Abstract
Introduction
Poor prognosis of gastric cancer (GC) has partly been a result of late diagnosis due to nonspecific symptoms in the early stages. The overall survival rate of patients with GC is quite low. Here, we presented the functional role and potential mechanism of long noncoding RNA STXBP5-AS1 in GC.
Materials and methods
CCK-8, scratch wound healing and Transwell assays were conducted to analyze proliferation, migration, and invasion of SGC7901 and MKN45 cells. Real-time polymerase chain reaction (qPCR) and Western blot assays were performed to investigate the relationship between STXBP5-AS1 and STXBP5. Finally, the correlation between STXBP5-AS1 and phosphorylated AKT1 (p-AKT1) was explored to reveal the potential mechanism of STXBP5-AS1 in GC. Western blot assays were performed to analyze phosphorylated AKT1 (p-AKT1) and AKT levels.
Results
Our results suggested that STXBP5-AS1 suppressed proliferation, migration, and invasion, and the upregulation of STXBP5-AS1 significantly repressed STXBP5 expression, and knockdown of STXBP5-AS1 promoted STXBP5 expression. In addition, the p-AKT1 level decreased when STXBP5-AS1 was overexpressed and the p-AKT1 level increased with STXBP5-AS1 knockdown in SGC7901 and MKN45 cells.
Conclusion
In summary, our results indicate that STXBP5-AS1 inhibits cell proliferation, migration, and invasion through PI3K/AKT in GC.
Keywords: long noncoding RNA, STXBP5-AS1, STXBP5, PI3K/AKT, GC
Introduction
Gastric cancer (GC) ranks as the fifth most frequently diagnosed cancer and the third leading cause of cancer-related deaths globally.1 It is often diagnosed at an advanced stage and is famous for undergoing malignant proliferation.2 Due to the lack of understanding about the pathogenesis, and absence of reliable markers, gastro-esophageal cancers are associated with delayed diagnosis.3 To improve the prognosis of GC, it is necessary to identify more potential biomarkers and reveal their molecular mechanisms.
lncRNAs are defined as transcripts longer than 200 nucleotides that do not encode proteins.4 To date, numerous studies have uncovered that lncRNAs play crucial roles in a wide array of tumors, including GC. For example, Farhangiyan et al showed that low expression of SOX2OT was related to poor diagnosis in GC.5 H19 has been considered a marker of the early stages of GC.6 Differentiation antagonizing non-protein coding RNA (DANCR) promotes migration and invasion in tumors.7 MAFG-AS1 has been shown to facilitate the migration and invasion of non-small cell lung carcinoma (NSCLC).8 Sun et al showed that GClnc1 affected cell proliferation, invasiveness, and metastasis in multiple GC models,9 and STXBP5-AS1 was involved in breast cancer10 and non-small-cell lung carcinoma.11 However, the association between STXBP5-AS1 and GC remains largely unknown.
STXBP5-AS1 was among the newly identified lncRNAs.10 According to the UCSC database (http://genome.ucsc.edu/), we found that STXBP5-AS1 is located on chromosome 6, ranging from 147,162,525 to 147,525,750 bp and STXBP5 is an antisense transcript of STXBP5-AS1. STXBP5-AS1 has been reported to play a role in breast cancer10 and inhibits migration and invasion in NSCLC.11 The significant finding is that STXBP5 was linked to GC.12 STXBP5 was associated with type 1 von Willebrand disease13 and found to participate in tumor processes.11 STXBP5 has also been identified as influenced by PI3K/AKT.11
In our study, using CCK-8, scratch wound healing and Transwell assays, we evaluated the effects of STXBP5-AS1 on GC cell proliferation, migration, and invasion. Additionally, qPCR and Western blot were performed to investigate the relationships between STXBP5-AS1, STXBP5, and p-AKT1. Our findings suggested that STXBP5-AS1 plays important role, and our study is expected to provide a theoretical basis for its role in GC.
Materials and methods
Cell culture
Human GC cell lines, SGC7901 and MKN45, were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA), and all cells were maintained in Dulbecco’s Modified Eagle’s Medium (DMEM) (Gibco, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA) in a humidified atmosphere of 5% (v/v) CO2 and 95% air at 37°C.
