Abstract
During synthesis, mRNA undergoes a number of modifications such as capping, splicing and polyadenylation. These processes are coupled with the orderly deposition of the TREX complex on the mRNA and subsequent recruitment of the NXF1-P15 heterodimer which stimulates the nuclear export of mature mRNAs. mRNAs also undergo a number of internal modifications, the most common of which is the N6‑methyladenosine (m6A) modification. In this review we discuss the recent evidence of coupling between the m6A modification, RNA processing and export.
Highlights
-
•
mRNA processing and export are coupled with the N6‑methyladenosine (m6A) modification of mRNA.
-
•
TREX and SR proteins cooperate with m6A writers and readers in mRNA export.
-
•
Ultimately the m6A modification leads to NXF1 recruitment and mRNA export
1. Introduction
DNA, RNA and proteins can all undergo post synthesis modifications allowing for tight regulation and diverse function. In regards to RNA, over 100 distinct chemical modifications have been identified [1]. The most abundant internal chemical modification present within eukaryotic mRNA is N6‑methyladenosine (m6A) [2]. Although the m6A modification of mRNA has been known for some time, recent advances in sequencing technologies have led to an explosion of new research. Not only discovering the precise distribution of m6A but also the protein players orchestrating its addition and subsequently carrying out a variety of m6A dependent functions [3,4]. The major effects of m6A addition on transcripts appear to be a reduction in their stability and influencing the splicing kinetics of nascent pre-mRNAs [[5], [6], [7]]. However, the addition of m6A also influences a variety of steps within the lifecycle of an mRNA. Nuclear processing, export and translation all appear altered with the addition of m6A [8]. Additionally, the m6A modification and its associated proteins have been found to regulate a variety of cell specific process. The removal of m6A can result in perturbation of a cells circadian rhythm, impair embryonic stem cell differentiation and disrupt Drosophila sex determination [[9], [10], [11], [12]]. In addition, m6A plays a role in the control of neurogenesis, DNA damage response pathways and X chromosome transcriptional inactivation [[13], [14], [15]].
The m6A modification appears to play a vital role in the expression of a certain subset of genes, influencing many aspects of their expression [8]. One of the key steps in gene expression is the export of a mature correctly packaged mRNA from the nucleus through the nuclear pore to the cytoplasm. The bulk of mRNA is exported from the nucleus in a Ran-independent manner via the Transcription Export (TREX) Complex and the nuclear heterodimeric export receptor NXF1-P15 [16]. RNA Polymerase II synthesises pre-mRNA that is then subject to multiple co- and post-transcriptional processing events, such as 5′ capping, splicing and 3′ polyadenylation [17]. Throughout the mRNA maturation process, members of the TREX complex are deposited on the mRNA and culminate in a messenger ribonucleoprotein complex (mRNP) containing the correct protein composition to bind and hand over the mRNA to the export receptor NXF1 [[18], [19], [20]]. m6A is found in pre-mRNA and some of the factors involved in this RNA modification are found in the nuclear speckles, sites where mRNA export factors reside in the nucleus [21]. Furthermore multiple members of the methylation complex have been found to interact with subunits of the TREX complex [22]. Therefore, it seems likely that this RNA modification might also influence the export of mRNAs harbouring this modification. This leads us to the purpose of this review, which is twofold. Firstly, we will discuss the recent m6A literature outlining the currently proposed pathway and also point out the inconsistencies present. Following this we will identify the evidence presented throughout the literature of an overlap between the m6A machinery and the nuclear mRNA export pathway.
2. m6A pathway
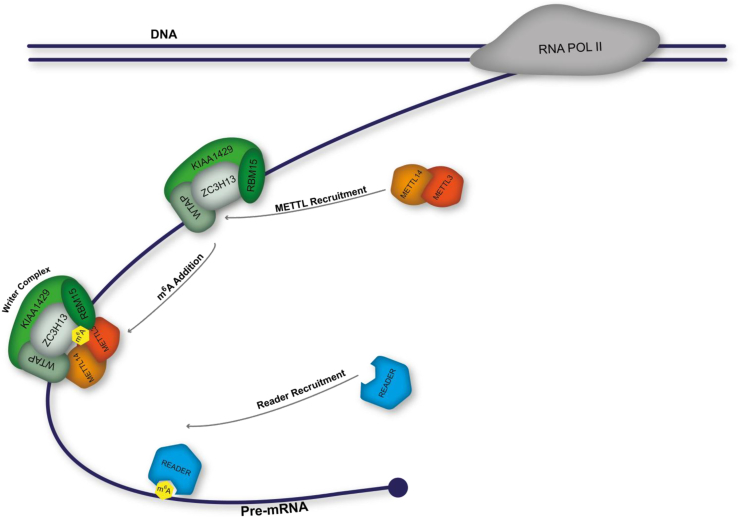
The transcriptome wide mapping of human m6A revealed widespread modification covering one third of the transcriptome confined to the consensus sequence of RRACU [2,3]. The majority of steady state m6A residing in the RRAC consensus sequence is deposited on target transcripts with a bias toward longer exons [2,23]. Recent advances within the field have also identified m6A within intronic regions of nascent pre-mRNA clustering around a consensus sequence with a SAG core [7]. The addition of m6A is carried out by the methyltransferase complex with its subunits often referred to as ‘writers’. There are also indications of demethylases, referred to as ‘erasers’, but this is a source of much debate and will be discussed in due course. The final class of proteins in the m6A pathway are ‘readers’. Readers are responsible for decoding the m6A mark and conducting further processes on a target mRNA, often bridging between two cellular pathways. Below we will discuss the m6A biogenesis pathway, describing its currently identified members and their roles. We will also take this opportunity to present the current inconsistencies within the literature. Fig. 1 illustrates the currently known m6A pathway.
Fig. 1.
The m6A pathway. The methylation is added to the target mRNA molecule in a co-transcriptional manner by the writer complex. The enzymatic activity of the writer complex is imparted by the METT3/METT14 heterodimer. Once a target RNA molecule has been methylated a reader can specifically bind the modification and impart further processing upon the molecule.
3. Writer complex
The methyltransferase writer complex is responsible for the co-transcriptional catalytic addition of m6A to target mRNAs [5,21,24]. The currently defined methyltransferase writer complex comprises adaptor proteins, RNA binding motif protein 15 (RBM15) and its paralogue RBM15B, responsible for initial recruitment of the complex to its target site on a pre-mRNA. The adaptors have also been implicated in an m6A dependent silencing mechanism for X-chromosome inactivation via XIST [13]. Regulatory members are responsible for the complex formation, these are Wilms' tumour 1-associating protein (WTAP) and KIAA1429 (also known as VIRMA) [21,23]. The recently characterised zinc finger CCCH domain-containing protein 13 (ZC3H13) has been found to act as a bridge between the adaptor RBM15 and WTAP [25]. The final member of the writer complex is the catalytic heterodimeric core consisting of Methyltransferase-like protein 3 (METTL3) and Methyltransferase-like protein 14 (METTL14) [24].
WTAP is a ubiquitously expressed protein identified in a yeast–two hybrid screen associating with splicing factors and was subsequently linked to mammalian cell cycle progression from G2 to M [26,27]. Additional mass spectrometry studies on the WTAP interactome revealed multiple methyltransferase complex members such as RBM15, KIAA1429 and METTL3/METTL14, although their role within the m6A pathway was not clarified at that time [28]. Subsequent work identified WTAP as a METTL3/METTL14 binding partner and responsible for the recruitment of the heterodimer to target transcripts facilitating methylation [21,29]. WTAP does not appear to harbour a catalytic domain, however its knockdown resulted in a 6.25-fold decrease in m6A transcriptome wide, this decrease appears to be as a result of reduced METTL3 recruitment [21,23].
Proteomic analysis of WTAP revealed another member of the regulatory component of the methyltransferase complex, KIAA1429 [23]. The knockdown of KIAA1429 results in a fourfold decrease in m6A levels transcriptome wide [23]. Further evidence for KIAA1429 association within the m6A pathway arose from studies on the Drosophila orthologue Virilizer. Two independent studies implicated Virilizer in the m6A dependent regulation of sex determination via alternative splicing [10,11]. KIAA1429 is responsible for recruitment of METTL3 and METTL14 along with WTAP. Furthermore, it associates with polyadenylation cleavage factors and play a role in Poly A site selection for certain m6A containing transcripts [30].
The protein ZC3H13 is the most recently identified member of the methyltransferase complex [25,31]. Knockout of ZC3H13 triggers an 80% reduction in m6A levels transcriptome wide, similar to that of a METTL3 knockout [25,31]. Interestingly ZC3H13 appears to be required for RBM15 and WTAP association, but not for the association between RBM15 and KIAA1429 [25]. ZC3H13 deficient Drosophila S2R+ cells display an increased intron retention rate as well as an increased usage of alternative 5′ splice sites [25]. Interestingly, the study by Knuckles et al. suggests the possibility of two compositionally distinct complexes; m6A-METTL-associated complex (MACOM), which comprises of RBM15, ZC3H13, WTAP, KIAA1429 and CBLL1 (HAKAI) and the m6A–METTL complex (MAC), comprising of METTL3 and METTL4. Together MACOM and MAC act to trigger the m6A modification. However, genetic studies in Drosophila suggest that MACOM has additional m6A independent functions [25].
