Abstract
The role of small noncoding RNAs, termed microRNAs (miRs), in development and disease has been recognized for many years. The number of miRs and regulated targets that reinforce a role for miRs in human disease and disease progression is ever-increasing. However, less is known about the involvement of miRs in steady-state, nondisease homeostatic pathways. In the kidney, much of the regulated ion transport is under the control of hormonal signaling. Evidence is emerging that miRs are involved in the hormonal regulation of kidney function and, particularly, in ion transport. In this short review, the production and intra- and extracellular signaling of miRs and the involvement of miRs in kidney disease are discussed. The discussion also focuses on the role of these small biological molecules in the homeostatic control of ion transport in the kidney. MiR regulation of and by corticosteroid hormones, in particular the mineralocorticoid hormone aldosterone, is considered. While information about the role of aldosterone-regulated miRs in the kidney is limited, an increase in the research in this area will undoubtedly highlight the involvement of miRs as central mediators of hormonal signaling in normal physiology.
Keywords: epithelial sodium channel, microRNAs, aldosterone, mineralocorticoids, steroid hormone, kidney, collecting duct
the short, noncoding pieces of RNA, typically 22 or 23 nucleotides (nt) long, termed microRNAs (miRs), were first identified in the early 1990s in Caenorhabditis elegans. It was discovered that a small RNA regulated C. elegans development; however, the gene product did not encode a protein, and thus a role for these small noncoding pieces of RNA was discovered (44, 78, 95). One of these C. elegans miRs, let-7, was subsequently found in a number of different organisms, including humans, and the exploration of these short RNA species took off (3, 43, 68). Since that time, an increasing number of miRs have been described in a range of organisms. With the advent of next-generation sequencing approaches, a further explosion in miR identification has occurred. The database miRBase lists >1,800 precursor miRs and ∼2,600 mature miR forms in humans, with >28,000 miRs identified for all organisms (21, 37, 38). For the majority of miRs investigated, the short RNA sequences bind to the 3′-untranslated region (UTR) of target mRNAs to cause degradation of the mRNA (56). This, in turn, downregulates steady-state protein levels. Therefore, as a general rule, it is considered that the action of miRs is a decrease in target protein expression (40). However, with the rapid growth in studies investigating miRs, exceptions to this established paradigm of miR action have emerged (22), e.g., where binding of miRs to target sequences results in protein upregulation. MiRs have been shown to bind to the 5′-UTR, rather than the 3′-UTR, of mRNA or bind directly to coding sequences of proteins to interrupt their translation. MiRs may also alter the regulation of RNA synthesis itself (20, 22, 32, 56). Whatever the mechanism of control, it is considered that, via direct or indirect mechanisms, miRs regulate the expression of ≥30% of all protein-coding genes (19). It has been suggested that ∼90% of all protein expression is regulated in some way by noncoding RNAs (this includes miRs, other short RNA species, and the long noncoding RNAs) (1).
The genomic sequences that produce the typical stem-loop, precursor miR structure are encoded throughout the genome. MiR sequences embedded in introns of protein-coding genes or in exon and intron sequences of non-protein-coding RNAs have been identified. In production, there is a diversity of strategies that transcribe miRs (described in numerous reviews including refs. 22, 35, 99). Some miR transcription is driven by its own promoter, resulting in the independent control of miR production (8, 55). Other miRs are contained within gene sequences and, therefore, share the promoters of the encoded gene, locking them into coregulation with their surrounding gene (26, 50). To add to this diversity of expression, miRs can be produced as a single immature species, or they can form part of a miR cluster where miR sequences are in close genomic proximity (12, 28). In miR clusters, a number of precursor family members are transcribed at the same time and then processed individually to mature miRs. The consequence of these numerous miR production strategies is an extensive diversity of miR species that are regulated in a variety of ways. This then allows for an intricate regulatory network to be established in cells to alter and fine-tune protein expression.
The canonical pathway producing miRs begins with miRs transcribed by RNA polymerase II into primary miRs (pri-miRs) (4, 45, 46). Pri-miRs are longer (>70-nt) stem-loop hairpin structures with an elongated polyadenylated tail (22, 23). The pri-miRs are trimmed in the nucleus in a complex that contains the ribonuclease III enzyme Drosha to produce a ∼70-nt precursor miR (pre-miR) (34, 45). This pre-miR is next exported to cytoplasm from the nucleus by the action of exportin-5 for further processing (33, 51). In the cytoplasm the ends of the stem-loop pre-miRs are cleaved by the enzyme Dicer to produce a ∼23-nt RNA double strand (22, 67). The complementary strands, termed the guide and passenger strands, are separated as the guide strand is loaded into a complex with Argonaute proteins for the final assembly into a mature miR silencing complex, with the passenger strand often degraded (13, 63).
As the miR field develops, exceptions to this canonical pathway are being described. For example, transcription by RNA polymerase III, as opposed to II, has been described in detail (22, 23). MiRs contained within coding gene introns can be excised by spliceosome action to generate pre-miRs directly from genomic sequences. MiRs produced in this manner have been termed mirtrons (5, 62, 76). MiRs can be generated from small-nucleolar, short-hairpin, short-interfering, or transfer RNA (8, 86). These sources of miRs do not require the action of Drosha for processing to pre-miRs. Similarly, examples of miR production without the need for Dicer processing (e.g., pre-miR-451) have emerged (11, 98). Again, this diversity of miR production permits a range of strategies that cells can employ to generate miRs, each of which can be regulated in different ways. The details of miR production by canonical and noncanonical pathways have been extensively reviewed (22, 56, 86, 99) and are not discussed in detail here. However, an appreciation of the elaborate and dynamic regulation of and by miRs should be noted when the role of this small molecule in biological processes is considered. For almost any example where a clear path has been identified for miR production, targeting, and gene product regulation, exceptions have been noted. It therefore remains a challenge to conclusively demonstrate the linear involvement of miRs in regulating a target gene product to demonstrate the physiological significance of miR action. However, these more labor-intensive efforts are underway in a number of fields, including investigations of miR involvement in ion channel regulation, the topic addressed by this review.
