Abstract
The integrative mobilizable elements of SGI1-family considerably contribute to the spread of resistance to critically important antibiotics among enteric bacteria. Even though many aspects of SGI1 mobilization by IncA and IncC plasmids have been explored, the basic transfer elements such as oriT and self-encoded mobilization proteins remain undiscovered. Here we describe the mobilization region of SGI1 that is well conserved throughout the family and carries the oriTSGI1 and two genes, mpsA and mpsB (originally annotated as S020 and S019, respectively) that are essential for the conjugative transfer of SGI1. OriTSGI1, which is located in the vicinity of the two mobilization genes proved to be a 125-bp GC-rich sequence with several important inverted repeat motifs. The mobilization proteins MpsA and MpsB are expressed from a bicistronic mRNA, although MpsB can be produced from its own mRNA as well. The protein structure predictions imply that MpsA belongs to the lambda tyrosine recombinase family, while MpsB resembles the N-terminal core DNA binding domains of these enzymes. The results suggest that MpsA may act as an atypical relaxase, which needs MpsB for SGI1 transfer. Although the helper plasmid-encoded relaxase proved not to be essential for SGI1 transfer, it appeared to be important to achieve the high transfer rate of the island observed with the IncA/IncC-SGI1 system.
Keywords: salmonella genomic island 1, integrative mobilizable element, IncA/C plasmids, origin of transfer (oriT), horizontal gene transfer, mobile genetic element (MGE), antibiotic resistance (AR)
Introduction
Conjugation is a widespread mechanism of horizontal gene transfer among naturally occurring plasmids and genomic islands. During conjugation, DNA is transferred from donor to recipient through a close cell-to-cell contact. Based on Gram-negative plasmid models, the process starts with the assembly of a multi-protein complex called the relaxosome around the origin of transfer (oriT). OriT is a cis-acting DNA region that is required for initiation of the transfer. The key enzyme of transfer initiation is the relaxase, which cuts either strand of oriT DNA at the nic site. The relaxase remains covalently bound to the 5′-end of the cleaved strand, which is subsequently delivered to the recipient across the membrane-associated DNA transport machinery, the type IV secretion system (T4SS). Initiation of conjugation often requires several auxiliary proteins implicated in the relaxosome (De La Cruz et al., 2010; Bellanger et al., 2014). The relaxosome is then recruited to the T4SS with the assistance of the membrane-associated coupling proteins (T4CP), which binds the cognate T4SS and the relaxosome complex through the relaxase or auxiliary proteins (Llosa and Alkorta, 2017). Based on phylogenetic analyses of relaxases, conjugative systems have been classified into eight major MOB families, however, numerous unclassified systems have also been reported (Garcillán-Barcia et al., 2009; Bellanger et al., 2014; Li et al., 2018). Some of these families include plasmids and mobile genomic islands (MGIs) as well (e.g., MOBH, MOBC, MOBP, MOBV, MOBT), indicating the relationship of the conjugative systems of the two groups of mobile elements.
In addition to resistance plasmids, MGIs are the major players in dissemination of multidrug resistance (MDR) phenotype among bacteria. MGIs provide selective advantages to their host by carrying resistance-, pathogenicity-, metabolic pathway- or symbiosis-related genes. After acquisition, MGIs integrate into the host chromosome by site-specific-, transposition- or homologous recombination to ensure their maintenance and vertical transmission (Bellanger et al., 2014). MGIs are classified into two major groups: integrative conjugative elements (ICEs, some of them were formerly known as conjugative transposons, (Liu et al., 2019) and integrative mobilizable elements (IMEs). In addition to the chromosomal excision/integration ability, ICEs encode for their own conjugation system and are fully autonomous in horizontal transfer (Carraro and Burrus, 2014), while IMEs have a limited set of transfer genes and thus require the presence of other conjugative helper elements to hijack their transfer systems (Douard et al., 2010, Daccord et al., 2013; Bellanger et al., 2014).
Many studies about MGI mobilization unravel their integration and excision reactions. However, the mechanism of their conjugation is less investigated, and their transfer genes are mostly identified by their similarities to those of conjugative plasmids. Like all conjugative and mobilizable plasmids, MGIs also carry their own oriT sequence (Bellanger et al., 2014). Most oriT sequences have been identified in conjugative plasmids, where they are often located adjacent to genes of the relaxosome components (e.g., in RP4, R388, R6K, pCW3, pIP501). Although conserved sequence motifs have been found in oriTs close to the nic site of IncPα, Ti, Ri, R64 and pTF-FC2 plasmids (Pansegrau and Lanka, 1991), oriTs are generally diverse sequences [oriTDB (Li et al., 2018)], which frequently contain inverted repeat (IR) motifs and AT-rich regions [F (Frost et al., 1994), R6K (Avila et al., 1996), IncPα (Fürste et al., 1989),(Pansegrau et al., 1994), pAD1 (Francia and Clewell, 2002), pAM373 (Francia and Clewell, 2002), pCW3 (Wisniewski et al., 2016) and IncA and IncC plasmids (Hegyi et al., 2017)]. Transfer systems of several ICEs have also been analyzed in details and the oriT has been identified in Tn916 (Jaworski and Clewell, 1995), Tn4445 (Smith and Parker, 1998), ICEBs1 (Lee and Grossman, 2007), SXT/R391 (Ceccarelli et al., 2008), ICEclc (Miyazaki and Van Der Meer, 2011), ICEhptfs4 (formerly referred as tfs4) (Grove et al., 2013) and the related IMEs mobilized by SXT (Daccord et al., 2010, 2013; Li et al., 2018). Although all oriTs contain several IR motifs, no extensive sequence similarities can be observed between oriTs of unrelated elements. In contrast, IMEs hijacking the same helper elements have similar oriTs to each other and to that of the helper (Daccord et al., 2010). Most conjugative plasmids and MGIs have a single oriT, however, in some cases two separate and functional oriTs have been identified [pAD1 (Francia et al., 2001; Francia and Clewell, 2002), R6K (Avila et al., 1996), ICEclc (Miyazaki and Van Der Meer, 2011)].
Among the few well-studied families of MGIs, the Salmonella genomic island 1 (SGI1) is one of the largest and most diverse IME family, which considerably contribute to spreading MDR and especially the resistance to critically important antibiotics such as extended-spectrum β-lactams or carbapenems. The prototype of SGI1 has been described in a multidrug resistant clone of Salmonella enterica serovar Typhimurium DT104 (Boyd et al., 2000), which emerged during the mid-1980s (Threlfall et al., 1994) and spread worldwide. The common multidrug resistance phenotype (ACSSuT) of this epidemic clone is conferred by the 42.4-kb SGI1, which contains 44 predicted open reading frames (ORFs) and carries a ca. 15 kb complex In4-type integron structure called In104 (Figure 1A). In104 is inserted near the 3′-end of the SGI1 backbone (Boyd et al., 2001) and flanked by 25-bp imperfect inverted repeats. In the prototype SGI1 this gene cluster contains two class 1 integron structures with gene cassettes aadA2 and blaCARB-2 (blaPSE-1), respectively, other resistance genes (tetA(G) and floR) occurring independently of the integrons, IS elements (ISCR3, IS6100) and some additional genes of unknown function. Loss, gain or exchange of antibiotic resistance genes have occurred mainly in In104 by exchanges of resistance gene cassettes, homologous recombinations or IS-induced rearrangements, which led to the emergence of known SGI1 variants (SGI1-A to SGI1-Z12, SGI2) identified in numerous S. enterica serovars, Proteus mirabilis strains and Morganella morganii (Levings et al., 2005, 2008; Doublet et al., 2008; Siebor and Neuwirth, 2011, 2014; Schultz et al., 2017). Recently, SGI1-related elements have been described in several other species such as PGI1 and PGI2 in P. mirabilis and AGI1 in Acinetobacter baumannii (Siebor and Neuwirth, 2014; Hamidian et al., 2015). Other uncharacterized SGI1-related elements are found in Aeromonas veronii, Vibrio cholerae (Johnson et al., 2015), Vibrio mimicus, Shewanella and Enterobacter spp. (Supplementary Figure S1).
FIGURE 1.
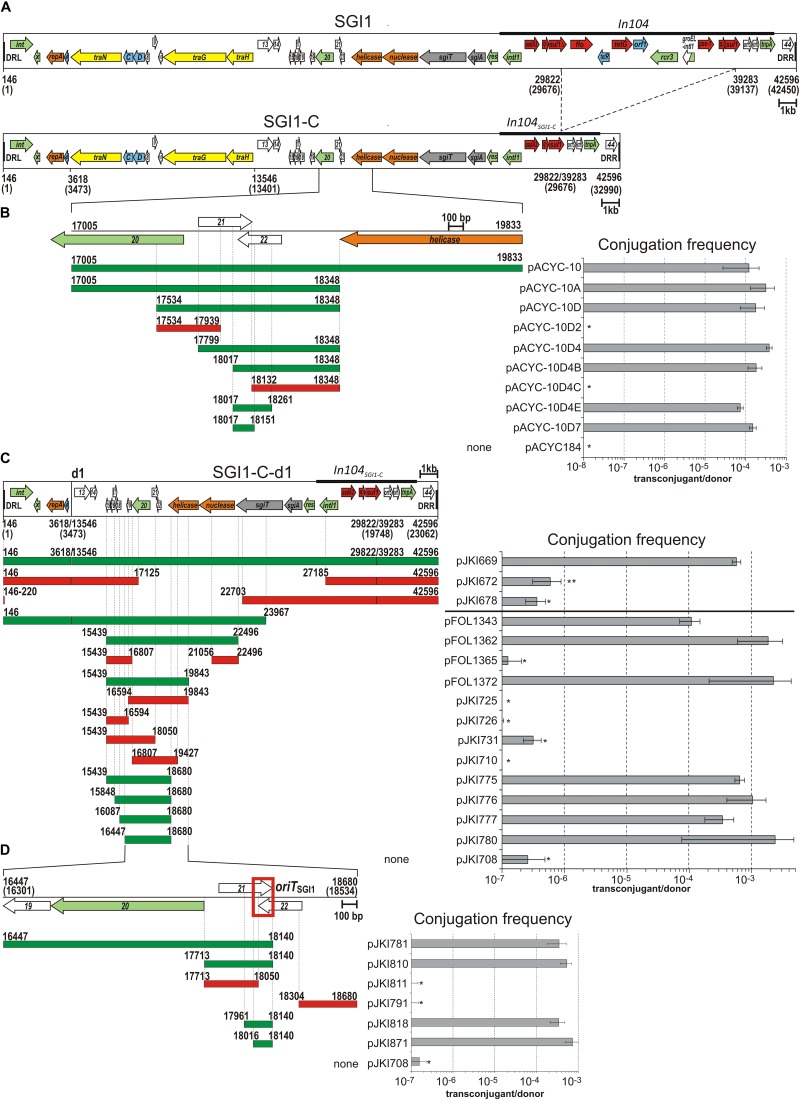
Identification of oriTSGI1. (A) Schematic map of SGI1 and SGI1-C. The annotated ORFs designated originally as S001-S044 are indicated by colored arrows: green – recombination, transposase; orange – replication; blue – transcription regulator; yellow – conjugation; gray – TA system; red – antibiotics resistance gene; white – unknown function. Names of genes with known function or identified homologs are indicated. Abbreviations: x - xis, C and D - flhDCSGI1, sgiT and sgiA – toxin and antitoxin gene of SGI1 TA system, qΔ – qacEΔ1, genes of unknown function are numbered according to their original numbering (e.g., “4” refers to S004, etc). Terminal direct repeats are shown as black boxes, In104 and its deletion derivative in SGI1-C are indicated. Coordinates are according to the published SGI1 sequence (AF261825) and refer to the ends of SGI1 (including DRs) and the endpoints of deletions led to the formation of SGI1-C and SGI1-C-d1 variants. The real coordinates of SGI1, SGI1-C and SGI1-C-d1 (taken into account of the missing A base from our SGI1 sequence at the 16338 bp position of AF261825) are indicated in brackets below the maps of SGI1 variants (5′-end of DRL was designated as the first position of SGI1). All maps are drawn to scale. (B) Identification of oriTSGI1 using the S. Agona 47SA97 (harboring R55 helper and the chromosomally integrated SGI1-CΔint) donor and the E. coli BM14 recipient strains for mating. The color-coded horizontal bars with coordinates represent the cloned SGI1 regions in the pACYC184-derivative plasmids: green – mobilizable; red – not mobilizable. The graph shows the transfer frequency of the plasmids. Frequency data were calculated as transconjugant/donor titres in each figure. Asterisk indicates that the transfer frequency was below the detection limit ( < 10-8/donor). (C) Identification of mobSGI1 region in SGI1-C-d1 variant. Conjugation frequency of SGI1-C-d1 variant carried by a p15A based vector and its p15A-based subclones was measured in the presence of the helper plasmid R55, but without chromosomal SGI1. Horizontal bars indicate the SGI1-derived sequences present in the test plasmids. For mobilization of the constructs carrying the SmR/SpR marker of SGI1-C-d1, TG1 donor and TG1Nal recipient, while for others, TG1Nal donor and TG2 recipient strains were used (separated by horizontal line). The asterisk indicates that the transfer frequency was close to or below the detection limits (<10-7/donor). The rare transconjugants obtained sporadically carried also the helper plasmid (co-transfer of the two plasmids was ∼100 %) suggesting an oriTSGI1-independent transfer mechanism (e.g., conduction, transfer of plasmids by cointegration with the helper). ∗∗In case of pJKI672, the NalRSmR colonies obtained with a frequency of ∼6 × 10-7 did not contain the transferred plasmid, thus were not real transconjugants. Other symbols are as in panels (A) and (B). (D) Identification of oriTSGI1 using mobSGI1-donor (harboring chromosomally integrated mobSGI1 and R55 helper) and TG2 recipient E. coli strains. The minimal fully mobilizable fragment designated as oriTSGI1 is indicated by red box, other symbols are as in panel (B).
