Abstract
Background
Long-tailed and pig-tailed macaque monkeys are natural hosts of Plasmodium knowlesi, which has been identified as a fifth malaria parasite infecting humans. In this study, we investigated possible infection by this Plasmodium parasite in macaque monkeys using a combination of polymerase chain reaction amplification and sequencing.
Methods
Forty-five blood samples were obtained in 2010 from macaques in northern Myanmar near Yunnan Province of China and investigated for possible infection with Plasmodium species using a nested polymerase chain reaction method for amplification of 18S SSU rRNA genes.
Results
Positive amplification was obtained from one monkey, and both sequence and phylogenetic analysis indicated that the parasite was of the Hepatocystis species lineage.
Conclusion
The results suggest that a combination of polymerase chain reaction amplification and sequence identification would be necessary for detection of Plasmodium knowlesi infection in both humans and its natural hosts.
Keywords: Plasmodium knowlesi, monkey, parasite, malaria
Background
In recent years, a fifth Plasmodium species, ie, P. knowlesi, has been identified as an invasive pathogen in humans,1 and cases of P. knowlesi infection have been concentrated in southeast Asian countries.2,3 We recently found that the prevalence of P. knowlesi in the border area between China and Myanmar was more than 20% in malaria-infected patients, and the parasite was frequently found in coinfection with either P. falciparum or P. vivax.4
Currently, species identification of P. knowlesi and differentiation from other malarial parasites has relied mainly on polymerase chain reaction (PCR) amplification of 18S SSU rRNA genes.1,5 Studies have indicated that it is difficult to differentiate P. knowlesi from other parasites by examination of blood smears.6 Further, apart from P. knowlesi, Hepatocystis species have been reported to be common pathogens in macaque monkeys.7–9 The genus of blood-parasitizing hemosporines (family Plasmodiidae) features gametocytes in red cells and cyst-like exoerythrocytic schizonts in the liver parenchyma. These are parasitic in old world primates, bats, and squirrels, but not in domestic animals.7 Hepatocystis species are transmitted by biting Culicoides species, such as C. adersi.8,9 Escalante et al first identified a species of Hepatocystis in a phylogeny of malarial parasites and found it clustered within the primate Plasmodium.10 Perkins and Schall suggested that Hepatocystis shared a common ancestor closer to primate malaria parasites than to avian parasites.11 Infected animals are asymptomatic and do not experience hemolysis.12 It was reported that Hepatocystis parasites do not produce the cyclical fever spikes typical of malaria in humans, but do cause anemia and visible merocyst formation, followed by scarring in the liver.7,13 Thus, it is clinically important to differentiate Hepatocystis parasites from other malaria parasites in both natural hosts and humans, for clinical diagnosis as well as for epidemiological clarification.
Systematic molecular studies provide an independent source of evidence for elucidating the origin of parasites. Studies found that partial gene conversion occurs among nonhomologous copies of the 18S SSU rRNA genes that are expressed during different parasite stages, which can generate misleading phylogenetic results.14 To overcome this limitation, we studied the cytochrome b gene independent of the mitochondrial genome, which is evolutionarily conserved.15 In this study, we investigated possible infection of Plasmodium parasite in macaque monkeys using a combination of PCR amplification and sequencing.
Materials and methods
Blood samples
Forty-five blood samples from macaque monkeys were collected in the border area between China and Myanmar. A 20–50 µL drop of finger-prick blood was spotted directly onto premarked filter paper which was allowed to dry in air and was then stored individually in plastic bags. A blood smear was made for each monkey and stained using Geimsa solution followed by microscopy. The study was performed with permission from the ethics committee of the Institute for Parasitic Disease Control of Yunnan province and the local administration authority in Myanmar.
DNA extraction
DNA templates were prepared from whole blood spots on filter paper according to a previously reported method.16 Each filter paper punch was soaked in 125 µL of methanol. After incubation at room temperature for 15 minutes, the methanol was removed and the samples were dried before adding 65 µL of distilled water. The punches were mashed using a new pipette tip for each punch and heated at 97°C for 15 minutes to elute the DNA.
