Highlights
-
•
Patterning of biosynthesis, catabolism and transport determines gibberellin distributions.
-
•
The first gibberellin transporters have only recently been discovered.
-
•
Novel methods are revealing spatiotemporal distributions of gibberellins in vivo.
-
•
Gibberellin gradients can be generated independently of biosynthesis patterns.
-
•
In some tissues, gibberellin levels correlate with cell elongation rates.
Abstract
The gibberellin phytohormones regulate growth and development throughout the plant lifecycle. Upstream regulation and downstream responses to gibberellins vary across cells and tissues, developmental stages, environmental conditions, and plant species. The spatiotemporal distribution of gibberellins is the result of an ensemble of biosynthetic, catabolic and transport activities, each of which can be targeted to influence gibberellin levels in space and time. Understanding gibberellin distributions has recently benefited from discovery of transport proteins capable of importing gibberellins as well as novel methods for detecting gibberellins with high spatiotemporal resolution. For example, a genetically-encoded fluorescent biosensor for gibberellins was deployed in Arabidopsis and revealed gibberellin gradients in rapidly elongating tissues. Although cellular accumulations of gibberellins are hypothesized to regulate cell growth in developing embryos, germinating seeds, elongating stems and roots, and developing floral organs, understanding the quantitative relationship between cellular gibberellin levels and cellular growth awaits further investigation. It is also unclear how spatiotemporal gibberellin distributions result from myriad endogenous and environmental factors directing an ensemble of known gibberellin enzymatic and transport steps.
Current Opinion in Plant Biology 2018, 47:9–15
This review comes from a themed issue on Growth and development
Edited by Adrienne H K Roeder and C Jill Harrison
For a complete overview see the Issue and the Editorial
Available online 30th August 2018
https://doi.org/10.1016/j.pbi.2018.08.001
1369-5266/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Plants use phytohormones — a suite of mobile small molecules — as potent regulators that coordinate and adjust development to suit environmental conditions. The gibberellin phytohormones were discovered nearly a century ago when delivery of a gibberellin was revealed to be the mechanism by which a fungal plant pathogen, Gibberella fujikuroi (now reclassified as Fusarium fujikuroi), tailors the growth of infected rice plants [1]. Plant breeders have since tailored the growth of cereal crops for dramatic yield increases through genetic manipulation of gibberellins or DELLA proteins involved in gibberellin signalling [2]. Farmers have even used direct application of gibberellins or gibberellin inhibitors to tailor the growth of crops, for example to increase the size of table grapes [3]. The success of these human and microbial strategies for regulating plant growth depends on the ability of gibberellins to influence plant growth in a variety of developmental contexts, and thus answering the question of where and when endogenous gibberellins influence plant development has been of great interest for several decades. This review will focus on recent developments as the spatiotemporal distribution of gibberellins has of late received significant attention.
Timely accumulation of gibberellins in specific tissues is relevant for organ size and morphology as well as for key developmental transitions. Gibberellin levels are low in dry mature seeds, but after imbibition and exposure to light [4], de novo gibberellin biosynthesis promotes germination, in part through stimulating cell division and expansion in the radicle [5]. Accumulation of gibberellins in the shoot apical meristem of short-day grown Arabidopsis is required for the transition to flowering, but gibberellin catabolism is subsequently required for normal development of the inflorescence [6]. Gibberellins are also mobile across cells and organs, for example in Pisum sativum, where growth of gibberellin mutant pea plant scions was complemented after grafting to wild-type root-stocks [7]. The spatiotemporal distribution of gibberellins influencing these and other aspects of plant development is the result of an ensemble of distinct and independently regulated enzymatic and transport activities.
Gibberellin biosynthesis, catabolism and transport
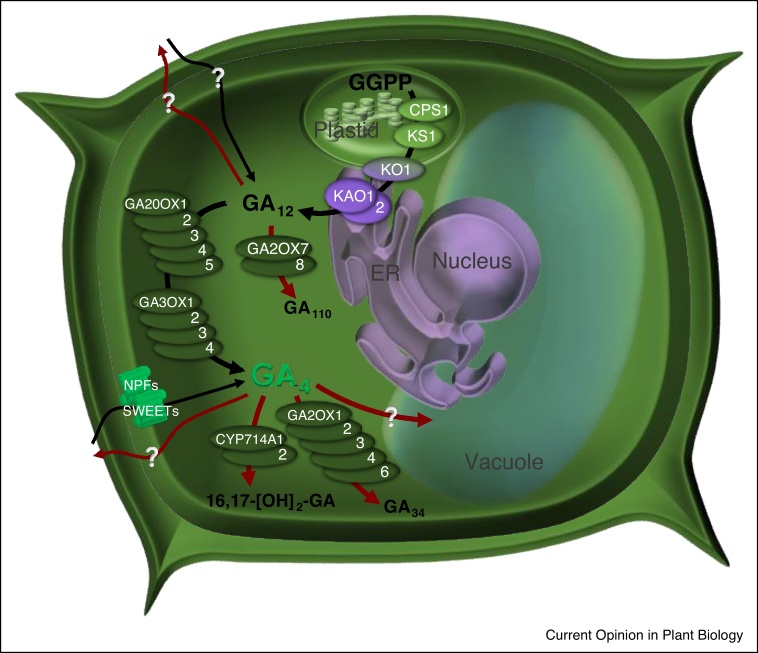
Gibberellin biosynthesis in plants proceeds in stages defined by three cellular compartments: plastids, endoplasmic reticulum (ER) and cytoplasm [8] (Figure 1). The first steps occur in the plastids where ent-copalyl diphosphate synthase and ent-kaur-16-ene synthase promote the conversion of geranylgeranyl diphosphate to ent-kaurene. In the ER, two membrane-associated cytochrome P450 monooxygenases (CYP), ent-kaurene oxidase and ent-kaurenoic acid oxidase, convert ent-kaurene to gibberellin A12 (GA12) though ent-kaurene oxidase also associates with the outer envelope of plastids thereby bridging plastid and ER steps of the pathway [9]. The last steps occur in the cytoplasm where enzymes belonging to GIBBERELLIN 20 OXIDASE (GA20ox), and GIBBERELLIN 3 OXIDASE (GA3ox) families promote the final conversions into bioactive GA1 and GA4. For individual family members of the five GA20ox and four GA3ox enzymes in Arabidopsis, promoter–reporter fusions driving β-glucuronidase (GUS) revealed highly localized expression domains, while genetic studies revealed overlapping functions [10, 11, 12, 13]. For example, AtGA3ox1 is expressed in stamen filaments and AtGA3ox2-4 are expressed in anthers, but mutation of AtGA3ox1 results in silique fertility defects only in combination with mutation of AtGA3ox3 [10,11].
