Abstract
Advances in mass spectrometry-based proteomics now permit analysis of complex cellular structures. Application to epidermis and its appendages (nail plate, hair shaft) has revealed a wealth of information about their protein profiles. The results confirm known site-specific differences in levels of certain keratins and add great depth to our knowledge of site specificity of scores of other proteins, thereby connecting anatomy and pathology. An example is the evident overlap in protein profiles of hair shaft and nail plate, helping rationalize their sharing of certain dystrophic syndromes distinct from epidermis. In addition, inter-individual differences in protein level are manifest as would be expected. This approach permits characterization of altered profiles as a result of disease, where the magnitude of perturbation can be quantified and monitored during treatment. Proteomic analysis has also clarified the nature of the isopeptide cross-linked residual insoluble material after vigorous extraction with protein denaturants, nearly intractable to analysis without fragmentation. These structures, including the cross-linked envelope of epidermal corneocytes, are comprised of hundreds of protein constituents, evidence for strengthening the terminal structure complementary to disulfide bonding. Along with other developing technologies, proteomic analysis is anticipated to find use in disease risk stratification, detection, diagnosis, and prognosis after the discovery phase and clinical validation.
Keywords: Cross-linked envelopes, Hair shaft, Keratins, Mass spectrometry, Transglutaminase
Introduction
Advances in genomics, proteomics, metabolomics and other technology-driven specialties provide a remarkable amount of detail on the fundamental mechanisms of life and are helping develop connections between anatomy and pathology. Mass spectrometry-enabled proteomics permits comparative profiling, characterization of post-translational modifications, elucidation of protein-protein interactions and networks and development of biomarkers of disease[1], especially in the last case integrated with other omics technologies.[2] For example, the depth of knowledge of the keratin gene superfamily, with distinct groups of keratin proteins found in a variety of epithelia, makes keratins a fundamental marker for all epithelia. We can now interrogate the protein content relatively noninvasively of the epidermis, hair shaft, and nail plate to understand not only their keratin compositions but also the wealth of other proteins located in them and how their structures differ in health and disease.[3] The results summarized below, from straightforward application of protein profiling to epidermal callus, hair shaft and nail plate,[4] provide a foundation for improved analysis of disease of the integument and monitoring of treatment.
Keratinocytes exhibit an intricate maturation program to yield mature corneocytes. These cells and the structures they form have presented a substantial challenge to characterization at the protein level due to the high density of disulfide bonding and the prevalence of transglutaminase-mediated isopeptide cross-linking. Traditional biochemical methods permitted isolation of keratins from corneocytes by solubilization with strong denaturants in the presence of reducing agents. The identities of proteins in the fraction (10–20%) that could not be solubilized remained mysterious until the advent of mass spectrometry-mediated analysis of complex protein mixtures. Reproducible fragmentation through tryptic proteolysis and the ability to match resulting fragments to a peptide database generated in silico has resulted in a dramatic advance in our information-gathering ability.
Nevertheless, several technical factors must be kept in mind to exploit this advance. Because keratinocytes, and especially corneocytes, are designed by nature to be tough, cohesive and resistant to their environment, the structures they form often need vigorous treatment with strong denaturants under reducing conditions, and the cysteine residues then are alkylated for stability. With care, the solubilizable protein can be separated from the heavily isopeptide cross-linked (insoluble) fraction if desired.[4] When the tryptic digests are matched in the peptide database to identify protein constituents, the presence of shared peptides needs attention to improve the quantitation. Thus peptides with the same sequence may be found in different proteins, not a rare phenomenon among closely related proteins such as the keratins, and can be distributed by a weighting process.[5] Spectral counting (weighted) is satisfactory for comparing relative amounts of a given protein among samples, but label free methods are becoming common for comparing amounts of different proteins.[6] Although the work described below has been performed in shotgun or discovery mode, a focus on given proteins for a specific purpose could speed up the analysis and sensitivity. As the sensitivity of mass spectrometry increases, more proteins are anticipated to be detected, increasing the need for scrupulous technique (e.g., sample processing in laminar flow hoods and, where possible, passage of reagent solutions through C18 solid phase extraction columns). Higher sensitivity brings with it more opportunity for detecting contamination, a particular risk for corneocyte samples inasmuch as shed corneocytes from epidermis and hair are a large fraction of dust in the human environment.
Hair shaft
Shotgun proteomics identified a score of keratins in the solubilized fraction of hair shaft protein. It also revealed a previously unappreciated complexity of the isopeptide cross-linked fraction that could not be solubilized with strong denaturants under reducing conditions. Some 300 proteins were identified that mapped to the various compartments of the cell.[7] In addition to comprising the overwhelming majority of protein in the solubilized fraction, keratins also appeared to comprise a majority of the protein in the insoluble fraction as well. Removal of the solubilized keratins, however, assisted detection of the remaining non-keratin proteins in the insoluble fraction.
