Alien species are often successful invaders in new environments, despite the introduction of a few isolates with a reduced genetic pool. This is called the genetic paradox of invasion. We found two mechanisms by which the invasive forest pathogen causing sudden oak and sudden larch death can evolve. Extensive mitotic recombination producing runs of homozygosity generates genotypic diversity even in the absence of sexual reproduction, and rapid turnover of genes in the non-core, or nonessential portion of genome not shared by all isolates, allows pathogenicity genes to evolve rapidly or be eliminated while retaining essential genes. Mitotic recombination events occur in genomic hot spots, resulting in similar ROH patterns in different isolates or groups; one ROH, independently generated in two different groups, was enriched in pathogenicity genes and may be a target for selection. This provides important insights into the evolution of invasive alien pathogens and their potential for adaptation and future persistence.
KEYWORDS: forest health, genome evolution, oomycetes, sudden larch death, sudden oak death, tree pathogen, clonality, mitotic recombination
ABSTRACT
Invasive alien species often have reduced genetic diversity and must adapt to new environments. Given the success of many invasions, this is sometimes called the genetic paradox of invasion. Phytophthora ramorum is invasive, limited to asexual reproduction within four lineages, and presumed clonal. It is responsible for sudden oak death in the United States, sudden larch death in Europe, and ramorum blight in North America and Europe. We sequenced the genomes of 107 isolates to determine how this pathogen can overcome the invasion paradox. Mitotic recombination (MR) associated with transposons and low gene density has generated runs of homozygosity (ROH) affecting 2,698 genes, resulting in novel genotypic diversity within the lineages. One ROH enriched in effectors was fixed in the NA1 lineage. An independent ROH affected the same scaffold in the EU1 lineage, suggesting an MR hot spot and a selection target. Differences in host infection between EU1 isolates with and without the ROH suggest that they may differ in aggressiveness. Non-core regions (not shared by all lineages) had signatures of accelerated evolution and were enriched in putative pathogenicity genes and transposons. There was a striking pattern of gene loss, including all effectors, in the non-core EU2 genome. Positive selection was observed in 8.0% of RxLR and 18.8% of Crinkler effector genes compared with 0.9% of the core eukaryotic gene set. We conclude that the P. ramorum lineages are diverging via a rapidly evolving non-core genome and that the invasive asexual lineages are not clonal, but display genotypic diversity caused by MR.
INTRODUCTION
Invasive alien tree pathogens are increasingly responsible for devastating forest disease epidemics. Successful pathogens can spread thousands of kilometers within a few decades, attacking naive hosts under new environmental conditions, causing ecosystem-wide change (1). However, there is a genetic paradox to many invasions: pathogen populations often undergo genetic bottlenecks as a result of founder events (2, 3), frequently accompanied by the elimination of sexual reproduction due to the absence of one mating type, resulting in the proliferation of asexual clones (4, 5). The ability to reproduce sexually is considered an important life trait that can impact a pathogen’s ability to overcome host resistance (6), adapt to new environments (7), and contribute to invasiveness (8). Sexual reproduction can generate genotypic diversity, produce novel gene combinations, rapidly disperse beneficial mutations, and purge deleterious ones (9, 10). In diploid species, it can speed up the rate of adaptation by generating the fittest homozygous genotypes among heterozygous isolates (11).
Cryptic diversity, or genotypic diversity in asexual populations, can increase the potential for adaptation. Mitotic recombination (MR) is one mechanism that can generate genotypic diversity, uncover beneficial mutations, and increase the potential for, and rate of, adaptation. In an asexually reproducing diploid, two rare mutational events must occur before a beneficial mutation can become fixed: one producing a heterozygous carrier and the second, at the same locus, converting the heterozygote to a homozygote (12). MR can accelerate this process (12). Another mechanism that could increase adaptive potential is a rapidly evolving non-core genome (13, 14). In some plant pathogens, effector genes that manipulate host processes are associated with gene-sparse regions enriched in repetitive sequences and transposable elements (TEs), which could facilitate rapid adaptation (13, 15–17).
The oomycete genus Phytophthora comprises some of the most destructive pathogens affecting crops and forests. Phytophthora ramorum Werres, De Cock & Man in 't Veld is an invasive pathogen with a remarkably broad host range responsible for the current sudden oak death epidemic in the western United States, the sudden larch death epidemic in the United Kingdom, and ramorum blight of trees and ornamental shrubs (18–20). Four divergent clonal lineages have spread in the United States, Europe, and Canada (21–25). NA1 is responsible for sudden oak death in the United States, NA1, NA2, and EU1 are responsible for ramorum blight in the United States and Canada, and EU1 and EU2 are responsible for sudden larch death and blight in Europe. These lineages appear reproductively isolated, and each comprises a single mating type; however, there is evidence for sexual reproduction in the ancestral source population (26–31). The lineages differ in morphology and aggressiveness and, despite their presumed clonality, exhibit considerable intralineage phenotypic variation (27, 30), raising questions about their adaptive potential.
To investigate genome evolution and the potential for adaptation in P. ramorum, we sequenced the genomes of 107 P. ramorum isolates in all lineages from a broad range of hosts and geographic origins. Although P. ramorum has only been found to reproduce asexually in its current known range, we uncovered a surprising level of cryptic diversity in these populations. We found evidence for MR generating extensive genotypic diversity within the lineages, generating runs of homozygosity (ROH) and fixing nonsynonymous changes in numerous genes. At the interlineage level, divergence is driven by a rapidly evolving non-core genome enriched in transposable elements and genes associated with host-pathogen interactions.
RESULTS
Genome-wide diversity in P. ramorum asexual lineages.
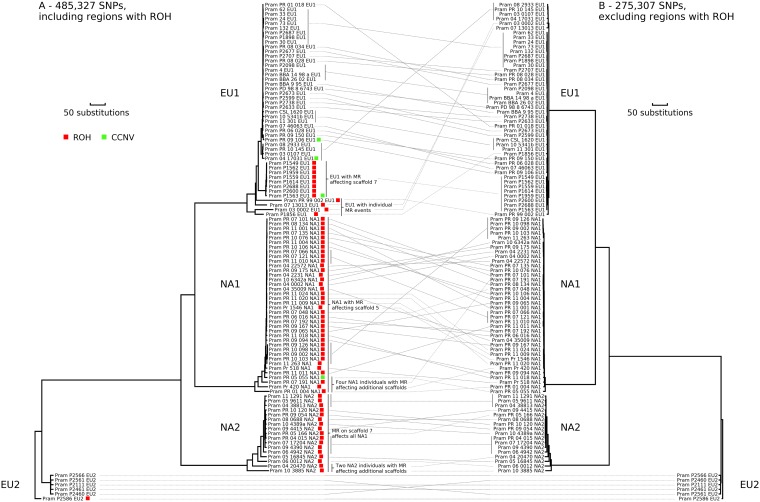
The lineages of P. ramorum clustered into four distinct clades in a neighbor-joining (NJ) tree (Fig. 1A) using 485,327 biallelic single nucleotide polymorphisms (SNPs). We estimate these lineages diverged 0.75 million years ago (MYA) for the split between EU1 and NA1 and 1.3 MYA for the split between EU2 and the other lineages (Text S1, “Divergence time between lineages”). We observed a high level of genotypic diversity within lineages, with some isolates diverging markedly from the main groups (Fig. 1A). The observed diversity resulted from extensive runs of homozygosity (ROH, also known as loss of heterozygosity [LOH] or copy-neutral LOH [cnLOH]), and to a lesser extent, plasticity in chromosome numbers (i.e., chromosomal copy number variants [CCNVs]) or aneuploidy. The most divergent isolates were characterized by ROH covering large portions of or entire scaffolds, ranging in size from 60 to 339 kb, which translated to 1 to 9% of the genome (see Table S1 in the supplemental material). ROH was found in all lineages, and in three lineages, ROH was shared either by the whole lineage or several clones. Eight EU1 isolates, all from the United Kingdom, had the same ROH, suggesting the emergence of a single genotype and subsequent spread (Text S1, “Detection of ROH and effects on genotype”). Four isolates exhibited CCNV, one of which also had ROH (see Fig. S1 and Text S1, “Genome-level copy number variation,” in the supplemental material). When phylogenetic analysis was performed on scaffolds not affected by these characteristics, only minor diversification was evident (Fig. 1B).
FIG 1.
Evolutionary relationships in Phytophthora ramorum. (A) Neighbor-joining tree constructed from Euclidean distances between isolates using 485,327 biallelic SNPs, including SNPs from run of homozygosity (ROH) regions. Isolates displaying ROH are indicated with red squares, and those having chromosomal copy number variants (CCNV) are indicated with green squares. (B) Neighbor-joining tree constructed from Euclidean distances between isolates using 275,307 biallelic SNPs, excluding SNPs from regions with ROH.
Additional materials, methods, and results. Download Text S1, DOCX file, 0.09 MB (95KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Read-depth analysis for chromosomal copy number variation in four Phytophthora ramorum isolates with CCNV. (A) Read-depth ratio of alleles at heterozygous sites over 10 scaffolds. For each scaffold, departure (χ2 test) of the read count for each allele averaged over all heterozygous sites from the null hypothesis of diploidy is represented as follows: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. (B) Distribution of read frequency for the minor allele at heterozygous sites for two scaffolds of interest: e.g., scaffold 11 for Pram_1563 has a distribution averaged around 0.5 typical of a 2n ploidy, distribution is centered around 0.33 for scaffold 2 in Pram_PR_09_106 and 0.25 for scaffold 1 in Pram_PR_04_17031, suggesting triploidy and tetraploidy, respectively. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Percentage of homozygous and heterozygous SNPs in Phytophthora ramorum isolates with runs of homozygosity (ROH) and representatives of the general population without ROH, as well as the combined length of scaffolds with ROH and percentage of genome affected in the scaffold. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Mitotic recombination is responsible for ROH.
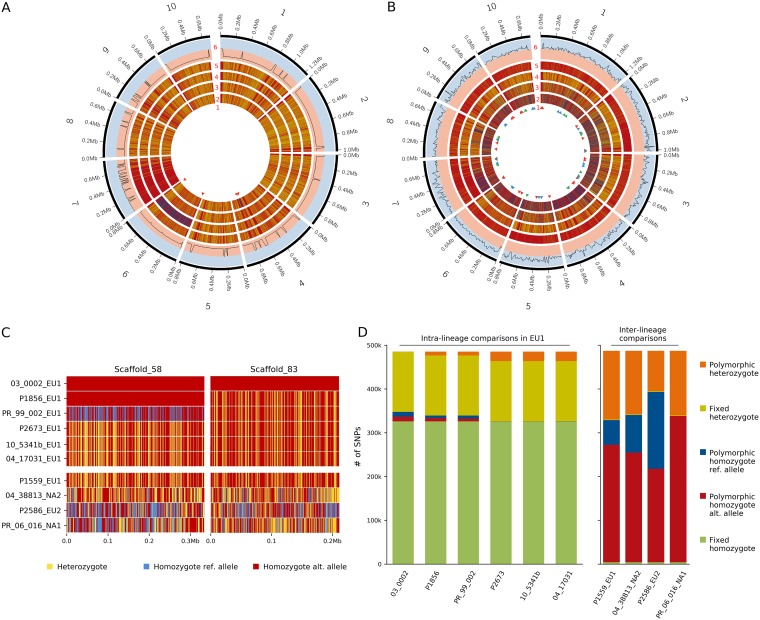
The SNP patterns, including fixed heterozygosity, the lack of polymorphic SNPs between isolates, and the negative within lineage inbreeding coefficient FIS throughout the genomes, are not consistent with those expected from sexual recombination and provide strong evidence of asexual reproduction within lineages (Fig. 2A, C, and D). We observed a limited number of chromosomal breakpoints along the genomes within lineages (Fig. 2A). These were predominantly confined to one or a few regions in a few isolates, resulting in ROH patterns in which all sites after the breakpoint were converted to homozygosity for either the reference or alternate allele relative to the reference genome (e.g., scaffold 4 in NA1 isolate Pram_PR_11_011 and scaffold 6 in NA1 isolate Pram_PR_420 in Fig. 2A and EU1 isolates 03_0002, PR_99_002, and P1856 in Fig. 2C). This pattern is consistent with MR caused by mitotic crossing over. In contrast, interlineage patterns suggest ancient sexual recombination. Alignments of isolates from different lineages reveal a high number of breakpoints, likely the result of ancient meiotic recombination, resulting in multiple short homozygous fragments scattered throughout the genomes (Fig. 2B). The numbers of polymorphic homozygous and heterozygous SNPs between isolates from the different lineages are also higher, and no large genome regions with fixed heterozygosity among isolates were observed (Fig. 2C and D). Furthermore, interlineage FIS was near equilibrium (Fig. 2B).