The catalytic transfer of a methyl group to nitrogen of an adenosine is carried out by a tight heterodimeric complex consisting of METTL3 and METTL14 [24]. The methylation donor was identified as S‑Adenosyl‑methionine (SAM) [24,32]. METTL3 was found to play a role in circadian rhythm regulation as knockdown of METTL3 results in a prolonged circadian period due to the reduced nuclear kinetics of specific clock genes [12]. Mechanistic details of methylation by the heterodimer were revealed by crystallographic studies [[33], [34], [35]]. Surprisingly METTL14 appears to have no catalytic activity, instead acting as a RNA binding scaffold to facilitate the METTL3 dependent m6A modification [33]. Initial studies revealed knockdown of either METTL3 or METTL14 did not yield a dramatic reduction in transcriptional m6A (compared with WTAP) indicating possible redundancy or that only low levels of the METTL3 enzyme are required for methylation to occur [23]. However recent publications have indicated the knockout of METTL3 results in a greater than 90% reduction in m6A levels [5].
The distribution of m6A within the human and mouse embryonic stem cell (ESC) transcriptome has revealed a unique methylation dependent regulatory mechanism. Core factors required for pluripotency were identified as METTL3 targets [36]. The m6A modification is thought to reduce the stability of specific mRNAs required for ESCs to move from a naïve to a primed state [37]. Chromatin associated zinc finger protein 217, ZFP217, is responsible for the regulation of core stem cells genes, and was found to associate with METTL3. High ZFP217 levels in ESCs result in the sequestering of METTL3 and therefore a reduction in methylation upon target mRNAs. Upon differentiation, ZFP217 levels decrease resulting in the release of METTL3 and its subsequent interaction with METTL14 [9].
4. Erasers
As well as writers the current literature suggests the existence of m6A demethylases, also known as erasers. With the potential to have the modification added and removed by the writers and erasers, akin to the epigenetic mechanisms regulating gene expression, the term epitranscriptomics was coined [38]. However, recent studies are starting to cast doubt on the once exciting concept of a general mechanism for m6A related epitranscriptomics [5,39].
Two possible demethylase enzymes/erasers have been discovered, FTO and ALKBH5, both are members of the ALKB subfamily of the superfamily of Fe(II)/2‑oxoglutarate dioxygenases [40,41]. The initial study connecting ALKBH5 and m6A revealed a demethylase activity with preference for single stranded RNA/DNA, unlike other ALK family members that can catalyse the demethylation of double stranded substrates [41]. A subsequent crystallographic study revealed the reason behind ALKBH5 substrate preference, a loop formation results in a steric clash with double stranded ribonucleic substrates therefore allowing only single stranded nucleic acids access to the catalytic site [42]. When assaying the enzymatic activity of ALKBH5, in vitro, using an m6A-RNA oligomer comprising the RRACU consensus only a 40% reduction in total m6A was observed post ALKBH5 treatment. Interestingly a non-consensus m6A-control saw a 20% decrease in m6A levels [41]. Further to this, the knockdown of ALKBH5 in HeLa cells resulted in a 9% increase in m6A transcriptome wide. One possibility for this modest effect could be redundancy within the pathway allowing for the cells to compensate for the loss. A greater effect on m6A levels was observed when over expressing ALKBH5 in HeLa cells as a ~29% reduction in m6A was detected [41]. The study by Zheng and colleagues showed that ALKBH5 deficient male mice were characterised by impaired fertility as a result of aberrant spermatogenesis and apoptosis in the testis [41]. ALKBH5 knockout results in increased m6A deposition at specific 3′ splice sites resulting in alternatively spliced 3′ UTRs and shorter transcripts with a decreased half-life [43].
Interestingly, ALKBH5 and its potential demethylase activity have been implicated in the tumour initiation of breast cancer stem cells [44]. Specifically, Zhang et al., identified the hypoxia dependent stabilisation of pluripotency factor mRNA of NANOG via ALKBH5 demethylation in its 3′ UTR [44]. However, a further study by the same group identified a known methyltransferase (METTL3) inhibitor ZNF217 [9], was expressed in the same hypoxia dependent manner within breast cancer stem cells and acted upon NANOG [45]. The subsequent study raises a level of uncertainty of the actions of ALKBH5 upon NANOG in this particular setting, as the observed reduction in methylation may be a consequence of the METTL3 inhibitor expression.
Another example of ALKBH5 mediated demethylation of m6A was identified with the FOXM1 transcript [46]. This study demonstrated the ALKBH5-FOXM1 pathway as a critical component for Glioblastoma Stem-like Cell proliferation and tumorigenesis. ALKBH5 was identified as acting upon FOXM1 transcript in a unique manner. FOXM1 transcript demethylation was promoted by the formation of an RNA-RNA duplex with anti-sense FOXM1 long non-coding RNA (FOXM1-AS) [46]. This duplex interaction promoting ALKBH5 catalytic activity upon FOXM1 transcript indicates a novel substrate recognition mechanism for the demethylase.
The second reported m6A demethylase is FTO. As with ALKBH5, the proposed role of FTO within the m6A pathway has changed since the first identification of its potential m6A demethylation activity [40]. Initial data suggested FTO harboured a preference toward 5′ m6A modifications [47,48]. However, recent work by Mauer et al. has identified the preferential target of FTO is the N62′‑O‑dimethyladenosine (m6Am) modification not m6A [39]. m6Am is located adjacent to the mRNA N7‑methylguanosine (m7G) cap and it was noted transcripts with this modification displayed an overall greater stability. To have the greatest catalytic activity on m6Am, it was shown FTO requires the presence of the m7G modification and triphosphate linker. Furthermore, FTO displays 100 fold greater catalytic activity for m6Am compared to m6A [39]. The increase in stability appears to be imparted upon select m6Am transcripts by a resistance to mRNA-decapping enzymes [39].
To add to the functional uncertainty regarding demethylases, the recent study by Ke et al. indicated that the vast majority of m6A is added to chromatin associated pre-mRNA and persists throughout the nucleoplasm and cytoplasm. This argues against frequent demethylation of steady state m6A RNA [5]. However the study does note a very small portion of m6A peaks present in the chromatin associated RNA were reduced in frequency in the nucleoplasm [5]. This suggests a potential demethylase acting prior to nuclear export on a small subset of mRNAs, but not the vast majority of m6A containing transcripts [5].
There is much work to be done on ALKBH5, and demethylases in general, to clarify their role within the m6A pathway. It appears from the current literature ALKBH5 only acts upon a small subset of mRNAs and although the m6A modification resides in the same consensus sequence, ALKBH5 may require a unique yet unidentified recognition method, exemplified in the FOXM1 study [46].
The case for global epitranscriptomic mRNA regulation is not as clear cut as initially thought. Further work is required to outline the specificities of the identified demethylases and possibility of identification of new ones.
5. Readers
The m6A readers carry out different functions once bound to the transcript (for an in-depth look at the m6A reader proteins consult Meyer and Jaffrey 2017 [49]). A variety of reader proteins have been discovered, often linking the m6A modification with other mRNA pathways. Two heterogeneous nuclear ribonucleoproteins (HNRNP) have been identified as nuclear located m6A readers, HNRNPA2B1 and HNRNPC [50,51]. Although they share similar characteristics, HNRNPs comprise a wide array of different domains and therefore carry out an array of functions [52]. HNRNPA2B1 has been identified as a direct nuclear reader of m6A and involved in the control and processing of a subset of pre-miRNAs [50]. The discovery occurred when an HNRNPA2B1 knockdown phenocopied the reduction in miRNA expression also observed in a METTL3 knockdown [50]. Post methylation HNRNPA2B1 binds to the methylated adenosine and bridges the initial interaction between pre-miRNA and the microprocessor complex, specifically DGCR8 [53,54]. HNRNPC was identified as an indirect reader of m6A [51]. The addition of an m6A within a stem loop structure resulted in a change in its thermostability, disrupting the secondary structure allowing HNRNPC-RNA interactions to occur. This was termed an m6A-dependent RNA switch and it is noteworthy that HNRNPC does not directly bind m6A but acts indirectly as a consequence of m6A addition. However, this switch only occurs on a subset of HNRNPC target RNAs, pre-mRNAs and lncRNAs and is said to influence splice site selection [51]. The catalogue of m6A readers is ever expanding, a recent global analysis identified new m6A readers but also proteins repelled by m6A [55]. This study identified FMR1 as a new reader of m6A, suggesting a link between the mRNA modification and an autism spectrum disorder [55].
