The number and diversity of β-lactamases are steadily increasing. The emergence of β-lactamases that hydrolyze carbapenems poses a significant threat to our antibiotic armamentarium. The explosion of OXA enzymes that are carbapenem hydrolyzers is a major challenge (carbapenem-hydrolyzing class D [CHD]). An urgent need exists to discover β-lactamase inhibitors with class D activity. The sulbactam-ETX2514 combination demonstrates the potential to become a treatment regimen of choice for Acinetobacter spp. producing class D β-lactamases.
KEYWORDS: Acinetobacter, DBO, diazabicyclooctanone, diazabicyclooctenone, ETX2514, sulbactam, β-lactamase inhibitor, beta-lactamases, beta-lactams
ABSTRACT
Multidrug-resistant (MDR) Acinetobacter spp. poses a significant therapeutic challenge in part due to the presence of chromosomally encoded β-lactamases, including class C Acinetobacter-derived cephalosporinases (ADC) and class D oxacillinases (OXA), as well as plasmid-mediated class A β-lactamases. Importantly, OXA-like β-lactamases represent a gap in the spectrum of inhibition by recently approved β-lactamase inhibitors such as avibactam and vaborbactam. ETX2514 is a novel, rationally designed, diazabicyclooctenone inhibitor that effectively targets class A, C, and D β-lactamases. We show that addition of ETX2514 significantly increased the susceptibility of clinical Acinetobacter baumannii isolates to sulbactam. AdeB and AdeJ were identified to be key efflux constituents for ETX2514 in A. baumannii. The combination of sulbactam and ETX2514 was efficacious against A. baumannii carrying blaTEM-1, blaADC-82, blaOXA-23, and blaOXA-66 in a neutropenic murine thigh infection model. We also show that, in vitro, ETX2514 inhibited ADC-7 (k2/Ki 1.0 ± 0.1 × 106 M−1 s−1) and OXA-58 (k2/Ki 2.5 ± 0.3 × 105 M−1 s−1). Cocrystallization of ETX2514 with OXA-24/40 revealed hydrogen bonding interactions between ETX2514 and residues R261, S219, and S128 of OXA-24/40 in addition to a chloride ion occupied in the active site. Further, the C3 methyl group of ETX2514 shifts the position of M223. In conclusion, the sulbactam-ETX2514 combination possesses a broadened inhibitory range to include class D β-lactamases as well as class A and C β-lactamases and is a promising therapeutic candidate for infections caused by MDR Acinetobacter spp.
INTRODUCTION
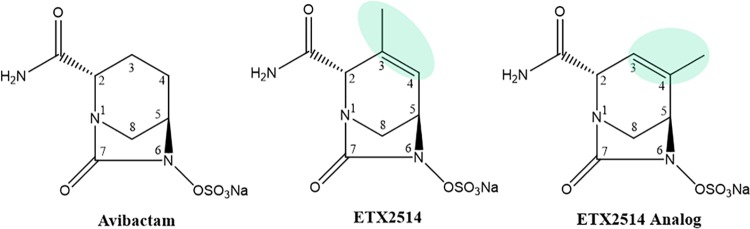
The discovery of ETX2514, a novel diazabicyclooctenone β-lactamase inhibitor that was derived from the diazabicyclooctanone (DBO), avibactam (Fig. 1), represents a major advance for non-β-lactam β-lactamase inhibitors (1, 2). ETX2514 effectively inhibits class A and C β-lactamases and a broad range of class D β-lactamases (1, 2), whereas the spectrum of most DBO inhibition had limited potency against class D OXA β-lactamases (e.g., OXA-48). The introduction of avibactam (Fig. 1) (3), the first commercial member of this inhibitor family, allowed successful inhibition of serine carbapenemases (e.g., KPC-2) and most class C cephalosporinases but with limited activity against class D β-lactamases (4). Avibactam inhibits the enterobacterial carbapenemase OXA-48 with a k2/Ki value of 1.4 ± 0.1 × 103 M−1 s−1 (4) but is much less effective at inhibiting other known OXA β-lactamases. For example, the k2/Ki value for OXA-10 obtained from Pseudomonas aeruginosa is 1.1 ± 0.1 × 101 M−1 s−1 (4). Similarly, the related DBO, relebactam, is also not effective at restoring susceptibility to strains carrying most OXA β-lactamases, with the exception of blaOXA-48 (5–7).
FIG 1.
Structures of the DBOs, avibactam, ETX2514, and ETX2514 analog compound 3.
Unlike other members in the DBO family, the structure of WCK 4234 consists of a nitrile R1 group, and its inhibitory activity extends to OXA-23 (k2/Ki value of 1.7 ± 0.2 × 104 M−1 s−1) and OXA-24/40 (k2/Ki value of 9.6 ± 1 × 103 M−1 s−1) (8). Notably, the acylation rates of ETX2514 for OXA-23 and OXA-24/40 are in a similar range (5 to 9 × 103 M−1 s−1) (2). Moreover, a diverse array of OXA β-lactamases (e.g., OXA-23, OXA-24/40, OXA-51, and OXA-58 families) are prevalent resistance determinants in Acinetobacter spp. (2, 9).
ETX2514 was rationally designed as a novel and more versatile inhibitor with increased reactivity and improved binding to β-lactamases, resulting from the addition of a double bond between C3 and C4 and a methyl group at the C3 position compared to avibactam (Fig. 1) (2). In addition to class D β-lactamases OXA-10, OXA-23, and OXA-24/40 inhibition (k2/Ki values in the range of 5 to 9 × 103 M−1 s−1), ETX2514 inhibits OXA-48 more robustly with an 8 ± 2 × 105 M−1 s−1 k2/Ki. (2). Further, ETX2514 inhibits class A β-lactamases (e.g., TEM-1, CTX-M-15, KPC-2, and SHV-5) and class C β-lactamases (e.g., AmpC and the Enterobacter cloacae AmpC, P99) with k2/Ki values ranging from 9 ± 2 × 105 to 1.4 ± 0.6 × 107 M−1 s−1 (2). The imipenem-ETX2514 combination is effective against Enterobacteriaceae and P. aeruginosa (10). Moreover, the sulbactam-ETX2514 combination extends efficacy to Acinetobacter baumannii (10, 11).
Carbapenem hydrolyzing OXA-like β-lactamases, including OXA-23-like, OXA-24/40-like, OXA-58-like, and OXA-143-like, have been identified in A. baumannii (12, 13). In the present study, we set out to test the susceptibility of a diverse panel of multidrug-resistant (MDR) A. baumannii isolates to sulbactam-ETX2514 as well as further biochemically and structurally characterize the ability of ETX2514 to inhibit class C ADC-7 (the AmpC ortholog in A. baumannii) and class D OXA-58 chromosomal β-lactamases which are highly prevalent in clinical isolates of Acinetobacter spp.
RESULTS AND DISCUSSION
Susceptibility testing of A. baumannii clinical strains.
Seventy-two well-characterized A. baumannii isolates from the Walter Reed Army Medical Center (WRAMC) were tested against sulbactam with and without ETX2514. Previous characterization of the genetic content of these strains revealed that the majority encoded class C (blaADC) genes (99%) and class D (blaOXA-69-like) genes (97%). Furthermore, 40% of strains carried a class A blaTEM gene, and 11% and 12.5% of the isolates carried class D blaOXA-23-like and blaOXA-58-like genes, respectively (Table 1) (14). Using the MIC breakpoint for ampicillin-sulbactam (S ≤ 8/4; I = 16/8, R ≥ 32/16 mg/liter) set by the Clinical and Laboratory Standards Institute (CLSI) (15), 47% (34/72) of the isolates were resistant to sulbactam alone (MIC ≥ 16 mg/liter), but only one isolate (1%) maintained an MIC higher than 8 mg/liter (i.e., AB054 MIC = 32 mg/liter) against sulbactam in combination with 4 mg/liter ETX2514 (Table 2). We rationalized the use of the breakpoint of sulbactam alone taken from the CLSI-defined breakpoints for the ampicillin-sulbactam combination, since ampicillin contributes little activity to sulbactam in Acinetobacter spp. (16). Seventeen of the 72 isolates (24%) were resistant to meropenem (R ≥ 8 according to CLSI [15]; Table 2).
TABLE 1.
Presence of β-lactamase gene in the Walter Reed Army Medical Center (WRAMC) isolates (14)
| β-Lactamase gene | Prevalence (%)a |
|---|---|
| blaADC | 99 (71/72) |
| blaOXA-69-like | 97 (70/72) |
| blaOXA-23-like | 11 (8/72) |
| blaOXA-58-like | 12.5 (9/72) |
| blaTEM | 40 (29/72) |
| blaSHV | 0 (0/72) |
| blaPER | 3 (2/72) |
The number of isolates with the β-lactamase gene and total number of isolates are shown within parentheses.
TABLE 2.