FIG 2.
Intralineage versus interlineage SNP patterns in Phytophthora ramorum. Shown is a representation of SNPs mapped on the NA1 JGI P. ramorum genome: heterozygous (yellow) and homozygous sites correspond to the reference (red) or alternate (blue) allele. (A) Intralineage diversity in NA1 (scaffolds 1 to 10). Track 1, triangle pairs with the same color indicate blocks delimited by two chromosomal breakpoints; tracks 2 to 5, NA1 isolates Pram_PR_11_011, Pram_PR_06_016, Pram_PR_420, and Pram_PR_09_065; track 6, subtracks show negative (red) and positive (blue) FIS. (B) Interlineage diversity (scaffolds 1 to 10). Tracks 1 and 6, same as in panel A; tracks 2 to 5, isolates Pram_P2586 (EU2), Pram_04_38813 (NA2), Pram_P1559 (EU1), and Pram_PR_06_016 (NA1). (C) Portions of two scaffolds with ROH. All homozygous SNPs were converted to homozygous reference for Pram_03_0002_EU1 and changed to an alternate or reference relative to Pram_03_0002_EU1 for the other isolates. Four EU1 isolates without ROH representing all other EU1 isolates sequenced in this study are shown for comparison as well as an isolate from each of the other lineages. In scaffold 58, the ROH was present in three isolates, two with the same haplotype (Pram_03_0002_EU1 and Pram_P1856_EU1) and the third with the opposite haplotype (Pram_PR_99_002_EU1). In scaffold 83, only one isolate (Pram_03_0002_EU1) has the ROH. (D) Comparison of fixed and polymorphic homozygous and heterozygous SNPs in the isolates in panel C, with intralineage comparison on the left and interlineage comparison on the right.
Protein-coding content differences within P. ramorum lineages.
The ROHs observed could result in altered protein-coding content if isolates with ROH lose one of two alleles with amino acid differences. We observed this pattern in 52% of the 5,172 genes located within scaffolds with ROHs (Table 1 and Text S1, “Detection of ROH and effects on genotype”). In 21% of those genes, there were five or more amino acid differences in isolates without ROH compared to those with ROH, and in one gene, 35 changes were observed (Table 1). Such differences have the potential to generate substantial phenotypic changes.
TABLE 1.
Number of Phytophthora ramorum proteins with amino acid differences between alleles retained and alleles lost through conversion to homozygosity after mitotic recombination
| Lineage | Isolate | Host, origin | No. of genes: |
|||
|---|---|---|---|---|---|---|
| In scaffolds with ROH |
With amino acid difference between alleles |
With allele loss due to ROH |
With ≥5 differences between alleles |
|||
| EU1 | Pram_03_0002 | Rhododendron sp., Canada | 1,587 | 893 | 880 | 202 |
| Pram_07_13013 | Rhododendron sp., Canada | 349 | 186 | 175 | 46 | |
| Pram_P1856 | Acer platanoides, UK | 879 | 516 | 507 | 128 | |
| Pram_PR_99_002 | Viburnum bodnantense, Germany | 875 | 514 | 501 | 127 | |
| EU2 | Pram_P2586 | Larix kaempferi, Scotland | 905 | 444 | 387 | 41 |
| NA1a | Pram_PR_01_004 | Lithocarpus densiflorus, USA | 641 | 328 | 298 | 47 |
| Pram_PR_07_191 | Camellia sp., USA | 204 | 108 | 55 | 9 | |
| Pram_PR_11_011 | Lithocarpus densiflorus, USA | 280 | 150 | 14 | 2 | |
| Pram_Pr_420 | Quercus agrifolia, USA | 220 | 147 | 146 | 36 | |
| NA2 | Pram_04_20470 | Rhododendron sp., Canada | 650 | 303 | 195 | 33 |
| Pram_10_3885 | Leucothoe fontanesiana, Canada | 1,202 | 638 | 615 | 104 | |
| NA1 scaffold_7 | All NA1 isolates | https://doi.org/10.5061/dryad.d81073k | 175 | 84 | 73 | 20 |
| NA1 scaffold_100 | All NA1 isolates | https://doi.org/10.5061/dryad.d81073k | 53 | 24 | 24 | 1 |
| EU1 scaffold_7b | P1559 | Rhododendron sp., UK | 175 | 94 | 63 | 15 |
| P1614 | Nothofagus sp., UK | |||||
| P2600 | Larix kaempferi, UK | |||||
| P1549 | Fagus sylvatica, UK | |||||
| P1562 | Quercus cerris, UK | |||||
| P1563 | Castanea sativa, UK | |||||
| P1959 | Quercus cerris, UK | |||||
| P2688 | Larix kaempferi, UK | |||||
| EU1 scaffold_100b | Shared by 8 isolates above |
Same as for scaffold 7 above | 53 | 25 | 24 | 2 |
| NA2 Scaffold_5 | All NA2 isolates | https://doi.org/10.5061/dryad.d81073k | 144 | 56 | 26 | 2 |
| Allc | All | 5,172 | 2,960 | 2,698 | 555 | |
Excludes scaffolds 7 and 100, which were shared by all NA1 isolates.
Shared by 8 isolates in the EU1 lineage.
Total across all four lineages and all affected isolates, where the same gene affected in two or more isolates is only counted once.
ROH can uncover beneficial mutations and be subject to selection.
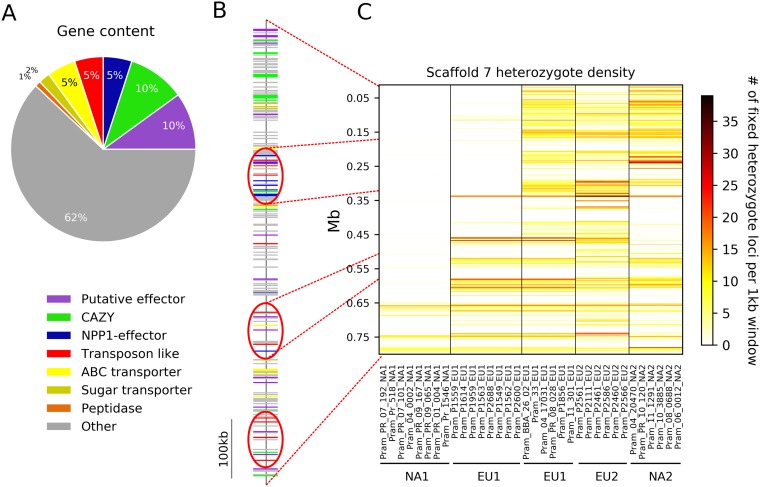
Scaffold 7 had the longest ROH, covering approximately 650 kb, which was shared by all 38 NA1 isolates (large stretch of red homozygous loci in Fig. 2A and shown in white in Fig. 3C). Interestingly, 8 (out of 46) EU1 isolates also have an ROH on scaffold 7, but this appears to be an independent MR event, suggesting an MR hot spot (Fig. 3). Protein-coding content differed on scaffold 7: it had the highest percentage of genes encoding secreted proteins (24.6%) compared to the 25 largest scaffolds (χ2 = 45.127; P < 0.001). Fifteen percent of the genes encode putative effectors, 5% of which are necrosis-inducing proteins (NPP1s). This represents the highest proportion of NPP1s on any scaffold and is significantly higher than expected across the genome (χ2 = 79.3; P < 0.001). The NPP1-encoding genes occur in a cluster within the homozygous region, as do most of the putative effectors (Fig. 3). Scaffold 7 also comprises several genes, such as those encoding carbohydrate-active enzymes, peptidases, and sugar transporters that act in some plant pathogens as virulence factors (Fig. 3 and Text S1, “Detection of ROH and effects on genotype”). An ROH affecting all NA2 isolates was also found on scaffold five (Fig. 1A and Text S1, “Detection of ROH and effects on genotype”).
FIG 3.
Comparison of heterozygote density, gene content, and gene location in scaffold 7 of Phytophthora ramorum. (A) Pie chart showing the percentage of each category of putative plant pathogenicity genes out of all genes on scaffold 7. (B) Gene map showing approximate location of genes on scaffold 7. Clusters of effectors, pathogenicity-related genes, and transposons are circled in red. (C) Heterozygote density of representative isolates of each lineage. The run of homozygosity (ROH) pattern displayed for the NA1 lineage is observed in all 38 NA1 isolates. EU1 isolates possessed two distinct patterns on scaffold 7: an ROH pattern shared by eight isolates and a normal pattern in the remaining 38 isolates.
Evidence of MR hot spots was also observed in other genome regions. EU1 isolates Pram_03_0002, Pram_PR_99_002, and Pram_P1856 share an ROH in at least nine scaffolds (scaffolds 24, 33, 41, 53, 58, 63, 69, 90, and 101 [e.g., Fig. 2C]); however, these genotypes appear to have been generated independently. Pram_PR_99_002 (isolated in Germany in 1999) had the alternate haplotype to Pram_P1856 (isolated in the United Kingdom in 2004) and Pram_03_0002 (isolated in Canada in 2003) in all scaffolds with shared ROH, suggesting these scaffolds lie on the same chromosome. In Pram_03_0002, a much larger portion of the genome was affected (71 scaffolds, totaling 5.9 Mb). ROHs were also observed in the same genome regions in different lineages, providing further evidence of MR hot spots. Overlapping ROH regions were found in Pram_04_20470_NA2 and Pram_PR_01_004_NA1; these isolates shared ROH in 20 scaffolds totaling approximately 3.1 Mb. Similarly, Pram_P2586_EU2 and Pram_PR_11_011_NA1 shared ROH of approximately 39 kbp on scaffold 4 and Pram_P2586_EU2 and Pram_PR_07_191_NA1 shared an ROH covering six scaffolds totaling approximately 0.87 Mb.
Phenotypic characterization of isolates affected by ROH.
EU1 isolates with ROH on scaffold 7 caused on average significantly larger lesions on larch than isolates without ROH (average = 57.4 mm and standard deviation [SD] = 22.2 mm versus average = 38.0 mm and SD = 13.5 mm; Kruskal-Wallis H = 4.412; P = 0.036) (see Fig. S2 and S3 in the supplemental material), but there was no significant difference in lesion sizes on Douglas fir after 4 weeks (average = 56.6 mm and SD = 14.0 versus average = 54.7 mm and SD = 14.9; P = 0.753). Eight weeks postinoculation, the trend was reversed on larch, but the differences were not significant (Text S1, “Effects of ROH on phenotype”). We did not observe loss of growth or infection ability in comparisons of isolates with and without ROH grown on sapwood agar or inoculated onto rhododendron leaves (Text S1, “Effects of ROH on phenotype”).
Lesion length on larch (top) and Douglas fir (middle) 4 and 8 weeks postinoculation of isolates with ROH on scaffold 7 compared to isolates without ROH. The pictures on the bottom represent lesion measured on larch for a no-ROH isolate (left) and a ROH isolate (right). Download FIG S2, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotyping of Phytophthora ramorum isolates with ROH on scaffolds 7 and 100 (n = 8) compared to isolates with no ROH (n = 8). (Top) Lesion length on rhododendron leaves; middle, growth (left) and sporulation (right) on minimum medium supplemented with larch sapwood extract; bottom, growth (left) and sporulation (right) on minimum medium supplemented with Douglas fir sapwood extract. The shaded area around the average represents ± standard deviation (SD). None of the differences were significant. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
TEs and low gene density are associated with mitotic recombination.
Transposable elements (TEs) were found in close proximity to chromosomal breakpoints in 10 out of 12 instances. This was considerably more frequent than in 100 randomly generated data sets; t = −60.16; P < 0.0001) (see Fig. S4A in the supplemental material). The chromosomal breakpoints occurred in regions with a lower gene density than the rest of the genome: intergenic distances surrounding breakpoints were significantly larger than the intergenic distances in random data sets (Kruskal-Wallis H = 38.34; P < 0.0001) (Fig. S4B).
Chromosomal breakpoints occur in regions enriched in transposable elements (TEs) and with less gene density. (A) Number of breakpoint regions (out of 12) with at least one TE (blue); this number is compared to a distribution of 100 × 12 regions randomly sampled along the Phytophthora ramorum reference genome. Statistically supported difference between the observed value and the random distribution is indicated by a one-sample t test. (B) Distribution of intergenic distances (represented by 5′ and 3′ distances between genes) in scaffolds with ROH (in blue) and random scaffolds without ROH (orange). Statistically supported difference between the two distributions is indicated according to a Kruskal-Wallis nonparametric test. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
The core and non-core genomes differ among Phytophthora ramorum lineages.
