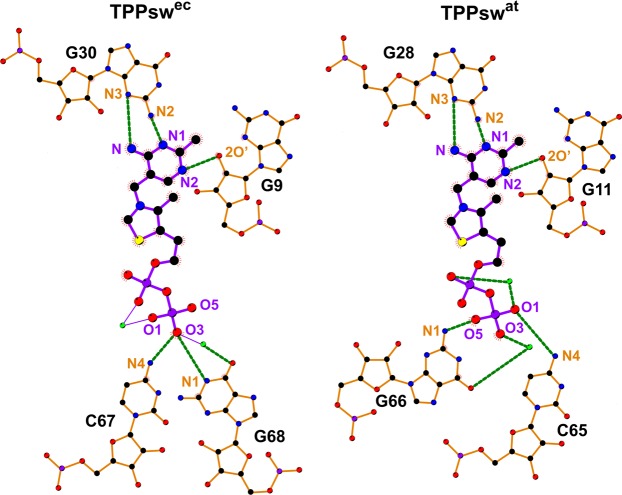
To verify the stability of TPP-RNA complexes during MD simulations, we calculated the H-bonds occupancy and the average distances between pairs of atoms involved in key interactions (Table 3). We confirmed the existence of similar interactions in all systems. The aminopyrimidine ring of TPP formed hydrogen bonds with N2 and N3 of G30(28) and the 2-OH′ of G9(11). Direct contacts to non-bridging oxygens of TPP β-phosphate were also formed via N4 of C67(65) and N1 of G68(66). All other pyrophosphate-RNA contacts were mediated through the two Mg2+ ions26,27 (Fig. 4).
Table 3.
Occupancy and distance of RNA-TPP interaction. The number in parenthesis indicates the distance standard deviation. “—” no RMSD values.
| Atoms | Occupancy (%) | Distance (Å) | |||
|---|---|---|---|---|---|
| RNA | TPP | TPPswec | TPPswat | TPPswec (SD) | TPPswat (SD) |
| G9(11) – 2O′ | TPP – N2 | 56 | 26 | 2.86 (0.19) | 3.03 (0.35) |
| G30(28) – N2 | TPP – N1 | 91 | 90 | 2.99 (0.14) | 3.06 (0.17) |
| G30(28) – N3 | TPP – N | 64 | 70 | 3.08 (0.17) | 3.02 (0.14) |
| C67(65) – N4 | TPP – O1 | 41 | 0 | 3.05 (0.40) | 5.18 (0.97) |
| C67(65) – N4 | TPP – O3 | 3 | 10 | 4.02 (0.33) | 4.67 (0.77) |
| C67(65) – N4 | TPP – O5 | 1 | 35 | 4.70 (0.70) | 4.08 (1.07) |
| G68(66) – N1 | TPP – O1 | 6 | 0 | 3.71 (0.51) | 4.17 (0.29) |
| G68(66) – N1 | TPP – O3 | 0 | 0 | 3.31 (0.25) | 3.24 (0.31) |
| G68(66) – N1 | TPP – O5 | 30 | 26 | 3.64 (0.62) | 3.65 (0.41) |
| Mg2+(1) | Mg2+(2) | — | — | 6.58 (0.97) | 3.35 (1.13) |
| Mg2+(1) | TPP – O3 | — | — | 1.85 (0.04) | 1.89 (0.06) |
| Mg2+(2) | TPP – O1 | — | — | 3.38 (1.53) | 1.90 (0.06) |
Figure 4.
2D representation of the RNA-TPP interaction of TPPswec and TPPswat crystallographic structures. TPP, RNA and Mg+2 ion are represented in purple, orange and green, respectively. C, N, O, P and S atoms are colored in black, blue, red, purple, and yellow, respectively. Green dashed lines indicate hydrogen bonds formation. The figure was generated using LigPlot+57.
Analysis of the distributions of the total number of RNA-TPP hydrogen bonds revealed that most TPPswec formed more interactions than TPPswat (Supplementary Fig. S2). The number of sampled structures presenting four or more hydrogen bonds was consistently higher in the TPPswec system than in TPPswat. The occupancy of the two hydrogen bonds formed between G30(28) and TPP was similar for both systems. Curiously, the occupancy of G9(11) for TPPswat was less than a half (26%) than for TPPswec (56%).
In the holo systems, the Mg2+(1) ion remained closer than 1.90 Å from O3 β-phosphate of TPP, interacting inespecifically with RNA. In TPPswat, the magnesium atom Mg2+(2) got closer to Mg2+(1) (3.35 ± 1.13 Å) and interacted strongly with O1 oxygen atom from TPP (1.90 ± 0.06 Å), leaving the O5 oxygen atom from β-phosphate available to interact with RNA, with consequent formation of a bifurcated H-bond. The O5 oxygen atom from TPP interacted with C65 e G66, with hydrogen bond occupancies of 35% and 26%, respectively.
Each TPPswec nucleotide involved in pyrophosphate recognition formed more than one hydrogen bond because they interacted with other oxygen atoms from TPP, being C67·TPP–O1 and G68·TPP–O5 examples of these interactions, with occupancies of 41% and 30%, respectively (Table 3).