Susceptibility of 72 Walter Reed Army Medical Center (WRAMC) A. baumannii isolatesa
| Strain | MIC (mg/liter) |
||
|---|---|---|---|
| Sulbactam | Sulbactam-ETX2514b | Meropenem | |
| AB001 | 16 | 2 | 1 |
| AB002 | 8 | 1 | 2 |
| AB003 | 8 | 1 | 2 |
| AB004 | 4 | 1 | 2 |
| AB005 | 4 | 1 | 1 |
| AB006 | 4 | 2 | 4 |
| AB007 | 4 | 1 | 0.5 |
| AB008 | 4 | 0.5 | 1 |
| AB009 | 4 | 1 | 1 |
| AB010 | 4 | 1 | 1 |
| AB011 | 8 | 1 | 1 |
| AB012 | 4 | 1 | 1 |
| AB013 | 4 | 1 | 1 |
| AB014 | 4 | 1 | 1 |
| AB015 | 4 | 1 | 2 |
| AB016 | 4 | 1 | 0.5 |
| AB017 | 32 | 1 | 0.5 |
| AB018 | 64 | 1 | 0.5 |
| AB019 | 16 | 1 | 0.25 |
| AB020 | 16 | 1 | 0.5 |
| AB021 | 16 | 1 | 0.5 |
| AB022 | 16 | 1 | 0.5 |
| AB023 | 16 | 1 | 2 |
| AB024 | 16 | 1 | 0.5 |
| AB025 | 16 | 1 | 0.5 |
| AB026 | 16 | 1 | 1 |
| AB027 | 16 | 2 | 0.5 |
| AB028 | 16 | 0.5 | 0.5 |
| AB029 | 16 | 0.5 | 0.5 |
| AB030 | 16 | 0.5 | 1 |
| AB031 | 16 | 0.25 | 1 |
| AB032 | 16 | 0.5 | 0.5 |
| AB033 | 64 | 0.5 | 1 |
| AB034 | 16 | 0.5 | 1 |
| AB035 | 16 | 0.5 | 0.25 |
| AB036 | 16 | 0.5 | 0.5 |
| AB037 | 64 | 0.5 | 0.25 |
| AB038 | 16 | 0.5 | 1 |
| AB039 | 4 | 0.5 | 1 |
| AB040 | 32 | 0.5 | 0.5 |
| AB041 | 16 | 0.5 | 0.5 |
| AB042 | 4 | 0.5 | 4 |
| AB043 | 4 | 1 | 16 |
| AB044 | 4 | 1 | 8 |
| AB045 | 2 | 0.5 | 8 |
| AB046 | 4 | 1 | 16 |
| AB047 | 4 | 1 | 8 |
| AB048 | 4 | 0.5 | 16 |
| AB049 | 4 | 0.5 | 32 |
| AB050 | 4 | 0.5 | 16 |
| AB051 | 2 | 1 | 1 |
| AB052 | 64 | 8 | 2 |
| AB053 | 32 | 8 | 2 |
| AB054 | 64 | 32 | 32 |
| AB055 | 16 | 4 | 8 |
| AB056 | 8 | 1 | 16 |
| AB057 | 16 | 1 | 32 |
| AB058 | 32 | 4 | 2 |
| AB059 | 32 | 1 | 32 |
| AB060 | 8 | 1 | 32 |
| AB061 | 8 | 2 | 32 |
| AB062 | 8 | 2 | 64 |
| AB063 | 32 | 1 | 0.5 |
| AB064 | 8 | 0.5 | 0.25 |
| AB065 | 8 | 2 | 32 |
| AB066 | 4 | 0.5 | 0.25 |
| AB067 | 4 | 0.5 | 0.25 |
| AB068 | 4 | 2 | 1 |
| AB069 | 4 | 1 | 1 |
| AB070 | 32 | 8 | 2 |
| AB071 | 8 | 2 | 0.5 |
| AB072 | 4 | 0.5 | 0.25 |
| MIC50 | 8 | 1 | 1 |
| MIC90 | 32 | 2 | 32 |
According to CLSI (15), ampicillin-sulbactam R ≥ 32/16 mg/liter and meropenem R ≥ 8 mg/liter.
The ETX2514 concentration was 4 mg/liter.
We also assessed 26 additional A. baumannii isolates that were carbapenem resistant (R > 8 mg/liter for meropenem and imipenem) (Table 3): 8 strains were collected from patients at the Brooke Army Medical Center, 10 strains tested were from patients at University Hospitals of Cleveland, and 8 isolates were collected at the Cleveland Clinic Foundation (CCF). ETX2514 combined with sulbactam was also extremely effective against this panel of A. baumannii isolates. The addition of ETX2514 lowered the MICs of sulbactam alone by 8- to 64-fold, resulting in MICs of ≤4 mg/liter for the sulbactam-ETX2514 combination against all 26 strains (Table 3).
TABLE 3.
Susceptibility of carbapenem-resistant A. baumannii isolatesa
| Strain | MIC (mg/liter) |
|||
|---|---|---|---|---|
| Sulbactam | Sulbactam-ETX2514b | Meropenem | Imipenem | |
| VA-432 | 16 | 2 | 64 | 32 |
| VA-433 | 16 | 1 | 32 | 16 |
| VA-434 | 16 | 2 | 64 | 64 |
| VA-435 | 16 | 2 | 64 | 32 |
| VA-436 | 16 | 2 | 64 | 64 |
| VA-437 | 8 | 2 | 32 | 32 |
| VA-438 | 8 | 1 | 32 | 16 |
| VA-505 | 8 | 1 | 32 | 32 |
| UH AB636 | 16 | 2 | >8 | >8 |
| UH AB667 | 32 | 4 | >8 | >8 |
| UH AB477 | 16 | 2 | >8 | >8 |
| UH AB491 | 16 | 2 | >8 | >8 |
| UH AB660 | 16 | 2 | >8 | >8 |
| UH AB202 | 16 | 2 | >8 | >8 |
| UH AB461 | 8 | 1 | >8 | >8 |
| UH AB489 | 16 | 1 | >8 | >8 |
| UH AB487 | 16 | 2 | >8 | >8 |
| UH AB490 | 16 | 2 | >8 | >8 |
| CCF 1MR | 32 | 2 | >8 | >8 |
| CCF 2MR | 16 | 0.25 | >8 | >8 |
| CCF 3MR | 4 | 0.5 | >8 | >8 |
| CCF 4MR | 16 | 1 | >8 | >8 |
| CCF 5MR | 16 | 2 | >8 | >8 |
| CCF 6MR | 16 | 2 | >8 | >8 |
| CCF 7MR | 32 | 4 | >8 | >8 |
| CCF 8MR | 8 | 0.5 | >8 | >8 |
According to CLSI guidelines (15), ampicillin-sulbactam R ≥ 32/16 mg/liter, meropenem R ≥ 8 mg/liter, and imipenem R ≥ 8 mg/liter.
The ETX2514 concentration was 4 mg/liter.
The genomic DNA from the four isolates with a sulbactam-ETX2514 MIC of ≥8 mg/liter was purified for whole-genome sequencing (WGS). All four strains contained a blaADC gene (three blaADC-25 genes and one blaADC-79 gene), blaOXA-66 (OXA-51-like), blaTEM-1, and a mutated adeJ (Acinetobacter drug efflux) gene (the homolog of P. aeruginosa mexB) (Table 4). Strains AB054 and AB070 additionally contained distinct pbp3 mutations. Strain AB070 also carried an additional blaOXA-69 gene (Table 4). Spontaneous resistance to sulbactam-ETX2514 was mapped previously to mutations in the active site of the PBP3 gene, but notably, those mutations and corresponding amino acid substitutions (S395F, S390T, T511S, and V505L) were not identified in these A. baumannii isolates (17). The H370Y mutation that was identified in PBP3 (ftsl gene) was reported previously and does not seem to contribute to carbapenem resistance (18), but this residue sits at a critical location, at the entrance of the active site, and therefore, could be involved in resistance of other classes of antibiotics. The A578T mutation in the ftsl gene is a second shell residue that possibly could affect antibiotic resistance through hydrogen bonding with the addition of the hydroxyl group on the threonine.
TABLE 4.
Presence of relevant genes (β-lactamases, PBP, and efflux gene mutations) in the four WRAMC isolates with sulbactam-ETX2514 MICs of ≥8 mg/liter
| Strain | Relevant genotype |
|---|---|
| AB052 | blaADC-25 blaOXA-66+ blaTEM-1+ adeJ[A326V]+ |
| AB053 | blaADC-25+ blaOXA-66+ bla TEM-1+ adeJ[A326V]+ |
| AB054 | blaADC-25+ blaOXA-66+ blaTEM-1+ adeJ[S1010R]+ pbp3[A578T]+ |
| AB070 | blaADC-79+ blaOXA-66+ blaOXA-69+ blaTEM-1+ adeJ[L670R]+ pbp3[H370Y]+ |
A summary of susceptibility profiles for all 98 A. baumannii strains tested against sulbactam and sulbactam-ETX2514 is presented in Fig. 2. The MIC90 of sulbactam against this collection was lowered from 32 mg/liter alone to 2 mg/liter in the presence of 4 mg/liter of ETX2514. These data closely resemble the MIC90 of 64 mg/liter for sulbactam alone and 4 mg/liter for sulbactam-ETX2514 in a panel of A. baumannii isolates in a previous study (2).