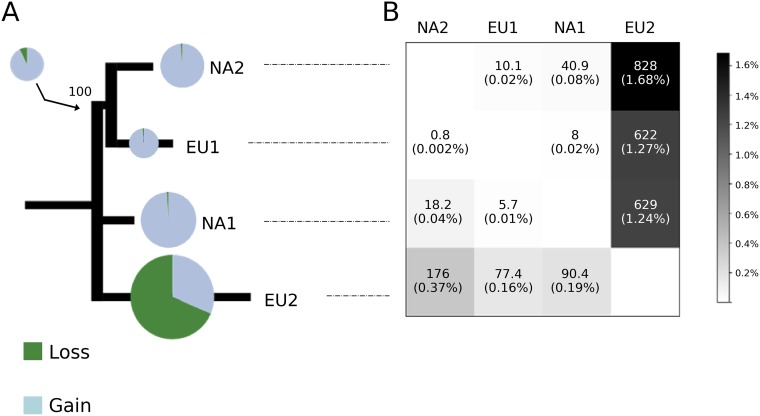
The P. ramorum lineages share a core genome representing 98.3 to 99.5% of the full content, leaving a non-core genome ranging from 0.24 Mbp (EU2) to 0.85 Mbp (NA2). EU2 had a non-core genome that was quite different from the other lineages. Relatively high proportions of its non-core genome (77 to 175.5 kbp, representing 0.16 to 0.37% of the total genome) were not shared with the genomes of the other lineages; similarly, substantial proportions of the NA1, NA2, and EU1 genomes (1.24, 1.27, and 1.68%, respectively) were not found in EU2 (Fig. 4). A neighbor-joining tree based on lineage similarity of non-core regions was topologically incongruent with the P. ramorum lineage phylogeny reconstructed from the core gene set (Fig. 4; see Fig. S5 in the supplemental material). The discordance between the two phylogenies suggests differences in the rate and extent of polymorphism among lineages in the non-core genome. Most of the presence/absence polymorphisms are due to losses in EU2 (68.6%), whereas they have resulted almost entirely from gains in the other three lineages (Fig. 4).
FIG 4.
Core and non-core genomes predicted in the four Phytophthora ramorum lineages. (A) Neighbor-joining tree based on genome similarity in terms of non-core regions (presence/absence of 100-bp non-core regions in the four lineages). Pie charts are proportional to the number of non-core regions that differed in each genome as predicted by CAFE, with expansions in blue and contractions in green. (B) Proportions of lineage non-core genome (rows) that are not found in the full genome content of the other three lineages (columns). For example, the top right corner indicates that 1.68% (i.e., 0.83 Mbp) of the whole genome of NA2 was not found in EU2, and the bottom left corner shows that 0.37% (0.18 Mbp) of the whole genome of EU2 was not found in NA2.
Gene family contraction and expansion in Phytophthora spp. (Left) A CAFE analysis was conducted on the 51,458 OrthoMCL gene families and the maximum likelihood phylogenetic tree obtained for the four P. ramorum lineages, P. lateralis, and five other Phytophthora species. The size of pie charts is proportional to the total number of gene families that changed relatively to a putative common ancestor; values in black at the node are bootstrap support for the RAxML tree. (Right) Neighbor-joining tree reconstructed from a distance matrix based on genome similarity in terms of gene content (presence/absence of 51,458 OrthoMCL gene families). Values above nodes are statistical bootstrap support obtained from 100 resamplings with replacement of gene families. The scale bar indicates 1% genomic dissimilarity (i.e., percentage of difference in terms of number of shared genes). Download FIG S5, TIF file, 0.7 MB (752.1KB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
A rapidly evolving non-core genome enriched in plant cell wall modification enzymes.
The nucleotide and gene compositions were notably distinct in the non-core and core genomes. The non-core genomes had a significantly lower average G+C content across the four lineages (49.4% versus 54.0%; t = 23.21, P < 0.01), a higher repeat content (46.3% versus 15.5%), an enrichment in TE-like genes (χ2 = 19.3; P < 0.001), four to seven times fewer genes per Mb, and more unannotated genes (https://doi.org/10.5061/dryad.d81073k). Genes in the non-core genome were shorter and had a lower G+C content than those in the core genome (average G+C content = 54.9% versus 57.9%; t = 5.14; P = 0.01) and genes in repetitive regions (57.3%; t = 3.87, P < 0.05) (https://doi.org/10.5061/dryad.d81073k). A large number of genes showed strong bias in codon usage (from 82% in NA2 to 100% in EU2), reflecting a preponderance of codons ending with T or A nucleotides (see Fig. S6 in the supplemental material). In addition, one-third to one-half of the genes found in non-core regions (35% in EU1 and EU2, 41% in NA1, and 50% in NA2) share paralogs in the core genome and have high mutation loads, as well as in several instances premature stop codons, suggesting they are degenerated copies (Text S1, “Core and non-core genomes”).
Principal-component analysis (PCA) of codon usage on protein-coding genes of the Phytophthora ramorum NA2 isolate 05-16845 showing a strong bias toward usage of codons ending with A or U. Results are shown for genes included in the core (top left), repetitive (top right), and non-core (bottom left) genomes. Color scales on the right indicate protein density, and the percentages in the top right corner are the proportions of genes found on the right part of the PC1 axis. The bottom right graph represents the contribution of each codon variable to the different components: codons ending with G or C are labelled in black, whereas those ending with A or U are in green. Download FIG S6, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
We did not observe significant enrichment in genes encoding secreted proteins (χ2 = 0.15; P = 0.98) or effectors in the non-core protein-coding genes (17 in EU2 to 44 in NA2). However, we observed presence/absence patterns for 13 effector-like proteins that are part of the RxLR effector family (Table 2) and for genes with functions related to degradation of the plant cell walls (Table 2); eight were degenerated copies of effectors found in the core genome. Remarkably, none of the 17 non-core protein models predicted for EU2 encode effector-like proteins (Table 2).
TABLE 2.
Protein family content predicted in the non-core genome of the four Phytophthora ramorum lineages
| Protein family | No. of effectors in lineagea
|
|||
|---|---|---|---|---|
| EU1 | EU2 | NA1 | NA2 | |
| Ester hydrolasesb | 1 (1) | |||
| Peptidasesb | 1 (1) | 2 | 4 (1) | |
| RxLR | 2 (2) | 1 | 2 (2) | |
| ABC transporter | 1 (1) | 1 | 1 | |
| Kinases | 1 (1) | |||
| Methyltransferases | 3 (3) | 1 | 1 (1) | 2 (2) |
| Glycoside transferasesb | 1 | |||
| Helicases | 2 | 1 (1) | ||
| Transposons | 3 | 6 | 8 (1) | 7 (1) |
| Other hypothetical proteins | 10 (4) | 5 (1) | 17 (5) | 22 (8) |
| Putative proteins without homologs | 5 (2) | 3 (3) | 2 (1) | 5 (1) |
| Total | 26 | 17 | 34 | 44 |
Results for putative effector families according to van Damme et al. (113) are in boldface, and numbers of models in a category that were predicted as putative effectors with EffectorP (114) are shown in parentheses.
Protein models with homologs potentially playing direct or indirect role in degradation of plant cell wall component.
A core P. ramorum proteome of 541 clusters (representing 2,197 proteins) not shared with any other Phytophthora species has likely been conserved since the lineages diverged (see Table S2 in the supplemental material). We identified only slight protein family expansion within each P. ramorum lineage followed by rapid divergence, resulting in proportions of 2.5 to 3.2% lineage-specific proteome (i.e., not shared with any other Phytophthora species or between P. ramorum lineages) (Fig. S5). Gene Ontology (GO) terms associated with peptidase activity and pectin and glucan modification were enriched among the unique protein models, suggesting that rapidly diverging proteins unique to each lineage are enriched in functions related to plant cell wall modification and/or degradation (see Table S3 in the supplemental material).
Shared and unique protein models found in Phytophthora ramorum lineages. Download Table S2, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Model-based gene set analysis for enriched Gene Ontology terms (the five most represented) in sets of proteins unique to each Phytophthora ramorum lineage. Download Table S3, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Evidence of host-driven adaptation in Phytophthora ramorum lineages.
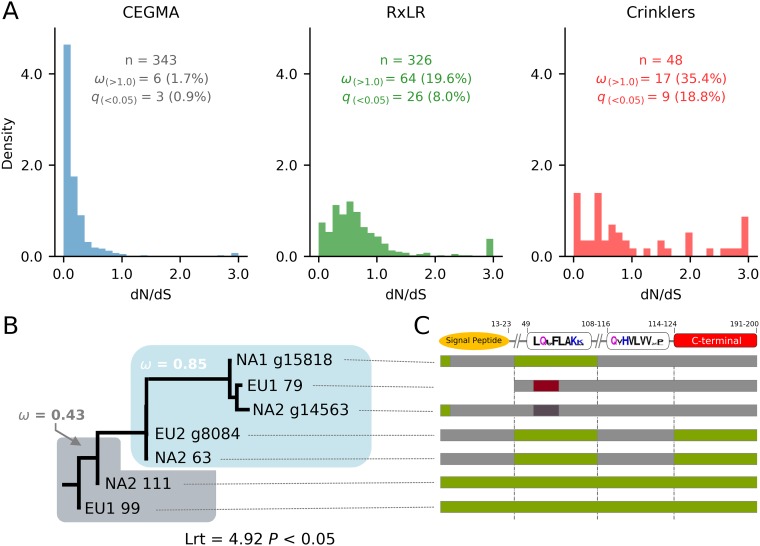
Significant positive selection (q value of <0.05) was observed in 8.0% of the RxLR genes and 18.8% of the Crinkler effector (CRN) genes compared to only 0.9% of the CEGMA (Core Eukaryotic Genes Mapping Approach) gene set and 3.3% in a random set of genes across the lineages (Fig. 5A). CEGMA (69.7%) and random genes (65.7%) were under strong significant negative selection (ratio of nonsynonymous to synonymous evolutionary changes [dN/dS ratio] of <0.3; q value of <0.05). The distributions of dN/dS ratios were different in the effector and CEGMA gene sets (F value = 376.93; P < 0.001) and in the effector and random gene sets (F value = 298.82; P < 0.001). We identified one CRN family that likely diversified within the prior evolutionary history of the P. ramorum lineages through duplication, recombination, and episodes of accelerated nucleotide evolution (Fig. 5B; Text S1, “Genes encoding effectors”).
FIG 5.
Positive selection on genes encoding effectors in Phytophthora ramorum. (A) Distribution of dN/dS values for the RxLR (255 genes), Crinkler (51 genes), and CEGMA (315 genes) gene sets. The number of alignments (n) with ω > 1.0 and significant dN > dS (FDR-adjusted q value of <0.05) are indicated for each gene set. (B) Phylogenetic relationships and evolutionary history of the CRN subfamily expanded in the P. ramorum EU1 and NA2 lineages (The designation after lineage names are gene names for isolates EU1 03-0002, NA1 PR_09_175 NA2 04-38813, and EU2 P2586). (C) Recombination blocks among sequences of the CRN subfamily expanded in the EU1 and NA2 lineages.
DISCUSSION
Phytophthora ramorum is an invasive pathogen that, despite being limited to asexual reproduction and a small effective population size, is spreading extensively in Europe and North America, causing heavy mortality on ecologically and economically valuable trees such as larch and oak species. The pathogen comprises four lineages that started diverging 1 MYA without signs of extant meiotic recombination within or between lineages (24, 26, 31–36). We highlight two mechanisms—MR and a rapidly evolving non-core genome—that could generate diversity and adaptive potential in this pathogen, allowing it to overcome the so-called “genetic paradox of invasion” (2, 3).
We uncovered extensive variation driven by MR causing ROH and potentially affecting hundreds of genes simultaneously. This appears to be an important mechanism fueling evolution in P. ramorum, producing ROH in isolates of all lineages and affecting 2,698 genes with nonsynonymous differences between alleles. One proposed advantage of MR is that it facilitates rapid evolution of asexual eukaryotic microorganisms facing a changing environment (37, 38). By reducing the time between the emergence of beneficial mutations in a heterozygote and the appearance of a homozygous mutant, it may increase the rate of adaptation to a level comparable to that in sexual populations (12). Conversion of genomic regions to homozygosity may result in the expression of new or recessive alleles and can improve the efficiency of selection on both beneficial and deleterious alleles (12).