Plasmid construction
pcDNA3.1 (+) vector was purchased from Thermo Fisher Scientific. STXBP5-AS1 DNA was obtained by PCR using cDNA, Pfu DNA polymerase and synthetic oligonucleotide primers incorporating restriction sites. PCR products were ligated into the pcDNA3.1 (+) vector according to the manufacturer’s protocol (Thermo Fisher Scientific) and then sequenced to confirm. Primer sequences were as follows (sequence from 5′ to 3′): STXBP5-AS1 Forward: GGAGTGGGAGTGTG AGGAGAA; Reverse: AAGATCCCTGTGGCAAAATCCC.
Cell transfection
pcDNA3.1 STXBP5-AS1 or pcDNA3.1 were separately transfected into cells using Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s protocol. Small interfering RNAs (siRNAs) for the STXBP5-AS1 and negative control (NC) siRNAs were purchased from Ribobio (Guangzhou, P.R. China). Cell transfection was performed using siRNAs, plasmids and Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. A random RNA duplex (Ribobio) was used as a negative control, and pcDNA3.1 was used as a negative control. The target sequences of siRNA were as follows: si-STXBP5-AS1: 5′-GCAAGTTGCTGAGTATTAT-3′ (sense).
RNA extraction and qPCR
Total RNA was extracted from the GC cells using the TRIzol Reagent (Thermo Fisher Scientific) according to the manufacturer’s protocol. cDNA synthesis was performed with 1 µg of total RNA using a cDNA Synthesis Kit (Takara, Ohtsu, Japan). Relative mRNA levels were determined by qPCR using an Applied Biosystems 7500 Fluorescent Quantitative PCR System (Applied Biosystems, Foster City, CA, USA), SYBR Premix Ex Taq (TaKaRa) and specific primers. The reaction mixtures started at 95°C for 30 seconds, followed by 40 amplification cycles of 95°C for 5 seconds and 60°C for 30 seconds. The quantification of gene expression was performed using the ΔΔCT calculation with CT as the threshold cycle. Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) was used as an endogenous control. The results represent at least three experiments. The sequences for qPCR are listed in Table 1.
Table 1.
The sequences used for qPCR
| Gene | Primer sequence (5′–3′)
|
|
|---|---|---|
|
| ||
| STXBP5-AS1 | Forward | AGGGACTTGCCTTGTCGCTGAT |
| Reverse | GAGATTTAGGTGGGGACGCTGC | |
| STXBP5 | Forward | GCTTTAAGGCTCTTTGGTCG |
| Reverse | GCGAATGTAGTATGGCAGGC | |
| GAPDH | Forward | ACGGATTTGGTCGTATTGGGCG |
| Reverse | GCTCCTGGAAGATGGTGATGGG | |
Abbreviation: GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
Cell proliferation assay
Cells were seeded at a density of 3,000 cells/well in 96-well culture plates and cultured for 18 hours prior to transfection. Subsequently, transfection experiments were performed using 20 nM siRNA, 1 µg plasmid and Lipofectamine 2000 (Thermo Fisher Scientific) according to the manufacturer’s instructions. From 0–3 days posttransfection, 20 µL/well CCK-8 solution was added (Beyotime Institute of Biotechnology, Haimen, P.R. China). The optical density was measured at a wavelength of 450 nm using a Thermo Fisher Scientific Multiskan FC.
Scratch wound healing assay
Cells were seeded in 6-well culture plates at a density of 1×105 cells/well and cultured for 18 hours prior to transfection. Cells were transfected with pcDNA STXBP5-AS1 or pcDNA3.1 and si-STXBP5-AS1 or si-NC. Cells were harvested 48 hours posttransfection. Uniform wounds were scraped. Images for the wound closures were viewed at 40× magnification by microscope and photographed 0, 24, 48 and 72 hours after scratching.