Figure 1.
Gibberellin biochemistry in Arabidopsis. Model of subcellular localization of gibberellin biosynthetic and catabolic pathways as well as putative intra/intercellular movements of gibberellins. Gibberellin accumulation and depletion steps are depicted with black and red arrows, respectively. Biosynthetic steps occur in the three cellular compartments (plastids, endoplasmic reticulum (ER) and cytoplasm); the corresponding enzymes are reported with the following nomenclature: ent-copalyl diphosphate synthase, CPS; ent-kaurene synthase, KS; ent-kaurene oxidase, KO; ent-kaurenoic acid oxidase, KAO; gibberellin 20-oxidase, GA20ox; gibberellin 3-oxidase, GA3ox. Gibberellin deactivation enzymes convert either precursors or bioactive gibberellins into inactive catabolites. Shown here are members of GA2ox and CYP714A families. Transporters belonging to NPF and SWEET families import extracellular GA4. Potential mechanisms for GA4 export and GA12 transport remain poorly understood.
Gibberellin can be inactivated through at least three distinct mechanisms as evidenced by biochemical and genetic studies in Arabidopsis and rice [14, 15, 16, 17]. Precursors and bioactive gibberellins are substrates of GA METHYL TRANSFERASE (GAMT), GIBBERELLIN 2 OXIDASE (GA2ox) and CYP714A catabolic enzymes and higher order mutants in the GA2ox and CYP714A classes display gibberellin overproduction phenotypes. As for Arabidopsis gibberellin biosynthetic genes, the eight AtGA2ox, two AtGAMT and two AtCYP714A genes exhibit localized expression patterns in various organs and developmental stages, potentially serving to modulate gibberellin spatiotemporal distributions initiated by patterns of gibberellin biosynthesis [14, 15, 16].
Gibberellin transport can also affect spatiotemporal distributions, because gibberellins are known to be mobile over short and long distances in plants. Gibberellin movements have been associated with inducing flowering in Arabidopsis [18] and sex determination in Lygodium japonicum ferns [19]. More recently, movements of gibberellins have been also discovered in cucumber, where GA9 moves from ovaries to sepal and petal tissues where it is converted to the bioactive GA4 [20]. In Arabidopsis, GA12 is the GA4 precursor associated with long-distance transport; it has been suggested that GA12 moves from root to shoot via the xylem and from photosynthetic source to sink tissues via the phloem [21,22•].
Intercellular and intercompartmental movement of gibberellins would depend on transmembrane transport activities that are only beginning to be characterized. As gibberellins are weak acids, an acid trap mechanism could contribute to their import from the low pH apoplasm into higher pH cytoplasm without the need for gibberellin transporters in the plasma membrane [23]. Nevertheless, in recent years several transporters belonging to the NITRATE TRANSPORTER 1/PEPTIDE TRANSPORTER FAMILY (NPF) proteins have been identified as gibberellin importers [24, 25, 26]. Using a yeast two-hybrid system expressing gibberellin receptors in combination with NPFs, it was possible to identify NPFs able to transport one or more gibberellins [24]. The Arabidopsis genome contains 53 genes encoding NPF proteins and most of those characterized were able to transport multiple hormone substrates in yeast in addition to any previously characterized nitrate or peptide transport activities [24]. In another example of multi-substrate transporters, the sugar transporters SWEET13 and SWEET14 were found to import gibberellin into yeast and oocyte cells and are involved in gibberellin mediated plant growth including seed, seedling and anther development [27•]. Importantly, no specific gibberellin exporters have been discovered [28], though the SWEETs were initially characterized as mediating sugar import and export [29].