Several genome regions appeared to be MR hot spots. Multiple ROH were observed in the same genome region in different lineages, in isolates from different continents, and in the same loci but affecting opposite chromosomes. Two separate MR events in lineages NA1 and EU1 affected a scaffold enriched in pathogenicity-related genes. This suggests that one or more genes in this region may be under selection that is maintaining or increasing the frequency of these genotypes. The EU1 ROH genotype appears to be spreading as it has been found on six different hosts since 2003 at multiple sites in the United Kingdom. This EU1 genotype may have arisen from an MR event in a single clone in the United Kingdom or through an introduction of an isolate (or isolates) with the ROH pattern. The NA1 genotype carrying ROH in this region also could have been introduced or originated early in the epidemic and subsequently spread. The likelihood of founder events in two different lineages, on different continents, having genotypes with ROH in the same genomic regions is low, unless the region is an MR hot spot or these genotypes are in high frequency or fixed in the founding populations.
We observed slight differences between EU1 isolates with and without ROH in inoculation experiments, but there were no clear differences of fitness under the conditions we tested. In its introduced state, P. ramorum is a generalist with over 100 hosts (23); therefore, we only tested fitness on a fraction of hosts. The changes induced by the ROH could provide genetic flexibility for interactions with a wide range of hosts and for survival in different environments. Further testing is needed to better understand the potential impact of the extensive variation uncovered. MR and ROH (or LOH) have been previously observed in P. ramorum and other Phytophthora species, but not to the extent reported here (24, 29, 39–49). Loss of heterozygosity (LOH) and aneuploidy have been found in the P. ramorum NA1 and EU1 lineages; in NA1, they have been associated with unstable non-wild-type phenotypes in isolates from “dead-end” hosts (31, 41, 47, 49). MR has been hypothesized to have caused rare mating-type switches in EU1 (31, 47, 49). In Phytophthora capsici, extensive LOH was found; although most changes occurred in noncoding regions, some isolates exhibited changes in virulence or mating type (48). The prevalence of MR in Phytophthora species suggests it is a salient characteristic of the Phytophthora genetic system.
The cause and importance of MR in adaptation in P. ramorum are unclear. There is evidence that MR can be caused by stress. Virulence differences and adaptation to environmental changes or stress have been associated with ROH in other species (38, 50). In Candida albicans, LOH increased proportionally with exposure to oxidative or heat stress and antifungal drugs (37). In Saccharomyces cerevesiae, a trade-off between outcrossing and MR may play a role in shaping genome architecture in response to nutrient stress (38). Stress induced by infection of bark in dead-end hosts has been implicated in changes in chromosome number in P. ramorum (47). The high frequency of MR in P. ramorum could be a response to environmental stressors such as interactions with novel hosts or exposure to fungicides in nurseries. The observed genotypic diversity could reflect phenotypes favored by selection. Alternatively, the very high rates of asexual reproduction during the current epidemics on highly susceptible hosts such as larch and tanoak may allow survival of novel MR genotypes that would not necessarily survive selection under nonepidemic conditions. It is possible that in a future postepidemic phase, when more susceptible host genotypes have been eliminated and selection on fitness components is more intense, some of these novel genotypes with lower fitness may be eliminated.
Rapid evolution in plant pathogens has been associated with high transposon content and activity (14–17, 51–54). Recombination between near-homologous copies of retrotransposons can create new genetic combinations (55). Stress-induced transposon activity has been proposed as an adaptive mechanism enabling pathogens facing new environmental conditions to overcome the invasion paradox (56). Previously, TEs have been associated with chromosomal breakpoints in non-wild-type P. ramorum isolates with LOH (47). We observed a high frequency of chromosomal breakpoints associated with transposons in regions with low gene density, suggesting that TE activity may be triggering MR in P. ramorum, thereby generating genotypic diversity, which is considered important in the successful establishment, persistence, and adaptability of invading populations (2, 57–59).
Pathogenicity genes in plant pathogens are often clustered in rapidly evolving, less-conserved, gene-poor genomic regions enriched in repeated elements or transposons (15, 16, 60–62). These regions generally contain effectors involved in host or ecological adaptation (55, 63). Rapid evolution of these regions can create lineage-specific or divergent non-core regions that vary between populations. The non-core genome of P. ramorum displays a distinct evolutionary trajectory compared to the core genome. It is also enriched in genes associated with plant-pathogen interactions and TE-like sequences, some of which are in tandem with effector genes. It is therefore likely to play an important role in the evolution and adaptive potential of the lineages. There was an extensive loss of effector loci in the non-core genome of the EU2 lineage likely caused by differences in host selection pressures. Selection pressure on effectors may be especially strong since a mismatch in effectors or host recognition is expected to have a high fitness cost (64). The evidence for positive selection and the evolutionary history of effector gene components provide further evidence of rapid evolution. Gene × environment and pathogenicity tests show that the EU2 lineage is adaptively different from EU1 and has higher pathogenicity on the bark of larch (28, 65). It is possible that the EU2 source population coevolved with different primary hosts than the other three lineages, making some effectors obsolete in EU2. The non-core genome appears to play an important role in the evolution of P. ramorum and in the wider adaptive potential of the lineages.
The non-core genome is distinct from the core genome in P. ramorum, having a lower GC content, a strong bias to codons ending in AT, numerous degenerated paralogs, and enrichment of transposons or transposon-like genes. Some ascomycete fungi possess genome defense mechanisms limiting the accumulation of transposable elements. The best-known mechanism is repeat-induced point (RIP) mutation, which inactivates repeated sequences by introducing point mutations in CpG sites, resulting in mutational loads favoring GC-to-AT changes (66). In Zymoseptoria tritici, sequences exclusive to isolates also showed a mutational load likely resulting from a genome defense mechanism similar to RIP (54). No RIP-like mechanism has been demonstrated in Phytophthora species or other oomycetes. We speculate, however, that Phytophthora species may have a defense mechanism, similar to RIP mutation, that can deactivate TEs and restrict genome reshuffling during low-stress periods or lead to a stable genome after periods of enhanced stress or episodic change (20).
The initial invasion “success” of an introduced pathogen such as P. ramorum is often seen in terms of ecological or economic damage that results from the availability of highly susceptible hosts and a lack of natural enemies in new environments. However, invasive pathogens may be limited to asexual reproduction and may face long-term extinction if they are unable to adapt to environmental changes, such as loss of susceptible hosts and the emergence of parasites or competitors. Hence, they may face a genetic paradox. In introduced diploid asexually reproducing organisms, some of the genetic diversity in the parental population is carried in the genome via heterozygosity. Excess heterozygosity can then build up via accumulated mutations—the so-called “Meselson effect” (67). Finally, mitotic recombination can produce different combinations of homozygous alleles, including the loss of deleterious alleles, increasing the potential for adaptation. MR may therefore be important in enhancing the adaptability of introduced asexual Phytophthora species. It could also enhance the pathogen’s ability to overcome more resistant host genotypes among surviving host isolates or those generated via breeding programs. Genomic plasticity, gene loss or gain, epigenetic variation, and effector evolution may further contribute to long-term success.
MATERIALS AND METHODS
Sample preparation and sequencing.
A total of 107 isolates (https://doi.org/10.5061/dryad.d81073k) were selected for genome resequencing. Representatives included the four lineages (38 NA1, 17 NA2, 46 EU1, and 6 EU2 isolates) and covered the chronological (1995 to 2012) and geographical (Canada, the United States, and Europe) ranges of the epidemic. Cultures were grown on peptone-dextrose agar (PDA) overlaid with a cellophane membrane (GE Healthcare Bio-Sciences Corp., Piscataway, NJ) at 20°C under a 12-h photoperiod for 7 to 10 days. DNA was extracted from ground mycelium using a chloroform method (68). DNA was quantified with a Qubit fluorometer (Life Technologies, Inc., Grand Island, NY).
Genome sequencing was done at Canada's Michael Smith Genome Sciences Centre (Vancouver Canada) using one of two methods for library construction (https://doi.org/10.5061/dryad.d81073k). The majority of libraries were constructed on an SPRI-TE robot (Beckman-Coulter, USA) according to the manufacturer’s instructions (SPRIworks Fragment Library System I kit, A84801) following fragmentation by Covaris E210 sonication for 30 s (duty cycle of 20%, intensity of 5) using 1 µg genomic DNA in a 96-well format. The library templates were quantified using a Qubit fluorometer. Five nanograms of template was PCR amplified using Phusion DNA polymerase (Thermo Fisher Scientific, Inc., USA) and Illumina’s PE indexed primer set, with cycle conditions of 98°C for 30 s, followed by 10 cycles of 98˚C for 15 s, 62˚C for 30 s, and 72˚C for 30 s and a final amplicon extension at 72°C for 5 min. For the remaining samples, the paired-end sequencing library preparation followed the BC Cancer Agency’s Genome Sciences Centre 96-well Genomic ∼350- to 450-bp insert Illumina Library Construction protocol using a Biomek FX robot (Beckman-Coulter, USA). The resulting PCR products from both methods were purified using Ampure XP SPRI beads and quantified with Caliper LabChip GX using the high-sensitivity assay (PerkinElmer, Inc., USA). PCR products of desired size range were purified using gel electrophoresis (8% PAGE or 1.5% Metaphor agarose gels in a custom-built robot). DNA quality was assessed and quantified using an Agilent DNA 1000 series II assay and Quant-iT double-stranded DNA (dsDNA) HS assay kit using a Qubit fluorometer (Invitrogen) and then diluted to 8 nM. The final concentration was verified by Quant-iT dsDNA HS assay prior to Illumina Sequencing. For sequencing, 100-bp paired-end tagged (PET) reads were prepared. The DNA of 12 isolates was pooled per lane and sequenced on the Illumina HiSeq 2000 (Illumina, Inc.). Reads were provided in BAM format.
Mapping and extracting SNPs.
PRINSEQ v0.20.3 (69) was used to filter BAM files for redundant reads and reads containing one or more N’s and to trim low-quality bases from the ends by discarding the last 10 nucleotides (for an average quality of <20 calculated on windows of 10 bp). Trimmed reads were mapped onto the P. ramorum reference genome (Pr-102 NA1 lineage; version 1.1; DOE Joint Genome Institute, Walnut Creek, CA) (70) with the Burrows-Wheeler Aligner (BWA) (71) using default parameters. Mapping and quality statistics were calculated using Qualimap v.0.7.1 (72). Variant sites were obtained in a mulitvcf format using the mpileup function of SAMtools and the Bayesian variant-calling models implemented in BCFtools (73). Two alleles were called—a reference allele corresponding to the JGI reference genome and an alternate allele corresponding to the most frequent non-reference allele across the reads of all 107 genomes—resulting in a biallelic data set. Single nucleotide polymorphisms (SNPs) were filtered using VCFtools (74) with parameters determined from testing to minimize false positives and maximize true positives (Text S1, “SNP extraction and filtering—testing SNP filtering parameters and false call rates”): maximum mean depth of 90 and minimum mean depth of 10, and for each SNP, a minimum quality of 30, a minimum depth of 10, a minimum mapping quality of 30, and a minimum distance to a gap of 10 bp. For each genotype, a minimum depth of 4 reads, a maximum of 240, and a minimum genotype quality of 20 were used. We then filtered for a minimum allele count of 2 (allele present in at least two isolates) and no missing data.
We determined if each SNP was fixed in all isolates within a lineage (but different between lineages) or if it was polymorphic in some of the isolates in a lineage. We then determined if the SNP was homozygous or heterozygous. SNPs were classified as (i) fixed homozygous (i.e., homozygous and fixed for one allele in one or more lineages and homozygous and fixed for another allele in another lineage or lineages), (ii) fixed heterozygous in one or more lineages and fixed homozygous in other lineage(s), (iii) polymorphic within lineages but homozygous for one allele in two or more isolates and homozygous for another allele in two or more isolates, or (iv) heterozygous in two or more isolates and homozygous in the other isolates.
De novo genome assemblies.
Two representative isolates of each lineage were assembled de novo (https://doi.org/10.5061/dryad.d81073k). PRINSEQ v0.20.3-filtered Illumina sequencing reads were assembled into contigs and prescaffolded using ABySS (75), with k values ranging from 32 to 96. Final scaffolding was completed with SSPACE v.3.0 (76), and the best assembly was selected based on genome size (∼50.0 Mb) and contiguity (best N50 and length of longest scaffold). Completeness of assemblies was assessed using BUSCO (Benchmarking Universal Single-Copy Orthologs) (77). Assemblies were repeat masked with RepeatMasker (A. F. A. Smit, R. Hubley, and P. Green, RepeatMasker at http://repeatmasker.org) using 2,101 Phytophthora repeats available in Repbase (78) and annotated using AUGUSTUS version 2.7 (79) trained with models from Phytophthora sojae, Phytophthora infestans, and P. ramorum (80). Predicted protein models were functionally annotated with Blast2GO (81) following homolog searches using BLASTp against NR (E value cutoff of 1e−05) and a protein domain search using Interproscan (82).