PCR amplification of 18S SSU rRNA gene
The DNA samples were initially analyzed using specific nested PCR assays as described elsewhere.5 The first PCR amplification (nest 1) using Plasmodium genus-specific primers for each sample was carried out in a 50 µL reaction mixture containing 2.5 mM of each primer (rPLU1 and rPLU5), 1 × PCR buffer, 2.5 mM of each dNTP, 1.25 U of Taq DNA polymerase (Promega, Madison, WI) and 0.5 µg of DNA template. Two microliters of nest 1 amplification was used as the template DNA in the second PCR amplification (nest 2) using the self-designed primers (CHN18-R 5′-TAA GGA TAA CTA CGG AAA AGC TGT-3′ and CHN18-F 5′-AAG ATT ACG ACG GTA TCT GA-3′). The PCR conditions were 4 minutes at 94°C, 30 seconds at 94°C, 30 seconds at 58°C, one minute at 72°C for 36 cycles, and a final 10-minute extension at 72°C. Amplicons were examined by 1% agarose gel electrophoresis and ethidium bromide staining.
DNA bands were removed from the gel, purified using the QIAquick gel extraction kit (Qiagen, Valencia, CA), and ligated to a T-cloning vector (Invitrogen, Carlsbad, CA) according to protocols provided by the manufacturers. The DNA sequences were determined from at least five plasmid clones and sequenced by BGI (Beijing, China).
PCR amplification of cytochrome b gene of Hepatocystis
To confirm further that the sequences amplified with the 18S SSU rRNA primers were derived from Hepatocystis, the cytochrome b fragments of Hepatocystis were amplified using nested PCR. The cytochrome b nested PCR primers were CHNb 1 (5′-GAG AAT TAT GGA GTG GAT GGT G-3′), CHNb 2 (5′-GTG GTA ATT GAC ATC CAA TCC-3′), CHNb 3 (5′-GGT GTT TCA GAT ATA TGC ATG C-3′), and CHNb 4 (5′-CAT CCA ATC CAT AAT AAA GCA TAG-3′), which were designed based on the Hepatocystis cytochrome b gene (GU930067.1). The PCR reaction was carried out in a total volume of 25 µL under the following conditions: 1 µM of each primer, ie, CHNb 1 and CHNb 2, 2.5 mM of each dNTP, 0.5 U of DNA polymerase (Promega), 0.1 µg DNA template, and 1 × reaction buffer. The PCR conditions were 4 minutes at 94°C, 30 seconds at 94°C, 30 seconds at 55°C, 90 seconds at 72°C for 36 cycles, and a final 10-minute extension at 72°C. Nested PCR was carried out using 1 µL of the PCR products and performed with CHNb 3 and CHNb 4 primers. The PCR conditions were 4 minutes at 94°C, 30 seconds at 94°C, 30 seconds at 58°C, 30 seconds at 72°C for 35 cycles, and a final seven-minute extension at 72°C. The PCR products was detected by agarose electrophoresis, and cloned and sequenced as described earlier.
Sequence analysis
Sequences were first blasted in the Genbank for identification. Homologous sequences were aligned using the CLUSTAL W program.17 Phylogenetic trees were reconstructed by the neighbor-joining method.18,19 Reliability of clustering patterns was tested by bootstrapping,20 and 1000 bootstrap samples were used.
Results
PCR amplification
In this study, 45 macaque blood samples were first investigated for possible infection of P. knowlesi by nested PCR amplification of 18S SSU rRNA. A positive amplification was obtained from one monkey (Figure 1A, lane 8). Further sequence identification found that the positive product might be generated from a closely related organism, ie, Hepatocystis species. For further clarification, primer sets were designed based on both 18S SSU rRNA and mitochondrial cytochrome b genes of Hepatocystis species. The PCR product of the secondary amplification of the 18S SSU rRNA gene was 961 bp4 (Figure 1B, lane 1). The size of the PCR product of the mitochondrial cytochrome b gene was 788 bp (Figure 1B, lane 2).