Movements of gibberellins were also followed in high spatiotemporal resolution by using fluorescently labelled GA molecules, that is, GA3 and GA4 linked to fluorescein (GA-Fl). Analyses of GA-FI showed highest accumulation of GA-Fl specifically in the vacuole of elongating endodermal cells of Arabidopsis roots, suggesting that GA-Fl movements are tightly regulated by patterning of gibberellin transporters [30]. Subsequently, mutations in the NPF3.1 gene, a member of the NPF family, were found to abolish GA3-Fl accumulation in the endodermal cells of the elongation zone [31•] and to result in developmental phenotypes under nitrate limitation [32], suggesting the patterning of NPF3.1 can affect tissue and condition specific GA distributions.
Measuring spatiotemporal distribution of gibberellins
Patterns of gibberellin biosynthesis, catabolism and transport integrate to determine gibberellin spatiotemporal distributions that then influence patterns of cell division and expansion underlying plant development (Figure 1). Thus, measuring gibberellin distributions with high spatiotemporal resolution is an important step towards understanding the role of gibberellins in plant development. Recent years have seen a variety of high-resolution analytic techniques ranging from indirect approaches such as tracking expression of gibberellin biosynthetic enzymes [33••] or signaling events downstream of gibberellin accumulation [34] to direct approaches such as high-resolution mass spectrometry [35,36] or genetically encoded fluorescent biosensors [37••].
In Arabidopsis embryos, a cell-level analysis of promoter–GUS fusions of gibberellin biosynthetic enzymes revealed highly localized expression in the tip of the radicle during early seed germination that corresponded to the expression domains of transcriptional targets of gibberellin signaling [33••]. An approach based on synthetic hormone activated Cas9-based repressors (HACRs) revealed an endosperm specific gibberellin distribution that corresponded with AtGA3ox4 expression in early seed development [34]. In the gibberellin HACRs, a gibberellin-targeted degron (i.e. the DELLA proteins REPRESSOR OF GA 1 (RGA1) or GIBBERELLIC ACID INSENSITIVE (GAI)) were fused to a transcriptional repressor protein such that gibberellin signaling would increase the expression of a reporter gene such as GFP [34]. Because the HACR approach is modular, it would be straightforward to test additional GA targeted DELLA proteins or indeed quantify other pathways dependent on regulated protein degradation. It will be interesting to determine how patterns of gibberellins reported by HACRs compare with the patterns of DELLA–GFP fusions that also indirectly report on gibberellins via a gibberellin induced degradation mechanism [38]. For example, pRGA:GFP-RGA reporters have long been used to track dynamics of gibberellin signaling and recently revealed a decrease in gibberellin signaling (GFP-RGA increase) in the Arabidopsis root division and elongation zones and an increase in gibberellin signaling (GFP-RGA decrease) in the epidermis of the differentiation zone under iron limitation [39]. In a more direct approach, derivatization-based ultra-high-performance liquid chromatography (UHPLC) coupled with electrospray ionization-tandem mass spectrometry (ESI-MS/MS) enables the detection of ultra-trace gibberellins in small organs, such as Arabidopsis flowers [35,36]. Interestingly, this method detected higher levels of bioactive GA4 in pistils as compared with other floral organs over three stages of flower development [36].
Recently, GIBBERELLIN PERCEPTION SENSOR 1 (GPS1), the first Förster Resonance Energy Transfer (FRET) biosensor for gibberellins, was developed for high-resolution measurements of gibberellin distributions in vivo [37••]. FRET biosensors are characterized by a FRET pair (a donor fluorescent protein and an acceptor fluorescent protein) linked to a sensory domain containing fusion proteins that directly interact with a ligand of interest. Upon ligand binding, the sensory domain undergoes conformational changes that are transmitted to the FRET pair resulting in a ligand dependent change in energy transfer between the donor and the acceptor. Once expressed in plants, such biosensors are measured by exciting the donor and measuring the fluorescence emission ratio of acceptor over donor, and thus FRET biosensor analysis is ratiometric and largely independent of the concentration of the biosensor itself [40,41]. The GPS1 sensory domain consists of the Arabidopsis gibberellin receptor AtGID1C linked via a 12-amino acid flexible linker to a 74-amino acid truncation of AtGAI and the GPS1 FRET pair consists of enhanced dimerization variants of Aphrodite (a codon diversified Venus) as FRET acceptor and Cerulean as FRET donor [37••]. GPS1 was found to report on increasing concentrations of gibberellins via an increase in fluorescence emission ratio and exhibit a high affinity and slow reversibility for GA4 (Kd approximately 20 nM).