FIG 2.
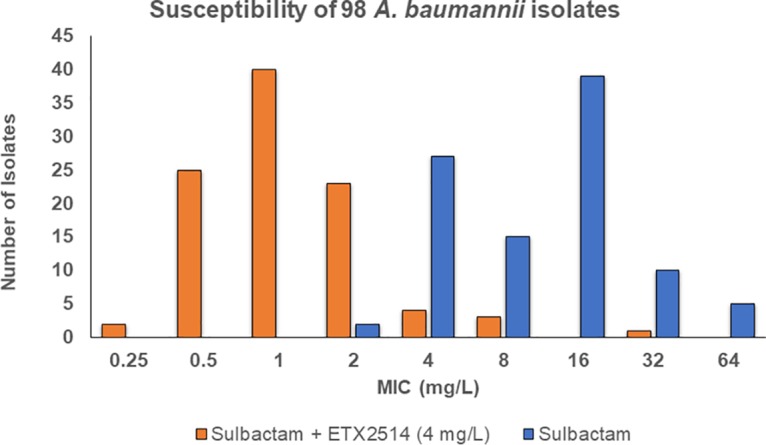
Susceptibility summary of 98 A. baumannii clinical isolates.
Susceptibility testing of Acinetobacter efflux knockout strains.
To further define the contribution of efflux to the antibacterial activity of sulbactam-ETX2514, the MICs of sulbactam with and without ETX2514 against four A. baumannii strains and four Acinetobacter nosocomialis strains with inactivated efflux constituents were compared to the respective wild-type parent of each species. The A. nosocomialis M2 strain was isolated from a patient in 1996 in Cleveland, Ohio (19). While the activity of sulbactam did not change upon addition of ETX2514 against any isogenic mutant tested compared to the parent strain, the addition of ETX2514 led to substantial decreases in sulbactam MICs in A. baumannii devoid of adeB and/or adeJ efflux pump genes, suggesting that ETX2514 is a substrate for these pumps (Table 5). These multidrug transporters are components of two out of the three most prevalent efflux pumps in A. baumannii, adeABC, adeFGH, and adeIJK (20). adeABC was the first resistance-nodulation-division (RND) efflux pump system characterized in A. baumannii (21, 22). A study in 2013 found that 10 out of 14 MDR clinical isolates overexpressed the adeABC operon, but not the other efflux systems (20). The toxic nature of high levels of adeIJK in multidrug-resistant A. baumannii predicts that overexpression of adeABC would be more prevalent than adeIJK in this species (23). In contrast to A. baumannii, susceptibility of A. nosocomialis strains was mostly affected by loss of the adeJ efflux pump (Table 5). Further investigation is needed to determine the relative effect on susceptibility to sulbactam-ETX2514 in Acinetobacter spp. overexpressing these efflux determinants.
TABLE 5.
Susceptibility of efflux knockouts in Acinetobacter
| Acinetobacter species (strain) | Efflux knockout |
MIC (mg/liter) |
Fold change |
|
|---|---|---|---|---|
| Sulbactam | Sulbactam-ETX2514 | |||
| A. baumannii (ATCC 17978) | WT | 1 | 1 | 0 |
| ΔadeB | 1 | 0.125 | 8 | |
| ΔadeJ | 1 | 0.125 | 8 | |
| ΔadeB/ΔadeJ | 1 | 0.125 | 8 | |
| A. nosocomialis (M2) | WT | 2 | 1 | 2 |
| ΔadeB | 2 | 1 | 2 | |
| ΔadeJ | 2 | 0.125 | 16 | |
| ΔadeB/ΔadeJ | 2 | 0.125 | 16 | |
Murine thigh infection model of A. baumannii.
The in vivo efficacy of the sulbactam-ETX2514 combination was evaluated in a neutropenic mouse thigh infection model using A. baumannii ARC5955 carrying blaTEM-1, blaADC-82, blaOXA-23, and blaOXA-66 (OXA-51-like) genes. This strain demonstrates poor susceptibility to sulbactam in vitro with an MIC of 64 mg/liter; however, susceptibility is restored in the presence of ETX2514 with a potentiated MIC of 4 mg/liter (24). As shown in Table 6, subcutaneous administration of sulbactam and ETX2514 individually every 3 h at doses of 75 and 50 mg/kg of body weight resulted in >2 log10 CFU/g of growth in 24 h (vehicle control = +3.39 log10 CFU/g). In combination at the same doses, however, >1-log CFU/g of bactericidal activity is realized relative to the CFU burden at the beginning of treatment. Increasing the ETX2514 dose to 200 mg/kg in combination with 50 mg/kg sulbactam resulted in a 1.5 log10 CFU/g reduction, exceeding the performance of colistin (−1.36 log10 CFU/g), which was used as a positive control for the study.
TABLE 6.
Change in log10 CFU/g burden in thigh tissue 24 h after initiation of therapy to CD-1 mice
| Group ID | Dose (mg/kg) | Route/regimen | Log10 CFU/g | SD | Change in log10 CFU/g |
|---|---|---|---|---|---|
| T = Rx | n/a | n/a | 6.6 | 0.23 | |
| 26-h infection control | Vehicle | SC/q3h | 10.0 | 0.19 | 3.4 |
| Sulbactam | 75 | SC/q3h | 9.0 | 0.22 | 2.5 |
| ETX2514 | 50 | SC/q3h | 8.8 | 0.31 | 2.2 |
| ETX2514-sulbactam | 12.5:75 | SC/q3h | 6.6 | 0.49 | 0.01 |
| ETX2514-sulbactam | 50:75 | SC/q3h | 5.6 | 0.34 | −1.0 |
| ETX2514-sulbactam | 200:75 | SC/q3h | 5.1 | 0.23 | −1.5 |
| Colistin | 40 | SC/q24h | 5.3 | 0.33 | −1.4 |
Kinetic analysis.
ADC-7 is acylated by ETX2514 very quickly with a rate constant of 1.0 ± 0.1 × 106 M−1 s−1 (Table 7). The acylation rate constant (k2/Ki) of ETX2514 for OXA-58 is appreciable, albeit four times lower than for ADC-7. In addition, the koff for ADC-7 (8 ± 1 × 10−4 s−1) was fivefold faster than that for OXA-58 (1.6 ± 0.3 × 10−4 s−1). Both β-lactamases shared the same Kd value (1 nM). The partition ratio (kcat/kinact) (1) was the same for ADC-7 and OXA-58, showing that the β-lactamases do not hydrolyze ETX2514 at an appreciable rate (Table 7). Due to its prevalence in clinical isolates of A. baumannii, we also purified and characterized OXA-66 (OXA-51-like). However, sulbactam was found to be a very poor substrate for this enzyme (data not shown), which also was only modestly inhibited by ETX2514 (Ki app 43 ± 4 µM, k2/K 600 ± 0.7 M−1 s−1).
TABLE 7.
Kinetic parameters of β-lactamase inhibition by ETX2514
| Drug |
Ki app (µM) |
k2/Ki (M−1 s−1) |
koff (residence time) (s−1) |
koff (residence time half-life) (min) |
Kd (nM) |
kcat/kinact |
|---|---|---|---|---|---|---|
| ADC-7 | 0.11 ± 0.01 | 1.0 ± 0.1 × 106 | 8 ± 1 × 10−4 | 14 ± 1 | 1 ± 0.1 | 1 |
| OXA-58 | 0.39 ± 0.04 | 2.5 ± 0.3 × 105 | 1.6 ± 0.3 × 10−4 | 72 ± 8 | 1 ± 0.2 | 1 |
Mass spectrometry of the binding of ETX2514 to β-lactamases.
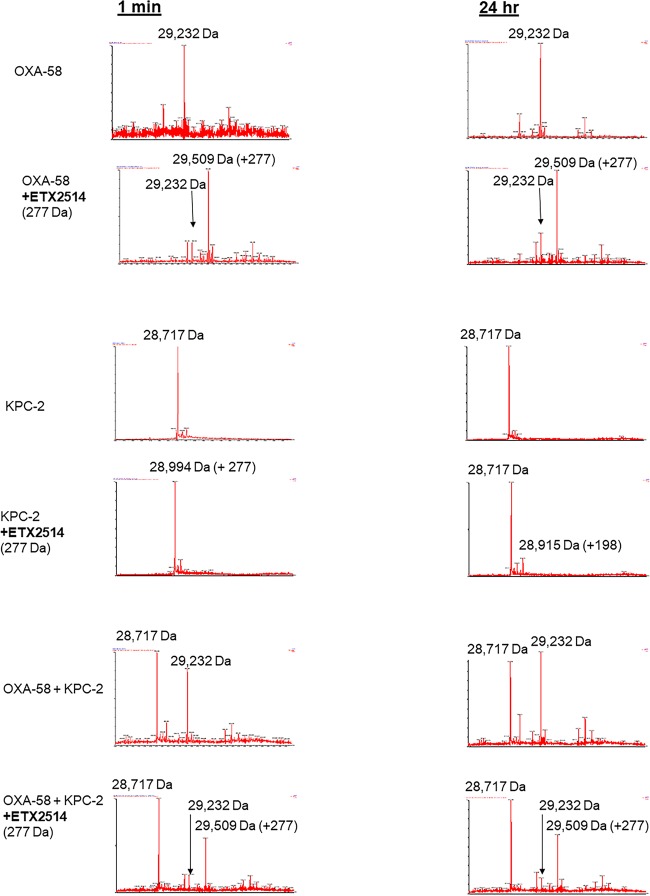
To identify intermediates in the reaction of ETX2514 with ADC-7 and OXA-58, the β-lactamases were incubated with ETX2514 at a 1:1 molar ratio for 5 min, 30 min, and 24 h prior to analysis by mass spectrometry (Fig. 3).
