Summary
Spermatogonial stem cells (SSCs) are essential for adult spermatogenesis. Recently, Wang et al. (2018), Guo et al. (2018), and Hermann et al. (2018) used single-cell RNA sequencing to define and molecular characterize human testicular cell populations, including spermatogonial subsets with characteristics of human SSCs.
The production of male gametes—or spermatogenesis—is critical for the propagation of all species. In mammals, spermatogenesis initiates in the testis at puberty and continues into late adulthood. As a testament to its endurance, children have been fathered by several male celebrities over the age of 65, including the likes of Pablo Picasso, Charlie Chaplin, and Mick Jagger. What makes this possible is the spermatogonial stem cell (SSC), a highly prolific stem cell that yields over 1,000 sperm every breath a man takes. SSCs do not directly generate sperm; instead, they form progenitors that undergo proliferative expansion followed by differentiation and meiosis to give rise to haploid cells that ultimately become sperm.
Our knowledge of spermatogenesis and SSCs has significantly expanded over the past several decades using rodents as models. These studies have provided valuable insights into key factors, including signaling pathways and transcriptional drivers essential for spermatogenesis. These past studies have also identified markers that label specific testicular cell subsets, however markers that exclusively label specific subsets, including SSCs, have been largely elusive. Another murky issue has been whether findings from rodents apply to humans. This has been particularly vexing given the evidence that human and mouse testes differ significantly in seminiferous epithelium organization, the pattern of SSC differentiation, SSC frequency, and sperm output per gram of tissue (Fayomi and Orwig, 2018).
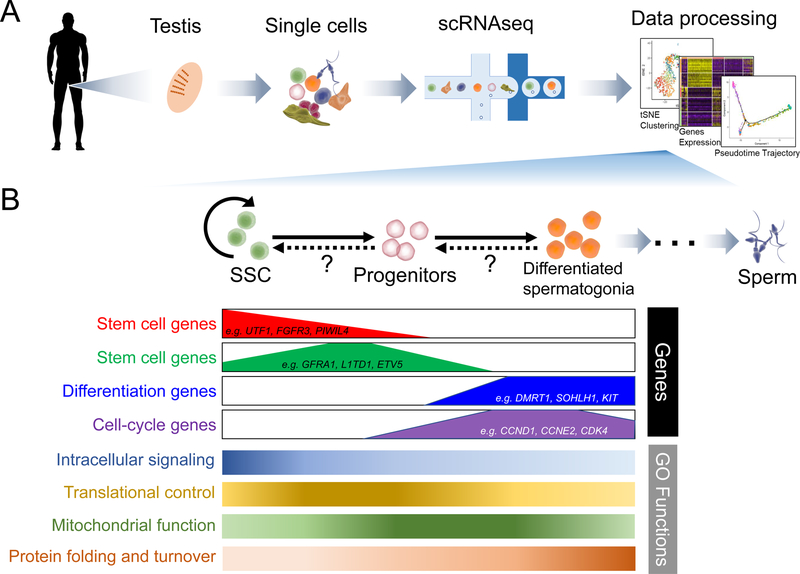
To address these questions head-on, several groups recently used different single-cell RNA-sequencing (scRNAseq) platforms to analyze dissociated cells (ranging from ~2800 to ~18,000) from the human testis (Figure 1) (Guo et al., 2018; Hermann et al., 2018; Wang et al., 2018). These studies independently identified all the known testicular germ cell types, including spermatogonia (diploid cells), spermatocytes (meiotic cells), and spermatids (haploid germ cells), as well as somatic cells (e.g., Sertoli cells, peritubular myoid cells, Leydig cells, and macrophages). More than one cell subset was identified from most known cell types, providing clear evidence for heterogeneity. In the case of spermatogonia, Wang et al., Guo et al., and Herman et al. identified 3, 5, and 10 distinct cell clusters (i.e., cell subsets), respectively. This suggested an astounding degree of human spermatogonial heterogeneity; and was particularly fascinating given that spermatogonia include SSCs. All 3 papers assigned a specific spermatogonial cell cluster as being SSC-enriched, based on previously defined mouse and human SSC markers, as well as Pseudotime trajectory analysis, which aligns the developmental order of cells based on transcriptome changes (Wu et al., 2017). Given this, we will refer to these cell clusters as “SSCs,” but it should be noted that none of the studies directly measured SSC activity (see more on this below). All 3 studies found some common characteristics of the SSC subset they each defined. For example, their cell-cycle gene expression pattern suggested that the SSCs were largely non-proliferative, while more differentiated spermatogonial cell clusters were dominated by proliferative cells. This agrees with previous studies using conventional approaches to measure cell proliferation in seminiferous tubules (Di Persio et al., 2017). Gene ontology analysis revealed that functions enriched in SSCs included genes involved in signaling pathways (BMP, FGF, and GDNF), transcription, as well as post-transcriptional regulation; e.g., RNA transport, translation, and RNA turnover (Figure 1).