YTHDF1/2/3 are cytoplasmic readers that recognise the m6A via an aromatic cage consisting of two conserved tryptophan residues, located in the YTH domain [[56], [57], [58], [59]]. Although all identified cytoplasmic readers contain a YTH-m6A RNA binding domain they differ in further domain composition allowing for an array of functions [60]. YTHDF1 is responsible for ribosomal loading onto m6A transcripts and subsequently their expression. This function is apparent as the interactome of YTHDF1 is heavily populated with translation initiation factors, notably eIF3 [61]. The YTHDF1 dependent translation initiation mechanism only occurs on a subset of m6A transcripts [61]. Conversely YTHDF2 destabilises its m6A-mRNA targets in the cytoplasm by directly recruiting them to the CCR4-NOT deadenylase complex [6]. The RNA-m6A-YTHDF2 complex can be located in the 3′ UTR or open reading frame and alters the lifetime of transcripts. This degradation mechanism is orchestrated by the N-terminus of YTHDF2 whilst its C-terminal YTH domain binds m6A [62]. This leads to the formation of a multidimensional cytoplasmic mechanism for select transcript expression via the m6A modification, as YTHDF1 promotes translation whilst YTHDF2 can induce transcript degradation. Cross talk between these proteins is evident as they share 50% of their targets [61]. Cellular response mechanisms upon stress have been found to involve m6A dependent mechanisms. One example is the heat shock induced nuclear translocation of YTHDF2, resulting in protection of specific mRNA targets subsequently allowing for their cytoplasmic expression [63]. A third member of the cytoplasmic YTH-domain subfamily, YTHDF3, has been implicated in both translation and degradation [64]. The current literature paints a methylation dependent regulatory pathway orchestrated by the cytoplasmic YTH containing proteins.
YTHDC1 (YT521-B) is a nuclear localised YTH domain containing protein initially described as a splicing associated factor [65]. The crystal structure of YTHDC1 revealed details of its m6A binding activity. As is the case with the previously discussed YTH containing proteins, YTHDC1 binds the modification via two tryptophan residues within an aromatic cage [59]. Interestingly 21% of all YTHDC1 bound transcripts are shared with YTHDF2 and 13% of all bound targets having a role in transcriptional regulation [59]. Recently the role of YTHDC1 in splicing regulation has been investigated in detail, providing mechanistic details of previously identified YTHDC1 knockdown phenotypes [65,66]. YTHDC1 was found to bind near splice sites and promote exon inclusion, working cooperatively with splicing factor SRSF3 [66]. Conversely Xiao et al. also suggest SRSF10 can bind at similar sites on mRNA, resulting in the blocking of YTHDC1 mRNA binding therefore resulting in exon skipping [66,67]. The proposed current competition model of YTHDC1, SRSF3 and SRSF10 may provide mechanistic reasoning for previous results showing exon inclusion is YTHDC1 dosage dependent, greater expression of YTHDC1 leads to an increase in exon inclusion [68,69].
Recent studies have built on the work performed by Xiao et al. to discover m6A is deposited within intronic regions and around splicing junctions. Loiloupi et al. [7] developed transient N‑6‑methyladenosine transcriptome sequencing (TNT-seq), a novel approach to identify the m6A landscape of nascent RNA. This method incorporated a bromouridine RNA isolation step prior to m6A RNA isolation, allowing for analysis of nascent RNA. The resulting data indicated up to 57% of m6A on nascent RNA resides within introns [7]. They concluded m6A deposition upon intronic sequences is associated with slowly processed introns and alternative splicing events, whereas m6A deposition close to splicing junctions promotes fast splicing. These data indicate m6A regulates the kinetics of splicing. It was also noted intronic m6A sits within a SAG core consensus and not the classical RRAC consensus [3,7]. The SAG core is reminiscent of the SRSF binding motifs consistent with the role of SRSF3/10 in m6A splicing regulation [66], although it does not reflect the YTHDC1 binding motif [59]. Conversely a study by Ke et al., demonstrates that whilst the vast majority of m6A is added to pre-mRNA prior to chromatin dissociation and splicing completion, very little occurs within introns [5]. Furthermore Ke et al. demonstrated, only ~10% of m6A sites were within 50 nucleotides of a 5′ or 3′ splice site and cells lacking METTL3 showed few splicing changes. Moreover, Ke et al. did not observe a universal demethylation event between chromatin associated RNA and nucleoplasm/cytoplasmic RNA. The discrepancy between these finding may be due to the RNA isolation and sequencing techniques applied: m6A-CLIP vs TNT-seq [5,7]. Further studies will be needed to define the complex pathways associated with m6A-related RNA biogenesis.
YTHDC1 also plays a role in mRNA export in concert with hypophosphorylated SR proteins and the major export receptor NXF1 [70]. This is the first study to offer mechanistic insight into the possible role of the m6A pathway and mRNA export. In order for us to outline the data presented throughout the literature on a possible overlap of the m6A pathway and mRNA export we will give a brief introduction on mRNA export and its key players.
6. mRNA export
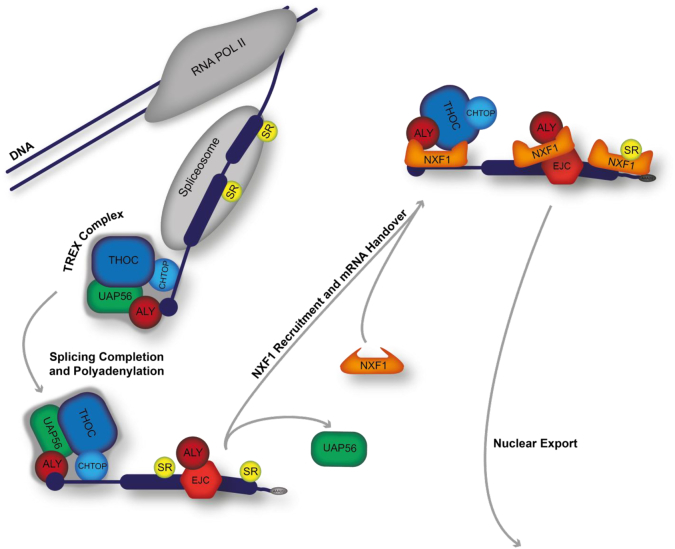
The bulk of mRNA nuclear export is mediated by the TREX complex and the heterodimeric nuclear export receptor NXF1-P15. TREX is deposited on the mRNA through a number of pre- and post-transcriptional maturation events. Fig. 2 gives an overview of the TREX-NXF1 mediated export pathway. The initial addition of the 5′ m7guanosine (m7G) cap to nascent pre-mRNAs protects them from the action of 5′-3′ exonucleases [17]. Moreover, the m7G cap confers binding of CBP80 and CBP20 that form the cap-binding complex (CBC). The action of CBC binding allows for deposition of TREX components onto the 5′ end of the mRNA [71,72]. mRNAs undergo co- and post-transcriptional splicing. The excision of introns and fusing of exons results in the formation of an exon junction complex (EJC) centred ~24 bases upstream from the exon-exon boundary. The EJC associates with TREX subunits whilst TREX member UAP56 is involved in spliceosome assembly [73,74].
Fig. 2.
The mRNA nuclear export pathway. The bulk of mRNA is exported into the cytoplasm by the TREX:NXF1 pathway. During transcription initial TREX complex members are deposited upon the mRNA. Further processing such as capping and splicing results in further TREX deposition. Once an mRNA has matured the export receptor NXF1 associates with the mRNA via TREX complex interactions. NXF1 guides the correctly packaged mRNA through the nuclear pore.
Polyadenylation of pre-mRNAs at the 3′ terminus is another maturation event that commits the mRNA for nuclear export. A hexanucleotide cleavage signal is bound by cleavage and polyadenylation factors responsible for removal of the downstream RNA. Post cleavage, Poly A polymerase catalyses the addition of adenosine repeats on the 3′ terminal cleaved mRNA. The adenosine repeats are then bound by PABPN1, protecting the mRNA from degradation by 3′-5′ exonuclease [75]. The combination of these processing events leads to the formation of a correctly packaged messenger ribonucleoparticle (mRNP) that contains all the licensing signals for nuclear pore translocation orchestrated by NXF1.
7. Transcription export complex
TREX is conserved from yeast to humans and is required for messenger ribonucleoprotein particles (mRNP) nuclear export [76,77]. Akin to the spliceosome, the TREX complex undergoes multiple conformational and compositional changes. At the core of the TREX complex sits the multimeric THO complex comprised of THOC1,2,3,5,6,7 and DEAD box RNA helicase, UAP56 [78,79]. Export adaptor subunits of TREX, ALYREF, UIF and LUZP4, aid in the loading and handover of the packaged mRNA to the nuclear export receptor NXF1 [20,80,81]. Coadaptor subunits of TREX, CHTOP and THOC5, also play a critical role in NXF1 loading [82].
ALYREF is recruited to the mRNP via UAP56 during splicing in an ATP dependent manner [19,78]. It contains two UAP56-binding motifs (UBM) at its N and C-termini. The core of the adaptor consists of an RNA recognition motif flanked by two disordered arginine rich regions which are the main RNA binding sites [74]. The arginine rich regions of ALYREF are also required for NXF1 binding [83]. UAP56 interacting factor (UIF) is a second export adaptor that contains a UBM. Although UIF interacts with ALYREF, the interaction is RNA-dependent, therefore UIF may exist in an alternative TREX complex populating the same mRNA as an ALYREF containing TREX. UIF recruitment to the mRNA differs to that of ALYREF. The FACT chromatin remodelling complex is required for the co-transcriptional loading of UIF but not ALYREF [80]. A third export adaptor, LUZP4, is a cancer testis antigen – a group of proteins normally restricted to testis which are commonly upregulated in cancer cells. LUZP4 displays the classical adaptor characteristics mentioned above, contains a UBM and NXF1s mRNA binding affinity increases upon association [81]. Members of the serine/arginine (SR) rich family of proteins can also act as mRNA export adaptors when in a hypophosphorylated state, this function is independent of their role within splicing [84].
