Abstract
Background:
Patients with peripheral artery disease (PAD) are at risk of major adverse cardiac and cerebrovascular events (MACCE). There are no readily available risk scores that can accurately identify which patients are most likely to sustain an event, making it difficult to identify those who might benefit from more aggressive intervention. Thus, we aimed to develop a novel predictive model – using machine learning methods on electronic health record (EHR) data – to identify which PAD patients are most likely to develop MACCE.
Methods and Results:
Data were derived from patients diagnosed with PAD at two tertiary care institutions. Predictive models were built using a common data model (CDM) that allowed for utilization of both structured (coded) and unstructured (text) data. Only data from time of entry into the health system up to PAD diagnosis were used for modeling. Models were developed and tested using nested cross-validation. A total of 7,686 patients were included in learning our predictive models. Utilizing almost 1,000 variables, our best predictive model accurately determined which PAD patients would go on to develop MACCE with an area under the curve (AUC) of 0.81 (95% Confidence Interval, 0.80-0.83).
Conclusions:
Machine learning algorithms applied to data in the EHR can learn models that accurately identify PAD patients at risk of future MACCE, highlighting the great potential of EHR to provide automated risk stratification for cardiovascular diseases. Common data models that can enable cross-institution research and technology development could potentially be an important aspect of widespread adoption of newer risk-stratification models.
Introduction
Ninety-six percent of acute care hospitals in the United States have adopted Electronic Health Record (EHR) technology as of 2015, up from just 9% in 2008 1. Though billing and administrative tasks have been the focus of EHR systems, the large amount of granular, observational patient data offer unprecedented opportunities for data mining and analysis. In particular, the combination of advanced statistical methods and data volume provide an opportunity for stakeholders to build new technologies that can automate portions of health care such as early detection of adverse drug reactions, predictions of in-hospital mortality, wound healing, and risk stratification for heart failure readmissions 2-5.
Risk stratification using EHR data, in particular, has been of increasing interest for many investigators 6. With only a limited number of well-validated risk stratification scores 7-10, the use of EHR data from tens of thousands of patients may produce predictive models that can more accurately discriminate between patients at high and low risk of disease development or progression. However, as Goldstein and investigators report in a meta-analysis of EHR-based risk stratification models, researchers often do not take full advantage of the diversity of data available in the EHR 6. Frequently, model development will only use a handful of variables pre-emptively identified by investigators, and/or only utilize coded data such as lab values or diagnosis codes. This limitation in data usage may unduly limit the accuracy of predictive models, especially where complex non-linear interactions are involved, even if the models are developed using data from thousands of patients.
In previous work, we demonstrated that using a database of patients with and without PAD, machine learning algorithms could accurately identify patients with PAD who were previously undiagnosed 11. Such approaches allow for the automatic identification of patients, require less active management by health care professionals directly, and can considerably shift the paradigm of health care delivery from a reactionary practice to a proactive one 12, 13. Though this prior work demonstrated the ability to identify undiagnosed PAD patients, the data were derived from a well-structured prospectively collected database. Patients included in our predictive models had complete data across hundreds of phenotypic and genomic variables and were prospectively followed for years with accurate ascertainment of outcomes from hours of chart review and patient interviews and phone calls.
Real-world EHR data do not come as neatly packaged. Despite the volume, data may be missing or mislabeled, and patients may drop out of the data set at varying lengths of time. In the current work, we evaluate the feasibility of machine learning algorithms in identifying risk of future adverse cardiovascular events in patients diagnosed with PAD using EHR data. Note that when finding undiagnosed PAD patients, the disease in question is already present (but is unrecognized and untreated). For risk stratification, we use the portion of the record up to PAD diagnosis to predict the probability of the event’s (MACCE) occurrence in the future--making it a much harder problem.
We hypothesized that using observational EHR data and supervised machine learning, we would be able to learn predictive models that could accurately identify which PAD patients would go on to have a major adverse cerebrovascular and cardiovascular event prior to their occurrence. Accurate identification of patients at risk offers the possibility of managing high-risk patients differently. Therefore, accurate prediction is the first-step towards personalized care, particularly when treatment alternatives exist 14.
Methods
Data Source
Data were derived from the EHR of two tertiary care hospitals. Only de-identified patient records were utilized and the requirement for informed consent was waived. To ensure data de-identification only pertinent clinical data without protected health information (PHI) were extracted from the overall clinical data warehouse. Specifically, we extracted non-negated terminology, codes and results for each patient and used unique patient identifier codes 15. Metadata including dates and locations of visits (e.g. inpatient, outpatient) were also retrieved and stored. Our Institutional Review Boards approved the study. Due to data privacy considerations the clinical data are not available to researchers outside of our institutions for purposes of reproducing the results. However, the software used in these analyses are publicly available 16, 17.
EHR data included all adults treated as outpatients and inpatients at Stanford Health Care (SHC) between 1995 and 2015 and between 1980 and 2015 at Mount Sinai School of Medicine (MSSM). Data included International Classification of Diseases, version 9 (ICD-9) codes, Current Procedural Terminology (CPT) codes, lab test values, prescription medications, vital signs and unstructured clinical notes. In order to combine and analyze these disparate data types across different institutions we chose to perform data standardization using the Observational Medical Outcomes Partnership Common Data Model (OMOP CDM), version 5 18. The common data model also allowed us to coerce enough structure into the EHR data to enable more efficient use of machine learning algorithms.
Common Data Model
A common data model allows for translation of different forms of observational data from different institutions into a cohesive database structure. This is achieved by defining broad concepts, their expected value units and rules as to how similar concepts will be mapped into one unifying concept. For example if “serum blood glucose” is used at one institution and “blood sugar” at another, they would fall into the broader category of “blood glucose measurement” in the common data model with units converted to “mg/dL” no matter what the initial units at each institution are. Common data models can thus allow for more rapid analysis of structured and unstructured data across multiple institutions.