Principal component analysis suggests different dominant motions in apo and holo states
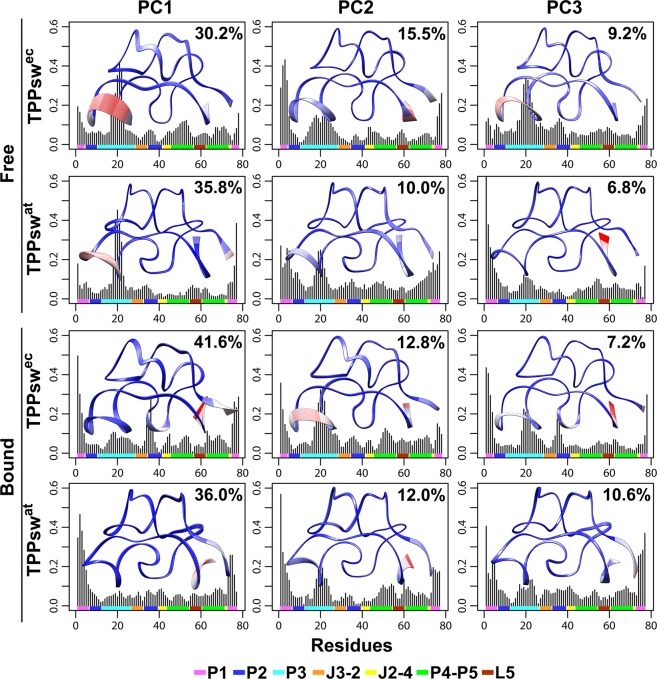
To identify statistically relevant motions of TPP riboswitches in solution, we performed principal component analysis (PCA) on the snapshots obtained from the MD trajectories. Overall, the first three components (named PC1, PC2, and PC3) captured the dominant motions, presenting the highest contributions to total fluctuations. The first three PCs accounted for 54.9% and 52.6% of the overall variance in apo TPPswec and TPPswat, respectively. In the holo states, the contribution of the first three PCs was slightly higher: 61.7% and 58.7% for TPPswec and TPPswat, respectively.
We compared the projections of the trajectories onto the subspace spanned by the first three principal components. RMSFs and structural projections along the three PCs showed that the four systems displayed a uniform and overlapping PC subspace (Fig. 5).
Figure 5.
RMSF contributed by the first three principal components. The fractional contribution of each PC to the overall variance is shown in the top right part of each graph. Interpolated structures obtained by displacements along each vector are displayed within each plot. Blue indicates overlapping regions with little or no motion. Red areas represent mobile regions. The secondary structure elements are given at the lower margin of the plots and colored according to the figure caption.
Inspection of the atomic fluctuations along PC1 revealed substantial higher flexibility of the P3 helix in the apo systems, confirming its stabilization by ligand binding. However, this region presented higher fluctuations along PC2 in the holo state in the TPPswec system, but not in TPPswat. Therefore, the stabilization of the most statistical relevant motions of P3 helix inducted by ligand binding was undoubtedly more pronounced in TPPswat, as lower fluctuations were noticed along both principal components. Furthermore, TPP binding resulted in the augmented flexibility of the P1 helix. In the holo TPPswec, the ligand promoted an increase in the flexibility of nucleotides 34–37 of the P2 helix that was unseen in TPPswat.
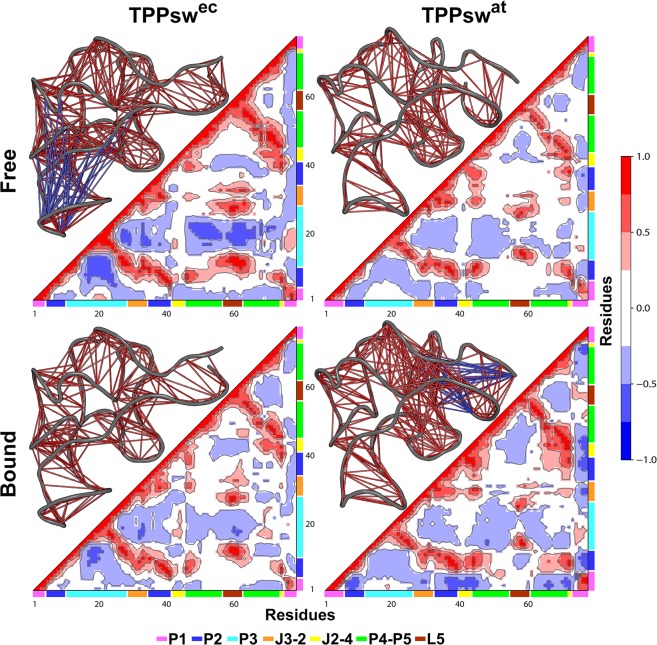
Correlation network analysis reveals distinct responses to ligand binding in TPPswec and TPPswat
We analyzed the correlations between pairs of nucleotides to investigate how TPP binding affects the dynamic couplings in TPPswec and TPPswa. We calculated the dynamic cross-correlation matrices (DCCM) for each simulated system, as described in the methods section. A similar cross-correlation pattern was observed for both systems in their apo states. However, both the extent of regions displaying anticorrelations and their magnitudes were greater in TPPswec, mainly in the P3–P4-P5–L5 region (Fig. 6).
Figure 6.
DCCMs of free and bound states of TPPswec and TPPswat. Next to each matrix, the corresponding 3D structures with lines connecting pairs of correlated residues are shown. For clarity sake, only the pairs presenting (|Cij|) > 0.6. are represented.