Figure 1.
(A) Guo et al., Hermann et al., and Wang et al. performed scRNAseq analysis to identify human testicular germ and somatic cell subsets. (B) Top: cartoon of major spermatogonial cell types and their developmental relationships. Bottom: developmental expression pattern of marker genes, as well as enriched gene ontology (GO) functions, as determined by scRNAseq analysis.
Future clinical application of SSCs depends on identifying human SSC-specific markers. In this regard, all 3 studies identified genes enriched in the respective human SSC subsets they defined. Reassuringly, many of these genes were previously shown to mark mouse and/or human SSCs (e.g., UTF1, ID4, GFRA1, and FGFR3). Novel SSC marker genes were also identified, many of which were verified as candidate SSC markers by in situ RNA hybridization and antibody studies, both of which showed a signal in the seminiferous tubule periphery, where SSCs reside (Figure 1). A controversial topic addressed by Guo et al. is whether the most primitive spermatogonia (i.e., SSCs) express high levels of the growth factor receptor and mouse SSC marker, GFRA1, and low levels of the transcription factor and human SSC marker, UTF1, as suggested by immunofluorescence analysis of whole mount testes (Di Persio et al., 2017). Pseudotime trajectory analysis suggested that instead primitive spermatogonia are GFRA1low UTF1high and progress to become GFRA1high UTF1low as they differentiate (Guo et al., 2018), which is consistent with their becoming proliferative and the finding by Wang et al. that they acquire the differentiation marker DMRT1 (Wang et al., 2018). It will be interesting to determine which of these models is correct, or whether, instead, there is actually no precursor-product relationship between these subsets; e.g., they may be distinct SSC types (e.g., reserve vs. active).
Like many stem cell types, SSCs not only self-renew but they differentiate into highly proliferative progenitors. The 3 scRNAseq studies illuminated some aspects of this process. For example, Hermann et al. found that differentiating spermatogonia first turned on genes involved in translational control, then mitochondrial function genes (suggesting a shift from glycolysis to oxidative phosphorylation upon initiation of proliferation), and finally genes encoding protein folding and protein turnover factors (the latter of which might help drive the shift to the next step of male germ cell progression: meiosis) (Figure 1) (Hermann et al., 2018). As a testament to the versatility of scRNAseq datasets, Guo et al. applied a new computational approach that infers developmental trajectories—called “RNA Velocity” (La Manno et al., 2018)—to obtain evidence that differentiating human spermatogonia not only move towards a more advanced state, but can reverse direction (i.e., de-differentiate) back towards SSCs (Guo et al., 2018), a phenomenon suggested—by other methods—to also occur in mice spermatogonia (Nakagawa et al., 2010).