Our text processing methodology allowed us to integrate unstructured clinical text into our common data model to utilize for predictive modeling. Details of our text processing pipeline are described in more detail in prior publications 19, 20, but briefly, we utilized a strategy of identifying known medical terms within each clinical note within a patient’s health record. We then mapped these terms to common medical concepts using standardized medical dictionaries 21, 22. Terms that are ambiguous are removed and terms that have similar meanings (e.g. type 2 diabetes, insulin-dependent diabetes) are collapsed into one overriding concept. We then analyze whether or not terms relate to the patient, or for instance family members and if the term is negated. Terms that are unambiguous, relate to the patient and are not negated are then tabulated for each patient (Online Figure 1). These terms are then further evaluated and integrated into the OMOP common data model along with coded data such as lab values, procedural codes, and diagnosis codes.
Study Population
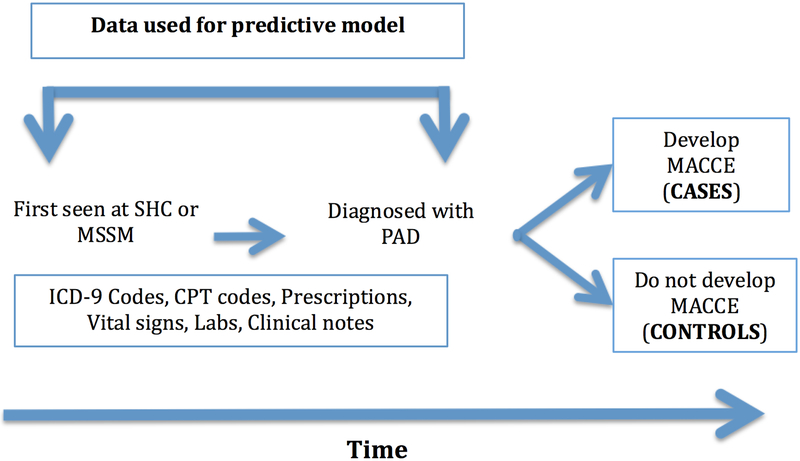
Patients older than 18 years of age treated at SHC and MSSM with a diagnosis of PAD were included in the initial patient data cohort. Age was defined at time of entry into the health system. Patients had to have either an ICD-9 code or an affirmative text mention of PAD to be included. Please see Online Table 1 for concept definitions for PAD. Previous validation of this methodology for PAD identification demonstrated a specificity of 98% and precision of 83% 23. Patients with MACCE including stroke, acute myocardial infarction, severe cardiac arrhythmias, or sudden cardiac death were also identified (Online Table 2) using previously vetted MACCE concepts and ICD-9 codes 23, 24. Positive example (“cases”) for learning the model were defined as patients with PAD who had a MACCE outcome at least 30 days after PAD diagnosis. Negative examples (“controls”) for the model were patients with a PAD diagnosis with no MACCE diagnosis prior to or after a PAD diagnosis. Data used in our prediction model included diagnosis codes, text data from notes, lab values, vital signs and prescriptions extracted from the time of first presentation to SHC or MSSM up to and including the time of PAD diagnosis (Figure 1). Thus, using the resulting model, a patient diagnosed with PAD could be risk stratified at time of diagnosis. Furthermore we did not pre-select cases or controls based on any prior evaluation of their risk of developing MACCE (e.g. age or smoking status).
Figure 1.
Data schematic. CPT – Current Procedural Terminology; ICD-9 – International Classification of Disease, version 9; MACCE – major adverse cardiac and cerebrovascular events; MSSM – Mt. Sinai Medical School of Medicine; PAD – peripheral artery disease; SHC – Stanford Health Care.
Patients were excluded if they had less than 1 year of data prior to PAD diagnosis to ensure enough data were available for predictive modeling. Since codes, notes and results data are all associated with specific encounter/visit dates (even during retrospective coding and billing procedures), ensuring that MACCE occurred on a different visit date than PAD diagnosis allowed us to determine that the events did not occur during the same visit. Patients were also excluded if less than 30 days of EHR data were available after PAD diagnosis to ensure that there was reasonable follow-up to exclude MACCE in control patients and also as another way to ensure MACCE and PAD diagnosis did not occur at or around the same time.
Missing data were not imputed in our data set. That is, if lab values, diagnosis codes or observations were not reported for a patient, we did not attempt to predict its value or likelihood of absence or presence. If variables were present for one patient but not the other, an encoding of 0 indicated the variable’s absence. The resulting data matrix, therefore, was relatively sparse. We did not impute data for two reasons. Our primary aim was to capture the performance of predictive modeling in real-world data, which will inherently include missing data. Secondly, there is no consensus as to which imputation methods are ideal for heterogeneous EHR data where data can be missing at random 25. Thus, our results are reflective of the natural health care utilization patterns of each institution. Please see the supplementary materials for further discussion on the handling of missing data.
Study Design & Statistical Analysis
The goal of our study was to evaluate whether machine learning algorithms could analyze thousands of data points from the EHR to identify whether a patient with a new diagnosis of PAD would go on to have MACCE prior to the event occurrence. Our approach was to combine data from SHC and MSSM to create a more generalizable predictive model.
We used penalized linear regression and random forest algorithms to build our predictive models. Both algorithms have the ability to automatically select variables in a data set that provide the most predictive power. That is, instead of identifying lab values or diagnosis codes that we felt were most likely to help predict the outcome of interest, we included all EHR data from every patient at an institution and let the algorithm identify the variables to keep. However, data pre-processing becomes important in optimizing performance of machine learning algorithms. Our data pre-processing included normalizing data by number of patient visits. That is, features (disease mentions, ICD-9 codes, lab values, etc) were tabulated, or averaged per patient then divided by the total number of visits over the course of the patient’s observation window. Doing so allowed us to both adjust the scale of each data point to improve machine learning algorithm performance and ensure numeric stability by adjusting for any skewed numerical values. Normalization also allowed us to account for differences in length of patient records and provide relative weights for different variables. That is, if certain variables occurred more frequently in a patient record than others, their weight in the predictive algorithm would be higher than a less frequently occurring variable.