It was observed that TPP binding stabilized the P3-L5 interaction in TPPswec, as evidenced by weaker anticorrelations, which are likely to be associated with the separation of these regions in the apo state. This result is in line with PCA revealing increased stability of P3 motions in the holo state (Fig. 5). Interestingly, no similar trend was observed in TPPswat. In TPPswat system, TPP binding resulted in increased anticorrelations at the P1-P2 helices, indicating a possible destabilization of interactions that ultimately resulted in a larger separation between them. In contrast, the dynamic coupling pattern in this region was not altered in TPPswec.
Next, we performed a correlation network analysis by constructing weighted graphs in which each residue was represented by a single node, and the weight of the connection between pairs of nodes was proportional to their respective previously calculated correlation coefficients. To quantify the relative importance of each residue in the network, we computed the betweenness centrality per nucleotide for each simulated system (Fig. 7). This metric is used to identify critical communication nodes over the network. Residues presenting high betweenness values are considered “bottlenecks” of information as they are found in the shortest communication paths28.
Figure 7.
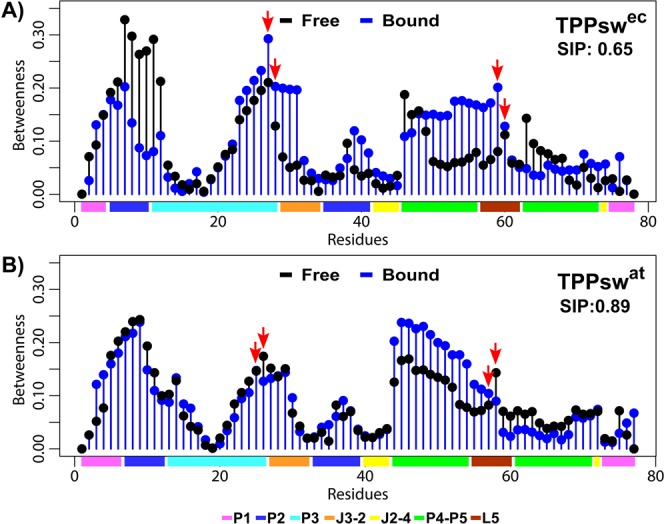
Betweenness centrality of the node for each residue of free and bound states. (A) TPPswec systems. (B) TPPswat systems. Red arrows indicate residues G27(25)–A59(57) involved between P3-L5 interaction. Square Inner Product (SIP) is shown above each graph.
We calculated the square inner product (SIP) to compare the overall similarity of the betweenness centrality profiles calculated for the apo and holo states. According to this analysis, high SIP values are associated with weak modulation of intramolecular communication as a consequence of the TPP binding. We obtained a higher SIP for TPPswat (0.89) than for TPPswec (0.65), thus reinforcing more noticeable TPP related effects in TPPswec systems. In both holo systems, we noted increased centrality values at the P4-P5 helices. Interestingly, the nucleotides G27, C28, and A59 were critical for P3–L5 interaction and displayed higher betweenness values in holo TPPswec, indicating that ligand binding favors an efficient communication through these nucleotides (Fig. 7). This feature was not observed in TPPswat, in which C26 and A58 centrality values were higher in the apo state.
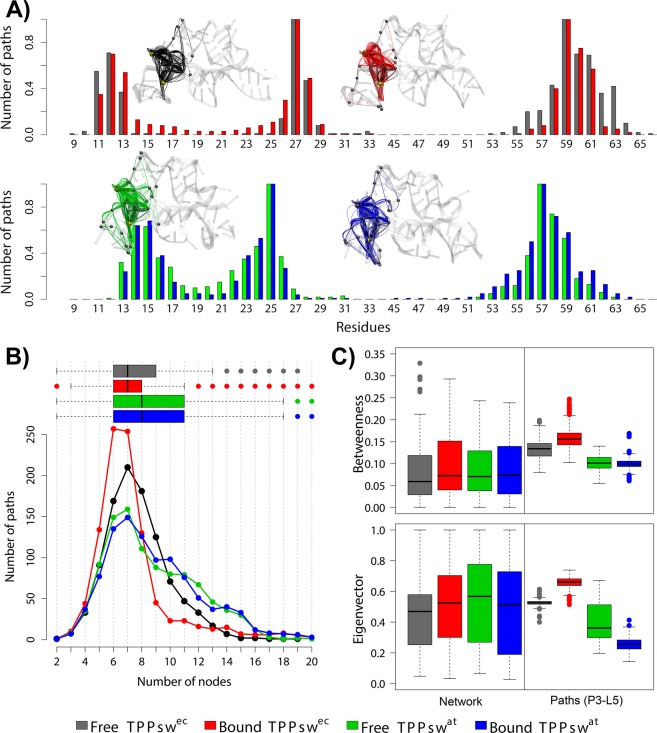
Communication pathways between P3-L5
To get a deeper understanding about the critical residues governing P3-L5 interactions, we computed the 1000 shortest paths between G27(25) [located in P3] and A59(57) [located in L5] (Fig. 8). The normalized node degeneracy metric reveals the percentage of paths accessing each node. We observed for all systems conserved critical residues for communication with degeneracy values > 0.35. While the majority of these residues belongs to L5 (U58(56), A60(58) and G61(59)), two of them (C12(14) and C28(26)) are located in P3 (Fig. 8A).