Application of this technology to inbred mouse strains did not permit establishment of “the” mouse hair shaft proteome. Rather, it revealed that strains differed substantially in their pelage hair proteomes and that single gene alterations could produce considerable alteration in the hair protein profile.[8] The corollary proposition that individuals in an outbred population would differ in hair protein profile was then substantiated for humans.[9] The variation among individuals within ancestral subpopulations appears greater than among subpopulations. A survey of hair samples from 10 human monozygotic twins showed that observed differences in hair protein profile among individuals has a genetic basis.[10] No clear evidence for male/female differences was obtained in this limited sample. However, the VSIG8 protein, recently found to be prominent in hair shaft and nail plate in humans[11] and mice[12] was seen only at very low levels in two of the 5 hair samples from female subjects (Supporting Figure S1). Using this information in a forensic context to distinguish among individuals seems feasible, although detection of genetically variant peptides in the hair shaft appears to offer a greater power of discrimination.[13]
The hair of AKR/J inbred mice provides an example of protein profiling as a useful complement to study of disease by genetic approaches. Superficially resembling human pili annulati, the hair appears softer than that from other inbred albino mouse strains. The hair shafts are similar externally but the inside appears to have bubbles, not unlike cooked human hair (“bubble hair”).[14] This abnormal mouse hair was termed “hair interior defect”.[15] When mapping the responsible mutation, the hair interior defect phenotype behaved like a simple autosomal recessive trait when affected mice were crossed with four unrelated strains, resulting in about 25% affected progeny. The fifth strain, which was the first one used, had only 14.62% affected progeny recovered. Mutations in sterol O-acyltransferase 1 (Soat1) result in hair interior defects in AKR/J mice, and linkage analysis indicated that at least 6 modifier genes affect this phenotype.[16] To help address the basis for the peculiar phenotype, proteomic analysis of the hair shafts showed that hair from AKR/J mice, in comparison with two other strains without the defect, was deficient in trichohyalin. This deficiency seemed responsible for the lack of proper orientation of cells in the medulla and thus its unusual appearance.[17]
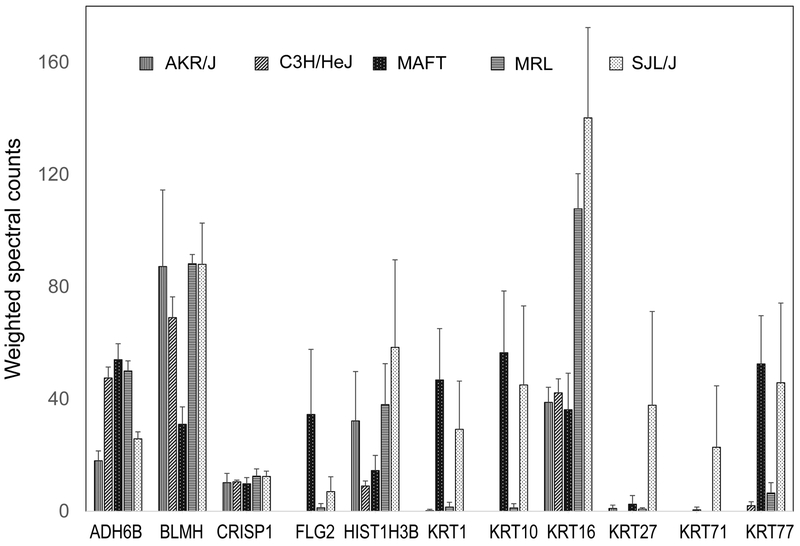
Alopecia areata is a cell mediated autoimmune disease directed against actively growing anagen stage hair follicles in humans, mice, rats, cattle, horses, and probably even chickens (growing feathers). Disruption of the hair follicle root sheaths results in a deformed hair shaft, commonly referred to as “exclamation point hair”. The defective hair shaft breaks off at or near the skin surface resulting in the clinical presentation of hair loss (alopecia). In fact, if the disruptive effect of the immune system is removed the hair grows back. The C3H/HeJ is a well-accepted mouse model for human alopecia areata.[18] Other inbred strains of mice also develop alopecia areata but usually far less frequently and less severely.[19, 20] Hair samples from AKR/J and 4 other mouse strains were analyzed to find whether a relation to incidence or severity of alopecia areata was discernable. MRL/MPJ and SJL/J are susceptible, and C3H/HeJ are susceptible as they age. Not susceptible are AKR/J and STOCK a/a Tmem79ma Flgft/J, a mouse model for atopic dermatitis with mutations in transmembrane protein 79 (Tmem79) and filaggrin (Flg) genes.[21, 22] Previous work had pointed to cysteine-rich secretory protein 1 (Crisp1), coding for a hair structural protein as a candidate gene for alopecia areata.[23] These 5 strains did not differ in their CRISP1 protein levels judging by mass spectrometry, but functionality of the protein was not assessed. Levels of several other proteins differed, notably keratins 1 (KRT1) and 10 (KRT10) (Figure 1). These were reported in human hair cuticle in some[9, 24] but not other reports,[25] while KRT10 has been found in sheep wool cuticle.[26] Figure 1 indicates they are detectable in the hair shaft of some (MAFT, SJL/J) but not other mouse strains.
Figure 1. Protein expression levels in 5 inbred mouse strains.
A sample from each of four mice per strain was processed essentially as previously described.[8] Numbers are weighted spectral counts of peptides, averages ± standard deviations. ADH6B, aldehyde dehydrogenase 6b; BLMH, bleomycin hydrolase; CRISP1, cysteine rich secretory protein 1; FLG2, filaggrin 2; HIST1H3B, histone 1H3B; KRT1, keratin 1; KRT10, keratin 10; KRT16, keratin 16; KRT27, keratin 27; KRT71, keratin 71; KRT77, keratin 77.