We performed nested cross-validation whereby model parameters and variable selection was done using an inner 5-fold cross-validation on a random sample of 75% of patients. The best-trained model was chosen based on the F-measure, balancing model precision and recall. Once the best model was selected, model performance was tested using an outer 5-fold cross validation using the remaining 25% of patients. Model performance was judged both on its ability to discriminate between patients with a high and low risk of an outcome (i.e. area under the receiver operating characteristic curve (AUC) and its calibration (i.e. ability to accurately quantify observed absolute risk). In addition to visual characterization of calibration we also calculated the model Brier Score 26. We chose AUC and calibration as metrics to assess the best model, as opposed to other metrics such as Akaike Information Criterion, since these metrics can provide robust results using linear and non-linear algorithms 27. The best performing model was chosen as the final model. We used the APHRODITE package from the OHDSI group 16 and R version 3.2.1 for all data analyses 17.
Results
At SHC we identified 9,729 patients with PAD. After applying exclusion criteria, there were a total of 3,577 patients remaining with 837 (23%) who had MACCE after PAD diagnosis and 2,740 controls. Median time from initial presentation to SHC to diagnosis of PAD was 6.2 years (± 4.7 years) for cases and median time of observation for control patients was 5.8 years (± 5.2 years). Median time to MACCE for cases after PAD diagnosis was 2.2 years (± 3 years).
The MSSM data set included 8,259 PAD patients. A total of 4,109 patients met inclusion criteria, of which 459 (12.6%) had MACCE after PAD diagnosis. Median time from initial presentation to MSSM to diagnosis of PAD was 5.3 years (± 3.4 years) for cases and median total observation time for controls was 4.4 years (± 3.4 years). Median time to MACCE for cases after PAD diagnosis was 2.1 years (± 3 years). Table 1 outlines differences in demographics, variable frequency, and outcomes across the two health care sites and between cases and controls.
Table 1.
Differences in demographic, clinical and outcome variables across institutions and between cases and controls.
| Stanford | Mt. Sinai | |||
|---|---|---|---|---|
| Cases (N = 837) | Controls (N = 2,740) |
Cases (N = 459) | Controls (N = 3,191) |
|
| Patients | ||||
| Age (mean, y ± SD) | 69.5 (± 13) | 67 (± 15) | 71 (± 12) | 71 (± 13) |
| Male (%) | 55% | 53% | 51% | 50% |
| Race/Ethnicity | ||||
| White | 70% | 65% | 31% | 35% |
| Black | 6% | 5% | 26% | 21% |
| Asian | 8% | 9% | 1% | 6% |
| Hispanic | 7% | 7% | 32% | 20% |
| Top 5 Diagnosis Codes | ||||
| Hypertension | Hypertension | Type II Diabetes | Hypertension | |
| Coronary artery disease | Type II Diabetes | Hypertension | Type II Diabetes | |
| Type II Diabetes | Hyperlipidemia | Hypercholesterolemia | Hyperlipidemia | |
| Hyperlipidemia | Coronary artery disease | Coronary artery disease | Coronary artery disease | |
| Atrial fibrillation | Back pain | Congestive heart failure | Depression | |
| Top 5 Medications | ||||
| Saline | Saline | Aspirin | Aspirin | |
| Tylenol | Tylenol | Insulin | Insulin | |
| Glucose | Glucose | Heparin | Heparin | |
| Potassium | Ondansetron | Saline | Saline | |
| Ondansetron | Potassium | Simvastatin | Docusate | |
| Top 5 Text mentions | ||||
| Assessment | Assessment | Assessment | Assessment | |
| Pain | Pain | Pain | Pain | |
| Review of systems | Review of systems | Normal sinus rhythm | Chief Complaint | |
| Procedure | Procedure | Review of systems | Review of systems | |
| Auscultation | Female | Chief complaint | Normal sinus rhythm | |
| Top 5 Labs | ||||
| Serum potassium | Erythrocyte mean corpuscular hemoglobin concentration | Erythrocyte mean corpuscular hemoglobin concentration | Erythrocyte mean corpuscular hemoglobin concentration | |
| Erythrocyte mean corpuscular hemoglobin concentration | Serum potassium | Complete blood count | Serum platelets | |