Phylogenetic reconstruction.
The multisample variant call format (VCF) file was converted to Plink format using VCFtools (74) and recoded for use in Adegenet (83) using Plink v1.07 (84). Euclidean distances were calculated in R and used to construct a neighbor-joining tree using APE (85). The analysis was repeated using a VCF that excluded all scaffolds where ROH was detected.
Detection of ROH.
The multisample VCF file was separated into independent files for each isolate, and files were searched for stretches of homozygous sites uninterrupted by heterozygotes using a custom Python script (Python 2.6). The length of these stretches was compared among isolates within a lineage to identify extensive ROH relative to the population and to identify scaffolds with extensive ROH. VCFtools (74) was used to extract the observed number of heterozygous and homozygous sites for each locus of isolates with ROH and compared to those without ROH, and percentages of homozygous sites for either allele and for heterozygous sites were calculated.
Effect of conversion to homozygosity on protein content.
Data from isolates with ROH were used to phase isolates without ROH using Beagle4 (86). Alternate genome sequences were generated using FastaAlternateReferenceMaker from the GATK suite (87), using the VCF file as the source of variants. We used BEDTools (88) to extract the genes, and two FASTA-formatted files per gene, one for each strand, were obtained for each isolate. Using a custom Python script, genes were translated to proteins and protein translations were compared for isolates with and without ROH to determine the number of amino acid changes between alleles for each group. Gene Ontology (GO) analysis was done on proteins in the ROH regions using the model-based gene set analysis (MGSA) approach in R (89, 90).
Identification of a pathogenicity gene hot spot.
Proteins that occurred in the ROH region in scaffold 7 were extracted from the reference genome (version 1.1) (70) and reannotated using Blast2GO (81). Putative effectors and other proteins potentially involved in pathogenicity (carbohydrate-active enzymes, some transporters, and peptidases) and transposons (or transposon-like elements) were counted and mapped by GenoPlotR in R (91). MGSA (89) was used to identify protein sets that were enriched in scaffold 7, and a χ2 test was used to test if putative effectors were located on scaffold 7 more often than expected.
Growth of isolates with ROH on Douglas fir and larch logs.
We assessed the pathogen’s ability to cause lesions on woody stems of two hosts: Pseudotsuga menziesii (Mirb.) Franco (Douglas fir) and Larix kaempferi (Lamb.) Carr. (Japanese larch) using eight EU1 isolates with ROH in scaffold 7 and eight EU1 isolates without ROH (https://doi.org/10.5061/dryad.d81073k). Three trees of each host between 10 and 25 cm in diameter were freshly felled from the University of British Columbia Malcolm Knapp Research Forest in Maple Ridge, British Columbia (49°15′48.72″N, 122°34′23.61″W). Logs were cut into bolts (0.5 to 0.7 m) and sealed with epoxy resin (Intergard 740; International Paint, Houston, TX). Four isolates were inoculated on each bolt as described in reference 33. There were three replicates for each isolate inoculated on a log bolt from a different tree. Logs were incubated at room temperature in a plastic bag for 8 weeks. The length and width at the longest and widest points of each lesion were measured 4 and 8 weeks postinoculation (wpi) and compared with a Kruskal-Wallis test (significance assessed at the 0.05 level).
Mitotic recombination breakpoints.
We searched for scaffolds with putative mitotic recombination breakpoints defined as regions where a switch from heterozygosity to homozygosity occurred. The 15-kbp region around each breakpoint was scanned for gene content using Blast2GO (81) annotations. Intergenic distances were calculated on full scaffolds containing potential breakpoints. For comparison, 100 data sets, each with 12 15-kbp genomic regions without potential breakpoints, ROH, or stretches of ambiguous nucleotides, were randomly generated. Gene content was determined for the 15-kbp regions in each data set, and intergenic distances were calculated on the full scaffolds for each random set. A nonparametric analysis of variance (Kruskal-Wallis test) was performed to compare the observed distribution of intergenic distances in scaffolds containing potential breakpoints versus the distribution of the randomized data sets.
OrthoMCL and evolution of gene family size.
Protein models obtained for de novo assemblies EU1 030002 (14,095 models), EU2 P2586 (14,028 models), NA1 PR09-175 (14,213 models), NA2 0438813 (14,186 models), and P. lateralis CBS_168.42 (17,463 models) were combined with 188,602 models from five sequenced Phytophthora genomes (15, 70, 92). A tentative clustering of one-to-one orthologs was carried out using OrthoMCL (93) (BLASTp search with an E value cutoff of 1e−05, coverage of at least 50% of the query sequence, and identity of at least 30%, as well as an OrthoMCL inflation value of 4) and then automatically aligned with MAFFT version 7.123b (94). The resulting 51,451 OrthoMCL clusters were then submitted to two filters to minimize the confounding effect of truncated proteins resulting from fragmented de novo assemblies and gene mispredictions as described in reference 95.
A neighbor-joining (NJ) tree representing genome content similarity was reconstructed by calculating a distance matrix based on the gene presence/absence matrix inferred from the OrthoMCL analysis in which the distances measured between pairs of taxa are inversely proportional to the number of genes they share (96). The tree was reconstructed by using the fneighbor program of the PHYLIP version 3.696 package, with 1,000 bootstrap replicate sampling columns of the presence/absence matrix. The maximum likelihood model of CAFE (Computational Analysis of gene Family Evolution) (96) was used to study gene family expansions/contractions while taking into account the one-to-one ortholog phylogeny reconstructed for Phytophthora spp. (Text S1, “Divergence time between lineages—phylogenetic analysis of one-to-one orthologs”).
Core and non-core genomes.
To identify core genomic regions (found in all lineages) and lineage-specific genomic regions, we used the mpileup function of SAMtools (73). We assessed coverage depth over 1-kb sliding windows of all 107 resequenced isolates over two de novo genome assemblies of each of the four lineages. A window was considered as non-core when a contiguous region representing more than 90% of the window size was missing in 50% of the isolates for the NA1, NA2, and EU1 lineages and the six isolates of the EU2 lineage of at least one of the four P. ramorum lineages. Lineage-specific or non-core genomic regions shared by two or three lineages were identified by reciprocal mapping to the de novo assemblies of each lineage. Groups of homologous non-core windows were identified with reciprocal BLASTn searches (E value of <1e−20) within and between lineages followed by graph clustering using a TCL implementation of the Deep-First Search algorithm (identity cutoff = 40%; overlap cutoff = 100 nucleotides [nt]) (97, 98).
A neighbor-joining tree was constructed from a distance matrix of similarity based on non-core genome regions (as done in Text S1, “Divergence time between lineages—phylogenetic analysis of one-to-one orthologs”). The extent of gene gain or loss was determined by attributing costs to gain and loss events and minimizing the total cost (maximum parsimony criterion [99]).
RxLR effectors.
RxLR protein annotation was performed on de novo assemblies and on the P. ramorum reference genome (version 1.1, DOE-JGI) (70) to evaluate optimal strategies for identifying and filtering candidate Avh (avirulence homolog) proteins. Avh effector annotation for each lineage was done in two parts to produce the most candidates, followed by a final manual inspection. First, genomic sequences were translated to a six-frame open reading frame using Emboss, (minimum length of 90) (100). Protein sequences were trimmed to the m-signal peptide. A database of previously identified Avh proteins was used to search de novo assemblies using Hmmer version 3.1b (101), which contained 9,779 proteins from 23 Phytophthora species genomes (P. cajani, P. europaea, P. foliorum, P. hibernalis, P. litchii, P. megakarya, P. melonis, P. parvispora, P. pistaciae, P. syringae, P. uliginosa, P. vignae, P. cinnamomi var. robiniae, Phytophthora taxon niederhauserii, P. pisi, P. cinnamomi, P. rubi, P. fragariae, P. palmivora, P. parasitica, P. sojae, P. ramorum, and P. infestans; provided by the Phytophthora Sequencing Consortium). Candidate proteins with an HMM score of <20, lacking both an RxLR motif and dEER motif, and duplicates were removed. The presence/location of a signal peptide cleavage site was predicted using Signal-P 3.0 (102), and those with a P value of ≥0.8 were retained.
In the second step, a multiple sequence alignment of candidates was created using MUSCLE version 3.8.31 (103) to build an HMM model in Hmmer. Candidate proteins were trimmed as described above, and those with an HMM score of <10 or lacking both the RxLR and dEER motifs were removed. Duplicates were removed, and only those with signal peptide scores of ≥0.8 were retained. This recursive search was repeated using a HMM database of Avh proteins from the first step and 370 previously identified in the P. ramorum genome (104). Candidates identified in both searches were combined, and duplicates were removed. Common motifs were identified using MEME version 4.9.1 (105) (minimum motif of 4 and maximum of 8).
Lastly, effector annotation was performed on all de novo assemblies and on the P. ramorum reference genome. Candidate proteins with an RxLR motif were subsequently categorized according to the actual RxLR sequence. The 370 Avh proteins previously identified from the P. ramorum genome were processed by MEME to generate a list of 312 that had an RxLR motif, which was used as a baseline for optimizing filtering to avoid exclusion of true positives.
CRNs.
The Crinkler protein effectors (CRNs) were identified using two approaches similar to those described by Haas et al. (15). A total of 552 previously reported CRNs from P. infestans (196 effectors), P. ramorum (19 effectors), P. sojae (100 effectors), and P. capsici (237 effectors) (CRNdb) were aligned with MAFFT (94). The recombination domain containing an LxKLAK motif in the first 60 amino acids (aa) of the alignment and the HVLVVVP motif were used to set and train two HMM models with the hmmbuild and hmmcalibrate commands of HMMER v3 (106). The whole proteomes predicted for the four P. ramorum lineages and P. lateralis (see Text S1, “Core and non-core genomes”) were searched for these two models with HMMER v3 (106) (cutoff E value of 1e−05). Candidate CRNs were then aligned with other CRNs of the CRNdb, before training a new HMM model based on full-length CRN sequences.
In the second approach, sequences of the CRNdb were searched against the genomes of P. lateralis and the four P. ramorum lineages using the TBLASTn algorithm (E value cutoff of 1e−04). The coordinates of matches were captured, and matches overlapping genes found in the first approach were removed. For the other matches, the corresponding DNA sequence was translated in ORFs using the EMBOSS package getorf (minimum size cutoff of 100 nt and a maximum size cutoff of 6,000 nt) (100). Predicted ORFs were submitted to an HMMsearch (−T 0) for the full-length CRN model developed in the previous approach. Additional editing was carried out on the positive hits by checking the presence of both LxKLAK and HVLVVVP domains. Both CRN sets were merged to generate a final nonoverlapping set of CRN-like proteins.
Analysis of positive selection on P. ramorum RxLR and CRN effectors.
Sequences of the RxLR and CRN protein data sets were individually clustered with OrthoMCL (93) (BLASTp E value cutoff of 1e−05, 50% coverage, 50% identity, and OrthoMCL inflation value = 1.5). Protein clusters were filtered for truncated proteins as described above. For each sequence cluster, tBLASTn was used to retrieve DNA sequence homologs in the P. ramorum de novo assemblies used for the RxLR search. To generate a set of “neutral” proteins for comparison, sequence homologs of the 458 proteins of the core eukaryotic CEGMA data set (107) were also retrieved by using the same tBLASTn approach. In addition, a random set of 500 proteins with sequence homologs was extracted and used for comparison. Each sequence set was aligned with MAFFT version 7.123b (94). Only sequence alignments of high quality (<5% ambiguous data) and at least 3 synonymous sites were retained to reduce statistical bias in the estimation of the dN/dS value (108). This resulted in 326, 48, 343, and 306 alignments for the RxLR, CRN, CEGMA, and random data sets, respectively. For each alignment, the average ω value (dN/dS) was estimated by fitting the sequence alignment with the basic maximum likelihood model M0 of Codeml (CodonFreq option set to F3X4) implemented in PAMLV4.0 (109). PhyML version 3.0 (110) was used to estimate branch lengths of the phylogenetic tree of the alignment, and used as starting values for Codeml. Values of dN and dS obtained for each branch of the phylogeny reestimated under model M0 of Codeml were extracted using a Python script, and the statistical significance of the difference between the average dN and dS was determined using a paired t test with a false-discovery rate (FDR)-adjusted P value (i.e., q value) for multiple testing (111). For statistical comparisons, RxLR and CRN data sets were grouped into effectors, a Box-Cox transformation was done on the full data set using the MASS package (112) in R (90), and the transformed data sets were compared by analysis of variance (ANOVA).