Nail plate
Proteomic analysis of nail plate showed that, like hair shaft, the overwhelming majority of the proteins solubilized by detergent under reducing conditions were keratins.[11] Also similar to hair shaft, keratins comprised a majority of the insoluble fraction as well, but numerous cytoplasmic, membrane and junctional proteins and histones were identified in addition. As indicated by previous work,[27, 28] the nail plate keratin composition was intermediate between epidermis and hair shaft containing some keratins abundant in epidermis but lacking in hair shaft and vice versa. The overlap of the nail plate and hair proteomes helps rationalize hair and nail dystrophies as seen in mice and humans with defects in the forkhead box N1 (FOXN1) gene.[29, 30] Illustrating the effect of programmed maturation on the corneocyte proteome, mice with a specific autophagy deficiency in the nail unit showed minimal effects on the content of keratin, keratin-associated or desmosomal proteins, but substantially higher levels of diverse enzymes and other proteins.[31]
Epidermis
Extension of shotgun proteomics to epidermis led to study of ichthyosis by sampling the stratum corneum with tape stripping. To provide a basis for comparison of afflicted skin at different sites, the dependence of the protein profile on bodily location was surveyed. Dependence on the site of sampling was observed generally, but the most dramatic contrast was between the palm and elsewhere (forearm, forehead, lower leg, upper back, abdomen) in content of keratin 9 and hornerin (HRNR).[32] Keratins, especially KRT9,[33] are well known to show anatomic site specificity.[34] Such differences, evident from transcriptomic studies, could affect susceptibility to infectious disease.[35]
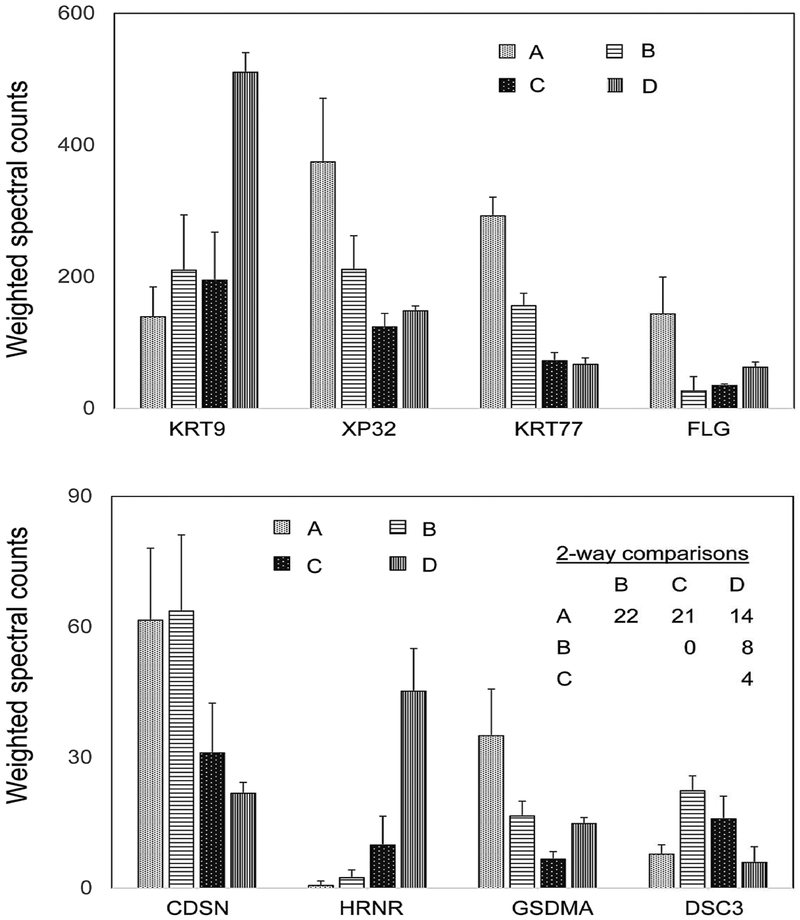
Samples were collected and compared to controls at the same sites from subjects with ichthyosis vulgaris (FLG deficiency), recessive X-linked ichthyosis (steroid sulfatase, STS, deficiency) and lamellar ichthyosis (transglutaminase 1, k polypeptide, TGM1, deficiency). Sampled lesions were distinguishable in appearance and in their protein profiles from each other and from control samples. Effects of mutations in the STS and gap junction protein beta 2 (GJB2) genes on appearance of ichthyosis vulgaris lesions and their protein profiles were also evident. The degree of departure from normal in the profile was parallel to that in the appearance. Indeed, this parallel was evident as well in normal and ichthyosis human epidermal samples grafted to mouse skin.[36] In this work, individual differences were clear in expression levels of certain proteins. This finding is also evident in a more recent survey of four individuals, where some individuals were distinguishable by protein profiling (Figure 2) in two way comparisons as previously performed with hair samples.[7]
Figure 2. Variation among individuals in expression of human epidermal callus proteins.
The data show the mean and standard deviation of weighted spectral counts from triplicate samples of each subject (A, B, C, D). The table of 2-way comparisons shows the numbers of proteins with significantly different weighted spectral counts. Protein expression was compared between subjects using a one-way ANOVA model in limma. KRT9, keratin 9; XP32, skin specific protein 32; KRT77, keratin 77; FLG, filaggrin; CDSN, corneodesmosin; HRNR, hornerin; GSDMA, gasdermin; DSC3, desmocollin 3.
A survey was undertaken analyzing the profiles of callus from the ball of the foot in individuals suffering from pachyonychia congenita. Gain of function mutations in keratins 6A, 6B, 6C, 16 or 17 lead to palmoplantar keratoderma, abnormalities in the nail unit and various other symptoms.[37, 38] Comparisons with samples from unaffected control subjects revealed the protein profiles were most altered in samples from individuals with KRT6 or KRT16 mutations, while those from KRT6C or KRT17 mutations displayed few protein alterations.[39] The degree of departure from normal generally fit the observed severity of the syndrome for the keratin gene categories. Although not clearly contributing to understanding the mechanism of pain generation, a debilitating symptom, the results do provide a quantitative basis for noninvasive monitoring of treatment effectiveness.