| Serum carbon dioxide | Serum Chloride | Serum platelets | Serum Hemoglobin | |
| Serum Chloride | Serum carbon dioxide | Serum potassium | Serum potassium | |
| Serum Sodium | Serum Sodium | Serum Sodium | Complete blood count | |
| MACCE | N = 837 | N = 459 | ||
| Myocardial Infarction (%) | 337 (40) | 250 (54) | ||
| Cardiac Arrest/Shock (%) | 155 (18) | 114 (25) | ||
| Cardiac arrhythmia* (%) | 143 (17) | 30 (6) | ||
| Stroke (%) | 182 (22) | 54 (12) | ||
| Sudden death (%) | 20 (2) | 11 (2) | ||
Ventricular fibrillation or ventricular tachycardia; MACCE – Major adverse cerebro-cardiovascular events
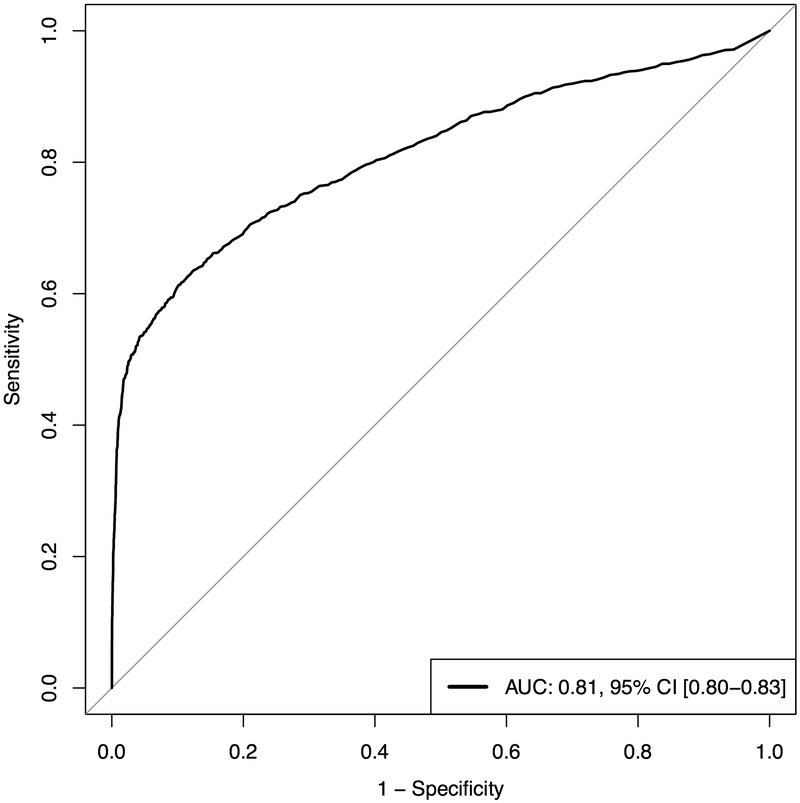
Compared to a penalized linear regression algorithm, random forest performed significantly better overall (AUC 0.69 [95% CI, 0.68-0.71] versus 0.81 [95% CI, 0.8-0.83], P < 0.001, respectively). Our final model was a random forest model that utilized 957 variables. Figure 2 demonstrates the average AUC and calibration as produced by 5-fold outer cross-validation on the test set. Our model had overall good calibration (Brier Score 0.10), though the model has a tendency to over-estimate risk for low-risk patients and under-estimate risk for high-risk patients. Sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) were 0.5, 0.96, 0.8, and 0.9, respectively. These values correspond to thresholds obtained from the AUC plot with a true positive rate as close to 1 and a false positive rate as close to 0 as possible. Online Figure 2 illustrates the precision-recall curve, which demonstrates the trade-offs involved in increasing model sensitivity or specificity.
Figure 2.
A. Area under receive operator curve plot for random forest predictive model. B. Calibration plot. AUC - area under the curve; CI – confidence interval. Confidence intervals computed using binomial test of proportions for each risk category.
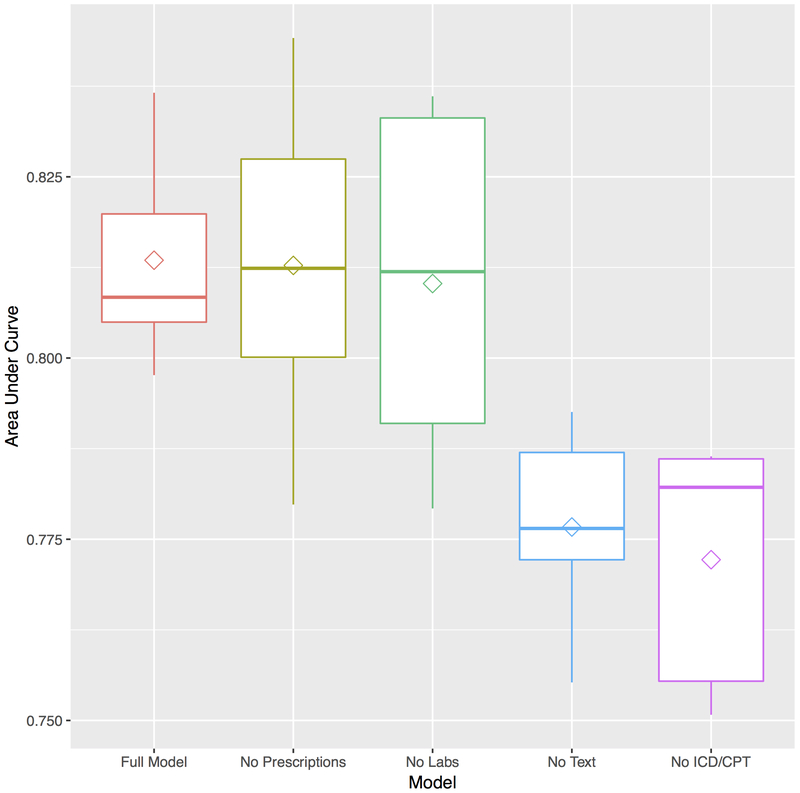
We performed sensitivity analysis on the best random forest model to evaluate how much each data type (unstructured text data, laboratory data, visit data (ICD-9 and CPT codes), and prescriptions) contributed to the accuracy of the final model (Figure 3). We found that unstructured text and coded data added significantly to model performance. Removal of text data resulted in a drop in AUC from 0.81 to 0.78, P = 0.002. Removal of coded data resulted in a decline in AUC to 0.77 (P = 0.0004). To evaluate whether these differences were clinically significant, we calculated the net reclassification index and found that on average 20% of patients would be reclassified as higher or lower risk with the addition of text or coded data. Removing lab and prescription data did not result in significant changes in model discrimination. Lastly, in Online Table 3, we provide the top 20 most important variables for each of the 5 outer cross-validated test sets. Given that our data pre-processing included normalization using the number of patient visits, in addition to other predictive characteristics of each variable, the frequency of a variable’s appearance may also contribute to their predictive importance.
Figure 3.
Box-plots of area under receiver operator curve plot by data type for random forest model. Removal of text and ICD/CPT coded data results in significant loss of model discrimination (P= 0.002) and (P = 0.0004), respectively. Diamond within box represents average model area under the curve while solid line represents median area under the curve.
Discussion
We have demonstrated that it is feasible to utilize real world EHR data to accurately identify PAD patients within a health system who are at high risk of developing subsequent MACCE. Our ability to build such predictive models is based on data integration via a common data model (OMOP CDM v5) and standard machine learning algorithms. Such a data analysis pipeline could potentially be deployed in real-time for personalized risk assessment for PAD patients; and thus guide personalized treatment planning.