Data availability.
Whole-genome sequencing (WGS) data for all of the isolates sequenced in this study have been deposited in the Sequence Read Archive under SRA accession no. PRJNA427329.
ACKNOWLEDGMENTS
We thank Brent Kronmiller (Center for Genome Research and Biocomputing, Oregon State University) and members of the Phytophthora Sequencing Consortium for making Avh sequences from 23 Phytophthora genomes available, Joan Webber of the UK Forestry Commission for providing isolates, Miranda Newton and Debbie Shearlaw of the Canadian Food Inspection Agency, Canada, for sample preparation, and Stéphanie Beauseigle and Hesther Yueh at UBC for managing the project.
This work was supported by Genome Canada, Genome British Columbia, Canadian Forest Service (Genomics Research and Development Initiative, GRDI), the Canadian Food Inspection Agency, and FPInnovations through the Large Scale Applied Research Projects (LSARP 2112 and LSARP 10106) and a Genomics Application Partnership (GAPP 6102) grant.
The authors declare they have no competing financial interests.
Footnotes
Citation Dale AL, Feau N, Everhart SE, Dhillon B, Wong B, Sheppard J, Bilodeau GJ, Brar A, Tabima JF, Shen D, Brasier CM, Tyler BM, Grünwald NJ, Hamelin RC. 2019. Mitotic recombination and rapid genome evolution in the invasive forest pathogen Phytophthora ramorum. mBio 10:e02452-18. https://doi.org/10.1128/mBio.02452-18.
REFERENCES
- 1.Loo JA. 2009. Ecological impacts of non-indigenous invasive fungi as forest pathogens. Biol Invasions 11:81–96. doi: 10.1007/s10530-008-9321-3. [DOI] [Google Scholar]
- 2.Gladieux P, Feurtey A, Hood ME, Snirc A, Clavel J, Dutech C, Roy M, Giraud T. 2015. The population biology of fungal invasions. Mol Ecol 24:1969–1986. doi: 10.1111/mec.13028. [DOI] [PubMed] [Google Scholar]
- 3.Pérez JE, Nirchio M, Alfonsi C, Muñoz C. 2006. The biology of invasions: the genetic adaptation paradox. Biol Invasions 8:1115–1121. doi: 10.1007/s10530-005-8281-0. [DOI] [Google Scholar]
- 4.Fry WE, Goodwin SB, Matuszak JM, Spielman LJ, Milgroom MG, Drenth A. 1992. Population genetics and intercontinental migrations of Phytophthora infestans. Annu Rev Phytopathol 30:107–130. doi: 10.1146/annurev.py.30.090192.000543. [DOI] [Google Scholar]
- 5.Garbelotto M. 2008. Molecular analysis to study invasions by forest pathogens: examples from Mediterranean ecosystems. Phytopathol Mediterr 47:183–203. [Google Scholar]
- 6.McDonald BA, Linde C. 2002. Pathogen population genetics, evolutionary potential, and durable resistance. Annu Rev Phytopathol 40:349–379. doi: 10.1146/annurev.phyto.40.120501.101443. [DOI] [PubMed] [Google Scholar]
- 7.North A, Pennanen J, Ovaskainen O, Laine AL. 2011. Local adaptation in a changing world: the roles of gene-flow, mutation, and sexual reproduction. Evolution 65:79–89. doi: 10.1111/j.1558-5646.2010.01107.x. [DOI] [PubMed] [Google Scholar]
- 8.Philibert A, Desprez-Loustau ML, Fabre B, Frey P, Halkett F, Husson C, Lung-Escarmant B, Marçais B, Robin C, Vacher C, Makowski D. 2011. Predicting invasion success of forest pathogenic fungi from species traits. J Appl Ecol 48:1381–1390. doi: 10.1111/j.1365-2664.2011.02039.x. [DOI] [Google Scholar]
- 9.Calo S, Billmyre RB, Heitman J. 2013. Generators of phenotypic diversity in the evolution of pathogenic microorganisms. PLoS Pathog 9:e1003181. doi: 10.1371/journal.ppat.1003181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Heitman J, Sun S, James TY. 2013. Evolution of fungal sexual reproduction. Mycologia 105:1–27. doi: 10.3852/12-253. [DOI] [PubMed] [Google Scholar]
- 11.Kirkpatrick M, Jenkins CD. 1989. Genetic segregation and the maintenance of sexual reproduction. Nature 339:300. doi: 10.1038/339300a0. [DOI] [PubMed] [Google Scholar]
- 12.Mandegar MA, Otto SP. 2007. Mitotic recombination counteracts the benefits of genetic segregation. Proc Biol Sci 274:1301–1307. doi: 10.1098/rspb.2007.0056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dong S, Raffaele S, Kamoun S. 2015. The two-speed genomes of filamentous pathogens: waltz with plants. Curr Opin Genet Dev 35:57–65. doi: 10.1016/j.gde.2015.09.001. [DOI] [PubMed] [Google Scholar]
- 14.Raffaele S, Kamoun S. 2012. Genome evolution in filamentous plant pathogens: why bigger can be better. Nat Rev Microbiol 10:417. doi: 10.1038/nrmicro2790. [DOI] [PubMed] [Google Scholar]
- 15.Haas BJ, Kamoun S, Zody MC, Jiang RHY, Handsaker RE, Cano LM, Grabherr M, Kodira CD, Raffaele S, Torto-Alalibo T, Bozkurt TO, Ah-Fong AMV, Alvarado L, Anderson VL, Armstrong MR, Avrova A, Baxter L, Beynon J, Boevink PC, Bollmann SR, Bos JIB, Bulone V, Cai G, Cakir C, Carrington JC, Chawner M, Conti L, Costanzo S, Ewan R, Fahlgren N, Fischbach MA, Fugelstad J, Gilroy EM, Gnerre S, Green PJ, Grenville-Briggs LJ, Griffith J, Grünwald NJ, Horn K, Horner NR, Hu CH, Huitema E, Jeong DH, Jones AME, Jones JDG, Jones RW, Karlsson EK, Kunjeti SG, Lamour K, Liu Z, et al. 2009. Genome sequence and analysis of the Irish potato famine pathogen Phytophthora infestans. Nature 461:393–398. doi: 10.1038/nature08358. [DOI] [PubMed] [Google Scholar]
- 16.Raffaele S, Farrer RA, Cano LM, Studholme DJ, MacLean D, Thines M, Jiang RHY, Zody MC, Kunjeti SG, Donofrio NM, Meyers BC, Nusbaum C, Kamoun S. 2010. Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330:1540–1543. doi: 10.1126/science.1193070. [DOI] [PubMed] [Google Scholar]
- 17.Raffaele S, Win J, Cano LM, Kamoun S. 2010. Analyses of genome architecture and gene expression reveal novel candidate virulence factors in the secretome of Phytophthora infestans. BMC Genomics 11:637. doi: 10.1186/1471-2164-11-637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Brasier C, Webber J. 2010. Sudden larch death. Science 329:824–825. doi: 10.1038/466824a. [DOI] [PubMed] [Google Scholar]
- 19.Grünwald NJ, Goss EM, Press CM. 2008. Phytophthora ramorum: a pathogen with a remarkably wide host range causing sudden oak death on oaks and ramorum blight on woody ornamentals. Mol Plant Pathol 9:729–740. doi: 10.1111/j.1364-3703.2008.00500.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rizzo DM, Garbelotto M, Davidson JM, Slaughter GW, Koike ST. 2002. Phytophthora ramorum as the cause of extensive mortality of Quercus spp. and Lithocarpus densiflorus in California. Plant Dis 86:205–214. doi: 10.1094/PDIS.2002.86.3.205. [DOI] [PubMed] [Google Scholar]
- 21.Goss EM, Larsen M, Vercauteren A, Werres S, Heungens K, Grünwald NJ. 2011. Phytophthora ramorum in Canada: evidence for migration within North America and from Europe. Phytopathology 101:166–171. doi: 10.1094/PHYTO-05-10-0133. [DOI] [PubMed] [Google Scholar]
- 22.Grünwald NJ, Goss EM, Ivors K, Garbelotto M, Martin FN, Prospero S, Hansen E, Bonants PJM, Hamelin RC, Chastagner G, Werres S, Rizzo DM, Abad G, Beales P, Bilodeau GJ, Blomquist CL, Brasier C, Brière SC, Chandelier A, Davidson JM, Denman S, Elliott M, Frankel SJ, Goheen EM, de Gruyter H, Heungens K, James D, Kanaskie A, McWilliams MG, Man in ‘T Veld W, Moralejo E, Osterbauer NK, Palm ME, Parke JL, Sierra AMP, Shamoun SF, Shishkoff N, Tooley PW, Vettraino AM, Webber J, Widmer TL. 2009. Standardizing the nomenclature for clonal lineages of the sudden oak death pathogen, Phytophthora ramorum. Phytopathology 99:792–795. doi: 10.1094/PHYTO-99-7-0792. [DOI] [PubMed] [Google Scholar]
- 23.Grünwald NJ, Garbelotto M, Goss EM, Heungens K, Prospero S. 2012. Emergence of the sudden oak death pathogen Phytophthora ramorum. Trends Microbiol 20:131–138. doi: 10.1016/j.tim.2011.12.006. [DOI] [PubMed] [Google Scholar]
- 24.Ivors KL, Garbelotto M, Vries IDE, Ruyter-Spira C, Hekkert BTE, Rosenzweig N, Bonants P. 2006. Microsatellite markers identify three lineages of Phytophthora ramorum in US nurseries, yet single lineages in US forest and European nursery populations. Mol Ecol 15:1493–1505. doi: 10.1111/j.1365-294X.2006.02864.x. [DOI] [PubMed] [Google Scholar]
- 25.Vercauteren A, De Dobbelaere I, Grünwald NJ, Bonants P, Van Bockstaele E, Maes M, Heungens K. 2010. Clonal expansion of the Belgian Phytophthora ramorum populations based on new microsatellite markers. Mol Ecol 19:92–107. doi: 10.1111/j.1365-294X.2009.04443.x. [DOI] [PubMed] [Google Scholar]
- 26.Boutet X, Vercauteren A, Heungens K, Laurent F, Chandelier A. 2010. Oospores progenies from Phytophthora ramorum. Fungal Biol 114:369–378. doi: 10.1016/j.funbio.2010.02.009. [DOI] [PubMed] [Google Scholar]
- 27.Brasier C, Kirk S, Rose J. 2006. Differences in phenotypic stability and adaptive variation between the main European and American lineages of Phytophthora ramorum. Proceedings of the 3rd International IUFRO Working Party, p 166–173. In Brasier C, Jung T, Oswald W (ed), Progress in research on Phytophthora diseases of forest trees. Forest Research, Farnham, United Kingdom. [Google Scholar]
- 28.Franceschini S, Webber JF, Sancisi-Frey S, Brasier CM. 2014. Gene × environment tests discriminate the new EU2 evolutionary lineage of Phytophthora ramorum and indicate that it is adaptively different. For Pathol 44:219–232. doi: 10.1111/efp.12085. [DOI] [Google Scholar]
- 29.Goss EM, Carbone I, Grünwald NJ. 2009. Ancient isolation and independent evolution of the three clonal lineages of the exotic sudden oak death pathogen Phytophthora ramorum. Mol Ecol 18:1161–1174. doi: 10.1111/j.1365-294X.2009.04089.x. [DOI] [PubMed] [Google Scholar]
- 30.Hüberli D, Garbelotto M. 2012. Phytophthora ramorum is a generalist plant pathogen with differences in virulence between isolates from infectious and dead-end hosts. For Pathol 42:8–13. doi: 10.1111/j.1439-0329.2011.00715.x. [DOI] [Google Scholar]
- 31.Vercauteren A, De Dobbelaere I, Van Bockstaele E, Maes M, Heungens K. 2011. Genotypic and phenotypic characterization of the European A2 isolates of Phytophthora ramorum. Eur J Plant Pathol 129:621–635. doi: 10.1007/s10658-010-9727-5. [DOI] [Google Scholar]
- 32.Brasier C, Kirk S. 2004. Production of gametangia by Phytophthora ramorum in vitro. Mycol Res 108:823–827. doi: 10.1017/S0953756204000565. [DOI] [PubMed] [Google Scholar]
- 33.Brasier CM, Kirk SA. 2001. Comparative aggressiveness of standard and variant hybrid alder phytophthoras, Phytophthora cambivora and other Phytophthora species on bark of Alnus, Quercus and other woody hosts. Plant Pathol 50:218–229. doi: 10.1046/j.1365-3059.2001.00553.x. [DOI] [Google Scholar]
- 34.Sansford C, Inman A, Baker R, Frankel S, de Gruyter J, Husson C, Kehlenbeck H, Kessel G, Moralejo E, Steeghs M, Webber J, Werres S. 2009. Risk analysis for Phytophthora ramorum, a newly recognised pathogen threat to Europe and the cause of sudden oak death in the USA (acronym—RAPRA). HAL https://hal.archives-ouvertes.fr/hal-01245979.