The SGI1-family islands are typical IMEs, as they integrate autonomously into or excise from the bacterial chromosome specifically at the 3′-end of trmE gene (formerly thdF and also called mnmE), but they do not encode all the genes necessary for their self-transfer, and require the conjugation system of plasmids of the incompatibility groups A or C for their horizontal transfer (Doublet et al., 2005, Daccord et al., 2010). The IncC family with hundreds of fully sequenced plasmids includes broad host range, single-copy conjugative plasmids implicated in MDR dissemination, while IncA group contains only one sequenced member, RA1, which has a backbone closely related to that of IncC plasmids, but proved to be compatible with them (Datta and Hedges, 1973; Ambrose et al., 2018). Their conjugative system, which is related to that of SXT/R391 ICEs (Wozniak et al., 2009; Poulin-Laprade et al., 2015), has been classified into the MOBH12 group (Garcillán-Barcia et al., 2009). SGI1 is mobilized by IncA and IncC (hereafter are referred as IncA/C) plasmids such as RA1, R55, R16a, IP40a, pVCR94 or pRMH760 (Douard et al., 2010; Kiss et al., 2012; Carraro et al., 2014; Harmer et al., 2016), however, other unrelated IMEs (e.g., MGIVmi1 of V. mimicus) have also been reported to utilize these plasmids as mobilization helpers (Carraro et al., 2014, 2016). Expression of the conjugation apparatus of IncA/C plasmids are controlled by the FlhDC-family master activator, AcaCD (Carraro et al., 2014), which is required for the transfer of both the plasmid and the mobilized IMEs (Carraro et al., 2014; Kiss et al., 2015). The plasmid-encoded AcaCD triggers the excision of SGI1 through the activation of expression of the SGI1-encoded recombination directionality factor Xis (Carraro et al., 2014; Kiss et al., 2015), but has no effect on the expression of the site-specific recombinase Int (Kiss et al., 2015) required for both the excision and integration (Doublet et al., 2005). After SGI1 transfer, the island efficiently integrates into the attB site in trmE.
Recent studies have indicated a more complex crosstalk between SGI1 and IncA/C plasmids. In addition to the xis promoter, four other AcaCD-responsive promoters have been identified on SGI1 (Carraro et al., 2014; Murányi et al., 2016). Moreover, the island also encodes its own functional FlhDC-family regulator, FlhDCSGI1 (Kiss et al., 2015). AcaCD and the closely related FlhDCSGI1 can activate the AcaCD-responsive promoters on both SGI1 and the conjugation regulon of IncA/C plasmids (Murányi et al., 2016). The SGI1-encoded transfer proteins TraN, TraG, and TraH, whose expression is also under AcaCD-control, interact with the plasmid encoded homologs and promote SGI1 transfer at the expense of the helper plasmid (Carraro et al., 2017). Furthermore, SGI1 and IncA/C plasmids encode for incompatibility functions impeding stable co-habitation of the partners in the same host (Harmer et al., 2016; Huguet et al., 2016). Destabilization of SGI1 by IncA/C plasmids seems to be based on triggering of SGI1 excision by AcaCD (Kiss et al., 2015), however, the similar effect of SGI1 on the helper plasmids appear more complex and poorly understood (Harmer et al., 2016).
Even though many aspects of the SGI1-IncA/C dual system have been studied, the basic transfer functions of SGI1, i.e., the oriT and the putative self-encoded mobilization proteins remain undiscovered. The three SGI1-encoded tra genes, traN, traG and traH, have an important but not essential role in the transfer of the island (Carraro et al., 2017; Kiss et al., 2012). Other ORFs of SGI1 do not show sequence similarity to known tra genes and relaxases, including that of IncA/C plasmids, thus, the implication of an atypical relaxase in SGI1 transfer can not be excluded. Similarly, oriT of SGI1 can not be identified on the basis of sequence homology with oriT of IncA/C plasmids (Hegyi et al., 2017), despite the fact that the plasmid-encoded relaxase is required for efficient SGI1 transfer. In the present work, we identify the mobilization region of SGI1 carrying the cis-acting oriT sequence and two predicted ORFs encoding proteins that are essential for the conjugative transfer of the island. The minimal oriT sequence, oriTSGI1, has been characterized, and the importance of its inverted repeat (IR) motifs for the transfer was examined. Comparative analysis of SGI1-related elements confirmed that the mobSGI1 region is well conserved throughout the family, strengthening its importance in the horizontal transfer of SGI1. The predicted structure and possible function of the two transfer proteins identified have also been discussed.
Materials and Methods
Microbial Techniques and DNA Procedures
Relevant features of the bacterial strains and plasmids are listed in Supplementary Tables S1, S2, respectively. Bacterial strains were maintained at -80°C in LB broth containing 30% glycerol and were routinely grown in Luria-Bertani (LB) broth at 37°C supplemented with the appropriate antibiotics used at a final concentration as follows: ampicillin (Ap) 150 μg/ml, chloramphenicol (Cm) 20 μg/ml, kanamycin (Km) 30 μg/ml, spectinomycin (Sp) 50 μg/ml, streptomycin (Sm) 50 μg/ml, nalidixic acid (Nal) 20 μg/ml, gentamicin (Gm) 25 μg/ml, tetracycline (Tc) 10 μg/ml, sodium azide (Az) 500 μg/ml. Standard molecular biology procedures were carried out according to Sambrook et al. (1989). Detailed methodology of plasmid constructions is described in Supplementary Methods. Test/colony PCRs were performed using Dream Taq polymerase (Thermo Fisher Scientific) as previously described (Kiss et al., 2012). The amplicons for cloning were amplified with Phusion (Thermo Fisher Scientific) or Pwo (Roche) polymerases and sequenced on ABI Prism 3100 Genetic Analyzer (PerkinElmer). Oligonucleotide primers used in this work are listed in Supplementary Table S3. Primers annealing to SGI1 or R55 were designed according to the published sequence of SGI1 (GenBank: AF261825) and R55 (GenBank: JQ010984).
The prototype SGI1 carries five functional resistance genes, which makes difficult to set up mating or KO mutagenesis experiments, thus for mating assays, cloning or mutagenesis we used the SmR/SpR, SulR derivative, SGI1-C (Figure 1A), which was previously shown to have the same mobilization properties as SGI1 (Kiss et al., 2012).
The β-galactosidase assay was performed in five replicates according to Miller (1972) except that the cultures were grown at 37°C to an OD600 ∼0.3 in LB both and diluted at a ratio of 1:1 with Z buffer.
Primer extension reactions were carried out as described (Murányi et al., 2016) using the β-gal tester plasmids pMSZ1017 and pMSZ948 carrying the upstream regions of S019 and S020, respectively.
Conjugation Assays
Deletion derivatives of SGI1-C-d1 in pJKI669 were generated by enzymatic digestion and re-ligation and introduced into Escherichia coli TG1/R55. Other smaller SGI1 backbone fragments were amplified by PCR, cloned into pJKI708 vector and transformed into TG1Nal/R55 strain. The resulting TG1 or TG1Nal donor strains harboring one of the test plasmids along with R55 helper were then used in standard mating assays with TG1Nal or TG2 recipients, respectively, as described previously (Kiss et al., 2012).
Mating with S. Agona donor strains were performed by mixing mid-log phase (OD600 = 0.5) cultures of the donor strains harboring SGI1 and R55 w/o pACYC184-derivative test plasmids and the sodium azide-resistant E. coli recipient strain BM14 in a ratio of 1:1. The mix was incubated overnight at 37°C without shaking and then cells were streaked on appropriate selective SS agar plates. Transconjugants were selected on sodium azide combined with antibiotics depending on the transferred element: SGI1 – Sm, R55 – Km and the pACYC184 derivatives – Tc. The transfer frequencies were calculated as the ratio of transconjugant and donor titers. Complementation assays for identification of promoter regions of S020 and S019 were carried out as described (Kiss et al., 2012), except that mating was done with TG90 recipient in 2YT plates for 6 h to reduce the growth of the donor over the recipient cells.
IncC plasmid R16a was used as helper plasmid in some mating assays due to its more suitable resistance markers. It has previously been shown to be as effective mobilizer of SGI1 as R55 (Douard et al., 2010). In mobilization tests of the S019 and S020 KO (Knock-Out) mutant or WT mobSGI1-containing plasmids (pJKI772, pJKI737, pFOL1372) or SGI1-C by R16aWT or R16aΔTraI helper plasmids, the E. coli donor strains TG1Nal or TG1Nal::SGI1-C were used with TG90 recipient. Mobilization of SGI1-CΔoriT and pMNI41 by R55ΔTn6187 helper plasmid was assayed by mating of E. coli TG1Nal donor and E. coli TG90 recipient starins. In case of low frequency transfer, rare transconjugants were detected by spreading 100 μl (instead of dropping 5 μl) of undiluted bacterial suspension obtained from the mating LB plates onto the appropriate selective plates.