Care for patients with chronic diseases such as PAD may be dramatically improved with use of automated, EHR-based technology. Patients with PAD often go undiagnosed in the primary care setting and are often not on optimal medical therapy 28-34. With annual costs of PAD-related care estimated to be over $4 billion, better screening and treatment strategies are needed, especially in an era where health care systems are being tasked with efficiently providing care for regional populations. Particularly in the case of PAD, better risk stratification would allow for more aggressive risk factor mitigation efforts that include prescription of anti-platelets, high dose cholesterol lowering medications, more aggressive blood pressure management, and enrollment in smoking cessation programs 30. What is more, with new Medicare reimbursement for supervised exercise programs and robust data showing improved outcomes with addition of rivaroxaban for stable PAD 35, automated solutions to identify high risk patients and increase awareness for new treatment options could substantially decrease cardiovascular morbidity and mortality.
To our knowledge there are no current risk scores for MACCE in PAD patients. While epidemiologically derived scores such as the Framingham Risk Score 36 (FRS) can be of good clinical utility in identifying discrete clinical risk factors, these scores are developed and validated in such a way that their predictive accuracy do not always translate to different clinical contexts. The FRS, for example, was not developed specifically for PAD patients and in our own experiments we have found that even a re-calibrated version of the FRS performs 20-25% worse than a machine learned risk prediction model for MACCE. Thus, an advantage to using EHR data for risk prediction is that such models can be fine-tuned to local populations and/or more specific disease states and risk stratification can be automated. As an illustration of this Arruda-Olson and colleagues describe their methodology of using EHR data to build a risk stratification model for 5-year mortality risk for PAD patients within their health system 37. By building a risk model specific to their patient population and extracting specific variables from the EHR they achieve good discrimination and calibration and are able to provide results in real-time.
Most published work evaluating EHR risk stratification models show mixed results 6. A majority of studies do not use longitudinal data, use relatively few predictive variables, with a median of 27 variables, and rarely develop multicenter models or validate models at different sites. A distinct strength of our work is that we used all variables available from the EHR, used longitudinal data from the patient’s health records and developed an accurate model by combining data from two distinct health care sites. In addition, we quantify the contribution of using features derived from unstructured and structured content and we set up the study as a true prediction problem to build a risk-model that can be used at the time of diagnosis.
Our study highlights the immense utility of common data models (CDMs). Despite utilizing data from different institutions representing two geographically distinct hospital systems, with different payer mixes, patient populations with varying prevalence of disease (Table 1), and different environmental risk factors, we were able to build a very good prediction model for MACCE in the PAD population. In general, CDMs allow researchers to combine observational data sources from multiple institutions, which allows for data standardization and the ability to create analytical tools that can be applied broadly. There are five main CDMs for EHR currently in use in the U.S. including OMOP 18, 38-42. While each CDM has a host of advantages and disadvantages 43, 44, they are key to enabling big data analysis in health care for tasks such as drug surveillance 44, 45, comparative effectiveness research 46, and reproducing EHR based research results 47. We show that a CDM is also useful for building risk predictive models.
Despite the accuracy of our findings there are limitations of our work. First, despite an AUC of 0.81, the model sensitivity (maximizing the true positive and minimizing false positive rates) is 0.5, with a specificity of 0.96. Thus, cases identified using our predictive model are likely to be truly high-risk patients, but many patients may be missed. Models can be adjusted to provide more sensitivity by adjusting threshold cut-offs. The precision recall curve (Online Figure 1) illustrates that increasing the sensitivity of the model to 100% would result in a drop in accuracy to less than 25%. This may be a reasonable trade-off in cases in which an intervention has a relatively low risk and/or low-cost (e.g. prescribing exercise therapy), but a higher specificity threshold would be warranted for higher risk interventions (e.g. prescribing anticoagulation that may significantly increase bleeding risk).
Another limitation is that use of data across multiple institutions can potentially lead to less accurate predictive models as local data storage and mapping practices may differ 48, as do patient populations. As detailed in Table 1 for instance, the demographic mix at the two care sites are quite different. Furthermore there are other clinical factors that may significantly vary across sites (e.g. severity of cardiovascular disease, number of comorbidities, etc) as Stanford has twice the rate of MACCE as MSSM. These population and data differences may require that models be trained locally to obtain higher accuracy.
On the other hand, use of data from multiple institutions helps improve model generalizability as models trained on one specific data set have the risk of being over fitted with little applicability to other sites. We believe that by using data from different institutions and by performing internal and external cross-validation we have appropriately balanced model accuracy and generalizability. Nevertheless, our models may suffer from issues such as over fitting and only further testing at other institutions would better clarify our model’s performance. What is more, in the effort to deliver “precision health” using institutional data some have advocated for data re-calibration at different sites to maximize predictive model accuracy 49. Need for re-calibration is not necessarily a failure of our approach to predictive modeling since others have found that re-calibrating more well-known epidemiologically derived risk scores provides better risk estimates for specific populations as well 50.
We also did not fully take into account missing data. Variables were included in the model if even 1 patient had a single value in the chart. This may have somewhat diminished our predictive accuracy, however a strength of this approach is that it represents the true nature of live EHR data with minimal transformations and no data imputations. Given that we are not engaged in inference questions, but rather focus on building a predictor for risk stratification 51, 52, we opt to use the data as they are. Another consideration is that we elected to define PAD and MACCE using diagnosis codes and term mentions since in our validation of this technique we achieve average specificity of 98% and precision of 90% 23. This methodology represents a more easily deployable solution to cohort building than natural language techniques, though there may be loss in sensitivity. Other techniques such as using an objective ABI measure could also be pursued, though ABIs are not always available for review since at tertiary care centers these exams are sometimes done at outside facilities and not adequately reported. Another limitation of our work is that given the fractured nature of health care in the U.S., it is possible that we did not capture all MACCE cases as events. While death data is integrated from the Social Security Death Index, other events may have occurred at other institutions without being reported to the index hospital of treatment. However, this issue would affect both the training and test sets of patients equally, and we still obtain reliable estimates of model performance. Such missing events do have the potential to reduce the overall performance of the model.