- 35.Van Poucke K, Franceschini S, Webber JF, Vercauteren A, Turner JA, McCracken AR, Heungens K, Brasier CM. 2012. Discovery of a fourth evolutionary lineage of Phytophthora ramorum: EU2. Fungal Biol 116:1178–1191. doi: 10.1016/j.funbio.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Werres S, Kaminski K. 2005. Characterisation of European and North American Phytophthora ramorum isolates due to their morphology and mating behaviour in vitro with heterothallic Phytophthora species. Mycol Res 109:860–871. doi: 10.1017/S0953756205003369. [DOI] [PubMed] [Google Scholar]
- 37.Forche A, Abbey D, Pisithkul T, Weinzierl MA, Ringstrom T, Bruck D, Petersen K, Berman J. 2011. Stress alters rates and types of loss of heterozygosity in Candida albicans. mBio 2:e00129-11. doi: 10.1128/mBio.00129-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Magwene PM, Kayıkçı Ö, Granek JA, Reininga JM, Scholl Z, Murray D. 2011. Outcrossing, mitotic recombination, and life-history trade-offs shape genome evolution in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A 108:1987–1992. doi: 10.1073/pnas.1012544108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamnanpunt J, Shan W, Tyler BM. 2001. High frequency mitotic gene conversion in genetic hybrids of the oomycete Phytophthora sojae. Proc Natl Acad Sci U S A 98:14530–14535. doi: 10.1073/pnas.251464498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobrowolski MP, Tommerup IC, Shearer BL, O’Brien PA. 2003. Three clonal lineages of Phytophthora cinnamomi in Australia revealed by microsatellites. Phytopathology 93:695–704. doi: 10.1094/PHYTO.2003.93.6.695. [DOI] [PubMed] [Google Scholar]
- 41.Elliott M, Yuzon J, Mathu Malar C, Tripathy S, Bui M, Chastagner GA, Coats K, Rizzo DM, Garbelotto M, Kasuga T. 2018. Characterization of phenotypic variation and genome aberrations observed among Phytophthora ramorum isolates from diverse hosts. BMC Genomics 19:320. doi: 10.1186/s12864-018-4709-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Goodwin SB, Cohen BA, Fry WE. 1994. Panglobal distribution of a single clonal lineage of the Irish potato famine fungus. Proc Natl Acad Sci U S A 91:11591–11595. doi: 10.1073/pnas.91.24.11591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goss EM, Press CM, Grünwald NJ. 2013. Evolution of RXLR-class effectors in the oomycete plant pathogen Phytophthora ramorum. PLoS One 8:e79347. doi: 10.1371/journal.pone.0079347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goss EM, Larsen M, Chastagner GA, Givens DR, Grünwald NJ. 2009. Population genetic analysis infers migration pathways of Phytophthora ramorum in US nurseries. PLoS Pathog 5:e1000583. doi: 10.1371/journal.ppat.1000583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hu J, Diao Y, Zhou Y, Lin D, Bi Y, Pang Z, Trout Fryxell R, Liu X, Lamour K. 2013. Loss of heterozygosity drives clonal diversity of Phytophthora capsici in China. PLoS One 8:e82691. doi: 10.1371/journal.pone.0082691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hüberli D, Tommerup IC, Dobrowolski MP, Calver MC, Hardy GE. 2001. Phenotypic variation in a clonal lineage of two Phytophthora cinnamomi populations from Western Australia. Mycol Res 105:1053–1064. doi: 10.1016/S0953-7562(08)61967-X. [DOI] [Google Scholar]
- 47.Kasuga T, Bui M, Bernhardt E, Swiecki T, Aram K, Cano LM, Webber J, Brasier C, Press C, Grünwald NJ, Rizzo DM, Garbelotto M. 2016. Host-induced aneuploidy and phenotypic diversification in the sudden oak death pathogen Phytophthora ramorum. BMC Genomics 17:385. doi: 10.1186/s12864-016-2717-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lamour KH, Mudge J, Gobena D, Hurtado-Gonzales OP, Schmutz J, Kuo A, Miller NA, Rice BJ, Raffaele S, Cano LM, Bharti AK, Donahoo RS, Finley S, Huitema E, Hulvey J, Platt D, Salamov A, Savidor A, Sharma R, Stam R, Storey D, Thines M, Win J, Haas BJ, Dinwiddie DL, Jenkins J, Knight JR, Affourtit JP, Han CS, Chertkov O, Lindquist EA, Detter C, Grigoriev IV, Kamoun S, Kingsmore SF. 2012. Genome sequencing and mapping reveal loss of heterozygosity as a mechanism for rapid adaptation in the vegetable pathogen Phytophthora capsici. Mol Plant Microbe Interact 25:1350–1360. doi: 10.1094/MPMI-02-12-0028-R. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Turner J, O'Neill P, Grant M, Mumford RA, Thwaites R, Studholme DJ. 2017. Genome sequences of 12 isolates of the EU1 lineage of Phytophthora ramorum, a fungus-like pathogen that causes extensive damage and mortality to a wide range of trees and other plants. Genomics Data 12:17–21. doi: 10.1016/j.gdata.2017.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.James TY, Litvintseva AP, Vilgalys R, Morgan JAT, Taylor JW, Fisher MC, Berger L, Weldon C, Du Preez L, Longcore JE. 2009. Rapid global expansion of the fungal disease chytridiomycosis into declining and healthy amphibian populations. PLoS Pathog 5:e1000458. doi: 10.1371/journal.ppat.1000458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jiang RHY, Tyler BM. 2012. Mechanisms and evolution of virulence in oomycetes. Annu Rev Phytopathol 50:295–318. doi: 10.1146/annurev-phyto-081211-172912. [DOI] [PubMed] [Google Scholar]
- 52.Judelson HS. 2012. Dynamics and innovations within oomycete genomes: insights into biology, pathology, and evolution. Eukaryot Cell 11:1304–1312. doi: 10.1128/EC.00155-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Moller M, Stukenbrock EH. 2017. Evolution and genome architecture in fungal plant pathogens. Nat Rev Microbiol 15:756–771. doi: 10.1038/nrmicro.2017.76. [DOI] [PubMed] [Google Scholar]
- 54.Plissonneau C, Stürchler A, Croll D. 2016. The evolution of orphan regions in genomes of a fungal pathogen of wheat. mBio 7:e01231-16. doi: 10.1128/mBio.01231-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.de Jonge R, Bolton MD, Kombrink A, van den Berg GC, Yadeta KA, Thomma BP. 2013. Extensive chromosomal reshuffling drives evolution of virulence in an asexual pathogen. Genome Res 23:1271–1282. doi: 10.1101/gr.152660.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stapley J, Santure AW, Dennis SR. 2015. Transposable elements as agents of rapid adaptation may explain the genetic paradox of invasive species. Mol Ecol 24:2241–2252. doi: 10.1111/mec.13089. [DOI] [PubMed] [Google Scholar]
- 57.Blackburn TM, Lockwood JL, Cassey P. 2015. The influence of numbers on invasion success. Mol Ecol 24:1942–1953. doi: 10.1111/mec.13075. [DOI] [PubMed] [Google Scholar]
- 58.Forsman A. 2014. Effects of genotypic and phenotypic variation on establishment are important for conservation, invasion, and infection biology. Proc Natl Acad Sci U S A 111:302–307. doi: 10.1073/pnas.1317745111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sakai AK, Allendorf FW, Holt JS, Lodge DM, Molofsky J, With KA, Baughman S, Cabin RJ, Cohen JE, Ellstrand NC, McCauley DE, O'Neil P, Parker IM, Thompson JN, Weller SG. 2001. The population biology of invasive species. Annu Rev Ecol Syst 32:305–332. doi: 10.1146/annurev.ecolsys.32.081501.114037. [DOI] [Google Scholar]
- 60.Bopp SER, Manary MJ, Bright AT, Johnston GL, Dharia NV, Luna FL, McCormack S, Plouffe D, McNamara CW, Walker JR, Fidock DA, Denchi EL, Winzeler EA. 2013. Mitotic evolution of Plasmodium falciparum shows a stable core genome but recombination in antigen families. PLoS Genet 9:e1003293. doi: 10.1371/journal.pgen.1003293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dutheil JY, Mannhaupt G, Schweizer G, Sieber CMK, Münsterkötter M, Güldener U, Schirawski J, Kahmann R. 2016. A tale of genome compartmentalization: the evolution of virulence clusters in smut fungi. Genome Biol Evol 8:681–704. doi: 10.1093/gbe/evw026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rouxel T, Grandaubert J, Hane JK, Hoede C, Van D, Wouw AP, Couloux A, Dominguez V, Anthouard V, Bally P, Bourras S, Cozijnsen AJ, Ciuffetti LM, Degrave A, Dilmaghani A, Duret L, Fudal I, Goodwin SB, Gout L, Glaser N, Linglin J, Kema GHJ, Lapalu N, Lawrence CB, May K, Meyer M, Ollivier B, Poulain J, Schoch CL, Simon A, Spatafora JW, Stachowiak A, Turgeon BG, Tyler BM, Vincent D, Weissenbach J, Amselem J, Quesneville H, Oliver RP, Wincker P, Balesdent MH, Howlett BJ. 2011. Effector diversification within compartments of the Leptosphaeria maculans genome affected by repeat-induced point mutations. Nat Commun 2:202. doi: 10.1038/ncomms1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hatta R, Ito K, Hosaki Y, Tanaka T, Tanaka A, Yamamoto M, Akimitsu K, Tsuge T. 2002. A conditionally dispensable chromosome controls host-specific pathogenicity in the fungal plant pathogen Alternaria alternata. Genetics 161:59–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Giraud T, Gladieux P, Gavrilets S. 2010. Linking the emergence of fungal plant diseases with ecological speciation. Trends Ecol Evol 25:387–395. doi: 10.1016/j.tree.2010.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Harris A. 2014. The epidemiology of Phytophthora ramorum on Larix in the UK. PhD thesis. Imperial College, London, United Kingdom. [Google Scholar]
- 66.Galagan JE, Selker EU. 2004. RIP: the evolutionary cost of genome defense. Trends Genet 20:417–423. doi: 10.1016/j.tig.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 67.Butlin R. 2002. The costs and benefits of sex: new insights from old asexual lineages. Nat Rev Genet 3:311–317. doi: 10.1038/nrg749. [DOI] [PubMed] [Google Scholar]
- 68.Möller EM, Bahnweg G, Sandermann H, Geiger HH. 1992. A simple and efficient protocol for isolation of high molecular weight DNA from filamentous fungi, fruit bodies, and infected plant tissues. Nucleic Acids Res 20:6115–6116. doi: 10.1093/nar/20.22.6115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Schmieder R, Edwards R. 2011. Quality control and preprocessing of metagenomic datasets. Bioinformatics 27:863–864. doi: 10.1093/bioinformatics/btr026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RHY, Aerts A, Arredondo FD, Baxter L, Bensasson D, Beynon JL, Chapman J, Damasceno CMB, Dorrance AE, Dou D, Dickerman AW, Dubchak IL, Garbelotto M, Gijzen M, Gordon SG, Govers F, Grunwald NJ, Huang W, Ivors KL, Jones RW, Kamoun S, Krampis K, Lamour KH, Lee M-K, McDonald WH, Medina M, Meijer HJG, Nordberg EK, Maclean DJ, Ospina GMD, Morris PF, Phuntumart V, Putnam NH, Rash S, Rose JKC, Sakihama Y, Salamov AA, Savidor A, Scheuring CF, Smith BM, Sobral BWS, Terry A, Torto ATA, Win J, Xu Z, Zhang H, et al. 2006. Phytophthora genome sequences uncover evolutionary origins and mechanisms of pathogenesis. Science 313:1261–1266. doi: 10.1126/science.1128796. [DOI] [PubMed] [Google Scholar]
- 71.Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.García-Alcalde F, Okonechnikov K, Carbonell J, Cruz LM, Götz S, Tarazona S, Dopazo J, Meyer TF, Conesa A. 2012. Qualimap: evaluating next-generation sequencing alignment data. Bioinformatics 28:2678–2679. doi: 10.1093/bioinformatics/bts503. [DOI] [PubMed] [Google Scholar]
- 73.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. 2009. The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Danecek P, Auton A, Abecasis G, Albers CA, Banks E, DePristo MA, Handsaker RE, Lunter G, Marth GT, Sherry ST, McVean G, Durbin R, 1000 Genomes Project Analysis Group. 2011. The variant call format and VCFtools. Bioinformatics 27:2156–2158. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Simpson JT, Wong K, Jackman SD, Schein JE, Jones SJ, Birol I. 2009. ABySS: a parallel assembler for short read sequence data structures assemblies and assembly algorithms. Genome Res 19:1117–1123. doi: 10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hunt M, Newbold C, Berriman M, Otto TD. 2014. A comprehensive evaluation of assembly scaffolding tools. Genome Biol 15:R42. doi: 10.1186/gb-2014-15-3-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. 2015. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 78.Jurka J, Kapitonov V, Pavlicek A, Klonowski P, Kohany O, Walichiewicz J. 2005. Repbase update, a database of eukaryotic repetitive elements. Cytogenet Genome Res 110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 79.Keller O, Kollmar M, Stanke M, Waack S. 2011. A novel hybrid gene prediction method employing protein multiple sequence alignments. Bioinformatics 27:757–763. doi: 10.1093/bioinformatics/btr010. [DOI] [PubMed] [Google Scholar]