MiRs in Kidney Disease
Even though the role of miRs in human disease is not the primary focus of this review, much of the interest in miRs arose because of their link to human diseases and their demonstrable role in disease progression (see Refs. 18, 27, 36, 59, and 70 as examples of thousands of publications). This is true for kidney pathophysiology and disease states, as well. Briefly, miRs have been shown to regulate or be involved in kidney development (7, 78, 89), kidney cancers, including renal cell carcinomas (29, 71, 94), diabetic nephropathy (10, 41, 89), polycystic kidney disease (66, 88), fibrotic kidney disease (14, 39, 61, 96), IgA nephropathy (10, 89), acute kidney injury (1, 93), and end-stage renal disease (24, 88, 89). Several studies have highlighted the role of miR-192, expressed in the kidney cortex, in the profibrotic progression of nephropathy (14, 30, 41). MiRs also play a role in kidney maintenance and normal kidney physiology, and conditional deletion of Dicer in podocytes is known to result in glomerular damage (24, 29, 100).
However, there are fewer reports of miR's involvement in the hormonal regulation of ion transport. Recent evidence, including a report by Edinger et al. (16), suggests that, here too, miRs may be important intermediaries in the signaling cascades that alter ion transport in the kidney.
MiRs and the Renin-Angiotensin-Aldosterone System Pathway
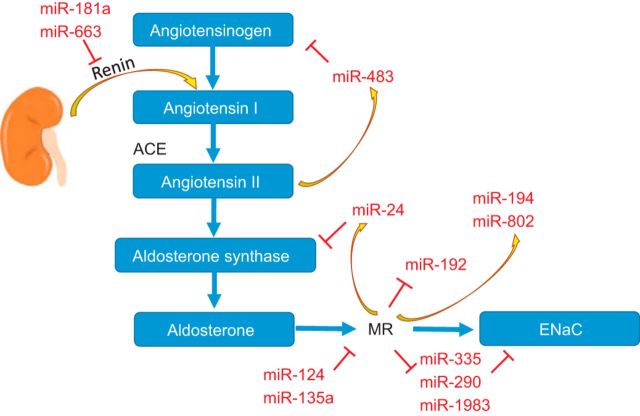
A well-established hormonal cascade is invoked in mammalian systems when Na+ reabsorption from the kidney nephron is required. Under conditions of low plasma Na+ or decreased blood volume, the renin-angiotensin (ANG)-aldosterone system (RAAS) is engaged as a homeostatic mechanism to increase Na+ uptake and expand blood volume (2, 9, 77, 81). A consequence of the increased Na+ reabsorption is water reabsorption, which leads to volume expansion. In this cascade, the final and major effector of Na+ balance is the mineralocorticoid hormone aldosterone (2). For ion channel regulation, aldosterone release is triggered in response to low plasma Na+ concentrations or elevated circulating K+ levels. The RAAS cascade is depicted in Fig. 1, but it is becoming apparent that, at each stage in this cascade, miRs play a role in the regulation of protein expression and, consequently, in the homeostatic regulation of electrolyte balance (see below).
Fig. 1.
Schematic representation of the renin-angiotensin-aldosterone system and points of microRNA (miR) regulation. Cascade of activation is indicated in the flow diagram. Role of miRs to regulate this cascade is illustrated by inhibitory (red) or stimulatory (yellow arrows) inputs. ACE, angiotensin-converting enzyme; ENaC, epithelial Na+ channel; MR, mineralocorticoid receptor.
Renin.
Aldosterone release is initiated by the production of renin in juxtaglomerular (JG) cells of the kidney, if they sense a decrease in renal perfusion. An indirect role for miRs in JG cell function was demonstrated with knockout of Dicer in JG precursors during kidney development to eliminate the production of all miRs (79). This resulted in a reduction of the number of the JG complexes in the mature kidney and a decrease in renin production. However, this decrease in renin production was the result of a developmental defect and failure to produce JG cells. Specific miRs involved in the regulation of JG cell development cannot be determined by Dicer1 knockout, as this would deplete the production of all miRs, and the specific miRs critical for JG cell development are still to be discovered. However, direct downregulation of renin expression has been demonstrated after increases in levels of miR-181a and miR-663, which were shown experimentally to bind to the 3′-UTR of renin (54). Marques et al. (54) reported that both of these miRs were repressed in samples obtained from hypertensive compared with normotensive subjects. As these miRs downregulate renin, the decreased levels of miR-181a and miR-633 were offered as a possible mechanism to account, in part, for the elevated blood pressure in the hypertensive patients (54). An increase in renin after release from the miR repression would lead to the eventual increase in aldosterone production, with associated hypernatremia and blood volume expansion.
Angiotensin.
Renin is responsible for the conversion of angiotensinogen to ANG I. Levels of angiotensinogen have been shown to be regulated by miRs. Specifically, Kemp et al. (31) reported that miR-483 was downregulated by ANG II to feed back and alter the expression of several RAAS components, including angiotensinogen. This miR also appeared to target additional members of the RAAS cascade, including the 3′-UTRs of ANG-converting enzymes 1 and 2 and ANG II receptor 2. The net result of repression of all these RAAS components by miR-483 would be downregulation of the RAAS cascade and reduction of aldosterone expression (31). However, aldosterone levels were not specifically measured in this study (31). The observation that miR-483 expression was altered by ANG II and that this miR, in turn, could target RAAS proteins provides an additional level of feedback control for the RAAS cascade and a new layer of regulation in homeostatic control of Na+ levels. In addition to alterations in miR levels, single-nucleotide polymorphisms (SNPs) in the target UTRs could also alter the regulation of the gene product if a canonical miR binding site is disrupted by the mutation. In a study investigating this possibility, Nossent et al. (60) identified SNPs in several RAAS genes that had the potential to alter miR binding. Disruption of miR binding would result in a derepression of the target protein and an upregulation of protein expression. Nossent et al. provided examples of such SNPs that were associated with increased blood pressure and linked to mutations in ANG II, ANG II receptors 1 and 2, renin, and the mineralocorticoid receptor (MR). While SNPs in protein coding sequences have long been identified as a source of sequence variation that could result in disease phenotypes, the possible alteration of miR binding in the UTRs is now also under consideration in an attempt to link pathophysiology to these genetic variations.