THOC5 and CHTOP have been identified as co-adaptor proteins involved in NXF1 binding and mRNA handover [82,85]. The co-adaptors bind to NXF1 co-operatively with adaptors. It is also worth noting that CHTOP and THOC5 bind to NXF1 in a mutually exclusive manner [82]. The presence of multiple adaptors and co-adaptors illustrates the dynamic changes and redundancy within the TREX complex dependent mRNA export pathway.
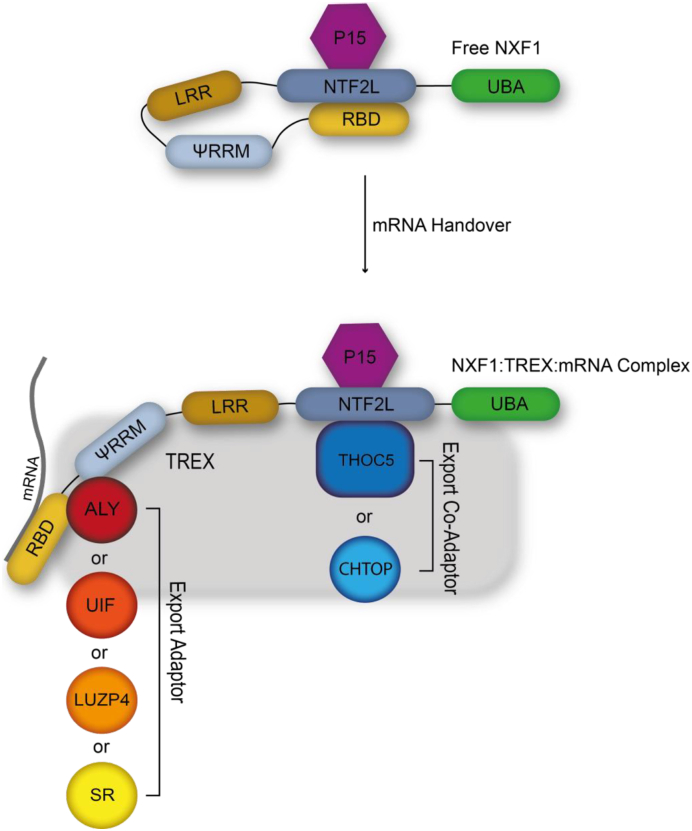
A key stage in export of the mRNP is the handover of the mRNA to the export receptor NXF1. NXF1 consists of five distinct domains (depicted in Fig. 3), the RNA binding domain (RBD), pseudo-RNA recognition motif (ΨRRM), leucine rich region (LRR), NTF2-like (NTF2L) domain and ubiquitin associated domain (UBA) [86]. The N-terminal arginine rich RBD is responsible for NXF1 RNA recognition, however it shows no sequence specificity [87]. To prevent the export of non-/incorrectly processed mRNA, NXF1 sequesters its own RNA binding activity via intramolecular association of the NTF2L and RBD domains [20]. Therefore, an mRNA is only handed to NXF1 once associated with the TREX complex. The mRNA handover is conducted via export adaptors binding the RBD and ΨRRM whilst export co-adaptors associate with the NTF2L domain [20]. The association of TREX components releases the NXF1 RBD, stabilising the NXF1-mRNA interaction. Subsequently NXF1-P15 escorts the mRNA to the nuclear pore where it interacts with nucleoporins via the NTF2L and Ubiquitin associated domains. P15 is essential for the stabilisation of NTF2L and its interaction with NPC members [88,89].
Fig. 3.
The NXF1 mRNA handover event. Once an mRNA has matured and is correctly packaged NXF1/P15 are recruited to initiate the final stages of the export pathway. Upon recruitment the mRNA binding domain (RBD) of NXF1 is released from its intramolecular interactions with the NTFL2 domain via export adaptor and co adapter binding. Once bound to the mRNA the NXF1:mRNA complex is guided through the nuclear pore into the cytoplasm.
8. The m6A pathway and nuclear export
The final section of this review will cover the present data suggesting an overlap between the m6A machinery and the TREX-NXF1 export pathway. Modifications of mRNA have previously been shown to be required for nuclear export, the most famous example being the requirement of the m7G cap and the accompanying CBC [74]. Evidence has also been presented implicating ALYREF in regulating the export of specific transcripts harbouring the internal mRNA modification m5C [90]. However, this study utilised RNA affinity chromatography to identify ALYREF and showed that it bound oligonucleotides bearing m5C with greater affinity than equivalent oligonucleotides lacking the methylation modification. This does not necessarily prove specificity for m5C for ALYREF since the hydrophobicity of the affinity column is also altered with the methyl group addition and so in the absence of a suitable control such as an m6A affinity column we cannot be certain that ALYREF specifically recognises m5C at present. It is not a stretch to think the m6A pathway may have links to export, as mRNA maturation events are closely coupled to TREX deposition and subsequent NXF1 dependent export [18,75,77]. A review of both the export and m6A literature reveals a number of possible links between the two pathways including associations between important regulatory players.
Historic work demonstrated the mRNA of Simian virus 40 (SV40) harboured m6A modifications which appear to be added by host machinery whilst progressing through its replication cycle [91]. An interesting observation from this early study was that cycloleucine treatment, a methylation inhibitor, inhibited SV40 mRNA export. It was noted the total amount of SV40 mRNA within the nucleus was unchanged however the cytoplasmic levels of the SV40 mRNA were dramatically reduced, indicating a defect in export upon inhibitor treatment [91]. Although it must be noted the use of cycloleucine will also inhibit 5′ Cap formation, which is also detrimental to mRNA export. More recent studies have demonstrated the role of the m6A and its reader in the replication of HIV-1 and ZIKA viruses [92,93]. Also of note, inhibition of S‑adenosylmethionine-dependent methyltransferase reactions by S‑Tubercidinylhomocysteine (STH), triggered a delay in mRNA export [94].
Further evidence of an overlap between mRNA export and m6A appear in more recent literature and is briefly summarised in Table 1 The knockdown of METTL3 caused a slowdown in the circadian clock [12]. One of the reasons behind this was the reduced export of mature mRNA of specific clock factors, Arntl and Per2 [12]. Therefore, the m6A modification was shown to be required for the efficient export of specific mature mRNAs. Conversely, it was demonstrated by Poly(A)+ RNA fluorescence in situ hybridisation that ALKBH5 knockdown causes excess cytoplasmic accumulation of mRNA, however this was looking at total Poly (A)+ RNA not just m6A containing and therefore may not be a trait limited to m6A containing RNA [41].
Table 1.
Interactions between the mRNA export machinery and components of the m6A pathway.
| m6A pathway member | Association with mRNA export | Refs. |
|---|---|---|
| METTL3 | Knockdown delayed nuclear export Associated with TREX |
[12] [22] |
| WTAP | Associated with TREX (Co-IP and Mass spec) Associated with SR-like proteins (BCLAF1/TRAP150) WTAP/KIAA1429 knockdown resulted in defective mRNA export for specific transcripts |
[22,28] [28] [22] |
| KIAA1429 | Associated with core TREX members WTAP/KIAA1429 knockdown resulted in defective mRNA export for specific transcripts |
[22,78] [22] |
| ZC3H13 | Associated with ALYREF and THOC5 | [78,98] |
| RBM15 | Knockdown results in an export defect Acts as an adaptor within NXF1/p15 pathway ALYREF and UAP56 binding partners |
[99] [100] [78,99,100] |
| YTHDC1 | Knockdown resulted in mRNA export defect of specific transcripts. Acts with SRSF3 to export mRNA via NXF1 TREX associated |
[22,70] [70] [22] |
| ALKBH5 | Knockdown results in increased PolyA+ RNA cytoplasmic accumulation. | [41] |
Yet another indication of the overlap between the TREX complex and the methylation machinery is demonstrated within a proteomic study of the export adaptor ALYREF [78]. KIAA1429, a key member of the methylation writer complex was found to associate with the export adaptor [78]. A possible association between the two complexes was also shown upon shotgun proteomic analysis of WTAP by mass spectrometry, identifying ALYREF and UAP56 as binding partners [28]. Further interesting binding partners were identified with WTAP that are suspected to be involved with the nuclear mRNA export process including: ERH which is known to associate with TREX [19] and the SR-like proteins BCLAF1 and THRAP3 [28]. BCLAF1 is implicated in mRNA export since its knockdown leads to an increase in cytoplasmic Poly(A)+ mRNA [95] similar to that observed upon ALKBH5 knockdown [41]. A recent study has shown that BCLAF1 and THRAP3 are required for the efficient splicing and export of DNA damage response mRNAs, though the precise molecular role in export remains to be determined [96]. The recently identified core member of the methyltransferase complex ZC3H13 also interacts with the TREX complex, being found to associate with ALYREF and THOC1 in multiple proteomics studies [78,97].