Cell invasion assay
Cells were treated with pcDNA STXBP5-AS1 or pcDNA3.1 and si-STXBP5-AS1 or si-NC, and Transwell assays were performed in 6.5 mm Transwell chambers with 8 µm pores (Corning Costar, Corning, NY, USA). The underside of each membrane was coated with 20% fetal bovine serum for 2 hours. Approximately 1×104 cells were seeded into the upper chambers in 100 µL of serum-free medium. Lower chambers contained 500 µL of medium with 10% fetal bovine serum. After cells were allowed to migrate, the medium in the upper chamber was aspirated, and cells on the upper side were removed with a cotton swab. Cells on the lower side of the membrane were stained using 0.1% crystal violet for 20 minutes at room temperature. The numbers of invaded cells in the lower chamber were calculated at 100× magnification using a microscope (Olympus Corporation, Tokyo, Japan). All experiments were performed at least three times independently.
Western blot assay
Cells were grown in 6-well plates and transfected with the indicated plasmids and siRNA using Lipofectamine 2000 (Thermo Fisher Scientific). Cells were washed with pre-cooling phosphate-buffered saline (PBS), and cells were collected and then lysed for 20 minutes in cold lysis buffer (Beyotime Institute of Biotechnology). Extracts were clarified by centrifugation (12,000 rpm/min) for 30 minutes at 4°C. Protein samples were resolved by electrophoresing on 10% polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred to polyvinylidene fluoride (PVDF). Membranes were blocked in Tris-buffered saline (TBS) containing 5% nonfat dry milk and 0.05% Tween-20 for 20 minutes with shaking and then incubated with primary antibody following the manufacturer’s instructions for 2 hours at room temperature. Samples were then washed with TBST (TBS-0.1% Tween-20) and incubated for 1 hour with horseradish peroxidase-conjugated secondary antibody (1:10,000 dilution). Protein bands were visualized using enhanced chemiluminescent substrate according to the manufacturer’s protocol (Millipore, Billerica, MA, USA). The primary antibodies were STXBP5, AKT1, p-AKT1 and GAPDH from ABclonal Biotechnology (Wuhan, P.R. China).
Statistical analyses
The data were expressed as the means ± SD. Differences were assessed by two-tailed Student’s t-test using GraphPad Prism 5.0 software (GraphPad Software, Inc. CA, USA). P<0.05 was considered statistically significant.
Results
STXBP5-AS1 attenuates the proliferation of GC cells
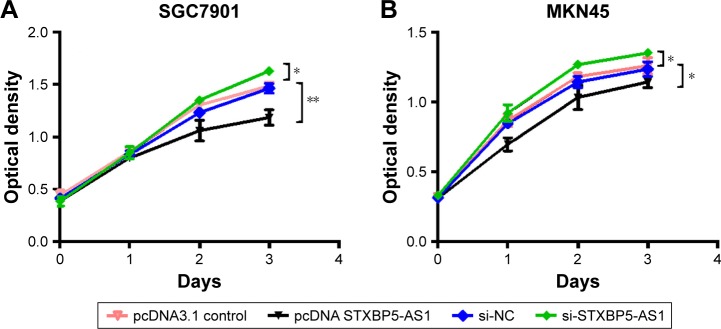
SGC7901 and MKN45 cells were transfected with pcDNA STXBP5-AS1 and pcDNA 3.1, and then a CCK-8 assay was performed to examine the effect of STXBP5-AS1 on proliferation. Compared with the corresponding pcDNA3.1 group, proliferation was inhibited in the pcDNA STXBP5-AS group (Figure 1A and B).
Figure 1.
STXBP5-AS1 attenuates the proliferation of GC cells.
Notes: CCK-8 assay indicated that STXBP5-AS1 inhibited the proliferation of SGC7901 (A) and MKN45 (B) cells. Error bars represent SD obtained from three independent experiments and all the data are shown as the mean ± SD; *P<0.05; **P<0.01.
Abbreviations: CCK-8, Cell Counting Kit-8; GC, gastric cancer; NC, negative control; si, small interfering RNA.
In contrast, si-STXBP5-AS1 and si-NC were transfected into SGC7901 and MKN45 cells, and a CCK-8 assay was conducted to further identify the effect of STXBP5-AS1 on proliferation. We observed that the proliferation properties of si-STXBP5-AS1 groups were promoted in SGC7901 and MKN45 cells compared with corresponding si-NC groups (Figure 1A and B). Taken together, STXBP5-AS1 attenuates the proliferation of GC cells.