Another potential limitation that warrants discussion is our use of a “black box” algorithm for predicting MACCE in PAD patients. In medical research our goals are often to understand causal pathways for diseases and outcomes or to predict their occurrence. While similar statistical algorithms can be applied to these separate modeling tasks (prediction or inference), models that are good predictors of disease are not always models from which we can derive understanding of the disease mechanisms 52, 53. Because we did not force the use of any particular variables, the random forest algorithm found any variable in the EHR that was most associated with risk of future MACCE. The upside of such a model is that it can be more accurate than a simple linear regression model. However, the fact that the variables used are not always explanatory can be a drawback of this methodology. For example, higher age is associated with MACCE in the general population, but might not be more associated with MACCE in the population with PAD. Therefore, when comparing equally aged PAD patients with equally aged non-PAD patients, age might not show up as “predictive”. Indeed in Online Table 3, many of the predictive variables used in the random forest model such as “assessment” or “chief complaint” cannot be used to understand why certain patients are at higher risk than others. Other, traditional epidemiologic approaches may be necessary to find causal links for disease risk in such cases where the variables used in a particular algorithm are not the most descriptive and may be more predictive of a pattern that produces increased risk rather than a specific, discrete risk factor. Even so, machine learning algorithms can be highly beneficial and may revolutionize our ability to deliver both precision and population health, especially with an understanding of the limitations of each algorithm 54. For instance, associations between clinical, demographic or imaging characteristics and patient outcomes are not always linear. Machine learning algorithms such as random forest are still able to identify non-linear signals that can accurately predict disease occurrence and outcomes 55. Such capabilities will become even more important as the depth and breadth of healthcare data grow. And while machine learning methodologies have many advantages, to truly improve patient care and outcomes, methods for teasing out causal relationships will remain an important part of the health care and biomedical armamentarium.
Conclusions
Machine learning algorithms using available data in the EHR can accurately predict which PAD patients are most likely to go on to develop MACCE. The use of a common data model enables de novo learning of accurate risk models across multiple sites. Such informatics approaches can be applied to the medical record in an automated fashion to risk stratify patients with vascular disease and identify those who might benefit from more aggressive disease management. Future evaluation of the prospective performance of machine learning techniques versus traditional risk scores could provide valuable understanding of the overall utility of a machine learning approach.
Supplementary Material
Clinical Perspective Summary.
What is known
Peripheral artery disease is a significant cause of cardiovascular disease morbidity and mortality.
Risk assessment and medical optimization can increase longevity and decrease cardiovascular events in these patients.
There are no currently well-validated methods for risk stratifying PAD patients.
Machine learning algorithms coupled with data from the electronic health record has potential to help clinicians rapidly risk stratify patients.
What this study adds
We present some of the latest methodologies for harnessing electronic health record data to predict patient outcomes.
Machine learning algorithms perform well in identifying which PAD patients will develop major adverse cardiac and cerebrovascular events in the future.
Acknowledgments
Sources of funding
Funding: National Heart, Lung, and Blood Institute Training Program in Mechanisms and Innovation in Vascular Disease (1T32HL098049-06) (EGR) and the National Health Institutes Grant 5R01HL12522402 (NJL).
Footnotes
Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Henry J, Pylypchuk Y, Searcy T & Patel V (May 2016). Adoption of Electronic Health Record Systems among U.S. Non-Federal Acute Care Hospitals: 2008-2015 ONC Data Brief, no.35. Office of the National Coordinator for Health Information Technology: Washington DC. [Google Scholar]
- 2.Zhao J, Henriksson A, Asker L and Bostrom H. Predictive modeling of structured electronic health records for adverse drug event detection. BMC medical informatics and decision making. 2015;15 Suppl 4:S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tabak YP, Sun X, Nunez CM and Johannes RS. Using electronic health record data to develop inpatient mortality predictive model: Acute Laboratory Risk of Mortality Score (ALaRMS). Journal of the American Medical Informatics Association : JAMIA. 2014;21:455–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jung K, Covington S, Sen CK, Januszyk M, Kirsner RS, Gurtner GC and Shah NH. Rapid identification of slow healing wounds. Wound repair and regeneration : official publication of the Wound Healing Society [and] the European Tissue Repair Society. 2016;24:181–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eapen ZJ, Liang L, Fonarow GC, Heidenreich PA, Curtis LH, Peterson ED and Hernandez AF. Validated, electronic health record deployable prediction models for assessing patient risk of 30-day rehospitalization and mortality in older heart failure patients. JACC Heart failure. 2013;1:245–51. [DOI] [PubMed] [Google Scholar]
- 6.Goldstein BA, Navar AM, Pencina MJ and Ioannidis JP. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. Journal of the American Medical Informatics Association : JAMIA. 2017;24:198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antman EM, Cohen M, Bernink PJ, McCabe CH, Horacek T, Papuchis G, Mautner B, Corbalan R, Radley D and Braunwald E. The TIMI risk score for unstable angina/non-ST elevation MI: A method for prognostication and therapeutic decision making. Jama. 2000;284:835–42. [DOI] [PubMed] [Google Scholar]