A mouse model for severe keratoderma was developed by inhibiting AP1 transcription factor action in the suprabasal epidermis through expression of a dominant negative jun proto-oncogene (Jun) construct, producing hyperplasia, hyperkeratosis, parakeratosis and impaired barrier function.[40] Perturbation of the protein profile was found to be severe as well.[41] Analysis of the disulfide and isopeptide cross-linked fraction revealed suppression of epidermal keratins, filaggrin, filaggrin family member 2, late cornifed envelope proteins and keratin-associated proteins but stimulation of hyperproliferation-associated keratins, desmosomal linker, small proline rich and S100 proteins. The results suggest the genetic modification reduced expression of late differentiation genes that was compensated by increased expression of early differentiation genes.
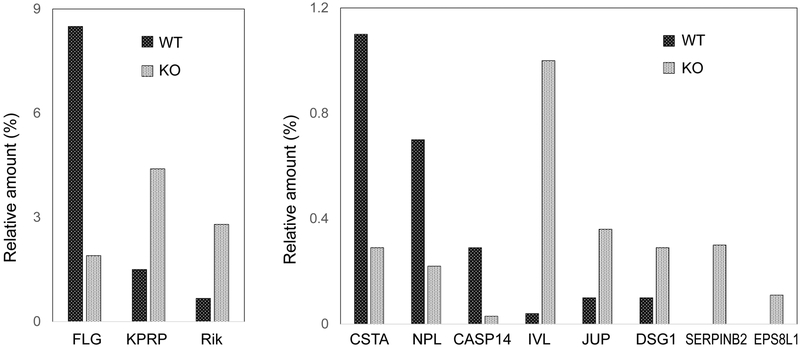
Although giving rise to a mild phenotype, effects of targeting a single gene are illustrated well by studies of a loricrin null mouse.[42] Proteomic analysis showed that keratins comprised ≈55% of the total protein of the cross-linked envelope in the control and null, with keratins 1 and 10 as primary constituents (30–40%).[43] In envelopes from the newborn (4 day) wild type mice, LOR was estimated as 11.5% of the total protein (Table 1). Its absence in the null mice led to altered incorporation of some 40 proteins into cross-linked envelopes, notably reductions of FLG, CSTA and CASP14 and increases in KPRP, IVL, JUP, DSG1 and EPS8L1 (Figure 3). SERPINB3 was reduced, while SERPINB2 was increased. Thus, the mild phenotype could be attributed to compensatory stimulation of alternate envelope proteins, some of which are encoded by NFE2L2 target genes such as Lce family members.[44]
Table 1.
Protein components of cross-linked envelopes in wild type (WT) and Lor knockout (KO) mice. Amounts were estimated by iBAQ calculations. Clusters of proteins (†) in some cases were used due to their shared peptides.
| %(WT) | Gene name | %(KO) Gene name | |
|---|---|---|---|
| 20.9 | Krt10 | 16.3 | Krt1 |
| 19.2 | Krt1 | 15.3 | Krt10 |
| 11.5 | Lor | 5.5 | Krt71 |
| 8.5 | Flg | 4.4 | Kprp |
| 3.2 | Flg2 | 4.1 | Hrnr |
| 3.0 | Krt28 | 3.8 | Flg2 |
| 2.8 | Hrnr | 3.2 | Krt27 |
| 2.2† | Krt13,14,15,16,17 | 2.8 | 2310050C09Rik |
| 2.1 | Krt27 | 2.8 | Sprr1a |
| 2.0 | Krt25 | 2.8† | Krt2,5,6a,6b,8,73,75,76,77,79 |
Figure 3. Change in envelope protein profile in loricrin null mice.
The epidermis of mouse skin, collected on day 4 after birth, was obtained by heat separation, extracted 4 times with SDS in the presence of dithioerythritol, processed for mass spectrometry and analyzed as described.[43] Relative protein amounts were estimated by iBAQ calculations. FLG, filaggrin; KPRP, keratinocyte proline rich protein; Rik, 2310050C09Rik; CSTA, cystatin A; NPL, N-acetylneuraminate pyruvate lyase; CASP14, caspase 14; IVL, involucrin; JUP, junctional plakoglobin; DSG1, desmoglein 1; SERPINB2, serine endopeptidase inhibitor family B member 2; EPS8L1, epidermal growth factor pathway substrate 8 like 1.
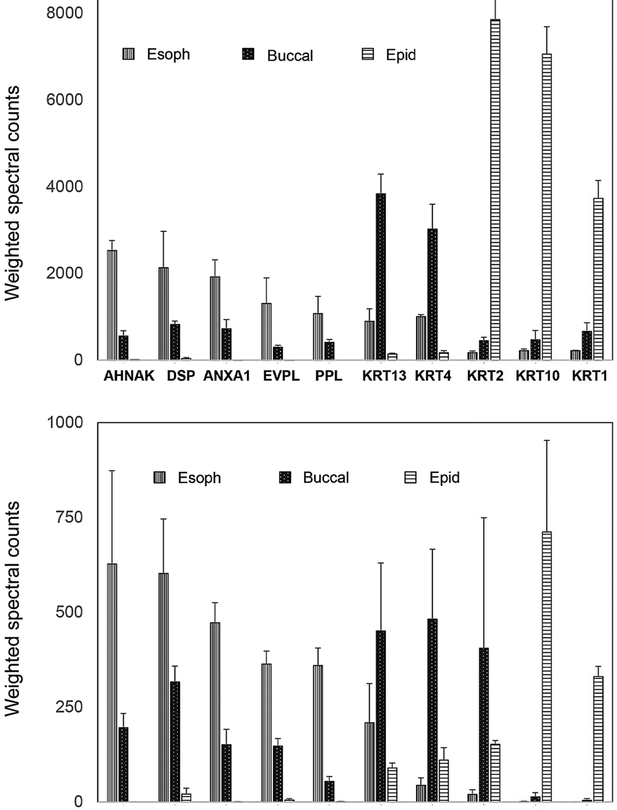
An extension of this work compared the profile of human epidermal callus to profiles from buccal cells obtained by cheek swab and from biopsy material from esophageal squamous epithelium. Figure 4 shows the striking variation observed among keratins, junctional and various other proteins. Statistical analysis showed that the majority of proteins were expressed at significantly different levels in comparisons of sample types (Supporting Figure S2). Site specificity of keratin expression is well known,[45] which these findings reflect. Some of the differences are potentially influenced by the different state of the sampled tissues. Unlike terminally differentiated epidermal callus samples, which are subjected to proteolytic remodeling, human esophageal specimens reflect the living interactive lining tissue. The sampled superficial buccal cells were of an intermediate state but closer to epidermal callus in terminal state. Efficiency of detection of proteins could be changed by the action of transglutaminase incorporating them into cross-linked envelopes, but envelopes comprise only ≈10% of the total cellular protein.