STXBP5-AS1 restrains the migration of GC cells
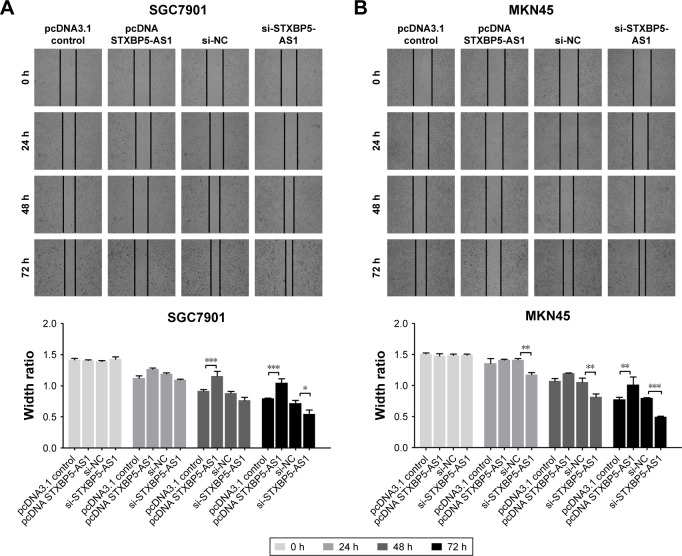
Next, SGC7901 and MKN45 cells were transfected with pcDNA STXBP5-AS1 and pcDNA 3.1, and then a scratch wound healing assay was performed to examine the effect of STXBP5-AS1 on migration. Migration was decreased in pcDNA STXBP5-AS1 groups but enhanced in pcDNA 3.1 groups (Figure 2A and B).
Figure 2.
STXBP5-AS1 restrains the migration of GC cells.
Notes: Scratch wound healing indicated that STXBP5-AS1 repressed the migration of SGC7901 (A) and MKN45 cells (B). *P<0.05; **P<0.01; ***P<0.001
Abbreviations: GC, gastric cancer; h, hours; NC, negative control; si, small interfering RNA.
SGC7901 and MKN45 cells were transfected with si-STXBP5-AS1 and si-NC and scratch wound healing assays were conducted separately. Cells transfected with si-STXBP5-AS1 had elevated cell migration, and in si-NC groups, cells had inhibited migration (Figure 2A and B). Collectively, these results suggested that STXBP5-AS1 restrains the migration of GC cells.
STXBP5-AS1 alleviates the invasion of GC cells
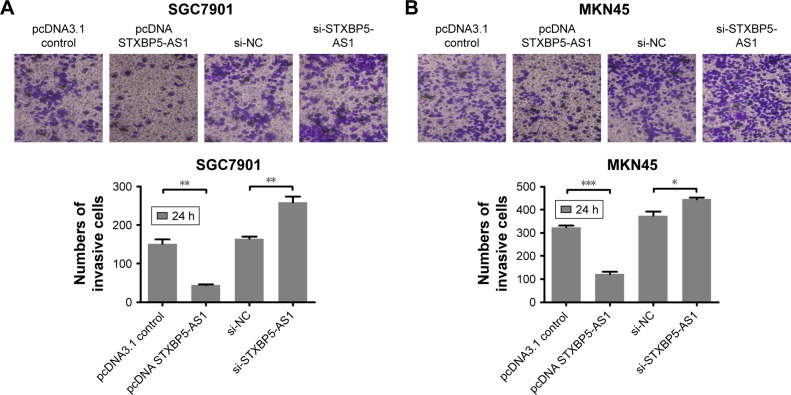
SGC7901 and MKN45 cells were transfected with pcDNA STXBP5-AS1 and pcDNA 3.1, and a transwell assay was conducted to investigate invasion. It showed that invasion was lower in pcDNA STXBP5-AS1 than pcDNA 3.1 groups (Figure 3A and B).
Figure 3.
STXBP5-AS1 alleviates the invasion of GC cells.
Notes: Transwell assay suggested that STXBP5-AS1 repressed the invasion of SGC7901 (A) and MKN45 (B) cells. *P<0.05; **P<0.01; ***P<0.001
Abbreviations: GC, gastric cancer; h, hours; NC, negative control; si, small interfering RNA.