- 8.D’Agostino RB Sr, Grundy S Sullivan LM, Wilson P and for the CHDRPG. Validation of the framingham coronary heart disease prediction scores: Results of a multiple ethnic groups investigation. Jama. 2001;286:180–187. [DOI] [PubMed] [Google Scholar]
- 9.Ntaios G, Lip GY, Makaritsis K, Papavasileiou V, Vemmou A, Koroboki E, Savvari P, Manios E, Milionis H and Vemmos K. CHADS(2), CHA(2)S(2)DS(2)-VASc, and long-term stroke outcome in patients without atrial fibrillation. Neurology. 2013;80:1009–17. [DOI] [PubMed] [Google Scholar]
- 10.Knaus WA, Wagner DP, Draper EA, Zimmerman JE, Bergner M, Bastos PG, Sirio CA, Murphy DJ, Lotring T, Damiano A and Harrell F Jr. The APACHE III prognostic system. Risk prediction of hospital mortality for critically ill hospitalized adults. Chest. 1991;100:1619–36. [DOI] [PubMed] [Google Scholar]
- 11.Ross EG, Shah NH, Dalman RL, Nead KT, Cooke JP and Leeper NJ. The use of machine learning for the identification of peripheral artery disease and future mortality risk. J Vasc Surg. 2016;64:1515–1522.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Banda JM, Halpern Y, Sontag D and Shah NH. Electronic phenotyping with APHRODITE and the Observational Health Sciences and Informatics (OHDSI) data network. AMIA Joint Summits on Translational Science proceedings AMIA Joint Summits on Translational Science. 2017;2017:48–57. [PMC free article] [PubMed] [Google Scholar]
- 13.Shah NH. Mining the ultimate phenome repository. Nature biotechnology. 2013;31:1095–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen JH and Asch SM. Machine Learning and Prediction in Medicine — Beyond the Peak of Inflated Expectations. New England Journal of Medicine. 2017;376:2507–2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.LePendu P, Iyer SV, Bauer-Mehren A, Harpaz R, Mortensen JM, Podchiyska T, Ferris TA and Shah NH. Pharmacovigilance using clinical notes. Clinical pharmacology and therapeutics. 2013;93:547–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Banda JM. APHRODITE (Automated PHenotype Routine for Observational Definition Identification Training and Evaluation) (2015). [computer program] Available at: https://github.com/OHDSI/Aphrodite. [Accessed 1 October 2016]
- 17.R: A language and environment for statistical computing. [computer program]. Vienna, Austria: R Foundation for Statistical Computing; 2017. [Google Scholar]
- 18.Makadia R and Ryan PB. Transforming the Premier Perspective Hospital Database into the Observational Medical Outcomes Partnership (OMOP) Common Data Model. EGEMS (Washington, DC). 2014;2:1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lependu P, Iyer SV, Fairon C and Shah NH. Annotation Analysis for Testing Drug Safety Signals using Unstructured Clinical Notes. Journal of biomedical semantics. 2012;3 Suppl 1:S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ross EG, Shah N and Leeper N. Statin Intensity or Achieved LDL? Practice-based Evidence for the Evaluation of New Cholesterol Treatment Guidelines. PloS one. 2016;11:e0154952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu ST, Liu H, Li D, Tao C, Musen MA, Chute CG and Shah NH. Unified Medical Language System term occurrences in clinical notes: a large-scale corpus analysis. Journal of the American Medical Informatics Association : JAMIA. 2012;19:e149–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parai GK, Jonquet C, Xu R, Musen MA and Shah NH. The Lexicon Builder Web service: Building Custom Lexicons from two hundred Biomedical Ontologies. AMIA Annual Symposium proceedings / AMIA Symposium AMIA Symposium. 2010;2010:587–91. [PMC free article] [PubMed] [Google Scholar]
- 23.Leeper NJ, Bauer-Mehren A, Iyer SV, Lependu P, Olson C and Shah NH. Practice-based evidence: profiling the safety of cilostazol by text-mining of clinical notes. PloS one. 2013;8:e63499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Agarwal V, Podchiyska T, Banda JM, Goel V, Leung TI, Minty EP, Sweeney TE, Gyang E and Shah NH. Learning statistical models of phenotypes using noisy labeled training data. Journal of the American Medical Informatics Association : JAMIA. 2016;23:1166–1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Beaulieu-Jones BK, Lavage DR, Snyder JW, Moore JH, Pendergrass SA and Bauer CR. Characterizing and Managing Missing Structured Data in Electronic Health Records: Data Analysis. JMIR medical informatics. 2018;6:e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gerds TA, Cai T and Schumacher M. The performance of risk prediction models. Biometrical journal Biometrische Zeitschrift. 2008;50:457–79. [DOI] [PubMed] [Google Scholar]
- 27.Steyerberg EW, Vickers AJ, Cook NR, Gerds T, Gonen M, Obuchowski N, Pencina MJ and Kattan MW. Assessing the performance of prediction models: a framework for some traditional and novel measures. Epidemiology (Cambridge, Mass). 2010;21:128–138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hirsch AT, Criqui MH, Treat-Jacobson D, Regensteiner JG, Creager MA, Olin JW, Krook SH, Hunninghake DB, Comerota AJ, Walsh ME, McDermott MM and Hiatt WR. Peripheral arterial disease detection, awareness, and treatment in primary care. Jama. 2001;286:1317–24. [DOI] [PubMed] [Google Scholar]
- 29.Lacroix P, Aboyans V, Voronin D, Le Guyader A, Cautres M and Laskar M. High prevalence of undiagnosed patients with peripheral arterial disease in patients hospitalised for non-vascular disorders. International journal of clinical practice. 2008;62:59–64. [DOI] [PubMed] [Google Scholar]
- 30.Hussain MA, Al-Omran M, Mamdani M, Eisenberg N, Premji A, Saldanha L, Wang X, Verma S and Lindsay TF. Efficacy of a Guideline-Recommended Risk-Reduction Program to Improve Cardiovascular and Limb Outcomes in Patients With Peripheral Arterial Disease. JAMA surgery. 2016;151:742–50. [DOI] [PubMed] [Google Scholar]