Targeted Gene KO Experiments
The PCR fragments for KO mutagenesis of S019, S020, S022 and oriTSGI1 were amplified from pKD3 template plasmid using primers delS019for-delS019rev, delS020for-delS020rev, delS022for-delS022rev, and deloriTfor-deloriTrev, respectively, (Supplementary Table S3). For promoting the gene replacement recombination between the targeted region and the respective KO PCR fragment, λ Red recombinase was expressed from the plasmid pKD46 or its TcR derivative plasmid pJKI842 using 1% L-arabinose as inductor at 30°C for 1.5 h. The CmR cassette was removed from the chromosomal KO alleles by expressing the FLP recombinase from the thermo-inducible expression plasmid pCP20 (Datsenko and Wanner, 2000), or by digestion with XbaI (present in FRT sites) followed by religation in the case of pFOL1372 derivatives. The plasmids having temperature-sensitive pSC101 replication system were maintained and cured at 30 and 42°C, respectively. In the resulting KO mutants, a short region was replaced with 83-bp sequence deriving from the PCR template plasmid pKD3. The replacements near the 5′-end of S019, S020, and S022 generated early stop codons in the ORFs.
The KO PCR fragment for the scarless deletion (Kolisnychenko et al., 2002) of Tn6187 in R55 was amplified from pJKI1023 template plasmid using primers R55-dTn6187ABfor- R55-dTn6187Crev. After the λ Red-induced recombination/gene replacement, the SmR marker gene was eliminated from the resulting R55ΔTn6187::SmR plasmid by DSB-stimulated recombinational repair process induced by I-SceI cleavage. Expression of I-SceI from pMSZ934 was induced by 30 μg/ml chlortetracycline (cTc) O/N at 30°C in LB+Cm+Ap. The SmS clones were selected by replica plating and the scarless site was amplified and sequenced. To demonstrate that deletion of Tn6187 did not affect the conjugation properties of R55, mating assay was conducted with TG1Nal donor and TG90Nal (TcR) recipient strains, where R55 and its ΔTn6187 derivative showed similar transfer frequencies (2.9 ± 1.0 × 10-1 and 2.5 ± 0.8 × 10-1, respectively).
Construction of mobSGI1-Donor Strain
For chromosomal integration of the 16447–18680 bp SGI1 regions, the pLOFKm (Herrero et al., 1990) derivative pJKI796 was introduced into E. coli S17-1 λpir strain, which allows the replication and transfer of R6K-based plasmids carrying the oriT of RK2. One of the S17-1 λpir/pJKI796 transformant colonies was used as donor in a standard mating (Kiss et al., 2012) with TG1Nal recipient. The miniTn10::mobSGI1-KmR chromosomal integrants were selected on LB+Nal+Km plates at 37°C O/N. KmRNalR transconjugants were streaked twice onto LB+Nal+Km plates and tested for ApS phenotype indicating the loss of plasmid backbone of pJKI796 (conservative transposition of the miniTn10 unit). Then, R55 was transferred into the resulting strain from TG1/R55 in a standard mating. Transconjugants were selected on LB+Nal+Cm plates at 37 °C O/N, and streaked twice on LB+Nal+Cm plates to get rid of donor contamination.
Bioinformatics
Sequence alignments were generated using the MultAlin interface1 (Corpet, 1988). Promoter motifs were predicted by BPROM2 (Solovyev and Salamov, 2011). For searching protein motifs MOTIF Search3 was used. Protein structure modeling was conducted using Phyre24 (Kelley et al., 2015), Swiss-Model5 (Arnold et al., 2006) and PSIPRED6 (Buchan et al., 2013). All homology searches were performed with the NCBI BLAST and DELTA-BLAST server7. SGI1-related elements were identified via a nucleotide BLAST search in GenBank using SGI1 backbone as query sequence, which was generated from the reference SGI1 sequence AF261825 by deletion of the flanking non-SGI1 sequences of DRs and In104 region along with one copy of the 5-bp direct repeat (DR associated with integron IRi) delimiting the In104 gene cluster. Inverted repeat motifs were detected using mFold8 server (Zuker, 2003).
Results
Identification and Functional Analysis of the SGI1 Transfer Origin
Localization of the oriT Sequence in SGI1
To determine the location of oriTSGI1, 14 different segments covering almost the entire SGI1-C backbone (excluding In104) were cloned into the non-mobilizable low-copy number vector pACYC184. These plasmids (pACYC184-1 to pACYC184-14) were introduced into the S. enterica serovar Agona strain 47SA97SGI1Δint harboring the IncC helper plasmid R55 and their mobilization were assessed in mating assays. Non-mobilizable chromosomal SGI1Δint was used to provide all (yet unknown) necessary trans mobilization factors and to prevent the mobilization of pACYC184 derivatives along with SGI1 via homologous recombination-mediated cointegration. Unlike other plasmids, pACYC-10 carrying SGI1 sequence from position 17005 to 19833 bp (Figure 1B) proved to be mobilizable at a comparable frequency (1.2 ± 1.0 × 10-4) to that of the wild-type (WT) SGI1-C (SGI1 positions are given according to the GenBank entry AF261825 used as reference sequence). However, pACYC-10 could not be mobilized in the absence of SGI1Δint in the donor strain (transfer frequency was below the 10-8 detection limit). This suggested that this SGI1 fragment (the region flanked by ORFs S020 and S023) carried at least the cis-acting oriT sequence of SGI1, while other mobilization functions provided in trans by SGI1Δint were also required for mobilization of pACYC-10.
To determine the minimal oriTSGI1, the fragment carried by pACYC-10 was progressively reduced in size and assessed for mobilization in similar assays (Figure 1B). The shortest mobilizable clone was pACYC-10D7 carrying a 135-bp SGI1 sequence extending from 18017 to 18151 bp, indicating that the fully functional oriTSGI1 overlaps the 3′-end of the predicted ORFs S021 and S022. The analysis of 10 transconjugants from each independent positive experiment confirmed the presence of the pACYC derivatives and the absence of the IncC helper plasmid, implying their conjugative mobilization in trans.
Minimal Mobilizable Region of SGI1
In parallel with the above approach, an alternative method was also applied to determine the entire region carrying all cis- and trans-acting mobilization functions of SGI1. The ca. 23 kb deletion derivative of SGI1-C (designated SGI1-C-d1, Figure 1C), which was previously shown to be mobilizable by R55 (Kiss et al., 2012) was cloned into a non-mobilizable p15A-based vector (plasmid pJKI669). Deletion derivatives and subclones of pJKI669 were used in mating assays in absence of chromosomal SGI1 in the donor strain. Only the plasmid constructs containing the intact S019-S022 region (position 16447 to 18680 bp) proved to be mobilizable in trans by R55 at a frequency comparable to the transfer of SGI1-C-d1 from pJKI669 (5.74 ± 0.93 × 10-4, Figure 1C). The shortest mobilizable construct, pJKI780, contained a 2.2 kb insert spanning SGI1 from the end of S019 to that of S023. Plasmids in which S019 (pJKI710, pJKI725), S020 (pJKI710, pJKI726) or S022 (pJKI731) were partially or entirely missing could not be mobilized. These results suggested that the 2.2 kb region cloned in pJKI780, hereafter called as mobSGI1, carries not only the oriTSGI1, but also all genes that are indispensable for SGI1 mobilization.
Identification of the Minimal oriTSGI1
Since further reduction of cloned mobSGI1 region impaired the transfer of the test plasmid, exact localization of oriTSGI1 and the determination of the minimal functional oriTSGI1 region required a helper strain which provides all mobilization factors in trans similarly, to the S. Agona strain 47SA97SGI1Δint applied previously for mobilization of pACYC184-derivatives. Thus, a donor strain containing the chromosomally integrated mobSGI1 region was constructed. The 16447–18680 bp mobSGI1 region was integrated into the chromosome of E. coli strain TG1Nal using a mini-Tn10 transposon (Herrero et al., 1990), and then the helper plasmid R55 was introduced. The resulting “mobSGI1-donor” strain was used to test the mobilization of plasmid subclones carrying progressively shortened fragments of mobSGI1 region by similar method applied above (see Figure 1C). The smallest SGI1 fragment permitting the mobilization of the test plasmid corresponded to positions 17961–18140 of SGI1 (pJKI818, Figure 1D). Based on the alternative methods (Figure 1B,D), we identified a 125-bp sequence corresponding to the 18016–18140 bp fragment of SGI1 (deduced from the overlap of inserts in pJKI818 and pACYC-10D7) as the minimal fully active oriTSGI1 (pJKI871, Figure 1D), which is localized in the 3′-end of ORF S021 including the overlapping part of S022.
Deletion of oriTSGI1 Causes Transfer-Deficiency
To further examine the oriTSGI1, we knocked out this segment from SGI1-C using the one-step gene inactivation method. In the conjugation assays, mobilization of SGI1-CΔoriT and a p15A-based plasmid carrying the 125-bp oriTSGI1 sequence (pMNI41) was assessed. For this assay, the R55ΔTn6187 helper plasmid was used due to its reduced antibiotic resistance spectrum (FloR/CmR, SulR). While the SGI1-CΔoriT mutant proved to be non-mobilizable (<7.3 ± 2.3 × 10-8), the plasmid pMNI41 was mobilized in trans at a frequency of 7.3 ± 2.9 × 10-3, which was comparable to the transfer rate of pJKI871 also containing oriTSGI1 (Figure 1D). This result indicated that deletion of oriTSGI1 sequence abolished the SGI1 transfer, while the functions required in trans for mobilization of oriTSGI1-carrying plasmid were not affected.
Functional Analysis of Mutations Affecting IR Motifs in oriTSGI1
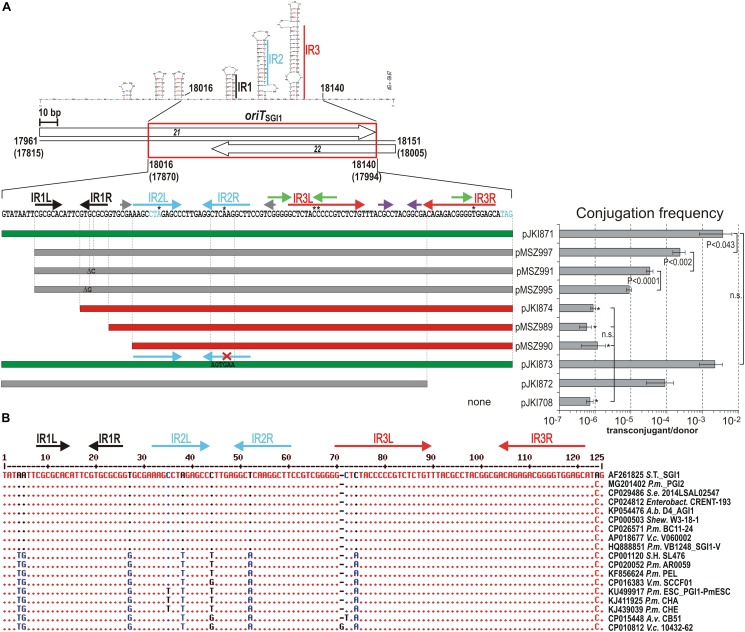
The oriTSGI1 sequence contains an array of inverted repeats (IRs, Figure 2A). For a more detailed analysis the oriT sequence, was further shortened or the repetitive motifs were mutated and the mobilization of the resulting oriTSGI1-derivatives was assayed using the mobSGI1 donor strain as described above. Mutations of the 7-bp perfect GC-rich inverse repeat IR1 resulted in significant reduction of the transfer frequency. Complete or partial deletion of IR1 entirely abolished the plasmid mobilization (pMSZ990, pMSZ989, and pJKI874; Figure 2A). Interestingly, deletion of the 7-bp AT rich tract located upstream of IR1 caused a 15-fold reduction of the transfer rate, which was further reduced by factors of 7 and 26 by the single base deletions in IR1R (compare pJKI871, pMSZ997, pMSZ995 and pMSZ991). Similarly, deletion of the right arm of the 19-bp imperfect repeat IR3 caused a 40-fold reduction of transfer rate (pJKI873 and pJKI872). In contrast, base changes introduced into the right arm of the 12-bp imperfect repeat IR2, abolishing the putative stem-loop structure, had no detectable effect on the transfer frequency. The two 6-bp GC-rich IRs located upstream of oriTSGI1 (17987–18017 bp, Figure 2A) seemed to have no or marginal role in oriT function, as their absence did not reduce the transfer frequency (compare pJKI818 and pJKI871, Figure 1D). The 125-bp oriTSGI1 sequence proved to be well conserved (95–100% identity) among the 63 SGI1-related IMEs found in public databases as only 11 divergent positions can be found in 17 elements (Figure 2B). These results confirmed that the fully functional oriTSGI1 is located between 18016 and 18140 bp of SGI1.