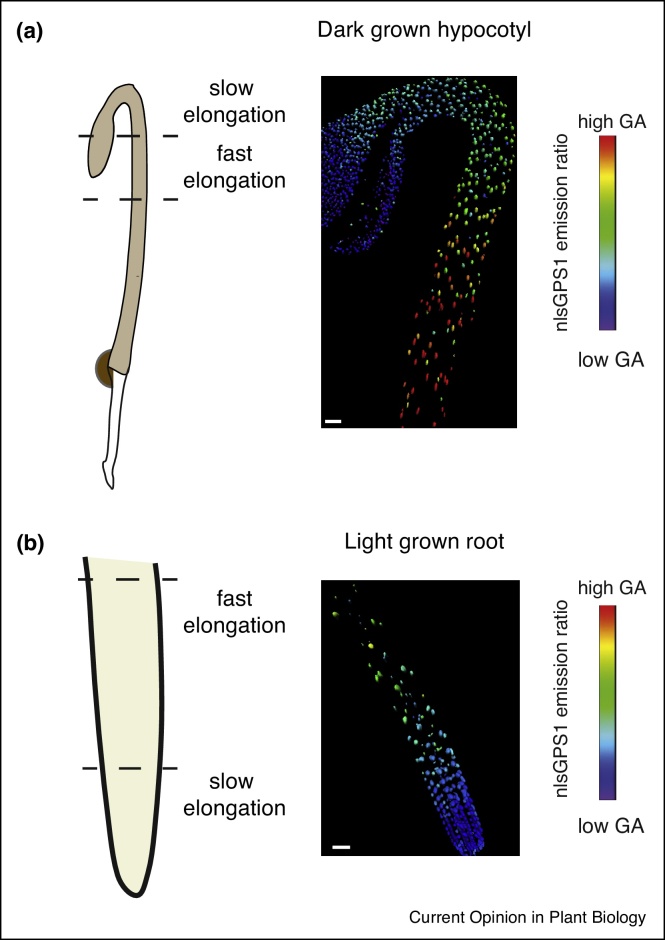
Using GPS1 targeted to nuclei, it was possible to monitor gibberellin levels in different Arabidopsis tissues including roots, hypocotyls and floral organs with cellular resolution [37••]. In roots, a gradient of endogenous gibberellin is formed, with high levels in the elongation zone grading to low levels in the division zone (Figure 2). A parallel result was found with exogenous gibberellin in that treatment with gibberellin resulted in faster accumulation in the elongation zone and slower accumulation in the division zone. The patterning of exogenous gibberellins in roots suggests that a gibberellin gradient can be generated by patterns of gibberellin catabolism and/or transport independently of patterns of gibberellin biosynthesis [37••], a finding that is supported by the observation that root growth defects of gibberellin biosynthetic mutants can be rescued with exogenous gibberellin [42]. Dark grown Arabidopsis hypocotyls also exhibited a gibberellin gradient with high levels in the basal hypocotyl grading to low levels in the apical hypocotyl and cotyledons (Figure 2). As GPS1 is genetically encoded and binds gibberellin directly, it will be possible to investigate gibberellin patterning in further Arabidopsis tissues amenable to fluorescence imaging as well as in further conditions and transformable plant species.
Figure 2.
Gibberellin gradients compared with cellular growth rate in dark grown hypocotyl and root tip. Analysis of nuclear localized GPS1 (nlsGPS1) expressed in Arabidopsis at three days post sowing as described in Ref. [37••]. Comparison of regions of cellular elongation with nlsGPS1 emission ratios in (a) a dark grown hypocotyl and (b) a light grown root. In both tissues, gibberellin levels correlate with cell elongation rate. Scale bars (a) 70 μm (b) 30 μm.
Spatiotemporal regulation of gibberellin and cellular growth
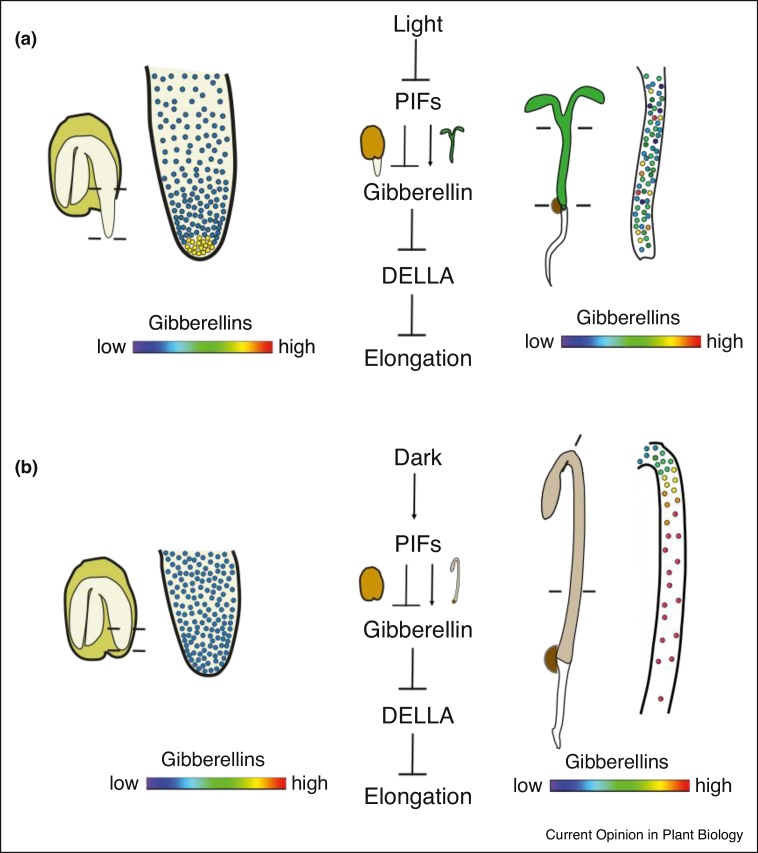
Many environmental and endogenous signals integrate to influence gibberellin enzymes and transport activities directing spatiotemporal distribution of gibberellins, which can then serve as a link between environmental conditions and plant development. For example, the light environment influences seed germination and hypocotyl elongation through modulation of gibberellins [43, 44, 45]. Interestingly, many of the same signaling modules are used to achieve light mediated stimulation of cell growth in the radicle versus light mediated inhibition of cell growth in hypocotyls (Figure 3). For example, PHYTOCHROME INTERACTING FACTOR (PIF) accumulation in darkness negatively regulates gibberellin accumulation in seeds [43,44] and positively regulates gibberellin accumulation in hypocotyls [37••]. The core signaling pathways connecting gibberellin accumulation to growth and germination are now well-characterized: in the presence of gibberellin in the cytoplasm and nucleus, gibberellin receptors (GIDs) bind gibberellins and this complex promotes the degradation of DELLA proteins that inhibit the expression of gibberellin mediated genes [47]. However, the quantitative relationship between gibberellin distributions and cellular growth remains unclear. In both roots and dark-grown hypocotyls, the spatiotemporal distribution of gibberellins detected with nlsGPS1 closely tracked the spatiotemporal distribution of absolute cell growth [37••], suggesting that local accumulation of gibberellin is important for directing cell growth. On the other hand, apparent gibberellin accumulation in the early radicle tip during seed germination was several cells removed from the domain of accelerated cell growth [33••]. Furthermore, in light grown hypocotyls expressing nlsGPS1 there was no overall correlation between nlsGPS1 emission ratios and cell length under the conditions tested ([37••], Figure 3).