Figure 8.
Shortest communication paths connecting G27(25) and A59(57) residues of free and bound TPPswec and TPPswat. (A) Normalized node degeneracy graph and visualization of sub-optimal paths in a correlation network. (B) Number of nodes per path. (C) Boxplot of betweenness and eigenvector centrality of the paths compared to the one corresponding to the complete network. Each system was colored according to the figure caption.
The distribution of node degeneracies obtained for TPPswec was narrower at the P3 region in the holo state, indicating that TPP binding restricted the presence of a few residues in the shortest paths. Whereas a larger number of residues are accessed in the apo state, the shortest paths were mostly formed by nucleotides 59–61 in the holo state. In contrast, the distributions obtained for both TPPswat states were very similar (Fig. 8A).
We calculated the number of nodes per path to further evaluate and characterize the influence of TPP binding on P3-L5 interactions (Fig. 8B). This analysis was based on the hypothesis that communication involving fewer nodes along the pathway is likely to be more efficient. Indeed, in holo TPPswec, the P3-L5 communication required fewer nodes (Fig. 8B). Interestingly, while TPP binding did not modify the global distributions of betweenness centralities obtained for both species (Fig. 8C left boxes), opposing trends were perceived concerning the average betweenness centrality calculated for the residues participating in shortest paths (Fig. 8C right boxes). However, TPPswec TPP binding resulted in increased centralities for the residues involved in shortest paths, leading to a slight decrease in average betweenness in TPPswat.
To support this analysis, we computed the eigenvector centralities for the overall network and the shortest paths (Fig. 8C lower boxes). Again, TPP binding resulted in higher centrality in the shortest paths only for TPPswec. The selection of specific P3 residues imposed by ligand binding in TPPswec resulted in a stronger communication along pathways accessing a selection of neighboring residues with high eigenvector centrality. In agreement with our previous analysis (Figs 6–8), a corresponding effect was not observed in TPPswat, which strongly suggests the weaker influence of TPP for an effective communication.
Discussion
Plant and bacterial TPP aptamers share similar core structures and bind to the same ligand. However, minor structural and dynamical differences between them could be found, specially concerning the behavior of P3 helix. Bacteria and archaea commonly have a P3a stem18, which is not observed in eukaryotic riboswitches. The eukaryotic P3 stem is significantly variable in length, sequence, and base pairings19. Particularly in plants, the length of distal P3 extension varies among TPP aptamer representatives of the same species, as observed in Physcomitrella patens8. The P3 distal portion is not required for ligand binding of L5-P3 interaction5,29, but might act as an anchor for the aptamer as it was already pointed out by Anthony et al.30. Also, the authors claim that the correct folding could help in the competition with other RNA structures with different regulation mechanisms.
However, despite that the P3 stem is significantly variable in length in plants, the TPP aptamer is structurally stable. This stability might lead to slower arm movement than the observed in the helix arm of E. coli TPP riboswitch31. Cross-correlation analysis corroborated this hypothesis because stronger negative correlations were noticed in the apo TPPswec involving substructures P3–P4-P5–L5 (Fig. 6). Our findings also suggest that communication pathways between P3-L5 may be different in E. coli and A. thaliana. The communication between P3-L5 in TPPswec can be very efficient in the holo state, while in TPPswat the corresponding effect was weakened, thus suggesting a slower response to TPP binding in plants than in bacteria (Fig. 8).
The A. thaliana crystallographic structure used as starting point for our simulations contains a shortened P3 stem formed by 14 nucleotides. On the other hand, the corresponding structure in E. coli is composed of 18 nucleotides. The P3 helix of TPPswat, although smaller than the one in TPPswec, showed no significant modifications in the presence of the ligand, indicating that the size of P3 can be oblivious to plants and its influence about slow folding can be negligible.
Guedich et al. wondered whether the slow TPPswat folding would be related to a single nucleotide. The authors concluded that U35, located on the P2 helix, is crucial for shaping a TPP-binding competent riboswitch32. In our analysis, the equivalent pyrimidine nucleotide in TPPswec is U36 (Fig. 1B). The magnitude of the fluctuations at this position was 2-fold higher in the holo state than in the apo TPPswec state (Fig. 3). In contrast, in the TPPswat system, similar fluctuations were perceived regardless of a ligand binding. Furthermore, PCA data also supported these outcomes by showing that the segment 34–37 of the holo TPPswec displayed the most significant motion amplitude along PC1 (Fig. 5).
Grounded on these findings, we hypothesize that different interactions found in the microenvironment surrounding nucleotide U36 of TPPswec (and U35 in TPPswat) are related to different TPP responses. In TPPswec, this nucleotide is neighbored at 3′ by a non-canonical A37-G9 base pair. A similar context is observed for U35 in TPPswat, which is delimited by a non-canonical G34-G11 base pair but on 5′ instead. Nucleotides G9 and G11 of TPPswec and TPPswat, respectively, form hydrogen bonds with the aminopyrimidine ring of TPP. Interestingly, our simulations have shown that hydrogen bond occupancy between G9(11) and N2 of TPP was less than a half for TPPswat (25.67%) than for TPPswec (55.58%). This suggests that slight differences in the environment may directly interfere the TPP-aptamer interaction stability.