Figure 4. Distinct protein profiles from human epidermal callus, esophageal epithelium and buccal swabs.
Shown are averages ± standard deviations of samples from three (esophageal), four (buccal) and 12 (epidermal) subjects. AHNAK, neuroblast differentiation associated protein; DSP, desmoplakin; ANXA1, annexin A1, EVPL, envoplakin; PPL, periplakin; KRT13, keratin 13; KRT4, keratin 4; KRT2, keratin 2; KRT10, keratin 10; KRT1, keratin 1; SPRR3, small proline rich protein 3; JUP, junctional plakoglobin; S100A11, soluble in 100% ammonium sulfate family A, number 11; PKP1, plakophilin 1; PLEC, pleckstrin; KRT6A, keratin 6A; KRT16, keratin 16; KRT76, keratin 76; KPRP, keratinocyte proline rich protein; FLG2, filaggrin 2.
Corneocyte cross-linked proteome
Mature keratinocytes of the epidermis, hair shaft and nail plate are remarkably resistant to physical damage. They consist largely of keratin intermediate filaments with abundant disulfide bonding and are bounded by a cross-linked protein envelope. In the epidermis, this envelope serves as a scaffold to which a lipid barrier is attached that prevents transepidermal water loss [46] and where glycerol originating from sebaceous glands maintains hydration.[47] Loss of TGM1 activity localized at the plasma membrane results in defective envelopes and thus deficient barrier function and manifests as a prominent cause of autosomal recessive congenital ichthyosis.[48, 49] Defects in genes affecting lipid processing also give rise to scaly skin.[46]
The mechanism of envelope formation has been studied intensively. In addition to TGM1 activity, the substrate proteins being cross-linked have received considerable attention. An inability to reverse the isopeptide bonding sufficiently to isolate individual protein components made such investigation quite difficult. The advent of mass spectrometric analysis of peptide fragments from complex protein structures, however, has provided considerable insight. Identification of at least 300 proteins from the isopeptide cross-linked fraction of hair shaft,[7] and similar results with nail plate,[11] suggest analogous complexity for epidermal corneocytes.
Exhaustive extraction of hair shaft, nail plate and epidermal callus with SDS under reducing conditions removes the protein held together by disulfide and minimal isopeptide bonding.[4] Analysis of the remainder (10–20% of the total protein), where an estimated 15–20% of the lysines participate in isopeptide bonding in epidermal keratinocytes,[50] revealed that a large fraction, roughly two-thirds, was keratin. Keratin participation in envelopes from epidermal callus had been reported,[51] and the ablation of keratin genes in mice provided clear evidence for their essentiality.[52]
Some proteins can serve as substrates for transglutaminase-mediated cross-linking through available glutamine residues, but most are capable of participating though their lysine residues. Thus, those with abundant glutamines (e.g., involucrin and small proline rich proteins) have been proposed to facilitate the incorporation of other proteins. In addition, protein-protein interactions could influence substrate availability leading up to cross-linking, for example, by increasing their representation in the vicinity of TGM1 at the plasma membrane. Moreover, the large array of proteins that participate suggests that flexibility in envelope composition is possible, particularly in the ability to compensate for missing components. Such considerations provide a rationale for the minimal effects of loricrin gene ablation on mouse epidermis.[42] They also explain the analogous lack of effect of involucrin gene ablation[53] and the mild phenotype observed when it is accompanied by ablation of envoplakin (Evpl) and periplakin (Ppl) genes as well.[54]
Future prospects
At present, shotgun proteomics offers an overview of protein level changes for the most prevalent several hundred proteins. With respect to the epidermis and appendages, it permits viewing the extent of perturbation, providing a quantitative description of visible changes. Application to related epithelia, including that of esophagus, appears feasible. Now that the anatomic site specificity of protein levels is appreciated, and applications to disease thus far indicate its potential usefulness, extension to analyzing environmental influences appears possible, including atmospheric pollutants[55] and well known therapeutic agents such as coal tar.[56] This direction has been explored in cultured human epidermal keratinocytes treated with the persistent halogenated aromatic contaminant 2,3,7,8-tetrachlorodibenzo-p-dioxin. The results provide a rationale for a hyperkeratotic response (in vivo) due to reduction in certain differentiation markers while stimulating cross-linked envelope formation.[57] Such experiments may be valuable in finding the influence of modifier genes in culture samples from different individuals.