Aldosterone production.
The next step in the cascade leading to aldosterone production is the action of the steroidogenic enzyme aldosterone synthase (AS), encoded by the gene CYP11B2, in the adrenal cortex. Aldosterone production via AS is stimulated by ANG II, and expression of AS can ultimately determine the amount of aldosterone released from the adrenals (25). In a study conducted by Robertson et al. (74) to establish whether miRs had a role in the regulation of AS, the miR-processing enzyme Dicer was depleted in adrenocortical (H295R) cells, and changes in AS mRNA levels were determined. The authors demonstrated that a global reduction of miRs by knockdown of Dicer1 resulted in increased expression of AS and the related 11β-hydrolase (CYP11B1) responsible for the production of cortisol (74). These data would then point to a chronic repression in AS by miRs under steady-state conditions. A number of candidate miRs were tested to determine which miR (or group of miRs) could be responsible for altering AS levels, and miR-24 was identified as the most likely candidate. Overexpression of miR-24 reduced the expression of AS and 11β-hydrolase, and depletion of miR-24 increased mRNA levels of both enzymes. Furthermore, a corresponding change in aldosterone production was observed in the H295R cells with the exogenous manipulation of miR-24 levels (74). Interestingly, in unrelated studies, miR-24 was upregulated by the action of aldosterone signaling via its MR (49). It is possible then that miR-24 could constitute a feedback signaling loop, repressing the expression of AS when aldosterone levels are elevated, just as suggested for ANG II and angiotensinogen in the case of miR-483. However, this idea remains to be tested.
Additional evidence for miR regulation of AS was obtained from an investigation of a polymorphism in the human AS gene 3′-UTR. A polymorphism at position +735 in the 3′-UTR of AS has been shown to alter binding of miR-766. Maharjan et al. (53) reported that individuals carrying the 735G allele would bind the miR-766, while those with a 735A variant would not. This then could alter the steady-state levels of AS and change aldosterone production, depending on the polymorphism. In support of this idea, the authors demonstrated that transient overexpression of miR-766 decreased AS levels in the human H295R cells, which carry the 735G allele of the CYP11B2 gene. In an effort to link the potential miR regulation of AS to changes in blood pressure, Maharjan et al. alluded to a study investigating populations at increased risk for salt-sensitive hypertension, where a −344T allele in AS was associated with high blood pressure. This SNP is in linkage disequilibrium with the 735G allele, presenting the possibility that alteration of AS expression, regulated by miRs, could eventually alter circulating aldosterone expression and contribute to misregulated blood pressure. However, a conclusive demonstration of this pathway has yet to emerge.
Mineralocorticoid receptor.
Release of aldosterone into the blood represents the final step in the RAAS pathway to produce the effector hormone responsible for changes in Na+ and volume regulation. Aldosterone enters target cells, where it binds to its MR encoded by the gene NR3C2. The bound MR translocates into the nucleus and binds to response elements on accessible chromosomes to initiate the transcription of aldosterone-induced genes. This is the classical pathway describing the action of the RAAS cascade and mode of aldosterone action. The specificity of aldosterone signaling is protected by the action of 11β-hydroxysteroid dehydrogenase (11β-HSD2), which prevents nonselective activation of MR by cortisol. However, accumulating evidence suggests that miRs have now infiltrated this long-established paradigm, and examples can be found for miRs altering steroid receptor expression and 11β-HSD2 action. MiR-124 and -135a were shown experimentally to bind to and alter the expression of MR (80). Similarly, Rezaei et al. (72) reported that miR-20a reduced the expression of 11β-HSD2 in rat models. They also demonstrated that miR-20a was differentially expressed in hypertensive vs. nonhypertensive rodent models. In addition, they found that miR-20a was slightly enriched in the cortical collecting duct (CCD) compared with proximal tubule segments (72). Of interest and with reference to the description provided above, the miR-23-24-27 cluster family was also seen to be enriched in the CCD, providing an additional line of evidence that miR-24 may be a central regulator of RAAS signaling and aldosterone action in the CCD, but again this will need to be validated experimentally.
Therefore, for each step in the RAAS signaling cascade, some involvement of miR regulation has been described. It is certain that, as miRs continue to be investigated, additional examples of physiological regulation of this pathway by miRs will emerge. The next question to be addressed is not miR regulation of aldosterone production but, rather, miR regulation by aldosterone and the impact on ion channel regulation in the kidney CCD.
Aldosterone Regulation of miRs
Profiling of miR levels in model systems or samples derived from human tissue previously involved the use of microarrays, complemented by quantitative PCR techniques to validate any changes observed in the array data. More recently, the advent of deep sequencing approaches has allowed assay of the entire library of small RNA species in one read, to gain an enormous volume of data from biological samples. The arduous task of assembling the datasets, validating initial findings, and determining potential miR target mRNAs has been a much more difficult process. Yet these efforts have been successful and produced an explosion of miR-related publications investigating target interactions and pathways. An investigation of miR regulation by steroid hormones has also begun. While all steroid hormones are not discussed here, it should be noted that estrogen, androgens, and progesterone have been shown to both induce and repress miR production in a number of different tissues (15). This topic has been addressed in several reviews (64, 65). For the corticosteroid hormones, however, there is little information on miR regulation, with the majority of studies focusing on glucocorticoid regulation, rather than mineralocorticoids (74, 91).