Finally, TPP riboswitches of Arabidopsis thaliana present subtler and slower regulation mechanisms than Escherichia coli30–32. Here, we have shown through molecular dynamics simulations and networking analysis that minor structural differences in the aptamer enable enhanced intramolecular communication in the presence of TPP in TPPswec, but not in TPPswat. Weaker responses to changes in the TPP concentration may be related to the autotrophic mode of nutrition, which demands the endogenous synthesis of thiamine. Unlike in plants, bacteria can grow under thiamine-rich conditions allowing them to satisfy their full demand for compounds like thiamine exogenously8. Taken together, our results provide new insights into RNA behavior of TPP riboswitch, which may have adapted in a different way to the different metabolic demands of each group of organisms to accomplish distinct TPP binding modulation.
Materials and Methods
Analysis of crystallographic structures
For this study, TPP riboswitch 3D-structures of Escherichia coli (TPPswec) and Arabidopsis thaliana (TPPswat) obtained by X-ray crystallography were selected from the Protein Data Bank (PDB)33. Both structures are bound to TPP and present high-resolution crystal, of 2.05 Å (PDB ID: 2GDI27) and 2.25 Å (PDB ID: 3D2G29), for TPPswec and TPPswat, respectively. Corresponding sequence and secondary structures information were also taken from the PDB files and analyzed with 3DNA software suite34. TPPswec and TPPswat sequences were then aligned using the SARA-Coffee mode of T-Coffee program35, and figures of sequence alignment were rendered using ALINE36. The secondary structure calculated on the basis of the PDB files of the crystallographic structures was compared through the SimTree server20. SimTree returns a similarity score and a mapping between the similar regions of the two structures in dot-bracket notation. Graphical representation of 2D and 3D structures were generated using VARNA37 and UCSF Chimera, respectively.
Molecular dynamics simulations
Molecular dynamics (MD) simulations were carried out using the GROMACS version 5.1.2 package38, and RNA interactions were represented using the amber 14sb39 force field with parmbsc0 + chiOL340. Bonded and Lennard-Jones molecular parameters for TPP have been obtained using the Generalized Amber force field (GAFF)41 and AM1-BCC42 tools while atomic partial charges were added using ANTECHAMBER43. ACPYPE44 program was employed to create a GROMACS compatible topology file. Electrostatic interactions were treated using the particle mesh Ewald (PME) algorithm with a cut-off of 10 Å.
MD trajectories were monitored to investigate possible differences in the dynamical behavior between apo and holo TPPswec and TPPswat. In the apo systems, TPP was removed from the X-ray crystal structure and replaced with solvent water. Two initially positioned magnesium ions in the crystal structure were kept in both the apo and holo systems and also contributed to neutralize the systems. These ions are essential for ligand binding and confer stability to the riboswitch as well45. Each system was simulated under periodic boundary conditions in a triclinic box whose dimensions were automatically defined considering a distance of 1 nm from the outermost RNA atoms in all cartesian directions. The simulation box was filled with TIP3P water molecules46.
Simulations were performed in three stages: (i) Energy minimization, (ii) thermalization and equilibration, and (iii) trajectory production.
Energy minimization procedure was performed through 5000 steps and a gradient tolerance <1.0 kJ mol−1 nm−1 of the steepest descent and conjugate-gradient algorithms. These steps were carried out with heavy atom restraints by applying a harmonic potential with a force constant of 1000 kJ mol−1 nm−2 for the steepest descent algorithm. Applications of the conjugate-gradient algorithm do not allow the application of restraints.
In the second phase, starting atomic velocities were assigned to all atoms of the system using a Maxwell-Boltzmann distribution, corresponding to an initial temperature of 20 K. Then, the systems were gradually heated up to 300 K over 500 picoseconds (ps) utilizing the Langevin thermostat. During this stage, all heavy atoms were harmonically restrained by applying a constant force of 1000 kJ mol−1 nm−2.
Systems were subsequently equilibrated during twenty successive 100 ps long equilibration simulations where position restraints progressively approached zero. After this period, the systems were simulated with no restraints all at 300 K for 1 µs (trajectory production). All simulations were performed in the NPT ensemble. The V-rescale thermostat and Berendsen barostat were used for temperature (300 K) and pressure control (1 atm), respectively.
Trajectory analysis
As in the trajectory analysis, we were interested only in the structural aspects of the systems, regardless the temporal correlation. Two independent MD simulations were concatenated, and trajectory analyses were conducted for crystal structures systems.
To investigate structural changes of the TPP aptamers, root-mean-square deviation (RMSD) values were calculated separately for the whole RNA and its substructures after fitting to their respective parts, taking the initial structure of the production dynamics as a reference. Hydrogen bond formation was defined using a geometric criterion with VMD software. It was considered a hit when the distance between two polar heavy atoms, with at least one hydrogen atom attached, was less than 3.5 Å using a D-Ĥ-A angle cutoff of 30°. Motif Identifier for Nucleic acids Trajectory (MINT)47 program was used to evaluate the number of Watson–Crick (WC)-edge and non-WC-edge hydrogen bonds (and their sum) per nucleotide throughout the simulations.