Figure 3.
Model of environmental regulation of gibberellin distribution and cell growth in seeds and hypocotyls. (a) Contrasting regulation of gibberellins by light in seeds (left) versus hypocotyls (right). In seeds, the absence of PIF1 protein allows for the accumulation of gibberellins and the inhibition of DELLAs, thereby inducing radicle cell elongation and germination. Accumulation of gibberellins might not correlate with cell elongation [33••]. In hypocotyls, the absence of PIF proteins instead reduces gibberellin levels and promotes DELLA activity, thereby reducing hypocotyl cell elongation. Accumulation of gibberellins again might not correlate with cell elongation [37••]. (b) Contrasting regulation of gibberellins by darkness in seeds (left) versus hypocotyls (right). In seeds, the presence of PIF1 protein reduces gibberellin levels and promotes DELLA activity, thereby inhibiting germination [43,44]. In hypocotyls, the presence of PIF proteins allows for the accumulation of gibberellins and the inhibition of DELLAs, thereby inducing hypocotyl cell elongation. Here, accumulation of gibberellins correlates with cell elongation [37••].
Conclusions
Fundamentally, the functional consequences of gibberellin spatiotemporal distributions can be considered as the combined effect of gibberellin accumulation in certain cells and gibberellin depletion in others. The need for gibberellins to accumulate is evident from numerous studies of gibberellin deficiency dating back to Gregor Mendel’s dwarf peas; the Le trait was caused by mutation of a GA3ox gene [11,13,46,48,49]. More recently, the relative importance of gibberellin signaling, that is, DELLA degradation, among specific root cell files was demonstrated using cell-type specific expression of non-degradable GAI [50,51••]. The need for gibberellins to be depleted from specific cells can be demonstrated by examining the phenotypes of wild-type Arabidopsis plants treated with exogenous gibberellins (e.g. [52]), gibberellin catabolism mutants [14,16], and higher-order mutants lacking all five DELLAs [53•]. Surprisingly for a growth regulatory hormone, each of these interventions results in broadly normal plant growth and development, perhaps indicating that gibberellin responses in Arabidopsis might be largely saturated under optimal growth conditions. Nevertheless, ectopically increased gibberellins and loss of DELLAs consistently leads to altered organ size and fertility defects that are presumably a consequence of disruptions in normal spatiotemporal patterning of gibberellins [14,16,52,53•]. In the future, it will be fascinating to resolve how patterns of gibberellin biosynthetic, catabolic, and transport activities contribute to spatiotemporal distributions of gibberellins. It would be interesting then to understand how altering gibberellin enzymes or transporters will redistribute gibberellins and reprogram plant development.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as
• of special interest
•• of outstanding interest
Acknowledgements
We gratefully acknowledge Bo Larsen for assistance with graphics and Jim Rowe for critical review of the manuscript. This work has received funding from the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement n° 759282).