RBM15 was also identified in the WTAP proteomics screen [28] which was later found to play a role in the m6A pathway [13]. Strikingly, RBM15 is also a bona fide member of the NXF1-P15 export pathway [[98], [99], [100]]. Knockdown of RBM15 results in cytoplasmic depletion and nuclear accumulation of mRNA [99]. RBM15 binds to NXF1, specifically NXF1s NTF2L domain [100]. RBM15 binding to NXF1 NTF2L domain suggests it may act as an export co-adaptor such as THOC5 and CHTOP, aiding in NXF1 loading on the mRNA [20]. RBM15 also associates with export adaptor ALYREF, but not with the core TREX component UAP56, suggesting RBM15 plays a role in the later stages of mRNA export [78,99,100]. Together these data showing multiple interactions between the core of the mRNA export machinery (TREX-NXF1) and proteins implicated in the m6A modification strongly suggests a functional connection between the two processes.
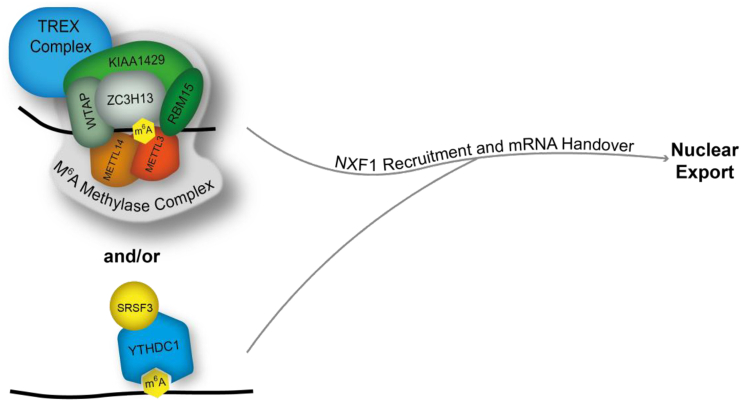
Two recent studies have now started to uncover the molecular mechanisms connecting the m6A modification and mRNA export, summarised in Fig. 4. In the first study, YTHDC1 was shown to interact with SRSF3 which can act as an mRNA export adaptor [70]. SRSF3 has previously been shown to bind NXF1 and act as a novel route for loading NXF1 onto the mRNP [84]. Knockdown of YTHDC1 or SRSF3 led to a nuclear export block for a common set of transcripts suggesting the two proteins act in the same pathway. Since SRSF3 binds NXF1 [84] it was proposed that YTHDC1, in recruiting NXF1, was able to trigger the selective export of m6A modified mRNAs [70]. Since SRSF3 only provides the adaptor function to unlock NXF1 and allow its stable binding to mRNAs, the co-adaptor function may be supplied by TREX. Alternatively, an intriguing possibility is that RBM15 might provide the co-adaptor function since it associates with both TREX and the m6A methylation machinery. Furthermore, it associates with NXF1 via the same domain used by co-adaptors, though this possibility remains to be tested. The second study showed that the TREX complex associates with the m6A writer machinery (WTAP, KIAA1429, METTL3, METTL14) and TREX deposition on specific m6A modified mRNAs requires the m6A writer complex [22]. The combined knockdown of WTAP/KIAA1429 led to an mRNA export block for a specific group of transcripts, which had largely been shown to be m6A modified in previous studies [22]. Moreover, this study showed that only a small proportion of mRNAs showed altered splicing following WTAP/KIAA1429 knockdown, confirming that the export block observed for many transcripts was not caused by defective splicing. This study further confirmed that YTHDC1 is required for the efficient export of specific mRNAs and demonstrated that knockdown of TREX in cells led to reduced levels of YTHDC1 associated with the mRNP, suggesting that TREX stabilises YTHDC1 binding to mRNA [22]. Together these two studies show that m6A plays an important role in mRNA export.
Fig. 4.
The m6A machinery and mRNA export. The m6A writer complex interacts with the TREX complex aiding in the export of specific mRNAs. The reader YTHDC1 associates with SRSF3. Both pathways may work in unison to recruit the export receptor NXF1/P15 to initiate nuclear export.
9. Concluding remarks
The recent explosion of m6A literature has revealed a number of exciting regulatory mechanisms that appear dependent on the addition of the modification. However, the literature does present a number of conflicting views on the role of m6A within splicing. There are some clear examples where m6A plays a role in splicing decisions, such as YTHDC1-m6A-SRSF3, HNRNPC-m6A-switch and Drosophila sex determination splicing mechanisms [11,51,66]. However, the recent identification of m6A not clustering near splice sites and knockout of METTL3 resulting in minimal splicing changes [5] does suggest that m6A regulation of splicing may be restricted to a limited set of transcripts. Furthermore the actions of the m6A proteins may be drastically different depending on cellular context, for example its role in spermatocytes compared to mRNA half-life in HeLa cells [5,41]. Going forward it will be important to begin to dissect the function for each member of the methyltransferase complex, as it may well be possible they have many roles within mRNA biogenesis, and these role may not all be linked with m6A deposition. An interesting example of this was presented above with ZC3H13 where it appears to exist in two separate complexes with one devoid of METTL3/14. The METTL3/14 lacking complex may harbour separate functions from m6A deposition, however knocking down ZC3H13, or WTAP/KIAA1429, will disrupt these functions along with m6A deposition and dissecting these two activities may be difficult [22,25]. Another example is RBM15 having export defects upon knockdown but also impairing m6A addition [13,100]. The question will be: are we seeing an export defect due to removing RBM15s interactions with NXF1 or due to the lack of m6A deposition or both? The dynamic aspect of the m6A modification may not be the modification itself but the proteins that are involved in its addition and subsequent binding.
Transparency document
Transparency document.
Acknowledgements
SW acknowledges support from the Biotechnology and Biological Sciences Research Council (BBSRC), UK via grants BB/N014839/1 and BB/N005430/1. SL was the recipient of a White Rose BBSRC doctoral training programme funded PhD studentship.
Footnotes
This article is part of a Special Issue entitled: mRNA modifications in gene expression control edited by Dr. Soller Matthias and Dr. Fray Rupert.
The Transparency document associated with this article can be found, in online version.
References
- 1.Dunin-Horkawicz S. MODOMICS: a database of RNA modification pathways. Nucleic Acids Res. 2006;34:D145–D149. doi: 10.1093/nar/gkj084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Meyer K.D., Saletore Y., Zumbo P., Elemento O., Mason C.E., Jaffrey S.R. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dominissini D., Moshitch-Moshkovitz S., Schwartz S., Salmon-Divon M., Ungar L., Osenberg S., Cesarkas K., Jacob-Hirsch J., Amariglio N., Kupiec M., Sorek R., Rechavi G. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 4.Desrosiers R., Friderici K., Rottman F. Identification of methylated nucleosides in messenger RNA from Novikoff hepatoma cells. Proc. Natl. Acad. Sci. 1974;71:3971–3975. doi: 10.1073/pnas.71.10.3971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ke S., Pandya-jones A., Saito Y., Fak J.J., Vågbø C.B., Geula S., Hanna J.H., Black D.L., Jr J.E.D., Darnell R.B. 2017. m6A mRNA Modifications Are Deposited in Nascent Pre-mRNA and Are Not Required for Splicing but Do Specify Cytoplasmic Turnover; pp. 990–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Du H., Zhao Y., He J., Zhang Y., Xi H., Liu M., Ma J., Wu L. YTHDF2 destabilizes m6A-containing RNA through direct recruitment of the CCR4–NOT deadenylase complex. Nat. Commun. 2016;7 doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Louloupi A., Ntini E., Conrad T., Ørom U.A.V. Transient N‑6‑methyladenosine transcriptome sequencing reveals a regulatory role of m6A in splicing efficiency. Cell Rep. 2018;23:3429–3437. doi: 10.1016/j.celrep.2018.05.077. [DOI] [PubMed] [Google Scholar]
- 8.Zhao B.S., Roundtree I.A., He C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2016;18:31–42. doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aguilo F., Zhang F., Sancho A., Fidalgo M., Di Cecilia S., Vashisht A., Lee D.F., Chen C.H., Rengasamy M., Andino B., Jahouh F., Roman A., Krig S.R., Wang R., Zhang W., Wohlschlegel J.A., Wang J., Walsh M.J. Coordination of m6A mRNA methylation and gene transcription by ZFP217 regulates pluripotency and reprogramming. Cell Stem Cell. 2015;17:689–704. doi: 10.1016/j.stem.2015.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Haussmann I.U., Bodi Z., Sanchez-Moran E., Mongan N.P., Archer N., Fray R.G., Soller M. m6A potentiates Sxl alternative pre-mRNA splicing for robust Drosophila sex determination. Nature. 2016;000 doi: 10.1038/nature20577. [DOI] [PubMed] [Google Scholar]