Principal components analysis
The study of large-scale domain motions is essential for characterizing the conformational dynamics of macromolecules. Functional motions are usually described by a few numbers of degrees of freedom that can be calculated using principal component analysis (PCA)48,49. PCA analysis was carried out for all systems using Bio3D50 library as implemented in R51. Rotational and translational motions were removed before calculation of the covariance matrix by least-squares superposition to the corresponding average structures. The 3 N × 3 N covariance matrices of C5′ atomic positions (Cartesian coordinates) were then calculated for each state. The conformations explored during the MD simulations were applied using hierarchical clustering in R (hclust) with the complete linkage method based on the PC1-PC2 subspace, where PC1 and PC2 denote the projections onto the two first eigenvectors.
Correlation network analysis
The cross-correlation and network analyses were carried out using the Bio3D and the igraph R packages52. Initially, the dynamic cross-correlation matrices (DCCM) were calculated separately for each simulation using as inputs the corresponding MD trajectory superimposed onto the initial structure. Then, each group of two matrices per riboswitch state was utilized to obtain a consensus matrix. A proximity/contact map filter was applied in the construction of the correlation network for residues that remained within 4.5 Å from one another for at least 75% of simulation time. Briefly, graphs were obtained considering C5′ atoms as nodes and the connection between nodes i and j were weighted using the absolute values of cross-correlations (C(i,j)) coefficients (Equation 1):
| 1 |
We also calculated the relative importance of each node for communication using centrality measures. According to the definition of the betweenness centrality (Equation 2), the relevance of a given node is defined by its presence in shortest communication paths connecting nodes over the entire network53.
| 2 |
According to the above equation, the betweenness centrality of a node depends on the total number of the shortest paths between nodes i and j that pass-through n ( and , which is the total number of shortest paths between nodes i and j (regardless of whether they cross or not through n).
Another measure of centrality can be given by the eigenvector (Equation 3) that accounts for the global relevance of each residue based on the connections with neighboring nodes. In other words, nodes with high eigenvector centrality are those connected to other central residues54.
| 3 |
The vector is obtained as a solution to the eigenvalue problem , where A is the adjacency matrix for the network graph G. More mathematical details can be found at Kolaczyk54.
The Square inner product (SIP) (Equation 4) was used to compare the overall similarity of the centrality profiles calculated for the systems. It varies between 0 and 1 and is defined as
| 4 |
where wA and wB are N-length vectors containing the fluctuation value for each atom in proteins A and B, respectively55.
The Yen’s algorithm56 was used to calculate the shortest pathways connecting two nodes in the network. Path lengths are defined as the sum of the edge weights connecting a pair of nodes in a given pathway. The first 1000 shortest paths were collected and employed to calculate the node degeneracy value, which represents the percentage of pathways from the overall ensemble in which a given node is present.
Supplementary information
Acknowledgements
This work was supported by the Fundação Carlos Chagas Filho de Amparo à Pesquisado Estado do Rio de Janeiro (FAPERJ) [E-26/111.401/2013 and E-26/203.428/2015 to E.R.C., E-26/203.229/2016 to F.P.); Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) [Papes VI 407741/2012-7 to E.R.C.]. This study was financed in part by the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior - Brasil (CAPES) - Finance Code 001 [Ph.D. scholarships to D.A. and N.A.N.J.].
Author Contributions
D.A. participated in the design research, performed calculations, analysed the results and wrote the article; N.A.N.J. contributed in the discussed results; M.G.S.C. analysed the results and helped writing the article; F.P. and E.R.C. participated in the design research, discussed results, helped writing the article. All authors reviewed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40875-1.
References
- 1.Edwards AL, Batey RT. Riboswitches: A common RNA regulatory element. Nat. Educ. 2010;3:9. [Google Scholar]
- 2.Breaker RR. Riboswitches and the RNA world. Cold Spring Harb. Perspect. Biol. 2012;4:a003566. doi: 10.1101/cshperspect.a003566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Miranda-Ríos J, Navarro M, Soberón M. A conserved RNA structure (thi box) is involved in regulation of thiamin biosynthetic gene expression in bacteria. Proc. Natl. Acad. Sci. USA. 2001;98:9736–41. doi: 10.1073/pnas.161168098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nudler E, Mironov AS. The riboswitch control of bacterial metabolism. Trends Biochem. Sci. 2004;29:11–17. doi: 10.1016/j.tibs.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 5.Thore S, Leibundgut M, Ban N. Structure of the eukaryotic thiamine pyrophosphate riboswitch with its regulatory ligand. Science (80-.). 2006;312:1208–1211. doi: 10.1126/science.1128451. [DOI] [PubMed] [Google Scholar]