- 31.McDermott MM, Kerwin DR, Liu K, Martin GJ, O’Brien E, Kaplan H and Greenland P. Prevalence and significance of unrecognized lower extremity peripheral arterial disease in general medicine practice*. Journal of general internal medicine. 2001;16:384–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olin JW and Sealove BA. Peripheral artery disease: current insight into the disease and its diagnosis and management. Mayo Clinic proceedings. 2010;85:678–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gonzalez-Clemente JM, Pinies JA, Calle-Pascual A, Saavedra A, Sanchez C, Bellido D, Martin-Folgueras T, Moraga I, Recasens A, Girbes J, Sanchez-Zamorano MA and Mauricio D. Cardiovascular risk factor management is poorer in diabetic patients with undiagnosed peripheral arterial disease than in those with known coronary heart disease or cerebrovascular disease. Results of a nationwide study in tertiary diabetes centres. Diabetic medicine : a journal of the British Diabetic Association. 2008;25:427–34. [DOI] [PubMed] [Google Scholar]
- 34.Pande RL, Perlstein TS, Beckman JA and Creager MA. Secondary prevention and mortality in peripheral artery disease: National Health and Nutrition Examination Study, 1999 to 2004. Circulation. 2011;124:17–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Anand SS, Bosch J, Eikelboom JW, et al. The Lancet. 2018;391:219–229. [DOI] [PubMed] [Google Scholar]
- 36.Hemann BA, Bimson WF and Taylor AJ. The Framingham Risk Score: an appraisal of its benefits and limitations. The American heart hospital journal. 2007;5:91–6. [DOI] [PubMed] [Google Scholar]
- 37.Arruda‐Olson Adelaide M, Afzal N, Priya Mallipeddi V, Said A, Moussa Pacha H, Moon S, Chaudhry Alisha P, Scott Christopher G, Bailey Kent R, Rooke Thom W, Wennberg Paul W, Kaggal Vinod C, Oderich Gustavo S, Kullo Iftikhar J, Nishimura Rick A, Chaudhry R and Liu H. Leveraging the Electronic Health Record to Create an Automated Real‐Time Prognostic Tool for Peripheral Arterial Disease. Journal of the American Heart Association. 2018;7:e009680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hripcsak G, Duke JD, Shah NH, Reich CG, Huser V, Schuemie MJ, Suchard MA, Park RW, Wong ICK, Rijnbeek PR, van der Lei J, Pratt N, Norén GN, Li YC, Stang PE, Madigan D and Ryan PB. Observational Health Data Sciences and Informatics (OHDSI): Opportunities for Observational Researchers. Studies in health technology and informatics. 2015;216:574–8. [PMC free article] [PubMed] [Google Scholar]
- 39.PCORnet. (2018). National Patient-Centered Clinical Research Network Common Data Model (CDM). Available at: http://www.pcornet.org/pcornet-common-data-model/ [Accessed 20 Nov 2017].
- 40.Ross TR, Ng D, Brown JS, Pardee R, Hornbrook MC, Hart G and Steiner JF. The HMO Research Network Virtual Data Warehouse: A Public Data Model to Support Collaboration. EGEMS (Washington, DC). 2014;2:1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sentinelinitiative.org. (2018). Distributed Database and Common Data Model ∣ Sentinel Initiative. Available at: https://www.sentinelinitiative.org/sentinel/data/distributed-database-common-data-model [Accessed 20 Nov. 2017].
- 42.Clinical Data Interchange Standards Consortium. (2018) CDISC Standards in the Clinical Research Process. Available at: https://www.cdisc.org/standards [Accessed 20 Nov 2017].
- 43.Garza M, Del Fiol G, Tenenbaum J, Walden A and Zozus MN. Evaluating common data models for use with a longitudinal community registry. Journal of biomedical informatics. 2016;64:333–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xu Y, Zhou X, Suehs BT, Hartzema AG, Kahn MG, Moride Y, Sauer BC, Liu Q, Moll K, Pasquale MK, Nair VP and Bate A. A Comparative Assessment of Observational Medical Outcomes Partnership and Mini-Sentinel Common Data Models and Analytics: Implications for Active Drug Safety Surveillance. Drug safety. 2015;38:749–65. [DOI] [PubMed] [Google Scholar]
- 45.Reisinger SJ, Ryan PB, O’Hara DJ, Powell GE, Painter JL, Pattishall EN and Morris JA. Development and evaluation of a common data model enabling active drug safety surveillance using disparate healthcare databases. Journal of the American Medical Informatics Association : JAMIA. 2010;17:652–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.FitzHenry F, Resnic F, Robbins S, Denton J, Nookala L, Meeker D, Ohno-Machado L and Matheny M. Creating a Common Data Model for Comparative Effectiveness with the Observational Medical Outcomes Partnership. Applied Clinical Informatics. 2015;6:536–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zozus MN, Richesson RL, Walden A, Tenenbaum JD and Hammond W. Research Reproducibility in Longitudinal Multi-Center Studies Using Data from Electronic Health Records. AMIA Summits on Translational Science Proceedings. 2016;2016:279–85. [PMC free article] [PubMed] [Google Scholar]
- 48.Matcho A, Ryan P, Fife D and Reich C. Fidelity Assessment of a Clinical Practice Research Datalink Conversion to the OMOP Common Data Model. Drug safety. 2014;37:945–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Celi LA, Tang RJ, Villarroel MC, Davidzon GA, Lester WT and Chueh HC. A Clinical Database-Driven Approach to Decision Support: Predicting Mortality Among Patients with Acute Kidney Injury. Journal of healthcare engineering. 2011;2:97–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yadlowsky S, Hayward RA, Sussman JB, McClelland RL, Min YI and Basu S. Clinical Implications of Revised Pooled Cohort Equations for Estimating Atherosclerotic Cardiovascular Disease Risk. Annals of internal medicine. 2018;169:20–29. [DOI] [PubMed] [Google Scholar]
- 51.Leek JT and Peng RD. Statistics. What is the question? Science (New York, NY). 2015;347:1314–5. [DOI] [PubMed] [Google Scholar]
- 52.Breiman L Statistical Modeling: The Two Cultures (with comments and a rejoinder by the author). 2001:199–231. [Google Scholar]
- 53.Bzdok D, Altman N and Krzywinski M. Statistics versus machine learning. Nature Methods. 2018;15:233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Verghese A, Shah NH and Harrington RA. What This Computer Needs Is a Physician: Humanism and Artificial Intelligence. Jama. 2018;319:19–20. [DOI] [PubMed] [Google Scholar]
- 55.Samad MD, Ulloa A, Wehner GJ, Jing L, Hartzel D, Good CW, Williams BA, Haggerty CM and Fornwalt BK. Predicting Survival From Large Echocardiography and Electronic Health Record Datasets: Optimization With Machine Learning. JACC: Cardiovascular Imaging. 2018. [Epub ahead of print], PMC6286869, DOI: 10.1016/j.jcmg.2018.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.