FIGURE 2.
Mutation and comparative analysis of oriTSGI1. (A) Functional analysis of oriTSGI1. The location of fully functional oriT is indicated by red box in the schematic map. The SGI1 coordinates are indicated as in Figure 1. Repeated motifs are shown above the sequence by color-coded arrows. Left and right arms of the inverted repeats longer than 6 bp are designated as IR1/2/3L and R, respectively. Asterisks above the sequence indicate mismatching bases in the IRs. The potential secondary structure of the region and the stem-forming IRs are also shown. Shortened or mutated subclones of oriT are represented by color-coded horizontal bars: green – wt transfer rate; gray – reduced transfer rate; red – not mobilizable. Single base deletions and the sequence of mutagenized IR2R are indicated in the respective bars. The transfer frequencies of the pJKI708-derivative constructs were measured using the mobSGI1-donor and TG2 recipient strains. Asterisk indicates that the transfer frequency was close to or below the detection limit. The rare TcRSmR colonies carried also the helper plasmid (co-transfer was ∼100 %) indicating that they did not result from in trans mobilization of the test plasmids. In all statistical evaluations paired t-test was used to calculate the significance of the differences. n.s. – not significant, other symbols are as in Figure 1A. (B) Comparison of the homologous sequences to oriTSGI1 in SGI1-related elements found in the GenBank database. The alignment shows the sequence divergence in the oriTSGI1 regions of 63 fully sequenced relatives of SGI1 (for the entire list see Supplementary Figure S1). The 17 oriT sequences having divergent positions are shown, while those identical to the reference sequence are represented by the respective region of the published SGI1 sequence AF261825. Short names of bacterial strains and the SGI1 variant/relative is indicated after the GenBank acc. number of the sequences. Abbreviations are as follows: A.b. – Acinetobacter baumannii; A.v. – Aeromonas veronii; Enterobact. - Enterobacter sp.; P.m. – Proteus mirabilis; S.e. – Salmonella enterica; Shew. – Shewanella sp.; S.T. – Salmonella Typhimurium; S.H. – Salmonella Heidelberg; V.c. – Vibrio cholerae; V.m. – Vibrio mimicus.
SGI1 Encodes Two Essential Mobilization Proteins
Identification of ORFs Required for SGI1 Mobilization
The mobSGI1 region containing 4 annotated ORFs S019, S020, S022, and S021 (latter is encoded on the upper strand) proved to carry all cis- and trans-acting elements that are required for mobilization of SGI1 (pJKI780 in Figure 1C or the mobSGI1-donor strain). Mobilization of plasmid pMNI41 from the TG1Nal::SGI1-CΔoriT donor strain, where both S021 and S022 are partially deleted due to the ΔoriT mutation suggested that the proteins encoded by the ORFs S021 and S022 are not required for in trans mobilization initiated at oriTSGI1. The ΔoriT mutation removes 40 and 29 amino acid (AA) residues from the C-terminus of the putative S021 and S022 proteins, respectively, which makes improbable that these proteins (if expressed at all) are still functional. To assess their role, independent KO mutations were generated in the 5′-end of both ORFs in mobSGI1-containing plasmids. As expected, transfer rates of both mutants were similar to the respective WT plasmid (2.7 ± 0.54 × 10-3 vs. 2.0 ± 0.32 × 10-3 for the S021 frameshift mutant pMSZ957/WT pMSZ949 and 6.1 ± 2.7 × 10-4 vs. 8.0 ± 1.7 × 10-4 for the S022 KO mutant pJKI774/WT pFOL1372) confirming that ORFs S021 and S022 do not encode proteins that are required for in trans mobilization of SGI1.
On the other hand, KO mutations of S019 and S020 had a deleterious effect on SGI1 conjugation, as the transfer frequency of both mutants dropped below 10-7 like that of the SGI1-CΔoriT mutant (Figure 3A). Interestingly, few transconjugant colonies could be obtained with a frequency of 0.9–2.9 × 10-8. Phenotypic and PCR analyses showed that the rare SGI1-CΔS020 and SGI1-CΔS019 transconjugants carried SGI1 predominantly alone, unlike SGI1-CΔoriT transconjugants, where the residual SGI1 transfer was tightly coupled to the conjugation of the helper plasmid. The absence of helper plasmid in the transconjugants suggested that the KO S020 and KO S019 mutants were trans-mobilized, albeit at a very low frequency. Similar result was obtained when the same KO mutations were introduced into the mobSGI1 region cloned in a p15A-based plasmid: the mobilization rates of the resulting KO S020 (pJKI737) and KO S019 (pJKI772) plasmids were 4 orders of magnitude lower than that of the WT control (pFOL1372) and were similar to the negative control (pJKI708). Using the mobSGI1-donor strain, WT transfer rates were observed with both KO mutant plasmids, indicating the effective trans-complementation by the chromosomally integrated mobSGI1 region (Figure 3B).
FIGURE 3.
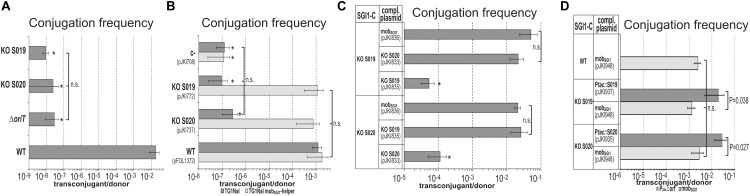
Mutation and complementation analysis of S019 and S020. (A) Transfer frequency of SGI1-C KO mutants. SGI1-CWT and the KO mutants were mobilized by the R55ΔTn6187 helper plasmid from TG1Nal strain into TG2 recipient. The asterisk indicates that the transfer frequency was close to the detection limit. In case of SGI1-CΔoriT the high rate of co-transfer with R55 among the sporadically occurring transconjugants refers to an oriTSGI1-independent way, while transconjugants of KO S019 and S020 SGI1-C mutants did not contain the helper plasmid and might derive via a yet unexplored way of in trans mobilization or lost the helper plasmid after conduction. (B) Transfer frequency of S019 and S020 KO mutant mobSGI1 regions cloned in pJKI708. In the mating assays TG1Nal/R55 or TG1Nal::mobSGI1/R55 strains containing the KO mutant or control plasmids were used as donor strains and TG2 was the recipient. Basal level of plasmid transfer (including the negative control plasmid pJKI708) indicated by asterisk did not result from regular in trans mobilization (high co-transfer rate with R55). (C) Trans-complementation of SGI1-CΔS019 and SGI1-CΔS020 by the respective KO mutant mobSGI1 regions. Donor E. coli strains TG1Nal::SGI1-CΔS019 and TG1Nal::SGI1-CΔS020 carried the helper plasmid R16a and one of the complementing plasmids containing the WT, KO S019 or KO S020 mobSGI1 region (pJKI836, pJKI835or pJKI833, respectively). ∗ Transconjugant frequency was below or around the detection limit. The relatively high detection limit was due to the lower donor titers compared to other matings (1.2–8.2 × 106 CFU/ml instead of the general 2–5 × 109 CFU/ml) in this experimental setup that was caused by the presence of two plasmids along with SGI1 in the donor strains. (D) Trans-complementation of SGI1-CΔS019 and SGI1-CΔS020 by expression of S019 or S020 proteins. Mating assays were performed using the recipient strain TG2 and donor strains TG1Nal::SGI1-CWT, TG1Nal::SGI1-CΔS019, or TG1Nal::SGI1-CΔS020 harboring the helper plasmid R16a and one of the complementing plasmids: pJKI948 contained the mobSGI1 region, while pJKI937 and pJKI935 expressed S019 and S020, respectively, from Ptac promoter. Protein expression was driven by leakage of the tac promoter (without IPTG induction).
Complementation of S020 and S019 KO Mutants
ORFs S020 and S019 are separated by a single TTG codon (beyond the stop codon of S020). To assess whether the two ORFs are translated into a single, biologically active fusion protein by a read-through mechanism or into two independent proteins, SGI1-CΔS020 and SGI1-CΔS019 mutants were complemented with plasmids carrying S020 and S019 KO mutant mobSGI1 regions. While neither SGI1-C mutant could be complemented by the respective KO mutant mobSGI1 region, SGI1-CΔS020 was complemented by S019 KO mobSGI1 region and vice versa as efficiently as was complemented by the WT mobSGI1 region (Figure 3C). This indicated that the two ORFs are translated into independent functional proteins.
Trans complementation of S020 and S019 KO mutant SGI1 was also carried out using expression vectors, which produced the two proteins under the control of Ptac promoter. These plasmids (pJKI935 and pJKI937 expressing S020 and S019, respectively) were used to complement the TG1Nal::SGI1-CΔS020 and TG1Nal::SGI1-CΔS019 donor strains harboring the IncC helper plasmid R16a. In similar mating assays used previously, both expression plasmids efficiently complemented the respective SGI1 mutants (Figure 3D). About tenfold higher rates of SGI1-transfer were observed with the non-mobilizable expression plasmids compared to the mobilizable control pJKI948, probably due to the lack of competition between SGI1 and the complementing plasmids. These data confirmed that ORFs S020 and S019 encode two essential transfer proteins, thus, they have been designated as mpsA and mpsB (mobilization protein of SGI1), respectively.