Subsequently, SGC7901 and MKN45 cells were transfected with si-STXBP5-AS1 and si-NC, and a transwell assay was conducted to further determine the role of STXBP5-AS1 on invasion. In the si-STXBP5-AS1 groups, invasion was increased compared to their corresponding si-NC groups (Figure 3A and B). Our results could further convey that STXBP5-AS1 weakened the proliferation, migration, and invasion of GC cells.
STXBP5-AS1 downregulates STXBP5 expression in GC cells
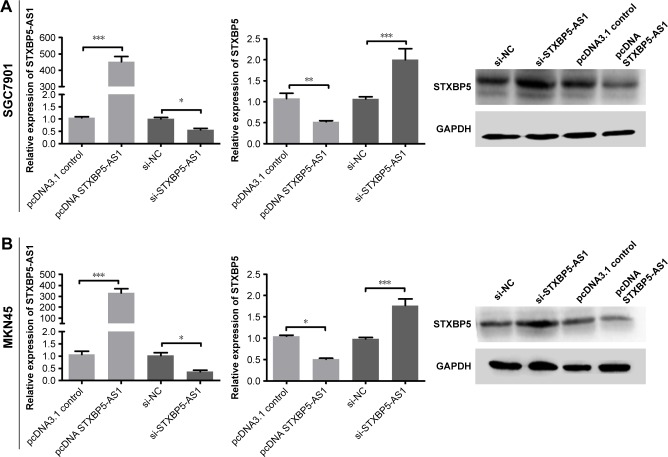
SGC7901 and MKN45 cells were transfected with pcDNA STXBP5-AS1 and pcDNA 3.1. qPCR, and a Western blot assay was conducted to analyze the association of STXBP5-AS1 and STXBP5. In pcDNA STXBP5-AS1 groups, STXBP5 expression was lower than in pcDNA 3.1 groups (Figure 4A and B).
Figure 4.
STXBP5-AS1 downregulates STXBP5 expression in GC cells.
Notes: Overexpression of STXBP5-AS1 restrained STXBP5 expression in SGC7901 (A) and MKN45 (B) cells, and its knockdown promoted STXBP5 expression. Error bars represent SD obtained from three independent experiments and all the data are shown as the mean ± SD. Differences between two groups were analyzed by Student’s t-test. *P<0.05; **P<0.01; ***P<0.001.
Abbreviations: GC, gastric cancer; NC, negative control; si, small interfering RNA.
In contrast, SGC7901 and MKN45 cells were transfected with si-STXBP5-AS1 and si-NC. qPCR, and a Western blot assay was conducted to further examine the association of STXBP5-AS1 and STXBP5. In si-STXBP5-AS1 groups, STXBP5 expression was higher than in si-NC groups (Figure 4A and B). Our results demonstrate that STXBP5-AS1 downregulates STXBP5 expression in GC cells.
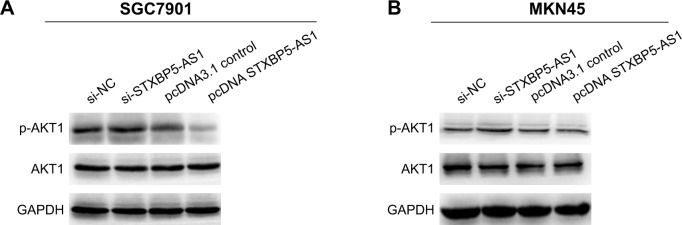
STXBP5-AS1 inhibits the PI3K/AKT signaling pathway in GC cells
To further explore the mechanism of STXBP5-AS1, SGC7901 and MKN45 cells were transfected with pcDNA STXBP5-AS1 and pcDNA 3.1. Western blot assays were conducted to analyze the association of STXBP5-AS1 and p-AKT1. We observed that p-AKT1 level was decreased in the pcDNA STXBP5-AS1 group compared to the pcDNA 3.1 group. STXBP5-AS1 had no significant effects on AKT1 (Figure 5A and B). Subsequently, SGC7901 and MKN45 cells were transfected with si-STXBP5-AS1 and si-NC. Western blot assays were conducted that showed that p-AKT1 was increased in si-STXBP5-AS1 groups compared to cells treated with si-NC. STXBP5-AS1 had no significant effects on AKT1 (Figure 5A and B). Conjointly, our findings imply that STXBP5-AS1 may constrain cell proliferation, migration, and invasion in GC cells through the PI3K/AKT signaling pathway.