- 6.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nat. Struct. Mol. Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 7.Fuchs RT, Grundy FJ, Henkin TM. The SMK box is a new SAM-binding RNA for translational regulation of SAM synthetase. Nat. Struct. Mol. Biol. 2006;13:226–233. doi: 10.1038/nsmb1059. [DOI] [PubMed] [Google Scholar]
- 8.Wachter A, et al. Riboswitch control of gene expression in plants by splicing and alternative 3′ end processing of mRNAs. Plant Cell Online. 2007;19:3437–3450. doi: 10.1105/tpc.107.053645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandal M, Breaker RR. Gene regulation by riboswitches. Nat. Rev. Mol. Cell Biol. 2004;5:451–463. doi: 10.1038/nrm1403. [DOI] [PubMed] [Google Scholar]
- 10.Tucker BJ, Breaker RR. Riboswitches as versatile gene control elements. Curr. Opin. Struct. Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 11.Winkler W, Nahvi A, Breaker RR. Thiamine derivatives bind messenger RNAs directly to regulate bacterial gene expression. Nature. 2002;419:952–956. doi: 10.1038/nature01145. [DOI] [PubMed] [Google Scholar]
- 12.Ontiveros-Palacios N, et al. Molecular basis of gene regulation by the THI-box riboswitch. Mol. Microbiol. 2008;67:793–803. doi: 10.1111/j.1365-2958.2007.06088.x. [DOI] [PubMed] [Google Scholar]
- 13.Kubodera T, et al. Thiamine-regulated gene expression of Aspergillus oryzae thiA requires splicing of the intron containing a riboswitch-like domain in the 5′-UTR. FEBS Lett. 2003;555:516–520. doi: 10.1016/S0014-5793(03)01335-8. [DOI] [PubMed] [Google Scholar]
- 14.Li S, Breaker RR. Eukaryotic TPP riboswitch regulation of alternative splicing involving long-distance base pairing. Nucleic Acids Res. 2013;41:3022–3031. doi: 10.1093/nar/gkt057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Croft MT, Moulin M, Webb ME, Smith AG. Thiamine biosynthesis in algae is regulated by riboswitches. Proc. Natl. Acad. Sci. 2007;104:20770–20775. doi: 10.1073/pnas.0705786105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bocobza, S. et al. Riboswitch-dependent gene regulation and its evolution in the plant kingdom Riboswitch-dependent gene regulation and its evolution in the plant kingdom. 2874–2879, 10.1101/gad.443907 (2007). [DOI] [PMC free article] [PubMed]
- 17.Nawrocki EP, et al. Rfam 12.0: Updates to the RNA families database. Nucleic Acids Res. 2015;43:D130–D137. doi: 10.1093/nar/gku1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rodionov DA, Vitreschak AG, Mironov AA, Gelfand MS. Comparative genomics of thiamin biosynthesis in procaryotes. New genes and regulatory mechanisms. J. Biol. Chem. 2002;277:48949–48959. doi: 10.1074/jbc.M208965200. [DOI] [PubMed] [Google Scholar]
- 19.Sudarsan, N., Barrick, J. E. & Breaker, R. R. Metabolite-binding RNA domains are present in the genes of eukaryotes. 644–647, 10.1261/rna.5090103.644 (2003). [DOI] [PMC free article] [PubMed]
- 20.Eden, E., Wallach, I. & Yakhini, Z. SimTree: A Tool for Computing Similarity Between RNA Secondary Structures, http://bioinfo.cs.technion.ac.il/SimTree/ (2005).
- 21.Gong Z, Zhao Y, Chen C, Duan Y, Xiao Y. Insights into ligand binding to preQ. 1 riboswitch aptamer from molecular dynamics simulations. PLoS One. 2014;9:1–12. doi: 10.1371/journal.pone.0092247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aytenfisu AH, Liberman JA, Wedekind JE, Mathews DH. Molecular mechanism for preQ. 1 -II riboswitch function revealed by molecular dynamics. Rna. 2015;21:1898–1907. doi: 10.1261/rna.051367.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang W, et al. Dynamics Correlation Network for Allosteric Switching of PreQ. 1 Riboswitch. Sci. Rep. 2016;6:31005. doi: 10.1038/srep31005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hanke CA, Gohlke H. Ligand-mediated and tertiary interactions cooperatively stabilize the P1 region in the guanine-sensing riboswitch. PLoS One. 2017;12:1–29. doi: 10.1371/journal.pone.0179271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lang K, Rieder R, Micura R. Ligand-induced folding of the thiM TPP riboswitch investigated by a structure-based fluorescence spectroscopic approach. Nucleic Acids Res. 2007;35:5370–5378. doi: 10.1093/nar/gkm580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kulshina N, Edwards TE, Ferré-D’Amaré AR. Thermodynamic analysis of ligand binding and ligand binding-induced tertiary structure formation by the thiamine pyrophosphate riboswitch. RNA. 2010;16:186–196. doi: 10.1261/rna.1847310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serganov A, Polonskaia A, Phan AT, Breaker RR, Patel DJ. Structural basis for gene regulation by a thiamine pyrophosphate-sensing riboswitch. Nature. 2006;441:1167–1171. doi: 10.1038/nature04740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sethi A, Eargle J, Black AA, Luthey-Schulten Z. Dynamical networks in tRNA:protein complexes. Proc. Natl. Acad. Sci. 2009;106:6620–6625. doi: 10.1073/pnas.0810961106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thore S, Frick C, Ban N. Structural basis of thiamine pyrophosphate analogues binding to the eukaryotic riboswitch. J. Am. Chem. Soc. 2008;130:8116–8117. doi: 10.1021/ja801708e. [DOI] [PubMed] [Google Scholar]