The Relaxase of IncC Helper Plasmid Is Not Essential for SGI1 Mobilization, but Increases Its Efficiency
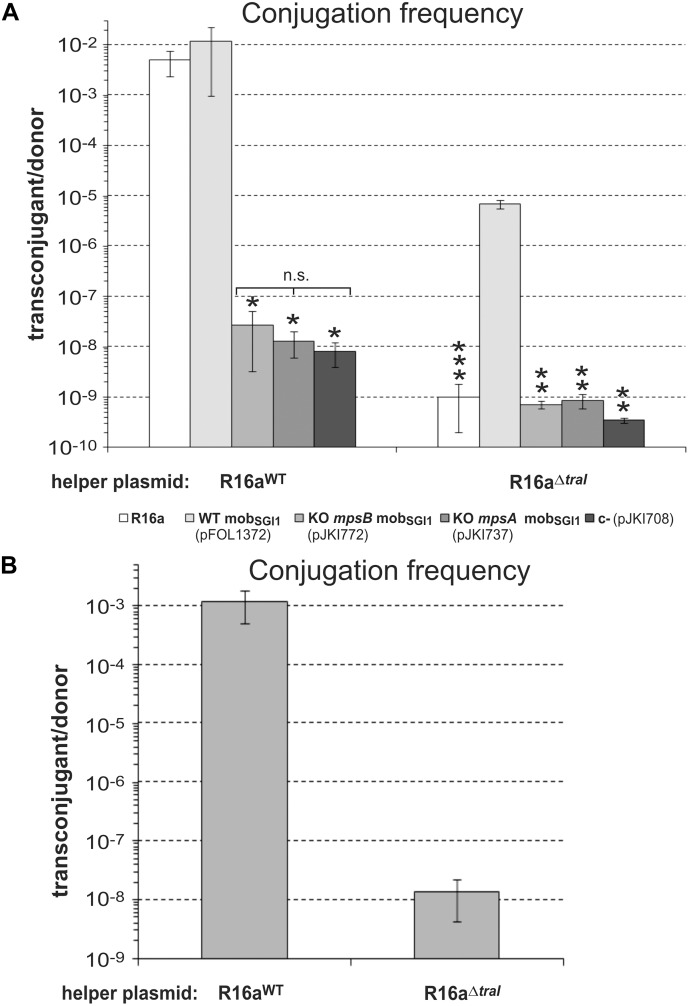
SGI1 exploits the transfer machinery of IncA/C plasmids in multiple manners (Kiss et al., 2015; Carraro et al., 2017), however, the role of the relaxase encoded by the traI gene of IncA/C plasmids in SGI1 mobilization has not yet been analyzed. To assess the possible cooperation between the relaxase and MpsAB proteins in mobilization of SGI1, p15A-based plasmids carrying the WT or mpsA or mpsB KO mutant mobSGI1 regions were compared in mating assays in the presence of the WT or the relaxase KO mutant helper plasmid, R16aWT and R16aΔTraI, respectively. R16aWT efficiently mobilized the plasmid containing the WT mobSGI1 region, while the two KO mutants (ΔmpsA and ΔmpsB) showed similar transfer rates to the negative control (Figure 4A). These data were in accordance with the previous results obtained with the helper plasmid R55 (Figure 3B) confirming that both mpsA and mpsB genes are required for transfer of the mobSGI1 region. Interestingly, the helper plasmid R16aΔTraI, which is unable to conjugate, mobilized the plasmid containing WT mobSGI1 region at a frequency of 6.9 ± 1.4 × 10-6, although with ca. 3 logs lower frequency compared to R16aWT (Figure 4A). Analysis of 10 transconjugants from each independent experiment proved that all were devoid of the helper plasmid and derived from trans-mobilization of the mobSGI1 plasmid. Moreover, no transconjugant at all was obtained in absence of either mpsA or mpsB. The R16aΔTraI was also able to mobilize the chromosomally integrated SGI1-C, although with 5 logs lower efficiency compared to R16aWT (Figure 4B). Transconjugants were proved to carry SGI1-C integrated at the attB site, and were devoid of the helper plasmid. These results indicate that mpsAB genes are indispensable, while the helper plasmid-encoded relaxase is also important for the efficient conjugative transfer initiated at oriTSGI1. However, mobilization rate of SGI1 remains detectable and significant in absence of the relaxase, suggesting that TraI provides important, but probably different functions in the initiation step of SGI1 transfer from the classical model.
FIGURE 4.
The role of mpsA, mpsB and the helper encoded relaxase traI in SGI1 mobilization. (A) Transfer frequency of the p15A-based plasmids, containing WT (pFOL1372), KO mpsB (KO S019, pJKI772) and KO mpsA (KO S020, pJKI737) mobSGI1 regions was assayed in the presence of the WT or relaxase KO mutant helper plasmid R16a. For mating, TG1Nal donor and TG90 recipient E. coli strains were used. The bars show means of 4 independent experiments except those representing the transfer rate of the helper plasmids. Since the conjugation frequency of the helper plasmids was the same independently of the test plasmids, the transfer rate of the helper plasmids R16aWT and R16aΔTraI correspond to the mean of their transfer frequencies in the 4 different settings that have been repeated 4 times. ∗Transconjugants indicated by an asterisk carried also the helper plasmid (co-transfer was ∼100 %) suggesting that the transfer of test plasmids did not derive from regular trans-mobilization. ∗∗Transfer frequency was below the detection limit (transconjugants were not obtained). ∗∗∗Transfer frequency of R16aΔTraI was close to the detection limit, 6 transconjugant colonies were obtained from five independent experiments (mean frequency was ≤ 1.9 × 10-9). These colonies did not contain test plasmid. (B) Mobilization of SGI1-CWT by the helper plasmids R16aWT and R16aΔTraI from TG1Nal::SGI1-C donor into TG90 recipient strain.
Identification of Promoter Regions Driving the Expression of mpsAB Genes
Based on the genetic context and the previous results, mpsAB genes were suspected to form a bicistronic operon. To localize the promoter region of this putative operon, the promoter activity of sequences located upstream of mpsA was measured in β-galactosidase assay. Two β-gal tester plasmids were constructed: the longer region fused to a promoterless lacZ gene extended from the start codon of mpsA to the end of S023 (pMSZ947), while the shorter one extended only to the end of the predicted ORF S022 (pMSZ948). The tester plasmids showed somewhat higher β-galactosidase activity (6.4 ± 1.5 and 4.2 ± 0.5 U, respectively) than the negative control plasmid pJKI990 (1.1 ± 0.4 U) suggesting that a weak promoter could drive the expression of the putative operon [for comparison Pint of SGI1 produced ∼350 U β-galactosidase in the same assay (Kiss et al., 2015)].
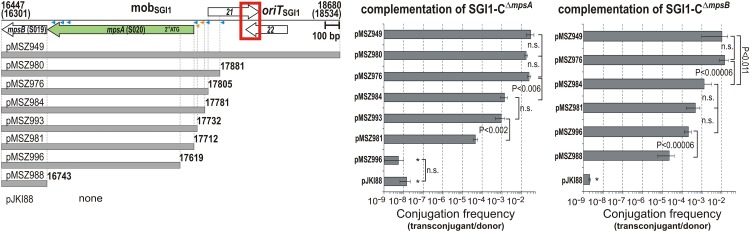
Then, a complementation method was applied to localize this weak promoter more exactly, where the mobilization rate of SGI1-CΔmpsA and SGI1-CΔmpsB mutants was examined in the presence of R55ΔTn 6187 helper and p15A-based complementing plasmids, which produced MpsA and MpsB proteins in trans depending on the intactness of the promoter in the cloned mobSGI1 fragments. Both SGI1-C mutants could be complemented to the WT transfer rate achieved with the whole mobSGI1 region (pMSZ949) by the fragments carrying mpsAB genes and at least the non-coding region between mpsA and S021 (pMSZ976, pMSZ980, Figure 5). This suggested that the PmpsAB promoter is located in the ca. 100 bp region upstream of the mpsA start codon. Then, primer extension experiments were conducted as previously described (Murányi et al., 2016) to identify the transcriptional start site (TSS) of mpsAB using plasmid pMSZ948. However, in repeated attempts, it failed to detect TSS, which again suggested a low transcription level of mpsAB.
FIGURE 5.
Identification of promoter regions driving expression of mpsA and mpsB. The fragments cloned into the complementing plasmids are shown below the schematic map of mobSGI1 region. Putative promoter elements predicted by BPROM or found by manual search are indicated by blue or yellow arrowheads, respectively. Other symbols are as in Figure 1B. Transfer frequency of SGI1-CΔS020 and SGI1-CΔS019 was measured in the presence of one of the complementing plasmids and the helper plasmid R55ΔTn6187. Plasmid pMSZ984 showed high st. deviations in three independent complementation assays carried out with 3–6 replicates with SGI1-CΔS019, thus the complementation efficiency of this plasmid could not exactly be determined. However, mean values appeared similar to that of pMSZ981 and pMSZ996. ∗Transconjugants resulting from oriTSGI1-independent transfer pathway.
Thus, the complementation method was further applied to localize the active promoters by progressively shortening the upstream region of mpsA in the complementing plasmids. Removing of the distal 25 bp (pMSZ984) resulted in a ca. 20-fold and 10-fold decrease of the transfer rate of SGI1-CΔmpsA and SGI1-CΔmpsB, respectively, compared to the shortest fully active plasmid pMSZ976 (Figure 5). The similarly, decreasing complementation efficiency observed with the two KO mutants supported that the two ORFs are expressed from a common promoter located upstream of mpsA. Interestingly, pMSZ993, carrying only 20 bp segment upstream of the mpsA start codon, achieved relatively efficient complementation of SGI1-CΔmpsA even though this segment was too short to contain an intact promoter. This raised the possibility that MpsA protein might be translated from the second, in-frame Met codon (17619 bp) instead of the originally identified start codon (17712 bp). This hypothesis was examined using two further complementing plasmids: pMSZ981 contained mpsAB without upstream sequences, while pMSZ996 carried a truncated mpsA beginning with the second Met codon. In both plasmids, the rrnB T1T2 terminators were inserted immediately upstream of the start codons to prevent any promoter activities from the plasmid backbone. The construct with truncated mpsA (pMSZ996) could not complement SGI1-CΔmpsA, while pMSZ981 containing only the coding sequence of mpsAB without promoter region had a residual activity in complementation of SGI1-CΔmpsA (ca. 540-fold reduction compared to the whole mobSGI1 region, pMSZ949, Figure 5). This result excluded that translation of MpsA protein starts from the in-frame Met codon and confirmed again that a very low level of MpsA expression is sufficient for complementation.
On the other hand, SGI1-CΔmpsB could be complemented by both constructs at a comparable level observed with pMSZ984, indicating that mpsB can also be expressed independently from PmpsAB. Interestingly, like SGI1-CΔmpsA, the transfer of SGI1-CΔmpsB could be complemented by a construct, which contained only the coding sequence of mpsB separated from the plasmid backbone by rrnB terminators. To detect TSS of mpsB, an analogous plasmid to pMSZ948 was constructed, which carried the 227 bp upstream region of mpsB (the 3′ part of mpsA) fused to the promoterless lacZ gene (pMSZ1017), and primer extension assay was carried out, but without positive result. These suggested again that low level of MpsB can ensure the complementation of SGI1-CΔmpsB (Figure 5). The complementation data indicated that the mpsAB genes can be transcribed into a bicistronic mRNA from a common promoter region PmpsAB, however, mpsB appeared to be expressed also from its own promoter PmpsB located in the 3′-end of mpsA.
The promoter search using BPROM server and additional thorough examination of the sequence revealed several putative promoters in the upstream regions of mpsA and mpsB (Figure 5). Their high divergence from the consensus of σ70 promoters and the low score values supported our earlier assumption that both the bicistronic mpsAB and mpsB mRNAs are synthesized at a low level from the two promoter regions.
Discussion
The boom in bacterial genome sequencing in the last decade highly contributed to the discovery of numerous MGIs of the SGI1 and related families gathering IMEs implicated in the spread of antimicrobial resistance among several Gram-negative pathogens such as S. enterica serovars, Proteus mirabilis strains and a few other species. Despite the evidence that SGI1-related elements are specifically mobilized by conjugative IncA/C plasmids, their own mobilization components such as oriT and self-encoded conjugation proteins have not yet been identified.