FIG 3.
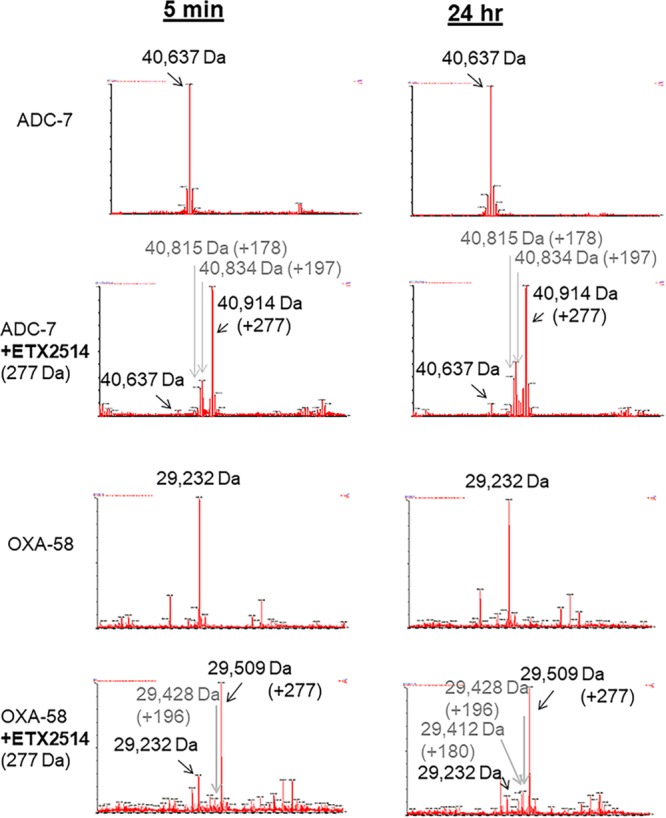
Mass spectra of ADC-7 and OXA-58 β-lactamases and ETX2514. Mass accuracy is within 5 Da.
Timed mass spectrometric analysis of ADC-7.
The ADC-7 β-lactamase was detected as a mass of 40,637 Da (Fig. 3). Incubation of ADC-7 with ETX2514 resulted in the expected mass addition of 277 Da (40,914 ± 5 Da), consistent with an adduct of the full ETX2514 molecule. A mass corresponding to the net loss of the sulfate group from ETX2514 bound to ADC-7 was also observed as a minor product at 40,834 ± 5 Da (40,914 – 80 Da). An additional mass at 40,815 Da corresponding to the loss of water from the desulfated acyl complex was determined. Interestingly, the intensity of the peaks corresponding to the desulfated acyl enzyme did not change significantly over 24 h. Desulfation seems to occur selectively for avibactam, but not for the relebactam or zidebactam DBOs, by the class A KPC-2 as was observed previously (4, 8, 25).
Timed mass spectrometric analysis of OXA-58.
The OXA-58 β-lactamase was detected as a mass of 29,232 Da ± 5 Da (Fig. 3). Incubation of OXA-58 with ETX2514 resulted in the expected mass addition of 277 Da (29,509 Da ± 5 Da). A mass corresponding to the net loss of the sulfate group from ETX2514 bound to ADC-7 was observed at 29,428 Da ± 5 Da (29,509 – 79 Da). An additional mass at 29,412 Da corresponding to the loss of water from the desulfated acyl complex was noted. The mass peaks corresponding to desulfated acyl complex were relatively small and did not dramatically change in intensity after 24 h.
Mass spectrometry of the ETX2514 acyl transfer between OXA-58 and KPC-2 β-lactamases.
To test the reversibility of the ETX2514 binding to OXA-58 and whether ETX2514 binds and dissociates as an intact compound or is modified, the class A KPC-2 β-lactamase was used as the acceptor β-lactamase to bind any ETX2514 that dissociated from the acylated OXA-58 (donor β-lactamase). Mass spectra of the reaction showed only the acylated OXA-58 with intact ETX2514 (+277 Da) from 15 s to 24 h (Fig. 4). ETX2514 did not transfer to KPC-2 under these conditions, which is consistent with a previous report of a lack of acyl exchange between ETX2514 acylated-OXA β-lactamases and CTX-M-15 up to 18 h (1). In the single β-lactamase control reaction using KPC-2 incubated with ETX2514, the intact (+277 Da) ETX2514 was detected bound to KPC-2 after a 15-s incubation, but at 24 h, only a mass addition of 197 Da was detected, corresponding to a desulfated ETX2514 (Fig. 4). The detection of desulfated ETX2514 bound to KPC-2 and the slow regeneration of unacylated KPC-2 due to ETX2514 were reported previously (1).
FIG 4.
Mass spectra of ETX2514 acyl transfer from OXA-58 using KPC-2 as the recipient β-lactamase. Mass accuracy is within 5 Da.
Crystallography.
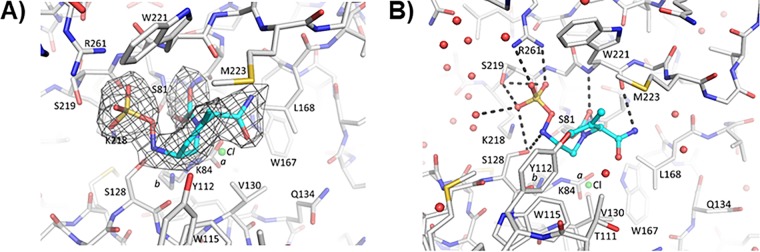
Herein is the first crystallization of OXA-24/40 with ETX2514. The crystal structure of the adduct of ETX2514 with a related carbapenem-hydrolyzing class D β-lactamase, OXA-24/40, revealed a well-ordered opened ring product of ETX2514 covalently bound to the catalytic serine S81 (Fig. 5). The K84 residue was observed to be partially carbamylated with an occupancy of 0.6 (Fig. 5); the occupancy of the noncarbamylated K84 side chain was 0.4, and a chloride ion (also 0.4 occupancy) was present instead of the carboxyl moiety. This was not surprising, as the active site lysine residue in OXA-24 has been shown to exist in equilibrium as carboxylated and noncarboxylated species (26). Further, the presence of the chloride ion was previously also observed for a related DBO inhibitor in complex with OXA-24/40 (27). The carbonyl oxygen of ETX2514 occupies the oxyanion hole formed by the backbone nitrogens of S81 and W221. The sulfate moiety of ETX2514 interacts with R261, S219, and S128. The nitrogen atom attached to the sulfate moiety interacts with S128, whereas the nitrogen atom of the amide moiety interacts with the backbone oxygen of W221. The latter direct protein interaction with the amide of ETX2514 was not observed for avibactam when complexed to OXA-24/40 (Fig. 6).
FIG 5.
ETX2514 in the active site of OXA-24/40. (A) Shown is an unbiased omit |Fo| − |Fc| electron density with ETX2514 removed from refinement and map calculations of OXA-24/40. The inhibitor is shown in blue carbon atom stick representation, whereas the protein is depicted in gray carbon atom stick representation. Electron density is contoured at the 3σ level. The K84 side chain was observed in two conformations; one carbamylated and one noncarbamylated (0.6 and 0.4 occupancy conformations labeled a and b, respectively). A chloride ion with 0.4 occupancy was also refined in the active site (green sphere labeled Cl). (B) Hydrogen bonds (dashed lines) between ETX2514 and OXA-24/40. Water molecules are depicted as red spheres, and the partially occupied chloride ion is depicted as a green sphere.
FIG 6.