Impact of Aldosterone-Regulated miR Channels and Transporter Function in the Kidney
A small number of studies of aldosterone regulation of Na+ and K+ transport by miRs have emerged. In one of the first of such studies, Elvira-Matelot et al. (17) identified miR-192 as a possible regulator of K+ secretion. Expression of this miR was shown to be inhibited in mice by maneuvers that increase circulating aldosterone levels, including K+ load, salt depletion, or chronic aldosterone infusion. One of the confirmed targets of miR-192 was the serine-threonine kinase, with no lysine (WNK1), specifically the long form (L-WNK1). Elvira-Matelot et al. showed that L-WNK1 protein expression increased without changes in mRNA levels. This suggests that posttranslation modifications may be responsible for the change in L-WNK1 expression, and it was confirmed that miR-192 was the effector (17). From a number of studies, the role of L-WNK and the kidney-specific form KS-WNK has been elucidated, and it is known that L-WNK1 is an important regulator of both K+ and Na+ transport (42, 84, 92, 97).
In a separate study, Lin et al. (48) described regulation of renal outer medullary K+ (ROMK) channels by miRs. They found that a change in K+ diet induced concomitant changes in miR-194 expression: a high-K+ diet increased miR-194 expression, and vice versa. A scaffold and regulatory protein, intersectin 1, was determined to be a target of this regulated miR. An increase in miR-194 reduced intersectin 1, and this, in turn, prevented the internalization of ROMK (48). Consequently, a longer membrane residency resulted in increased K+ transport. In an earlier study, Lin et al. (47) identified another miR that was regulated by changes in K+ diet, namely, miR-802. In this case, the target protein was caveolin-1 (47). Similar to the miR-194 example, a high-K+ diet induced miR-802 production, which inhibited caveolin-1 expression and reduced ROMK internalization. This then provides two separate mechanisms to regulate K+ transport, but with similar outcomes. It is likely that a number of miRs acting though different pathways, but working in concert, may converge to alter ROMK expression at the plasma surface and, ultimately, increase K+ transport in the distal kidney nephron.
For Na+ regulation, there is little information. The alteration of WNK expression by miR-192 would play into regulation of the Na+-Cl− cotransporter and the epithelial Na+ channel (ENaC), as WNKs are known regulators of these transporters. However, direct regulation of ENaC function by miRs was only recently demonstrated by our group as a possible intermediary component of the aldosterone signaling cascade (16). To our knowledge, our report represents the first example of this mode of regulation for ENaC in the kidney (16). Regulation of ENaC by miRs has been recently reported for miR-16 in alveolar epithelial cells (87) and regulation of miR-101 and miR-199 by ENaC in endometrial cells (85). Neither of these reports focused on miR regulation by hormones.
In an effort to understand the role of aldosterone in miR regulation in the kidney, Edinger et al. (16) stimulated a model mouse CCD (mCCD) cell line with aldosterone and profiled miR expression using microarrays. A small subset of miRs was significantly up- and downregulated. The initial description focused on the downregulated miRs. The array expression data were validated using quantitative PCR to confirm that the identified miRs were regulated by aldosterone in the mCCD cells and in isolated CCD from mouse kidney (for mice fed low-Na+ diets). An in silico approach was used to identify targets for the regulated miRs, namely, miR-335, -290, and -1983. One of these, ankyrin-G (Ank-G), was verified as a target for the downregulated miRs and shown to alter ENaC activity. Expression and cellular localization of Ank-G were altered by aldosterone, and the total protein levels of Ank-G were regulated by the downregulated miRs directly (16). Ongoing studies are investigating the mechanism of Ank-G's action on ENaC in the CCD. However, just as for ROMK, it appears that the change in membrane surface residency may account, in part, for the upregulation observed with increased Ank-G expression (unpublished observations).
Our focus has recently shifted to those miRs that are upregulated by aldosterone. Of interest, the miR cluster family miR-23-24-27 was significantly upregulated by aldosterone (Ref. 16 and unpublished data). This is the same group of miRs that has been implicated at several points in the RAAS cascade (see above). It will therefore be of interest to verify whether these miRs also play a role in the terminal regulation of Na+ transport in the kidney. Validation of such a role would offer an extensive feedback network to exquisitely regulate the RAAS cascade, and these studies are ongoing.
MiR Regulation of Other Kidney Transporters and Regulatory Proteins
To conclude this description of miRs and hormonal regulation in the kidney, a number of unconnected examples of miR regulation of kidney transporters are provided. Investigating the role of miRs in epithelial transport, Mladinov et al. (57) examined miR expression in epithelial cells in proximal vs. distal kidney nephron segments in rodents. Their work described 55 miRs that were differentially expressed in each segment. Of these, miR-192 was enriched in proximal tubules, and its expression was decreased when animals were fed a low-Na+ diet (which stimulates aldosterone release) (57). This finding is similar to the reports investigating regulation of ROMK function by miR-192 (see above). However, Mladinov et al. found that miR targeted the β1-subunit of the Na+-K+-ATPase. Knockdown of miR-192 increased β1-subunit expression, and regulation was suggested to occur through the 5′-UTR, rather than the 3′-UTR (noncanonical pathway). Other differentially regulated miRs were also demonstrated to interact with the 3′-UTR of transporters, including ROMK2 (miR-16, -195, and -382) and the Na+-K+-Cl− cotransporter (miR-16 and -195). The addition of another validated miR-192 target further highlights the nascent status of the miR field. It is almost certain that, with further investigation, additional targets of kidney-specific or enriched miRs will be uncovered. Understanding the miR interactome will obviously take some time.
The glucocorticoid-induced leucine zipper (GILZ) is known to regulate the function of ENaC in the distal kidney nephron. As its name suggests, this protein is induced by the action of glucocorticoids, as well as aldosterone (6, 58, 73, 82, 83). In a study investigating the glucocorticoid receptor, Vreugdenhil et al. (91) demonstrated that miR-18 and -124a were regulated by dexamethasone and that these miRs altered the expression of GILZ. The action of aldosterone was not considered in these studies. The data for aldosterone stimulation of CCD cells reported by Edinger et al. (16) showed no significant alteration in expression of these two miRs, and it could be that it is a glucocorticoid receptor-specific regulation.