References
- 1.Hedden P., Sponsel V. A century of gibberellin research. J Plant Growth Regul. 2015;34:740–760. doi: 10.1007/s00344-015-9546-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hedden P. The genes of the Green Revolution. Trends Genet. 2003;19:5–9. doi: 10.1016/s0168-9525(02)00009-4. [DOI] [PubMed] [Google Scholar]
- 3.Weaver R.J. Effect of gibberellic acid on fruit set and berry enlargement in seedless grapes of Vitis vinifera. Nature. 1958;181:851–852. [Google Scholar]
- 4.Graeber K., Nakabayashi K., Miatton E., Leubner-Metzger G., Soppe W.J. Molecular mechanisms of seed dormancy. Plant Cell Environ. 2012;35:1769–1786. doi: 10.1111/j.1365-3040.2012.02542.x. [DOI] [PubMed] [Google Scholar]
- 5.Holdsworth M.J., Bentsink L., Soppe W.J. Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 2008;179:33–54. doi: 10.1111/j.1469-8137.2008.02437.x. [DOI] [PubMed] [Google Scholar]
- 6.Yamaguchi N., Winter C.M., Wu M.F., Kanno Y., Yamaguchi A., Seo M., Wagner D. Gibberellin acts positively then negatively to control onset of flower formation in Arabidopsis. Science. 2014;344:638–641. doi: 10.1126/science.1250498. [DOI] [PubMed] [Google Scholar]
- 7.Proebsting W.M., Hedden P., Lewis M.J., Croker S.J., Proebsting L.N. Gibberellin concentration and transport in genetic lines of pea: effects of grafting. Plant Physiol. 1992;100:1354–1360. doi: 10.1104/pp.100.3.1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi S. Gibberellin metabolism and its regulation. Annu Rev Plant Biol. 2008;59:225–251. doi: 10.1146/annurev.arplant.59.032607.092804. [DOI] [PubMed] [Google Scholar]
- 9.Helliwell C.A., Sullivan J.A., Mould R.M., Gray J.C., Peacock W.J., Dennis E.S. A plastid envelope location of Arabidopsis ent-kaurene oxidase links the plastid and endoplasmic reticulum steps of the gibberellin biosynthesis pathway. Plant J. 2001;28:201–208. doi: 10.1046/j.1365-313x.2001.01150.x. [DOI] [PubMed] [Google Scholar]
- 10.Hu J., Mitchum M.G., Barnaby N., Ayele B.T., Ogawa M., Nam E., Lai W.C., Hanada A., Alonso J.M., Ecker J.R. Potential sites of bioactive gibberellin production during reproductive growth in Arabidopsis. Plant Cell. 2008;20:320–336. doi: 10.1105/tpc.107.057752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mitchum M.G., Yamaguchi S., Hanada A., Kuwahara A., Yoshioka Y., Kato T., Tabata S., Kamiya Y., Sun T.P. Distinct and overlapping roles of two gibberellin 3-oxidases in Arabidopsis development. Plant J. 2006;45:804–818. doi: 10.1111/j.1365-313X.2005.02642.x. [DOI] [PubMed] [Google Scholar]
- 12.Plackett A.R., Powers S.J., Fernandez-Garcia N., Urbanova T., Takebayashi Y., Seo M., Jikumaru Y., Benlloch R., Nilsson O., Ruiz-Rivero O. Analysis of the developmental roles of the Arabidopsis gibberellin 20-oxidases demonstrates that GA20ox1, -2, and -3 are the dominant paralogs. Plant Cell. 2012;24:941–960. doi: 10.1105/tpc.111.095109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieu I., Ruiz-Rivero O., Fernandez-Garcia N., Griffiths J., Powers S.J., Gong F., Linhartova T., Eriksson S., Nilsson O., Thomas S.G. The gibberellin biosynthetic genes AtGA20ox1 and AtGA20ox2 act, partially redundantly, to promote growth and development throughout the Arabidopsis life cycle. Plant J. 2008;53:488–504. doi: 10.1111/j.1365-313X.2007.03356.x. [DOI] [PubMed] [Google Scholar]
- 14.Rieu I., Eriksson S., Powers S.J., Gong F., Griffiths J., Woolley L., Benlloch R., Nilsson O., Thomas S.G., Hedden P. Genetic analysis reveals that C19-GA 2-oxidation is a major gibberellin inactivation pathway in Arabidopsis. Plant Cell. 2008;20:2420–2436. doi: 10.1105/tpc.108.058818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Varbanova M., Yamaguchi S., Yang Y., McKelvey K., Hanada A., Borochov R., Yu F., Jikumaru Y., Ross J., Cortes D. Methylation of gibberellins by Arabidopsis GAMT1 and GAMT2. Plant Cell. 2007;19:32–45. doi: 10.1105/tpc.106.044602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang Y., Zhang B., Yan D., Dong W., Yang W., Li Q., Zeng L., Wang J., Wang L., Hicks L.M. Two Arabidopsis cytochrome P450 monooxygenases, CYP714A1 and CYP714A2, function redundantly in plant development through gibberellin deactivation. Plant J. 2011;67:342–353. doi: 10.1111/j.1365-313X.2011.04596.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhu Y., Nomura T., Xu Y., Zhang Y., Peng Y., Mao B., Hanada A., Zhou H., Wang R., Li P. ELONGATED UPPERMOST INTERNODE encodes a cytochrome P450 monooxygenase that epoxidizes gibberellins in a novel deactivation reaction in rice. Plant Cell. 2006;18:442–456. doi: 10.1105/tpc.105.038455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Eriksson S., Bohlenius H., Moritz T., Nilsson O. GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell. 2006;18:2172–2181. doi: 10.1105/tpc.106.042317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka J., Yano K., Aya K., Hirano K., Takehara S., Koketsu E., Ordonio R.L., Park S.H., Nakajima M., Ueguchi-Tanaka M. Antheridiogen determines sex in ferns via a spatiotemporally split gibberellin synthesis pathway. Science. 2014;346:469–473. doi: 10.1126/science.1259923. [DOI] [PubMed] [Google Scholar]
- 20.Pimenta Lange M.J., Lange T. Ovary-derived precursor gibberellin A9 is essential for female flower development in cucumber. Development. 2016;143:4425–4429. doi: 10.1242/dev.135947. [DOI] [PubMed] [Google Scholar]
- 21.Hoad G.V., Bowen M.R. Evidence for gibberellin-like substances in phloem exudate of higher plants. Planta. 1968;82:22–32. doi: 10.1007/BF00384695. [DOI] [PubMed] [Google Scholar]
- 22•.Regnault T., Daviere J.M., Wild M., Sakvarelidze-Achard L., Heintz D., Carrera Bergua E., Lopez Diaz I., Gong F., Hedden P., Achard P. The gibberellin precursor GA12 acts as a long-distance growth signal in Arabidopsis. Nat Plants. 2015;1:15073. doi: 10.1038/nplants.2015.73. [DOI] [PubMed] [Google Scholar]; Characterisation of GA12 as a key long-distance transport form of gibberellin in Arabidopsis.