Figure 5.
STXBP5-AS1 inhibits the PI3K/AKT signaling pathway in GC cells.
Notes: (A, B) Western blot was conducted to examine the association of STXBP5-AS1 and p-AKT1. Significant reduction in p-AKT1 level was observed in pcDNA STXBP5-AS1 groups compared to groups transfected with pcDNA 3.1. Then, the increase in p-AKT1 was found in the si-STXBP5-AS1 group compared to the si-NC group.
Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GC, gastric cancer; NC, negative control; si, small interfering RNA.
Discussion
GC, the most common cause of death from cancer, is a tremendous burden worldwide with high morbidity and high mortality.14 Patients with GC commonly have a poor prognosis owing to its invasiveness and distant metastasis.15 Over the past few years, much effort has gone into exploring biological functions in GC. Researchers found that lncRNAs play critical roles in growth, metastasis and prognosis in a variety of tumors, such as, lncRNA AWPPH promotes the growth of triple-negative breast cancer by upregulating FZD7.16 MEG3 inhibits metastasis of GC via the p53 signaling pathway.17 Overexpression of DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer.18 Certainly, it has been identified that a few lncRNAs were also related to GC,5,6,9,19,20 whereas the role of lncRNAs in GC remains largely unclear.
STXBP5-AS1 is a recently reported anti-sense RNA, and researchers found that STXBP5-AS1 was associated with OS in patients with breast cancer.10 STXBP5-AS1 inhibits cell proliferation, migration, and invasion in NSCLC.11 Based on UCSC, we noticed that STXBP5-AS1 is an anti-sense of STXBP5. STXBP5 is involved in docking and fusion of synaptic vesicles with the presynaptic plasma membrane. It is widely known that a variety of vesicles secreted by several types of cells have roles in different processes related to cancer biology. For example, exosomes from lung cancer promote cell migration.21 The presence of tumor-derived cilia vesicles enhanced glioma cell proliferation.22 STXBP5 recently was found to have effects in NSCLC.11
Based on our study, the results showed that STXBP5-AS1 could influence the p-AKT1 levels, which is likely to be associated with PI3K/AKT signaling pathway. In fact, the PI3K/AKT signaling pathway is critically involved in tumor cellular functions.23,24 It is notable that lncRNAs have also been identified to participate in proliferation, migration, metastasis, and invasion via regulating the PI3K/AKT pathway in GC, such as ADAMTS9-AS2,25 CRNDE,26 HOTAIR,27 and HULC.28
In our study, we conducted CCK-8, scratch wound healing and transwell assays to analyze proliferation, migration and invasion, respectively. Our results suggested that STXBP5-AS1 inhibits SGC7901 and MKN45 cell proliferation, migration, and invasion. Then, using qPCR and Western blot assay, we found that in the pcDNA STXBP5-AS1 group, STXBP5 expression was lower than the pcDNA 3.1 group. In the si-STXBP5-AS1 group, STXBP5 expression was higher than in the si-NC group. Our results suggested that STXBP5-AS1 downregulates STXBP5 expression in GC cells. Subsequently, the potential mechanism was further affirmed by Western blot assay results, indicating that p-AKT1 level was decreased in cells transfected with pcDNA STXBP5-AS1 and significantly increased in the si-STXBP5-AS1 group. Overall, our findings collectively demonstrated that STXBP5-AS1 inhibits cell proliferation, migration, and invasion in GC via blocking the PI3K/AKT signaling pathway.
Conclusion
This study may provide a fresh look at the functional role and potential mechanism of STXBP5-AS1 in GC, but there were some limitations in our study. More specific mechanisms need to be clarified than we currently understand, such as the exact regulatory relationship between STXBP5-AS1 and STXBP5 and more potential events associated with STXBP5-AS1 in GC. Such findings will enable further follow-up on the basis of the findings that provide a new understanding of STXBP5-AS1 in GC.