For a study investigating miRs induced by osmotic stress, a transgenic mouse model overexpressing miR-466a was generated; this model was previously linked to osmotic stress pathways. In mice with overexpression of this miR, expression for a range of transporters and proteins in the kidney linked to osmoregulation, including aquaporins 1, 2, and 3, urea transporters 1, 2, and 3, the serum and glucocorticoid kinase 1 (SGK1), and osmotic response element-binding proteins, was reduced (52). SGK1 is a known regulator of ENaC (69, 75, 90). These mice also demonstrated an impaired ability to appropriately concentrate urine and effectively osmoregulate.
Conclusions
While much of the attention generated in the miR field has arisen due to their link to diseases and disease progression, it is becoming increasingly apparent that miRs may be involved in many of the long-characterized homeostatic physiological processes and pathways that regulate ion transport. This seems certainly to be the case for hormonal signaling linked to ion transport regulation in the kidney. While there are relatively few examples of miRs involved in aldosterone signaling, we, along with colleagues in the field, are beginning to identify miR's role here as well. The significance of miR action has yet to be fully appreciated. While the tools may not be available to dissect all the actions of miRs in aldosterone signaling, it is certain that additional examples of miR regulation will be uncovered. Over time, it is very possible that we may come to consider miRs as central players in signaling cascades, rather than merely a cellular mechanism to fine-tune protein levels. Differential regulation and expression of miRs, already noted in different nephron segments, hint at the possibility that miRs could be the site of signal integration. MiRs could therefore be the source of contextual regulation of hormonal inputs, to elicit an appropriate tissue- or cell-specific response and be responsible for the pleotropic actions noted, for example, in aldosterone signaling.
GRANTS
Studies described in the review that emerged from our lab were supported by the Carl W. Gottschalk Research Scholar Grant from the American Society of Nephrology.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author.
AUTHOR CONTRIBUTIONS
M.B.B. developed the concept and designed the research; M.B.B. drafted the manuscript; M.B.B. approved the final version of the manuscript.
ACKNOWLEDGMENTS
My thanks go to the current and past members of the lab for assistance and support and to all the colleagues whom I have had the privilege of working with.
M. B. Butterworth is the recipient of the New Investigator Award from the Cell and Molecular Physiology section of the American Physiological Society.
REFERENCES
- 1.Aguado-Fraile E, Ramos E, Conde E, Rodriguez M, Liano F, Garcia-Bermejo ML. MicroRNAs in the kidney: novel biomarkers of acute kidney injury. Nefrologia : 826–834, 2013. [DOI] [PubMed] [Google Scholar]
- 2.Atlas SA. The renin-angiotensin aldosterone system: pathophysiological role and pharmacologic inhibition. J Manag Care Pharm : 9–20, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Axtell MJ, Bartel DP. Antiquity of microRNAs and their targets in land plants. Plant Cell : 1658–1673, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, function. Cell : 281–297, 2004. [DOI] [PubMed] [Google Scholar]
- 5.Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell : 328–336, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bhalla V, Soundararajan R, Pao AC, Li H, Pearce D. Disinhibitory pathways for control of sodium transport: regulation of ENaC by SGK1 and GILZ. Am J Physiol Renal Physiol : F714–F721, 2006. [DOI] [PubMed] [Google Scholar]
- 7.Bhatt K, Mi QS, Dong Z. MicroRNAs in kidneys: biogenesis, regulation, and pathophysiological roles. Am J Physiol Renal Physiol : F602–F610, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Breving K, Esquela-Kerscher A. The complexities of microRNA regulation: mirandering around the rules. Int J Biochem Cell Biol : 1316–1329, 2010. [DOI] [PubMed] [Google Scholar]
- 9.Campese VM, Park J. The kidney and hypertension: over 70 years of research. J Nephrol : 691–698, 2006. [PubMed] [Google Scholar]
- 10.Chandrasekaran K, Karolina DS, Sepramaniam S, Armugam A, Wintour EM, Bertram JF, Jeyaseelan K. Role of microRNAs in kidney homeostasis and disease. Kidney Int : 617–627, 2012. [DOI] [PubMed] [Google Scholar]
- 11.Cheloufi S, Dos Santos CO, Chong MM, Hannon GJ. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature : 584–589, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chhabra R, Dubey R, Saini N. Cooperative and individualistic functions of the microRNAs in the miR-23a-27a-24-2 cluster and its implication in human diseases. Mol Cancer : 232, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Choe J, Cho H, Lee HC, Kim YK. MicroRNA/Argonaute 2 regulates nonsense-mediated messenger RNA decay. EMBO Rep : 380–386, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chung AC, Huang XR, Meng X, Lan HY. miR-192 mediates TGFβ/Smad3-driven renal fibrosis. J Am Soc Nephrol : 1317–1325, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cochrane DR, Cittelly DM, Richer JK. Steroid receptors and microRNAs: relationships revealed. Steroids : 1–10, 2011. [DOI] [PubMed] [Google Scholar]