Figure 1.
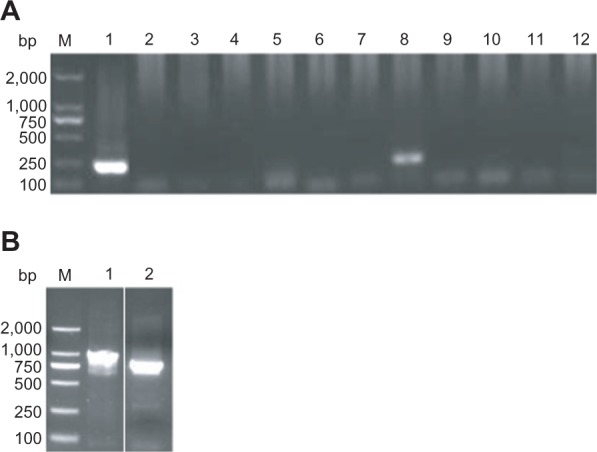
Polymerase chain reaction amplification of 18S SSU rRNA and cytochrome b genes. (A) Nested polymerase chain reaction amplification of the 18S SSU rRNA gene using the primers rPLU3 and rPLU4 matched to all pathogenic Plasmodium species. Lane 1 is a positive control. Lanes 2–11 are products of amplification of 10 monkey blood samples, and lane 12 is a negative control without DNA. (B) The 18S SSU rRNA and the cytochrome b genes were amplified by nested polymerase chain reaction with specific primers to the sequences of Hepatocystis species. Lanes 1 and 2 were the amplicons of 18S SSU rRNA using the primers CHN18-R and CHN18-F and the cytochrome b genes with the primers CHNb 3 and CHNb 4, respectively.
T18S SSU rRNA sequences and cytochrome b derived from Hepatocystis
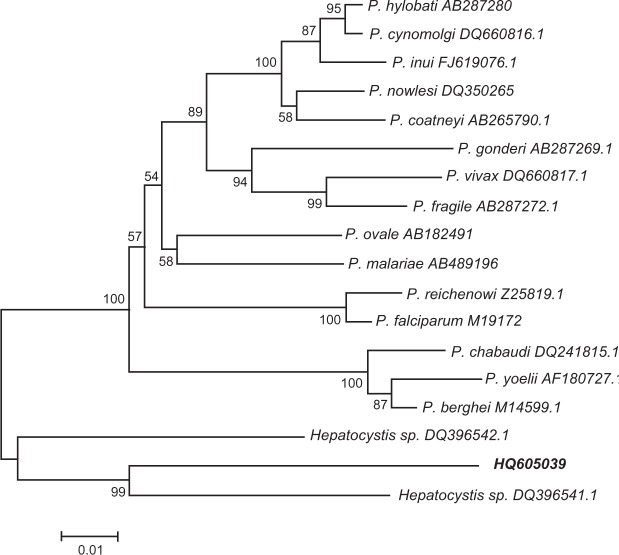
Sequences of the PCR products were blasted against the sequences in GenBank (http://blast.ncbi.nlm.nih.gov/). Phylogenetic analysis of the 18S SSU rRNA gene (GenBank HQ605039) and those of published malaria parasites showed that the sequence was more related to Hepatocystis species than to Plasmodium species (Figure 2).
Figure 2.
Phylogenetic analysis based on the 18S SSU rRNA genes of malaria and Hepatocystis species using the neighbour-joining method. HQ605039 was the sequence newly obtained in this study.