Use of mouse models can greatly assist elucidating the genetic basis for disease, including genodermatoses, and effects of modifier genes. Clinical subtypes of so-called “single gene based diseases” are now amenable to study. For example, a number of single gene mutations that occur or were created on a variety of inbred mouse backgrounds develop few to no lesions when crossed with the FVB/NJ strain, suggesting that identification of the gene(s) accounting for this phenomenon would be extremely useful biomarkers for disease prognosis if the discovery is transferable to human skin disease. As an indication of the profound effects on proteomic profiles, a single gene mutation on one background may give an outcome quite distinct from a second allelic mutation in the same gene on a different inbred mouse background. Such effects are magnified for humans (outbred) where each individual is unique. In combination with transcriptomics, metabolomics, lipidomics and other evolving and emerging technologies, including genetically modified organisms (e.g., CRISPR), proteomic approaches can help pinpoint the primary modifier gene(s) and protein(s) involved or elaborate on the molecular networks that direct the variety of clinical outcomes that patients experience. Newer technologies will increase the complexity of our understanding of pathophysiology. However, as profound changes or subtle differential expression variations correlate better with specific disease diagnoses, diagnostic and therapeutic accuracy will improve along with the prognostic abilities of the dermatologist.
Supplementary Material
Supporting Figure S1. Variation in VSIG8 protein level in hair samples from monozygotic twin pairs. Illustrated are the averages and standard deviations of each twin pair (pooled for the two individuals, each sampled in quadruplicate) for 5 female and 5 male pairs. Spectral counts were not significantly different within twin pairs. The average VSIG8 level from the 5 male pairs was the same as from the highest three female pairs (88 ± 22), but the level in two female pairs was significantly lower (5 ± 4).
Supporting Figure S2. Comparison of buccal, epidermal and esophageal protein profiles. The multidimensional scaling plot of all the samples shows log fold change distances between samples. Prior to analysis, proteins averaging less than 1 weighted spectral count were filtered. Analyses were conducted using the limma-voom Bioconductor pipeline[58] with limma version 3.32.10, edgeR version 3.18.1, and R version 3.4.4, originally developed for RNA sequencing data. Protein expression was compared between sample types using a one-way ANOVA model in limma, with standard errors of log fold changes adjusted to account for within-subject correlations. Illustrated is a compilation of buccal (4 subjects, b1-b4)), epidermal (4 subjects in triplicate, B1-B3, M1-M3, S1-S3, W1-W3) and esophageal samples (3 subjects, he1-he3).
Acknowledgments
We thank Dr. Young Moo Lee for initiating this work at the University of California Davis. We thank the National Institute of Arthritis, Musculoskeletal, and Skin Diseases (NIH grants R21- AR063781 and R01AR056635), the National Center for Advancing Translational Sciences (NH grant UL1 TR001860) and the USDA(NIFA)/University of California Agricultural Experiment Station for financial support of this work.
Abbreviations:
- *CASP14
caspase 14
- CRISP1
cysteine-rich secretory protein 1
- CRISPR
clustered regularly interspaced short palindromic repeats
- CSTA
cystatin A1
- DSG1
desmoglein 1
- EPS8L1
epidermal growth factor receptor kinase substrate 8-like protein 1
- FLG
filaggrin
- EVPL
envoplakin
- FOXN1
forkhead box N1
- GABPA (formerly NRF2)
GA repeat binding protein alpha
- GJB2
gap junction protein beta 2
- HRNR
hornerin
- IVL
involucrin
- KPRP
keratinocyte expressed proline-rich
- KRT
keratin
- JUN
jun proto-oncogene
- LCE
late cornified envelope protein
- LOR
loricrin
- NFE2L2
nuclear factor erythroid derived 2 like 2
- PPL
periplakin
- SERPINB2/3
serine endopeptidase inhibitor clade B member 2/3
- SOAT1
sterol O-acyltransferase 1
- STS
steroid sulfatase
- SDS
sodium dodecyl sulfate
- TGM1
transglutaminase 1
- TMEM79
transmembrane protein 79.
Footnotes
Conflict of interest
Dr. Sundberg has sponsored research projects with Biocon, Bioniz, Curadim, Takeda and Theravance, none of which are related to the work presented here. The other authors state they have no conflicts of interest.
Gene symbols are in italics, capital letters for human genes, first letter capitalized, remainder in lower case for mouse genes. Proteins are all in capital letters, not italicized, for both species.
This is the peer reviewed version of the following article: Corneocyte Proteomics: Applications to Skin Biology and Dermatology; Exp Dermatol. 2018 Aug.27(8):931–938, which has been published in final form at https://doi.org/10.1111/exd.13756. This article may be used for non-commercial purposes in accordance with Wiley Terms and Conditions for Use of Self-Archived Versions.