Footnotes
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Nanki K, Toshimitsu K, Takano A, et al. Divergent routes toward Wnt and R-spondin niche Independency during human gastric carcinogenesis. Cell. 2018;174(4):e817, 856–869. doi: 10.1016/j.cell.2018.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Catalano V, Labianca R, Beretta GD, Gatta G, de Braud F, van Cutsem E. Gastric cancer. Crit Rev Oncol Hematol. 2009;71(2):127–164. doi: 10.1016/j.critrevonc.2009.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Leung WK, M-Shiang WU, Kakugawa Y, et al. Screening for gastric cancer in Asia: current evidence and practice. Lancet Oncol. 2008;9(3):279–287. doi: 10.1016/S1470-2045(08)70072-X. [DOI] [PubMed] [Google Scholar]
- 4.Perkel JM. Visiting “noncodarnia”. BioTechniques. 2013;54(6):301, 303–304. doi: 10.2144/000114037. [DOI] [PubMed] [Google Scholar]
- 5.Farhangian P, Jahandoost S, Mowla SJ, Khalili M. Differential expression of long non-coding RNA SOX2OT in gastric adenocarcinoma. Cancer Biomark. 2018;23(2):221–225. doi: 10.3233/CBM-181325. [DOI] [PubMed] [Google Scholar]
- 6.Yörüker EE, Keskin M, Kulle CB, Holdenrieder S, Gezer U. Diagnostic and prognostic value of circulating lncRNA H19 in gastric cancer. Biomed Rep. 2018;9(2):181–186. doi: 10.3892/br.2018.1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao Z, Li H, Du B, et al. LncRNA DANCR promotes migration and invasion through suppression of lncRNA-LET in gastric cancer cells. Biosci Rep. 2017;37(6):BSR20171070. doi: 10.1042/BSR20171070. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Liu YY, Li B, Guo H. LncRNA MAFG-AS1 facilitates the migration and invasion of NSCLC cell via sponging miR-339-5p from MMP15. Cell Biol Int. 2019 Jan 1; doi: 10.1002/cbin.11092. Epub. [DOI] [PubMed] [Google Scholar]
- 9.Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6(7):784–801. doi: 10.1158/2159-8290.CD-15-0921. [DOI] [PubMed] [Google Scholar]
- 10.Guo W, Wang Q, Zhan Y, et al. Transcriptome sequencing uncovers a three-long noncoding RNA signature in predicting breast cancer survival. Sci Rep. 2016;6(1):27931. doi: 10.1038/srep27931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Huang J, Xie N, Huang H, Yao J, Hu W. Long noncoding RNA STXBP5-AS1 inhibits cell proliferation, migration, and invasion via preventing the PI3K/Akt against STXBP5 expression in non-small-cell lung carcinoma. J Cell Biochem. 2018 Nov 18; doi: 10.1002/jcb.28023. Epub. [DOI] [PubMed] [Google Scholar]
- 12.Lee B, Yoon K, Lee S, et al. Homozygous deletions at 3p22, 5p14, 6q15, and 9p21 result in aberrant expression of tumor suppressor genes in gastric cancer. Genes Chromosomes Cancer. 2015;54(3):142–155. doi: 10.1002/gcc.22226. [DOI] [PubMed] [Google Scholar]
- 13.Lind-Halldén C, Manderstedt E, Carlberg D, Lethagen S, Halldén C. Genetic variation in the syntaxin-binding protein STXBP5 in type 1 von Willebrand disease patients. Thromb Haemost. 2018;118(8):1382–1389. doi: 10.1055/s-0038-1661352. [DOI] [PubMed] [Google Scholar]