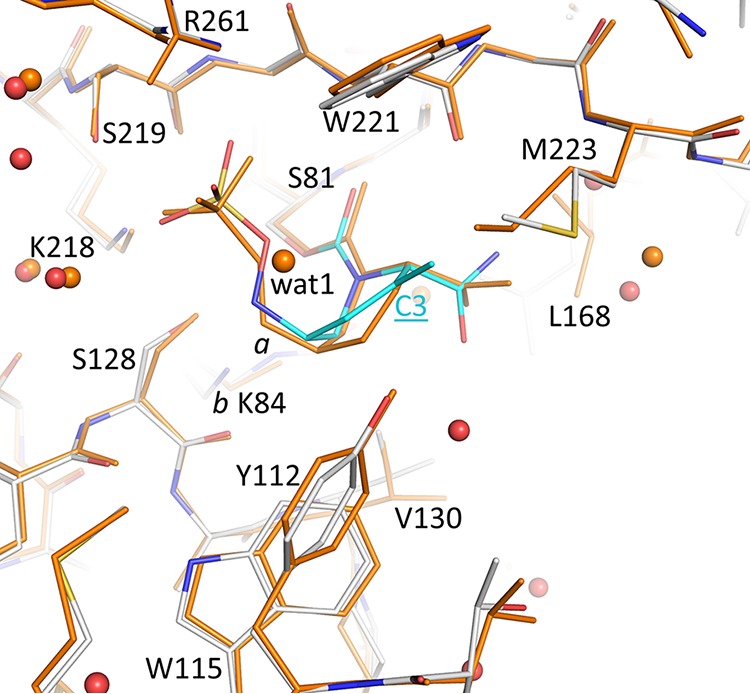
Superposition of ETX2514 and avibactam OXA-24/40 complexes. The avibactam OXA-24/40 complex (PDB ID 4wm9 [38]) is colored orange; the ETX2514 complex is colored as described in the legend to Fig. 5. The C3 atom for ETX2514 is labeled; the two conformations of K84 are labeled a and b. Crystallographic water molecules observed in the two structures are shown as spheres (colored red and orange for the ETX2514 and avibactam OXA-24/40 complexes, respectively).
Interestingly, the tetrahydropyridine ring of ETX2514 adopts a chair-like conformation similar to what was observed for avibactam as well as the ETX2514 analog compound 3 (2) (Fig. 1, 6, and 7). The IC50s of ETX2514 and compound 3 for OXA 24/40 are 0.19 µM and 0.36 µM, respectively, compared to that of avibactam at 16 µM (2). The presence of the extra methyl at the C3 position in ETX2514, compared to compound 3, shifts the side chain of M223 to potentially form tighter van der Waals interactions (Fig. 7). In addition to increasing the chemical reactivity of the DBO core, the double bond in ETX2514, not present in avibactam, gives the six-membered ring in ETX2514 a more planar character to present the C3 methyl group at the right orientation (Fig. 5 and 6). An additional difference between the ETX2514 and avibactam OXA-24/40 crystal structure complexes is the presence of a water molecule near the sulfate of avibactam which was not observed in the ETX2514 complex (“wat1” in Fig. 6). A likely reason for the absence of this water in the ETX2514 complex is that it would be situated at 4.1 Å from the methyl group attached to the C3 ring atom; this environment would probably be too hydrophobic to allow a water molecule to stably cluster at this position in the ETX2514 complex. The possibility of water occupancy differences adjacent to the sulfate of avibactam/ETX2514 in OXA-24/40 complexes was previously also observed in molecular dynamics simulations (2). A water molecule present at this position in close proximity to the ETX2514 sulfate moiety could affect the deacylation rate, and thus efficacy, since that step involves movement of the sulfate moiety to position its bonded nitrogen atom in close proximity to the carbonyl carbon of the bicyclic ring.
FIG 7.
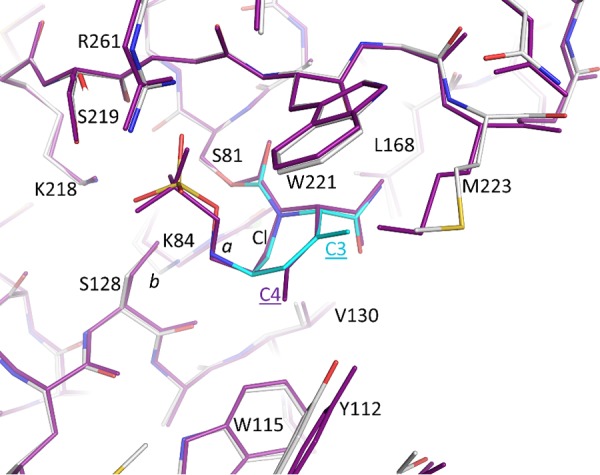
Superpositioning of ETX2514 and compound 3 OXA-24/40 complexes. The compound 3 OXA-24/40 complex (PDB ID 5vfd [2]) is colored magenta; the ETX2514 complex is colored as in Fig. 5. The differentiating methyl substituents on the DBO ring are labeled C4 (for compound 3) and C3 for ETX2514.
Conclusions.
In the present study, we show that sulbactam-ETX2514 effectively restores sulbactam susceptibility in A. baumannii clinical isolates. Only 4 isolates out of 98 tested against the sulbactam-ETX2514 combination demonstrated an MIC of ≥8 mg/liter. In addition, sulbactam-ETX2514 demonstrates robust efficacy in a neutropenic thigh model against A. baumannii ARC5955, a strain that expresses β-lactamases associated with sulbactam degradation (28). Equivalent exposure of ETX2514 and sulbactam in this murine study has been achieved in first-in-human trials suggesting that the combination has the potential to work therapeutically in a clinical setting. Using whole-genome sequencing, mutations in the adeJ efflux pump and/or pbp3 were identified as potential contributors to the elevated MIC values. Indeed, susceptibility testing of A. baumannii isolates lacking individual or a combination of efflux pumps against sulbactam with and without ETX2514 showed increased susceptibility of the strains that were missing adeB and/or adeJ efflux pumps.
Our findings further show that ETX2514 efficiently inhibits ADC-7 and OXA-58 β-lactamases. Inactivation of the class D β-lactamases by a DBO was also recently shown with WCK 4234 that inactivates OXA-23, OXA-24/40, and OXA-51 β-lactamases (27). ETX2514 acylates ADC-7 with a rate constant of 1.0 ± 0.1 × 106 M−1 s−1 and deacylates with a rate constant of 8 ± 1 × 10−4 s−1. A small but consistent amount of desulfated acyl complex was observed with ADC-7 and OXA-58. Further, acyl transfer experiments show that ETX2514 does not transfer from acylated OXA-58 to KPC-2. This behavior mimics the interactions of ETX2514 with other OXA β-lactamases (1). Our study is the first to show ETX2514 cocrystallized with OXA-24/40. OXA-24/40 residues R261, S219, and S128 interact with ETX2514 in the active site, and ETX2514 displaces M223. A chloride ion was also identified in the active site in the ETX2514-OXA-24/40 structure. The ability of ETX2514 to efficiently inhibit the class D OXA carbapenemases in addition to the class A and C β-lactamases makes sulbactam-ETX2514 a promising antibiotic combination for the treatment of Acinetobacter infections, for which it is has recently completed phase 2 clinical testing (ClinicalTrials.gov identifier NCT03445195).
MATERIALS AND METHODS
Critical reagents.
ETX2514 powder was provided by Entasis Therapeutics. Sulbactam was purchased from AstaTech. Nitrocefin (catalog no. BR0063G) was purchased from Oxoid.
Susceptibility testing.
Strains were phenotypically characterized using Mueller-Hinton agar dilution MICs according to the criteria of the CLSI, as previously described (15). A Steer’s Replicator was used to obtain MICs using sulbactam, ETX2514, and sulbactam-ETX2514. In testing, the sulbactam concentration was varied, while ETX2514 was held at a constant concentration of 4 mg/liter. All MIC measurements were performed at least three times, and the mode of the replicates is reported.
For susceptibility testing against carbapenems, MICs were determined by broth microdilution in 96-well plates customized by ThermoFisher Scientific (Cleveland, OH) or TREK Diagnostic Systems (Cleveland, OH), according to CLSI guidelines.
ADC-7 β-lactamase purification.
The ADC-7 β-lactamase was purified from Escherichia coli BL21(DE3) cells carrying the pET-24a(+)blaADC-7 plasmid. Cells were grown in super optimal broth (SOB) at 37°C to an approximate optical density at λ 600 nm (OD600) of 0.6 to 0.8 and induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for a minimum of 3 h to express the β-lactamase. The cells were pelleted and frozen at −20°C for ≥12 h prior to lysis in 50 mM Tris-HCl buffer, pH 7.4, containing 40 mg/ml lysozyme, 0.1 mM magnesium sulfate, and 250 U benzonase nuclease. The supernatant was further purified by preparative isoelectric focusing (pIEF), eluted from the Sephadex 10 mM phosphate-buffered saline (PBS) at pH 7.4, and polishing steps were completed on a HiLoad 16/600 Superdex 75 pg gel filtration column (GE Healthcare). The final sample of protein was concentrated using centrifugal filter units with a molecular weight cutoff of 10,000 daltons (Millipore). The purity of the protein was assessed by quadrupole time of flight (Q-TOF) mass spectrometry and Coomassie blue-stained SDS-PAGE.