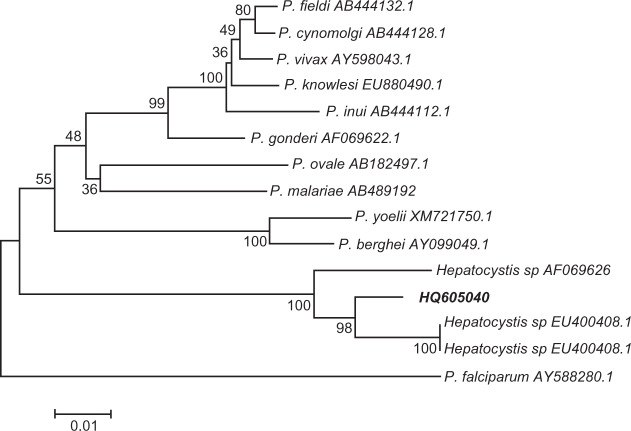
The PCR product of cytochrome b gene was analyzed to confirm that the 18S SSU rRNA sequence was truly of Hepatocystis origin. The 788 bp PCR product was sequenced and only one sequence was obtained. A blast search indicated that the sequence has a high similarity with that of Hepatocystis species lineage. A phylogenetic tree was constructed with the novel sequence (HQ605040) and related cytochrome b sequences from Hepatocystis species isolated from Myanmar macaques and other malaria parasites. The sequence clearly clustered with that of Hepatocystis species (Figure 3).
Figure 3.
Phylogenetic analysis based on the mitochondrial cytochrome b genes of malaria and Hepatocystis species using the neighbour-joining method. HQ605040 was newly obtained in this study.
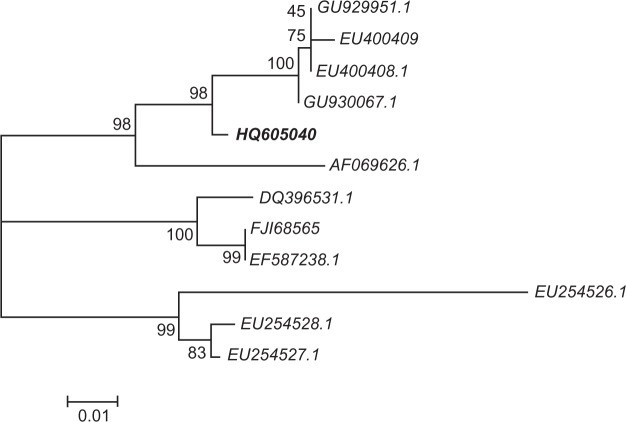
Further phylogenetic analysis of the cytochrome b sequences (see Table 1) showed that these sequences form three distinct clusters (Figure 4). A cluster with 98% bootstrap indicated that our sequence (HQ605040) formed a clade of primate species (GU929951, EU400409, EU400408, GU930067) from Southeast Asian and East Africa (AF069626, Figure 4). The bat Hepatocystis species fell into two divergent clades. The first contained three parasite sequences from Pteropus (FJ168565, EF587238, DQ396531) from Southeast Asia. The other species of Rousettus (EU254526) from the same region formed a sister group to the Hepatocystis species in Rousettus (EU254527, EU254528) from West Africa (Guinea, Figure 4).
Table 1.
Sequence accession numbers and origin of the mitochondrial cytochrome b gene identified in Hepatocystis species, which was used in the phylogenetic analysis of Figure 4
| GenBank accession | Geographic local source | Parasite host |
|---|---|---|
| GU929951 | Southeast Asian | Primate |
| EU400409 | Thailand | Primate |
| EU400408 | Thailand | Primate |
| GU930067 | Thailand | Primate |
| AF069626 | Ethiopia | Primate |
| HQ605040 | Myanmar | Primate |
| FJ168565 | Asian | Pteropus |
| EF587238 | Malaysia | Pteropus |
| DQ396531 | Malaysia | Pteropus |
| EU254526 | Singapore | Rousettus |
| EU254527 | Guinea | Rousettus |
| EU254528 | Guinea | Rousettus |
Figure 4.
Phylogeny of Hepatocystis species using mitochondrial cytochrome b genes. Numbers on branches are percentages of 1000 bootstrap supporting a given branch. Sequences from Hepatocystis species are listed along with their respective GenBank accession numbers. HQ605040 was newly obtained in this study.