In this study, the essential SGI1 conjugative mobilization functions were found to be clustered in the 2.2-kb mobSGI1 region (Figure 1C,D) carrying the oriTSGI1 and two essential genes (ORFs S020 and S019) named mpsA and mpsB. The fully active minimal oriT sequence has been localized in the vicinity of the mpsAB genes between 18016 and 18140 bp positions (GenBank accession AF261825), which includes the overlapping 3′ parts of two small predicted ORFs S021 and S022. Like many oriT regions identified to date, the oriTSGI1 also contains inverted repeat motifs. Three long IR motifs ( ≥ 7 bp, IRs1-3) have been found in the 125-bp minimal oriTSGI1. The most important one is the GC-rich 7-bp perfect IR1 (Figure 2A), suggesting its functional implication in binding relaxosome proteins and that the potential nic-site is probably located near this motif. Different methodologies have been successfully applied to identify nic-sites of several well-known conjugative plasmids and ICEs (Pansegrau et al., 1993; Llosa et al., 1995; Zechner et al., 1997; Núñez and De La Cruz, 2001; Varsaki et al., 2003; Lee and Grossman, 2007; Chen et al., 2010; Tsvetkova et al., 2010; Grove et al., 2013). However, the nic-site of one of the best studied ICEs, SXT, and the related IncA/C plasmids could not be defined yet [V. Burrus personal communication, (Hegyi et al., 2017)]. Although many efforts have been made to identify the nic-site in oriTSGI1 applying biochemical (primer extension, RACE PCR, in vitro nicking assay) and in vivo (interrupted mating) methods, all failed to give convincing results to date (data not shown).
In contrast to most oriT regions, oriTSGI1 is a GC-rich (60%) sequence compared to the SGI1 backbone (44%). Similar high GC-content (61%) can be seen in both oriTs of ICEclc element (Miyazaki and Van Der Meer, 2011), however, in this case the entire ICE has also high (62.5%) GC-content.
The largest part or the entire mobSGI1 region is present in all the 63 fully sequenced SGI1-related IMEs (Supplementary Figure S1). In the distant relatives (see bottom of Supplementary Figure S1), the 3′-end of mpsB homologs are more divergent and the distal part (ca. 210 bp at the 3′-end of mobSGI1 region) of the non-coding region adjacent to S023 along with ORF S023 are missing (e.g., in AGI1 or PGI2). The nucleotide identity of the mobSGI1-homolog regions to the corresponding SGI1 reference varies from 85 to 100%. The 125-bp oriTSGI1 sequence is highly conserved (95–100% identity) among the SGI1-related IMEs as only 11 divergent positions can be found in 17 out of the 63 elements (Figure 2B). Four and three divergent positions occur in the IR2L-space-IR2R region and in the 5′-end of IR3L, respectively, while the GC-rich 7-bp perfect IR1 is fully conserved. These observations are congruent with the conclusion of the experimental results, strengthening the importance of this region in SGI1 mobilization.
OriTSGI1 does not show striking similarity to that of IncA/C plasmids (oriTA/C), either in genetic context, sequence motifs or potential secondary structures (Supplementary Figures S2A,B). OriTA/C locates in an intergenic region between divergent genes and overlaps the promoter of one of them, mobI, which encodes a plasmid-specific transfer factor that is indispensable for conjugation of IncA/C plasmids, but not required for SGI1 mobilization (Hegyi et al., 2017). Although oriTA/C also contains several IRs, these are shorter and share no sequence similarity to those of oriTSGI1. Furthermore, the most important part of oriTA/C is a 14-bp direct repeat motif, which has no similar counterparts in oriTSGI1. Other striking difference is the GC-content: while oriTSGI1 is rather a GC-rich sequence, oriTA/C has a low GC content (37.4%). The only similarity with oriTA/C as well as with oriTs of many other mobilizable elements is the location of oriTSGI1 in the vicinity of mobilization genes.
Two pivotal SGI1-encoded transfer genes, ORFs S020 and S019, have been identified and were renamed here as mpsA and mpsB, respectively. Unlike mpsAB genes, the two other small predicted ORFs of mobSGI1, S021 and S022, do not appear to encode proteins that are involved in transfer process. Complementation experiments showed that mpsA and mpsB separated by a single codon beyond the stop codon of mspA are translated into independent polypeptides, in part, from a bicistronic mRNA, however, MpsB protein can also be expressed from its own mRNA (Figure 3C,D, 5). Interestingly, the sequential reduction of the upstream region of mpsA caused gradual decrease in complementation efficiencies of the respective KO mutant, suggesting the presence of at least two different functional promoters in this region. The sequence-based prediction, which identified several putative promoters in the intergenic region between mpsA and S021 and in the 5′ part of S021 (Figure 5) supported this assumption. By BPROM prediction three potential promoters located upstream of mpsB (in the 3′ part of mpsA) were found approving that mpsB has also its own promoter region. All of these promoter-like sequences show low similarity to the σ70 consensus. The weak promoter activity observed in β-gal assays, the negative results of primer extension assays to detect TSSs and the fact that plasmid constructs carrying mpsB or mpsAB coding sequences without their upstream regions could complement the respective KO mutants indicated that very low amount of MpsA and MpsB proteins are sufficient to reach detectable or even WT transfer rates.
Despite the crucial role of oriTSGI1, MpsA and MpsB proteins in SGI1 mobilization, a very low transfer frequency was observed with each SGI1-C KO mutants (around 10-8/donor, see Figure 3A, 4A) and mobSGI1-plasmids (Figure 3B) suggesting that the helper plasmid is somehow able to mobilize SGI1 at a very low level independently of cis- and trans-acting mobilization factors of SGI1. In case of deletion of oriTSGI1, the majority (≥ 97%) of the rare transconjugants contained also the helper plasmid suggesting that the transfer of SGI1 or the test plasmids was not independent of the transfer of the helper plasmid. The most plausible explanation for this phenomenon is that co-integrates was formed with the helper plasmid. The low transfer rate and the lack of extensive homology between SGI1-C derivatives or mobSGI1-plasmids and the helper plasmids suggest the involvement of a recA-independent illegitimate recombination process. Further support for a recA-independent mechanism may be that similar transfer rates were observed with the oriTSGI1-free pJKI708 and its derivatives when the recA- E. coli TG2 was used as donor strain (Figure 1C). A co-integrate based transfer mechanism has been reported previously (Hegyi et al., 2017), however, in that case the high transfer frequency was the consequence of an efficient site-specific recombination. The sporadically occurring helper-free transconjugants obtained with mpsA or mpsB KO mobSGI1-plasmids (≤3%, Figure 3B, 4A) and the generally helper-free SGI1-CΔmpsA or SGI1-CΔmpsB transconjugants (Figure 3A) may represent a yet unexplored way of inefficient mobilization by IncA/C plasmids in absence of the major transfer proteins MpsA and MpsB.
Both the homology searches and the structure predictions advocate that MpsA protein belongs to the Tyr-recombinase/integrase superfamily of DNA breaking-rejoining enzymes. The 126–225 AA tract of MpsA shows high homology to the DNA_BRE_C conserved C-terminal catalytic domain that is characteristic of site-specific integrases, bacterial recombinases XerD/C and type IB topoisomerases.
Phyre2 modeling confirmed that MpsA is related to the Tyr-recombinase family, as the best templates with known 3D-structure were XerH (Fold library id.: c5jjvA) and Cre recombinase (Fold library id.: c1ma7A) with 15–13% sequence identity, 91–94% coverage and 100% confidence. All the other hits showing > 90% coverage were also site-specific Tyr-recombinases (XerC,D; phage integrases). Similar result was obtained from Swiss-Model. Based on AA sequence alignments with related recombinases, the conserved catalytic nucleophile tyrosine residue was predicted at the C-terminus of MpsA protein (Y319). In addition to the Y319, DELTA-BLAST allowed to identify the R162, H247, R250, H251 in MpsA matching with the catalytic residues of the active sites of bacteriophage Hp1 Integrase (1AIH_A), Cre (2CRX_B), λ Integrase (1AE9_A) and IntI4 (2A3V_A). The mpsA gene appears to be well conserved among the 63 SGI1-related elements as only 19 show some sequence divergence compared to the reference SGI1 (99–87% identity). Although these base variations cause several changes in the predicted amino acid sequences (Supplementary Figure S3A), the only case when MpsA protein is apparently truncated and possibly inactive is the Aeromonas veronii CB51.
The mpsB gene encoding for the other essential mobilization factor of SGI1 is also similarly, conserved as only 17 of 63 elements show some DNA sequence variations in this gene (99–76% identity). These changes cause AA substitutions at 18 positions of the predicted MpsB proteins, however, truncation of the protein occurs only in V. mimicus SCCF01 and incomplete protein can be expressed in A. veronii CB51 (Supplementary Figure S3B). Using MotifFinder, the 98-AA-long MpsB protein was predicted to contain a phage integrase N-terminal SAM-4 like domain (PF13495, Pfam database), which was also supported by the best Phyre2 modeling based on the lambda integrase-like N-terminal domain template (Fold library id.: d1f44a1) and by Swiss-model where, following two eukaryotic proteins spectrin and plectin, the best hits were the N-terminal domains of XerH and Cre. Based on these data and PSIPRED secondary structure prediction (Buchan et al., 2013), MpsB appears to be an independent domain-like protein containing 4 α-helices of which the last three (20–82 AA) are related to a portion of the N-terminal core binding domain of lambda integrases (Swalla et al., 2003). This raises the possibility that MpsB is involved in DNA binding at oriTSGI1, however, a protein-recruiting function into the initiation complex cannot be ruled out either.
SGI1 exploits the transfer machinery of IncA/C plasmids in multiple manners (Kiss et al., 2015; Carraro et al., 2017), however, the role of the plasmid-borne relaxase TraI in SGI1 mobilization has not yet been analyzed. The mobilization assays using R16ΔTraI helper plasmid provided an unexpected result. While conjugation of the helper plasmid appears strictly TraI-dependent, the WT mobSGI1-plasmid and SGI1-C proved to be mobilizable in absence of TraI (Figure 4), albeit with orders of magnitude less efficiently. Considering the striking difference between oriTSGI1 and oriTA/C (Supplementary Figure S2), it is unlikely that similar relaxosome complexes are formed at these oriTs. The fact that SGI1 mobilization was found to be possible in absence of the helper-encoded relaxase suggests that other SGI1-encoded proteins ensure the essential relaxase functions, at least in part, in the SGI1 relaxosome at oriTSGI1.