Focus Theme issue “Skin barrier”
References
- [1].Altelaar AFM, Munoz J, Heck AJR, Nat. Rev. Genet 2013, 14, 35. [DOI] [PubMed] [Google Scholar]
- [2].Karczewski KJ, Snyder MP, Nat. Rev. Genet 2018, 19, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Hammers CM, Tang HY, Chen J, Emtenani S, Zheng Q, Stanley JR, J. Invest. Dermatol 2018, 138, 1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Rice RH, Int. J. Cosmetic Sci 2011, 62, 229. [PMC free article] [PubMed] [Google Scholar]
- [5].Zhang Y, Wen Z, Washburn MP, Florens L, Analyt. Chem 2015, 87, 4749. [DOI] [PubMed] [Google Scholar]
- [6].Dowle AA, Wilson J, Thomas JR, J. Proteome Res 2016, 15, 3550. [DOI] [PubMed] [Google Scholar]
- [7].Lee YJ, Rice RH, Lee YM, Molec. Cell Proteom 2006, 5, 789. [DOI] [PubMed] [Google Scholar]
- [8].Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS, Sundberg JP, PLoS One 2012, 7, e51956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Laatsch CN, Durbin-Johnson BP, Rocke DM, Mukwana S, Newland AB, Flagler MJ, Davis MG, Eigenheer RA, Phinney BS, Rice RH, PeerJ 2014, 2, e506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Wu P-W, Mason KE, Durbin-Johnson BP, Salemi M, Phinney BS, Rocke DM, Parker GJ, Rice RH, Proteomics 2017, 17, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Rice RH, Xia Y, Alvarado RJ, Phinney BS, J. Proteome Res 2010, 9, 6752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Rice RH, Phillips MA, Sundberg JP, J. Invest. Dermatol 2011, 131, 1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Parker GJ, Leppert T, Anex DS, Hilmer JK, Matsunami N, Baird L, Stevens J, Parsawar K, Durbin-Johnson BP, Rocke DM, Nelson C, Fairbanks DJ, Wilson AS, Rice RH, Woodward SR, Bothner B, Hart H, Leppert M, PLoS One 2016, 11(9), e0160653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Whiting DA, Howsden FL, Canfield Publishing, Cedar Grove, NJ, 1996 [Google Scholar]
- [15].Giehl KA, Potter CS, Wu B, Silva KA, Rowe L, Awgulewitsch A, Sundberg JP, Clin. Exp. Dermatol 2009, 34, 509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wu B, Potter CS, Silva KA, Liang Y, Reinholdt LG, Alley LM, Rowe LB, Roopenian DC, Awgulewitsch A, Sundberg JP, J. Invest. Dermatol 2010, 130, 2666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Rice RH, Rocke DM, Tsai H-S, Lee YJ, Silva KA, Sundberg JP, J. Invest. Dermatol 2009, 129, 2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pratt CH, King LEJ, Messenger AG, Christiano AM, Sundberg JP, Nature Reviews Disease Primers 2017, 3, 17011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].McElwee KJ, Boggess D, Miller J, King LEJ, Sundberg JP, J. Invest. Dermatol. Symp. Proc 1999, 4, 202. [DOI] [PubMed] [Google Scholar]
- [20].Sundberg JP, Chevallier L, Silva KA, Kennedy VE, Sundberg BA, Li Q, Uitto J, King LEJ, Berndt A, J. Invest. Dermatol 2014, 134, 279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Saunders SP, Goh CS, Brown SJ, Palmer CN, Porter RM, Cole C, Campbell LE, Gierlinski M, Barton G, Schneider G, Balmain A, Prescott AR, Weidinger S, Baurecht H, Kabesch M, Gieger C, Lee YA, Tavendale R, Mukhopadhyay S, Turner SW, Madhok VB, Sullivan FM, Relton C, Burn J, Meggitt S, Smith CH, Allen MA, Barker JN, Reynolds NJ, Cordell HJ, Irvine AD, McLean WH, Sandilands A, Fallon PG, Allergy Clin J. Immunol. 2013, 132, 1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Sasaki T, Shiohama A, Kubo A, Kawasaki H, Ishida-Yamamoto A, Yamada T, Hachiya T, Shimizu A, Okano H, Kudoh J, Amagai M, Allergy Clin J. Immunol. 2013, 132, 1111. [DOI] [PubMed] [Google Scholar]
- [23].Sundberg JP, Awgulewitsch A, Pruett ND, Potter CS, Silva KA, Stearns TM, Sundberg BA, Muñoz MW, Cuasnicu PS, King LEJ, Rice RH, Exp. Molec. Pathol 2014, 97, 525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Stark HJ, Breitkreutz D, Limat A, Ryle CM, Roop D, Leigh I, Fusenig N, Eur. J. Cell Biol 1990, 52, 359. [PubMed] [Google Scholar]
- [25].Langbein L, Yoshida H, Praetzel-Wunder S, Parry DA, Schweizer J, J. Invest. Dermatol 2010, 130, 55. [DOI] [PubMed] [Google Scholar]
- [26].Koehn H, Clerens S, Deb-Choudhury S, Morton J, Dyer JM, Plowman JE, J. Proteome Res 2010, 9, 2920. [DOI] [PubMed] [Google Scholar]
- [27].De Berker D, Wojnarowska F, Sviland L, Westgate GE, Dawber RP, Leigh IM, Br. J. Dermatol 2000, 142, 89. [DOI] [PubMed] [Google Scholar]
- [28].Perrin C, Langbein L, Schweizer J, Br. J. Dermatol 2004, 151, 362. [DOI] [PubMed] [Google Scholar]
- [29].Mecklenburg L, Paus R, Halata Z, Bechtold LS, Fleckman P, Sundberg JP, J. Invest. Dermatol 2004, 123, 1001. [DOI] [PubMed] [Google Scholar]
- [30].Auricchio L, Adriani M, Frank J, Busiello R, Christiano A, Pignata C, Arch. Dermatol 2005, 141, 647. [DOI] [PubMed] [Google Scholar]
- [31].Jaeger J, Sukseree S, Zhong S, Mlitz V, Tschachler E, Rice RH, Eckhart L, J. Invest. Dermatol 2018, 138, S228 [Google Scholar]
- [32].Rice RH, Bradshaw KM, Durbin-Johnson BP, Rocke DM, Eigenheer RA, Phinney BS, Schmuth M, Gruber R, PLoS One 2013, 8(10), e75355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Knapp AC, Franke WW, Heid H, Hatzfeld M, Jorcano J, Moll R, J. Cell Biol 1986, 103, 657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Swensson O, Langbein L, R MJ, Stevens HP, Leigh IM, McLean WHI, Lane EB, Eady RAJ, Br. J. Dermatol 1998, 139, 767. [DOI] [PubMed] [Google Scholar]