- 23.Kramer E.M. How far can a molecule of weak acid travel in the apoplast or xylem? Plant Physiol. 2006;141:1233–1236. doi: 10.1104/pp.106.083790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chiba Y., Shimizu T., Miyakawa S., Kanno Y., Koshiba T., Kamiya Y., Seo M. Identification of Arabidopsis thaliana NRT1/PTR FAMILY (NPF) proteins capable of transporting plant hormones. J Plant Res. 2015;128:679–686. doi: 10.1007/s10265-015-0710-2. [DOI] [PubMed] [Google Scholar]
- 25.Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., Koshiba T., Kamiya Y., Seo M. Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc Natl Acad Sci U S A. 2012;109:9653–9658. doi: 10.1073/pnas.1203567109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Saito H., Oikawa T., Hamamoto S., Ishimaru Y., Kanamori-Sato M., Sasaki-Sekimoto Y., Utsumi T., Chen J., Kanno Y., Masuda S. The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat Commun. 2015;6:6095. doi: 10.1038/ncomms7095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27•.Kanno Y., Oikawa T., Chiba Y., Ishimaru Y., Shimizu T., Sano N., Koshiba T., Kamiya Y., Ueda M., Seo M. AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 2016;7:13245. doi: 10.1038/ncomms13245. [DOI] [PMC free article] [PubMed] [Google Scholar]; Characterisation of gibberellin transporters whose mutation results in gibberellin mobility as well as growth and development phenotypes.
- 28.Binenbaum J., Weinstain R., Shani E. Gibberellin localization and transport in plants. Trends Plant Sci. 2018;23:410–421. doi: 10.1016/j.tplants.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 29.Chen L.Q., Hou B.H., Lalonde S., Takanaga H., Hartung M.L., Qu X.Q., Guo W.J., Kim J.G., Underwood W., Chaudhuri B. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shani E., Weinstain R., Zhang Y., Castillejo C., Kaiserli E., Chory J., Tsien R.Y., Estelle M. Gibberellins accumulate in the elongating endodermal cells of Arabidopsis root. Proc Natl Acad Sci U S A. 2013;110:4834–4839. doi: 10.1073/pnas.1300436110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31•.Tal I., Zhang Y., Jorgensen M.E., Pisanty O., Barbosa I.C., Zourelidou M., Regnault T., Crocoll C., Erik Olsen C., Weinstain R. The Arabidopsis NPF3 protein is a GA transporter. Nat Commun. 2016;7:11486. doi: 10.1038/ncomms11486. [DOI] [PMC free article] [PubMed] [Google Scholar]; Characterisation of a gibberellin transporter whose mutation abolished GA-Fl accumulation in roots.
- 32.David L.C., Berquin P., Kanno Y., Seo M., Daniel-Vedele F., Ferrario-Mery S. N availability modulates the role of NPF3.1, a gibberellin transporter, in GA-mediated phenotypes in Arabidopsis. Planta. 2016;244:1315–1328. doi: 10.1007/s00425-016-2588-1. [DOI] [PubMed] [Google Scholar]
- 33••.Bassel G.W., Stamm P., Mosca G., Barbier de Reuille P., Gibbs D.J., Winter R., Janka A., Holdsworth M.J., Smith R.S. Mechanical constraints imposed by 3D cellular geometry and arrangement modulate growth patterns in the Arabidopsis embryo. Proc Natl Acad Sci U S A. 2014;111:8685–8690. doi: 10.1073/pnas.1404616111. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes and models an apparent physical separation of gibberellin signaling and cell growth responses in the germinating radicle.
- 34.Khakhar A., Leydon A.R., Lemmex A.C., Klavins E., Nemhauser J.L. Synthetic hormone-responsive transcription factors can monitor and re-program plant development. Elife. 2018;7 doi: 10.7554/eLife.34702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li D., Guo Z., Chen Y. Direct derivatization and quantitation of ultra-trace gibberellins in sub-milligram fresh plant organs. Mol Plant. 2016;9:175–177. doi: 10.1016/j.molp.2015.07.001. [DOI] [PubMed] [Google Scholar]
- 36.Li D., Guo Z., Liu C., Li J., Xu W., Chen Y. Quantification of near-attomole gibberellins in floral organs dissected from a single Arabidopsis thaliana flower. Plant J. 2017;91:547–557. doi: 10.1111/tpj.13580. [DOI] [PubMed] [Google Scholar]
- 37••.Rizza A., Walia A., Lanquar V., Frommer W.B., Jones A.M. In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants. 2017;3:803–813. doi: 10.1038/s41477-017-0021-9. [DOI] [PubMed] [Google Scholar]; Describes the invention of a high-resolution fluorescent biosensor for gibberellins and its use to discover gibberellin gradientsin vivo.