- 11.Lence T., Akhtar J., Bayer M., Schmid K., Spindler L., Ho C.H., Kreim N., Andrade-Navarro M.A., Poeck B., Helm M., Roignant J.-Y. m6A modulates neuronal functions and sex determination in Drosophila. Nature. 2016 doi: 10.1038/nature20568. [DOI] [PubMed] [Google Scholar]
- 12.Fustin J.-M., Doi M., Yamaguchi Y., Hida H., Nishimura S., Yoshida M., Isagawa T., Morioka M.S., Kakeya H., Manabe I., Okamura H. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell. 2013;155:793–806. doi: 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Patil D.P., Chen C.-K., Pickering B.F., Chow A., Jackson C., Guttman M., Jaffrey S.R. m6A RNA methylation promotes XIST-mediated transcriptional repression. Nature. 2016;537:1–25. doi: 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xiang Y., Laurent B., Hsu C.-H., Nachtergaele S., Lu Z., Sheng W., Xu C., Chen H., Ouyang J., Wang S., Ling D., Hsu P.-H., Zou L., Jambhekar A., He C., Shi Y. RNA m6A methylation regulates the ultraviolet-induced DNA damage response. Nature. 2017;543:573–576. doi: 10.1038/nature21671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoon K.J., Ringeling F.R., Vissers C., Jacob F., Pokrass M., Jimenez-Cyrus D., Su Y., Kim N.S., Zhu Y., Zheng L., Kim S., Wang X., Doré L.C., Jin P., Regot S., Zhuang X., Canzar S., He C., li Ming G., Song H. Temporal control of mammalian cortical neurogenesis by m6A methylation. Cell. 2017;171:877–889.e17. doi: 10.1016/j.cell.2017.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heath C.G., Viphakone N., Wilson S.A. The role of TREX in gene expression and disease. Biochem. J. 2016;473:2911–2935. doi: 10.1042/BCJ20160010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shatkin A., Manley J. The ends of the affair: capping and polyadenylation. Nat. Struct. Biol. 2000;7:1–5. doi: 10.1038/79583. [DOI] [PubMed] [Google Scholar]
- 18.Masuda S. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dufu K., Livingstone M.J., Seebacher J., Gygi S.P., Wilson S. a, Reed R. ATP is required for interactions between UAP56 and two conserved mRNA export proteins, Aly and CIP29, to assemble the TREX complex. Genes Dev. 2010;24:2043–2053. doi: 10.1101/gad.1898610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Viphakone N., Hautbergue G.M., Walsh M., Chang C.-T., Holland A., Folco E.G., Reed R., Wilson S.A. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat. Commun. 2012;3 doi: 10.1038/ncomms2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ping X.-L., Sun B.-F., Wang L., Xiao W., Yang X., Wang W.-J., Adhikari S., Shi Y., Lv Y., Chen Y.-S., Zhao X., Li A., Yang Y., Dahal U., Lou X.-M., Liu X., Huang J., Yuan W.-P., Zhu X.-F., Cheng T., Zhao Y.-L., Wang X., Danielsen J.M.R., Liu F., Yang Y.-G. Mammalian WTAP is a regulatory subunit of the RNA N6‑methyladenosine methyltransferase. Cell Res. 2014;24:177–189. doi: 10.1038/cr.2014.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lesbirel Simon, Viphakone Nicolas, Parker Matthew, Parker Jacob, Heath Catherine, Sudbury I., Wilson S.A. The m6A‑methylase complex recruits TREX and regulates mRNA export. Sci. Rep. 2018;8(13827):1–12. doi: 10.1038/s41598-018-32310-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz S., Mumbach M.R., Jovanovic M., Wang T., Maciag K., Bushkin G.G., Mertins P., Ter-Ovanesyan D., Habib N., Cacchiarelli D., Sanjana N.E., Freinkman E., Pacold M.E., Satija R., Mikkelsen T.S., Hacohen N., Zhang F., Carr S.A., Lander E.S., Regev A. Perturbation of m6A writers reveals two distinct classes of mRNA methylation at internal and 5′ sites. Cell Rep. 2014;8:284–296. doi: 10.1016/j.celrep.2014.05.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liu J., Yue Y., Han D., Wang X., Fu Y., Zhang L., Jia G., Yu M., Lu Z., Deng X., Dai Q., Chen W., He C. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6‑adenosine methylation. Nat. Chem. Biol. 2014;10:93–95. doi: 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knuckles P., Lence T., Haussmann I.U., Jacob D., Kreim N., Carl S.H., Masiello I., Hares T., Villaseñor R., Hess D., Andrade-Navarro M.A., Biggiogera M., Helm M., Soller M., Bühler M., Roignant J.-Y. Zc3h13/Flacc is required for adenosine methylation by bridging the mRNA-binding factor Rbm15/Spenito to the m6A machinery component Wtap/Fl(2)d. Genes Dev. 2018:1–15. doi: 10.1101/gad.309146.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Little N.A., Hastie N.D., Davies R.C. Identification of WTAP, a novel Wilms' tumour 1-associating protein. Hum. Mol. Genet. 2000;9:2231–2239. doi: 10.1093/oxfordjournals.hmg.a018914. [DOI] [PubMed] [Google Scholar]
- 27.Horiuchi K., Umetani M., Minami T., Okayama H., Takada S., Yamamoto M., Aburatani H., Reid P.C., Housman D.E., Hamakubo T., Kodama T. Wilms' tumor 1-associating protein regulates G2/M transition through stabilization of cyclin A2 mRNA. Proc. Natl. Acad. Sci. U. S. A. 2006;103:17278–17283. doi: 10.1073/pnas.0608357103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Horiuchi K., Kawamura T., Iwanari H., Ohashi R., Naito M., Kodama T., Hamakubo T. Identification of Wilms' tumor 1-associating protein complex and its role in alternative splicing and the cell cycle. J. Biol. Chem. 2013;288:33292–33302. doi: 10.1074/jbc.M113.500397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schöller E., Weichmann F., Treiber T., Ringle S., Treiber N., Flatley A., Feederle R., Bruckmann A., Meister G. Interactions, localization and phosphorylation of the m6A generating METTL3-METTL14-WTAP complex. RNA. 2018 doi: 10.1261/rna.064063.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu J., Yue Y., Liu J., Cui X., Cao J., Luo G., Zhang Z., Cheng T., Gao M., Shu X., Ma H., Wang F., Wang X., Shen B., Wang Y., Feng X., He C. VIRMA mediates preferential m6A mRNA methylation in 3′UTR and near stop codon and associates with alternative polyadenylation. Cell Discov. 2018;4 doi: 10.1038/s41421-018-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wen J., Lv R., Ma H., Shen H., He C., Wang J., Jiao F., Liu H., Yang P., Tan L., Lan F., Shi Y.G., He C., Shi Y., Diao J. Zc3h13 regulates nuclear RNA m6A methylation and mouse embryonic stem cell self-renewal. Mol. Cell. 2018;69:1028–1038.e6. doi: 10.1016/j.molcel.2018.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bokar J.A., Shambaugh M.E., Polayes D., Matera A.G., Rottman F.M. Purification and cDNA cloning of the AdoMet-binding subunit of the human mRNA (N6‑adenosine)‑methyltransferase. RNA. 1997;3:1233–1247. [PMC free article] [PubMed] [Google Scholar]
- 33.Wang P., Doxtader K.A., Nam Y. Structural basis for cooperative function of Mettl3 and Mettl14 methyltransferases. Mol. Cell. 2016;63:306–317. doi: 10.1016/j.molcel.2016.05.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Śledź P., Jinek M. Structural insights into the molecular mechanism of the m6A writer complex. elife. 2016;5:1–16. doi: 10.7554/eLife.18434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wang X., Feng J., Xue Y., Guan Z., Zhang D., Liu Z., Gong Z., Wang Q., Huang J., Tang C., Zou T., Yin P. Structural basis of N6‑adenosine methylation by the METTL3–METTL14 complex. Nature. 2016:1–15. doi: 10.1038/nature18298. [DOI] [PubMed] [Google Scholar]