- 14.Fitzmaurice C, Dicker D, Pain A, et al. The global burden of cancer 2013. JAMA. 2015;1(4):505–527. doi: 10.1001/jamaoncol.2015.0735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang Y, Zhang J, Hou L, et al. LncRNA AK023391 promotes tumorigenesis and invasion of gastric cancer through activation of the PI3K/Akt signaling pathway. J Exp Clin Cancer Res. 2017;36(1):194. doi: 10.1186/s13046-017-0666-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang K, Li X, Song C, Li M. LncRNA AWPPH promotes the growth of triple-negative breast cancer by up-regulating frizzled homolog 7 (FZD7) Biosci Rep. 2018;38(6):BSR20181223. doi: 10.1042/BSR20181223. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 17.Wei GH, Wang X. lncRNA MEG3 inhibit proliferation and metastasis of gastric cancer via p53 signaling pathway. Eur Rev Med Pharmacol Sci. 2017;21(17):3850–3856. [PubMed] [Google Scholar]
- 18.Liu Y, Zhang M, Liang L, Li J, Chen YX. Overexpression of lncRNA DANCR is associated with advanced tumor progression and poor prognosis in patients with colorectal cancer. Int J Clin Exp Pathol. 2015;8(9):11480–11484. [PMC free article] [PubMed] [Google Scholar]
- 19.Yan J, Huang W, Huang X, Xiang W, Ye C, Liu J. A negative feedback loop between long noncoding RNA NBAT1 and SOX9 inhibits the malignant progression of gastric cancer cells. Biosci Rep. 2018;38(6):BSR20180882. doi: 10.1042/BSR20180882. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 20.Luo Y, Liang M, Yao W, et al. Functional role of lncRNA LOC101927497 in N-methyl-N′-nitro-N-nitrosoguanidine-induced malignantly transformed human gastric epithelial cells. Life Sci. 2018;193:93–103. doi: 10.1016/j.lfs.2017.12.007. [DOI] [PubMed] [Google Scholar]
- 21.Lukic A, Wahlund CJE, Gómez C, et al. Exosomes and cells from lung cancer pleural exudates transform LTC4 to LTD4, promoting cell migration and survival via CysLT1. Cancer Lett. 2019;444:1–8. doi: 10.1016/j.canlet.2018.11.033. [DOI] [PubMed] [Google Scholar]
- 22.Hoang-Minh LB, Dutra-Clarke M, Breunig JJ, Sarkisian MR. Glioma cell proliferation is enhanced in the presence of tumor-derived cilia vesicles. Cilia. 2018;7:6. doi: 10.1186/s13630-018-0060-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Engelman JA. Targeting PI3K signalling in cancer: opportunities, challenges and limitations. Nat Rev Cancer. 2009;9(8):550–562. doi: 10.1038/nrc2664. [DOI] [PubMed] [Google Scholar]
- 24.Thorpe LM, Yuzugullu H, Zhao JJ. PI3K in cancer: divergent roles of isoforms, modes of activation and therapeutic targeting. Nat Rev Cancer. 2015;15(1):7–24. doi: 10.1038/nrc3860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao B, Liu C, Yang G. Down-regulation of lncRNA ADAMTS9-AS2 contributes to gastric cancer development via activation of PI3K/Akt pathway. Biomed Pharmacother. 2018;107:185–193. doi: 10.1016/j.biopha.2018.06.146. [DOI] [PubMed] [Google Scholar]
- 26.Du DX, Lian DB, Amin BH, Yan W. Long non-coding RNA CRNDE is a novel tumor promoter by modulating PI3K/Akt signal pathways in human gastric cancer. Eur Rev Med Pharmacol Sci. 2017;21(23):5392–5398. doi: 10.26355/eurrev_201712_13925. [DOI] [PubMed] [Google Scholar]
- 27.Cheng C, Qin Y, Zhi Q, Wang J, Qin C. Knockdown of long non-coding RNA HOTAIR inhibits cisplatin resistance of gastric cancer cells through inhibiting the PI3K/Akt and Wnt/β-catenin signaling pathways by up-regulating miR-34a. Int J Biol Macromol. 2018;107(Pt B):2620–2629. doi: 10.1016/j.ijbiomac.2017.10.154. [DOI] [PubMed] [Google Scholar]
- 28.Feng H, Wei B, Zhang Y. Long non-coding RNA HULC promotes proliferation, migration and invasion of pancreatic cancer cells by down-regulating microRNA-15a. Int J Biol Macromol. 2019;126:891–898. doi: 10.1016/j.ijbiomac.2018.12.238. [DOI] [PubMed] [Google Scholar]