OXA-24/40 β-lactamase purification.
OXA-24/40 was expressed and purified from E. coli BL21(DE3) cells expressing blaOXA-24/40 pET24 (+) as previously described (27). Briefly, cells were grown in SOB with 50 µg/ml kanamycin and induced with IPTG. Cells were centrifuged to pellets and frozen at −20°C at least overnight. Cells were lysed in 50 mM Tris-HCl (pH 7.4) with 40 µg/ml lysozyme, 1 mM MgSO4, and 250 U benzonuclease, and debris was cleared by centrifugation. Lysate was processed by an overnight preparative isoelectric focusing (pIEF), and a nitrocefin was used to identify the region of the β-lactamase. OXA-24/40 eluted from the pIEF was further purified on a HiLoad 16/60 Superdex 75 column (GE Healthcare Life Science).
OXA-58 and OXA-66 β-lactamase purification.
The OXA-58 (H22-L280) and OXA-66 (P27-L274) β-lactamase genes were cloned into pET30a downstream of the pelB signal sequence and transformed into E. coli Rosetta2 (DE3) pLysS cells. Cells were grown in 1 liter of LB to an OD600 of approximately 0.6 at 37°C. The culture was cooled in an ice water bath to room temperature and then induced with 0.5 mM isopropyl-β-D-1-thiogalactopyranoside (IPTG) for 16 h at 23°C. The cells were pelleted at 6,000 × g for 30 min, and the pellets were stored at −20°C. Cell pellets were resuspended in 50 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (pH 7) with a protease inhibitor. For OXA-66, DNase and magnesium chloride were also added. Cells were subjected to two rounds of lysis using a French press at 16,000 lb/in2, and the lysates were cleared by centrifugation at 12,000 × g for 20 min. β-Lactamases were purified from the resulting supernatant using a cation exchange column in 50 mM HEPES (pH 7) and eluted by the addition of a linear gradient of sodium chloride of up to 1 M. Further purification of OXA-58 was conducted using a HiLoad 16/600 Superdex 200 gel filtration column preequilibrated with 50 mM HEPES buffer (pH 7) with 100 mM sodium chloride for OXA-58 and 50 mM sodium phosphate (pH 7) for OXA-66. The final stock concentrations of OXA-58 and OXA-66 were 1 mM and 0.12 mM, respectively.
β-Lactamase steady-state kinetic analysis.
Kinetic parameters at 482 nm for nitrocefin and 297 nm for imipenem were determined using an Agilent (Santa Clara, CA) 8453 diode array spectrophotometer using a 1-cm-path-length cuvette. All reactions with ADC-7 were conducted in 10 mM PBS at pH 7.4 at room temperature. Stock solutions of OXA-58 and OXA-66 were diluted accordingly in 50 mM sodium phosphate (pH 7.2). Reactions with OXA-58 and OXA-66 were conducted in 50 mM sodium phosphate (pH 7.2) and 20 mM sodium bicarbonate at ambient temperature.
The Km and Vmax for ADC-7 and OXA-58 β-lactamases and nitrocefin (NCF) were determined by maintaining the enzyme at low nanomolar concentrations and using NCF in excess molar concentration to establish pseudo-first-order kinetics. The resulting data were fit to the Henri Michaelis-Menten equation using Origin 9.1 (OriginLab, Northhampton, MA).
The mechanism of inactivation of β-lactamases by ETX2514 is represented according to the following scheme which is based on previous work with β-lactamases and avibactam (3, 29):
Ki app.
Determination of Ki apparent (Ki app) was described previously (30). Ki app was determined for β-lactamases using a direct competition assay under steady-state conditions. ADC-7 β-lactamase (0.74 nM) was mixed with 100 μM NCF (Km = 20 µM) and 100 to 400 nM ETX2514, and the reaction velocity was monitored for the first 10 s of the reaction. The reaction velocity of OXA-58 β-lactamase (2 nM), 85 μM NCF (Km = 17 μM), and 750 to 2,000 nM ETX2514 was monitored for the first 10 s. OXA-66 β-lactamase (380 nM) was mixed with 300 μM imipenem (Km = 60 μM) and 150 to 300 μM ETX2514 and monitored for the first 20 s. Data were linearized using a Dixon plot of inverse initial steady-state velocities (1/v0) versus inhibitor concentration ([I]). The observed Ki app was determined by dividing the value for the y-axis intercept by the slope of the line. The data were corrected to account for the substrate concentration and affinity for the β-lactamase using equation 1.
| (1) |
where [S] is the concentration of the substrate.
k2/Ki.
Progress curves of β-lactamases mixed with increasing concentrations of ETX2514 using 100 µM NCF (OXA-58 and ADC-7) (85 µM NCF for OXA-58) or 300 µM imipenem (OXA-66) as a reporter substrate were fit to equation 2 to obtain kobs values.
| (2) |
where V0 is the initial velocity, Vf is the final velocity, and A0 is the initial absorbance.
The kobs data were plotted against the concentration of ETX2514. The acylation rate was obtained by correcting the value obtained for the slope of the line (k2/Ki) for the use of the reporter substrate (NCF or imipenem [IMI]) (equation 3).
| (3) |
kcat/kinact.
Partition ratios at 24 h for ADC-7 and OXA-58 with ETX2514 were obtained by incubating each β-lactamase with increasing concentrations of ETX2514. The ratio of inhibitor to enzyme (I:E) necessary to inhibit the hydrolysis of NCF by >95% was determined.
koff.
To determine the off-rate of ETX2514 for ADC-7, the β-lactamase (1 µM) was incubated with 825 nM ETX2514 for 5 min, diluted 1:10,000, and reacted with 100 µM NCF. OXA-58 β-lactamase (1 µM) was incubated with 2.9 µM ETX2514 for 5 min, diluted 1:1,000, and reacted with 85 µM NCF. Progress curves of NCF hydrolysis were collected for 1 h, and the data were fit to a single exponential decay equation to obtain koff. β-lactamase alone and ETX2514 alone were used as controls.
Electrospray ionization mass spectrometry of β-lactamases.
To discern the nature of the intermediates of inactivation by ETX2514 in the reaction pathway with β-lactamases, electrospray ionization (ESI) mass spectrometry (MS) was performed on a Waters Synapt G2-Si quadrupole-time-of-flight mass spectrometer. The Synapt G2-Si mass spectrometer was calibrated with a sodium iodide solution using a mass range of 50 to 2000 m/z. Five micrograms of each β-lactamase (ADC-7 and OXA-58) were incubated with ETX2514 at a 1:1 molar ratio for 5 min, 30 min, or 24 h at room temperature in 10 mM phosphate-buffered saline (PBS), pH 7.4. Reactions were quenched with the addition of 0.1% formic acid in water and 1% acetonitrile. The samples were analyzed using Q-TOF coupled to a Waters Acquity H class ultraperformance liquid chromatograph (UPLC) with an Acquity UPLC BEH C18 column (1.7 µm; 2.1 by 100 mm). The mobile phase consisted of 0.1% formic acid in water. The β-lactamase and β-lactamase–ETX2514 complexes were eluted using a gradient with final conditions of 15% mobile phase and 85% organic phase (100% acetonitrile with 0.1% formic acid). The tune settings for each of the data run was as follows: capillary voltage at 3.5 kV, sampling cone at 35, source offset at 35, source temperature at 100°C, desolvation temperature at 500°C, cone gas at 100 liters/h, desolvation gas at 800 liters/h, and nebulizer bar at 6.0. The protein peaks were deconvoluted using the MaxEnt1 program in MassLynx v4.1 software.
OXA-58 acyl-transfer experiment.
To characterize the acyl transfer, the OXA-58 β-lactamase (10 µM) was incubated with 10 µM ETX2514 for 1 min. Immediately thereafter, the ability of the acylated OXA-58 to donate the acyl group to a second recipient β-lactamase was tested by the addition of a 10 µM concentration of the class A KPC-2 β-lactamase. The koff and k2/Ki of KPC-2 were reported previously as 1.0 ± 0.1 × 10−3 s−1 (1) and (9.3 ± 0.6 × 105 M−1 s−1 (2). The dual β-lactamase reaction was terminated after 15 s, 1 min, 30 min, 1 h, 2 h, 18 h, and 24 h and prepared for mass spectrometry as described above.
Preparation of whole-genome DNA from A. baumannii isolates.
The genomic DNA from four A. baumannii isolates (AB052, AB053, AB054, and AB070) with sulbactam ETX2514 MICs of ≥8 mg/liter was isolated using the MasterPure Gram-positive DNA purification kit (Epicentre). Whole-genome sequencing was performed as previously described (31).
Cocrystallization of OXA-24/40 with ETX2514.