- 36.Batista P.J., Molinie B., Wang J., Qu K., Zhang J., Li L., Bouley D.M., Lujan E., Haddad B., Daneshvar K., Carter A.C., Flynn R.A., Zhou C., Lim K.-S.S., Dedon P., Wernig M., Mullen A.C., Xing Y., Giallourakis C.C., Chang H.Y. m6A RNA modification controls cell fate transition in mammalian embryonic stem cells. Cell Stem Cell. 2014;15:707–719. doi: 10.1016/j.stem.2014.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Geula S., Moshitch-Moshkovitz S., Dominissini D., Mansour A.A., Kol N., Salmon-Divon M., Hershkovitz V., Peer E., Mor N., Manor Y.S., Ben-Haim M.S., Eyal E., Yunger S., Pinto Y., Jaitin D.A., Viukov S., Rais Y., Krupalnik V., Chomsky E., Zerbib M., Maza I., Rechavi Y., Massarwa R., Hanna S., Amit I., Levanon E.Y., Amariglio N., Stern-Ginossar N., Novershtern N., Rechavi G., Hanna J.H. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 38.Saletore Y., Chen-Kiang S., Mason C.E. Novel RNA regulatory mechanisms revealed in the epitranscriptome. RNA Biol. 2013;10:342–346. doi: 10.4161/rna.23812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mauer J., Luo X., Blanjoie A., Jiao X., Grozhik A.V., Patil D.P., Linder B., Pickering B.F., Vasseur J.J., Chen Q., Gross S.S., Elemento O., Debart F., Kiledjian M., Jaffrey S.R. Reversible methylation of m6Amin the 5′ cap controls mRNA stability. Nature. 2017;541:371–375. doi: 10.1038/nature21022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jia G., Fu Y., Zhao X., Dai Q., Zheng G., Yang Y., Yi C., Lindahl T., Pan T., Yang Y.G., He C. N6‑methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat. Chem. Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng G., Dahl J.A., Niu Y., Fedorcsak P., Huang C.-M., Li C.J., Vågbø C.B., Shi Y., Wang W.-L., Song S.-H., Lu Z., Bosmans R.P.G., Dai Q., Hao Y.-J., Yang X., Zhao W.-M., Tong W.-M., Wang X.-J., Bogdan F., Furu K., Fu Y., Jia G., Zhao X., Liu J., Krokan H.E., Klungland A., Yang Y.-G., He C. ALKBH5 is a mammalian RNA demethylase that impacts RNA metabolism and mouse fertility. Mol. Cell. 2013;49:18–29. doi: 10.1016/j.molcel.2012.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xu C., Liu K., Tempel W., Demetriades M., Aik W., Schofield C.J., Min J. Structures of human ALKBH5 demethylase reveal a unique binding mode for specific single-stranded N6‑methyladenosine RNA demethylation. J. Biol. Chem. 2014;289:17299–17311. doi: 10.1074/jbc.M114.550350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tang C., Klukovich R., Peng H., Wang Z., Yu T., Zhang Y., Zheng H., Klungland A., Yan W. ALKBH5-dependent m6A demethylation controls splicing and stability of long 3′-UTR mRNAs in male germ cells. Proc. Natl. Acad. Sci. 2017 doi: 10.1073/pnas.1717794115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang C., Samanta D., Lu H., Bullen J.W., Zhang H., Chen I., He X., Semenza G.L. Hypoxia induces the breast cancer stem cell phenotype by HIF-dependent and ALKBH5-mediated m6A‑demethylation of NANOG mRNA. Proc. Natl. Acad. Sci. 2016;113:E2047–E2056. doi: 10.1073/pnas.1602883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang C., Zhi W.I., Lu H., Samanta D., Chen I., Gabrielson E., Semenza G.L. Hypoxia-inducible factors regulate pluripotency factor expression by ZNF217- and ALKBH5-mediated modulation of RNA methylation in breast cancer cells. Oncotarget. 2016;7 doi: 10.18632/oncotarget.11743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang S., Zhao B.S., Zhou A., Lin K., Zheng S., Lu Z., Chen Y., Sulman E.P., Xie K., Bögler O., Majumder S., He C., Huang S. m6A demethylase ALKBH5 maintains tumorigenicity of glioblastoma stem-like cells by sustaining FOXM1 expression and cell proliferation program. Cancer Cell. 2017;31:591–606.e6. doi: 10.1016/j.ccell.2017.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Huang Y., Yan J., Li Q., Li J., Gong S., Zhou H., Gan J., Jiang H., Jia G.-F., Luo C., Yang C.-G. Meclofenamic acid selectively inhibits FTO demethylation of m6A over ALKBH5. Nucleic Acids Res. 2015;43:373–384. doi: 10.1093/nar/gku1276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhao X., Yang Y., Sun B.-F., Shi Y., Yang X., Xiao W., Hao Y.-J., Ping X.-L., Chen Y.-S., Wang W.-J., Jin K.-X., Wang X., Huang C.-M., Fu Y., Ge X.-M., Song S.-H., Jeong H.S., Yanagisawa H., Niu Y., Jia G.-F., Wu W., Tong W.-M., Okamoto A., He C., Danielsen J.M.R., Wang X.-J., Yang Y.-G. FTO-dependent demethylation of N6‑methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Meyer K.D., Jaffrey S.R. Rethinking m6A readers, writers, and erasers. Annu. Rev. Cell Dev. Biol. 2017;33:319–342. doi: 10.1146/annurev-cellbio-100616-060758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Alarcón C.R., Goodarzi H., Lee H., Liu X., Tavazoie S., Tavazoie S.F. HNRNPA2B1 is a mediator of m(6)A-dependent nuclear RNA processing events. Cell. 2015;162:1299–1308. doi: 10.1016/j.cell.2015.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Liu N., Dai Q., Zheng G., He C., Parisien M., Pan T. N(6)‑methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han S.P., Tang Y.H., Smith R. Functional diversity of the hnRNPs: past, present and perspectives. Biochem. J. 2010;430:379–392. doi: 10.1042/BJ20100396. [DOI] [PubMed] [Google Scholar]
- 53.Berulava T., Rahmann S., Rademacher K., Klein-Hitpass L., Horsthemke B. N6‑adenosine methylation in MiRNAs. PLoS One. 2015;10 doi: 10.1371/journal.pone.0118438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Alarcón C.R., Lee H., Goodarzi H., Halberg N., Tavazoie S.F. N6‑methyladenosine marks primary microRNAs for processing. Nature. 2015;519:482–485. doi: 10.1038/nature14281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Edupuganti R.R., Geiger S., Lindeboom R.G.H., Shi H., Hsu P.J., Lu Z., Wang S.Y., Baltissen M.P.A., Jansen P.W.T.C., Rossa M., Müller M., Stunnenberg H.G., He C., Carell T., Vermeulen M. N6‑methyladenosine (m6A) recruits and repels proteins to regulate mRNA homeostasis. Nat. Struct. Mol. Biol. 2017 doi: 10.1038/nsmb.3462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xu C., Liu K., Ahmed H., Loppnau P., Schapira M., Min J. Structural basis for the discriminative recognition of N6‑methyladenosine RNA by the human YT521-B homology domain family of proteins. J. Biol. Chem. 2015;290:24902–24913. doi: 10.1074/jbc.M115.680389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li F., Zhao D., Wu J., Shi Y. Structure of the YTH domain of human YTHDF2 in complex with an m6A mononucleotide reveals an aromatic cage for m6A recognition. Cell Res. 2014;24:1490–1492. doi: 10.1038/cr.2014.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang Z., Theler D., Kaminska K.H., Hiller M., De La Grange P., Pudimat R., Rafalska I., Heinrich B., Bujnick J.M., Allain F.H.T., Stamm S. The YTH domain is a novel RNA binding domain. J. Biol. Chem. 2010;285:14701–14710. doi: 10.1074/jbc.M110.104711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xu C., Wang X., Liu K., Roundtree I.A., Tempel W., Li Y., Lu Z., He C., Min J. Structural basis for selective binding of m6A RNA by the YTHDC1 YTH domain. Nat. Chem. Biol. 2014;10:927–929. doi: 10.1038/nchembio.1654. [DOI] [PubMed] [Google Scholar]
- 60.Roundtree I.A., Evans M.E., Pan T., He C. Dynamic RNA modifications in gene expression regulation. Cell. 2017;169:1187–1200. doi: 10.1016/j.cell.2017.05.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wang X., Zhao B.S., Roundtree I.A., Lu Z., Han D., Ma H., Weng X., Chen K., Shi H., He C. N6‑methyladenosine modulates messenger RNA translation efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee K.-M., Tarn W.-Y. TRAP150 activates splicing in composite terminal exons. Nucleic Acids Res. 2014;42:12822–12832. doi: 10.1093/nar/gku963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou J., Wan J., Gao X., Zhang X., Jaffrey S.R., Qian S.-B. Dynamic m(6)A mRNA methylation directs translational control of heat shock response. Nature. 2015 doi: 10.1038/nature15377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Shi H., Wang X., Lu Z., Zhao B.S., Ma H., Hsu P.J., He C. YTHDF3 facilitates translation and decay of N6‑methyladenosine-modified RNA. Cell Res. 2017;27:315–328. doi: 10.1038/cr.2017.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hartmann A.M., Nayler O., Schwaiger F.W., Obermeier A., Stamm S. The interaction and colocalization of Sam68 with the splicing-associated factor YT521-B in nuclear dots is regulated by the Src family kinase p59(fyn) Mol. Biol. Cell. 1999;10:3909–3926. doi: 10.1091/mbc.10.11.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Xiao W., Adhikari S., Dahal U., Chen Y.-S., Hao Y.-J., Sun B.-F., Sun H.-Y., Li A., Ping X.-L., Lai W.-Y., Wang X., Ma H.-L., Huang C.-M., Yang Y., Huang N., Jiang G.-B., Wang H.-L., Zhou Q., Wang X.-J., Zhao Y.-L., Yang Y.-G. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Mol. Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 67.Roundtree I.A., He C. Nuclear m6A reader YTHDC1 regulates mRNA splicing. Trends Genet. 2016;32:320–321. doi: 10.1016/j.tig.2016.03.006. [DOI] [PubMed] [Google Scholar]
- 68.Rafalska I., Zhang Z., Benderska N., Wolff H., Hartmann A.M., Brack-Werner R., Stamm S. The intranuclear localization and function of YT521-B is regulated by tyrosine phosphorylation. Hum. Mol. Genet. 2004;13:1535–1549. doi: 10.1093/hmg/ddh167. [DOI] [PubMed] [Google Scholar]