- 38.Silverstone A.L., Jung H.S., Dill A., Kawaide H., Kamiya Y., Sun T.P. Repressing a repressor: gibberellin-induced rapid reduction of the RGA protein in Arabidopsis. Plant Cell. 2001;13:1555–1566. doi: 10.1105/TPC.010047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wild M., Daviere J.M., Regnault T., Sakvarelidze-Achard L., Carrera E., Lopez Diaz I., Cayrel A., Dubeaux G., Vert G., Achard P. Tissue-specific regulation of gibberellin signaling fine-tunes Arabidopsis iron-deficiency responses. Dev Cell. 2016;37:190–200. doi: 10.1016/j.devcel.2016.03.022. [DOI] [PubMed] [Google Scholar]
- 40.Jones A.M., Danielson J.A., Manojkumar S.N., Lanquar V., Grossmann G., Frommer W.B. Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife. 2014;3:e01741. doi: 10.7554/eLife.01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Walia A., Waadt R., Jones A.M. Genetically encoded biosensors in plants: pathways to discovery. Annu Rev Plant Biol. 2018;69:497–524. doi: 10.1146/annurev-arplant-042817-040104. [DOI] [PubMed] [Google Scholar]
- 42.Band L.R., Ubeda-Tomas S., Dyson R.J., Middleton A.M., Hodgman T.C., Owen M.R., Jensen O.E., Bennett M.J., King J.R. Growth-induced hormone dilution can explain the dynamics of plant root cell elongation. Proc Natl Acad Sci U S A. 2012;109:7577–7582. doi: 10.1073/pnas.1113632109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gabriele S., Rizza A., Martone J., Circelli P., Costantino P., Vittorioso P. The Dof protein DAG1 mediates PIL5 activity on seed germination by negatively regulating GA biosynthetic gene AtGA3ox1. Plant J. 2010;61:312–323. doi: 10.1111/j.1365-313X.2009.04055.x. [DOI] [PubMed] [Google Scholar]
- 44.Kim D.H., Yamaguchi S., Lim S., Oh E., Park J., Hanada A., Kamiya Y., Choi G. SOMNUS, a CCCH-type zinc finger protein in Arabidopsis, negatively regulates light-dependent seed germination downstream of PIL5. Plant Cell. 2008;20:1260–1277. doi: 10.1105/tpc.108.058859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reid J.B., Botwright N.A., Smith J.J., O’Neill D.P., Kerckhoffs L.H. Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol. 2002;128:734–741. doi: 10.1104/pp.010607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yamaguchi S., Kamiya Y., Sun T. Distinct cell-specific expression patterns of early and late gibberellin biosynthetic genes during Arabidopsis seed germination. Plant J. 2001;28:443–453. doi: 10.1046/j.1365-313x.2001.01168.x. [DOI] [PubMed] [Google Scholar]
- 47.Sun T.P. Gibberellin-GID1-DELLA: a pivotal regulatory module for plant growth and development. Plant Physiol. 2010;154:567–570. doi: 10.1104/pp.110.161554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lester D.R., Ross J.J., Davies P.J., Reid J.B. Mendel’s stem length gene (Le) encodes a gibberellin 3 beta-hydroxylase. Plant Cell. 1997;9:1435–1443. doi: 10.1105/tpc.9.8.1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Martin D.N., Proebsting W.M., Hedden P. Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci U S A. 1997;94:8907–8911. doi: 10.1073/pnas.94.16.8907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ubeda-Tomas S., Federici F., Casimiro I., Beemster G.T., Bhalerao R., Swarup R., Doerner P., Haseloff J., Bennett M.J. Gibberellin signaling in the endodermis controls Arabidopsis root meristem size. Curr Biol: CB. 2009;19:1194–1199. doi: 10.1016/j.cub.2009.06.023. [DOI] [PubMed] [Google Scholar]
- 51••.Ubeda-Tomas S., Swarup R., Coates J., Swarup K., Laplaze L., Beemster G.T., Hedden P., Bhalerao R., Bennett M.J. Root growth in Arabidopsis requires gibberellin/DELLA signalling in the endodermis. Nat Cell Biol. 2008;10:625–628. doi: 10.1038/ncb1726. [DOI] [PubMed] [Google Scholar]; Describes the effects of disturbing gibberellin signaling in specific root cell types.
- 52.Plackett A.R.G., Powers S.J., Phillips A.L., Wilson Z.A., Hedden P., Thomas S.G. The early inflorescence of Arabidopsis thaliana demonstrates positional effects in floral organ growth and meristem patterning. Plant Reprod. 2018;31:171–191. doi: 10.1007/s00497-017-0320-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53•.Fuentes S., Ljung K., Sorefan K., Alvey E., Harberd N.P., Ostergaard L. Fruit growth in Arabidopsis occurs via DELLA-dependent and DELLA-independent gibberellin responses. Plant Cell. 2012;24:3982–3996. doi: 10.1105/tpc.112.103192. [DOI] [PMC free article] [PubMed] [Google Scholar]; Describes phenotypes of a della quintuple mutant.