Discussion
In this study, 45 macaque monkeys were investigated for possible infection of Plasmodium species using the classical PCR method by amplification of the 18S SSU rRNA genes. The blood samples were first examined on-spot by microscopy and no parasite was observed. Surprisingly, a positive amplification was obtained from one sample using the primers specific for Plasmodium species. The molecular size of the PCR product was as expected (Figure 1A). Since P. knowlesi was detected earlier in human blood samples in the same region,4 we suspected that the amplified sequence might be derived from P. knowlesi. A blast search with the sequence against the sequences in the Genbank indicated that the sequence was more related to that of Hepatocystis species, although it had high similarity to the 18S SSU rRNA sequences of Plasmodium species (data not shown). A literature search indicated that Hepatocystis has indeed been found in the monkey in the region before.21,22
The result of the 18S SSU rRNA sequences was further confirmed by amplification of mitochondrial cytochrome b gene with a nested PCR. The sequence of the amplified product clustered within that of Hepatocystis. Thus, the parasite identified in the macaque monkey belonged to the genus of Hepatocystis. Importantly, only one sequence was obtained with the second set of primers, indicating only one species existed in the samples.
Our results are reliable for three reasons. First, nested PCR methods have proved to be more specific and sensitive than conventional microscopy, and nested PCR assays are extremely valuable tools for obtaining accurate epidemiologic data.17 Second, the 18S SSU rRNA and cytochrome b gene from the mitochondrial genome have been regarded as reference sequences for the detection of Hepatocystis,8,22 and we applied the sequence signatures of both 18S SSU rRNA and the cytochrome b gene to identify the parasite. Third, phylogenetic analysis based on the two genes supported a close phylogenetic relationship of both genes with that from the Hepatocystis species lineage. Finally, our data suggest that both PCR and deep sequence analysis are necessary for correct identification of primate malaria parasites which may have a similar genome sequence signature to that of Hepatocystis species.
Conclusion
In this study, the possible prevalence of P. knowlesi in 45 long-tailed macaque monkeys in northern Myanmar was investigated using Plasmodium genus-specific primers followed by sequencing of the PCR product. Positive amplification was obtained from one sample. Sequence comparison indicated that the sequence had high similarity to that of Hepatocystis species. Further confirmation with primers to both 18S SSU rRNA and mitochondrial cytochrome b genes suggested that the monkey was infected by Hepatocystis species, but not P. knowlesi. Thus, the results suggest that a combination of PCR amplification and sequence identification is necessary for correct detection of P. knowlesi infection in both humans and its natural hosts.
Acknowledgments
This study was carried out with support to QC from the National Basic Research Program of China (973 Program, 2007CB513100), the National Science and Technology Project (2008zc-10004-011), the Chinese Natural Scientific Foundation (81171592, 81130033), and the institutional grants to QC from Institute of Pathogen Biology (IPB, CAMS) and Jilin University, China.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Singh B, Sung LK, Matusop A, et al. A large focus of naturally acquired Plasmodium knowlesi infections in human beings. Lancet. 2004;363:1017–1024. doi: 10.1016/S0140-6736(04)15836-4. [DOI] [PubMed] [Google Scholar]
- 2.Baird JK. Malaria zoonoses. Travel Med Infect Dis. 2009;7:269–277. doi: 10.1016/j.tmaid.2009.06.004. [DOI] [PubMed] [Google Scholar]