- 16.Edinger RS, Coronnello C, Bodnar AJ, Laframboise WA, Benos PV, Ho J, Johnson JP, Butterworth MB. Aldosterone regulates microRNAs in the cortical collecting duct to alter sodium transport. J Am Soc Nephrol : 2445–2457, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Elvira-Matelot E, Zhou XO, Farman N, Beaurain G, Henrion-Caude A, Hadchouel J, Jeunemaitre X. Regulation of WNK1 expression by miR-192 and aldosterone. J Am Soc Nephrol : 1724–1731, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Erson AE, Petty EM. MicroRNAs in development and disease. Clin Genet : 296–306, 2008. [DOI] [PubMed] [Google Scholar]
- 19.Filipowicz W, Bhattacharyya SN, Sonenberg N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat Rev Genet : 102–114, 2008. [DOI] [PubMed] [Google Scholar]
- 20.Flynt AS, Greimann JC, Chung WJ, Lima CD, Lai EC. MicroRNA biogenesis via splicing and exosome-mediated trimming in Drosophila. Mol Cell : 900–907, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S. Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res : D154–D158, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol : 509–524, 2014. [DOI] [PubMed] [Google Scholar]
- 23.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev : 3016–3027, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harvey SJ, Jarad G, Cunningham J, Goldberg S, Schermer B, Harfe BD, McManus MT, Benzing T, Miner JH. Podocyte-specific deletion of dicer alters cytoskeletal dynamics and causes glomerular disease. J Am Soc Nephrol : 2150–2158, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hattangady NG, Olala LO, Bollag WB, Rainey WE. Acute and chronic regulation of aldosterone production. Mol Cell Endocrinol : 151–162, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hinske LC, Galante PA, Kuo WP, Ohno-Machado L. A potential role for intragenic miRNAs on their hosts' interactome. BMC Genomics : 533, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microRNA expression in human heart disease. Physiol Genomics : 367–373, 2007. [DOI] [PubMed] [Google Scholar]
- 28.Janga SC, Vallabhaneni S. MicroRNAs as post-transcriptional machines and their interplay with cellular networks. Adv Exp Med Biol : 59–74, 2011. [DOI] [PubMed] [Google Scholar]
- 29.Kato M, Arce L, Natarajan R. MicroRNAs and their role in progressive kidney diseases. Clin J Am Soc Nephrol : 1255–1266, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kato M, Zhang J, Wang M, Lanting L, Yuan H, Rossi JJ, Natarajan R. MicroRNA-192 in diabetic kidney glomeruli and its function in TGFβ-induced collagen expression via inhibition of E-box repressors. Proc Natl Acad Sci USA : 3432–3437, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kemp JR, Unal H, Desnoyer R, Yue H, Bhatnagar A, Karnik SS. Angiotensin II-regulated microRNA 483-3p directly targets multiple components of the renin-angiotensin system. J Mol Cell Cardiol : 25–39, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nat Rev Mol Cell Biol : 376–385, 2005. [DOI] [PubMed] [Google Scholar]
- 33.Kim VN. MicroRNA precursors in motion: exportin-5 mediates their nuclear export. Trends Cell Biol : 156–159, 2004. [DOI] [PubMed] [Google Scholar]
- 34.Kim VN. Small RNAs: classification, biogenesis, function. Mol Cells : 1–15, 2005. [PubMed] [Google Scholar]
- 35.Kim YK, Kim VN. Processing of intronic microRNAs. EMBO J : 775–783, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kluiver J, Kroesen BJ, Poppema S, van den Berg A. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia : 1931–1936, 2006. [DOI] [PubMed] [Google Scholar]
- 37.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res : D68–D73, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kozomara A, Griffiths-Jones S. miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res : D152–D157, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kriegel AJ, Fang Y, Liu Y, Tian Z, Mladinov D, Matus IR, Ding X, Greene AS, Liang M. MicroRNA-target pairs in human renal epithelial cells treated with transforming growth factor β1: a novel role of miR-382. Nucleic Acids Res : 8338–8347, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet : 597–610, 2010. [DOI] [PubMed] [Google Scholar]
- 41.Krupa A, Jenkins R, Luo DD, Lewis A, Phillips A, Fraser D. Loss of microRNA-192 promotes fibrogenesis in diabetic nephropathy. J Am Soc Nephrol : 438–447, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lang F, Capasso G, Schwab M, Waldegger S. Renal tubular transport and the genetic basis of hypertensive disease. Clin Exp Nephrol : 91–99, 2005. [DOI] [PubMed] [Google Scholar]
- 43.Lau NC, Lim LP, Weinstein EG, Bartel DP. An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science : 858–862, 2001. [DOI] [PubMed] [Google Scholar]
- 44.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell : 843–854, 1993. [DOI] [PubMed] [Google Scholar]
- 45.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, Kim VN. The nuclear RNase III Drosha initiates microRNA processing. Nature : 415–419, 2003. [DOI] [PubMed] [Google Scholar]
- 46.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J : 4663–4670, 2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lin DH, Yue P, Pan C, Sun P, Wang WH. MicroRNA 802 stimulates ROMK channels by suppressing caveolin-1. J Am Soc Nephrol : 1087–1098, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lin DH, Yue P, Zhang C, Wang WH. MicroRNA-194 (miR-194) regulates ROMK channel activity by targeting intersectin 1. Am J Physiol Renal Physiol : F53–F60, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci USA : 12103–12108, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu N, Williams AH, Kim Y, McAnally J, Bezprozvannaya S, Sutherland LB, Richardson JA, Bassel-Duby R, Olson EN. An intragenic MEF2-dependent enhancer directs muscle-specific expression of microRNAs 1 and 133. Proc Natl Acad Sci USA : 20844–20849, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lund E, Guttinger S, Calado A, Dahlberg JE, Kutay U. Nuclear export of microRNA precursors. Science : 95–98, 2004. [DOI] [PubMed] [Google Scholar]