The OXA-24/40 β-lactamase (5 µM) was incubated with ETX2514 compound (250 µM) in 10 mM Tris-HCl (pH 7.8) buffer at 4°C overnight. Subsequently, this reaction mixture was purified over a Superdex 75 size exclusion column in buffer containing 10 mM Tris-HCl (pH 7.8) and 0.15 mM ETX2514. The eluted protein was concentrated to 7 mg/ml and crystallized using the Classic Suite crystallization screening kit (Qiagen) with a 1:1 ratio of protein to reservoir solution in the drop. Best crystals were obtained in a crystallization condition consisting of 0.1 M HEPES sodium salt (pH 7.5), 10% (vol/vol) isopropanol, and 20% (wt/vol) PEG 4000. Crystals were frozen after 3 days of setting up the crystallization experiments.
X-ray data collection and refinement.
Data for the ETX2514–OXA-24/40 complex were collected at the SSRL beamline and processed using AutoXDS scripts (32, 33) (Table 8). The β-lactamase structure was refined using the Refmac (34) and Coot programs (35). The starting protein coordinates for the OXA-24/40 starting structure was PDB identifier (ID) 3mbz. The parameter and topology files for ETX2514 were generated using the PRODRG program (36). The coordinates were checked using the structure validation program PROCHECK (37) and found to have no outliers in the Ramachandran plot. Coordinates and structure factors of the OXA-24–ETX2514 complex have been deposited in Protein Data Bank (PDB ID 6MPQ).
TABLE 8.
Data collection and refinement statistics for ETX2514 complex with OXA-24/40
| Data collection | OXA-24/40 + ETX2514 |
|---|---|
| Wavelength (Å) | 0.97946 |
| Data range (outer shell) (Å) | 1.95–37.3 (1.95–2.0) |
| Space group | P41212 |
| Cell dimensions (Å) | 102.6 102.6 87.0 90° 90° 90° |
| Completeness (outer shell) (%) | 99.6 (99.3) |
| Unique reflections (outer shell) | 34,248 (2,350) |
| Total no. of observations (outer shell) | 226,430 |
| Average multiplicity (outer shell) | 6.6 (6.5) |
| Mean I/sd(I) (outer shell) | 11.3 (2.4) |
| Rmerge (%) | 10.5 (79.1) |
| Refinement | |
| Resolution (Å) | 1.95–37.3 |
| Rwork | 0.187 |
| Rfree | 0.210 |
| No. of molecules (a.s.u.) | 1 |
| Ligands | 1 ETX2514, 1 Cl |
| No. of water molecules | 192 |
| RMSD bond length (Å) | 0.012 |
| RMSD bond angles (°) | 1.51 |
Murine thigh infection model.
All animal procedures were performed at Neosome LLC under the supervision of an IACUC utilizing policies consistent with OLAW standards for the ethical treatment and welfare of animals. Following 5 days of acclimation to the facility, Crl:CD-1 mice (n = 3/treatment group) were rendered transiently neutropenic via two doses of cyclophosphamide administered intraperitoneally on days −4 and −1 at doses of 150 mg/kg and 100 mg/kg, respectively. Following overnight plate culture, some of the bacterial colonies were resuspended in sterile saline and adjusted to deliver 1.0 × 107 CFU/mouse into each thigh of mice via intramuscular injection (100 µl). At 2 h following inoculation, mice were dosed subcutaneously q3h separately with vehicle (control), ETX2514, sulbactam, and combinations of ETX2514 and sulbactam. Colistin was also administered to a separate group of animals at 40 mg/kg q24h as a positive therapy control. Animals were euthanized 24 h after the initiation of therapy, thighs were excised aseptically, weighed, and homogenized in 2 ml of sterile saline. Homogenates were then serially diluted, plated on growth media, and incubated overnight at 37°C, and CFU were enumerated the following day. Bacterial burden was expressed as log10 CFU/g of tissue, and the change in burden over the 24-h course of therapy was considered as a measure of efficacy.
ACKNOWLEDGMENTS
Research reported in this publication was supported in part by a research grant from Entasis Therapeutics (R.A.B. and F.V.D.A.), by funds and/or facilities provided by the Cleveland Department of Veterans Affairs to K.M.P.-W. and R.A.B., the Veterans Affairs Merit Review Program Awards 1I01BX002872 (K.M.P.-W.) and 1I01BX001974 (R.A.B.) from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development, the Geriatric Research Education and Clinical Center VISN 10 (R.A.B.), and the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under awards R01AI100560, R01AI063517, and R01AI072219 to R.A.B.
P.N.R. is supported by VA Merit Award I01 BX001725 and Research Career Scientist Award IK6BX004470, both from the Department of Veterans Affairs.
We thank Neosome Life Sciences for executing the in vivo efficacy studies.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health, the U.S. Department of Veterans Affairs, or the U.S. Government.
Footnotes
Citation Barnes MD, Kumar V, Bethel CR, Moussa SH, O’Donnell J, Rutter JD, Good CE, Hujer KM, Hujer AM, Marshall SH, Kreiswirth BN, Richter SS, Rather PN, Jacobs MR, Papp-Wallace KM, van den Akker F, Bonomo RA. 2019. Targeting multidrug-resistant Acinetobacter spp.: sulbactam and the diazabicyclooctenone β-lactamase inhibitor ETX2514 as a novel therapeutic agent. mBio 10:e00159-19. https://doi.org/10.1128/mBio.00159-19.
REFERENCES
- 1.Shapiro AB, Gao N, Jahic H, Carter NM, Chen A, Miller AA. 2017. Reversibility of covalent, broad-spectrum serine β-lactamase inhibition by the diazabicyclooctenone ETX2514. ACS Infect Dis 3:833–844. doi: 10.1021/acsinfecdis.7b00113. [DOI] [PubMed] [Google Scholar]
- 2.Durand-Reville TF, Guler S, Comita-Prevoir J, Chen B, Bifulco N, Huynh H, Lahiri S, Shapiro AB, McLeod SM, Carter NM, Moussa SH, Velez-Vega C, Olivier NB, McLaughlin R, Gao N, Thresher J, Palmer T, Andrews B, Giacobbe RA, Newman JV, Ehmann DE, de Jonge B, O'Donnell J, Mueller JP, Tommasi RA, Miller AA. 2017. ETX2514 is a broad-spectrum β-lactamase inhibitor for the treatment of drug-resistant Gram-negative bacteria including Acinetobacter baumannii. Nat Microbiol 2:17104. doi: 10.1038/nmicrobiol.2017.104. [DOI] [PubMed] [Google Scholar]
- 3.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Kern G, Walkup GK, Fisher SL. 2012. Avibactam is a covalent, reversible, non-β-lactam β-lactamase inhibitor. Proc Natl Acad Sci U S A 109:11663–11668. doi: 10.1073/pnas.1205073109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ehmann DE, Jahic H, Ross PL, Gu RF, Hu J, Durand-Reville TF, Lahiri S, Thresher J, Livchak S, Gao N, Palmer T, Walkup GK, Fisher SL. 2013. Kinetics of avibactam inhibition against Class A, C, and D β-lactamases. J Biol Chem 288:27960–27971. doi: 10.1074/jbc.M113.485979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blizzard TA, Chen H, Kim S, Wu J, Bodner R, Gude C, Imbriglio J, Young K, Park YW, Ogawa A, Raghoobar S, Hairston N, Painter RE, Wisniewski D, Scapin G, Fitzgerald P, Sharma N, Lu J, Ha S, Hermes J, Hammond ML. 2014. Discovery of MK-7655, a beta-lactamase inhibitor for combination with PrimaxinR. Bioorg Med Chem Lett 24:780–785. doi: 10.1016/j.bmcl.2013.12.101. [DOI] [PubMed] [Google Scholar]
- 6.Lapuebla A, Abdallah M, Olafisoye O, Cortes C, Urban C, Landman D, Quale J. 2015. Activity of imipenem with relebactam against Gram-negative pathogens from New York City. Antimicrob Agents Chemother 59:5029–5031. doi: 10.1128/AAC.00830-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haidar G, Clancy CJ, Chen L, Samanta P, Shields RK, Kreiswirth BN, Nguyen MH. 2017. Identifying spectra of activity and therapeutic niches for ceftazidime-avibactam and imipenem-relebactam against carbapenem-resistant Enterobacteriaceae. Antimicrob Agents Chemother 61:e00642-17. doi: 10.1128/AAC.00642-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moya B, Barcelo IM, Bhagwat S, Patel M, Bou G, Papp-Wallace KM, Bonomo RA, Oliver A. 2017. WCK 5107 (Zidebactam) and WCK 5153 are novel inhibitors of PBP2 showing potent “β-lactam enhancer” activity against Pseudomonas aeruginosa, including multidrug-resistant metallo-β-lactamase-producing high-risk clones. Antimicrob Agents Chemother 61:e02529-16. doi: 10.1128/AAC.02529-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mushtaq S, Warner M, Livermore DM. 2010. In vitro activity of ceftazidime+NXL104 against Pseudomonas aeruginosa and other non-fermenters. J Antimicrob Chemother 65:2376–2381. doi: 10.1093/jac/dkq306. [DOI] [PubMed] [Google Scholar]