A critical question for the future is to determine SSC frequency in the spermatogonial subsets defined by the 3 groups by purifying these subsets using specific markers and then assaying them for “SSC activity” via the xeno-transplanation assay (Fayomi and Orwig, 2018). This experiment may lead to surprises. An assumption by the field is that SSCs have enriched expression for known SSC marker genes. As a case in point, Guo et al. defined “SSC subsets” through scRNAseq analysis of human spermatogonia purified using a marker—SSEA4—known to enrich for SSCs (Guo et al., 2017), but their subsequent analysis using unfractionated testes revealed the existence of a more primitive spermatogonial cell cluster that lacks SSEA4 expression (state “0”) that is a better candidate to be SSC-enriched (Guo et al., 2018). Another typical assumption by the field is that SSCs are most enriched in the least advanced spermatogonial subset. In this regard, it is notable that Hermann et al. identified a spermatogonial subset that was less differentiated (based on pseudotime trajectory analysis) than the subset they defined as the SSC subset (based on known markers) (Hermann et al., 2018). This novel primitive cell subset—which was found to be conserved in both mice and humans—might be a vestigial SSC precursor related to primordial germ cells. Another important future question is: can we still rely on mice to reflect what occurs in humans? Recent studies using scRNAseq to analyze mice testicular cells (Green et al., 2018; Hermann et al., 2018) suggest the answer is “yes,” based on the many similarities they found between mouse and human spermatogenesis. However, functional comparative studies will be needed to determine the extent to which mice and human SSCs (and spermatogenesis in general) overlap. Areas of overlap will justify performing studies in mice to address in vivo questions about SSCs not easily evaluated in humans.
Footnotes
DECLARATION OF INTEREST
The authors declare no competing interests.
REFERENCES:
- Di Persio S, Saracino R, Fera S, Muciaccia B, Esposito V, Boitani C, Berloco BP, Nudo F, Spadetta G, Stefanini M, et al. (2017). Spermatogonial kinetics in humans. Development 144, 3430–3439. [DOI] [PubMed] [Google Scholar]
- Fayomi AP, and Orwig KE (2018). Spermatogonial stem cells and spermatogenesis in mice, monkeys and men. Stem Cell Res 29, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green CD, Ma Q, Manske GL, Shami AN, Zheng X, Marini S, Moritz L, Sultan C, Gurczynski SJ, Moore BB, et al. (2018). A Comprehensive Roadmap of Murine Spermatogenesis Defined by Single-Cell RNA-Seq. Dev Cell 46, 651–667 e610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J, Grow EJ, Mlcochova H, Maher GJ, Lindskog C, Nie X, Guo Y, Takei Y, Yun J, Cai L, et al. (2018). The adult human testis transcriptional cell atlas. Cell Res [DOI] [PMC free article] [PubMed]
- Guo J, Grow EJ, Yi C, Mlcochova H, Maher GJ, Lindskog C, Murphy PJ, Wike CL, Carrell DT, Goriely A, et al. (2017). Chromatin and Single-Cell RNA-Seq Profiling Reveal Dynamic Signaling and Metabolic Transitions during Human Spermatogonial Stem Cell Development. Cell Stem Cell 21, 533–546 e536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hermann BP, Cheng K, Singh A, Roa-De La Cruz L, Mutoji KN, Chen IC, Gildersleeve H, Lehle JD, Mayo M, Westernstroer B, et al. (2018). The Mammalian Spermatogenesis Single-Cell Transcriptome, from Spermatogonial Stem Cells to Spermatids. Cell Rep 25, 1650–1667 e1658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lonnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Sharma M, Nabeshima Y, Braun RE, and Yoshida S (2010). Functional hierarchy and reversibility within the murine spermatogenic stem cell compartment. Science 328, 62–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Liu X, Chang G, Chen Y, An G, Yan L, Gao S, Xu Y, Cui Y, Dong J, et al. (2018). Single-Cell RNA Sequencing Analysis Reveals Sequential Cell Fate Transition during Human Spermatogenesis. Cell Stem Cell [DOI] [PubMed]
- Wu AR, Wang J, Streets AM, and Huang Y (2017). Single-Cell Transcriptional Analysis. Annu Rev Anal Chem (Palo Alto Calif) 10, 439–462. [DOI] [PubMed] [Google Scholar]