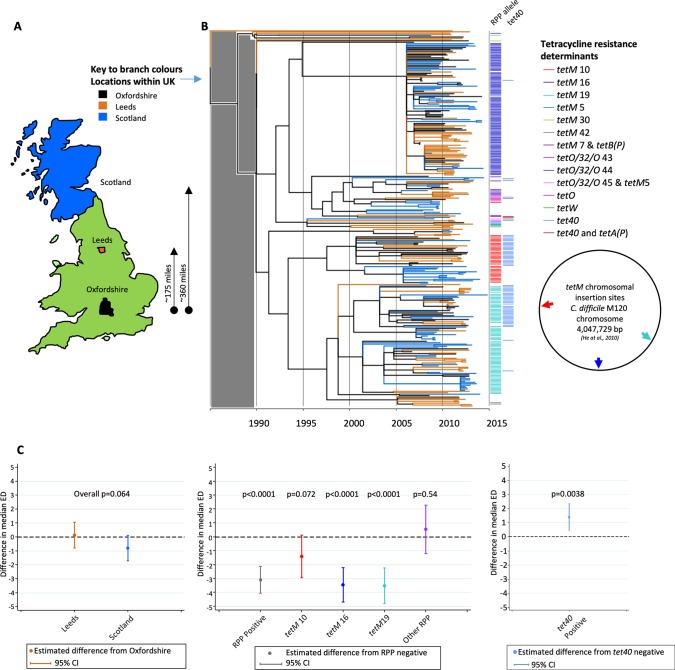
FIG 2.
United Kingdom-representative, time-scaled RT078 phylogeny revealing a lack of geographic structure but strong structuring of tetracycline resistance. (A) Map showing the areas of the United Kingdom from which the RT078 C. difficile genomes were obtained. (B) Time-scaled ClonalFrameML phylogeny constructed using genomes from United Kingdom C. difficile isolates, comprising Oxfordshire (n = 78), Leeds (n = 104), and Scottish (n = 110) isolates. Branch colors, as defined for panel A, denote the location of each genome. Colored bars to the right of the phylogeny indicate the presence of tetracycline resistance determinants; ribosomal protection protein (RPP) allele sequences detected within each genome were assigned numbers to identify distinct nucleotide sequences of tetM, tetO/32/O, tetO, or tetW. To the right of the phylogeny, the chromosomal locations of the three most prevalent tetM alleles (designated tetM 10, 16, and 19) relative to the RT078 M120 genome (NCBI reference sequence NC_017174.1) are shown. All phylogenies included in this study are directly comparable post-1990, i.e., in the time frame of RT078 emergence; the gray shaded block over the region corresponding to the time period prior to that date indicates that region is not scaled identically and should not be used for comparisons. (C) The extent to which RT078 clonal expansions are associated with geographic structure and tetracycline resistance (ribosomal protection proteins and efflux pumps) was determined using two-sided quantile regression. (Left) Differences in median evolutionary distinctiveness scores compared to Oxfordshire samples. A lower evolutionary distinctiveness value indicates a larger proportion of close relatives in the tree. The P values indicate the overall significance of geographic location in the evolutionary distinctiveness score. (Center) Differences in median evolutionary distinctiveness scores for samples with ribosomal protection proteins detected compared to ribosomal protection protein-negative samples, overall and for each of the three putative tetM-associated clonal expansions. A lower evolutionary distinctiveness value indicates a larger proportion of close relatives in the tree. The P values indicate the significance of gene presence in the evolutionary distinctiveness score. (Right) Differences in median evolutionary distinctiveness scores for samples with tetracycline efflux pumps [tet40 and tetA(P)] detected compared to efflux pump-negative samples. A lower evolutionary distinctiveness value indicates a larger proportion of close relatives in the tree. The P value indicates the significance of gene presence in the evolutionary distinctiveness score.