- 52.Luo Y, Liu Y, Liu M, Wei J, Zhang Y, Hou J, Huang W, Wang T, Li X, He Y, Ding F, Yuan L, Cai J, Zheng F, Yang JY. Sfmbt2 10th intron-hosted miR-466(a/e)-3p are important epigenetic regulators of Nfat5 signaling, osmoregulation and urine concentration in mice. Biochim Biophys Acta : 97–106, 2014. [DOI] [PubMed] [Google Scholar]
- 53.Maharjan S, Mopidevi B, Kaw MK, Puri N, Kumar A. Human aldosterone synthase gene polymorphism promotes miRNA binding and regulates gene expression. Physiol Genomics : 860–865, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Marques FZ, Campain AE, Tomaszewski M, Zukowska-Szczechowska E, Yang YH, Charchar FJ, Morris BJ. Gene expression profiling reveals renin mRNA overexpression in human hypertensive kidneys and a role for microRNAs. Hypertension : 1093–1098, 2011. [DOI] [PubMed] [Google Scholar]
- 55.Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J, Calabrese JM, Dennis LM, Volkert TL, Gupta S, Love J, Hannett N, Sharp PA, Bartel DP, Jaenisch R, Young RA. Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell : 521–533, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miyoshi K, Miyoshi T, Siomi H. Many ways to generate microRNA-like small RNAs: non-canonical pathways for microRNA production. Mol Genet Genomics : 95–103, 2010. [DOI] [PubMed] [Google Scholar]
- 57.Mladinov D, Liu Y, Mattson DL, Liang M. MicroRNAs contribute to the maintenance of cell-type-specific physiological characteristics: miR-192 targets Na+/K+-ATPase-β1. Nucleic Acids Res : 1273–1283, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Muller OG, Parnova RG, Centeno G, Rossier BC, Firsov D, Horisberger JD. Mineralocorticoid effects in the kidney: correlation between α-ENaC, GILZ, and Sgk-1 mRNA expression and urinary excretion of Na+ and K+. J Am Soc Nephrol : 1107–1115, 2003. [DOI] [PubMed] [Google Scholar]
- 59.Negrini M, Nicoloso MS, Calin GA. MicroRNAs and cancer—new paradigms in molecular oncology. Curr Opin Cell Biol : 470–479, 2009. [DOI] [PubMed] [Google Scholar]
- 60.Nossent AY, Hansen JL, Doggen C, Quax PH, Sheikh SP, Rosendaal FR. SNPs in microRNA binding sites in 3′-UTRs of RAAS genes influence arterial blood pressure and risk of myocardial infarction. Am J Hypertens : 999–1006, 2011. [DOI] [PubMed] [Google Scholar]
- 61.Oba S, Kumano S, Suzuki E, Nishimatsu H, Takahashi M, Takamori H, Kasuya M, Ogawa Y, Sato K, Kimura K, Homma Y, Hirata Y, Fujita T. miR-200b precursor can ameliorate renal tubulointerstitial fibrosis. PLos One : e13614, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Okamura K, Chung WJ, Lai EC. The long and short of inverted repeat genes in animals: microRNAs, mirtrons and hairpin RNAs. Cell Cycle : 2840–2845, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev : 1655–1666, 2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ottaviani S, de Giorgio A, Harding V, Stebbing J, Castellano L. Noncoding RNAs and the control of hormonal signaling via nuclear receptor regulation. J Mol Endocrinol : R61–R70, 2014. [DOI] [PubMed] [Google Scholar]
- 65.Pandey DP, Picard D. Multidirectional interplay between nuclear receptors and microRNAs. Curr Opin Pharmacol : 637–642, 2010. [DOI] [PubMed] [Google Scholar]
- 66.Pandey P, Brors B, Srivastava PK, Bott A, Boehn SN, Groene HJ, Gretz N. Microarray-based approach identifies microRNAs and their target functional patterns in polycystic kidney disease. BMC Genomics : 624, 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Park JE, Heo I, Tian Y, Simanshu DK, Chang H, Jee D, Patel DJ, Kim VN. Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature : 201–205, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Pasquinelli AE, Reinhart BJ, Slack F, Martindale MQ, Kuroda MI, Maller B, Hayward DC, Ball EE, Degnan B, Muller P, Spring J, Srinivasan A, Fishman M, Finnerty J, Corbo J, Levine M, Leahy P, Davidson E, Ruvkun G. Conservation of the sequence and temporal expression of let-7 heterochronic regulatory RNA. Nature : 86–89, 2000. [DOI] [PubMed] [Google Scholar]
- 69.Pearce D, Kleyman TR. Salt, sodium channels, SGK1. J Clin Invest : 592–595, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Perron MP, Provost P. Protein components of the microRNA pathway and human diseases. Methods Mol Biol : 369–385, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Powers MP, Alvarez K, Kim HJ, Monzon FA. Molecular classification of adult renal epithelial neoplasms using microRNA expression and virtual karyotyping. Diagn Mol Pathol : 63–70, 2011. [DOI] [PubMed] [Google Scholar]
- 72.Rezaei M, Andrieu T, Neuenschwander S, Bruggmann R, Mordasini D, Frey FJ, Vogt B, Frey BM. Regulation of 11β-hydroxysteroid dehydrogenase type 2 by microRNA. Hypertension : 860–866, 2014. [DOI] [PubMed] [Google Scholar]
- 73.Robert-Nicoud M, Flahaut M, Elalouf JM, Nicod M, Salinas M, Bens M, Doucet A, Wincker P, Artiguenave F, Horisberger JD, Vandewalle A, Rossier BC, Firsov D. Transcriptome of a mouse kidney cortical collecting duct cell line: effects of aldosterone and vasopressin. Proc Natl Acad Sci USA : 2712–2716, 2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Robertson S, MacKenzie SM, Alvarez-Madrazo S, Diver LA, Lin J, Stewart PM, Fraser R, Connell JM, Davies E. MicroRNA-24 is a novel regulator of aldosterone and cortisol production in the human adrenal cortex. Hypertension : 572–578, 2013. [DOI] [PubMed] [Google Scholar]