- 3.Cox-Singh J, Singh B. Knowlesi malaria: newly emergent and of public health importance? Trends Parasitol. 2008;24:406–410. doi: 10.1016/j.pt.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jiang N, Chang Q, Sun X, et al. Co-infections with Plasmodium knowlesi and other malaria parasites, Myanmar. Emerg Infect Dis. 2010;16:1476–1478. doi: 10.3201/eid1609.100339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh B, Bobogare A, Cox-Singh J, Snounou G, Abdullah MS, Rahman HA. A genus- and species-specific nested polymerase chain reaction malaria detection assay for epidemiologic studies. Am J Trop Med Hyg. 1999;60:687–692. doi: 10.4269/ajtmh.1999.60.687. [DOI] [PubMed] [Google Scholar]
- 6.Lee KS, Cox-Singh J, Brooke G, Matusop A, Singh B. Plasmodium knowlesi from archival blood films: Further evidence that human infections are widely distributed and not newly emergent in Malaysian Borneo. Int J Parasitol. 2009;39:1125–1128. doi: 10.1016/j.ijpara.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garnham PCC. Malaria Parasites and Other Haemosporidia. Oxford, UK: Blackwell Scientific; 1966. [Google Scholar]
- 8.Seethamchai S, Putaporntip C, Malaivijitnond S, Cui L, Jongwutiwes S. Malaria and Hepatocystis species in wild macaques, southern Thailand. Am J Trop Med Hyg. 2008;78:646–653. [PubMed] [Google Scholar]
- 9.Garnham PC, Heisch RB, Minter DM. Culicoides adersi Ingram and Macfie, 1923, a presumed vector of hepatocystis (=Plasmodium) kochi (Laveran, 1899) Nature. 1961;190:737–741. [Google Scholar]
- 10.Escalante AA, Freeland DE, Collins WE, Lal AA. The evolution of primate malaria parasites based on the gene encoding cytochrome b from the linear mitochondrial genome. Proc Natl Acad Sci U S A. 1998;95:8124–8129. doi: 10.1073/pnas.95.14.8124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Perkins SL, Schall JJ. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J Parasitol. 2002;88:972–978. doi: 10.1645/0022-3395(2002)088[0972:AMPOMP]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 12.Zeiss CJ, Shomer N. Hepatocystosis in a baboon (Papio anubis) Contemp Top Lab Anim Sci. 2001;40:41–42. [PubMed] [Google Scholar]
- 13.Tung J, Primus A, Bouley AJ, Severson TF, Alberts SC, Wray GA. Evolution of a malaria resistance gene in wild primates. Nature. 2009;460:388–391. doi: 10.1038/nature08149. [DOI] [PubMed] [Google Scholar]
- 14.Escalante AA, Goldman IF, De Rijk P, et al. Phylogenetic study of the genus Plasmodium based on the secondary structure-based alignment of the small subunit ribosomal RNA. Mol Biochem Parasitol. 1997;90:317–321. doi: 10.1016/s0166-6851(97)00121-7. [DOI] [PubMed] [Google Scholar]
- 15.Milinkovitch MC, Meyer A, Powell JR. Phylogeny of all major groups of cetaceans based on DNA sequences from three mitochondrial genes. Mol Biol Evol. 1994;11:939–948. doi: 10.1093/oxfordjournals.molbev.a040164. [DOI] [PubMed] [Google Scholar]
- 16.Bereczky S, Martensson A, Gil JP, Farnert A. Short report: Rapid DNA extraction from archive blood spots on filter paper for genotyping of Plasmodium falciparum. Am J Trop Med Hyg. 2005;72:249–251. [PubMed] [Google Scholar]
- 17.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr Protoc Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2: Unit 2.3. [DOI] [PubMed] [Google Scholar]
- 18.Saitou N, Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 19.Nei M, Kumar S. Molecular Evolution and Phylogenetics. Oxford, UK: Oxford University Press; 2000. [Google Scholar]
- 20.Felsenstein J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution. 1985;39:783–791. doi: 10.1111/j.1558-5646.1985.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 21.Takenaka T, Hashimoto K, Gotou H, Matsumoto S, Nishikawa T. Studies on Hepatocystis spp. in rhesus monkeys from Yunnan, China. Jikken Dobutsu. 1990;39:273–279. doi: 10.1538/expanim1978.39.2_273. Japanese. [DOI] [PubMed] [Google Scholar]
- 22.Putaporntip C, Jongwutiwes S, Thongaree S, Seethamchai S, Grynberg P, Hughes AL. Ecology of malaria parasites infecting Southeast Asian macaques: evidence from cytochrome b sequences. Mol Ecol. 2010;19:3466–3476. doi: 10.1111/j.1365-294X.2010.04756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]