The IncA/C relaxase TraI, although significantly increases the SGI1 transfer by a yet unknown mechanism, seems unable to mobilize oriTSGI1 in absence of MpsA or MpsB. Its role might be the unwinding of donor DNA or delivery of the relaxosome to T4SS by recruiting the coupling protein. Since the components of the initiation complex of IncA/C plasmids are only predictable through their homologies to those of well-studied model systems, the functions of TraI and involvement of other IncA/C-encoded proteins in the plasmid transfer and SGI1 mobilization need further investigations. In addition to the DNA nicking domain, other functional domains of relaxases, such as helicase domains (Llosa et al., 1996; Cheng et al., 2011; Alperi et al., 2013) and translocation signals, which are required for binding by the cognate coupling proteins and delivery of the relaxase-DNA complex to the T4SS (Lang et al., 2010), have been identified in several conjugation systems. Since the helicase domain seems to be the peculiarity of the IncW/N/F family relaxases (Llosa et al., 1996) and such domain have not yet been identified in TraI of IncA/C plasmids, the most possible way by which TraI can enhance SGI1 mobilization is to facilitate the transport of the relaxosome complex to or even through the T4SS. In addition to TraI, some conjugation systems require several auxiliary relaxosome components. In the case of IncPα plasmid RP4, TraJ protein is responsible for sequence-specific recognition of oriT and directing TraI to the origin of transfer (Ziegelin et al., 1989), TraH stabilizes the oriT-DNA complex through specific protein-protein interactions (Pansegrau et al., 1990) and TraK stimulates the relaxation by wrapping DNA near oriT (Ziegelin et al., 1992). In IncW plasmid R388, TrwA binds specifically to oriT and stimulates the ATPase activity of the coupling protein TrwB, which is indispensable to link the relaxosome complex to T4SS (Cabezón et al., 2015). Similar auxiliary factors are not yet known in the conjugative apparatus of IncA/C plasmids. Besides the structural components and the assembly factors of T4SS, only TraI and the indispensable transfer factor MobI have been described in IncA/C transfer. Although the exact function of MobI is not clear, it appears not to be involved in SGI1 mobilization (Hegyi et al., 2017).
Based on the results presented here we suggest that MpsA functions as an atypical relaxase of SGI1, which needs MpsB for binding and/or nicking oriTSGI1, but does not require TraI for the initial step of SGI1 relaxosome formation. This explanation is strengthened by the recent description of the atypical relaxase TcpM of plasmid pCW3 from Clostridium perfringens that appears related to tyrosine recombinases (our Phyre2 modeling showed the best homology of TcpM with XerH) without sequence similarity to known relaxases (Wisniewski et al., 2016). The fact that mpsB KO mutant mobSGI1-plasmid could not be mobilized even by a WT helper plasmid (Figure 4) suggests that MpsB rather has a role in the initiation of transfer (maybe in binding oriTSGI1 together with MpsA), than recruiting additional proteins like TraI. The relaxase TraI might be involved in unwinding the donor DNA or more possibly in the transport of the relaxosome to T4SS. The key elements and an accessory partner of the SGI1 relaxosome have been here identified, however, the understanding their exact role in the conjugative DNA processes needs further investigations.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
JK, BD, AC, and FO conceived the project. JK, MS, AH, GD, KP, BD, IN, and FO designed and carried out the experiments and analyzed the data. JK and BD performed the bioinformatic analyses, JK prepared the figures. JK, BD, AC, and AH wrote the manuscript. All the authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Erika Sztánáné-Keresztúri and Mária Turai for the excellent technical assistance.
Funding. This work was supported by the Hungarian Scientific Research Fund K 105635 and NKFI K 128203 to JK and by public funds from the French National Institute of Agricultural Research to BD. HA was supported by a fellowship of Campus France. GD was supported by a Ph.D. fellowship from Ministère de l’Enseignement Supérieur et de la Recherche, France. IN was supported by the Ph.D. fellowship of Eötvös Loránd University, Budapest, Hungary.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.00457/full#supplementary-material
References
- Alperi A., Larrea D., Fernández-González E., Dehio C., Zechner E. L., Llosa M. (2013). A translocation motif in relaxase trwc specifically affects recruitment by its conjugative type IV secretion system. J. Bacteriol. 195 4999–5006. 10.1128/JB.00367-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose S. J., Harmer C. J., Hall R. M. (2018). Compatibility and entry exclusion of IncA and IncC plasmids revisited: IncA and IncC plasmids are compatible. Plasmid 9 7–12. 10.1016/j.plasmid.2018.02.002 [DOI] [PubMed] [Google Scholar]
- Arnold K., Bordoli L., Kopp J., Schwede T. (2006). The SWISS-MODEL workspace: a web-based environment for protein structure homology modelling. Bioinformatics 22 195–201. 10.1093/bioinformatics/bti770 [DOI] [PubMed] [Google Scholar]
- Avila P., Núñez B., de la Cruz F. (1996). Plasmid R6K contains two functional oriTs which can assemble simultaneously in relaxosomes in vivo. J. Mol. Biol. 261 135–143. 10.1006/jmbi.1996.0447 [DOI] [PubMed] [Google Scholar]
- Bellanger X., Payot S., Leblond-Bourget N., Guédon G. (2014). Conjugative and mobilizable genomic islands in bacteria: evolution and diversity. FEMS Microbiol. Rev. 38 720–760. 10.1111/1574-6976.12058 [DOI] [PubMed] [Google Scholar]
- Boyd D., Peters G. A., Cloeckaert A., Boumedine K. S., Chaslus-Dancla E., Imberechts H., et al. (2001). Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183 5725–5732. 10.1128/JB.183.19.5725-5732.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyd D. A., Peters G. A., Ng L., Mulvey M. R. (2000). Partial characterization of a genomic island associated with the multidrug resistance region of Salmonella enterica Typhymurium. FEMS Microbiol. Lett. 189 285–291. 10.1111/j.1574-6968.2000.tb09245.x [DOI] [PubMed] [Google Scholar]
- Buchan D. W. A., Minneci F., Nugent T. C. O., Bryson K., Jones D. T. (2013). Scalable web services for the PSIPRED protein analysis workbench. Nucleic Acids Res. 41 W349–W357. 10.1093/nar/gkt381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cabezón E., Ripoll-Rozada J., Peña A., de la Cruz F., Arechaga I. (2015). Towards an integrated model of bacterial conjugation. FEMS Microbiol. Rev. 39 81–95. 10.1111/1574-6976.12085 [DOI] [PubMed] [Google Scholar]
- Carraro N., Burrus V. (2014). Biology of three ICE families: SXT/R391, ICEBs1, and ICESt1/ICESt3. Microbiol. Spectr. 2 1–20. 10.1128/microbiolspec.MDNA3-0008-2014 [DOI] [PubMed] [Google Scholar]
- Carraro N., Durand R., Rivard N., Anquetil C., Barrette C., Humbert M., et al. (2017). Salmonella genomic island 1 (SGI1) reshapes the mating apparatus of IncC conjugative plasmids to promote self-propagation. PLoS Genet. 13:e1006705. 10.1371/journal.pgen.1006705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N., Matteau D., Luo P., Rodrigue S., Burrus V. (2014). The master activator of IncA/C conjugative plasmids stimulates genomic islands and multidrug resistance dissemination. PLoS Genet. 10:e1004714. 10.1371/journal.pgen.1004714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carraro N., Rivard N., Ceccarelli D., Colwell R. R., Burrus V. (2016). IncA/C conjugative plasmids mobilize a new family of multidrug resistance islands in clinical Vibrio cholerae non-O1/non-O139 isolates from Haiti. mBio 7:e00509–16. 10.1128/mBio.00509-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ceccarelli D., Daccord A., René M., Burrus V. (2008). Identification of the origin of transfer (oriT) and a new gene required for mobilization of the SXT/R391 family of integrating conjugative elements. J. Bacteriol. 190 5328–5338. 10.1128/JB.00150-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Pappas D. L., Galli D. M. (2010). Mapping of the nick site on conjugative plasmid pVT745 by interrupted mating. Plasmid 63 136–142. 10.1016/j.plasmid.2010.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng Y., McNamara D. E., Miley M. J., Nash R. P., Redinbo M. R. (2011). Functional characterization of the multidomain F plasmid TraI relaxase-helicase. J. Biol. Chem. 286 12670–12682. 10.1074/jbc.M110.207563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corpet F. (1988). Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16 10881–10890. 10.1093/nar/16.22.10881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccord A., Ceccarelli D., Burrus V. (2010). Integrating conjugative elements of the SXT/R391 family trigger the excision and drive the mobilization of a new class of Vibrio genomic islands. Mol. Microbiol. 78 576–588. 10.1111/j.1365-2958.2010.07364.x [DOI] [PubMed] [Google Scholar]
- Daccord A., Ceccarelli D., Rodrigue S., Burrus V. (2013). Comparative analysis of mobilizable genomic islands. J. Bacteriol. 195 606–614. 10.1128/JB.01985-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko K. A., Wanner B. L. (2000). One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U.S.A. 97 6640–6645. 10.1073/pnas.120163297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta N., Hedges R. W. (1973). R factors of compatibility group A. J. Gen. Microbiol. 74 335–337. 10.1099/00221287-74-2-335 [DOI] [PubMed] [Google Scholar]
- De La Cruz F., Frost L. S., Meyer R. J., Zechner E. L. (2010). Conjugative DNA metabolism in Gram-negative bacteria. FEMS Microbiol. Rev. 34 18–40. 10.1111/j.1574-6976.2009.00195.x [DOI] [PubMed] [Google Scholar]
- Douard G., Praud K., Cloeckaert A., Doublet B. (2010). The Salmonella genomic island 1 is specifically mobilized in trans by the IncA/C multidrug resistance plasmid family. PLoS One 5:e15302. 10.1371/journal.pone.0015302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doublet B., Boyd D., Mulvey M. R., Cloeckaert A. (2005). The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55 1911–1924. 10.1111/j.1365-2958.2005.04520.x [DOI] [PubMed] [Google Scholar]
- Doublet B., Praud K., Bertrand S., Collard J.-M., Weill F.-X., Cloeckaert A. (2008). Novel insertion sequence- and transposon-mediated genetic rearrangements in genomic island SGI1 of Salmonella enterica serovar Kentucky. Antimicrob. Agents Chemother. 52 3745–3754. 10.1128/AAC.00525-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia M. V., Clewell D. B. (2002). Transfer origins in the conjugative Enterococcus faecalis plasmids pAD1 and pAM373: identification of the pAD1 nic site, a specific relaxase and a possible TraG-like protein. Mol. Microbiol. 45 375–395. 10.1046/j.1365-2958.2002.03007.x [DOI] [PubMed] [Google Scholar]
- Francia M. V., Haas W., Wirth R., Samberger E., Muscholl-Silberhorn A., Gilmore M. S., et al. (2001). Completion of the nucleotide sequence of the Enterococcus faecalis conjugative virulence plasmid pAD1 and identification of a second transfer origin. Plasmid 46 117–127. 10.1006/plas.2001.1533 [DOI] [PubMed] [Google Scholar]
- Frost L. S., Ippen-Ihler K., Skurray R. A. (1994). Analysis of the sequence and gene products of the transfer region of the F sex factor. Microbiol. Rev. 5 162–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fürste J. P., Pansegrau W., Ziegelin G., Kröger M., Lanka E. (1989). Conjugative transfer of promiscuous IncP plasmids: interaction of plasmid-encoded products with the transfer origin. Proc. Natl. Acad. Sci. U.S.A. 86 1771–1775. 10.1073/pnas.86.6.1771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcillán-Barcia M. P., Francia M. V., De La Cruz F. (2009). The diversity of conjugative relaxases and its application in plasmid classification. FEMS Microbiol. Rev. 33 657–687. 10.1111/j.1574-6976.2009.00168.x [DOI] [PubMed] [Google Scholar]