- 75.Rotin D. Regulation of the epithelial sodium channel (ENaC) by accessory proteins. Curr Opin Nephrol Hypertens : 529–534, 2000. [DOI] [PubMed] [Google Scholar]
- 76.Ruby JG, Jan CH, Bartel DP. Intronic microRNA precursors that bypass Drosha processing. Nature : 83–86, 2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruster C, Wolf G. Renin-angiotensin-aldosterone system and progression of renal disease. J Am Soc Nephrol : 2985–2991, 2006. [DOI] [PubMed] [Google Scholar]
- 78.Saal S, Harvey SJ. MicroRNAs and the kidney: coming of age. Curr Opin Nephrol Hypertens : 317–323, 2009. [DOI] [PubMed] [Google Scholar]
- 79.Sequeira-Lopez ML, Weatherford ET, Borges GR, Monteagudo MC, Pentz ES, Harfe BD, Carretero O, Sigmund CD, Gomez RA. The microRNA-processing enzyme dicer maintains juxtaglomerular cells. J Am Soc Nephrol : 460–467, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sober S, Laan M, Annilo T. MicroRNAs miR-124 and miR-135a are potential regulators of the mineralocorticoid receptor gene (NR3C2) expression. Biochem Biophys Res Commun : 727–732, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Soldner A, Spahn-Langguth H, Mutschler E. The renin-angiotensin-aldosterone system: focus on its distinct role in arterial hypertension and its various inhibitors as a therapeutic strategy to effectively lower blood pressure. Pharmazie : 783–799, 1996. [PubMed] [Google Scholar]
- 82.Soundararajan R, Melters D, Shih IC, Wang J, Pearce D. Epithelial sodium channel regulated by differential composition of a signaling complex. Proc Natl Acad Sci USA : 7804–7809, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Soundararajan R, Zhang TT, Wang J, Vandewalle A, Pearce D. A novel role for glucocorticoid-induced leucine zipper protein in epithelial sodium channel-mediated sodium transport. J Biol Chem : 39970–39981, 2005. [DOI] [PubMed] [Google Scholar]
- 84.Subramanya AR, Yang CL, McCormick JA, Ellison DH. WNK kinases regulate sodium chloride and potassium transport by the aldosterone-sensitive distal nephron. Kidney Int : 630–634, 2006. [DOI] [PubMed] [Google Scholar]
- 85.Sun X, Ruan YC, Guo J, Chen H, Tsang LL, Zhang X, Jiang X, Chan HC. Regulation of miR-101/miR-199a-3p by the epithelial sodium channel during embryo implantation: involvement of CREB phosphorylation. Reproduction : 559–568, 2014. [DOI] [PubMed] [Google Scholar]
- 86.Suzuki HI, Miyazono K. Emerging complexity of microRNA generation cascades. J Biochem : 15–25, 2011. [DOI] [PubMed] [Google Scholar]
- 87.Tamarapu Parthasarathy P, Galam L, Huynh B, Yunus A, Abuelenen T, Castillo A, Kollongod Ramanathan G, Cox R Jr, Kolliputi N. MicroRNA 16 modulates epithelial sodium channel in human alveolar epithelial cells. Biochem Biophys Res Commun : 203–208, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tan YC, Blumenfeld J, Rennert H. Autosomal dominant polycystic kidney disease: genetics, mutations and microRNAs. Biochim Biophys Acta : 1202–1212, 2011. [DOI] [PubMed] [Google Scholar]
- 89.Trionfini P, Benigni A, Remuzzi G. MicroRNAs in kidney physiology and disease. Nat Rev Nephrol : 23–33, 2015. [DOI] [PubMed] [Google Scholar]
- 90.Verrey F, Loffing J, Zecevic M, Heitzmann D, Staub O. SGK1: aldosterone-induced relay of Na+ transport regulation in distal kidney nephron cells. Cell Physiol Biochem : 21–28, 2003. [DOI] [PubMed] [Google Scholar]
- 91.Vreugdenhil E, Verissimo CS, Mariman R, Kamphorst JT, Barbosa JS, Zweers T, Champagne DL, Schouten T, Meijer OC, de Kloet ER, Fitzsimons CP. MicroRNA 18 and 124a down-regulate the glucocorticoid receptor: implications for glucocorticoid responsiveness in the brain. Endocrinology : 2220–2228, 2009. [DOI] [PubMed] [Google Scholar]
- 92.Wade JB, Fang L, Liu J, Li D, Yang CL, Subramanya AR, Maouyo D, Mason A, Ellison DH, Welling PA. WNK1 kinase isoform switch regulates renal potassium excretion. Proc Natl Acad Sci USA : 8558–8563, 2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wei Q, Bhatt K, He HZ, Mi QS, Haase VH, Dong Z. Targeted deletion of Dicer from proximal tubules protects against renal ischemia-reperfusion injury. J Am Soc Nephrol : 756–761, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wei Q, Mi QS, Dong Z. The regulation and function of microRNAs in kidney diseases. IUBMB Life : 602–614, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell : 855–862, 1993. [DOI] [PubMed] [Google Scholar]
- 96.Xiong M, Jiang L, Zhou Y, Qiu W, Fang L, Tan R, Wen P, Yang J. The miR-200 family regulates TGF-β1-induced renal tubular epithelial-to-mesenchymal transition through Smad pathway by targeting ZEB1 and ZEB2 expression. Am J Physiol Renal Physiol : F369–F379, 2012. [DOI] [PubMed] [Google Scholar]
- 97.Yang CL, Zhu X, Wang Z, Subramanya AR, Ellison DH. Mechanisms of WNK1 and WNK4 interaction in the regulation of thiazide-sensitive NaCl cotransport. J Clin Invest : 1379–1387, 2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Yang JS, Lai EC. Dicer-independent, Ago2-mediated microRNA biogenesis in vertebrates. Cell Cycle : 4455–4460, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Zhang W, Gao S, Zhou X, Xia J, Chellappan P, Zhang X, Jin H. Multiple distinct small RNAs originate from the same microRNA precursors. Genome Biol : R81, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Zhdanova O, Srivastava S, Di L, Li Z, Tchelebi L, Dworkin S, Johnstone DB, Zavadil J, Chong MM, Littman DR, Holzman LB, Barisoni L, Skolnik EY. The inducible deletion of Drosha and microRNAs in mature podocytes results in a collapsing glomerulopathy. Kidney Int : 719–730, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]