- 10.Shapiro A, Guler S, Carter N, Comita-Prevoir J, McLeod S, deJonge B, McLaughlin R, Huynh H, Gao N, Prince D, Ferguson A, Velez C, Durand-Réville T, Miller A, Mueller J, Tommasi R. 2016. ETX2514, a novel, rationally designed inhibitor of class A, C and D β-lactamases, for the treatment of Gram-negative infections, abstr LB-024. ASM Microbe 2016, Boston, MA American Society for Microbiology, Washington, DC. [Google Scholar]
- 11.O'Donnell J, Newman J, Tanudra A, Chen A, De Jonge B, Shankaran H, Mueller J, Tommasi R. 2016. In vitro and in vivo efficacy of the novel β-lactamase inhibitor ETX2514 combined with sulbactam against multidrug resistant Acinetobacter baumannii, abstr LB-117. ASM Microbe 2016, Boston, MA American Society for Microbiology, Washington, DC. [Google Scholar]
- 12.Nowak P, Paluchowska P. 2016. Acinetobacter baumannii: biology and drug resistance - role of carbapenemases. Folia Histochem Cytobiol 54:61–74. doi: 10.5603/FHC.a2016.0009. [DOI] [PubMed] [Google Scholar]
- 13.Evans BA, Amyes SG. 2014. OXA beta-lactamases. Clin Microbiol Rev 27:241–263. doi: 10.1128/CMR.00117-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hujer KM, Hujer AM, Hulten EA, Bajaksouzian S, Adams JM, Donskey CJ, Ecker DJ, Massire C, Eshoo MW, Sampath R, Thomson JM, Rather PN, Craft DW, Fishbain JT, Ewell AJ, Jacobs MR, Paterson DL, Bonomo RA. 2006. Analysis of antibiotic resistance genes in multidrug-resistant Acinetobacter sp. isolates from military and civilian patients treated at the Walter Reed Army Medical Center. Antimicrob Agents Chemother 50:4114–4123. doi: 10.1128/AAC.00778-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Clinical and Laboratory Standards Institute. 2017. Performance standards for antimicrobial susceptibility testing; twenty-seventh informational supplement. Clinical and Laboratory Standards Institute, Wayne, PA. [Google Scholar]
- 16.Corbella X, Ariza J, Ardanuy C, Vuelta M, Tubau F, Sora M, Pujol M, Gudiol F. 1998. Efficacy of sulbactam alone and in combination with ampicillin in nosocomial infections caused by multiresistant Acinetobacter baumannii. J Antimicrob Chemother 42:793–802. doi: 10.1093/jac/42.6.793. [DOI] [PubMed] [Google Scholar]
- 17.McLeod SM, Shapiro AB, Moussa SH, Johnstone M, McLaughlin RE, de Jonge BLM, Miller AA. 2018. Frequency and mechanism of spontaneous resistance to sulbactam combined with the novel beta-lactamase inhibitor ETX2514 in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 62:e01576-17. doi: 10.1128/AAC.01576-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cayô R, Rodríguez M-C, Espinal P, Fernández-Cuenca F, Ocampo-Sosa AA, Pascual A, Ayala JA, Vila J, Martínez-Martínez L. 2011. Analysis of genes encoding penicillin-binding proteins in clinical isolates of Acinetobacter baumannii. Antimicrob Agents Chemother 55:5907–5913. doi: 10.1128/AAC.00459-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Carruthers MD, Harding CM, Baker BD, Bonomo RA, Hujer KM, Rather PN, Munson RS Jr.. 2013. Draft genome sequence of the clinical isolate Acinetobacter nosocomialis strain M2. Genome Announc 1:e00906-13. doi: 10.1128/genomeA.00906-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yoon EJ, Courvalin P, Grillot-Courvalin C. 2013. RND-type efflux pumps in multidrug-resistant clinical isolates of Acinetobacter baumannii: major role for AdeABC overexpression and AdeRS mutations. Antimicrob Agents Chemother 57:2989–2995. doi: 10.1128/AAC.02556-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Magnet S, Courvalin P, Lambert T. 2001. Resistance-nodulation-cell division-type efflux pump involved in aminoglycoside resistance in Acinetobacter baumannii strain BM4454. Antimicrob Agents Chemother 45:3375–3380. doi: 10.1128/AAC.45.12.3375-3380.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Marchand I, Damier-Piolle L, Courvalin P, Lambert T. 2004. Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system. Antimicrob Agents Chemother 48:3298–3304. doi: 10.1128/AAC.48.9.3298-3304.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Coyne S, Courvalin P, Perichon B. 2011. Efflux-mediated antibiotic resistance in Acinetobacter spp. Antimicrob Agents Chemother 55:947–953. doi: 10.1128/AAC.01388-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shapiro A, Moussa S, Tommasi R, Mueller J, O'Donnell J, Miller A. 2017. Restoration of sulbactam activity against multidrug-resistant Acinetobacter baumannii by the novel, broad spectrum beta-lactamase inhibitor ETX2514 correlates with beta-lactamase inhibition in vitro and in vivo. OS0562. 27th ECCMID, Vienna, Austria European Society of Clinical Microbiology and Infectious Diseases, Basel, Switzerland. [Google Scholar]
- 25.Barnes MD, Winkler ML, Taracila MA, Page MGP, Desarbre E, Kreiswirth BN, Shields RK, Nguyen MH, Clancy CJ, Spellberg B, Papp-Wallace KM, Bonomo RA. 2017. Klebsiella pneumoniae carbapenemase-2 (KPC-2), substitutions at Ambler position Asp179, and resistance to ceftazidime-avibactam: unique antibiotic-resistant phenotypes emerge from β-lactamase protein engineering. mBio 8:e00528-17. doi: 10.1128/mBio.00528-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Che T, Bethel CR, Pusztai-Carey M, Bonomo RA, Carey PR. 2014. The different inhibition mechanisms of OXA-1 and OXA-24 beta-lactamases are determined by the stability of active site carboxylated lysine. J Biol Chem 289:6152–6164. doi: 10.1074/jbc.M113.533562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Papp-Wallace KM, Nguyen NQ, Jacobs MR, Bethel CR, Barnes MD, Kumar V, Bajaksouzian S, Rudin SD, Rather PN, Bhavsar S, Ravikumar T, Deshpande PK, Patil V, Yeole R, Bhagwat SS, Patel MV, van den Akker F, Bonomo RA. 2018. Strategic approaches to overcome resistance against Gram-negative pathogens using beta-lactamase inhibitors and beta-lactam enhancers: activity of three novel diazabicyclooctanes, WCK 5153, zidebactam (WCK 5107), and WCK 4234. J Med Chem 61:4067–4086. doi: 10.1021/acs.jmedchem.8b00091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapiro AB. 2017. Kinetics of sulbactam hydrolysis by beta-lactamases, and kinetics of beta-lactamase inhibition by sulbactam. Antimicrob Agents Chemother 61:e01612-17. doi: 10.1128/AAC.01612-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morrison JF, Walsh CT. 1988. The behavior and significance of slow-binding enzyme inhibitors. Adv Enzymol Relat Areas Mol Biol 61:201–301. [DOI] [PubMed] [Google Scholar]
- 30.Papp-Wallace KM, Winkler ML, Gatta JA, Taracila MA, Chilakala S, Xu Y, Johnson JK, Bonomo RA. 2014. Reclaiming the efficacy of β-lactam-β-lactamase inhibitor combinations: avibactam restores the susceptibility of CMY-2-producing Escherichia coli to ceftazidime. Antimicrob Agents Chemother 58:4290–4297. doi: 10.1128/AAC.02625-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Penwell WF, Shapiro AB, Giacobbe RA, Gu RF, Gao N, Thresher J, McLaughlin RE, Huband MD, DeJonge BL, Ehmann DE, Miller AA. 2015. Molecular mechanisms of sulbactam antibacterial activity and resistance determinants in Acinetobacter baumannii. Antimicrob Agents Chemother 59:1680–1689. doi: 10.1128/AAC.04808-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gonzalez A, Tsay Y. 2010. A quick XDS tutorial for SSRL. http://smb.slac.stanford.edu/facilities/software/xds/.
- 33.Kabsch W. 2010. Xds. Acta Crystallogr D Biol Crystallogr 66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 35.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 36.Schuttelkopf AW, van Aalten DM. 2004. PRODRG: a tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr 60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 37.Laskowski RA, MacArthur MW, Moss DS, Thornton JM. 2001. PROCHECK - a program to check the sterochemical quality of protein structures. J Appl Crystallogr 26:283–291. doi: 10.1107/S0021889892009944. [DOI] [Google Scholar]
- 38.Lahiri SD, Mangani S, Jahic H, Benvenuti M, Durand-Reville TF, De Luca F, Ehmann DE, Rossolini GM, Alm RA, Docquier JD. 2015. Molecular basis of selective inhibition and slow reversibility of avibactam against class D carbapenemases: a structure-guided study of OXA-24 and OXA-48. ACS Chem Biol 10:591–600. doi: 10.1021/cb500703p. [DOI] [PubMed] [Google Scholar]