- Grove J. I., Alandiyjany M. N., Delahay R. M. (2013). Site-specific relaxase activity of a VirD2-like protein encoded within the tfs4 genomic island of helicobacter pylori. J. Biol. Chem. 288 26385–26396. 10.1074/jbc.M113.496430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamidian M., Holt K. E., Hall R. M. (2015). Genomic resistance island AGI1 carrying a complex class 1 integron in a multiply antibiotic-resistant ST25 Acinetobacter baumannii isolate. J. Antimicrob. Chemother. 70 2519–2523. 10.1093/jac/dkv137 [DOI] [PubMed] [Google Scholar]
- Harmer C. J., Hamidian M., Ambrose S. J., Hall R. M. (2016). Destabilization of IncA and IncC plasmids by SGI1 and SGI2 type Salmonella genomic islands. Plasmid 8 51–57. 10.1016/j.plasmid.2016.09.003 [DOI] [PubMed] [Google Scholar]
- Hegyi A., Szabó M., Olasz F., Kiss J. (2017). Identification of oriT and a recombination hot spot in the IncA/C plasmid backbone. Sci. Rep. 7:10595. 10.1038/s41598-017-11097-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrero M., De Lorenzo V., Timmis K. N. (1990). Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J. Bacteriol. 172 6557–6567. 10.1128/jb.172.11.6557-6567.1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huguet K. T., Gonnet M., Doublet B., Cloeckaert A. (2016). A toxin antitoxin system promotes the maintenance of the IncA/C-mobilizable Salmonella genomic Island 1. Sci. Rep. 6:32285. 10.1038/srep32285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaworski D. D., Clewell D. B. (1995). A functional origin of transfer (oriT) on the conjugative transposon Tn916. J. Bacteriol. 177 6644–6651. 10.1128/jb.177.22.6644-6651.1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson S. L., Khiani A., Bishop-Lilly K. A., Chapman C., Patel M., Verratti K., et al. (2015). Complete genome assemblies for two single-chromosome Vibrio cholerae isolates, strains 1154-74 (Serogroup O49) and 10432-62 (Serogroup O27). Genome Announc. 3:e00462–15. 10.1128/genomeA.00462-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley L. A., Mezulis S., Yates C. M., Wass M. N., Sternberg M. J. E. (2015). The Phyre2 web portal for protein modeling, prediction and analysis. Nat. Protoc. 10 845–858. 10.1038/nprot.2015.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J., Nagy B., Olasz F. (2012). Stability, entrapment and variant formation of Salmonella genomic island 1. PLoS One 7:e32497. 10.1371/journal.pone.0032497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss J., Papp P. P., Szabó M., Farkas T., Murányi G., Szakállas E., et al. (2015). The master regulator of IncA/C plasmids is recognized by the Salmonella genomic Island SGI1 as a signal for excision and conjugal transfer. Nucleic Acids Res. 43 8735–8745. 10.1093/nar/gkv758 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolisnychenko V., Plunkett G., Herring C. D., Fehér T., Pósfai J., Blattner F. R., et al. (2002). Engineering a reduced Escherichia coli genome. Genome Res. 12 640–647. 10.1101/gr.217202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang S., Gruber K., Mihajlovic S., Arnold R., Gruber C. J., Steinlechner S., et al. (2010). Molecular recognition determinants for type IV secretion of diverse families of conjugative relaxases. Mol. Microbiol. 78 1539–1555. 10.1111/j.1365-2958.2010.07423.x [DOI] [PubMed] [Google Scholar]
- Lee C. A., Grossman A. D. (2007). Identification of the origin of transfer (oriT) and DNA relaxase required for conjugation of the integrative and conjugative element ICEBs1 of Bacillus subtilis. J. Bacteriol. 189 7254–7261. 10.1128/JB.00932-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings R. S., Djordjevic S. P., Hall R. M. (2008). SGI2, a relative of Salmonella genomic island SGI1 with an independent origin. Antimicrob. Agents Chemother. 52 2529–2537. 10.1128/AAC.00189-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levings R. S., Lightfoot D., Partridge S. R., Hall R. M., Djordjevic S. P. (2005). The genomic Island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187 4401–4409. 10.1128/JB.187.13.4401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Xie Y., Liu M., Tai C., Sun J., Deng Z., et al. (2018). OriTfinder: a web-based tool for the identification of origin of transfers in DNA sequences of bacterial mobile genetic elements. Nucleic Acids Res. 46 W229–W234. 10.1093/nar/gky352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu M., Li X., Xie Y., Bi D., Sun J., Li J., et al. (2019). ICEberg 2.0: an updated database of bacterial integrative and conjugative elements. Nucleic Acids Res. 47 D660–D665. 10.1093/nar/gky1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llosa M., Alkorta I. (2017). Coupling proteins in type IV secretion. Curr. Top. Microbiol. Immunol. 413 143–168. 10.1007/978-3-319-75241-9_6 [DOI] [PubMed] [Google Scholar]
- Llosa M., Grandoso G., de la Cruz F. (1995). Nicking activity of TrwC directed against the origin of transfer of the IncW plasmid R388. J. Mol. Biol. 246 54–62. 10.1006/jmbi.1994.0065 [DOI] [PubMed] [Google Scholar]
- Llosa M., Grandoso G., Hernando M. A., De La Cruz F. (1996). Functional domains in protein TrwC of plasmid R388: dissected DNA strand transferase and DNA helicase activities reconstitute protein function. J. Mol. Biol. 264 56–67. 10.1006/jmbi.1996.0623 [DOI] [PubMed] [Google Scholar]
- Miller J. H. (1972). Experiments in Molecular Genetics. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Miyazaki R., Van Der Meer J. R. (2011). A dual functional origin of transfer in the ICEclc genomic island of Pseudomonas knackmussii B13. Mol. Microbiol. 79 743–758. 10.1111/j.1365-2958.2010.07484.x [DOI] [PubMed] [Google Scholar]
- Murányi G., Szabó M., Olasz F., Kiss J. (2016). Determination and analysis of the putative AcaCD-responsive promoters of Salmonella genomic island 1. PLoS One 11:e0164561. 10.1371/journal.pone.0164561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Núñez B., De La Cruz F. (2001). Two atypical mobilization proteins are involved in plasmid CloDF13 relaxation. Mol. Microbiol. 39 1088–1099. 10.1046/j.1365-2958.2001.02308.x [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Balzer D., Kruft V., Lurz R., Lanka E. (1990). In vitro assembly of relaxosomes at the transfer origin of plasmid RP4. Proc. Natl. Acad. Sci. U.S.A. 87 6555–6559. 10.1073/pnas.87.17.6555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E. (1991). Common sequence motifs in DNA relaxases and nick regions from a variety of DNA transfer systems. Nucleic Acids Res. 19 3455. 10.1093/nar/19.12.3455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pansegrau W., Lanka E., Barth P. T., Figurski D. H., Guiney D. G., Haas D., et al. (1994). Complete nucleotide sequence of birmingham IncPα plasmids. J. Mol. Biol. 239 623–663. 10.1006/jmbi.1994.1404 [DOI] [PubMed] [Google Scholar]
- Pansegrau W., Schröder W., Lanka E. (1993). Relaxase (TraI) of IncP alpha plasmid RP4 catalyzes a site-specific cleaving-joining reaction of single-stranded DNA. Proc. Natl. Acad. Sci. U.S.A. 90 2925–2929. 10.1073/pnas.90.7.2925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulin-Laprade D., Carraro N., Burrus V. (2015). The extended regulatory networks of SXT/R391 integrative and conjugative elements and IncA/C conjugative plasmids. Front. Microbiol. 6:837. 10.3389/fmicb.2015.00837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J., Fritsch E. F., Maniatis T. (1989). Molecular Cloning: A Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press. [Google Scholar]
- Schultz E., Barraud O., Madec J.-Y., Haenni M., Cloeckaert A., Ploy M.-C., et al. (2017). Multidrug resistance Salmonella genomic Island 1 in a Morganella morganii subsp. morganii human clinical isolate from France. mSphere 2:e00118–17. 10.1128/mSphere.00118-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siebor E., Neuwirth C. (2011). The new variant of Salmonella genomic island 1 (SGI1-V) from a Proteus mirabilis French clinical isolate harbours blaVEB-6 and qnrA1 in the multiple antibiotic resistance region. J. Antimicrob. Chemother. 66 2513–2520. 10.1093/jac/dkr335 [DOI] [PubMed] [Google Scholar]
- Siebor E., Neuwirth C. (2014). Proteus genomic island 1 (PGI1), a new resistance genomic island from two Proteus mirabilis French clinical isolates. J. Antimicrob. Chemother. 69 3216–3220. 10.1093/jac/dku314 [DOI] [PubMed] [Google Scholar]
- Smith C. J., Parker A. C. (1998). The transfer origin for Bacteroides mobilizable transposon Tn4555 is related to a plasmid family from gram-positive bacteria. J. Bacteriol. 180 435–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solovyev V., Salamov A. (2011). “Automatic annotation of microbial genomes and metagenomic sequences,” in Metagenomics and Its Applications in Agriculture, Biomedicine and Environmental Studies, ed. Li R. W. (Hauppauge, NY: Nova Science Publishers; ), 61–78. [Google Scholar]
- Swalla B. M., Gumport R. I., Gardner J. F. (2003). Conservation of structure and function among tyrosine recombinases: homology-based modeling of the lambda integrase core-binding domain. Nucleic Acids Res. 31 805–818. 10.1093/nar/gkg142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Threlfall E. J., Frost J. A., Ward L. R., Rowe B. (1994). Epidemic in cattle and humans of Salmonella Typhimurium DT 104 with chromosomally integrated multiple drug resistance. Vet. Rec. 134:577. 10.1136/vr.134.22.577 [DOI] [PubMed] [Google Scholar]
- Tsvetkova K., Marvaud J. C., Lambert T. (2010). Analysis of the mobilization functions of the vancomycin resistance transposon Tn1549, a member of a new family of conjugative elements. J. Bacteriol. 192 702–713. 10.1128/JB.00680-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsaki A., Lucas M., Afendra A. S., Drainas C., De La Cruz F. (2003). Genetic and biochemical characterization of MbeA, the relaxase involved in plasmid ColE1 conjugative mobilization. Mol. Microbiol. 48 481–493. 10.1046/j.1365-2958.2003.03441.x [DOI] [PubMed] [Google Scholar]
- Wisniewski J. A., Traore D. A., Bannam T. L., Lyras D., Whisstock J. C., Rood J. I. (2016). TcpM: a novel relaxase that mediates transfer of large conjugative plasmids from Clostridium perfringens. Mol. Microbiol. 99 884–896. 10.1111/mmi.13270 [DOI] [PubMed] [Google Scholar]
- Wozniak R. A., Fouts D. E., Spagnoletti M., Colombo M. M., Ceccarelli D., Garriss G., et al. (2009). Comparative ICE genomics: insights into the evolution of the SXT/R391 family of ICEs. PLoS Genet. 5:e1000786. 10.1371/journal.pgen.1000786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner E. L., Prüger H., Grohmann E., Espinosa M., Högenauer G. (1997). Specific cleavage of chromosomal and plasmid DNA strands in gram-positive and gram-negative bacteria can be detected with nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 94 7435–7440. 10.1073/pnas.94.14.7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegelin G., Furste J. P., Lanka E. (1989). TraJ protein of plasmid RP4 binds to a 19-base pair invert sequence repetition within the transfer origin. J. Biol. Chem. 264 11989–11994. [PubMed] [Google Scholar]
- Ziegelin G., Pansegrau W., Lurz R., Lanka E. (1992). TraK protein of conjugative plasmid RP4 forms a specialized nucleoprotein complex with the transfer origin. J. Biol. Chem. 267 17279–17286. [PubMed] [Google Scholar]
- Zuker M. (2003). Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31 3406–3415. 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.