- 80.Grigoriev IV, Nordberg H, Shabalov I, Aerts A, Cantor M, Goodstein D, Kuo A, Minovitsky S, Nikitin R, Ohm RA, Otillar R, Poliakov A, Ratnere I, Riley R, Smirnova T, Rokhsar D, Dubchak I. 2012. The genome portal of the Department of Energy Joint Genome Institute. Nucleic Acids Res 40:D26–D32. doi: 10.1093/nar/gkr947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. 2005. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- 82.Zdobnov EM, Apweiler R. 2001. InterProScan—an integration platform for the signature-recognition methods in InterPro. Bioinformatics 17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 83.Jombart T. 2008. Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405. doi: 10.1093/bioinformatics/btn129. [DOI] [PubMed] [Google Scholar]
- 84.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. 2007. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Paradis E, Claude J, Strimmer K. 2004. APE: analyses of phylogenetics and evolution in R language. Bioinformatics 20:289–290. doi: 10.1093/bioinformatics/btg412. [DOI] [PubMed] [Google Scholar]
- 86.Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet 81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bauer S, Robinson PN, Gagneur J. 2011. Model-based gene set analysis for Bioconductor. Bioinformatics 27:1882–1883. doi: 10.1093/bioinformatics/btr296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.R Core Team. 2017. R: a language and environment for statistical computing. 2.13. R Foundation for Statistical Computing, Vienna, Austria. [Google Scholar]
- 91.Guy L, Roat Kultima J, Andersson SGE. 2010. genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26:2334–2335. doi: 10.1093/bioinformatics/btq413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Feau N, Taylor G, Dale AL, Dhillon B, Bilodeau GJ, Birol I, Jones SJ, Hamelin RC. 2016. Genome sequences of six Phytophthora species threatening forest ecosystems. Genomics Data 10:85–88. doi: 10.1016/j.gdata.2016.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li L, Stoeckert CJ, Roos DS. 2003. OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res 13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Katoh K, Standley DM. 2013. MAFFT Multiple Sequence Alignment Software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Feau N, Beauseigle S, Bergeron M-J, Bilodeau GJ, Birol I, Cervantes-Arango S, Dhillon B, Dale AL, Herath P, Jones SJM, Lamarche J, Ojeda DI, Sakalidis ML, Taylor G, Tsui CKM, Uzunovic A, Yueh H, Tanguay P, Hamelin RC. 2018. Genome-Enhanced Detection and Identification (GEDI) of plant pathogens. PeerJ 6:e4392. doi: 10.7717/peerj.4392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Snel B, Bork P, Huynen MA. 1999. Genome phylogeny based on gene content. Nat Genet 21:108–110. doi: 10.1038/5052. [DOI] [PubMed] [Google Scholar]
- 97.Kozik A, Chan B, Michemore R. 2003. BLAST parser, distance matrix file and protein sequence clustering. How to obtain a distance matrix file from BLAST search results and use it for sequence clustering analysis. http://cgpdb.ucdavis.edu/BlastParser/Blast_Parser.html.
- 98.Joly DL, Feau N, Tanguay P, Hamelin RC. 2010. Comparative analysis of secreted protein evolution using expressed sequence tags from four poplar leaf rusts (Melampsora spp.). BMC Genomics 11:422. doi: 10.1186/1471-2164-11-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mirkin BG, Fenner TI, Galperin MY, Koonin EV. 2003. Algorithms for computing parsimonious evolutionary scenarios for genome evolution, the last universal common ancestor and dominance of horizontal gene transfer in the evolution of prokaryotes. BMC Evol Biol 3:2. doi: 10.1186/1471-2148-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Rice P, Longden I, Bleasby A. 2000. EMBOSS: the European Molecular Biology Open Software Suite. Trends Genet 16:276–277. doi: 10.1016/S0168-9525(00)02024-2. [DOI] [PubMed] [Google Scholar]
- 101.Wheeler TJ, Eddy SR. 2013. nhmmer: DNA homology search with profile HMMs. Bioinformatics 29:2487–2489. doi: 10.1093/bioinformatics/btt403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J Mol Biol 340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 103.Edgar RC. 2004. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics 5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jiang RHY, Tripathy S, Govers F, Tyler BM. 2008. RXLR effector reservoir in two Phytophthora species is dominated by a single rapidly evolving superfamily with more than 700 members. Proc Natl Acad Sci 105:4874–4879. doi: 10.1073/pnas.0709303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Bailey TL, Williams N, Misleh C, Li WW. 2006. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res 34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Eddy SR. 2008. A probabilistic model of local sequence alignment that simplifies statistical significance estimation. PLoS Comput Biol 4:e1000069. doi: 10.1371/journal.pcbi.1000069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Parra G, Bradnam K, Ning Z, Keane T, Korf I. 2009. Assessing the gene space in draft genomes. Nucleic Acids Res 37:289–297. doi: 10.1093/nar/gkn916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Stoletzki N, Eyre-Walker A. 2011. The positive correlation between dN/dS and dS in mammals is due to runs of adjacent substitutions. Mol Biol Evol 28:1371–1380. doi: 10.1093/molbev/msq320. [DOI] [PubMed] [Google Scholar]
- 109.Yang Z, Nielsen R. 1998. Synonymous and nonsynonymous rate variation in nuclear genes of mammals. J Mol Evol 46:409–418. doi: 10.1007/PL00006320. [DOI] [PubMed] [Google Scholar]
- 110.Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O. 2010. New algorithms and methods to astimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 111.Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci U S A 100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Venables WN, Ripley BD. 2002. Modern applied statistics with S, 4th ed Springer, New York, NY. [Google Scholar]
- 113.van Damme M, Cano LM, Oliva R, Schornack S, Segretin ME, Kamoun S, Raffaele S. 2012. Evolutionary and functional dynamics of Oomycetes effector genes, p 426 In Martin F, Kamoun S (ed), Effectors in plant-microbe interactions. John Wiley & Sons, Inc., Chichester, United Kingdom. [Google Scholar]
- 114.Sperschneider J, Gardiner DM, Dodds PN, Tini F, Covarelli L, Singh KB, Manners JM, Taylor JM. 2015. EffectorP: predicting fungal effector proteins from secretomes using machine learning. New Phytol 210:743–761. doi: 10.1111/nph.13794. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional materials, methods, and results. Download Text S1, DOCX file, 0.09 MB (95KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Read-depth analysis for chromosomal copy number variation in four Phytophthora ramorum isolates with CCNV. (A) Read-depth ratio of alleles at heterozygous sites over 10 scaffolds. For each scaffold, departure (χ2 test) of the read count for each allele averaged over all heterozygous sites from the null hypothesis of diploidy is represented as follows: *, P ≤ 0.05; **, P ≤ 0.01; and ***, P ≤ 0.001. (B) Distribution of read frequency for the minor allele at heterozygous sites for two scaffolds of interest: e.g., scaffold 11 for Pram_1563 has a distribution averaged around 0.5 typical of a 2n ploidy, distribution is centered around 0.33 for scaffold 2 in Pram_PR_09_106 and 0.25 for scaffold 1 in Pram_PR_04_17031, suggesting triploidy and tetraploidy, respectively. Download FIG S1, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Percentage of homozygous and heterozygous SNPs in Phytophthora ramorum isolates with runs of homozygosity (ROH) and representatives of the general population without ROH, as well as the combined length of scaffolds with ROH and percentage of genome affected in the scaffold. Download Table S1, DOCX file, 0.02 MB (19.6KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Lesion length on larch (top) and Douglas fir (middle) 4 and 8 weeks postinoculation of isolates with ROH on scaffold 7 compared to isolates without ROH. The pictures on the bottom represent lesion measured on larch for a no-ROH isolate (left) and a ROH isolate (right). Download FIG S2, TIF file, 1.8 MB (1.8MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Phenotyping of Phytophthora ramorum isolates with ROH on scaffolds 7 and 100 (n = 8) compared to isolates with no ROH (n = 8). (Top) Lesion length on rhododendron leaves; middle, growth (left) and sporulation (right) on minimum medium supplemented with larch sapwood extract; bottom, growth (left) and sporulation (right) on minimum medium supplemented with Douglas fir sapwood extract. The shaded area around the average represents ± standard deviation (SD). None of the differences were significant. Download FIG S3, TIF file, 1.3 MB (1.3MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Chromosomal breakpoints occur in regions enriched in transposable elements (TEs) and with less gene density. (A) Number of breakpoint regions (out of 12) with at least one TE (blue); this number is compared to a distribution of 100 × 12 regions randomly sampled along the Phytophthora ramorum reference genome. Statistically supported difference between the observed value and the random distribution is indicated by a one-sample t test. (B) Distribution of intergenic distances (represented by 5′ and 3′ distances between genes) in scaffolds with ROH (in blue) and random scaffolds without ROH (orange). Statistically supported difference between the two distributions is indicated according to a Kruskal-Wallis nonparametric test. Download FIG S4, TIF file, 1.1 MB (1.1MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Gene family contraction and expansion in Phytophthora spp. (Left) A CAFE analysis was conducted on the 51,458 OrthoMCL gene families and the maximum likelihood phylogenetic tree obtained for the four P. ramorum lineages, P. lateralis, and five other Phytophthora species. The size of pie charts is proportional to the total number of gene families that changed relatively to a putative common ancestor; values in black at the node are bootstrap support for the RAxML tree. (Right) Neighbor-joining tree reconstructed from a distance matrix based on genome similarity in terms of gene content (presence/absence of 51,458 OrthoMCL gene families). Values above nodes are statistical bootstrap support obtained from 100 resamplings with replacement of gene families. The scale bar indicates 1% genomic dissimilarity (i.e., percentage of difference in terms of number of shared genes). Download FIG S5, TIF file, 0.7 MB (752.1KB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Principal-component analysis (PCA) of codon usage on protein-coding genes of the Phytophthora ramorum NA2 isolate 05-16845 showing a strong bias toward usage of codons ending with A or U. Results are shown for genes included in the core (top left), repetitive (top right), and non-core (bottom left) genomes. Color scales on the right indicate protein density, and the percentages in the top right corner are the proportions of genes found on the right part of the PC1 axis. The bottom right graph represents the contribution of each codon variable to the different components: codons ending with G or C are labelled in black, whereas those ending with A or U are in green. Download FIG S6, TIF file, 1.2 MB (1.2MB, tif) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Shared and unique protein models found in Phytophthora ramorum lineages. Download Table S2, DOCX file, 0.01 MB (15KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Model-based gene set analysis for enriched Gene Ontology terms (the five most represented) in sets of proteins unique to each Phytophthora ramorum lineage. Download Table S3, DOCX file, 0.01 MB (15.2KB, docx) .
Copyright © 2019 Dale et al.
This content is distributed under the terms of the Creative Commons Attribution 4.0 International license.
Data Availability Statement
Whole-genome sequencing (WGS) data for all of the isolates sequenced in this study have been deposited in the Sequence Read Archive under SRA accession no. PRJNA427329.