- 30.Anthony PC, Perez CF, Garcia-Garcia C, Block SM. Folding energy landscape of the thiamine pyrophosphate riboswitch aptamer. Proc. Natl. Acad. Sci. 2012;109:1485–1489. doi: 10.1073/pnas.1115045109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Duesterberg VK, Fischer-Hwang IT, Perez CF, Hogan DW, Block SM. Observation of long-range tertiary interactions during ligand binding by the TPP riboswitch aptamer. Elife. 2015;4:1–17. doi: 10.7554/eLife.12362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guedich S, et al. Quantitative and predictive model of kinetic regulation by E. coli TPP riboswitches. RNA Biol. 2016;13:373–390. doi: 10.1080/15476286.2016.1142040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lu X-J, Olson WK. 3DNA: a versatile, integrated software system for the analysis, rebuilding and visualization of three-dimensional nucleic-acid structures. Nat. Protoc. 2008;3:1213–1227. doi: 10.1038/nprot.2008.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Di Tommaso P, et al. SARA-Coffee web server, a tool for the computation of RNA sequence and structure multiple alignments. Nucleic Acids Res. 2014;42:W356–60. doi: 10.1093/nar/gku459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bond CS, Schüttelkopf AW. ALINE: A WYSIWYG protein-sequence alignment editor for publication-quality alignments. Acta Crystallogr. Sect. D Biol. Crystallogr. 2009;65:510–512. doi: 10.1107/S0907444909007835. [DOI] [PubMed] [Google Scholar]
- 37.Blin G, Denise A, Dulucq S, Herrbach C, Touzet H. VARNA: Interactive drawing and editing of the RNA secondary structure. IEEE/ACM Trans. Comput. Biol. Bioinforma. 2010;7:309–322. doi: 10.1109/TCBB.2008.28. [DOI] [PubMed] [Google Scholar]
- 38.Abraham MJ, et al. Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX. 2015;1–2:19–25. doi: 10.1016/j.softx.2015.06.001. [DOI] [Google Scholar]
- 39.Maier, J. A. et al. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. J. Chem. Theory Comput. 10.1021/acs.jctc.5b00255 (2015). [DOI] [PMC free article] [PubMed]
- 40.Pérez, A. et al. Refinement of the AMBER force field for nucleic acids: Improving the description of α/γ conformers. Biophys. J. 10.1529/biophysj.106.097782 (2007). [DOI] [PMC free article] [PubMed]
- 41.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J. Comput. Chem. 2004;25:1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 42.Jakalian A, Jack DB, Bayly CI. Fast, efficient generation of high-quality atomic charges. AM1-BCC model: II. Parameterization and validation. J. Comput. Chem. 2002;23:1623–1641. doi: 10.1002/jcc.10128. [DOI] [PubMed] [Google Scholar]
- 43.Wang J, Wang W, Kollman PA, Case DA. Automatic atom type and bond type perception in molecular mechanical calculations. J. Mol. Graph. Model. 2006;25:247–260. doi: 10.1016/j.jmgm.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 44.Sousa da Silva AW, Vranken WF. ACPYPE - AnteChamber PYthon Parser interfacE. BMC Res. Notes. 2012;5:367. doi: 10.1186/1756-0500-5-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leipply D, Draper DE. Effects of Mg2+ on the Free Energy Landscape for Folding a Purine Riboswitch RNA. Biochemistry. 2011;50:2790–2799. doi: 10.1021/bi101948k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 47.Górska A, Jasiński M, Trylska J. MINT: Software to identify motifs and short-range interactions in trajectories of nucleic acids. Nucleic Acids Res. 2015;43(17):e114. doi: 10.1093/nar/gkv559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wold S, Esbensen K, Geladi P. Principal component analysis. Chemom. Intell. Lab. Syst. 1987;2:37–52. doi: 10.1016/0169-7439(87)80084-9. [DOI] [Google Scholar]
- 49.Abdi H, Williams LJ. Principal component analysis. Wiley Interdisciplinary Reviews: Computational Statistics. 2010;2:433–459. doi: 10.1002/wics.101. [DOI] [Google Scholar]
- 50.Grant BJ, Rodrigues APC, ElSawy KM, McCammon JA, Caves LSD. Bio3d: an R package for the comparative analysis of protein structures. Bioinformatics. 2006;22:2695–2696. doi: 10.1093/bioinformatics/btl461. [DOI] [PubMed] [Google Scholar]
- 51.R Development Core Team. R: A Language and Environment for Statistical Computing. R Found. Stat. Comput. Vienna, Austria0, {ISBN} 3-900051-07-0 (2016).
- 52.Csárdi G, Nepusz T. The igraph software package for complex network research. InterJournal, Complex Systems. 2006;1695:1–9. [Google Scholar]
- 53.Kolaczyk, E. D. & Csárdi, G. Statistical Analysis of Network Data with R., 10.1007/978-0-387-88146-1 (2014).
- 54.Kolaczyk, E. D. Statistical Analysis of Network Data. Learning26 (2009).
- 55.Skjærven L, Yao X-Q, Scarabelli G, Grant BJ. Integrating protein structural dynamics and evolutionary analysis with Bio3D. BMC Bioinformatics. 2014;15:399. doi: 10.1186/s12859-014-0399-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yen JY. Finding the K Shortest Loopless Paths in a Network. Manage. Sci. 1971;17:712–716. doi: 10.1287/mnsc.17.11.712. [DOI] [Google Scholar]
- 57.Laskowski, R. A. & Swindells, M. B. LigPlot+: Multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 10.1021/ci200227u (2011). [DOI] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.