- 69.Nayler O., Hartmann A.M., Stamm S. The ER repeat protein YT521-B localizes to a novel subnuclear compartment. J. Cell Biol. 2000;150:949–961. doi: 10.1083/jcb.150.5.949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Roundtree I.A., Luo G.-Z., Zhang Z., Wang X., Zhou T., Cui Y., Sha J., Huang X., Guerrero L., Xie P., He E., Shen B., He C. YTHDC1 mediates nuclear export of N6‑methyladenosine methylated mRNAs. elife. 2017;6 doi: 10.7554/eLife.31311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Reed R., Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. http://www.sciencedirect.com/science/article/pii/S009286740200627X [DOI] [PubMed] [Google Scholar]
- 72.Cheng H., Dufu K., Lee C.-S., Hsu J., Dias A., Reed R. Human mRNA export machinery recruited to the 5′ end of mRNA. Cell. 2006;127:1389–1400. doi: 10.1016/j.cell.2006.10.044. [DOI] [PubMed] [Google Scholar]
- 73.Luo M.L., Zhou Z., Magni K., Christoforides C., Rappsilber J., Mann M., Reed R. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413:644–647. doi: 10.1038/35098106. [DOI] [PubMed] [Google Scholar]
- 74.Gromadzka A.M., Steckelberg A.L., Singh K.K., Hofmann K., Gehring N.H. A short conserved motif in ALYREF directs cap- and EJC-dependent assembly of export complexes on spliced mRNAs. Nucleic Acids Res. 2016;44:2348–2361. doi: 10.1093/nar/gkw009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Libri D., Dower K., Boulay J., Thomsen R., Rosbash M., Jensen T.H. Interactions between mRNA export commitment, 3′-end quality control, and nuclear degradation. Mol. Cell. Biol. 2002;22:8254–8266. doi: 10.1128/MCB.22.23.8254-8266.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Reed R., Hurt E. A conserved mRNA export machinery coupled to pre-mRNA splicing. Cell. 2002;108:523–531. doi: 10.1016/s0092-8674(02)00627-x. [DOI] [PubMed] [Google Scholar]
- 77.Strässer K., Masuda S., Mason P., Pfannstiel J., Oppizzi M., Rodriguez-Navarro S., Rondón A., Aguilera A., Struhl K., Reed R., Hurt E. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417:304–308. doi: 10.1038/nature746. [DOI] [PubMed] [Google Scholar]
- 78.Masuda S., Das R., Cheng H., Hurt E., Dorman N., Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19:1512–1517. doi: 10.1101/gad.1302205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chi B., Wang Q., Wu G., Tan M., Wang L., Shi M., Chang X., Cheng H. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013;41:1294–1306. doi: 10.1093/nar/gks1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Hautbergue G.M., Hung M.-L.L., Walsh M.J., Snijders A.P.L., Te Chang C.-T., Jones R., Ponting C.P., Dickman M.J., Wilson S.A. UIF, a new mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr. Biol. 2009;19:1918–1924. doi: 10.1016/j.cub.2009.09.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Viphakone N., Cumberbatch M.G., Livingstone M.J., Heath P.R., Dickman M.J., Catto J.W., Wilson S.A. Luzp4 defines a new mRNA export pathway in cancer cells. Nucleic Acids Res. 2015;43:2353–2366. doi: 10.1093/nar/gkv070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Chang C.-T., Hautbergue G.M., Viphakone N., Dijk T.B., Philipsen S., Wilson S.A., van Dijk T.B., Chang C.-T., Hautbergue G.M., Viphakone N., Dijk T.B., Philipsen S., Wilson S.A., Dijk T. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013;32:473–486. doi: 10.1038/emboj.2012.342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hautbergue G., Hung M., Golovanov A., Lian L., Wilson S. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc. Natl. Acad. Sci. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Müller-McNicoll M., Botti V., de Jesus Domingues A.M., Brandl H., Schwich O.D., Steiner M.C., Curk T., Poser I., Zarnack K., Neugebauer K.M. SR proteins are NXF1 adaptors that link alternative RNA processing to mRNA export. Genes Dev. 2016;30:553–566. doi: 10.1101/gad.276477.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Katahira J., Yoneda Y., Katahira J., Yoneda Y. Roles of the TREX complex in nuclear export of mRNA. RNA Biol. 2009;6:149–152. doi: 10.4161/rna.6.2.8046. http://eutils.ncbi.nlm.nih.gov/entrez/eutils/elink.fcgi?dbfrom=pubmed&id=19229134&retmode=ref&cmd=prlinks [DOI] [PubMed] [Google Scholar]
- 86.Liker E., Fernandez E., Izaurralde E., Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zolotukhin A.S., Tan W., Bear J., Smulevitch S., Felber B.K. U2AF participates in the binding of TAP (NXF1) to mRNA. J. Biol. Chem. 2002;277:3935–3942. doi: 10.1074/jbc.M107598200. [DOI] [PubMed] [Google Scholar]
- 88.Fribourg S., Braun I.C., Izaurralde E., Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol. Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 89.Braun I.C., Herold A., Rode M., Conti E., Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J. Biol. Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 90.Yang X., Yang Y., Sun B.F., Chen Y.S., Xu J.W., Lai W.Y., Li A., Wang X., Bhattarai D.P., Xiao W., Sun H.Y., Zhu Q., Ma H.L., Adhikari S., Sun M., Hao Y.J., Zhang B., Huang C.M., Huang N., Bin Jiang G., Zhao Y.L., Wang H.L., Sun Y.P., Yang Y.G. 5‑Methylcytosine promotes mRNA export-NSUN2 as the methyltransferase and ALYREF as an m5C reader. Cell Res. 2017;27:606–625. doi: 10.1038/cr.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Finkel D., Groner Y. Methylations of adenosine residues (m6A) in pre-mRNA are important for formation of late simian virus 40 mRNAs. Virology. 1983;131:409–425. doi: 10.1016/0042-6822(83)90508-1. [DOI] [PubMed] [Google Scholar]
- 92.Lichinchi G., Zhao B.S., Wu Y., Lu Z., Qin Y., He C., Rana T.M. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe. 2016;20:666–673. doi: 10.1016/j.chom.2016.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kennedy E.M., Bogerd H.P., Kornepati A.V.R., Kang D., Ghoshal D., Marshall J.B., Poling B.C., Tsai K., Gokhale N.S., Horner S.M., Cullen B.R. Posttranscriptional m6A editing of HIV-1 mRNAs enhances viral gene expression. Cell Host Microbe. 2016;19:675–685. doi: 10.1016/j.chom.2016.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Camper S.A., Albers R.J., Coward J.K., Rottman F.M. Effect of undermethylation on mRNA cytoplasmic appearance and half-life. Mol. Cell. Biol. 1984;4:538–543. doi: 10.1128/mcb.4.3.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Varia S., Potabathula D., Deng Z., Bubulya A., Bubulya P. a. Btf and TRAP150 have distinct roles in regulating subcellular mRNA distribution. Nucleus. 2013;4:229–240. doi: 10.4161/nucl.25187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Vohhodina J., Barros E.M., Savage A.L., Liberante F.G., Manti L., Bankhead P., Cosgrove N., Madden A.F., Harkin D.P., Savage K.I. The RNA processing factors THRAP3 and BCLAF1 promote the DNA damage response through selective mRNA splicing and nuclear export. Nucleic Acids Res. 2017;45:12816–12833. doi: 10.1093/nar/gkx1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hein M.Y., Hubner N.C., Poser I., Cox J., Nagaraj N., Toyoda Y., Gak I.A., Weisswange I., Mansfeld J., Buchholz F., Hyman A.A., Mann M. A human Interactome in three quantitative dimensions organized by stoichiometries and abundances. Cell. 2015;163:712–723. doi: 10.1016/j.cell.2015.09.053. [DOI] [PubMed] [Google Scholar]
- 98.Lindtner S., Zolotukhin A.S., Uranishi H., Bear J., Kulkarni V., Smulevitch S., Samiotaki M., Panayotou G., Felber B.K., Pavlakis G.N. RNA-binding motif protein 15 binds to the RNA transport element RTE and provides a direct link to the NXF1 export pathway. J. Biol. Chem. 2006;281:36915–36928. doi: 10.1074/jbc.M608745200. [DOI] [PubMed] [Google Scholar]
- 99.Zolotukhin A.S., Uranishi H., Lindtner S., Bear J., Pavlakis G.N., Felber B.K. Nuclear export factor RBM15 facilitates the access of DBP5 to mRNA. Nucleic Acids Res. 2009;37:7151–7162. doi: 10.1093/nar/gkp782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Uranishi H., Zolotukhin A.S., Lindtner S., Warming S., Zhang G.M., Bear J., Copeland N.G., Jenkins N.A., Pavlakis G.N., Felber B.K. The RNA-binding motif protein 15B (RBM15B/OTT3) acts as cofactor of the nuclear export receptor NXF1. J. Biol. Chem. 2009;284:26106–26116. doi: 10.1074/jbc.M109.040113. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Transparency document.