- [35].Sundberg J, Stearns TM, Joh J, Proctor M, Ingle A, Silva KA, Dadras SS, Jenson AB, Ghim SJ, PLoS One 2014, 9(12), e113582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Aufenvenne K, Rice RH, Hausser I, Oji V, Hennies HC, Rio MD, Traupe H, Larcher F, J. Invest. Dermatol 2012, 132, 1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Leachman SA, Kaspar RL, Fleckman P, Florell SR, Smith FJ, McLean WH, Lunny DP, Milstone LM, van Steensel MA, Munro CS, O’Toole EA, Celebi JT, Kansky A, Lane EB, J. Invest. Dermatol. Symp. Proc 2005, 10, 3. [DOI] [PubMed] [Google Scholar]
- [38].Eliason MJ, Leachman SA, Feng BJ, Schwartz ME, Hansen CD, J. Am. Acad. Dermatol 2012, 67, 680. [DOI] [PubMed] [Google Scholar]
- [39].Rice RH, Durbin-Johnson BP, Salemi M, Schwartz ME, Rocke DM, Phinney BS, J. Proteomics 2017, 165, 132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rorke EA, Adhikary G, Young CA, Roop DR, Eckert RL, J. Invest. Dermatol 2015, 135, 170. [DOI] [PMC free article] [PubMed] [Google Scholar] [Research Misconduct Found]
- [41].Rorke E, Gautam A, Young C, Rice R, Elias P, Crumrine D, Meyer J, Blumenberg M, Eckert R, Cell Death Dis 2015, 6, e1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Koch PJ, de Viragh PA, Scharer E, Bundman D, Longley MA, Bickenbach J, Kawachi Y, Suga Y, Zhou Z, Huber M, Hohl D, Kartasova T, Jarnik M, Steven AC, Roop DR, J. Cell Biol 2000, 151, 389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Rice RH, Durbin-Johnson BP, Ishitsuka YI, Salemi M, Phinney BS, Rocke DM, Roop DR, J. Proteome Res 2016, 15, 2560. [DOI] [PubMed] [Google Scholar]
- [44].Ishitsuka Y, Huebner AJ, Rice RH, Zhou Z, Koch PJ, Jarnik M, Speransk VV, Steven AC, Roop DR, J. Invest. Dermatol 2016, 136, 1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Moll R, Divo M, Langbein L, Histochem. Cell Biol 2008, 129, 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Akiyama M, J. Dermatol. Sci 2017, 88, 3. [DOI] [PubMed] [Google Scholar]
- [47].Fluhr JW, Mao-Qiang M, Brown BE, Wertz PW, Crumrine D, Sundberg JP, Feingold KR, Elias PM, J. Invest. Dermatol 2003, 120, 728. [DOI] [PubMed] [Google Scholar]
- [48].Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SPM, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D, Science 1995, 267, 525. [DOI] [PubMed] [Google Scholar]
- [49].Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ, Nat. Genet 1995, 9, 279. [DOI] [PubMed] [Google Scholar]
- [50].Rice RH, Green H, Cell 1977, 11, 417. [DOI] [PubMed] [Google Scholar]
- [51].Candi E, Tarcsa E, DiGiovanna JJ, Compton JG, Marekov LN, Steinert PM, Proc. Natl. Acad. Sci. USA 1998, 95, 2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Kumar V, Bouameur J-E, Bär J, Rice RH, Hornig-Do H-T, Roop DR, Schwarz N, Brodesser S, Thiering S, Leube RE, Wiesner RJ, Brazel CB, Heller S, Binder H, Löffler-Wirth H, Seibel P, Magin TM, J. Cell Biol 2015, 211, 1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Djian P, Easley K, Green H, J. Cell Biol 2000, 151, 381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Sevilla LM, Nachat R, Groot KR, Klement JF, Uitto J, Djian P, Määttä A, Watt FM, J. Cell Biol 2007, 179, 1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rajagopalan P, Jain AP, Nanjappa V, Patel K, Mangalaparthi KK, Babu N, Cavusoglu N, Roy N, Soeur J, Breton L, Pandey A, Gowda H, Chatterjee A, Misra N, J. Dermatol. Sci 2018, in press, [DOI] [PubMed] [Google Scholar]
- [56].van den Bogaard EH, Bergboer JGM, Vonk-Bergers M, van Vlijmen-Willems IMJJ, Hato SV, van der Valk PGM, Schröder JM, Joosten I, Zeeuwen PLJM, J JS, J. Clin. Invest 2013, 123, 917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hu Q, Rice RH, Qin Q, Phinney BS, Eigenheer RA, Bao W, Zhao B, J. Proteome Res 2013, 12, 5340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK, Nucl. Acids Res. 2015, 43, doi: 10.1093/nar/gkv007 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Figure S1. Variation in VSIG8 protein level in hair samples from monozygotic twin pairs. Illustrated are the averages and standard deviations of each twin pair (pooled for the two individuals, each sampled in quadruplicate) for 5 female and 5 male pairs. Spectral counts were not significantly different within twin pairs. The average VSIG8 level from the 5 male pairs was the same as from the highest three female pairs (88 ± 22), but the level in two female pairs was significantly lower (5 ± 4).
Supporting Figure S2. Comparison of buccal, epidermal and esophageal protein profiles. The multidimensional scaling plot of all the samples shows log fold change distances between samples. Prior to analysis, proteins averaging less than 1 weighted spectral count were filtered. Analyses were conducted using the limma-voom Bioconductor pipeline[58] with limma version 3.32.10, edgeR version 3.18.1, and R version 3.4.4, originally developed for RNA sequencing data. Protein expression was compared between sample types using a one-way ANOVA model in limma, with standard errors of log fold changes adjusted to account for within-subject correlations. Illustrated is a compilation of buccal (4 subjects, b1-b4)), epidermal (4 subjects in triplicate, B1-B3, M1-M3, S1-S3, W1-W3) and esophageal samples (3 subjects, he1-he3).